Abstract
The psychostimulant methylphenidate (Ritalin) is used in conjunction with selective serotonin reuptake inhibitors (SSRIs) in the treatment of medical conditions such as attention-deficit hyperactivity disorder with anxiety/depression comorbidity and major depression. Co-exposure also occurs in patients on SSRIs that use psychostimulant “cognitive enhancers”. Methylphenidate is a dopamine/norepinephrine reuptake inhibitor that produces altered gene expression in the forebrain; these effects partly mimic gene regulation by cocaine (dopamine/norepinephrine/serotonin reuptake inhibitor). We investigated whether the addition of SSRIs (fluoxetine or citalopram; 5 mg/kg) modified gene regulation by methylphenidate (2–5 mg/kg) in the striatum and cortex of adolescent rats. Our results show that SSRIs potentiate methylphenidate-induced expression of the transcription factors zif 268 and c-fos in the striatum, rendering these molecular changes more cocaine-like. Present throughout most of the striatum, this potentiation was most robust in its sensorimotor parts. The methylphenidate + SSRI combination also enhanced behavioral stereotypies, consistent with dysfunction in sensorimotor striatal circuits. In so far as such gene regulation is implicated in psychostimulant addiction, our findings suggest that SSRIs may enhance the addiction liability of methylphenidate.
Keywords: psychostimulant, immediate-early gene, caudate-putamen, rat
INTRODUCTION
Use of the psychostimulant methylphenidate (Ritalin), both in the treatment of attention-deficit hyperactivity disorder (ADHD) and as a “cognitive enhancer” in the healthy, has increased considerably over the past decades (Kollins et al., 2001; Swanson & Volkow, 2008; Bogle & Smith, 2009). While it remains controversial whether the medical use of psychostimulants is completely safe (Kollins, 2008; Wilens et al., 2008), especially in children and adolescents (Carlezon & Konradi, 2004; Andersen, 2005), one aspect of such drug treatments is often overlooked: potential drug interactions. Methylphenidate is frequently administered together with selective serotonin reuptake inhibitors (SSRIs). This concomitant therapy is used, for example, to treat ADHD with anxiety/depression comorbidity (Safer et al., 2003; Bhatara et al., 2004), as a high percentage of ADHD patients is also diagnosed with major depressive or bipolar disorder (Kollins, 2008). Methylphenidate plus SSRI concomitant therapies are also employed in depression, for example, as augmentation therapy in major depressive disorder (e.g., Nelson, 2007; Ishii et al., 2008; Ravindran et al., 2008), as acceleration treatment with SSRIs (e.g., Lavretsky et al., 2003), or to treat sexual dysfunction (e.g., Csoka et al., 2008). In addition to such clinical co-administration, it is presently unknown how much uncontrolled methylphenidate + SSRI co-exposure occurs due to “cognitive enhancer” use (Greely et al., 2008) by patients on SSRIs.
Concerns regarding potential harmful consequences of methylphenidate + SSRI co-exposure are related to the neurochemical effects of these drugs. The mode of action of methylphenidate overlaps with that of others psychostimulants. Similar to cocaine, methylphenidate blocks the dopamine (and norepinephrine) transporter, thus, indirectly producing excessive dopamine receptor stimulation and ensuing changes in gene regulation in dopamine target areas such as the striatum (for review, see Yano & Steiner, 2007). Among the many genes affected by methylphenidate (Adriani et al., 2006a; Adriani et al., 2006b), those encoding transcription factors (immediate-early genes, IEGs) such as zif 268 and c-fos (Lin et al., 1996; Brandon & Steiner, 2003; Chase et al., 2003; Yano & Steiner, 2005b) are of special interest, as they regulate the expression of effector genes and are thus implicated in neuroplasticity underlying psychostimulant addiction (Hyman & Nestler, 1996; Berke & Hyman, 2000).
However, methylphenidate differs from cocaine in that it has a much lower affinity for the serotonin transporter and does not produce serotonin overflow (Kuczenski & Segal, 1997; Borycz et al., 2008; for review, see Yano & Steiner, 2007). This may explain why not all gene regulation effects of cocaine are mimicked by methylphenidate (Yano & Steiner, 2007). Evidence indicates that serotonin interacts with dopamine to modify striatal gene regulation by psychostimulants. For example, interruption of the serotonin transmission by transmitter depletion (Bhat & Baraban, 1993) or receptor deletion (Lucas et al., 1997) reduces IEG induction by cocaine.
We here determied whether concomitant treatment with SSRIs, which elevate extracellular serotonin levels, enhances methylphenidate-induced gene regulation and produces more cocaine-like effects. We investigated, in adolescent rats, the effects of the SSRIs fluoxetine and citalopram on gene regulation in the striatum and cortex. We assessed the expression of the two transcription factors/IEGs zif 268 and c-fos to allow for a more sensitive two-marker correlation analysis of drug effects (Yano & Steiner, 2005a,b). Our results show that these SSRIs potentiate methylphenidate-induced gene expression preferentially in the sensorimotor striatum. A partial account of our findings has been presented in a brief report (Steiner et al., 2010).
MATERIALS AND METHODS
Subjects
Male Sprague–Dawley rats (35-day old at the time of the drug treatment; Harlan, Madison, WI, USA) were housed 2 per cage under standard laboratory conditions (12:12h light/dark cycle; lights on at 07:00h) with food and water available ad libitum. Experiments were performed between 13:00 and 17:00h. Prior to the drug treatment, the rats were allowed one week of acclimation during which they were repeatedly handled. All procedures met the NIH guidelines for the care and use of laboratory animals and were approved by the Rosalind Franklin University Animal Care and Use Committee.
Drug treatments
Rats received a single intraperitoneal injection of vehicle (V), methylphenidate HCl (2 mg/kg, MP2, or 5 mg/kg, MP5; in 0.02% ascorbic acid, 1 ml/kg; Sigma, St. Louis, MO, USA), fluoxetine HCl (5 mg/kg, FLX; Sigma), or methylphenidate plus fluoxetine (MP2+FLX or MP5+FLX) (n=5–7 each). Other groups were treated with citalopram HBr (5 mg/kg, CIT; Sigma), methylphenidate (5 mg/kg) plus citalopram (MP5+CIT) or cocaine HCl (25 mg/kg; Sigma). After the injection, the rat was placed in an open-field apparatus (43 × 43 cm), and locomotion (ambulatory distance) and stereotypy (“stereotypy 2”) counts were measured for 40 min with an activity monitoring system (Truscan, Coulbourn Instruments, Allentown, PA, USA). These “stereotypy” counts reflect local, repetitive movements (e.g., head bobbing, focused sniffing).
Tissue preparation and in situ hybridization histochemistry
The rats were killed with CO2 40 min after the injection. The brain was rapidly removed, frozen in isopentane cooled on dry ice and then stored at −30 °C until cryostat sectioning. Coronal sections (12 µm) were thaw-mounted onto glass slides (Superfrost/Plus, Daigger, Wheeling, IL, USA), dried on a slide warmer and stored at −30 °C. In preparation for the in situ hybridization histochemistry, the sections were fixed in 4% paraformaldehyde/0.9% saline for 10 min at room temperature, incubated in a fresh solution of 0.25% acetic anhydride in 0.1 M triethanolamine/0.9% saline (pH 8.0) for 10 min, dehydrated, defatted for 2 × 5 min in chloroform, rehydrated, and air-dried. The slides were then stored at −30 °C until hybridization.
Oligonucleotide probes (48-mers; Invitrogen, Rockville, MD, USA) were labeled with [35S]-dATP as described earlier (Steiner & Kitai, 2000). The probes had the following sequence: zif 268, complementary to bases 352–399, GenBank accession number M18416; c-fos, bases 207–254, X06769. One hundred µl of hybridization buffer containing labeled probe (~3 × 106 cpm) was added to each slide. The sections were coverslipped and incubated at 37 °C overnight. After incubation, the slides were first rinsed in four washes of 1X saline citrate (150 mM sodium chloride, 15 mM sodium citrate), and then washed 3 times 20 min each in 2X saline citrate/50% formamide at 40 °C, followed by 2 washes of 30 min each in 1X saline citrate at room temperature. After a brief water rinse, the sections were air-dried and then apposed to X-ray film (BioMax MR-2, Kodak) for 5–9 days.
Analysis of autoradiograms
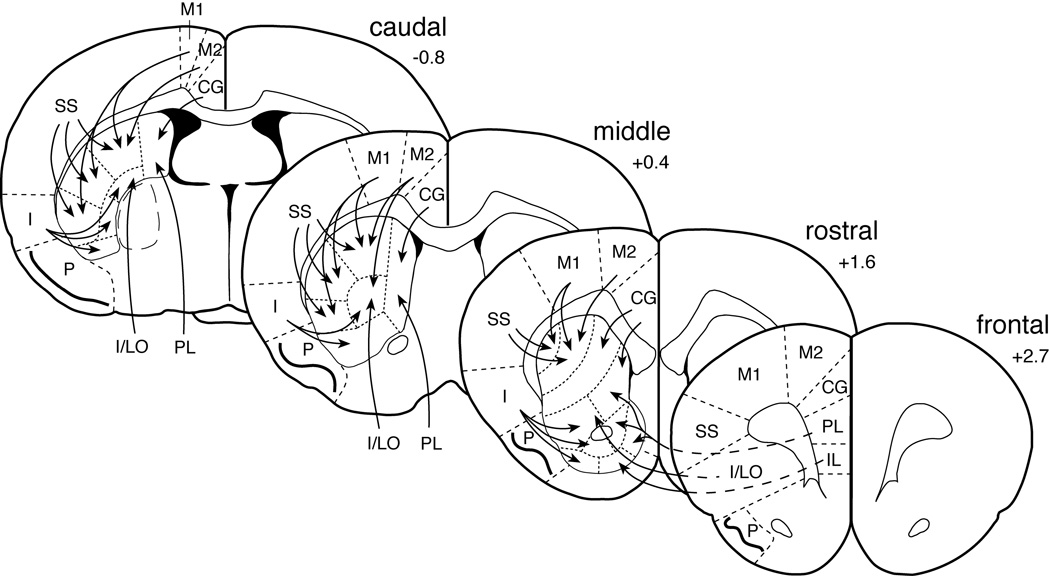
Gene expression in the cortex was assessed in sections from 4 rostrocaudal levels (Fig. 1): frontal, approximately at +2.7 mm relative to bregma (Paxinos & Watson, 1998); rostral, +1.6; middle, +0.4; caudal, −0.8. Levels of mRNA were measured in a total of 22 cortical regions (from medial to lateral; Paxinos & Watson, 1998): cingulate (CG), medial agranular (M2), motor (M1), somatosensory (SS) and insular (I) cortex on frontal to caudal levels, and infralimbic (IL), prelimbic (PL) and insular/lateral orbital cortex (I/LO) on the frontal level. Striatal gene expression was determined on rostral, middle and caudal levels in a total of 23 sectors mostly defined by their predominant cortical inputs (Fig. 1; see Willuhn et al., 2003). Eighteen of these sectors represented the caudate-putamen: medial (m), dorsomedial (dm), dorsal (d), dorsolateral (dl), ventrolateral (vl), ventral (v), central (c), dorsal central (dc), ventral central (vc); and 5 the nucleus accumbens: medial core (mC), lateral core (lC), medial shell (mS), ventral shell (vS) and lateral shell (lS) (Yano & Steiner, 2005a).
Figure 1.
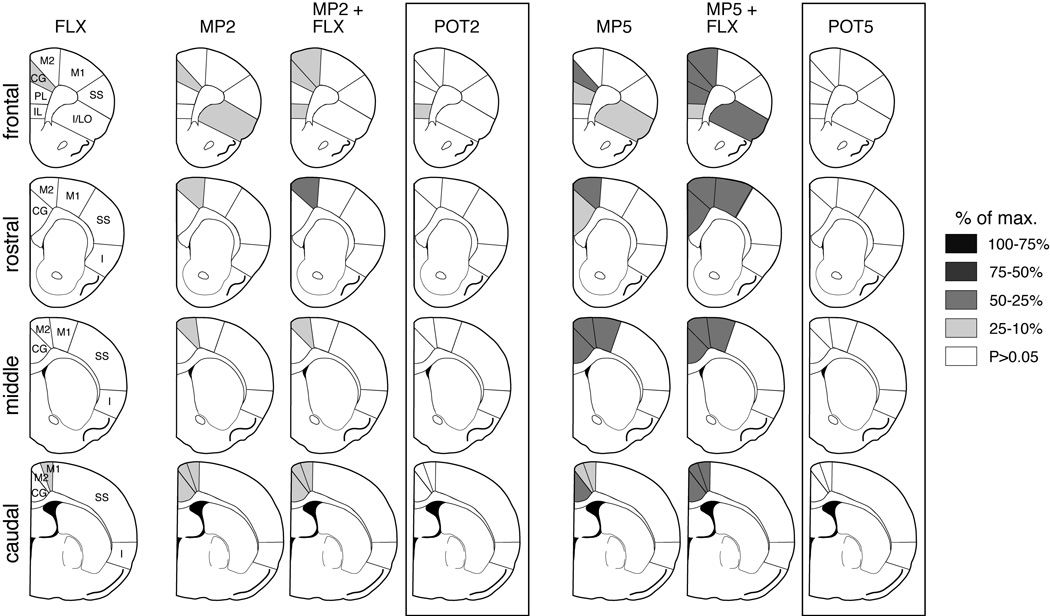
Schematic illustration of the 23 striatal and 22 cortical regions used to measure gene expression. The predominant cortical inputs to these striatal sectors are indicated by arrows (simplified; see Willuhn et al., 2003, for discussion). Gene expression was assessed on 4 rostrocaudal levels: frontal, rostral, middle, and caudal (ranging from +2.7 to −0.8 mm relative to bregma; Paxinos & Watson, 1998). Cortical areas (from medial to lateral): CG, cingulate; M2, medial agranular; M1, motor; SS, somatosensory; I, insular; P, piriform; IL, infralimbic; PL, prelimbic, I/LO, insular/lateral orbital. For striatal areas, see Figure 6.
Hybridization signals on film autoradiograms were measured by densitometry (NIH Image; Wayne Rasband, NIMH, Bethesda, MD, USA). The films were captured using a light table (Northern Light, Imaging Research, St. Catharines, Ontario, Canada) and a Sony CCD camera (Imaging Research). The “mean density” value of a region of interest was measured by placing a template over the captured image. Mean densities were corrected for background by subtracting mean density values measured over white matter (corpus callosum). Values from corresponding regions in the two hemispheres were then averaged. The illustrations of film autoradiograms displayed in Figures 3, 5 and 10 are computer-generated images, and are contrast-enhanced where necessary. Maximal hybridization signal is black.
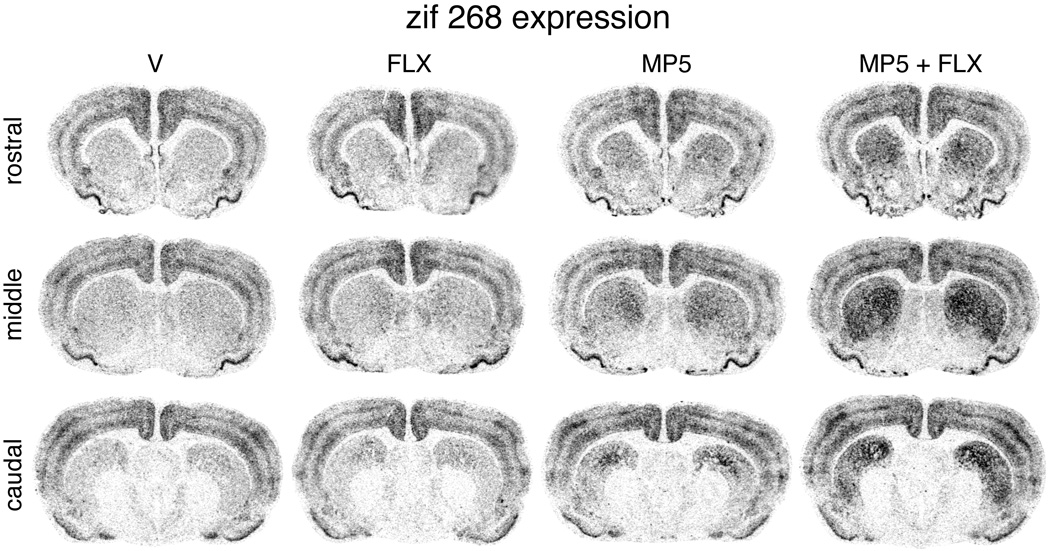
Figure 3.
Fluoxetine potentiates methylphenidate-induced zif 268 expression in the striatum. Illustrations of film autoradiograms depict zif 268 expression in coronal sections from the rostral, middle and caudal striatum in rats treated with vehicle (V), fluoxetine (5 mg/kg; FLX), methylphenidate (5 mg/kg; MP5), or methylphenidate + fluoxetine (MP5+FLX). The maximal hybridization signal is black.
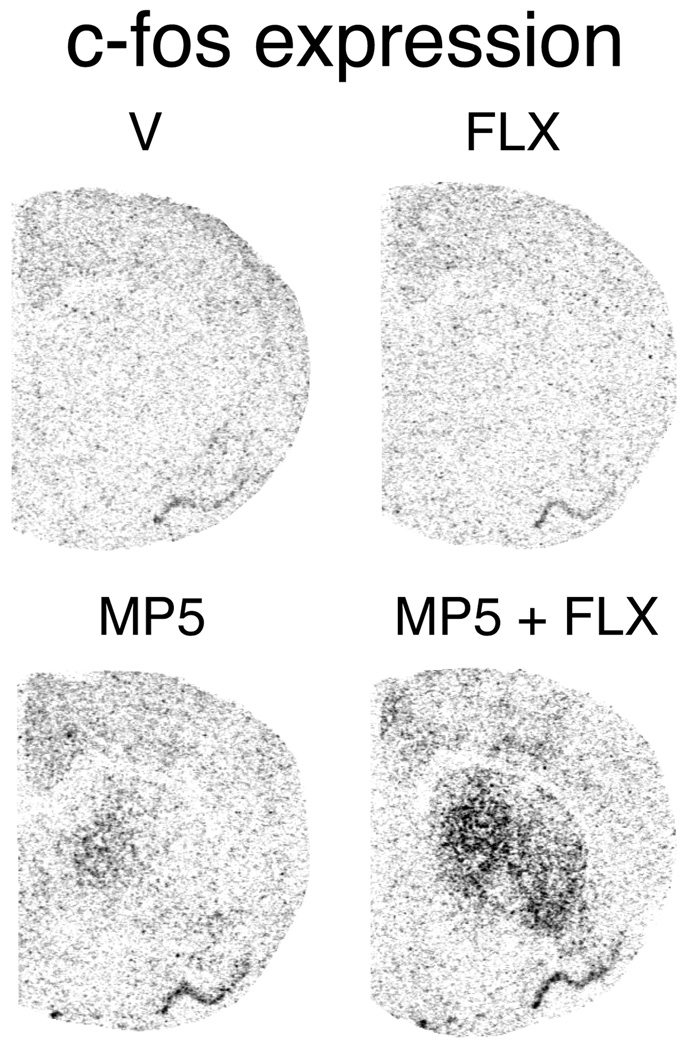
Figure 5.
Fluoxetine potentiates methylphenidate-induced c-fos expression in the striatum. Illustrations of film autoradiograms depict c-fos expression in coronal sections from the middle striatum in rats that received vehicle (V), fluoxetine (5 mg/kg; FLX), methylphenidate (5 mg/kg; MP5), or methylphenidate + fluoxetine (MP5+FLX). The maximal hybridization signal is black.
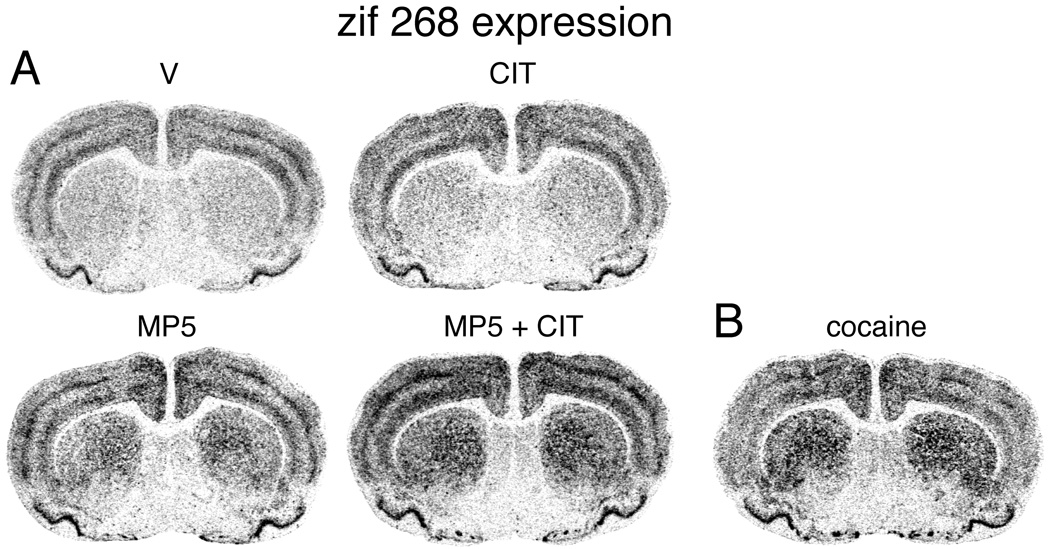
Figure 10.
Citalopram potentiates methylphenidate-induced zif 268 expression in the striatum. (A) Illustrations of film autoradiograms depict zif 268 expression in coronal sections from the middle striatum in rats treated with vehicle (V), citalopram (5 mg/kg; CIT), methylphenidate (5 mg/kg; MP5), or methylphenidate + citalopram (MP5+CIT). (B) For comparison, zif 268 induction by cocaine (25 mg/kg) is also shown. The maximal hybridization signal is black.
Statistics
Treatment effects were determined by two- and three-factor ANOVA with methylphenidate (0, 2, 5 mg/kg) and fluoxetine or citalopram (0, 5 mg/kg) as between-subject variables. Newman-Keuls post hoc tests were used to describe differences between individual groups (Statistica, StatSoft, Tulsa, OK, USA). For illustrations of topographies (maps), the change in gene expression in a given region was expressed as the percentage of the maximal change in the striatum observed for a particular probe (% max.). It was of interest to determine whether the regional distribution in the striatum of the changes in gene expression was similar for c-fos and zif 268. Thus, these changes in the 23 striatal sectors were compared by Pearson correlations. Also, our previous studies showed that such two-marker correlation analyses are more sensitive than ANOVAs to measure threshold drug effects, because trends contribute to correlations (Yano & Steiner, 2005a,b). For these analyses, the values were normalized relative to the maximal change observed for the 5 mg/kg methylphenidate treatment.
RESULTS
Effects of fluoxetine on methylphenidate-induced open-field behavior
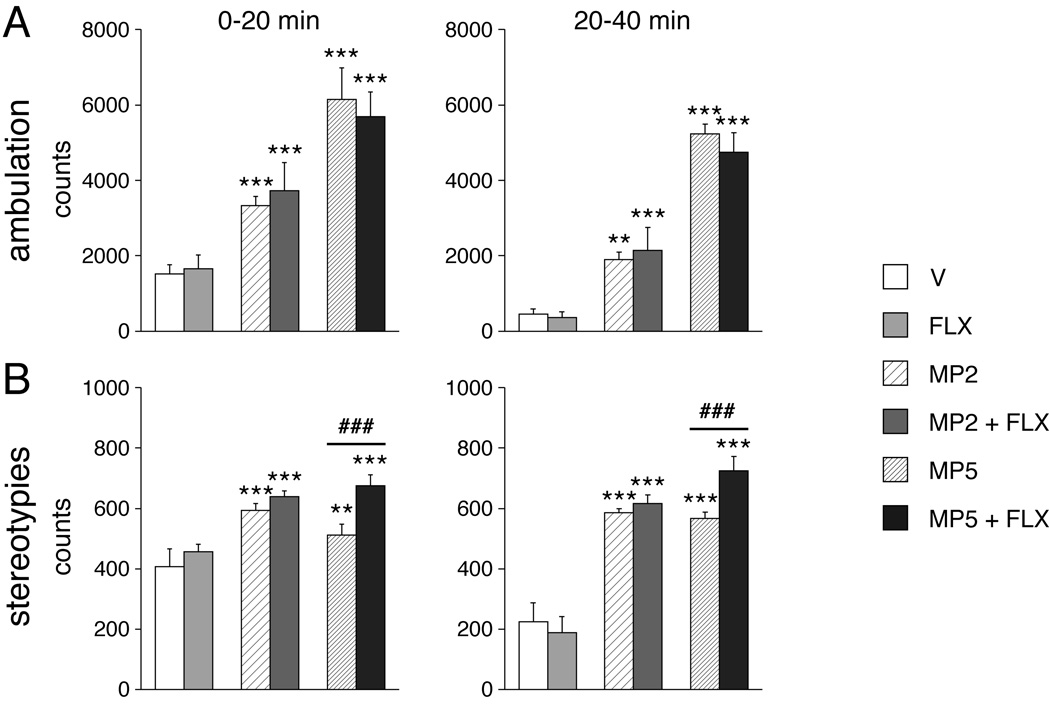
Administration of methylphenidate increased ambulation (MP2 vs. V, P<0.01; MP5 vs. V, P<0.001) and stereotypy counts (MP2 vs. V, P<0.001; MP5 vs. V, P<0.01) in the first (0–20 min) and second half (20–40 min) of the test (Fig. 2). Fluoxetine alone had no effect on these two parameters (FLX vs. V, P>0.05). The methylphenidate + fluoxetine combinations increased ambulation counts in a manner similar to methylphenidate alone (MP2+FLX vs. MP2 and MP5+FLX vs. MP5, P>0.05) and more than fluoxetine alone (MP2+FLX vs. FLX and MP5+FLX vs. FLX, P<0.001). As with methylphenidate alone, the methylphenidate + fluoxetine combinations induced stereotypies (MP2+FLX vs. V or FLX, and MP5+FLX vs. V or FLX, P<0.001). This increase in stereotypy counts was similar in the MP2 and MP2+FLX groups (P>0.05). However, the MP5+FLX group showed higher stereotypy counts than the MP5 alone group (P<0.001; Fig. 2) early and late in the test, demonstrating that fluoxetine potentiated methylphenidate-induced stereotypies for this higher methylphenidate dose.
Figure 2.
Drug effects on open-field behavior. Ambulation (A) and stereotypy counts (B) are shown for animals that received a systemic injection of vehicle (V), fluoxetine (5 mg/kg; FLX), methylphenidate (2 or 5 mg/kg; MP2, MP5), or methylphenidate + fluoxetine combinations (MP2+FLX and MP5+FLX) (n=5–7) and were tested for 40 min in a novel open field. Fluoxetine selectively potentiated methylphenidate (5 mg/kg)-induced stereotypies. ** P<0.01, *** P<0.001, vs. respective control group (V or FLX); ### P<0.001, MP5+FLX vs. MP5 (potentiation).
Effects of fluoxetine on methylphenidate-induced gene expression in the striatum
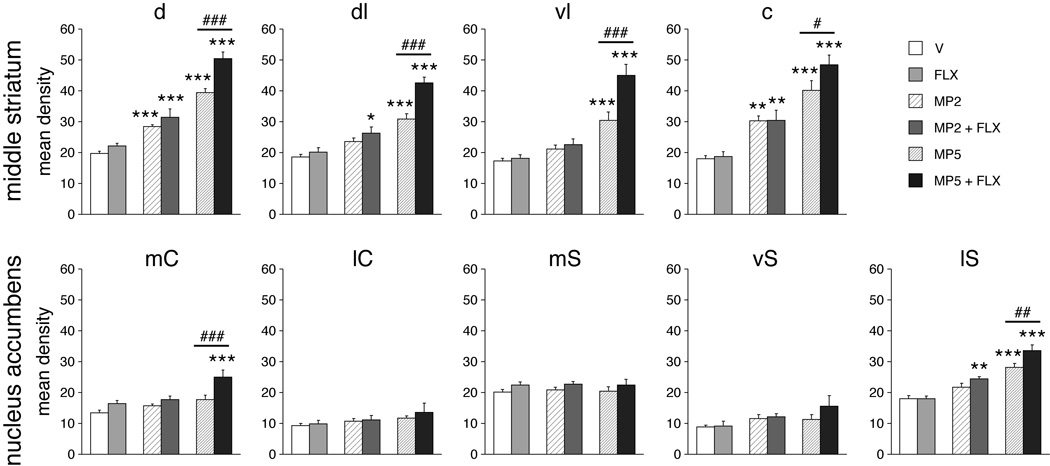
Administration of methylphenidate alone induced a dose-dependent increase in zif 268 and c-fos expression in the striatum on all three rostrocaudal levels (Figs. 3–6 and Tables 1 and 2), consistent with our previous findings (Brandon & Steiner, 2003; Yano & Steiner, 2005a,b). For zif 268, a significant increase in expression was observed in five (MP2) and 17 (MP5) of the 23 striatal sectors, and for c-fos, in three (MP2) and 11 (MP5) sectors (P<0.05 vs. V) (Fig. 6). Gene regulation varied considerably between different striatal regions. For both zif 268 and c-fos, the most robust increase was observed on middle and caudal striatal levels, in dorsal/central and medial sectors (Figs. 3–6) that receive sensorimotor and cingulate cortical inputs (Fig. 1). In contrast, the nucleus accumbens displayed more modest drug effects. No statistically significant changes in gene expression were seen with 2 mg/kg of methylphenidate alone (P>0.05 vs. V). The 5 mg/kg dose significantly increased zif 268 expression in the lateral shell only (MP5 vs. V, P<0.001, Figs. 4 and 6). In order to compare the regional patterns of methylphenidate-induced zif 268 and c-fos expression across the 23 striatal sectors, we performed a correlation analysis. This analysis confirmed that the regional distribution of increases (vs. vehicle-treated controls) was highly correlated between zif 268 and c-fos expression (zif 268 x c-fos: MP2, r=0.844, P<0.001; MP5, r=0.931, P<0.001; not shown).
Figure 6.
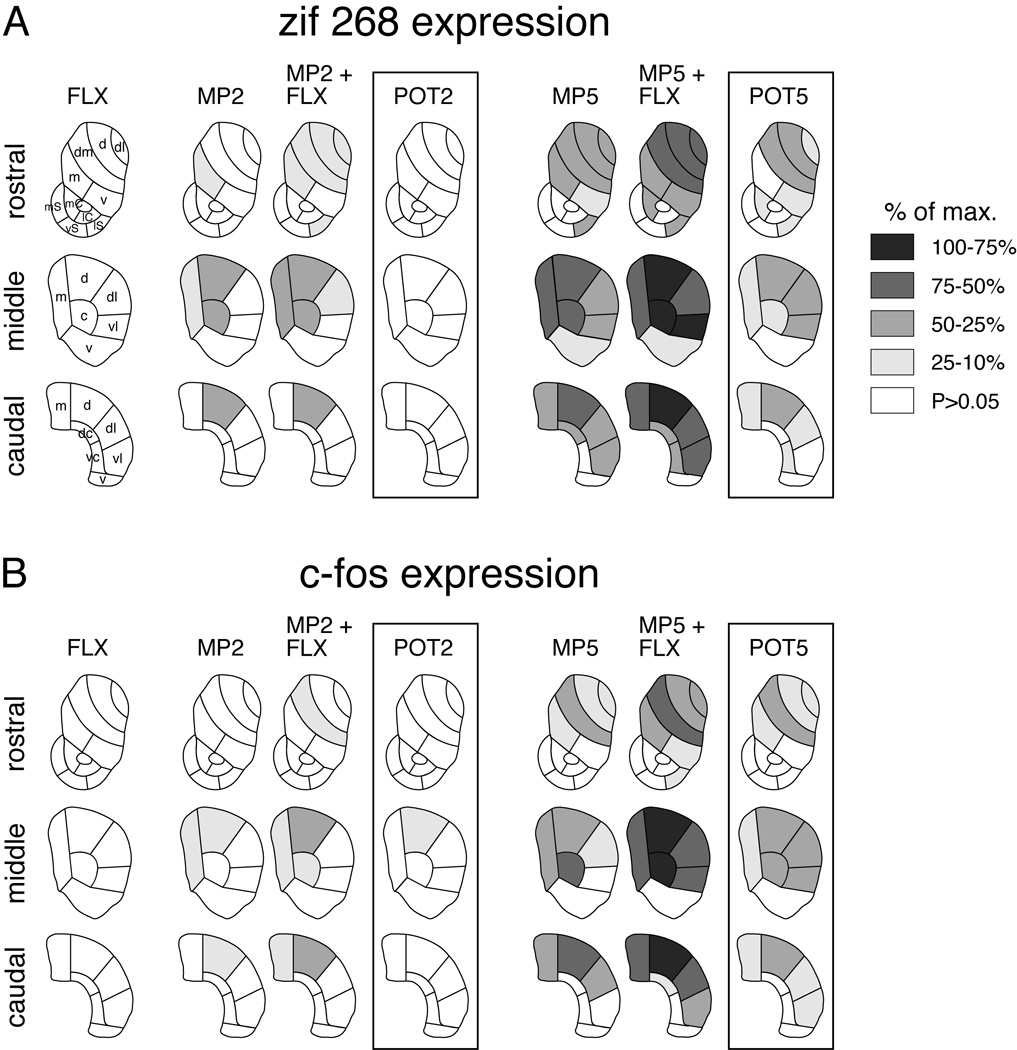
Topography of fluoxetine-potentiated gene regulation by methylphenidate. Maps depict the distribution of zif 268 (A) and c-fos expression (B) in the rostral, middle and caudal striatum after an injection of fluoxetine (5 mg/kg; FLX), methylphenidate (2 or 5 mg/kg; MP2, MP5), or methylphenidate + fluoxetine combinations (MP2+FLX, MP5+FLX). The potentiation (POT) denotes the difference between methylphenidate + fluoxetine and methylphenidate alone. The data are normalized relative to the maximal increase observed in the striatum (% of max.). Sectors with significant differences vs. vehicle-treated controls (P<0.05) are coded as indicated. Sectors without significant effects are in white. Abbreviations: caudate-putamen: c, central; d, dorsal; dc, dorsal central; dl, dorsolateral; dm, dorsomedial; m, medial; v, ventral; vc, ventral central; vl, ventrolateral; nucleus accumbens: mC, medial core; lC, lateral core; mS, medial shell; vS, ventral shell; lS, lateral shell.
Table 1.
Effects of fluoxetine, methylphenidate and methylphenidate + fluoxetine on zif 268 expression in the striatum.
| V | FLX | MP2 | MP2 + FLXFLX |
MP5 | MP5 + FLX |
||
|---|---|---|---|---|---|---|---|
| rostral | dl | 18.9±0.8 | 20.4±1.1 | 22.8±1.0 | 24.4±1.6 | 29.0±0.9*** | 36.8±2.2***,### |
| d | 19.9±0.8 | 23.3±1.4 | 24.5±0.9 | 27.5±1.2 | 31.2±0.8*** | 39.9±2.6***,### | |
| dm | 20.4±0.7 | 23.5±0.9 | 24.8±0.5 | 28.5±1.3 | 34.5±0.9*** | 43.9±3.1***,### | |
| m | 17.9±0.5 | 20.7±1.3 | 22.7±0.9* | 25.4±0.7* | 31.6±1.8*** | 34.3±1.8*** | |
| v | 16.2±0.9 | 17.7±1.4 | 18.8±0.7 | 20.4±0.8 | 21.9±1.3* | 27.7±1.8***,## | |
| mC | 13.4±0.9 | 16.4±1.0 | 15.7±0.7 | 17.7±1.1 | 17.8±1.4 | 25.0±2.4***,### | |
| lC | 9.3±0.7 | 9.9±1.1 | 10.7±0.9 | 11.2±1.4 | 11.8±0.7 | 13.6±3.0 | |
| mS | 20.2±0.8 | 22.5±1.0 | 20.9±0.8 | 22.7±0.9 | 20.5±1.5 | 22.5±1.8 | |
| vS | 8.8±0.7 | 9.2±1.4 | 11.6±1.2 | 12.1±1.0 | 11.3±1.5 | 15.6±3.4 | |
| lS | 18.0±1.0 | 18.1±0.8 | 21.7±1.3 | 24.5±0.7** | 28.2±1.2*** | 33.6±1.9***,## | |
| middle | m | 19.0±1.1 | 20.9±0.9 | 26.4±1.2** | 27.8±2.3** | 36.3±1.7*** | 42.6±1.2***,## |
| d | 19.8±0.7 | 22.2±0.9 | 28.4±0.6*** | 31.4±2.7*** | 39.5±1.3*** | 50.4±2.1***,### | |
| dl | 18.6±0.9 | 20.2±1.4 | 23.6±1.1 | 26.3±2.1* | 30.9±1.6*** | 42.6±1.9***,### | |
| vl | 17.3±0.9 | 18.1±1.2 | 21.2±1.2 | 22.6±1.8 | 30.4±2.7*** | 45.0±3.5***,### | |
| v | 13.1±0.6 | 13.6±0.9 | 14.9±1.0 | 14.7±1.2 | 19.1±1.7** | 20.6±1.4** | |
| c | 18.0±1.1 | 18.7±1.6 | 30.3±1.6** | 30.4±3.3** | 40.1±3.2*** | 48.4±3.1***,# | |
| caudal | m | 15.5±1.2 | 15.8±1.3 | 19.8±1.5 | 20.0±1.4 | 25.3±2.5** | 33.4±2.5***,## |
| d | 16.3±0.9 | 18.0±1.1 | 26.3±1.0*** | 28.3±2.1*** | 37.4±1.1*** | 49.9±2.3***,### | |
| dl | 13.6±1.4 | 15.6±1.4 | 19.1±1.4 | 20.2±1.4 | 28.4±1.2*** | 34.5±2.9***,# | |
| vl | 13.2±1.1 | 15.7±1.6 | 14.9±1.1 | 16.1±1.2 | 26.3±3.5** | 32.0±4.0*** | |
| v | 11.6±1.1 | 13.5±0.8 | 11.6±0.9 | 10.9±0.9 | 13.2±1.6 | 15.5±1.5 | |
| vc | 8.6±0.9 | 10.5±0.9 | 10.4±0.8 | 10.4±0.9 | 15.2±2.8 | 21.6±3.5***,# | |
| dc | 9.8±1.3 | 11.6±0.7 | 12.0±1.3 | 11.2±1.6 | 20.4±2.4** | 25.3±3.2*** |
Mean density values (mean±SEM) measured in different striatal sectors on rostral, middle and caudal levels for rats that received an injection of vehicle (V), fluoxetine (5 mg/kg; FLX), methylphenidate (2 or 5 mg/kg; MP2, MP5), or methylphenidate + fluoxetine combinations (MP2+FLX, MP5+FLX). Abbreviations: nucleus accumbens: lC, lateral core; mC, medial core; lS, lateral shell; mS, medial shell; vS, ventral shell; caudate-putamen: c, central; d, dorsal; dc, dorsal central; dl, dorsolateral; dm, dorsomedial; m, medial; v, ventral; vc, ventral central; vl, ventrolateral.
P<0.05,
P<0.01,
P<0.001, vs. respective control group (V or FLX);
P<0.05,
P<0.01,
P<0.001, MP+FLX vs. MP (potentiation).
Table 2.
Effects of fluoxetine, methylphenidate and methylphenidate + fluoxetine on c-fos expression in the striatum.
| V | FLX | MP2 | MP2 + FLX |
MP5 | MP5 + FLX |
||
|---|---|---|---|---|---|---|---|
| rostral | dl | 2.2±0.6 | 3.4±0.7 | 4.3±0.7 | 5.2±0.8 | 8.7±0.6*** | 13.0±1.9***,## |
| d | 2.8±0.4 | 3.6±0.4 | 5.2±0.6 | 6.1±1.2 | 8.8±0.4** | 14.8±2.0***,### | |
| dm | 3.2±0.6 | 3.9±0.3 | 5.8±0.5 | 7.5±1.1* | 11.0±0.6*** | 19.1±2.1***,### | |
| m | 3.7±0.5 | 4.8±0.6 | 6.1±0.8 | 7.1±0.7 | 8.0±1.2* | 12.4±1.5***,## | |
| v | 2.3±0.6 | 2.1±0.5 | 4.2±0.8 | 2.7±0.4 | 4.2±0.8 | 7.2±1.7** | |
| mC | 5.1±1.1 | 3.7±0.7 | 5.7±0.6 | 5.5±1.3 | 4.3±1.4 | 4.8±1.7 | |
| lC | 2.1±0.6 | 1.7±0.8 | 3.7±1.4 | 2.5±1.4 | 1.3±0.8 | 2.3±2.4 | |
| mS | 5.2±0.9 | 4.3±0.9 | 6.2±1.0 | 7.4±1.1 | 4.5±1.1 | 5.4±1.3 | |
| vS | 2.1±1.0 | 1.4±0.8 | 3.3±1.6 | 1.4±1.2 | 2.0±1.1 | 1.9±2.2 | |
| lS | 3.7±0.8 | 2.7±0.6 | 5.8±0.9 | 6.1±0.8 | 8.2±1.6 | 11.3±2.0*** | |
| middle | m | 4.1±0.5 | 3.5±1.1 | 7.3±1.0* | 8.3±1.2* | 13.3±1.1*** | 20.9±1.0***,### |
| d | 3.1±0.3 | 2.9±0.9 | 7.0±0.6* | 11.0±2.4***# | 17.1±0.6*** | 32.3±2.2***,### | |
| dl | 2.5±0.6 | 1.6±0.8 | 3.8±0.4 | 4.7±1.1 | 8.7±0.8** | 20.2±1.9***,### | |
| vl | 3.4±1.1 | 2.0±0.9 | 3.9±0.6 | 4.1±1.6 | 8.5±1.0 | 22.5±3.4***,### | |
| v | 4.2±1.1 | 2.6±1.0 | 4.8±0.7 | 2.9±1.4 | 5.6±1.8 | 8.5±1.2* | |
| c | 2.9±0.7 | 1.3±0.7 | 10.3±1.1 | 9.8±2.4* | 18.9±1.5*** | 27.9±4.6***,## | |
| caudal | m | 3.2±0.5 | 3.9±0.6 | 7.9±1.0 | 8.8±1.5 | 12.5±1.9*** | 20.2±2.5***,### |
| d | 3.8±0.3 | 4.2±0.5 | 11.1±1.3** | 11.9±2.0** | 20.2±1.5*** | 34.9±2.7***,### | |
| dl | 2.4±0.4 | 1.6±0.6 | 6.3±1.3 | 5.9±0.7 | 10.9±1.6*** | 18.3±2.6***,### | |
| vl | 1.8±0.7 | 1.7±0.7 | 3.2±1.2 | 3.4±0.9 | 6.4±2.6 | 13.1±2.5***,## | |
| v | 2.6±0.6 | 1.8±0.6 | 3.7±1.2 | 1.9±1.0 | 1.8±1.6 | 4.7±1.3 | |
| vc | 2.1±0.7 | 1.3±0.7 | 3.1±1.2 | 1.4±0.9 | 2.3±2.1 | 6.0±1.2 | |
| dc | 1.5±0.5 | 2.2±0.5 | 5.0±1.3 | 2.0±0.8 | 4.5±3.0 | 8.4±1.9* |
Mean density values (mean±SEM) measured in different striatal sectors on rostral, middle and caudal levels for rats that received an injection of vehicle (V), fluoxetine (5 mg/kg; FLX), methylphenidate (2 or 5 mg/kg; MP2, MP5), or methylphenidate + fluoxetine combinations (MP2+FLX, MP5+FLX). Abbreviations: nucleus accumbens: lC, lateral core; mC, medial core; lS, lateral shell; mS, medial shell; vS, ventral shell; caudate-putamen: c, central; d, dorsal; dc, dorsal central; dl, dorsolateral; dm, dorsomedial; m, medial; v, ventral; vc, ventral central; vl, ventrolateral.
P<0.05,
P<0.01,
P<0.001, vs. respective control group (V or FLX);
P<0.05,
P<0.01,
P<0.001, MP+FLX vs. MP (potentiation).
Figure 4.
Fluoxetine potentiation of methylphenidate-induced zif 268 expression in specific striatal sectors. Mean density values (mean±SEM) for zif 268 expression in rats that received an injection of vehicle (V), fluoxetine (5 mg/kg; FLX), methylphenidate (2 or 5 mg/kg; MP2, MP5), or methylphenidate + fluoxetine combinations (MP2+FLX and MP5+FLX) (n=5–7) are depicted for 4 middle striatal sectors (top) and the 5 sectors of the nucleus accumbens (bottom). Abbreviations: caudate-putamen: d, dorsal; dl, dorsolateral; vl, ventrolateral; c, central; nucleus accumbens: mC, medial core; lC, lateral core; mS, medial shell; vS, ventral shell; lS, lateral shell. * P<0.05, ** P<0.01, *** P<0.001 vs. respective control group (V or FLX); # P<0.05, ## P<0.01, ### P<0.001, MP+FLX vs. MP (potentiation).
In contrast to methylphenidate, fluoxetine (5 mg/kg) alone did not modify gene expression, neither in the caudate-putamen nor in the nucleus accumbens (Figs. 3–6). None of the 23 sectors showed significant changes in zif 268 or c-fos expression (P>0.05 vs. V, Figs. 4 and 6, Tables 1 and 2).
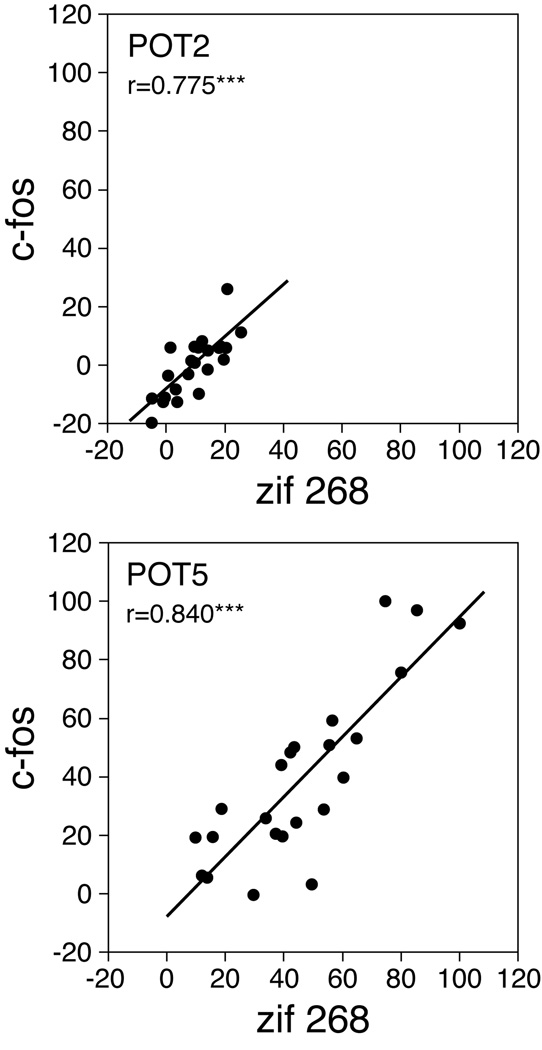
However, when given in conjunction with methylphenidate, fluoxetine potentiated methylphenidate-induced IEG expression in the striatum. Correlation analyses show that the regional distribution of this potentiation (POT; i.e., the difference between MP+FLX and MP) was similar for zif 268 and c-fos expression (zif 268 x c-fos: methylphenidate 2 mg/kg, POT2, r=0.775, P<0.001; 5 mg/kg, POT5, r=0.840, P<0.001; Fig. 7). Moreover, despite relatively modest potentiation for 2 mg/kg of methylphenidate, principally the same sectors were affected as for 5 mg/kg (POT2 x POT5: zif 268, r=0.659, P<0.05; c-fos, r=0.759, P<0.01; not shown).
Figure 7.
The potentiation of gene induction displays a similar regional distribution in the striatum for zif 268 and c-fos. Scatterplots show the correlations between zif 268 and c-fos potentiation for 2 mg/kg (r=0.775, POT2, top) and 5 mg/kg (r=0.840, POT5, bottom) of methylphenidate in the 23 striatal sectors. The values are expressed as the percentages of the maximal value in the 5 mg/kg group. *** P<0.001.
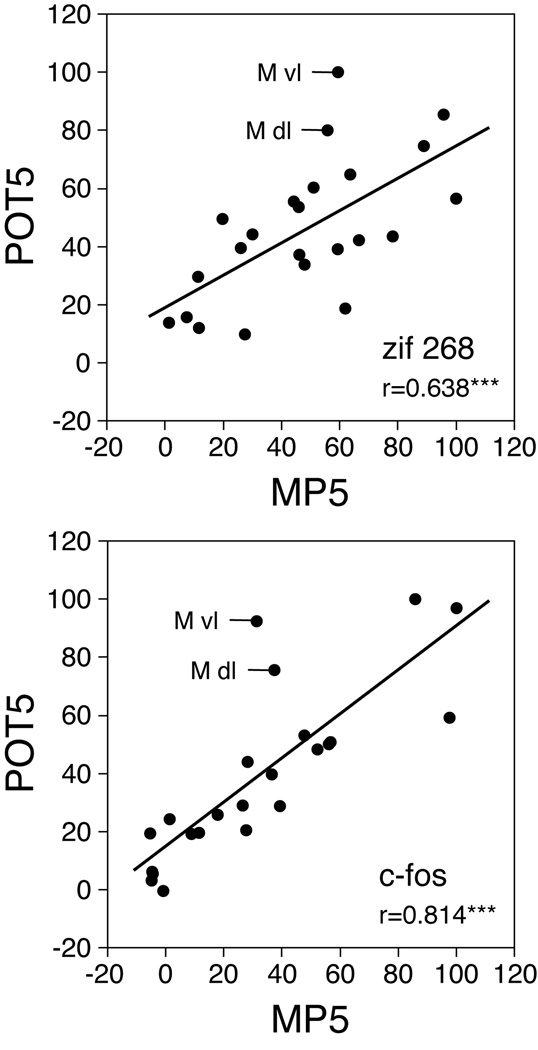
The fluoxetine potentiation was reflected in a higher proportion of the 23 striatal sectors displaying significantly increased zif 268 and c-fos expression after the methylphenidate + fluoxetine treatment, compared with methylphenidate alone (zif 268: MP2+FLX vs. MP2, 10 sectors vs. 5 sectors; MP5+FLX vs. MP5, 19 vs. 17; c-fos: 6 vs. 3 and 16 vs. 11; Fig. 6). Direct statistical comparisons showed that, for the 2 mg/kg dose of methylphenidate, c-fos induction was significantly more robust in the MP2+FLX group than in the MP2 group in one sector (middle level, dorsal sector; Fig. 6, POT2). For the 5 mg/kg dose, the potentiation (MP5+FLX vs. MP5, POT5) was statistically significant in 15 and 13 of the 23 striatal sectors, for zif 268 and c-fos, respectively. Further analysis showed that the magnitude of the fluoxetine potentiation was principally related to the magnitude of gene induction produced by methylphenidate alone (5 mg/kg, MP5 x POT5: zif 268, r=0.638, P<001; c-fos, r=0.814, P<001; Fig. 8; 2 mg/kg: P>0.05; not shown). However, a more pronounced potentiation than predicted by the methylphenidate response was seen in the dorsolateral and ventrolateral (sensorimotor) sectors of the middle striatum for both zif 268 and c-fos expression (Fig. 8). This finding confirms that the lateral (sensorimotor) striatum displayed a more pronounced fluoxetine potentiation of gene regulation than the medial (associative) striatum; this is also apparent in the potentiation maps (Fig. 6).
Figure 8.
Relationship between the gene induction by methylphenidate alone and the fluoxetine potentiation in the different striatal sectors. Scatterplots depict the correlations between gene induction by 5 mg/kg methylphenidate (MP5) and the fluoxetine potentiation (POT5) in the 23 sectors, for zif 268 (r=0.638, top) and c-fos (r=0.814, bottom). The values are expressed as the percentages of the maximal value in each group. Abbreviations: M vl, middle level ventrolateral sector; M dl, middle level dorsolateral sector. *** P<0.001.
Fluoxetine also potentiated methylphenidate-induced gene expression in selective regions of the nucleus accumbens, predominantly in the lateral part of the shell (Figs. 4 and 6). After MP2+FLX, but not MP2 alone, zif 268 expression was significantly increased in the lateral shell, as was c-fos expression after MP5+FLX, but not MP5 alone. For zif 268, a significant potentiation in MP5+FLX vs. MP5 animals (POT5) was seen in the lateral shell as well as in the medial core (Fig. 6).
Effects of fluoxetine on methylphenidate-induced zif 268 expression in the cortex
Administration of methylphenidate alone induced a dose-dependent up-regulation of zif 268 expression in the cortex on all four rostrocaudal levels (Fig. 9, Table 3). A statistically significant increase in zif 268 mRNA levels was observed in 7 (MP2) and 11 (MP5) of the 22 cortical areas (P<0.05 vs. V). However, this effect was restricted to dorsomedial cortical regions, including the cingulate, medial agranular and motor cortex (mainly on rostral to caudal levels), as well as the prelimbic and insular/lateral orbital cortex (frontal level). These are mostly limbic and associative areas. In contrast, the somatosensory cortex and insular cortex (except frontal level) were not affected by methylphenidate on any rostrocaudal levels (MP2 or MP5 vs. V, P>0.05).
Figure 9.
Effects of methylphenidate + fluoxetine combination treatment on zif 268 expression in the cortex. Maps show the distribution of zif 268 expression in the cortex on frontal, rostral, middle and caudal levels after an injection of fluoxetine (5 mg/kg; FLX), methylphenidate (2 or 5 mg/kg; MP2, MP5), or methylphenidate + fluoxetine combinations (MP2+FLX, MP5+FLX). The potentiation (POT) denotes the difference between methylphenidate + fluoxetine and methylphenidate alone. The data are normalized relative to the maximal increase observed in the striatum (% of max.). Sectors with significant differences vs. vehicle-treated controls (P<0.05) are coded as indicated. Sectors without significant effects are in white. Abbreviations: CG, cingulate; M2, medial agranular; M1, motor; SS, somatosensory; I, insular; IL, infralimbic; PL, prelimbic, I/LO, insular/lateral orbital.
Table 3.
Effects of fluoxetine, methylphenidate and methylphenidate + fluoxetine on zif 268 expression in the cortex.
| V | FLX | MP5 | MP5 + FLX |
||
|---|---|---|---|---|---|
| frontal | IL | 24.9±1.2 | 28.1±1.2 | 29.9±2.0 | 33.2±1.2 |
| PL | 38.2±2.3 | 41.9±1.7 | 46.3±2.9* | 48.4±1.2 | |
| CG | 39.4±1.8 | 45.5±2.3* | 51.5±2.5*** | 50.5±1.1 | |
| M2 | 26.1±1.9 | 32.0±2.7 | 33.4±1.8 | 36.8±0.7 | |
| M1 | 18.5±2.5 | 23.0±3.4 | 19.6±2.1 | 23.9±2.0 | |
| SS | 18.6±1.9 | 22.6±3.5 | 20.0±2.0 | 23.2±2.2 | |
| I/LO | 33.4±0.9 | 36.4±1.8 | 40.3±1.5** | 39.8±0.8 | |
| rostral | CG | 42.3±1.6 | 43.6±2.9 | 50.4±1.6* | 50.8±1.4 |
| M2 | 37.3±1.9 | 39.7±2.1 | 46.1±1.6** | 51.2±1.4*** | |
| M1 | 25.9±1.9 | 28.6±2.6 | 32.7±1.6 | 37.8±1.3* | |
| SS | 23.3±2.5 | 26.1±4.4 | 25.8±2.2 | 31.0±2.6 | |
| I | 22.9±0.7 | 26.1±2.3 | 27.7±2.1 | 30.0±2.0 | |
| middle | CG | 38.0±1.7 | 41.1±2.8 | 46.6±2.0* | 46.8±1.7 |
| M2 | 33.4±1.7 | 40.4±2.8 | 46.3±2.1** | 45.3±1.2 | |
| M1 | 28.1±1.5 | 31.6±1.7 | 36.6±1.8** | 35.8±1.9 | |
| SS | 25.4±2.1 | 28.0±3.5 | 28.2±1.7 | 31.0±2.6 | |
| I | 20.3±1.1 | 23.2±2.5 | 22.7±2.6 | 24.0±1.9 | |
| caudal | CG | 41.9±2.0 | 43.9±2.7 | 52.6±1.5** | 52.5±0.7* |
| M2 | 37.5±1.5 | 40.6±1.6 | 45.2±1.7** | 47.1±1.6* | |
| M1 | 32.5±1.3 | 38.6±0.8** | 40.4±1.6*** | 42.8±0.8 | |
| SS | 28.3±2.2 | 31.1±2.9 | 33.2±2.2 | 36.2±1.8 | |
| I | 18.0±1.1 | 17.5±1.7 | 17.6±1.3 | 18.0±1.1 |
Mean density values (mean±SEM) measured in different cortical areas on frontal, rostral, middle and caudal levels for rats that received an injection of vehicle (V), fluoxetine (5 mg/kg; FLX), methylphenidate (5 mg/kg; MP5), or methylphenidate + fluoxetine (MP5+FLX). Abbreviations: CG, cingulate; I, insular; IL, infralimbic; LO, lateral orbital; M1, motor; M2, medial agranular; PL, prelimbic; SS, somatosensory.
P<0.05,
P<0.01,
P<0.001, vs. respective control group (V or FLX).
Fluoxetine given alone tended to increase zif 268 mRNA levels in many cortical areas, but this effect was statistically significant only in the cingulate (frontal level) and motor cortex (caudal level) (FLX vs. V, P<0.05 and P<0.01, respectively, Fig. 9 and Table 3). After the methylphenidate + fluoxetine treatment, significantly enhanced zif 268 expression was found in eight (MP2+FLX) and 14 (MP5+FLX) of the 22 cortical areas (as compared to 7 and 11, respectively, for methylphenidate only, see above). However, the methylphenidate + fluoxetine treatment did not produce significantly higher zif 268 mRNA levels than methylphenidate alone in any cortical area (MP+FLX vs. MP; P>0.05), except in the infralimbic cortex on the frontal level for 2 mg/kg of methylphenidate (Fig. 9; MP2+FLX vs. MP2; P<0.01; MP5+FLX vs. MP5; P>0.05). Therefore, in contrast to the striatum, this dose of fluoxetine did not robustly potentiate cortical gene regulation by methylphenidate.
Our earlier findings (Yano & Steiner, 2005a; Cotterly et al., 2007) showed that there is a positive correlation between psychostimulant-induced gene expression in cortical areas and gene induction in the striatal sectors targeted by these cortical areas, indicating coordinated molecular changes in cortical and striatal nodes of corticostriatal circuits. We assessed whether such a relationship existed in the present study and whether it was affected by the present SSRI treatments. Thus, drug-induced increases in zif 268 expression in cortical areas were compared with those in their respective striatal target sectors (see Fig. 1). When a striatal sector received input from more than one cortical area, the values of these cortical areas were averaged. Our results show that, overall, methylphenidate-induced zif 268 expression in the cortical areas was positively correlated with that in their connected 23 striatal sectors (MP2, r=0.493, P<0.05; MP5, r=0.436, P<0.05), confirming our earlier findings (Yano & Steiner, 2005a; Cotterly et al., 2007). This effect was more robust when only the 18 sectors of the caudate-putamen were included (MP2, r=0.556, P<0.05; MP5, r=0.676, P<0.01). The addition of fluoxetine to methylphenidate weakened this correlation (23 sectors: MP2+FLX, r=0.326, P>0.05; MP5+FLX, r=0.307, P>0.05; 18 sectors: MP2+FLX, r=0.503, P<0.05; MP5+FLX, r=0.567, P<0.05).
Effects of citalopram on methylphenidate-induced IEG expression in the striatum
We also assessed whether the potentiation of methylphenidate-induced gene regulation generalized to other SSRIs. Our results demonstrate that this is the case. Administration of the SSRI citalopram (5 mg/kg) together with methylphenidate (5 mg/kg) potentiated methylphenidate-induced expression of zif 268 (Fig. 10) and c-fos (not shown) in the striatum. The regional distribution of this potentiation was similar to that produced by fluoxetine.
DISCUSSION
In these studies, we demonstrate that concomitant administration of serotonin reuptake inhibitors (fluoxetine, citalopram) robustly potentiates gene regulation by the psychostimulant and dopamine reuptake blocker methylphenidate, consistent with the notion that serotonin facilitates dopamine-mediated gene regulation (see Yano & Steiner, 2007). Our findings show enhanced induction of IEG transcription factors (zif 268 and c-fos) in the striatum, with the most robust effects occurring in sensorimotor parts, which mediate motor learning/habit formation and are implicated in compulsive aspects of drug taking (see below), and more modest effects in the nucleus accumbens, which participates in reward processes. These molecular changes were associated with selective potentiation of motor stereotypies, which are thought to reflect dysfunction in sensorimotor striatal circuits and may be related to compulsive behavior.
SSRIs potentiate methylphenidate-induced IEG expression in the striatum and nucleus accumbens
Activation of transcription factors by psychostimulants regulates the expression of effector genes and is thus critical for many forms of long-term neuroplasticity related to addiction. The effects of methylphenidate on zif 268 and c-fos expression emerged with 2 mg/kg (i.p.) and were more pronounced with 5 mg/kg, consistent with previous findings (e.g., Brandon & Steiner, 2003; Yano & Steiner, 2005b; see also Yano & Steiner, 2007). Our present results show that 5 mg/kg of fluoxetine, which by itself had no effect on gene expression, potentiated gene regulation by both 2 and 5 mg/kg of methylphenidate, with a more robust potentiation for the higher methylphenidate dose. We further confirmed this SSRI potentiation of methylphenidate-induced gene regulation with a second SSRI, citalopram (5 mg/kg). These findings highlight the ability of SSRIs to potentiate psychostimulant-induced molecular changes.
Acute IEG induction by psychostimulants is predictive of altered gene regulation after repeated psychostimulant treatments (Brandon & Steiner, 2003; Willuhn et al., 2003; Cotterly et al., 2007; Unal et al., 2009) and thus serves as a marker to identify brain regions prone to such neuroplasticity induced by repeated treatments. In so far as such molecular changes underlie addiction (Hyman & Nestler, 1996; Berke & Hyman, 2000; Nestler, 2001), it is of interest to compare the propensity of methylphenidate to alter gene regulation with that of psychostimulants such as cocaine (Yano & Steiner, 2007). While similar to cocaine effects in many ways, methylphenidate effects after acute and repeated treatment also display distinct differences (for review, see Yano & Steiner, 2007). For example, previous mapping studies revealed, and the present results confirmed, that methylphenidate-induced zif 268 and c-fos expression is most pronounced on middle-to-caudal striatal levels (Brandon & Steiner, 2003; Yano & Steiner, 2005a,b; Cotterly et al., 2007), whereas cocaine-induced gene regulation peaks on more caudal levels (Willuhn et al., 2003; Unal et al., 2009). As to the medial-lateral distribution, methylphenidate-induced gene regulation is most robust in medial and central striatal regions (associative striatum) (see above), again confirmed here, whereas cocaine prominently involves the lateral (sensorimotor) striatum as well (Willuhn et al., 2003; Unal et al., 2009).
Our present findings demonstrate that the SSRI potentiation of methylphenidate-induced gene regulation occurs on all rostrocaudal levels of the striatum and, generally, is directly related to the magnitude of gene induction by methylphenidate alone. Thus, the addition of the SSRI did not produce a shift in the rostrocaudal distribution of gene regulation. However, this was not the case for the medial-lateral distribution. Our correlation analysis shows that two striatal sectors, the dorsolateral and ventrolateral (sensorimotor) sectors on the middle level, displayed a more pronounced potentiation than predicted by the gene response to methylphenidate alone. This preferential potentiation in these two sensorimotor sectors shifted the regional distribution to also include the lateral striatum. In this respect, SSRI-potentiated gene regulation by methylphenidate resembles more gene regulation induced by cocaine (Willuhn et al., 2003; Unal et al., 2009). As these preferentially affected lateral striatal regions subserve habit formation (Packard & Knowlton, 2002), it will be important to determine whether such concomitant SSRI + methylphenidate treatment facilitates drug taking habits/addiction, similar to cocaine (see also below).
Our earlier studies showed that, in addition to the dorsal striatum, methylphenidate-induced gene regulation also occurs in the nucleus accumbens, although to a more modest extent (Brandon & Steiner, 2003; Yano & Steiner, 2005a,b; Cotterly et al., 2007). Consistent with these earlier findings, the most robust gene regulation in the nucleus accumbens in the present study was seen in the lateral part of the shell. Fluoxetine also produced a statistically significant potentiation of methylphenidate-induced zif 268 expression in the lateral shell (as well as in the medial core), but had no effect on c-fos expression. These regional effects also mimic those of cocaine (Unal et al., 2009). The functional consequences of these changes in specific nucleus accumbens subregions remain to be determined.
Mechanisms that may mediate the SSRI potentiation
A number of mechanisms may account for the SSRI potentiation of methylphenidate-induced gene regulation as described in the present study. It is unlikely that metabolic interactions between methylphenidate and SSRIs contributed to the observed effects. The principal metabolic pathway for methylphenidate is deesterification by carboxylesterases (e.g., Sun et al., 2004; Zhu et al., 2008), and, to our knowledge, there is no evidence for an inhibition of carboxylesterases by SSRIs. Conversely, the metabolism of SSRIs involves mostly demethylation by liver enzymes (cytochrome P450 system; Sandson et al., 2005). While there is some evidence that methylphenidate can inhibit such liver enzymes (Le Nedelec & Rosengren, 2002), it is unclear whether the specific P450 isozymes that metabolize fluoxetine and citalopram (Sandson et al., 2005) are affected. In addition, a recent study failed to find altered pharmacokinetics for methylphenidate (10 mg/kg) after the addition of citalopram (5 mg/kg), while this drug combination facilitated dopamine overflow in the prefrontal cortex (Weikop et al., 2007). Furthermore, it would be difficult to envision how systemic drug interactions could result in a potentiation of gene regulation with the distinct regional variations as observed here.
More likely, this potentiation is mediated by systems-level interactions between the dopamine and serotonin neurotransmissions (e.g., Bhat & Baraban, 1993; Gardier et al., 2000). The effects of psychostimulants (including methylphenidate; Yano et al., 2006) on gene expression in the striatum are principally mediated by the activation of dopamine receptors (for reviews, see Steiner & Gerfen, 1998; Yano & Steiner, 2007), but are also dependent on cortical (glutamate) inputs (e.g., Wang & McGinty, 1996; Steiner, 2010). Both glutamate and dopamine inputs are modulated by serotonin. For example, serotonin and agonists are well-known to enhance activity of the mesostriatal and mesolimbic/cortical dopamine pathways, by complex interactions in both the dopamine terminal regions (e.g., Benloucif & Galloway, 1991; Benloucif et al., 1993; Balcioglu & Wurtman, 1998; Bubar et al., 2003) as well as in the somatodendritic areas in the midbrain (for reviews, see Muller & Huston, 2006; Weikop et al., 2007; Bubar & Cunningham, 2008). Consistent with these findings, a facilitatory role for serotonin in dopamine/glutamate-mediated gene regulation in the striatum has been shown before (Bhat & Baraban, 1993; Torres & Rivier, 1993; Guerra et al., 1998; Wirtshafter & Cook, 1998; Gardier et al., 2000; Horner et al., 2005). Therefore, the present SSRI potentiation of methylphenidate-induced gene regulation in the striatum could reflect increased cortical input to the striatum and/or potentiated dopamine action that occurs with enhanced serotonin activity in the striatum and/or other brain areas.
Since drugs were administered systemically, we can not conclude which of the above local mechanisms played a role in this SSRI potentiation. However, our further analysis suggests that enhanced cortical input is not a main determinant. Our previous work showed that acute administration of methylphenidate produces coordinated IEG induction in cortical neurons and their striatal targets (Yano & Steiner, 2005a; Cotterly et al., 2007), which suggests enhanced activity in specific corticostriatal circuits. In order to assess a possible contribution of enhanced cortical activity, we thus compared zif 268 induction between specific cortical areas and their striatal target sectors (see Willuhn et al., 2003). Our results confirmed coordinated up-regulation of zif 268 expression between cortical areas and functionally related striatal sectors for both doses of methylphenidate used. However, the co-administration of fluoxetine with methylphenidate disrupted this coordinated response, mainly because fluoxetine robustly potentiated gene induction in the striatum, but not in the cortex. This dissociation suggests that the potentiated gene regulation in the striatum is not likely a consequence of enhanced activity in corticostriatal projections.
A host of serotonin receptor subtypes are known to mediate serotonin/dopamine interactions (for reviews, see Muller & Huston, 2006; Bubar & Cunningham, 2008) and to regulate striatal gene expression (Keefe & Horner, 2010), and could thus conceivably contribute to the SSRI potentiation. Interestingly, a recent study provided evidence for a role of the 5-HT1B receptor subtype in fluoxetine-potentiated methylphenidate effects (Borycz et al., 2008). This study showed that adjunct treatment with fluoxetine facilitated the stimulatory effects of methylphenidate on locomotor activity (Borycz et al., 2008). Moreover, this behavioral potentiation by fluoxetine was inhibited by a selective 5-HT1B receptor antagonist and mimicked by a 5-HT1B receptor agonist (Borycz et al. 2008). 5-HT1B receptors have previously also been shown to induced IEG expression (Wirtshafter & Cook, 1998) and facilitate cocaine-induced gene regulation (Lucas et al., 1997; Castanon et al., 2000) in the striatum. Therefore, the 5-HT1B serotonin receptor subtype may be one of the mediators of the SSRI potentiation of striatal gene regulation and could thus be a potential target for preventing this effect. Future studies with local administration of selective serotonin receptor agents will have to determine the relevant serotonin receptors in the striatum and/or other brain regions.
Stereotypies: Behavioral correlates of the SSRI-potentiated striatal gene regulation
In our study, the fluoxetine potentiation of methylphenidate-induced gene regulation in the striatum was accompanied by potentiation of behavioral stereotypies. Similar to the gene regulation effects, this behavioral potentiation was dose-dependent; it emerged with the lower methylphenidate dose (2 mg/kg; statistically not significant) and was very robust with the higher dose (5 mg/kg). While this behavioral effect is principally consistent with the above findings by Borycz et al. (2008), we did not see potentiated locomotor activity (ambulation) in our study. Different drug doses for fluoxetine or methylphenidate (10 mg/kg each in Borycz et al., 2008) and/or other experimental variables (e.g., non-habituated animals in our study vs. habituated animals in the study by Borycz et al.) may account for these differences.
Previous work has related behavioral stereotypies to drug-induced dysfunction and molecular changes in sensorimotor striatal circuits (for reviews, see Graybiel et al., 2000; Steiner, 2010). “Stereotypies” denotes a compulsive repetition of specific behavioral elements without apparent purpose (Randrup & Munkvad, 1967; Ellinwood & Balster, 1974). Sensorimotor striatal circuits are critical for selection and switching of motor actions (and thoughts) (e.g., Mink, 1996; Redgrave et al., 1999), and behavioral stereotypies have been interpreted as a switching dysfunction (Redgrave & Gurney, 2006). It has also been pointed out that such stereotypies share both phenomenological characteristics (Randrup & Munkvad, 1967) and neuronal substrates (Graybiel et al., 2000) with motor compulsions in humans. It is presently unclear whether stereotypies are mechanistically related to compulsions; however, stereotypies may to some degree reflect similar basal ganglia output deficiencies (see Steiner, 2010). Our findings thus suggest that one of the functional consequences of concomitant methylphenidate plus SSRI treatment is an enhanced liability for compulsive-like behavior.
Functional implications of SSRI-potentiated gene regulation by psychostimulants
A wealth of literature demonstrates that the most robust molecular changes induced by psychostimulants such as cocaine (Willuhn et al., 2003; Unal et al., 2009), amphetamine (Badiani et al., 1998; Uslaner et al., 2003) and methylphenidate (Yano & Steiner, 2005b; Cotterly et al., 2007) occur in the dorsal/lateral striatum that includes sensorimotor circuits (for reviews, see Berke & Hyman, 2000; Steiner, 2010). Our present findings show that the SSRI potentiation of methylphenidate-induced gene regulation preferentially occurs in the sensorimotor striatum. The sensorimotor striatum is known to mediate stimulus-response (SR) learning/habit formation (Graybiel, 1995; Packard & Knowlton, 2002), and it has been proposed that such drug-induced molecular changes may contribute to aberrant habit formation and compulsive aspects of drug taking (Berke & Hyman, 2000; Everitt & Robbins, 2005). Indeed, previous studies showed that the sensorimotor striatum is critical for relapse to cocaine seeking in the self-administration paradigm in animals (Vanderschuren et al., 2005; Fuchs et al., 2006; See et al., 2007).
Methylphenidate is also self-administered (Kollins et al., 2001), and pretreatment with this psychostimulant facilitates subsequent self-administration of cocaine (Brandon et al., 2001) in animal models of addiction, both signifying a certain addiction liability for methylphenidate. Given that the SSRI potentiation of striatal gene regulation is paralleled by more pronounced compulsive-like behavior, future studies will have to determine whether methylphenidate+SSRI combinations enhance drug seeking/taking tendencies and/or facilitate relapse.
Conclusions
Our study demonstrates that SSRIs potentiate gene regulation effects of methylphenidate in the striatum, with the most robust effects in sensorimotor parts. Such molecular changes are implicated in psychostimulant addiction. These findings thus suggest that such concomitant drug exposure, for example, during medical treatments or in patients on SSRIs who also use methylphenidate as a cognitive enhancer or for recreational purposes, may increase the liability for drug abuse disorder.
Acknowledgments
This work was supported by National Institutes of Health Grants DA011261 (HS) and DA020654 (MM). We thank Joel Beverley for excellent technical assistance.
References
- Adriani W, Leo D, Greco D, Rea M, di Porzio U, Laviola G, Perrone-Capano C. Methylphenidate administration to adolescent rats determines plastic changes in reward-related behavior and striatal gene expression. Neuropsychopharmacology. 2006a;31:1946–1956. doi: 10.1038/sj.npp.1300962. [DOI] [PubMed] [Google Scholar]
- Adriani W, Leo D, Guarino M, Natoli A, Di Consiglio E, De Angelis G, Traina E, Testai E, Perrone-Capano C, Laviola G. Short-term effects of adolescent methylphenidate exposure on brain striatal gene expression and sexual/endocrine parameters in male rats. Ann. N. Y. Acad. Sci. 2006b;1074:52–73. doi: 10.1196/annals.1369.005. [DOI] [PubMed] [Google Scholar]
- Andersen SL. Stimulants and the developing brain. Trends Pharmacol. Sci. 2005;26:237–243. doi: 10.1016/j.tips.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Badiani A, Oates MM, Day HE, Watson SJ, Akil H, Robinson TE. Amphetamine-induced behavior, dopamine release, and c-fos mRNA expression: modulation by environmental novelty. J. Neurosci. 1998;18:10579–10593. doi: 10.1523/JNEUROSCI.18-24-10579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcioglu A, Wurtman RJ. Dexfenfluramine enhances striatal dopamine release in conscious rats via a serotoninergic mechanism. J. Pharmacol. Exp. Ther. 1998;284:991–997. [PubMed] [Google Scholar]
- Benloucif S, Galloway MP. Facilitation of dopamine release in vivo by serotonin agonists: studies with microdialysis. Eur. J. Pharmacol. 1991;200:1–8. doi: 10.1016/0014-2999(91)90658-d. [DOI] [PubMed] [Google Scholar]
- Benloucif S, Keegan MJ, Galloway MP. Serotonin-facilitated dopamine release in vivo: pharmacological characterization. J. Pharmacol. Exp. Ther. 1993;265:373–377. [PubMed] [Google Scholar]
- Berke JD, Hyman SE. Addiction, dopamine, and the molecular mechanisms of memory. Neuron. 2000;25:515–532. doi: 10.1016/s0896-6273(00)81056-9. [DOI] [PubMed] [Google Scholar]
- Bhat RV, Baraban JM. Activation of transcription factor genes in striatum by cocaine: role of both serotonin and dopamine systems. J. Pharmacol. Exp. Ther. 1993;267:496–505. [PubMed] [Google Scholar]
- Bhatara V, Feil M, Hoagwood K, Vitiello B, Zima B. National trends in concomitant psychotropic medication with stimulants in pediatric visits: practice versus knowledge. J. Atten. Disord. 2004;7:217–226. doi: 10.1177/108705470400700404. [DOI] [PubMed] [Google Scholar]
- Bogle KE, Smith BH. Illicit methylphenidate use: a review of prevalence, availability, pharmacology, and consequences. Curr. Drug Abuse Rev. 2009;2:157–176. doi: 10.2174/1874473710902020157. [DOI] [PubMed] [Google Scholar]
- Borycz J, Zapata A, Quiroz C, Volkow ND, Ferré S. 5-HT(1B) receptor-mediated serotoninergic modulation of methylphenidate-induced locomotor activation in rats. Neuropsychopharmacology. 2008;33:619–626. doi: 10.1038/sj.npp.1301445. [DOI] [PubMed] [Google Scholar]
- Brandon CL, Marinelli M, Baker LK, White FJ. Enhanced reactivity and vulnerability to cocaine following methylphenidate treatment in adolescent rats. Neuropsychopharmacology. 2001;25:651–661. doi: 10.1016/S0893-133X(01)00281-0. [DOI] [PubMed] [Google Scholar]
- Brandon CL, Steiner H. Repeated methylphenidate treatment in adolescent rats alters gene regulation in the striatum. Eur. J. Neurosci. 2003;18:1584–1592. doi: 10.1046/j.1460-9568.2003.02892.x. [DOI] [PubMed] [Google Scholar]
- Bubar MJ, Cunningham KA. Prospects for serotonin 5-HT2R pharmacotherapy in psychostimulant abuse. Prog. Brain Res. 2008;172:319–346. doi: 10.1016/S0079-6123(08)00916-3. [DOI] [PubMed] [Google Scholar]
- Bubar MJ, McMahon LR, De Deurwaerdère P, Spampinato U, Cunningham KA. Selective serotonin reuptake inhibitors enhance cocaine-induced locomotor activity and dopamine release in the nucleus accumbens. Neuropharmacology. 2003;44:342–353. doi: 10.1016/s0028-3908(02)00381-7. [DOI] [PubMed] [Google Scholar]
- Carlezon WAJ, Konradi C. Understanding the neurobiological consequences of early exposure to psychotropic drugs: linking behavior with molecules. Neuropharmacology. 2004;47:47–60. doi: 10.1016/j.neuropharm.2004.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanon N, Scearce-Levie K, Lucas JJ, Rocha B, Hen R. Modulation of the effects of cocaine by 5-HT1B receptors: a comparison of knockouts and antagonists. Pharmacol. Biochem. Behav. 2000;67:559–566. doi: 10.1016/s0091-3057(00)00389-0. [DOI] [PubMed] [Google Scholar]
- Chase TD, Brown RE, Carrey N, Wilkinson M. Daily methylphenidate administration attenuates c-fos expression in the striatum of prepubertal rats. Neuroreport. 2003;14:769–772. doi: 10.1097/00001756-200304150-00022. [DOI] [PubMed] [Google Scholar]
- Cotterly L, Beverley JA, Yano M, Steiner H. Dysregulation of gene induction in corticostriatal circuits after repeated methylphenidate treatment in adolescent rats: Differential effects on zif 268 and homer 1a. Eur. J. Neurosci. 2007;25:3617–3628. doi: 10.1111/j.1460-9568.2007.05570.x. [DOI] [PubMed] [Google Scholar]
- Csoka A, Bahrick A, Mehtonen OP. Persistent sexual dysfunction after discontinuation of selective serotonin reuptake inhibitors. J. Sex. Med. 2008;5:227–233. doi: 10.1111/j.1743-6109.2007.00630.x. [DOI] [PubMed] [Google Scholar]
- Ellinwood EH, Jr, Balster RL. Rating the behavioral effects of amphetamine. Eur. J. Pharmacol. 1974;28:35–41. doi: 10.1016/0014-2999(74)90109-5. [DOI] [PubMed] [Google Scholar]
- Everitt BJ, Robbins TW. Neural systems of reinforcement for drug addiction: from actions to habits to compulsion. Nat. Neurosci. 2005;8:1481–1489. doi: 10.1038/nn1579. [DOI] [PubMed] [Google Scholar]
- Fuchs RA, Branham RK, See RE. Different neural substrates mediate cocaine seeking after abstinence versus extinction training: A critical role for the dorsolateral caudate-putamen. J. Neuroscience. 2006;26:3584–3588. doi: 10.1523/JNEUROSCI.5146-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardier AM, Moratalla R, Cuellar B, Sacerdote M, Guibert B, Lebrec H, Graybiel AM. Interaction between the serotoninergic and dopaminergic systems in d-fenfluramine-induced activation of c-fos and jun B genes in rat striatal neurons. J. Neurochem. 2000;74:1363–1373. doi: 10.1046/j.1471-4159.2000.0741363.x. [DOI] [PubMed] [Google Scholar]
- Graybiel AM. Building action repertoires: memory and learning functions of the basal ganglia. Curr. Opin. Neurobiol. 1995;5:733–741. doi: 10.1016/0959-4388(95)80100-6. [DOI] [PubMed] [Google Scholar]
- Graybiel AM, Canales JJ, Capper-Loup C. Levodopa-induced dyskinesias and dopamine-dependent stereotypies: a new hypothesis. Trends Neurosci. 2000;23:S71–S77. doi: 10.1016/s1471-1931(00)00027-6. [DOI] [PubMed] [Google Scholar]
- Greely H, Sahakian B, Harris J, Kessler RC, Gazzaniga M, Campbell P, Farah MJ. Towards responsible use of cognitive-enhancing drugs by the healthy. Nature. 2008;456:702–705. doi: 10.1038/456702a. [DOI] [PubMed] [Google Scholar]
- Guerra MJ, Liste I, Labandeira-Garcia JL. Interaction between the serotonergic, dopaminergic, and glutamatergic systems in fenfluramine-induced Fos expression in striatal neurons. Synapse. 1998;28:71–82. doi: 10.1002/(SICI)1098-2396(199801)28:1<71::AID-SYN9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Horner KA, Adams DH, Hanson GR, Keefe KA. Blockade of stimulant-induced preprodynorphin mRNA expression in the striatal matrix by serotonin depletion. Neuroscience. 2005;131:67–77. doi: 10.1016/j.neuroscience.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Hyman SE, Nestler EJ. Initiation and adaptation: a paradigm for understanding psychotropic drug action. Am. J. Psychiatry. 1996;153:151–162. doi: 10.1176/ajp.153.2.151. [DOI] [PubMed] [Google Scholar]
- Ishii M, Tatsuzawa Y, Yoshino A, Nomura S. Serotonin syndrome induced by augmentation of SSRI with methylphenidate. Psychiatry Clin. Neurosci. 2008;62:246. doi: 10.1111/j.1440-1819.2008.01767.x. [DOI] [PubMed] [Google Scholar]
- Keefe KA, Horner KA. Neurotransmitter regulation of basal ganglia gene expression. In: Steiner H, Tseng KY, editors. Handbook of Basal Ganglia Structure and Function. London: Academic Press/Elsevier; 2010. pp. 461–490. [Google Scholar]
- Kollins SH. ADHD, Substance Use Disorders, and Psychostimulant Treatment: Current Literature and Treatment Guidelines. J. Atten. Disord. 2008 doi: 10.1177/1087054707311654. X, epub. [DOI] [PubMed] [Google Scholar]
- Kollins SH, MacDonald EK, Rush CR. Assessing the abuse potential of methylphenidate in nonhuman and human subjects: a review. Pharmacol. Biochem. Behav. 2001;68:611–627. doi: 10.1016/s0091-3057(01)00464-6. [DOI] [PubMed] [Google Scholar]
- Kuczenski R, Segal DS. Effects of methylphenidate on extracellular dopamine, serotonin, and norepinephrine: comparison with amphetamine. J. Neurochem. 1997;68:2032–2037. doi: 10.1046/j.1471-4159.1997.68052032.x. [DOI] [PubMed] [Google Scholar]
- Lavretsky H, Kim MD, Kumar A, Reynolds CF. Combined treatment with methylphenidate and citalopram for accelerated response in the elderly: an open trial. J. Clin. Psychiatry. 2003;64:1410–1414. doi: 10.4088/jcp.v64n1202. [DOI] [PubMed] [Google Scholar]
- Le Nedelec MJ, Rosengren RJ. Methylphenidate inhibits cytochrome P450 in the Swiss Webster mouse. Hum. Exp. Toxicol. 2002;21:273–280. doi: 10.1191/0960327102ht245oa. [DOI] [PubMed] [Google Scholar]
- Lin JS, Hou Y, Jouvet M. Potential brain neuronal targets for amphetamine-, methylphenidate-, and modafinil-induced wakefulness, evidenced by c-fos immunocytochemistry in the cat. Proc. Natl. Acad. Sci. USA. 1996;93:14128–14133. doi: 10.1073/pnas.93.24.14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas JJ, Segu L, Hen R. 5-Hydroxytryptamine1B receptors modulate the effect of cocaine on c-fos expression: converging evidence using 5-hydroxytryptamine1B knockout mice and the 5-hydroxytryptamine1B/1D antagonist GR127935. Mol. Pharmacol. 1997;51:755–763. doi: 10.1124/mol.51.5.755. [DOI] [PubMed] [Google Scholar]
- Mink JW. The basal ganglia: focused selection and inhibition of competing motor programs. Prog. Neurobiol. 1996;50:381–425. doi: 10.1016/s0301-0082(96)00042-1. [DOI] [PubMed] [Google Scholar]
- Muller CP, Huston JP. Determining the region-specific contributions of 5-HT receptors to the psychostimulant effects of cocaine. Trends Pharmacol. Sci. 2006;27:105–112. doi: 10.1016/j.tips.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Nelson JC. Augmentation strategies in the treatment of major depressive disorder. Recent findings and current status of augmentation strategies. CNS Spectr. 2007;12 Suppl 22:6–9. doi: 10.1017/s1092852900016011. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nat. Rev. Neurosci. 2001;2:119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- Packard MG, Knowlton BJ. Learning and memory functions of the basal ganglia. Annu. Rev. Neurosci. 2002;25:563–593. doi: 10.1146/annurev.neuro.25.112701.142937. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. New York: Academic Press; 1998. [Google Scholar]
- Randrup A, Munkvad I. Stereotyped activities produced by amphetamine in several animal species and man. Psychopharmacologia. 1967;11:300–310. doi: 10.1007/BF00404607. [DOI] [PubMed] [Google Scholar]
- Ravindran AV, Kennedy SH, O’Donovan MC, Fallu A, Camacho F, Binder CE. Osmotic-release oral system methylphenidate augmentation of antidepressant monotherapy in major depressive disorder: results of a double-blind, randomized, placebo-controlled trial. J. Clin. Psychiatry. 2008;69:87–94. doi: 10.4088/jcp.v69n0112. [DOI] [PubMed] [Google Scholar]
- Redgrave P, Gurney K. The short-latency dopamine signal: a role in discovering novel actions? Nat. Rev. Neurosci. 2006;7:967–975. doi: 10.1038/nrn2022. [DOI] [PubMed] [Google Scholar]
- Redgrave P, Prescott TJ, Gurney K. The basal ganglia: a vertebrate solution to the selection problem? Neuroscience. 1999;89:1009–1023. doi: 10.1016/s0306-4522(98)00319-4. [DOI] [PubMed] [Google Scholar]
- Safer DJ, Zito JM, DosReis S. Concomitant psychotropic medication for youths. Am. J. Psychiatry. 2003;160:438–449. doi: 10.1176/appi.ajp.160.3.438. [DOI] [PubMed] [Google Scholar]
- Sandson NB, Armstrong SC, Cozza KL. An overview of psychotropic drug-drug interactions. Psychosomatics. 2005;46:464–494. doi: 10.1176/appi.psy.46.5.464. [DOI] [PubMed] [Google Scholar]
- See RE, Elliott JC, Feltenstein MW. The role of dorsal vs ventral striatal pathways in cocaine-seeking behavior after prolonged abstinence in rats. Psychopharmacology. 2007;194:321–331. doi: 10.1007/s00213-007-0850-8. [DOI] [PubMed] [Google Scholar]
- Steiner H. Psychostimulant-induced gene regulation in corticostriatal circuits. In: Steiner H, Tseng KY, editors. Handbook of Basal Ganglia Structure and Function. London: Academic Press/Elsevier; 2010. pp. 501–525. [Google Scholar]
- Steiner H, Gerfen CR. Role of dynorphin and enkephalin in the regulation of striatal output pathways and behavior. Exp. Brain Res. 1998;123:60–76. doi: 10.1007/s002210050545. [DOI] [PubMed] [Google Scholar]
- Steiner H, Kitai ST. Regulation of rat cortex function by D1 dopamine receptors in the striatum. J. Neurosci. 2000;20:5449–5460. doi: 10.1523/JNEUROSCI.20-14-05449.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner H, Van Waes V, Marinelli M. Fluoxetine potentiates methylphenidate-induced gene regulation in addiction-related brain regions: Concerns for use of cognitive enhancers? Biol. Psychiatry. 2010;67:592–594. doi: 10.1016/j.biopsych.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Murry DJ, Sanghani SP, Davis WI, Kedishvili NY, Zou Q, Hurley TD, Bosron WF. Methylphenidate is stereoselectively hydrolyzed by human carboxylesterase CES1A1. J. Pharmacol. Exp. Ther. 2004;310:469–476. doi: 10.1124/jpet.104.067116. [DOI] [PubMed] [Google Scholar]
- Swanson JM, Volkow ND. Increasing use of stimulants warns of potential abuse. Nature. 2008;453:586. doi: 10.1038/453586a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres G, Rivier C. Cocaine-induced expression of striatal c-fos in the rat is inhibited by NMDA receptor antagonists. Brain Res. Bull. 1993;30:173–176. doi: 10.1016/0361-9230(93)90055-g. [DOI] [PubMed] [Google Scholar]
- Unal CT, Beverley JA, Willuhn I, Steiner H. Long-lasting dysregulation of gene expression in corticostriatal circuits after repeated cocaine treatment in adult rats: Effects on zif 268 and homer 1a. Eur. J. Neurosci. 2009;29 doi: 10.1111/j.1460-9568.2009.06691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uslaner JM, Crombag HS, Ferguson SM, Robinson TE. Cocaine-induced psychomotor activity is associated with its ability to induce c-fos mRNA expression in the subthalamic nucleus: effects of dose and repeated treatment. Eur. J. Neurosci. 2003;17:2180–2186. doi: 10.1046/j.1460-9568.2003.02638.x. [DOI] [PubMed] [Google Scholar]
- Vanderschuren LJ, Di Ciano P, Everitt BJ. Involvement of the dorsal striatum in cue-controlled cocaine seeking. J. Neurosci. 2005;25:8665–8670. doi: 10.1523/JNEUROSCI.0925-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JQ, McGinty JF. Glutamatergic and cholinergic regulation of immediate-early gene and neuropeptide gene expression in the striatum. In: Merchant KM, editor. Pharmacological Regulation of Gene Expression in the CNS. Boca Raton: CRC; 1996. pp. 81–113. [Google Scholar]
- Weikop P, Yoshitake T, Kehr J. Differential effects of adjunctive methylphenidate and citalopram on extracellular levels of serotonin, noradrenaline and dopamine in the rat brain. Eur. Neuropsychopharmacol. 2007;17:658–671. doi: 10.1016/j.euroneuro.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Wilens TE, Adler LA, Adams J, Sgambati S, Rotrosen J, Sawtelle R, Utzinger L, Fusillo S. Misuse and diversion of stimulants prescribed for ADHD: a systematic review of the literature. J. Am. Acad. Child Adolesc. Psychiatry. 2008;47:21–31. doi: 10.1097/chi.0b013e31815a56f1. [DOI] [PubMed] [Google Scholar]
- Willuhn I, Sun W, Steiner H. Topography of cocaine-induced gene regulation in the rat striatum: Relationship to cortical inputs and role of behavioural context. Eur. J. Neurosci. 2003;17:1053–1066. doi: 10.1046/j.1460-9568.2003.02525.x. [DOI] [PubMed] [Google Scholar]
- Wirtshafter D, Cook DF. Serotonin-1B agonists induce compartmentally organized striatal Fos expression in rats. Neuroreport. 1998;9:1217–1221. doi: 10.1097/00001756-199804200-00047. [DOI] [PubMed] [Google Scholar]
- Yano M, Beverley JA, Steiner H. Inhibition of methylphenidate-induced gene expression in the striatum by local blockade of D1 dopamine receptors: Interhemispheric effects. Neuroscience. 2006;140:699–709. doi: 10.1016/j.neuroscience.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Yano M, Steiner H. Methylphenidate (Ritalin) induces Homer 1a and zif 268 expression in specific corticostriatal circuits. Neuroscience. 2005a;132:855–865. doi: 10.1016/j.neuroscience.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Yano M, Steiner H. Topography of methylphenidate (Ritalin)-induced gene regulation in the striatum: differential effects on c-fos, substance P and opioid peptides. Neuropsychopharmacology. 2005b;30:901–915. doi: 10.1038/sj.npp.1300613. [DOI] [PubMed] [Google Scholar]
- Yano M, Steiner H. Methylphenidate and cocaine: the same effects on gene regulation? Trends Pharmacol. Sci. 2007;28:588–596. doi: 10.1016/j.tips.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Zhu HJ, Patrick KS, Yuan HJ, Wang JS, Donovan JL, DeVane CL, Malcolm R, Johnson JA, Youngblood GL, Sweet DH, Langaee TY, Markowitz JS. Two CES1 gene mutations lead to dysfunctional carboxylesterase 1 activity in man: clinical significance and molecular basis. Am. J. Hum. Genet. 2008;82:1241–1248. doi: 10.1016/j.ajhg.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]