Abstract
Infections as a result of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) are an issue of increasing global healthcare concern. In Europe, this principally involves strains of multi-locus sequence type clonal complex 80 sequence type 80 with methicillin resistance in a staphylococcal chromosomal cassette (SCCmec) type IV arrangement (CC80:ST80-IV). As with other CA-MRSA strains, CC80:ST80-IV isolates tend to appear uniform when analysed by common molecular typing methods (e.g. pulsed field gel electrophoresis, multi-locus sequence type, SCCmec). To explore whether DNA sequence-based differences exist, we compared the genetic composition of six CC80:ST80-IV isolates of diverse chronological and geographic origin (i.e. Denmark and the Middle East) using an Affymetrix high-density microarray that was previously used to analyse CA-MRSA USA300 isolates. The results revealed a high degree of homology despite the diversity in isolation date and origin, with isolate differences primarily in conserved hypothetical open reading frames and intergenic sequences, but also including regions of known function. This included the confirmed loss of SCCmec recombinase genes in two Danish isolates representing potentially new SCCmec types. Microarray analysis grouped the six isolates into three relatedness pairs, also identified by pulsed field gel electrophoresis, which were consistent with both the clinical and molecular data.
Keywords: Community, microarray, ST80, Staphylococcus aureus
Introduction
Although healthcare-associated (HA) methicillin-resistant Staphylococcus aureus (MRSA) have been a subject of longstanding clinical concern, infections as a result of community-associated (CA) MRSA have now become an intense focus of interest and investigation [1–3]. Although CA-MRSA are globally distributed, specific strains continue to exhibit geographic predominance. In the USA, this is typified by isolates that are multi-locus sequence type (MLST) clonal complex 8, sequence type 8, staphylococcal cassette chromosome (SCCmec) type IV (CC8:ST8-IV), exhibiting the USA300 pulsed-field gel electrophoresis (PFGE) profile [4,5]. In Europe, the most common CA-MRSA strain is CC80:ST80-IV [6,7]. In Denmark, CC80:ST80-IV isolates are the predominant cause of CA-MRSA infections, with epidemiological studies [7,8] showing a large proportion of patients with family relationships in the Middle East. Individuals colonized when traveling abroad or visiting such high endemic areas have been suspected as likely sources of CC80:ST80-IV importation because the overall proportion of MRSA in Denmark is very low (approximately 0.1%) [7,9]. However, establishing direct transmission routes is challenging because of the conserved nature of CC80:ST80-IV genomic (e.g. PFGE) profiles [7], similar to other CA-MRSA, such as USA300 [4,10]. Although at least 17 PFGE subtypes have been identified in the Danish CC80:ST80-IV collection, there has been no specific association between specific subtypes and infections acquired domestically vs. those most likely acquired abroad [7]. Concern regarding the increased incidence of infections as a result of CA-MRSA has prompted investigations regarding their genetic composition, to better understand their potential for virulence and epidemic spread. In this context, microarrays have been a powerful tool for assessing the genomic presence or absence of important loci (e.g. regulatory, resistance, virulence or adhesion). For example, analysis of USA300 (ST8) in comparison to CA-MRSA USA400 (ST1) and HA-MRSA USA100 (ST5) and 500 (ST8) using a high-density microarray (i.e. 7775 loci) revealed a high degree of relatedness, especially between USA300 and USA500, with a set of 20 known or hypothetical genes unique to USA300 [4].
Past microarray analysis of CC80:ST80-IV by Monecke et al. [11,12] used 100 and 87 probes, respectively (e.g. genes for resistance and virulence) to compare isolates (12 from Germany, five from the UK and two from Switzerland) with a variety of S. aureus strains encoding the Panton–Valentine leukocidin (PVL), including the sequenced S. aureus USA400 strain MW2. These analyses revealed a diverse origin for pandemic PVL-positive strains but differences between CC80:ST80-IV isolates in only four plasmid-born antibiotic-resistance loci. Similarly, a recent study by Monecke et al. [13] comparing eight German CC80:ST80-IV isolates using 157 probes for resistance and virulence revealed differences only for plasmid-associated antibiotic resistance genes.
The S. aureus Affymetrix high-density microarray represents a powerful tool for genomic comparison because the 7775 loci include not only resistance determinants, toxins, virulence regulators and cell surface factors, but also hypothetical genes and intergenic sequences from the published S. aureus N315, Mu50, COL and NCTC8325 genomes [4,14]. Sung et al. [15] noted the importance of mobile genetic elements (e.g. plasmids, transposable elements and bacteriophages) in strain differentiation. In addition, as our understanding of microbial genomic organization, gene structure and function increases, sequences initially considered to be unimportant are finding new significance (e.g. phenol-soluble modulins) [16,17]. Thus, the present study aimed to use the S. aureus Affymetrix high-density microarray to investigate inter-relationships between six CC80:ST80-IV isolates obtained from 1997–2003, including one of the earliest (i.e. ‘ancestral’) entries in the Danish database, as well as isolates with possible ties to the Middle East (Lebanon and Egypt).
Materials and Methods
Bacterial isolates and susceptibility testing
Subsequent to 1988, Danish regional clinical microbiology departments have systematically referred all MRSA isolates to Statens Serum Institut. Based on hospital discharge summaries or notes from outpatient clinics and physicians, all patients with MRSA infections were evaluated for the potential origin of infection according to previously described criteria [9,18]. A total of 294 CC80:ST80-IV cases were registered (1988–2004), most of which were CA originating in Denmark. However, a large proportion of cases had family relationships in the Middle East [7]. For array investigation, six isolates spanning a 6-year period were chosen including an isolate from a patient infected during hospitalization in Egypt and one from a patient born in Lebanon. Four isolates caused CA infections (1198, 1200, 1202 and 1209) and one caused a health care-associated community-onset infection (1201). The remaining isolate (1199) was a surveillance culture (1199) from a patient transferred to Denmark from an Egyptian hospital with no record of earlier hospitalization for approximately 2 years. The isolates were unrelated as determined by epidemiological information. Susceptibility to cefoxitin, penicillin, streptomycin, tetracycline, erythromycin, clindamycin, fusidic acid, norfloxacin, kanamycin, rifampicin and linezolid was assessed using Neosensitabs® (Rosco, Taastrup, Denmark) on Danish Blood agar (SSI, Copenhagen, Denmark) [9]. Suspected methicillin/oxacillin resistance, predicted by cefoxitin test results, and fusidic acid resistance, was confirmed by detecting the mecA and fusB genes, respectively [19,20].
PFGE and PCR
Molecular characterization by PFGE and PCR analysis for the presence of the PVL genes, protein A gene (spa), accessory global regulator (agr), SCCmec and MLST were performed as described previously [7]. SCCmec typing was primarily conducted using the multiplex PCR method of Oliveira and de Lencastre [19] with additional ccr recombinase and mec typing as outlined by Kondo et al. [21] and Milheirico [22].
Microarray analysis
The S. aureus CC80:ST80-IV isolates were analysed using a commercially available S. aureus Affymetrix GeneChip® (Affymetrix, Santa Clara, CA, USA) as described previously [4,14]. Chromosomal DNA was interrogated for the presence or absence of the 7775 loci on the GeneChip®, which included resistance determinants, exoenzymes, exo- or enterotoxins and a variety of virulence regulators and cell surface factors from the S. aureus N315, Mu50, COL and NCTC8325 published genomes. Chromosomal DNA was purified from each of the CC80:ST80-IV isolates, fragmented, and biotinylated at the 3′ end [4,14]. Labelled DNA (1.5 µg) was hybridized to a GeneChip® and adjusted ‘present’ and ‘absent’ determinations were made for each array locus with an average of 20 probe sets per open reading frame (ORF) or intergenic region [4,14]. For adjusted calls, raw values were log transformed and normalized by dividing each value by the chip mean. Cut-off values for p calls were ≤0.89 = absent; ≥0.981 = present; and 0.9–0.98 = marginal.
Results
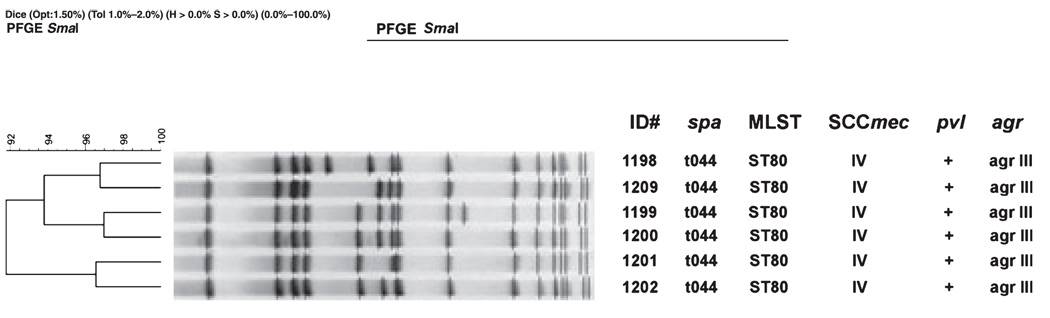
As shown in Table 1, the CC80:ST80-IV isolates were primarily associated with skin and soft tissue infections and were chosen to represent differences in year of isolation, potential geographic origin and antimicrobial susceptibility. Initial genotypic characterization (Fig. 1) revealed the expected homogeneity for spa, MLST, SCCmec (see below), PVL and agr type. Minor variations (>90% similarity) in PFGE patterns were consistent with published CC80:ST80-IV profiles. However, PFGE identified three subgroup pairs (approximately 96% relatedness) that linked the two Middle East isolates (1199 and 1200) cultured in 2001, Danish isolates 1201 and 1202 (cultured in 2003 and 2001, respectively) and Danish isolates 1198 and 1209, which were isolated in 1997 (i.e, one of the earliest CC80:ST80-IV in the database) and 2001, respectively.
TABLE 1.
CC80:ST80-IV isolates examined
| Isolate number |
Year of isolation |
Presumed country of origin |
Age of patient (years) |
Infection/screeninga | Antibiotic resistance patternb |
|---|---|---|---|---|---|
| 1198 | 1997 | Denmark | 21 | Skin and soft tissue infection (CA) | Ox, P, K, F |
| 1199 | 2001 | Egypt | 43 | Screening in Denmark after heart attack abroad (S) | Ox, P, S, T, K, F |
| 1200 | 2001 | Lebanon | 5 | Scratches infected during vacation in Lebanon (CA) | Ox, P, S, T, K, F |
| 1201 | 2003 | Denmark | 20 | Folliculitis at thigh, knee, and eyelid (HACO) | Ox, P, T, E, Cli, F |
| 1202 | 2001 | Denmark | 19 | Inflammation in axilla (CA) | Ox, P, T, F |
| 1209 | 2001 | Denmark | 10 | Skin abscesses (CA) | Ox, P, S, T, K, F |
CA, community-associated; S, surveillance; HACO, healthcare-associated community-onset.
antibiotic resistance detected against: Ox, oxacillin; P, penicillin; S, streptomycin; T, tetracycline; E, erythromycin; Cli, clindamycin; F, fusidic acid; K, kanamycin.
FIG. 1.
A summary of CC80:ST80-IV isolate molecular characteristics by pulsed-field gel electrophoresis (PFGE) and analysis by PCR for spa, MLST, SCCmec, pvl, and agr.
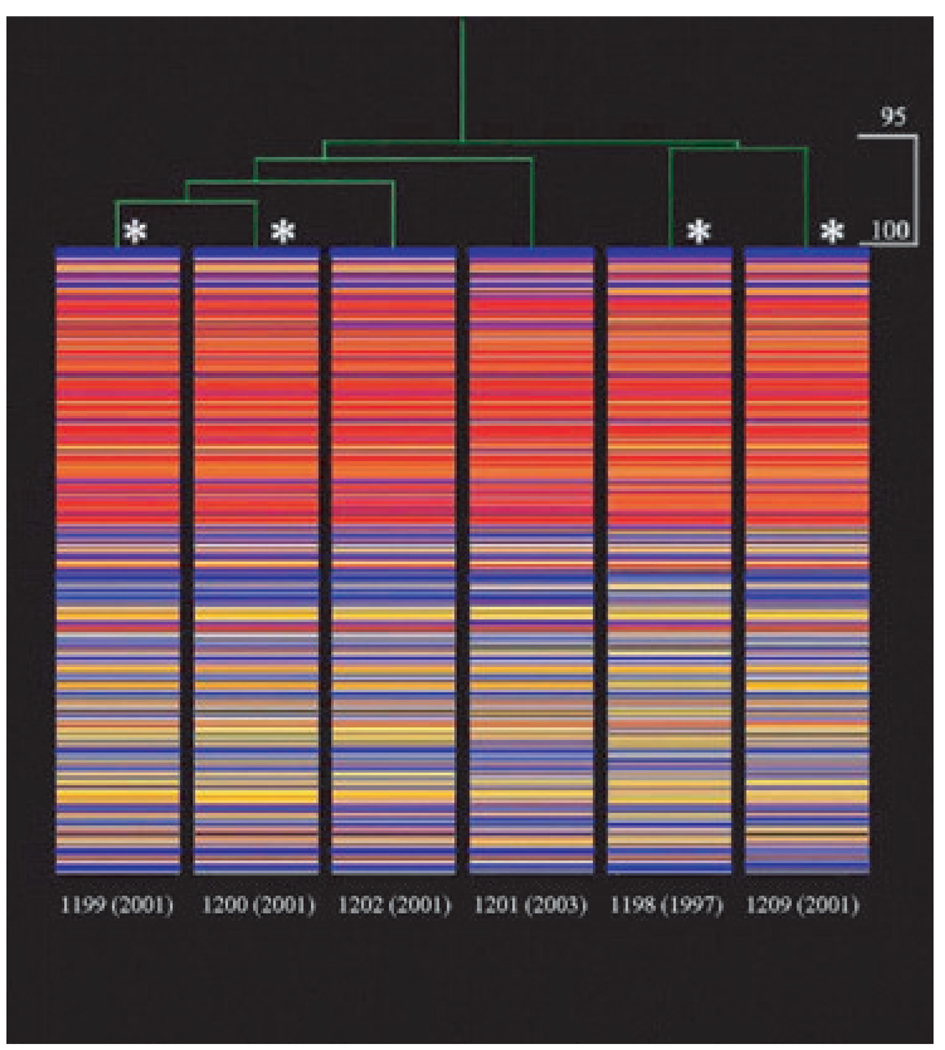
Overall, when analysed on the S. aureus Affymetrix Gene-Chip, DNA from the isolates hybridized to an average of 58% of the 7775 loci (i.e. 4489 ± 174). As shown in Fig. 2, although 95% related, the isolates appeared to group in pairs (i.e. 1199/1200, 1201/1202 and 1198/1209) similar to that seen by PFGE. Differences were primarily in conserved hypothetical ORFs and intergenic sequences as seen by pairwise analysis (Table 2), indicating the number of the 7775 queried loci present in one isolate but absent from another. This comparison confirmed that isolates 1199 and 1200 were the most similar, with only 73 instances (18 plus 55) where a locus found in one isolate was absent from the other. Isolate pairs 1201/1202 and 1198/1209 had 354 and 326 instances of nonshared loci, respectively. The inter-relationships were even more clearly seen with intergenic sequences removed from the analysis. As shown in Table 2, isolate pairs 1199/1200 shared all remaining 3514 ORFs, followed by isolate pair 1198/1209 with only 15 instances of nonshared loci. Isolate pair 1201 /1202 was more distantly related but lacked the SCCmec-associated recombinase (ccr) genes but retained mecA (see below).
FIG. 2.
Dendrogram (top) with heat map (beneath) for all loci that were analysed in each isolate. The dendrogram illustrates relatedness based on the signal intensity of each locus across all isolates. Within the heat map, each locus (total = 7775) is shown vertically for each strain. Red indicates high signal intensity; yellow indicates marginal signal intensity, and blue indicates low signal intensity. The order of loci is identical for all strains. For adjusted calls, raw values were log transformed and normalized by dividing each value by the chip mean. Cut-off values for p calls were ≤0.89 = absent; ≥0.981 = present; and 0.9–0.98 = marginal. Asterisks indicate isolates that especially clustered together by pairwise comparison (Table 2).
TABLE 2.
Pairwise comparison of CC80:ST80-IV isolates for the number of 7775 queried loci and the 3514 open reading frames (total loci minus intergenic sequences; shown in parentheses) that were present in one isolate but absent from another
| Loci present |
||||||
|---|---|---|---|---|---|---|
| Loci absent | 1199 | 1200 | 1202 | 1201 | 1198 | 1209 |
| 1199 | 0 (0) | 55 (0) | 63 (16) | 99 (3) | 145 (47) | 142 (46) |
| 1200 | 18 (0) | 0 (0) | 44 (17) | 80 (3) | 125 (47) | 130 (44) |
| 1202 | 65 (11) | 79 (11) | 0 (0) | 79 (3) | 145 (59) | 146 (58) |
| 1201 | 275 (22) | 337 (22) | 275 (23) | 0 (0) | 206 (64) | 211 (63) |
| 1198 | 366 (17) | 449 (17) | 383 (28) | 239 (10) | 0 (0) | 175 (7) |
| 1209 | 364 (18) | 425 (19) | 364 (38) | 204 (10) | 151 (8) | 0 (0) |
A summary of differences between the CC80:ST80-IV isolates (not including intergenic regions) is shown for 82 representative loci in Table 3. In many instances, these appeared to relate to variation in bacteriophage carriage. For example, the isolates were identical for 20 of 21 probes associated with PVL-encoding bacteriophages, with most being similar to the Mu50 array sequences. However, only 1198 and 1209 contained the bacteriophage-associated SA1789 sequence. Isolate 1198 contained at least one bacteriophage and various hypothetical genes not found in isolates 1199 and 1200. Isolate 1198 did not contain the epidermin immunity factor (epiG) gene or hypothetical protein SA0848, found in all other isolates. Isolate 1201 lacked hypothetical protein SA0406, the SAA0001 replication initiation protein repC, the SA0002 tetracycline resistance protein, the SAA0003 plasmid recombination-mobilization protein pre, and uniquely contained hypothetical protein SA2487. Isolate 1202 contained a unique set of bacteriophage-associated adjacent genes (COL SA1573–1586) encoding replication protein, integrase and various hypothetical ORFs. Isolate 1209, most similar to isolate 1198, was unique in lacking virulence genes sdrD and sdrE and hypothetical proteins SA0397, SA0753 and SA1346. As noted above, although initially characterized as SCCmec IV based on the Oliveira and de Lencastre multiplex PCR protocol [19], further SCCmec subtyping using the strategy of Kondo et al. [21] and Milheirico [22] revealed that isolates 1201 and 1202 lacked the SCCmec recombinase (ccr), whereas all isolates contained the SCCmec IVc J1 sequence (data not shown). All isolates were positive for mecA, ΔmecR1, and ΨIS1272 SCCmec IV probes. Within loci that varied among the CC80:ST80-IV (Table 4), the unique adjacent genes (COL SA1573–1586) in isolate 1202, as mentioned above, were also found in USA300 (CC8:ST8-IV). Of the 57 most variable loci (i.e. either present or absent in two to three of the six isolates), the majority (37/57; 65%) were absent in both USA300 and USA400 (Table 3) [4]. For loci of known function, CC80:ST80-IV isolates were generally similar to USA300 and USA400. However, for 19 loci of known function that varied between USA300 and USA400 [4], the CC80:ST80-IV isolates were more similar to USA400. These differences included agr, capsule type, the presence in USA300 of a complete ebh gene, fosfomycin resistance, and assorted extracellular virulence determinants (e.g. exotoxin 3) not found in USA400 or CC80:ST80-IV (data not shown).
TABLE 3.
GeneChip® loci which varied between the CC80:ST80-IV isolates
| Results for: | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genes | ORF no. | Descriptiona | 1198 | 1199 | 1200 | 1201 | 1202 | 1209 | Comments |
| MW1419 | Conserved hypothetical protein (MSSA476, MRSA252, MW2) | + | − | − | − | − | + | USA300− USA400− | |
| SAR395a | Conserved hypothetical protein (MRSA252) | − | + | + | + | + | − | USA300+ USA400− | |
| SAV0876 | Conserved hypothetical protein similar to phage phi ETA protein (MU50) |
+ | − | − | − | − | + | USA300− USA400+ | |
| SAV0880 | Hypothetical protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0884 | Phage phi 11-like int gene activator protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0884 | Phage phi 11-like int gene activator protein (MU50) | − | − | − | − | − | − | USA300− USA400− | |
| SAV0885 | Phage phi 11-like terminase protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0886 | Similar to phage terminase large subunit (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0887 | Phage phi MU50B/phi11-like portal protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0888 | Phage phi MU50B/phi11-like head protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0890 | Hypothetical phage phi MU50B/phi11-like protein (MU50) | − | − | − | − | − | − | USA300− USA400− | |
| SAV0890 | Hypothetical phage phi MU50B/phi11-like protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0891 | Hypothetical phage phi11 head protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0892 | Phage phi MU50B/phi11-like head protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0893 | Phage phi MU50B/phi11-like protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0894 | Phage phi MU50B/phi11-like protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0896 | Phage phi MU50B/phi11-like protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0897 | Phage phi MU50B/phi11-like protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0898 | Similar to phage phi MU50 tail protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0899 | Hypothetical phage phi11 protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0900 | Hypothetical phage phi 11 protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0901 | Hypothetical phage minor tail subunit protein (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0902 | Conserved phi ETA orf 54-like protein (Mu50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0903 | Conserved phi ETA orf 55-like protein (Mu50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0904 | phi ETA orf 56-like protein (Mu50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0905 | phiETA ORF57-like protein (Mu50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0906 | phiETA ORF58-like protein (Mu50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0907 | Conserved phage phiETA ORF59-like protein (Mu50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0909 | Phage phi11 cell wall hydrolyase (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0910 | Phage phi11 tail fiber (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV0913 | Amidase (MU50) | + | − | − | − | − | + | USA300− USA400− | |
| SAV1977 | phi PV83-like protein (Mu50) | + | − | − | − | − | + | USA300+ USA400− | |
| SAS1087 | Conserved hypothetical protein (MSSA476) | − | + | + | + | + | − | USA300+ USA400+ | |
| SA0043 | Hypothetical protein pathogenicity island SaPIn1 | + | + | + | − | − | + | USA300+ USA400+ | |
| SA0044 | Conserved hypothetical protein | + | + | + | − | − | + | USA300− USA400− | |
| SA0022 | Hypothetical protein (N315) | + | − | − | + | − | + | USA300− USA400− | |
| ccrB | SA0057 | Cassette chromosome recombinase B (N315) | + | + | + | − | − | + | USA300+ USA400+ |
| ccrA | SA0058 | Cassette chromosome recombinase A (N315) | + | + | + | − | − | + | USA300+ USA400+ |
| SA0059 | Putative membrane protein (N315) | + | + | + | − | − | + | USA300+ USA400+ | |
| SA0061 | Hypothetical protein (N315) | + | + | + | − | − | + | USA300− USA400− | |
| SA1591 | Arsenical resistance operon repressor homologue (N315) | + | − | − | − | − | + | USA300− USA400− | |
| SA0321 | Prophage L54a, Cro-related protein | + | − | − | − | − | + | USA300− USA400− | |
| SA0322 | Putative prophage L54a repressor protein | + | − | − | − | − | + | USA300− USA400− | |
| SA0331 | Hypothetical protein | − | + | + | − | + | − | USA300− USA400− | |
| SA0359 | Hypothetical protein | + | − | − | − | − | + | USA300− USA400− | |
| SA0360 | Conserved hypothetical protein | + | − | − | − | − | + | USA300− USA400− | |
| SA0397 | Conserved hypothetical protein | + | + | + | + | + | − | USA300− USA400− | |
| SA0406 | Hypothetical protein | + | + | + | − | + | + | USA300+ USA400− | |
| sdrD | SA0520 | Virulence gene (N315) | + | + | + | + | + | − | USA300+ USA400+ |
| sdrE | SA0610 | Virulence gene | + | + | + | + | + | − | USA300+ USA400+ |
| SA0753 | Conserved hypothetical protein | + | + | + | + | + | − | USA300+ USA400+ | |
| SA0848 | Hypothetical protein | − | + | + | + | + | + | USA300+ USA400− | |
| SA0865 | Hypothetical protein | − | + | + | − | + | − | USA300+ USA400+ | |
| SA0871 | Putatative acetyltransferase | − | + | + | − | + | − | USA300+ USA400+ | |
| SA0881 | Putative thioredoxin | − | + | + | + | + | − | USA300+ USA400+ | |
| SA0984 | Conserved hypothetical protein | − | + | + | − | + | − | USA300+ USA400+ | |
| SA1345 | Hypothetical protein | − | + | + | − | − | + | USA300+ USA400− | |
| SA1346 | Conserved hypothetical protein | + | + | + | + | + | − | USA300+ USA400+ | |
| SA1527 | Conserved hypothetical protein | + | − | − | + | − | − | USA300+ USA400− | |
| SA1573 | Integrase/recombinase/transposase (phage) | − | − | − | − | + | − | USA300+ USA400− | |
| SA1575 | Hypothetical protein | − | − | − | − | + | − | USA300+ USA400− | |
| SA1576 | Hypothetical protein, similar to secretory antigen precursor SsaA |
− | − | − | − | + | − | USA300+ USA400− | |
| SA1577 | Conserved hypothetical protein | − | − | − | − | + | − | USA300+ USA400− | |
| FtsK/SpoIIIE | SA1578 | Cell division protein | − | − | − | − | + | − | USA300+ USA400− |
| SA1579 | Conserved putative conjugative transposon protein | − | − | − | − | + | − | USA300+ USA400− | |
| SA1580 | Hypothetical protein | − | − | − | − | + | − | USA300+ USA400− | |
| SA1581 | Conserved hypothetical protein | − | − | − | − | + | − | USA300+ USA400− | |
| SA1582 | Conserved hypothetical protein | − | − | − | − | + | − | USA300+ USA400− | |
| SA1583 | Replication initiation protein (transposon) | − | − | − | − | + | − | USA300+ USA400− | |
| SA1584 | Hypothetical protein | − | − | − | − | + | − | USA300+ USA400− | |
| SA1585 | Conserved hypothetical protein | − | − | − | − | + | − | USA300+ USA400− | |
| SA1586 | Hypothetical protein | − | − | − | − | + | − | USA300+ USA400− | |
| SA1598 | Conserved hypothetical protein (competence) | − | + | + | − | + | − | USA300+ USA400+ | |
| SA1789 | Hypothetical protein phage phi PVL | + | − | − | − | − | + | USA300− USA400− | |
| SA1829 | Hypothetical protein | − | + | + | − | + | − | USA300+ USA400+ | |
| SA1839 | IS200-like transposase | − | + | + | − | + | − | USA300+ USA400+ | |
| epiG | SA1871 | Epidermin immunity protein F | − | + | + | + | + | + | USA300+ USA400+ |
| SA2307 | Hypothetical protein | − | + | + | − | + | − | USA300+ USA400+ | |
| SA2327 | Hypothetical protein (N315) | − | + | + | + | + | − | USA300+ USA400+ | |
| SA2338 | Hypothetical protein | − | + | + | + | + | + | USA300+ USA400+ | |
| SA2487 | Conserved hypothetical protein | − | − | − | + | − | − | USA300+ USA400+ | |
| repC | SAA0001 | Replication initiation protein | − | + | + | − | + | + | USA300+ USA400+ |
| tetR | SAA0002 | Tetracycline resistance protein | + | + | + | − | + | + | USA300+ USA400+ |
| pre | SAA0003 | Plasmid recombination/mobilization protein | + | + | + | − | + | + | USA300+ USA400+ |
Unless otherwise noted, poen reading frame (ORF) numbers and descriptions refer to Staphylococcus aureus strain COL. Hypothetical proteins are sequences lacking a homologue in the National Center for Biotechnology Information NR database; conserved hypothetical proteins have a homologue, although of no known function.
Discussion
As noted above, CA-MRSA strains are generally characterized by phenotypic and genotypic homogeneity which, coupled with their ability to spread, complicates the epidemiological picture. Currently, there is no accurate way to determine whether the multiple isolates that one wishes to compare represent the spread of a single or limited number of organisms vs. introduction from multiple independent sources. This dilemma is potentially more problematic with a higher MRSA prevalence. Because Denmark represents an environment of low MRSA prevalence, we were interested in comparatively analysing CC80:ST80-IV isolates chosen to represent different years of isolation and the probability of different geographic origin.
Although genetic exchange is known to occur between S. aureus strains, the high degree of genomic relatedness (95%) among the CC80:ST80-IV examined in the present study supports a model of clonal expansion leading to genomic uniformity, despite differences in time and geography. However, upon closer examination, subtle differences were observed, leading to potentially interesting ‘sub-type’ inter-relationships. For example, the most similar isolates were from outside of Denmark (1199 and 1200), both cultured in 2001 but from different locations (i.e. Lebanon and Egypt). Because of the small number of isolates examined, it is unclear whether clustering of the Lebanese and Egyptian isolates apart from the isolates of Danish origin is significant, as are the conclusions regarding possible transmission from the Middle East to Denmark. However, CC80:ST80-IV was recently shown to constitute 55% of all MRSA in a large hospital in Lebanon, which may support this hypothesis (Tokajian, et al., 13th International Symposium on Staphylococci and Staphylococcal Infections, 2008, abstract P655). In addition, Denmark is a country of low MRSA endemicity (approximately 0.1%) [23], with only a single case of CC80:ST80-IV hospital transmission being documented to date [7]. Therefore, the acquisition of MRSA in these two patients before leaving Denmark for Lebanon and Egypt would appear to be unlikely. Interesting inter-relationships were also noted among the Danish isolates. Although cultured over a 4-year time span (1997 vs. 2001), isolates 1198 and 1209 were the second most highly related pair. The final pair of isolates (1201 and 1202), found in Denmark over a 2-year period, were somewhat more distantly related but shared the interesting loss of SCCmec-associated recombinase genes at the same times as retaining mecA, ΔmecR1 and ΨIS1272, and the SCCmec IVc specific J1 region. Recent studies have reported both S. aureus [24] and Staphylococcus epidermidis [25] isolates positive for mecA but negative for ccr by PCR analysis. However, whether this is a result of the absence of ccr or sequence divergence influencing primer recognition remains unknown. Thus, to our knowledge, this is the first report of such a deletion that would clearly affect the mobility/excision of SCCmec in these isolates. The multi-year observation of these isolates suggests that they may represent a stable CC80:ST80-IV subpopulation (e.g. a SCCmec IV variant or potentially new type), the frequency and significance of which is currently unknown. Regarding PVL, two recent studies [26,27] reported interesting sequence differences related to specific MRSA strains and geography. Microarray analysis indicated that all six CC80:ST80-IV isolates appeared to carry the same PVL-associated bacteriophage, similar to that of S. aureus strain Mu50. As with issues related to SCCmec differences, additional sequence-based studies of PVL could provide potentially interesting information regarding possible isolate sub-type inter-relationships.
It is reassuring to note that both PFGE and microarray analysis identified the same relatedness pairs, which fit well with the overall clinical and molecular data, grouping the Middle East and ccr-deleted pairs from the remaining Danish isolates. This suggests that minor differences in both PFGE and microarray analysis of genomically uniform strains such as CC80:ST80-IV may have the potential for clinical and epidemiological significance. However, conclusions regarding these observations must be tempered by the small number of isolates analysed. As with other CA-MRSA, the conserved nature of CC80:ST80-IV isolates has limited the usefulness of current molecular approaches for epidemiological investigation. Nevertheless, as with recent USA300 genomic analyses [28,29], the data obtained in the present study suggest that differences such as single-nucleotide polymorphisms and divergence in hypothetical ORFs and intergenic regions may ultimately provide a useful sequence-based foundation for discerning epidemiologically meaningful inter-relationships. This is further supported by the recently demonstrated ability of a small number of single-nucleotide polymorphisms to provide meaningful typing data in the highly conserved genomic background of Bacillus anthracis [30]. However, the potential usefulness of microarray data for the epidemiological analysis of CC80:ST80-IV clearly requires further evaluation with isolates of known relatedness. Although microarray analysis revealed differences between the CC80:ST80-IV isolates primarily in conserved hypothetical ORFs and intergenic sequences, these differences are worth noting because the unknown function of such loci does not automatically equate with unimportance. For example, approximately 80% of the USA300 genome represents a coding sequence that includes numerous conserved hypothetical proteins whose role is yet to be determined [28]. In S. epidermidis, phenol-soluble modulins, now known to be important virulence factors [16], were initially poorly annotated in genomic sequences [17]. Thus, sequence-based comparisons (i.e. differences and similarities) of CA-MRSA strains such as CC80:ST80-IV hold potential promise as a means of uncovering relevant and important information that will hopefully lead to a better understanding of both the pathogenicity and epidemiology of these increasingly important pathogens.
Footnotes
Transparency Declaration
The authors declare that they have no conflicting interests in relation to this work.
References
- 1.Deurenberg RH, Vink C, Kalenic S, et al. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2007;13:222–235. doi: 10.1111/j.1469-0691.2006.01573.x. [DOI] [PubMed] [Google Scholar]
- 2.Humphreys H. National guidelines for the control and prevention of methicillin-resistant Staphylococcus aureus—what do they tell us? Clin Microbiol Infect. 2007;13:846–853. doi: 10.1111/j.1469-0691.2007.01766.x. [DOI] [PubMed] [Google Scholar]
- 3.Harbarth S. Control of endemic methicillin-resistant Staphylococcus aureus-recent advances and future challenges. Clin Microbiol Infect. 2006;12:1154–1162. doi: 10.1111/j.1469-0691.2006.01572.x. [DOI] [PubMed] [Google Scholar]
- 4.Tenover FC, McDougal LK, Goering RV, et al. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol. 2006;44:108–118. doi: 10.1128/JCM.44.1.108-118.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson JK, Khoie T, Shurland S, et al. Skin and soft tissue infections caused by methicillin-resistant Staphylococcus aureus USA300 clone. Emerg Infect Dis. 2007;13:1195–1200. doi: 10.3201/eid1308.061575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tristan A, Bes M, Meugnier H, et al. Global distribution of Panton–Valentine leukocidin—positive methicillin-resistant Staphylococcus aureus, 2006. Emerg Infect Dis. 2007;13:594–600. doi: 10.3201/eid1304.061316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsen AR, Bocher S, Stegger M, et al. Epidemiology of European community-associated methicillin-resistant Staphylococcus aureus clonal complex 80 type IV strains isolated in Denmark from 1993 to 2004. J Clin Microbiol. 2008;46:62–68. doi: 10.1128/JCM.01381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urth T, Juul G, Skov R, et al. Spread of a methicillin-resistant Staphylococcus aureus ST80-IV clone in a Danish community. Infect Control Hosp Epidemiol. 2005;26:144–149. doi: 10.1086/502518. [DOI] [PubMed] [Google Scholar]
- 9.Larsen AR, Stegger M, Bocher S, et al. Emergence and characterization of community associated methicillin-resistant Staphyloccocus aureus infections in Denmark, 1999–2006. J Clin Microbiol. 2009;47:73–78. doi: 10.1128/JCM.01557-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kazakova SV, Hageman JC, Matava M, et al. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N Engl J Med. 2005;352:468–475. doi: 10.1056/NEJMoa042859. [DOI] [PubMed] [Google Scholar]
- 11.Monecke S, Slickers P, Hotzel H, et al. Microarray-based characterisation of a Panton–Valentine leukocidin-positive community-acquired strain of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2006;12:718–728. doi: 10.1111/j.1469-0691.2006.01420.x. [DOI] [PubMed] [Google Scholar]
- 12.Monecke S, Berger-Bachi B, Coombs G, et al. Comparative genomics and DNA array-based genotyping of pandemic Staphylococcus aureus strains encoding Panton–Valentine leukocidin. Clin Microbiol Infect. 2007;13:236–249. doi: 10.1111/j.1469-0691.2006.01635.x. [DOI] [PubMed] [Google Scholar]
- 13.Monecke S, Jatzwauk L, Weber S, et al. DNA microarray-based genotyping of methicillin-resistant Staphylococcus aureus strains from Eastern Saxony. Clin Microbiol Infect. 2008;14:534–545. doi: 10.1111/j.1469-0691.2008.01986.x. [DOI] [PubMed] [Google Scholar]
- 14.Dunman PM, Mounts W, McAleese F, et al. Uses of Staphylococcus aureus GeneChips in genotyping and genetic composition analysis. J Clin Microbiol. 2004;42:4275–4283. doi: 10.1128/JCM.42.9.4275-4283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sung JM, Lloyd DH, Lindsay JA. Staphylococcus aureus host specificity: comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology. 2008;154:1949–1959. doi: 10.1099/mic.0.2007/015289-0. [DOI] [PubMed] [Google Scholar]
- 16.Klingenberg C, Ronnestad A, Anderson AS, et al. Persistent strains of coagulase-negative staphylococci in a neonatal intensive care unit: virulence factors and invasiveness. Clin Microbiol Infect. 2007;13:1100–1111. doi: 10.1111/j.1469-0691.2007.01818.x. [DOI] [PubMed] [Google Scholar]
- 17.Yao Y, Sturdevant DE, Otto M. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J Infect Dis. 2005;191:289–298. doi: 10.1086/426945. [DOI] [PubMed] [Google Scholar]
- 18.Salgado CD, Farr BM, Calfee DP. Community-acquired methicillin-resistant Staphylococcus aureus: a meta-analysis of prevalence and risk factors. Clin Infect Dis. 2003;36:131–139. doi: 10.1086/345436. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Neill AJ, Larsen AR, Henriksen AS, et al. A fusidic acid-resistant epidemic strain of Staphylococcus aureus carries the fusB determinant, whereas fusA mutations are prevalent in other resistant isolates. Antimicrob Agents Chemother. 2004;48:3594–3597. doi: 10.1128/AAC.48.9.3594-3597.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo Y, Ito T, Ma XX, et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51:264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milheirico C, Oliveira DC, de Lencastre H. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J Antimicrob Chemother. 2007;60:42–48. doi: 10.1093/jac/dkm112. [DOI] [PubMed] [Google Scholar]
- 23.Benfield T, Espersen F, Frimodt-Moller N, et al. Increasing incidence but decreasing in-hospital mortality of adult Staphylococcus aureus bacteraemia between 1981 and 2000. Clin Microbiol Infect. 2007;13:257–263. doi: 10.1111/j.1469-0691.2006.01589.x. [DOI] [PubMed] [Google Scholar]
- 24.Petrelli D, Repetto A, D’Ercole S, et al. Analysis of meticillin-susceptible and meticillin-resistant biofilm-forming Staphylococcus aureus from catheter infections isolated in a large Italian hospital. J Med Microbiol. 2008;57:364–372. doi: 10.1099/jmm.0.47621-0. [DOI] [PubMed] [Google Scholar]
- 25.Ibrahem S, Salmenlinna S, Virolainen A, et al. Carriage of methicillin-resistant staphylococci and their SCCmec types in a long term care facility. J Clin Microbiol. 2008;47:32–37. doi: 10.1128/JCM.01085-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolter DJ, Chatterjee A, Varman M, et al. Isolation and characterization of an epidemic methicillin-resistant Staphylococcus aureus 15 variant in the central United States. J Clin Microbiol. 2008;46:3548–3549. doi: 10.1128/JCM.00985-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Hara FP, Guex N, Word JM, et al. A geographic variant of the Staphylococcus aureus Panton–Valentine leukocidin toxin and the origin of community-associated methicillin-resistant S. aureus USA300. J Infect Dis. 2008;197:187–194. doi: 10.1086/524684. [DOI] [PubMed] [Google Scholar]
- 28.Highlander SK, Hulten KG, Qin X, et al. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol. 2007;7:99–113. doi: 10.1186/1471-2180-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennedy AD, Otto M, Braughton KR, et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci USA. 2008;105:1327–1332. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okinaka RT, Henrie M, Hill KK, et al. Single nucleotide polymorphism typing of Bacillus anthracis from Sverdlovsk tissue. Emerg Infect Dis. 2008;14:653–656. doi: 10.3201/eid1404.070984. [DOI] [PMC free article] [PubMed] [Google Scholar]