Abstract
In plants, the amino acids tyrosine and phenylalanine are synthesized from arogenate by arogenate dehydrogenase and arogenate dehydratase, respectively, with the relative flux to each being tightly controlled. Here the characterization of a maize opaque endosperm mutant (mto140), which also shows retarded vegetative growth, is described The opaque phenotype co-segregates with a Mutator transposon insertion in an arogenate dehydrogenase gene (zmAroDH-1) and this led to the characterization of the four-member family of maize arogenate dehydrogenase genes (zmAroDH-1–zmAroDH-4) which share highly similar sequences. A Mutator insertion at an equivalent position in AroDH-3, the most closely related family member to AroDH-1, is also associated with opaque endosperm and stunted vegetative growth phenotypes. Overlapping but differential expression patterns as well as subtle mutant effects on the accumulation of tyrosine and phenylalanine in endosperm, embryo, and leaf tissues suggest that the functional redundancy of this gene family provides metabolic plasticity for the synthesis of these important amino acids. mto140/arodh-1 seeds shows a general reduction in zein storage protein accumulation and an elevated lysine phenotype typical of other opaque endosperm mutants, but it is distinct because it does not result from quantitative or qualitative defects in the accumulation of specific zeins but rather from a disruption in amino acid biosynthesis.
Keywords: Arogenate dehydrogenase, maize endosperm, opaque mutant, tyrosine, zein
Introduction
The development of a vitreous endosperm at kernel maturity is a valuable trait in maize, since it affords resistance to insect and disease damage as well as seed durability during harvest, transportation, and storage (Gibbon et al., 2003). Many of the grains’ milling and processing characteristics also depend on this trait (Fox and Manley, 2009). There is an inverse correlation between vitreous endosperm and kernel protein nutritional quality (Mertz et al., 1964). There is evidence to support the hypothesis that the zein storage proteins and their packaging into endoplasmic reticulum-localized protein bodies are central to this trait. For example, the vitreous portion of the kernel contains much more zein than the soft, starchy interior (Dombrink-Kurtzman and Bietz, 1993), and environmental conditions that reduce zein synthesis, such as nitrogen deprivation, result in kernels that are soft and starchy (Tsai et al., 1978). Perhaps the most compelling evidence for this relationship comes from the molecular characterization of several maize mutants that have a starchy, opaque kernel and are caused by either quantitative or qualitative alterations in zein proteins. For example, the opaque2 (o2) mutant accumulates low levels of α-zeins and has protein bodies that are much smaller than the wild type. O2 encodes a transcription factor that regulates several endosperm-expressed genes, in particular those encoding 22 kDa α-zeins (Schmidt et al., 1990). Several other opaque mutants such as floury2, De-B30, and Mucronate have structural defects in different types of zein proteins which result in improper zein packaging, protein body deformation, and the ‘unfolded protein response’ (UPR) (Lending and Larkins, 1992; Kim et al., 2004). Since the UPR causes a general translational repression of zeins, these mutants show an increase in lysine though less pronounced than in o2. The floury1 (fl1) mutation is somewhat different, since it does not affect the amount or composition of zein proteins but rather results in the abnormal placement of α-zeins within the protein body (Holding et al., 2007).
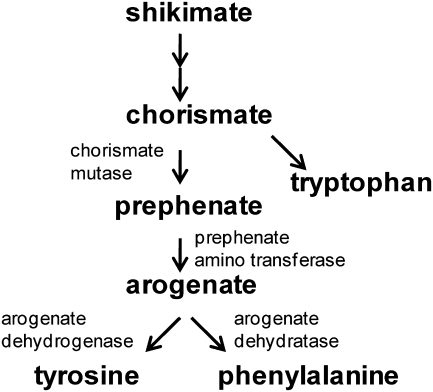
The characterization of additional starchy endosperm mutants can further our understanding of the mechanisms that lead to the formation of vitreous endosperm, and shed light on general processes of seed metabolism. Here a novel opaque mutant which co-segregates with a Mutator insertion in an arogenate dehydrogenase gene is described. This gene functions in the final steps of the aromatic amino acid pathway that produces tyrosine (Fig. 1). The arogenate route to tyrosine and phenylalanine is one of two distinct pathways that exist in plants, yeast, and bacteria (Stenmark et al., 1974). It involves the conversion of arogenate into tyrosine by arogenate dehydrogenase and into phenylalanine by arogenate dehydratase. The other route involves the transformation of prephenate into p-hydroxyphenyl pyruvate or phenylpyruvate, which are then transaminated into tyrosine or phenylalanine, respectively. Bacteria and yeast use various combinations of the above pathways, often resulting in complex feedback regulation (Legrand et al., 2006). Most plants exclusively use the arogenate pathway to synthesize tyrosine and phenylalanine, which are essential for protein synthesis and many phenylpropanoid compounds. The partitioning between tyrosine and phenylalanine is controlled by the sensitivity of arogenate dehydrogenase to feedback inhibition by tyrosine (Rippert and Matringe, 2002a, b). Control of the end-product is critical given that phenylalanine is the primary substrate for lignin biosynthesis. In this study, the identification and characterization of mutants in AroDH-1 and AroDH-3 genes which exhibit non-lethal opaque endosperm phenotypes are described, and this is discussed in the context of the redundantly expressed arogenate dehydrogenase gene family.
Fig. 1.
Aromatic amino acid pathway in plants.
Materials and methods
Cloning of arodh mutants
AroDH-1 was cloned using selective amplification of insertion-flanking fragments (SAIFF; Muszynski et al., 2006) according to the conditions previously described (Holding et al., 2007). A DNA fragment segregating with the mutant phenotype was identified with the +2 primer BfaIntGCA. For co-segregation analysis of the 50 additional mutant DNA samples, a gene-specific primer (5′-CCTTCCTCAACATCTTCGAGCGCGAGG-3′) was used with the TIR primer to amplify an ∼200 bp fragment.
For the TUSC (Trait Utility Screening Collection) screen, primers flanking the insertion site in arodh-1, DO133334 (5′-CACAAGCAAAGATGCTGTCGTCTCCAC-3′) and DO133335 (5′-GCCGTCGAAGAGCCTCTTCTTGAC-3′), were used in conjunction with the Mu TIR primer, DO9242 (5′-AGAGAAGCCAACGCCAWCGCCTCYATTTCGTC-3′). MU-flanking PCR fragments were cloned into the TOPO vector (Invitrogen) and sequenced using M13 F and R primers.
Phylogenetic analysis
AroDH-1, -2, -3, and -4 proteins were used to search the plant portion of Uniprot and the sorghum gene models. Seventeen complete and non-redundant sequences were identified in Uniprot and three additional sorghum proteins were identified in the sorghum set. The 24 protein sequences were aligned with ClustalW, trimmed at both the N- and C-termini to remove non-homologous regions, and a phylogenetic tree was built using the Phylogenator application (builds a consensus Neighbor–Joining tree using phylip and adds branch lengths using maximum likelihood).
Expression analysis
Six 20 DAP (days after pollination) kernels were dissected from each of three biological replicate ears taken from greenhouse-grown, true breeding arodh-1 mutant and wild-type plants. Total RNA was extracted from endosperm and embryo tissue from the six-kernel pools, as well as leaf tissue taken from the corresponding plants. The RNA extraction protocol was as previously described (Holding et al., 2008). cDNA was synthesized from 1 μg samples of DNase I-treated total RNA using IScript plus (Bio-Rad, Hercules, CA, USA), according to the manufacturer's instructions, and cDNAs were diluted 10-fold in water. For conventional RT-PCR, cDNAs were amplified using GC polymerase according to the manufacturer's instructions including a 3 min extension step at 68 °C. Primers were designed for 68–70 °C annealing temperatures as follows: AroDH-1 F, 5′-GGAGTTCCCCAAGAACCTGCTGCTCAGCTCCC-3′; AroDH-1 R, 5′-CCGGCAGCCCTCGCGCTCGAAGATGTT-3′; AroDH-2 F, 5′-GGAGGGCCCGGCGGAGCGCACG-3′; AroDH-2 R, 5′-CATTCTTCTGCTCTCCCGACTCGTCGCCGTC-3′; AroDH-3 F, 5′-GACGGACCACTCGGCGGCGGCGTCC-3′; AroDH-3 R, 5′-GCTCGAACACCAGCGGCAGGCCGTCCC-3′; AroDH-4 F, 5′-CAATATCAATGCCACAGAGCAGCTCGACAGGCTGG-3′; and AroDH-4 R, 5′-GTCGATAAGTGGTGCGCTGCAGGAGAACTGGAC-3′.
Real-time PCR primers were designed to amplify a 100–200 bp region using the primer 3 software available at http://frodo.wi.mit.edu/. All primers were designed for a 60 °C annealing temperature, and their sequences are as follows: AroDH-1 F, 5′-AATTCTCCTTGCCATTGTATTTGT-3′; AroDH-1R, 5′-TCATCCCTCAACTGTAAAACTGAA-3′; AroDH-2F, 5′-CTTGCATGCTTGTTTGATCTTATC-3′; AroDH-2R, 5′-TGATGATATTCTTGTCATGTCACAGTT-3′; AroDH-3F, 5′-CATCCTCCCAATGTCATATGTTTA-3′; and AroDH-3R, 5′-ACTGCCGCTTTATTCATTCTCTAC-3′.
A My iQ Real-time PCR thermocycler (Bio-Rad) was used with the following program: 95 °C for 5 min, followed by 45 cycles of 95 °C (10 s), and 60 °C (10 s) with 20 C s−1 ramp rates. Melting curves were obtained by heating from 65 °C to 95 °C with a 0.1 C s−1 ramp rate. Expression levels of AroDH-2 and -3 genes were calculated relative to the reference (AroDH-1) for each tissue type (Fig. 4B). Tissue-specific expression levels for each gene were calculated relative to the reference (endosperm expression level) for each gene (Fig. 4C). The internal control gene was glyceraldehyde phosphate dehydrogenase (GAPDH) which showed minimal variation between samples. For each expression calculation, the average crossing temperature (Ct) value was determined for three biological replicate samples of arodh-1 and the wild type for each tissue type. Relative expression was calculated using the following equation, where X=subject gene, C=control gene (GAPDH), W=average Ct of three reference samples, and G=average Ct of three subject samples: 2[(WX–WC)–(GX–GC)]. The final fold change values shown are the mean of three independent experiments (±SD).
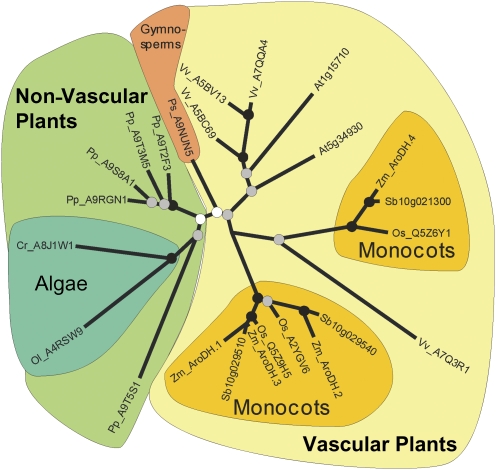
Fig. 4.
Phylogenic analysis of maize and other plant AroDH genes. Genus-species codes were prepended to the accessions and names: At, Arabidopsis thaliana; Cr, Chlamydomonas reinhardtii; Os, Oryza sativa; Ol, Ostreococcus lucimarinus; Pp, Physcomitrella patens; Ps, Picea sitchensis; Sb, Sorghum bicolor; Vv, Vitis vinifera; and Zm, Zea mays. Coloured backgrounds denote taxonomic groups as follows: light green designates non-vascular plants (moss and algae); light yellow designates vascular plants (gymnosperms and angiosperms); blue-green designates algae (Chlamydomonas and Ostreococcus); peach designates gymnosperms (Picea); and gold designates monocots (Orzya, Sorghum, and Zea).
Amino acid profiling
For analysis of metabolites including free amino acids in leaf tissues, aerial parts of 10 seedlings, grown in the greenhouse for 14 d, for each genotype were frozen in liquid N2 and lyophilized before being processed for metabolic profiling. Metabolites were extracted from three lyophilized leaf discs of each biological replicate, which were weighed and homogenized in 500 ml of chloroform:methanol:water (2:5:2, v/v/v) containing 0.015 mg of ribitol internal standard and subsequently rotated at 4 °C for 30 min. Samples were centrifuged at 1454 g for 15 min at 4 °C and 300 ml aliquots of the supernatant were lyophilized. Samples were resuspended in 50 μl of 20 mg ml−1 methoxyamine hydrochloride in pyridine and incubated with shaking at 30 °C for 90 min to form methoxyamine derivatives. To each sample, 80 ml of N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA) was added to each sample to form trimethylsilyl derivatives. The MSTFA delivery to individual samples was performed by the gas chromatograph autosampler 30 min prior to injection, to minimize differences in the state of derivatization. Trimethylsilyl derivatives were separated by gas chromatography on a Restek 30 m×0.25 mm i.d.×0.25 μm film thickness Rtx®-5Sil MS column with a 10 m integra guard column. Injections (1 ml) were made with a 1:10 split ratio using a CTC Combi PAL autosampler. The Agilent 6890N gas chromatograph was programmed for an initial temperature of 80 °C for 5 min, increased to 350 °C at 18 °C min−1 where it was held for 2 min before being cooled rapidly to 80 °C in preparation for the next run. The injector and transfer line temperatures were 230 °C and 250 °C, respectively, and the source temperature was 200 °C. Helium was used as the carrier gas with a constant flow rate of 1 ml min−1 maintained by electronic pressure control. Data acquisition was performed on a Leco Pegasus III time-of-flight mass spectrometer with an acquisition rate of 10 spectra s−1 in the mass range of m/z 45–600. An electron beam of 70 eV was used to generate spectra. Detector voltage was 1725 V. An instrument autotune for mass calibration using PFTBA (perfluorotributylamine) was performed prior to each GC sequence. GC/TOF-MS data were acquired with Leco ChromaTof software. Peaks were identified using a baseline offset of 0.5, a peak width of 2 s, a signal:noise of 10, and a smoothing factor of five data points. Metabolites were identified based on matching to a reference library created from authentic standards. Metabolites were quantified based on baseline-corrected heights of specified m/z values. Metabolite quantifications were normalized for internal standard (ribitol) amount and sample dry weight, and expressed as means ±SEM. Metabolite fingerprints, represented as an m/z value×retention time matrix, were also normalized for internal standard amount and sample dry weight. Metabolite fingerprint data were processed with Genedata Refiner MS software.
For analysis of protein-bound amino acids in mature endosperm, pools of 10 vitreous and opaque kernels were taken from each of four wild-type and mutant ears segregating for arodh-1. All ears were derived from the same parent and grown in the same row in the field in Tucson, Arizona. The embryos were removed with a small hand-held drill and the remaining endosperm and pericarp portions of the kernel were ground to a fine powder with a wigglebug dental amalgamator; traces of residual moisture were removed by lyophilization. The amino acids were profiled at the Pioneer Hi-Bred Metabolomics Facility using standard targeted analysis procedures. In brief, 100 mg aliquots of each sample were subjected to acid hydrolysis in 6 N HCl (except for tryptophan analysis, for which samples were hydrolysed in 4.2 N NaOH) followed by derivatization with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, reversed-phase HPLC separation of the resulting fluorescent amino acid derivates, and identification and quantification of amino acid derivatives using internal (α-aminobutyric acid) and external standards. The cumulative quantity of all protein amino acids detected was used as a proxy to calculate the protein content of each analysed tissue. For developing endosperm and embryo measurements, 20 DAP ears from three greenhouse-grown sibling wild-type and homozygous mutant plants (derived from the same segregating ear) were frozen in liquid nitrogen. Parent plants were genotyped using primers DO133334 and DO133335 for the wild-type allele and DO133335 and the MU internal primer, DO9242, for the Mu insertion allele. Kernels stored at –80 °C were defrosted enough to dissect out the endosperm and embryo before being immediately refrozen in liquid nitrogen and lyophilized. Pooled lyophilized tissues were ground to a fine powder using a 2000 Geno/Grinder ball mill homogenizer. Amino acid profiles were generated as described above, and the values were calculated relative to tissue dry weight rather than total protein. This is because the mutation affects the abundance of several aromatic amino acids and, consequently, overall protein synthesis. Expressing values relative to dry weight shows the changes in amino acids, whereas expression relative to total protein tends to mask them.
Results
Identification of maize arogenate dehydrogenase genes
Mutator-tagged opaque 140 (mto140) was one of several opaque endosperm mutants identified during a screen of Pioneer Hi-Bred's TUSC (Bensen et al., 1995). The mutation was introgressed into W64A (BC6) at the University of Arizona. mto140 grows more slowly than wild-type plants in the greenhouse (Fig. 2A), and has severely retarded growth and reduced seed production under Arizona field conditions (data not shown). The mature kernel phenotype manifests a marked reduction in the thickness of the vitreous endosperm layer (Fig. 2B) and has a reduced capacity to transmit light (opacity), as shown in Fig. 2C. Like some opaque mutants, mto140 has reduced zein protein accumulation, but, unlike o2, it affects accumulation of all zein classes (Fig. 2E), perhaps being indicative of a general effect on protein synthesis. Similar to the opaque5 (o5) and opaque 9 (o9) mutants, mto140 expresses a viridescent seedling phenotype (data not shown).
Fig. 2.
arodh-1 and arodh-3 mutant phenotypes. (A) Phenotype of greenhouse-grown wild-type and arodh-1 isogenic plants (left three, AroDH-1/AroDH-1; right three, arodh-1/arodh-1). (B) Endosperm phenotypes shown in cracked kernels (left, WT; centre, arodh-1; right, arodh-3). (C) Illuminated kernels from an AroDH-1 segregating ear shown on a light box (opaque kernels on the bottom row). (D) Illuminated kernels from an AroDH-3 segregating ear shown on a light box (opaque kernels on the bottom row). (E) SDS–PAGE separation of the zein fraction from vitreous and opaque kernels of an AroDH-1 segregating ear.
To identify the Mu insertion responsible for the mutant phenotype of mto140, SAIFF (Muszynski et al., 2006) was performed. Single kernels from 10 true breeding homozygous mutant ears and 10 true breeding non-segregating ears (derived from a single segregating ear) were germinated and DNA extracted from leaf tissue. Perfect co-segregation with the mutant phenotype was observed for a PCR product of ∼1150 bp. This DNA fragment was sequenced, revealing 1106 bp of flanking sequence between the Mu end and the adaptor primer sequence. PCR amplification with a gene-specific primer facing the Mu insertion and the Mu TIR primer (see Materials and methods) confirmed perfect co-segregation of the Mu insertion with the opaque phenotype in 50 additional mutants tested. The SAIFF sequence matched a 1477 bp cDNA (GenBank accession no. BT085368.1) with a 1080 bp open reading frame encoding 360 amino acids. Comparison with maize genomic sequences revealed the absence of introns in the gene. Blast N and P searches showed a high degree of identity with plant arogenate dehydrogenases, so this gene was designated zmAroDH-1 and the mutant was designated zmarodh-1. The Mu insertion site is 895 bp downstream of the ATG start codon and is predicted to interrupt the protein in its C-terminal domain (Fig. 3), which is predicted to be involved in dimerization (Legrand et al., 2006) (see Discussion).
Fig. 3.
Amino acid sequence of the maize genes encoding arogenate dehydrogenase. (A) Highly conserved region of AroDH-1 and -3 open reading frames showing the Mutator ‘hotspot’. (B) ClustalW amino acid alignment of predicted maize arogenate dehydrogenase proteins. The black shading with white letters indicates residues conserved in all four members, whereas grey shading indicates conservation between two or more of the family members. Solid white line boxes indicate 100% conservation with AroDH genes from other plants, bacteria, and yeast, whereas dashed white boxes indicate close to 100% conservation (Legrand et al., 2006). An asterisk under the conserved histidine residue at position 207 is predicted to be an essential part of the active site (Legrand et al., 2006) and the black arrow at position 223 shows the predicted start of the α-helix-rich, C-terminal dimerization domain (Legrand et al., 2006). The region corresponding to mutator insertion sites indicated in AroDH-1, -2 and -3 is shown with arrows.
Because of the similar opaque kernel and vegetative phenotypes of arodh-1 and o5 and o9, which map to chromosome 7 and 5, respectively, allelism tests were performed with these mutants potentially to uncover other alleles of zmarodh-1. For this purpose, homozygous plants of o5 and o9 were crossed with homozygous W64A arodh-1 parents. However, all kernels of the resulting hemizygous o5/+, arodh-1 /+ ears and of the o9/+, arodh-1 /+ ears showed a vitreous (i.e. wild-type) endosperm phenotype, indicating that neither o5 nor o9 is allelic to arodh-1.
A reverse genetics screen of the TUSC population was then conducted to identify additional mutations in AroDH-1, and two additional putative mutant alleles were found. Upon sequencing the Mu insertion sites in these genes, it was found that although the regions containing the primer-binding sites were identical, the Mu-flanking sequences were less than a perfect match with AroDH-1. An analysis of the B73 draft maize genome sequence indicated that mutant alleles of two very closely related gene family members had been isolated (AroDH-2 and AroDH-3; Fig. 3). The computational analysis of the B73 maize genome revealed a small family of maize arogenate dehydrogenase genes, consisting of (at least) four members (AroDH-1–AroDH-4). The DNA and protein sequence similarities shown in Fig. 3 and Table 1 demonstrate that AroDH-1 and AroDH-3 are the most closely related genes. A search of Pioneer's in silico transcript profiling data obtained through massively parallel signature sequencing (MPSS) revealed that AroDH-1 and AroDH-3 are the dominantly expressed genes in most maize tissues, including endosperm and leaf, whereas AroDH-2 and AroDH-4 are expressed in only a few tissues and at much lower levels (Supplementary Table S1 available at JXB online). Notably, AroDH-2 expression was not found in endosperm by MPSS, but it was detected at a low level using both quantitative and non-quantitative RT-PCR (see below). Consistent with AroDH-2 playing little or no functional role in the endosperm, the Mu insertion mutant arodh-2 (Fig. 3) resulted in no kernel phenotype, i.e. the kernels were vitreous. Moreover, an adult plant phenotype was also not observed. In contrast, the arodh-3 mutant (Fig. 3) had a strong opaque kernel phenotype (Fig. 2B, D). Like arodh-1, arodh-3 mutant plants also showed slightly retarded growth in the greenhouse (data not shown). Interestingly, in arodh-3 the Mu transposon was found in virtually the same position as in arodh-1 in a sequence that is almost perfectly conserved between the two genes (Fig. 3). However, despite analogous insertions in these highly similar family members and the similar phenotypes, since there are not multiple mutant alleles in each gene, it may not be formally concluded that these insertions cause the observed opaque kernel and vegetative phenotypes.
Table 1.
Nucleotide and amino acid similarities in the maize AroDH family
| AroDH-1 | AroDH-2 | AroDH-3 | AroDH-4 | |
| Accession no. | BT085368.1 | LOC100284089 | LOC100281078 | BT068692.1 |
| Transcript (bp) | 1477 | 1417 | 1336 | 1327 |
| Protein (amino acids) | 360 | 406 | 349 | 392 |
| ID to AroDH-1 mRNA (coverage and max ID) | 73% and 85% | 83% and 93% | 65% and 71% | |
| ID to AroDH-1 protein (identities and positives) | 67% and 77%a | 88% and 91%a | 62% and 76%a | |
| ID to AroDH-2 mRNA (coverage and max ID) | 66% and 86% | 58% and 69% | ||
| ID to AroDH-2 protein (coverage and max ID) | 67% and 75%a | 52% and 66%a | ||
| ID to AroDH-3 mRNA (coverage and max ID) | 67% and 70% | |||
| ID to AroDH-3 protein (coverage and max ID) | 61% and 76%a |
Upstream and downstream divergent regions not included in the comparison.
The AroDH-1 and AroDH-2 genes are found on chromosome 5 in opposite orientations (head to head), only ∼20 kb apart, indicating that they arose from a gene duplication event. Despite the fact that AroDH-3 is located on chromosome 6, it is more similar to AroDH-1 than to AroDH-2, suggesting that it arose from a duplication of an AroDH-1/-3 progenitor by a more recent event than that giving rise to the neighbouring AroDH-1 and AroDH-2 genes. Phylogenetic analysis (Fig. 4) of AroDH-1 and AroDH-3 suggests that these genes emerged relatively recently as paralogues and are therefore unlikely to be the result of the ancient tetraploidization event that has been postulated to have occurred ∼11 Mya in the lineage that led to the Zea genus (Gaut and Doebley, 1997). Figure 4 also shows that the more ancient AroDH-1–AroDH-2 split is shared by other grasses, including sorghum, where the orthologues show the same head to head orientation. The locations, short arm of chromosome 5 for AroDH-1 and AroDH-2 and short arm of chromosome 6 for AroDH-3, are not syntenous based on SyMap (Soderlund et al., 2006). In addition, the neighbouring genes are not similar and there is no evidence for a remnant AroDH-2-like sequence near AroDH-3. AroDH-1, AroDH-2, and AroDH-3 are each encoded by single exons. The AroDH-4 gene, located on chromosome 9, is more divergent and consists of five exons. The phylogeny of the arogenate dehydrogenase genes from various plant species indicates that monocot genes form two distinct clades with a deep division between them. Consistent with the gene structures, AroDH-1, AroDH-2, and AroDH-3 from maize are found in one of the two monocot clades and AroDH-4 is found in the other.
Expression of maize arogenate dehydrogenase genes
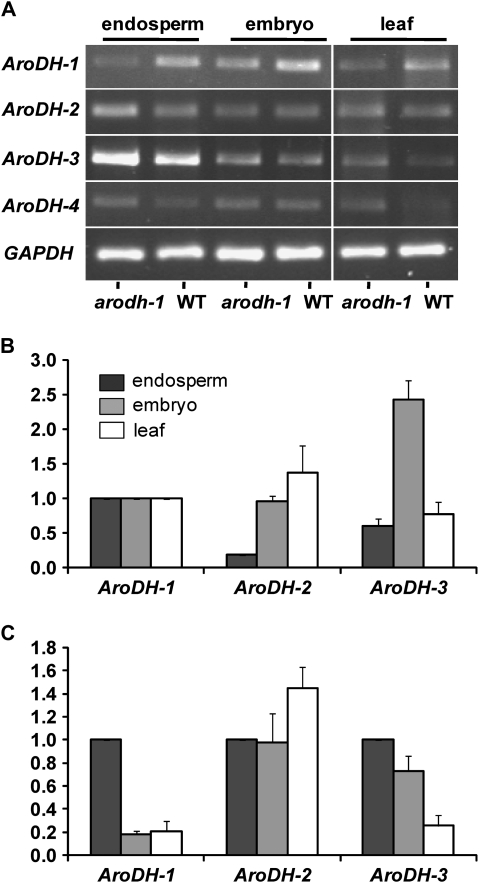
Given that the Mu insertion in arodh-1 occurs in a region encoding the C-terminal domain of the protein and that the mutant phenotype is non-lethal, it was of interest to determine whether a transcript could be detected. A truncated protein could retain some enzymatic function, since the predicted active site (Legrand et al., 2006) is intact, and only the predicted dimerization domain (Legrand et al., 2006) is disrupted. Also, since the maize arogenate dehydrogenase family is predicted to have redundant function, it was of interest to determine whether a compensatory increase in the expression of other gene family members could be detected in the arodh1 mutant. Using RT-PCR, expression of AroDH-1, AroDH-2, AroDH-3 and AroDH-4 was detected in endosperm, embryo, and leaf tissues of both arodh-1 and wild-type plants (Fig. 5A). Primers upstream of the Mu insertion where the genes are very GC rich were designed, necessitating use of GC polymerase (Clontech) and high annealing temperatures for the PCRs. Since ethidium bromide staining of stationary phase RT-PCR products is not suitable for detecting quantitative differences in transcript abundance, it was not possible to draw conclusions on the relative expression of AroDH family members from this analysis. Therefore, real-time RT-PCR was used to quantify expression of the AroDH genes in these tissues. In this case, primers were designed to the divergent 3′-untranslated regions so annealing temperatures compatible with the real-time PCR were possible. Despite several different designs, AroDH-4 primers were not suitable for real-time PCR, since they gave multiple products. The expression of each gene was measured in the arodh-1 background and it was expressed relative to the wild type for each tissue type. This analysis showed that, as expected, no transcript is produced downstream of the Mu insertion site in arodh-1 and, furthermore, that AroDH-2 and AroDH-3 expression is not increased in arodh-1 mutant tissues (not shown). Wild-type endosperm, embryo, and leaf RNAs were then used to address the relative expression levels of AroDH-2 and AroDH-3, in comparison with AroDH-1, and to determine the tissue specificity of each gene family member. In Fig. 5B the expression of AroDH-2 and AroDH-3 in each tissue relative to the AroDH-1 expression level for each tissue type is shown. In endosperm, AroDH-2 is expressed at 18% the level of AroDH-1, and AroDH-3 at 60% of that level. AroDH-2 has the highest relative expression of the three genes in the leaf tissue, and AroDH-3 has the highest relative expression of the three genes in the embryo tissue. In Fig. 5C the expression of each gene in embryo and leaf tissue relative to its own endosperm expression level is shown to demonstrate the tissue specificity of each gene. These data show that AroDH-1 is predominantly an endosperm-expressed gene, whereas AroDH-2 shows little tissue specificity, although it is most highly expressed in the leaf. AroDH-3 expression is biased towards the seed and shows a greater relative embryo expression than does AroDH-1.
Fig. 5.
Expression of AroDH family genes in maize tissues. (A) RT-PCR analysis of AroDH-1–AroDH-4 in arodh-1 mutant and wild-type tissues. (B) Real-time qRT-PCR analysis of AroDH-2 and AroDH-3 showing their expression relative to AroDH-1 expression in endosperm, embryo, and leaf tissues (shown as 1 relative expression unit). (C) Real-time qRT-PCR analysis of embryo and leaf expression relative to endosperm expression for AroDH-1, AroDH-2, and AroDH-3 (each gene relative to its own endosperm expression level; 1 relative expression unit). The key (black=endosperm, grey=embryo, white=leaf) for B also applies to C.
Amino acid profiling
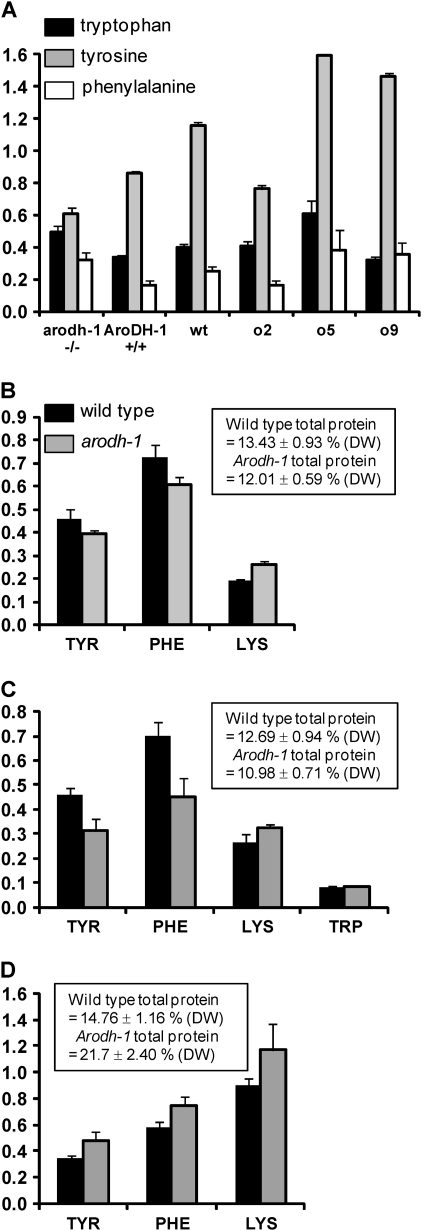
The arodh-1 mutant displayed a stunted vegetative growth phenotype under field conditions that is similar to, although less severe than, the phenotype of two other pleiotropic opaque mutants, o5 and o9. Since it was determined by test crossing that arodh-1 is not allelic to either o5 or o9 (see above), it was hypothesized that the three gene products may be linked in metabolic networks and that the mutants could therefore share similar metabolite profiles. To investigate this question, and in particular if the common growth phenotypes could be linked to a disruption to the aromatic amino acid pathway in arodh-1, metabolomics analysis was conducted on aerial plant material of these three mutants and on W64Ao2 and W64A wild type as controls. Based on the compiled data set of relative metabolite concentrations (Supplementary Table S2 at JXB online) a principle component analysis (PCA) was performed. This analysis (Supplementary Fig. S1 at JXB online) showed two major clusters. One cluster contained the o5 and o9 samples and the second contained the W64A+, o2, and arodh-1 samples. PCA therefore seems to confirm the suspected phenotypic similarity of the o5 and o9 mutants (both mutants were previously shown to exhibit similar transcript profiling characteristics; Hunter et al., 2002), but on the same grounds seems not to support a metabolic relationship between either O5 or O9 and the AroDH-1 gene product. The metabolomics analysis revealed the levels of aromatic amino acids in seedling leaf tissue. Figure 6A shows the levels of tryptophan, tyrosine, and phenylalanine in arodh-1 mutant tissue in comparison with three control tissues: AroDH-1 (wild-type isoline of arodh-1), W64A o2 (as an opaque kernel control), and W64A+ wild type, as well as W64Ao5 and W64Ao9. Of the three amino acids, tryptophan was the least variable, although a small increase was observed in arodh-1 and o5 compared with the controls, possibly indicative of an alteration in the flux through the pathway. Tyrosine showed reduced accumulation in arodh-1, suggesting that the mutation has an effect on the synthesis of this amino acid in leaf tissue. Phenylalanine accumulation was increased ∼2-fold in arodh-1 compared with the control tissues, possibly suggesting that reduced flux from arogenate to tyrosine causes a diversion from arogenate to phenylalanine. However, the phenylalanine increase may also reflect a general increase of all free amino acids except tyrosine in arodh-1 (Supplementary Table S2 at JXB online), and be symptomatic of a general reduction in protein synthesis. o5 and o9 both showed an even more pronounced general increase in leaf free amino acids, including tyrosine (Supplementary Table S2), suggesting that their reduced level of zein synthesis in the endosperm (Hunter et al., 2002) is part of a general reduction in protein synthesis in the whole plant. Interestingly, both o5 and o9 showed a 2-fold decrease in shikimate (Supplementary Table S2), an upstream precursor of the aromatic amino acids. It is not known if this represents a decrease in metabolites upstream of shikimate or an increase in turnover of downstream products (such as phenylalanine entering the phenylpropanoid pathway). o5 and o9 show highly similar metabolic profiles, suggesting that their functions may be closely related.
Fig. 6.
Amino acid profiles of arodh-1 mutant and wild-type tissues. (A) Seedling leaf tissue. Relative levels of free amino acids tryptophan (black bars), tyrosine (grey bars), and phenylalanine (white bars) extrapolated from metabolomics data of seedling leaf tissue. The y-axis shows relative amounts derived from peak heights of quantitative m/z values. Bars show standard errors of 10 biological replicates for each genotype. (B–D) Protein-bound amino acid content in seeds. The key in B also refers to C and D. Values shown are percentages of endosperm dry weight ±SD (three biological replicates). Insert boxes showing total protein are also expressed as the percentage of dry weight; tryptophan data were only available for developing endosperm samples. B, mature endosperm; C, 20 DAP developing endosperm; D, 20 DAP developing embryo.
Next accumulation of protein-bound amino acids in mature endosperm was measured. The levels of most protein-bound amino acids were similar between the wild type and arodh-1 although there were prominent reductions in glutamic acid, alanine, proline, and leucine in mutant endosperm (Supplementary Fig. S2A at JXB online). These amino acids are common in zeins, and are also reduced in opaque2 kernels, although to a greater extent than in arodh-1. There was a slight reduction in tyrosine and phenylalanine in mature arodh-1 endosperm (Fig. 6B). These reductions may simply reflect the slight reduction in total endosperm protein but, in the case of tyrosine, it could also be the result of a reduction in endosperm AroDH activity. Both of these amino acids are required for zein synthesis, and their reduction could contribute to the lower level of zeins. There was a 12% reduction in total endosperm protein in arodh-1 (see box in Fig. 6B), which reflects the drop in zein proteins and the failure of non-zein proteins to compensate completely. In common with other reduced zein mutants, such as opaque2, mature arodh-1 endosperm also has significantly higher levels of lysine (37% increase over wild type). Also in common with other opaque mutants, and perhaps symptomatic of reduced endosperm protein synthesis, general increases in free amino acids were detected in arodh-1 (data not shown).
In light of the existence of a redundantly expressed AroDH gene family and the metabolic plasticity of plant tissues, the amino acid profiles of developing seed tissues were also investigated to see if the effects of the mutation were more pronounced before seed maturity. In general, the amino acid profiles of developing endosperm were very similar to those of mature endosperm (Supplementary Fig. S2B at JXB online). However, while similar to mature endosperm, the reduced zein synthesis caused a 13.5% drop in total protein (box in Fig. 6C). Figure 6C shows that tyrosine and phenylalanine were reduced to a greater extent than in mature endosperm, possibly demonstrating an effect of their reduced biosynthesis. At this stage of endosperm development, though zeins are rapidly accumulating, they have not reached their final levels. Consequently, if the lower protein-bound tyrosine and phenylalanine levels were simply secondary effects of reduced zein synthesis, their reduced accumulation would be expected to be less pronounced than in mature endosperm. Lysine showed a 22% increase in developing arodh-1 endosperm (Fig. 6C), although this was less pronounced than in mature endosperm. In this analysis, it was also possible to measure tryptophan separately, which showed no significant difference between the wild type and arodh-1 (Fig. 6C). In o2 (Mertz et al., 1964) and low-zein transgenic lines (RJ, unpublished data), increases in lysine and tryptophan are closely correlated. The absence of an increase in tryptophan in arodh-1 suggests further evidence of alterations in the regulation and output of the aromatic amino acid pathway.
In developing arodh-1 embryos, there was a global increase in all protein-bound amino acids (Supplementary Fig. S2C at JXB online), and total protein was 47% greater than in wild-type embryos (see insert in Fig. 6D). This increase suggests that transport of nitrogenous compounds from endosperm to embryo is increased in response to reduced endosperm protein synthesis, which in turn triggers embryo protein synthesis. This compensatory effect extends to tyrosine and phenylalanine, both of which were increased in arodh-1 embryo tissue (Fig. 6D). Compared with AroDH-1, both AroDH-2 and AroDH-3 have greater embryo than endosperm expression in the wild type (Fig. 5), suggesting that in arodh-1 they are able to compensate for the lower endosperm levels of tyrosine, by increasing its abundance in the embryo. Consequently, this is a good demonstration of how a redundant gene family can maintain overall organ (seed in this case) homeostasis.
Discussion
The gene AroDH-1 gene is a member of a small family of at least four maize genes encoding arogenate dehydrogenase, the enzyme that synthesizes tyrosine from arogenate. There is high sequence conservation between gene family members (Table 1), with AroDH-1 and AroDH-3 exhibiting the greatest similarity (93% and 91% at the DNA and protein levels), suggesting that they resulted from gene duplication events of an ancestral gene (Fig. 4). The phylogenetic tree positions of AroDH-1 and AroDH-3 support the hypothesis that these genes emerged relatively recently as paralogues and are therefore unlikely to be the result of the ancient tetraploidization event that has been postulated to have occurred ∼11 Mya in the lineage that led to the Zea genus (Gaut and Doebley, 1997). The Mu transposon insertions in AroDH-1 and AroDH-3 occurred in virtually identical positions within their most highly conserved regions, suggesting either that Mu insertions at other sites result in lethal phenotypes and therefore escaped detection, or that the corresponding nucleotide sequence is a ‘hotspot’ for transposition. A transposition hotspot was also observed among different alleles of the fl1 mutant (Holding et al., 2007), and the existence of this phenomenon for genes located on different maize chromosomes suggests that it is governed at least in part at the sequence level, rather than at the level of chromosome architecture. The fact that Mu insertions at the same position in arodh-1 or arodh-3 result in the same opaque kernel phenotype supports the conclusion that the genes responsible for these mutant phenotypes have been identified. However, the absence of additional alleles of either arodh-1 or arodh-3 mutants means that it cannot be formally concluded that these insertions cause the observed opaque kernel and vegetative phenotypes.
The functional domain organization of TyrA synthase (AroDH) from Synechocystis was determined by a combination of biochemical and molecular modelling approaches (Legrand et al., 2006). The N-terminal region adopts a β-sheet structure and contains the NADP-binding and catalytic domains. NADP increases the affinity of the enzyme for arogenate. The C-terminal region is largely α-helical in structure and forms a dimerization interface. The enzyme exists largely as a homodimer with minor amounts of homotetramer. Dimerization of Arabidopsis arogenate dehydrogenase (TyrAAT2) was also shown to occur (Rippert and Matringe, 2002b). By aligning arogenate dehydrogenate amino acid sequences from several plant, bacteria, and yeast species, it was shown that although overall sequence conservation is low, there are key similarities in terms of globally conserved residues predicted to be essential in the active site and secondary structure characteristics (Legrand et al., 2006). All enzymes compared exhibit common NADP-binding domains and α-helical C-termini. The Mu transposons in arodh-1 and arodh-3 are inserted in virtually identical positions within the C-terminal region predicted to be involved in dimerization (annotated in Fig. 3). It is not known whether dimerization occurs nor whether the enzymes form homo- or heterodimers (which seems likely considering their amino acid similarity), but it is likely that the maize arodh-1 and arodh-3 proteins exhibit reduced or abolished dimerization activity. It could also be that the putative monomeric proteins encoded by the mutant genes retain some enzymatic activity. Discussions about different functional domains in AroDH proteins are based on the assumptions that their transcripts are translated into functional proteins.
All four AroDH gene family members are expressed in endosperm, embryo, and leaf tissues (Fig. 5A), although different tissues show variable expression, suggesting some degree of functional specialization (Fig. 5B, C). For example, AroDH-1 and AroDH-3 appear to encode the major endosperm-expressed forms, while AroDH-2 has the highest leaf expression. AroDH-1 and AroDH-3 are expressed at much lower levels in leaves than in endosperm (Fig. 5C). Although aerial plant growth of arodh-1 and arodh-3 mutants is severely reduced under Arizona field conditions, and moderately reduced under greenhouse conditions, they are still capable of completing their life cycle. This non-lethality may be attributable to the greater relative importance of AroDH-2 in leaf tissue, as indicated by it being the predominant leaf transcript. In leaf tissue, the arodh-1 mutant displays a slight decrease in tyrosine and a slight increase in phenylalanine (Fig. 6A), suggesting that this gene has some leaf function. AroDH-2 is expressed at a low level in the endosperm. Consistent with this, a Mu insertion in AroDH-2 in the region encoding the N-terminal NADP-binding and catalytic domains (annotated in Fig. 3) (Legrand et al., 2006) produced no apparent seed phenotype, suggesting that any AroDH-2 function in the endosperm may be dispensable. Future work will address the likely functional redundancy of AroDH-1, -2, and -3 by generating double and triple mutants.
A general increase in protein-bound amino acids and total protein was found in arodh-1 embryos. The arodh-1 mutant embryos also showed increased tyrosine levels (Fig. 6D). This suggests that while AroDH-1 is expressed to some extent in the embryo (Fig. 5), the combined activities of AroDH-2 and AroDH-3 completely mask any deficiency. Consistent with this, arodh-1 is not embryo lethal. Embryo expression of AroDH-3 is more substantial relative to AroDH-1 embryo expression (Fig. 5B) and has a greater expression relative to its own endosperm than does AroDH-1 (Fig. 5C). Moreover, the observed general increase in embryo proteins probably serves as a seed homeostasis mechanism to compensate for the general reduction in amino acid incorporation into endosperm proteins, mainly zeins. An overall deficiency in a single AroDH family member does not affect the general increase in all protein-bound amino acids in embryo tissue.
The majority of the maize opaque endosperm mutants that have been characterized result from quantitative or qualitative effects on specific zein proteins. However, a general reduction in zein synthesis occurs in a number of these mutants, including o5 and o9 (Hunter et al., 2002), and this is perhaps the cause of the starchy kernel phenotype of arodh-1 and arodh-3. The opaque6 mutant, which is allelic with the proline deficiency mutant, pro1 (Tonelli et al., 1986), is an example of a starchy endosperm phenotype that is associated with a defect in amino acid biosynthesis. Total free amino acid data were generated from leaf tissue, whereas the endosperm data showed total protein-bound amino acids. While it is difficult to relate these data sets adequately, especially given the differential activity of the AroDH family members in these tissues, they do suggest a disruption in tyrosine synthesis in the arodh-1 mutant. The metabolic profiling of arodh-1 leaf tissue showed that most free amino acids were increased (Supplementary Table S2 at JXB online), suggesting a reduction in incorporation into proteins. However, tyrosine was the only free amino acid that was reduced in abundance. This supports the hypothesis of a partial reduction in total arogenate dehydrogenase activity. By inference from their relative expression levels, other AroDH family members, especially AroDH-2, probably provide functional redundancy in leaf tissue. The reduction of tyrosine in the protein-bound endosperm fraction is only minor but may also result, in part, from partial blockage at arogenate dehydrogenase. However, since the endosperm protein-bound tyrosine reduction is less pronounced than is observed when measuring leaf free amino acids, the difference could be masked by a more general reduction in protein-bound amino acids in this tissue. The minor effect on tyrosine in endosperm proteins in arodh-1 also probably demonstrates the functional redundancy of AroDH-1 and AroDH-3 in this tissue, as indicated by their expression level (Fig. 5B, C).
Since zein proteins contain substantial amounts of tyrosine, a reduction in the abundance or activity of arogenate dehydrogenase could reduce the rate of zein synthesis, thereby leading to an opaque kernel phenotype. However, it is surprising that an apparently small reduction in tyrosine could result in major changes at the protein level. This suggests there may be other unidentified consequences of the mutation that contribute to the endosperm phenotype. Since the aromatic amino acid pathway is involved in production of many important molecules, including auxin and lignin, its perturbation could have multiple effects, making it difficult to predict the consequence of alterations in amino acid flux.
The functional redundancy of the AroDH gene family complicates attempts to explain the kernel phenotype. Extrapolating from gene expression levels, it is hypothesized that AroDH-1 and AroDH-3 are the major functional endosperm genes, although enzymatic data to support this are not available. It appears that functional enzymes encoded by both genes are necessary to maintain zein synthesis and support the formation of vitreous endosperm. This could be because both enzymes are required for an optimal level of amino acid biosynthesis or that a heterodimer composed of both enzymes is important in the endosperm.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Plot of principle component analysis (PCA) scores of leaf tissue metabolite profiles obtained from W64A arodh-1, W64A AroDH-1 wild-type isoline, W64Ao2, W64Ao5, W64o9, and W64A wild type. PCA was performed on the m/z value×retention time matrix of metabolite fingerprinting data following Z-scoring using the Eigenvector PLS Toolbox software. Group boundaries for samples of each genotype are shown with solid lines. The W64Ao5 and W64Ao9 sample groups form a cluster that is distinct from W64Aarodh-1, which overlaps with sample groups of W64A wild type and W64Ao2.
Figure S2. Protein-bound amino acid profiles of arodh-1 mutant and wild-type tissues. (A) Mature endosperm; (B) 20 DAP developing endosperm; (C) 20 DAP developing embryo. The key in A also refers to B and C. Values shown are percentages of endosperm dry weight ±SD (three biological replicates). Insert boxes showing total protein are also expressed as the percentage dry weight; tryptophan data were only available for developing endosperm samples.
Table S1. Expression levels from massively parallel signature sequencing (MPSS) analysis of zmAroDH genes in wild-type B73 maize tissues. Seed tissues are from 22 DAP kernels, and black bars represent no detectible expression
Table S2. Metabolic profiling of arodh-1 and other similar opaque mutants. Relative abundances of known metabolites for the 10 biological replicate leaf samples each for W64A arodh-1, W64A AroDH-1 wild-type isoline, W64Ao2, W64Ao5, W64o9, and W64A wild type. Data were generated from signature mass (m/z value)/retention time combinations for each metabolite from the fingerprint data. Data were normalized for both internal standard signal and sample dry weight. Worksheet 1 shows raw data, worksheet 2 shows averages, and worksheet 3 shows values normalized to the AroDH-1 wild-type isoline (values set at 1). Colours represent a heat map to illustrate commonalities in metabolite abundance between genotypes, where red means levels lower than the AroDH-1 wild-type isoline (yellow) and green means higher levels. Zero abundance is represented by deep red, between 0 and 1 by shades between red and yellow, between 1 and 2 by shades between yellow and green, and >2 by dark green. Bold lines show all amino acids except cysteine.
Supplementary Material
Acknowledgments
We thank Craig Coleman, Dwight Bostwick, and Brenda Hunter for their participation in the identification and introgression of mto140. This research was supported by the Department of Agronomy and Horticulture of the University of Nebraska, grants to BAL from DOE (DE-96ER20242), NSF (DBI-00776760), and Pioneer Hi-Bred International a Du Pont Company.
References
- Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP. Cloning and characterization of the maize AN1 gene. The Plant Cell. 1995;7:75–84. doi: 10.1105/tpc.7.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrink-Kurtzman MA, Bietz JA. Zein composition in hard and soft endosperm of maize. Cereal Chemistry. 1993;70:105–108. [Google Scholar]
- Fox G, Manley M. Hardness methods for testing maize kernels. Journal of Agricultural and Food Chemistry. 2009;57:5647–5657. doi: 10.1021/jf900623w. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proceedings of the National Academy of Sciences, USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon BC, Wang XL, Larkins BA. Altered starch structure is associated with endosperm modification in quality protein maize. Proceedings of the National Academy of Sciences, USA. 2003;100:15329–15334. doi: 10.1073/pnas.2136854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding DR, Hunter BG, Chung T, Gibbon BC, Ford CF, Bharti AK, Messing J, Hamaker BR, Larkins BA. Genetic analysis of opaque2 modifier loci in quality protein maize. Theoretical and Applied Genetics. 2008;117:157–170. doi: 10.1007/s00122-008-0762-y. [DOI] [PubMed] [Google Scholar]
- Holding DR, Otegui MS, Li BL, Meeley RB, Dam T, Hunter BG, Jung R, Larkins BA. The maize floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. The Plant Cell. 2007;19:2569–2582. doi: 10.1105/tpc.107.053538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter BG, Beatty MK, Singletary GW, Hamaker BR, Dilkes BP, Larkins BA, Jung R. Maize opaque endosperm mutations create extensive changes in patterns of gene expression. The Plant Cell. 2002;14:2591–2612. doi: 10.1105/tpc.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CS, Hunter BG, Kraft J, Boston RS, Yans S, Jung R, Larkins BA. A defective signal peptide in a 19-kD alpha-zein protein causes the unfolded protein response and an opaque endosperm phenotype in the maize De*-B30 mutant. Plant Physiology. 2004;134:380–387. doi: 10.1104/pp.103.031310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand P, Dumas R, Seux M, Rippert P, Ravelli R, Ferrer JL, Matringe M. Biochemical characterization and crystal structure of Synechocystis arogenate dehydrogenase provide insights into catalytic reaction. Structure. 2006;14:767–776. doi: 10.1016/j.str.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Lending CR, Larkins BA. Effect of the floury-2 locus on protein body formation during maize endosperm development. Protoplasma. 1992;171:123–133. [Google Scholar]
- Mertz ET, Nelson OE, Bates LS. Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science. 1964;145:279–280. doi: 10.1126/science.145.3629.279. [DOI] [PubMed] [Google Scholar]
- Muszynski MG, Dam T, Li B, Shirbroun DM, Hou Z, Bruggemann E, Archibald R, Ananiev EV, Danilevskaya ON. Delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiology. 2006;142:1523–1536. doi: 10.1104/pp.106.088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippert P, Matringe M. Molecular and biochemical characterization of an Arabidopsis thaliana arogenate dehydrogenase with two highly similar and active protein domains. Plant Molecular Biology. 2002a;48:361–368. doi: 10.1023/a:1014018926676. [DOI] [PubMed] [Google Scholar]
- Rippert P, Matringe M. Purification and kinetic analysis of the two recombinant arogenate dehydrogenase isoforms of Arabidopsis thaliana. European Journal of Biochemistry. 2002b;269:4753–4761. doi: 10.1046/j.1432-1033.2002.03172.x. [DOI] [PubMed] [Google Scholar]
- Schmidt RJ, Burr FA, Aukerman MJ, Burr B. Maize regulatory gene opaque-2 encodes a protein with a leucine-zipper motif that binds to zein DNA. Proceedings of the National Academy of Sciences, USA. 1990;87:46–50. doi: 10.1073/pnas.87.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C, Nelson W, Shoemaker A, Paterson A. SyMAP: a system for discovering and viewing syntenic regions of FPC maps. Genome Research. 2006;16:1159–1168. doi: 10.1101/gr.5396706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark SL, Pierson DL, Jensen RA, Glover GI. Blue-green bacteria synthesize l-tyrosine by the pretyrosine pathway. Nature. 1974;247:290–292. doi: 10.1038/247290a0. [DOI] [PubMed] [Google Scholar]
- Tonelli C, Gavazzi G, Manzocchi L, Di Fonzo N, Soave C. Opaque 6 allelic to pro1 mutant. Maize Newsletter. 1986;60:100. [Google Scholar]
- Tsai CY, Huber DM, Warren HL. Relationship of kernel sink for N to maize productivity. Crop Science. 1978;18:399–404. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.