SUMMARY
RNA import into mammalian mitochondria is considered essential for replication, transcription, and translation of the mitochondrial genome but the pathway(s) and factors that control this import are poorly understood. Previously, we localized polynucleotide phosphorylase (PNPASE), a 3′ → 5′ exoribonuclease and poly-A polymerase, in the mitochondrial intermembrane space, a location lacking resident RNAs. Here, we show a new role for PNPASE in regulating the import of nuclear-encoded RNAs into the mitochondrial matrix. PNPASE reduction impaired mitochondrial RNA processing and polycistronic transcripts accumulated. Augmented import of RNase P, 5S rRNA, and MRP RNAs depended on PNPASE expression and PNPASE–imported RNA interactions were identified. PNPASE RNA processing and import activities were separable and a mitochondrial RNA targeting signal was isolated that enabled RNA import in a PNPASE-dependent manner. Combined, these data strongly support an unanticipated role for PNPASE in mediating the translocation of RNAs into mitochondria.
INTRODUCTION
Much is understood about the mechanisms that regulate nuclear-encoded protein import into mitochondria (Chacinska et al., 2009). By contrast, much less is known about the factors that regulate mitochondrial RNA import. Almost every organism with mitochondria imports tRNAs and aminoacyl-tRNA synthetases (Alfonzo and Soll, 2009; Duchene et al., 2009). The number of imported tRNAs ranges from one in yeast to all in trypanosomes, with mammalian mitochondria importing several different tRNAs both in vitro and in vivo (Kolesnikova et al., 2004; Rubio et al., 2008). Recently, microRNAs were isolated from mitochondria (Kren et al., 2009) and the nuclear-encoded 5S rRNA was identified as one of the most abundant RNAs in human mitochondria (Entelis et al., 2001; Smirnov et al., 2008).
RNase MRP and RNase P enzyme complexes localize and function in mammalian mitochondria and may contain RNAs that are encoded within the nucleus. RNase MRP functions as a site-specific endoribonuclease involved in primer RNA cleavage during the replication of mitochondrial DNA (Chang and Clayton, 1987). Mammalian RNase P functions in the processing of tRNAs during the maturation of mitochondrial transcripts that encode oxidative phosphorylation (OXPHOS) components. Large polycistronic RNA transcripts are generated from the heavy and light strand promoters in mammalian mitochondria (Bonawitz et al., 2006). tRNAs often separate the coding regions for OXPHOS subunits and are processed and removed by RNase P (Doersen et al., 1985). Earlier studies showed that the RNA component of RNase P was imported into mammalian mitochondria (Doersen et al., 1985) and the RNase P RNA and processing activity has been co-purified from mitochondria (Puranam and Attardi, 2001). By contrast, RNase P RNA is encoded in the mitochondrial genome in Saccharomyces cerevisiae (Hollingsworth and Martin, 1986). Recently, an additional RNase P enzyme, consisting of three protein subunits, has been purified from human mitochondria. This alternative RNase P enzyme processes single tRNA 5’ precursor sequences in vitro without an RNA component (Holzmann et al., 2008; Walker and Engelke, 2008). The identification of two enzymes with RNase P activity in mammalian mitochondria warrants further investigation.
Critical open questions about RNA import into mammalian mitochondria include the selectivity of RNAs, the factor(s) targeting RNA from the cytosol, and the translocation pathway(s) across the mitochondrial membranes (Duchene et al., 2009). Different import pathways have been proposed for a subset of precursors in different species and the details of the components involved remain ill-defined. For example, the TOM and TIM protein import complexes have been implicated in the import of tRNALys in yeast mitochondria (Tarassov et al., 1995). By contrast, a protein import pathway-independent mechanism may exist and involve a 600 kDa multi-subunit RNA import complex (RIC) in Leishmania (Mukherjee et al., 2007). Thus, inconsistencies among RNA import systems suggest a critical need to learn more about the nature of the RNAs that are imported and the factor(s) mediating this import.
Previously we showed an unexpected location in the mitochondrial intermembrane space (IMS) for mammalian polynucleotide phosphorylase (PNPASE) (Chen et al., 2007; Chen et al., 2006; Rainey et al., 2006), a 3′ → 5′ exoribonuclease and poly-A polymerase that uses phosphorolysis to degrade RNA (Yehudai-Resheff et al., 2001). This was a surprise because Instead we expected that PNPASE would instead localize in the RNA-abundant mitochondrial matrix. Therefore, initial studies on mammalian PNPASE focused on a general role in maintaining mitochondrial homeostasis, potentially by regulating adenine nucleotide levels (Chen et al., 2006; Portnoy et al., 2008). Here we show that PNPASE has a central role in augmenting the import of small RNA components required for DNA replication and RNA processing into the mitochondrial matrix. We suggest that PNPASE regulates adenine nucleotide levels and mitochondrial homeostasis at least partly by regulating RNA import to control the abundance of the electron transport chain (ETC) components.
RESULTS
PNPASE Forms a Trimer in Yeast and Mammalian Mitochondria
To examine PNPASE in the IMS, a co-immunoprecipitation (IP) assay was performed to identify potential binding partners. A 6XHis-Protein-C (HisPC) tag was added to the C-terminus of PNPASE and stable PNPASE-HisPC expressing HEK293 cells were generated. The PNPASE-HisPC protein was detected in isolated mitochondria by immunoblot (Figure S1A). PNPASE-HisPC was isolated from mitochondria using sequential purification over Ni2+ and Protein-C affinity columns, followed by elution, and co-purifying proteins were identified by Sypro Ruby staining (Figure S1B) and liquid chromatography-tandem mass spectrometry (LC-MS/MS) (data not shown). All of the identified bands originated from PNPASE, suggesting that PNPASE lacks partner proteins in vivo. Bands of lower molecular weight than the ~85 kDa of PNPASE monomers were likely degradation products. The assembly state of PNPASE was also investigated. Mitochondria from yeast expressing human PNPASE (Rainey et al., 2006) were detergent solubilized and separated on blue-native (BN) gels. Immunoblot showed PNPASE in a complex of ~240 kDa (Figure S1C), similar to the trimeric complex of endogenous mouse hepatocyte PNPASE (Chen et al., 2006) and bacterially-expressed human PNPASE (French et al., 2007). PNPASE-HisPC isolated from HEK293 mitochondria also migrated in a similarly-sized complex (Figure S1D). Overall, PNPASE assembled identically in yeast and mammalian mitochondria into a homo-oligomeric complex, consisting of a trimer or a ‘dimer of trimers’ (Symmons et al., 2002). These results strongly suggest that PNPASE assembles and may function similarly in yeast and mammalian mitochondria.
Pnpt1 Knockout Cells Show Altered Mitochondrial Morphology and Impaired Respiration
We used several approaches to determine the function of PNPASE in mitochondria. First, the gene encoding PNPASE (Pnpt1) was knocked out (KO) in C57BL/6 mice (Figure S2, S3). Homozygous Pnpt1neo-flox mice, in which exon 2 was flanked by loxP recombination sites, were viable and fertile. A complete KO of Pnpt1 exon 2 was generated by crossing CMVCRE expressing mice with Pnpt1WT/neo-flox heterozygotes followed by inter-crossing the Pnpt1WT/KO progeny. Pnpt1KO/KO mice were embryonic lethal (Figure 1A).
Figure 1. Deletion of Pnpt1 in Hepatocytes Impairs Mitochondrial Function.
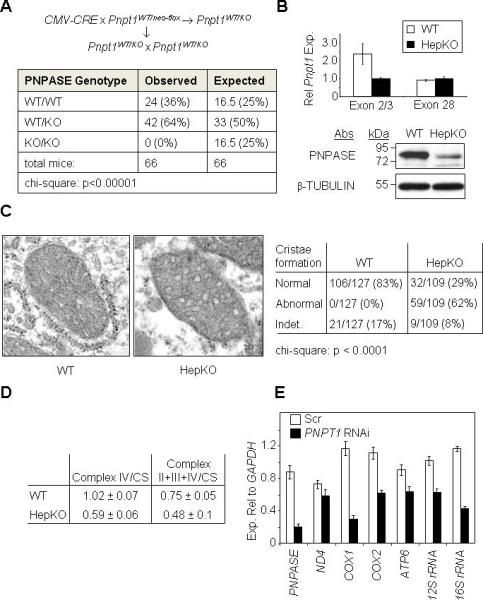
(A) Breeding strategy (top) and results (bottom) for attempting to generate a PNPASE KO mouse.
(B) Hepatocyte-specific Pnpt1 KO (HepKO) in 4-week old mice. Top: QPCR for liver Pnpt1 expression using an exon 2 – exon 3 primer pair versus a primer pair within exon 28. Bottom: PNPASE immunoblot from 4-week old WT and HepKO mouse livers.
(C) HepKO mitochondria have altered cristae. Left: TEM of 6-week old littermate livers shows circular, smooth HepKO IM cristae in contrast to linear, stacked cristae of WT mitochondria. Right: Analysis of cristae morphology in which a single normal cristae within a mitochondrion was scored as normal. Indet = indeterminate.
(D) Decreased respiration in isolated HepKO mitochondria. Oxygen consumption (nmol/min/mg protein) for ETC complexes IV and II+III+IV was measured using an O2 electrode. Mitochondrial mass was determined by citrate synthase (CS) activity using a spectrophotometer. Respiratory activities are shown normalized to CS activity.
(E) Decreased mature mtRNAs in HEK293 cells with RNAi to PNPT1. Transcripts were quantified relative to cytosolic GAPDH expression by QPCR from HEK293 cells 7d post-infection (nadir) with scramble (Scr) or PNPT1 RNAi retroviral constructs. See also Figure S1, S2, and S3.
Our prior cell line studies showed that RNAi targeting PNPASE reduced ATP production by OXPHOS and slowed cell growth (Chen et al., 2006). Recently, a viable liver KO of the COX10 gene, which is required for cytochrome c oxidase and ETC function, was produced, suggesting that targeted disruption of Pnpt1 in hepatocytes might be tolerated (Diaz et al., 2008). Therefore, a liver-specific KO (HepKO) of Pnpt1 was generated by the cross AlbCRE/WT/Pnpt1neo-flox/neo-flox× AlbWT/WT/Pnpt1neo-flox/neo-flox, which produced fertile progeny at the expected frequency (Figure S2C). Quantitative real-time PCR (QPCR) from HepKO liver showed reduced Pnpt1 transcripts containing targeted exon 2 compared with those containing untargeted exon 28 (Figure 1B). PNPASE protein expression was also markedly reduced in HepKO liver compared with sex-matched littermate WT liver (Figure 1B). The expression of Pnpt1 transcripts and PNPASE protein in HepKO liver likely arises from heterogeneous liver elements. Also, the AlbCRE deleting strain is hepatocyte-specific but incomplete (Postic and Magnuson, 2000). The physiology of these mice will be reported elsewhere.
The ultrastructure of HepKO liver mitochondria was investigated by transmission electron microscopy (TEM). Rather than displaying ordered, linear cristae with convolutions as in WT mitochondria, the HepKO mitochondria showed disordered circular and smooth IM cristae (Figure 1C), similar to mitochondria that are impaired for OXPHOS (Mandel et al., 2001) and to Pnpt1 RNAi mammalian cell lines (Chen et al., 2006). These and prior results suggesting that reduced PNPASE caused a decrease in ATP production prompted the evaluation of O2 consumption from HepKO liver mitochondria. Oxygen electrode studies showed a ~1.5 – 2-fold decrease in the activity of Complex IV and Complexes II+III+IV when normalized to citrate synthase activity in HepKO compared to WT mitochondria (Figure 1D). Combined, these data establish an in vivo role for PNPASE in mitochondrial morphogenesis and respiration through an unknown mechanism.
PNPASE is Required for the Processing of Mitochondrial RNA Transcripts
The data showing decreased respiration in HepKO mitochondria suggested a reduction in functional ETC complexes. Therefore, RNA processing and translation were examined in cells with decreased PNPASE. HEK293 cells with >75% reduced PNPASE expression were generated by RNAi, followed by mitochondrial RNA (mtRNA) transcript quantification using QPCR normalized to cytosolic GAPDH RNA. All mtRNAs tested were reduced in Pnpt1 RNAi cells compared to WT cells (Figure 1E). Proteins translated from mtRNAs were decreased in HEK293 Pnpt1 RNAi cells (data not shown) and HepKO liver cells (Figure 2A), strongly suggesting that a decrease in functional ETC complexes was responsible for decreased respiration.
Figure 2. HepKO Liver Mitochondria Do Not Efficiently Process mtRNA Precursors.
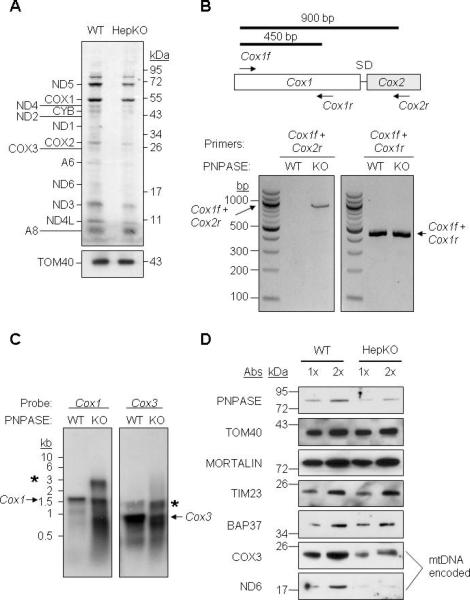
(A) In organello protein synthesis. WT and HepKO mitochondria (100 μg) were treated with micrococcal nuclease S7, and in organello translation was performed using [35S]-MET. TOM40 immunoblot shows equivalent mitochondria in each assay.
(B) RNA was isolated from WT and HepKO liver mitochondria followed by DNase I treatment to remove contaminating DNA. RT-PCR was performed for Cox1 and Cox2 with primers shown in the schematic (upper) and separated on a 1.5% agarose gel.
(C) Northern blot of mtRNA from WT and HepKO mouse liver mitochondria using a Cox1 or Cox3 DNA probe. * marks larger precursor mtRNAs and the arrow shows the mature mtRNA.
(D) Steady-state expression of nuclear- and mitochondrial-encoded proteins in WT and HepKO liver mitochondria. Equivalent nuclear-encoded protein expression shows that HepKO reduced mitochondria-encoded protein expression was not due to differing mitochondrial content between WT and HepKO liver cells. See also Figure S4.
The processing of polycistronic mtRNAs was investigated because reduced PNPASE could cause an accumulation of large precursor transcripts, resulting in reduced ETC proteins. Transcript processing requires RNase P excision of the tRNAs between ETC gene coding regions. Primers were designed to test processing between adjacent Cox1 and Cox2 transcripts that are separated by tRNAser and tRNAasp (Figure 2B). The primer set Cox1f and Cox1r generates a 450-bp fragment whereas the primer pair Cox1f and Cox2r generates a 900-bp fragment when tRNAser and tRNAasp are not excised from large precursor transcripts. Isolated mitochondrial RNAs were examined by RT-PCR. A 900-bp fragment was detected from HepKO but not from WT liver mitochondria (Figure 2B). Similar results were obtained using the same primers in PNPASE KO mouse embryonic fibroblasts (MEFs) (Figure S3, S4A). To query RNA processing at a second site, primers were generated for adjacent Cox2 and Atp8/6 loci, separated by tRNAlys. Again, polycistronic transcripts accumulated in the PNPASE KO MEFs (Figure S4B). The sizes of Cox1 and Cox3 transcripts were investigated using specific probes and Northern blot (Figure 2C). In addition to the mature Cox1 and Cox3 transcripts, a range of larger precursor transcripts was seen in HepKO liver cells. Also, the mature 0.9-kb Cox3 transcript was more abundant in WT than HepKO liver. The abundance of mitochondria-encoded COX3 and ND6 proteins was determined (Figure 2D). In HepKO liver mitochondria, the steady-state abundance of PNPASE was decreased by ~2-fold compared to the WT, similar to a ~2-fold decrease for COX3 and ND6 proteins. Controls TOM40, MORTALIN, TIM23, and BAP37 showed that the amount of nuclear-encoded mitochondrial proteins, and therefore the mitochondrial mass, was similar between HepKO and sex-matched WT littermate liver cells. Thus, the processing of polycistronic mtRNAs was impaired in mitochondria with reduced PNPASE, resulting in fewer mature mtRNAs and reduced ETC complexes.
RNase P RNA Binds to PNPASE and May Function in PNPASE-Dependent mtRNA Processing
Although an in vitro form of the RNase P enzyme can function without nucleic acid (Holzmann et al., 2008; Walker and Engelke, 2008), the RNA component of RNase P localizes to mitochondria (Puranam and Attardi, 2001) and has not been excluded from mtRNA processing in vivo (Holzmann et al., 2008). Therefore, the abundance of RNase P RNA in HepKO liver mitochondria was determined by RT-PCR and QPCR (Figure 3A). Reproducibly, RNase P RNA was decreased by ~75% in HepKO versus WT liver mitochondria, suggesting that PNPASE may help import and/or stabilize RNase P RNA. Therefore, we determined whether RNase P RNA directly bound to PNPASE in HEK293 cells stably expressing dual-tagged PNPASE-HisPC. Isolated mitochondria were treated with nuclease and tagged PNPASE was purified. RNase P RNA was amplified by RT-PCR and co-purified with PNPASE (Figure 3B, lane 4). Importantly, control IMS-localized TIM23-HisPC in stably-expressing HEK293 cells did not bind RNase P RNA (Figure 3B, lane 2). PNPASE also did not adventitiously bind RNA because the RNA transcripts for Cox1, GAPDH, mitochondrial 12S rRNA and mitochondrial tRNAtrp were not bound to PNPASE (Figure 3B). Furthermore, PNPASE-HisPC, but not TIM23-HisPC, bound in vitro transcribed and imported RNase P and MRP RNAs, but not control mitochondrial RNA, in cross linking IP assays (Figure S5A) Thus, RNase P and MRP RNA bound specifically to PNPASE.
Figure 3. RNase P RNA Binds to PNPASE and May Function in PNPASE-Dependent tRNA Processing.
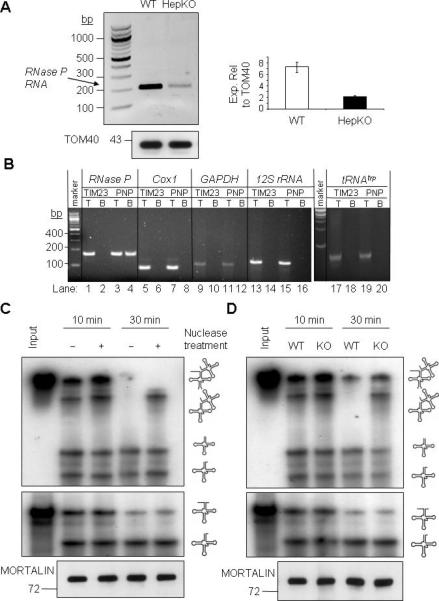
(A) Left- RNA was isolated from WT and HepKO liver mitochondria following nuclease treatment. RT-PCR was performed with primers that amplify nuclear-encoded RNase P RNA (212-bp). Right- QPCR analysis of RNase P RNA expression relative to TOM40 protein in isolated mitochondria.
(B) PNPASE-HisPC (PNP) or TIM23-HisPC (TIM23) was purified from stably-transfected HEK293 cells. Candidate interacting RNAs that co-purified in the final eluate with PNPASE-HisPC and TIM23-HisPC were identified by primer-specific RT-PCR. T is the total lysate (0.3% of the reaction) before mitochondrial purification and B is the bound fraction. Note that only RNase P RNA bound to PNPASE-HisPC (lane 4).
(C) Mitoplast extract (10 μg) was prepared from nuclease-treated, intact WT liver mitochondria. The extract was then treated with nuclease (+), as indicated, and then inactivated with EDTA and EGTA. The nuclease-treated or untreated extract was incubated with abutted tRNAs (tRNAHistRNASer) or a single tRNA (tRNALys) at 25°C for 10 or 30 min. RNA was separated on an urea-acrylamide gel and detected by autoradiography. A MORTALIN immunoblot shows equivalent mitoplast extract in each assay.
(D) Mitoplast extract was prepared from nuclease-treated, intact WT or HepKO liver mitochondria. The enzymatic assay was performed as described for panel C. See also Figure S5.
A protein-only RNase P complex was shown to process the 5’-terminus of a single mitochondrial tRNAlys (Holzmann et al., 2008; Walker and Engelke, 2008). However, most mammalian mitochondrial tRNA genes are grouped together in the mitochondrial genome and lack an intervening pre-sequence, causing them to be expressed with the ETC genes in polycistronic transcripts. Therefore, we examined whether protein-only RNase P can efficiently process paired mitochondrial tRNAs, as must occur in vivo. As done previously (Holzmann et al., 2008), mitoplast lysates were untreated or treated with micrococcal nuclease, followed by inactivation with EGTA and EDTA. tRNAlys or abutting tRNAs, tRNAhistRNAser, were then added to the lysates. Nuclease-treated mitoplast lysates cleaved the 5’-precursor sequence of the single and abutting tRNA substrates as efficiently as the lysates without treatment, as shown before (Figure 3C). By contrast, nuclease-treated lysates were impaired in cleaving the two abutting tRNAs into individual tRNAs, strongly suggesting an additional nucleic acid component is required for efficient processing. Interestingly, mitoplast lysates from HepKO liver showed the same defect on abutting tRNA maturation as the nuclease-treated WT mitoplast lysates (Figure 3D). Furthermore, the in vivo processing and separation of an endogenous paired tRNAhistRNAser substrate was inhibited in HepKO compared to WT liver mitochondria, whereas a linked 12s rRNA-tRNAval substrate was processed equivalently (Figure S5B). These results, combined with prior data (Puranam and Attardi, 2001), suggest protein-only and RNase P RNA-containing RNase P complexes coexist in mitochondria, with efficient tRNA processing requiring PNPASE-dependent RNase P RNA.
PNPASE Augments the Import of RNase P, 5S rRNA, and MRP RNAs into Yeast Mitochondria
Similar PNPASE complexes in yeast and mammalian mitochondria (Chen et al., 2006) (Figure S1) suggest yeast as a model for studying the import of nuclear-encoded RNAs. Importantly, added human PNPASE did not alter yeast mitochondrial morphology, rate of proliferation, or extent of cell death, supporting this choice (Figure S6). Mitochondria isolated from WT yeast or yeast expressing human PNPASE were incubated with in vitro transcribed human RNase P RNA in import buffer. The reaction was treated with nuclease to remove non-imported RNA followed by RNA isolation and RT-PCR. RNase P RNA abundance was increased in mitochondria containing PNPASE compared to WT mitochondria (Figure 4A). This RNA increase was specific for certain RNAs because cytosolic GAPDH RNA was not increased in the same mitochondria (Figure 4B). Osmotic shock was used to identify the location of the imported RNase P RNA (Koehler et al., 1998). Mitochondria were incubated in hypotonic buffer to rupture the outer membrane and the mitoplasts (P, pellet fraction that contains the matrix and IM) and the supernatant (S, contains the soluble IMS contents) were separated by centrifugation (Figure 4C). RNase P RNA was detected by RT-PCR and was localized in the mitochondrial matrix. Detergent exposed the matrix to verify that the nuclease degraded the RNase P RNA. To confirm that osmotic shock did not disrupt the IM, antibodies against cytochrome b2 (cyt b2; IMS) and α-ketoglutarate dehydrogenase (KDH; matrix) showed that cyt b2 was sensitive to protease in the IMS, but KDH was resistant to protease until the IM was lysed with Triton X-100. Thus, RNase P RNA import was augmented, or RNase P RNA was stabilized, in the yeast mitochondrial matrix when exogenous PNPASE was present in the IMS.
Figure 4. PNPASE Augments RNase P, 5S rRNA, and MRP RNA Import into Yeast Mitochondria.
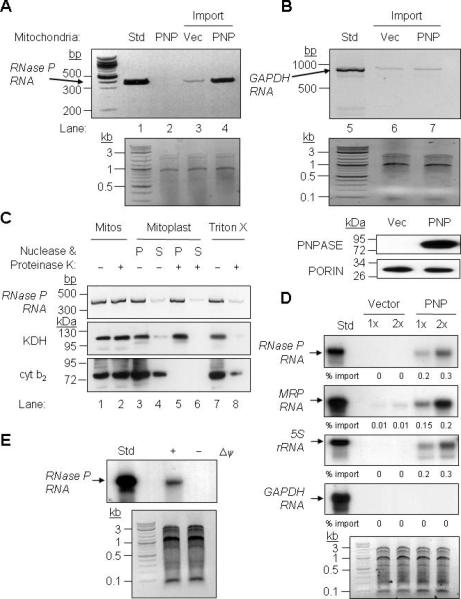
(A) Upper- In vitro transcribed human RNase P RNA was incubated with yeast mitochondria expressing human PNPT1 (PNP) or an empty vector (Vec) control. Non-imported RNA was digested with nuclease and the imported RNA was detected by RT-PCR. PNPT1-expressing mitochondria without added RNase P RNA was included as a specificity control for import and RT-PCR (lane 2 – Std, 1% of the reaction). Lower- Control showing equivalent total mitochondrial nucleic acid in each reaction.
(B) Upper- As in panel A, with cytosolic human GAPDH RNA used as a substrate. Middle- Control showing equivalent total mitochondrial nucleic acid in each reaction. Lower- Western blot showing PNPASE expression and PORIN immunoblot showing equivalent mitochondria in each import assay.
(C) After import as in panel A, mitochondria were subjected to osmotic shock, fractionated by centrifugation into soluble (S) and pellet (P) fractions, followed by proteinase K and nuclease additions where indicated. The pellet fraction was solubilized with Triton X-100 to expose the matrix. Localization was determined by RT-PCR for RNase P RNA and immunoblot for KDH (matrix) and cyt b2 (IMS) proteins.
(D) Upper- Radiolabeled RNase P, MRP, 5S rRNA, and GAPDH human RNAs were in vitro transcribed and then incubated with yeast mitochondria expressing PNPASE or an empty vector control. Non-imported RNA was digested with nuclease, followed by RNA isolation, separation on a urea acrylamide gel, and autoradiography. Import reactions were repeated with 1X and 2X amounts of RNA. Lower- Control showing equivalent total mitochondrial nucleic acid in each reaction.
(E) Upper- As in panel A except that the mitochondrial membrane potential (Δψ) was dissipated prior to import. Lower- Control showing equivalent total mitochondrial nucleic acid in each reaction.
To confirm the RT-PCR results and assay other imported RNAs, we performed the in vitro RNA import assay with yeast mitochondria and radiolabeled human RNAs (Figure 4D). Two different RNA volumes were used and the imported RNA was isolated and separated on a urea-acrylamide gel followed by autoradiography. RNase P, 5S rRNA, and MRP RNAs showed augmented import or stability in mitochondria expressing PNPASE relative to control mitochondria (Figure 4D). Again, this increase was RNA-type specific as PNPASE did not augment GAPDH RNA levels. When the mitochondrial membrane potential was dissipated, the RNase P RNA level was not increased in this assay system (Figure 4E).
PNPASE Mutations that Inactivate RNA Processing do not Affect RNA Import or Stability
Specific point mutations in conserved regions of PNPASE impair its RNA exonuclease or 3’ poly-A polymerase activity in biochemical assays (Portnoy et al., 2008), although the effects of these mutations on PNPASE in vivo are unknown. To determine whether the RNA import or stabilization activity of PNPASE was separable from its RNA processing activities, RNase P RNA import was studied when different PNPASE mutants were expressed in yeast mitochondria (Figure 5A). The point mutants generated and tested were based on results from Schuster and colleagues (Portnoy et al., 2008). Mutants D135G and S484A lacked poly-A polymerase and RNA degradation activities in vitro. Mutant D544G and double mutant R445E/R446E showed enhanced in vitro poly-A polymerase activity but compromised degradation activity. Of the four mutants, PNPASE S484A and R445E/R446E supported the import or stabilization of RNase P RNA, whereas mutants D135G and D544G were defective in this function (Figure 5A). The abundance of WT and the four mutant PNPASE proteins were similar between yeast strains and all of the PNPASE proteins assembled into ~240 kDa complexes without impairment (Figure 5A, lower panel). This contrasts with the expectation that mutant D135G would fail to form a trimeric complex from prior studies (Portnoy et al., 2008). Therefore, the mitochondrial RNA import or stabilization function of PNPASE is separable from its poly-A polymerase or exoribonuclease activities.
Figure 5. PNPASE Mutations that Inactivate RNA Processing do not Affect RNA Import or Stability.
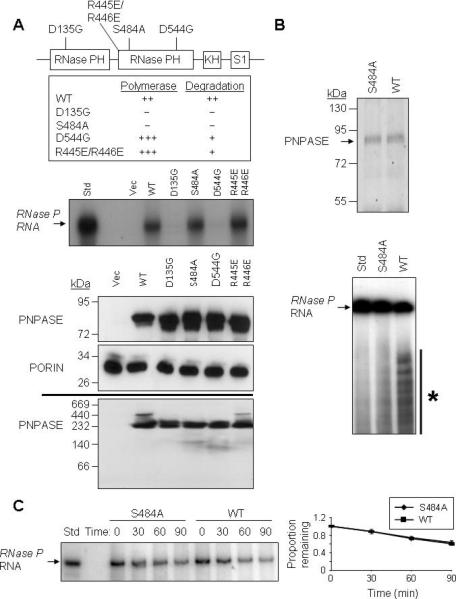
(A) Upper- Schematic for the positions of point mutations made in the PNPASE protein. Listed are the in vitro effects of mutations on 3’ polymerase and RNA degrading activities, from (Portnoy et al., 2008). Middle- Import reactions were performed as in Figure 4A. Radiolabeled RNase P RNA was incubated with isolated yeast mitochondria expressing an empty vector or the listed PNPASE constructs. Lower panels- Immunoblot of WT and point mutant PNPASE yeast transfectants used in panel A import assay. A PORIN immunoblot confirms the co-localization of PNPASE WT and mutants in yeast mitochondria. The assembly state of WT and point mutant PNPASE was determined by solubilization with 1% digitonin and separation on a 6-16% BN gel, followed by PNPASE immunoblot.
(B) Upper- WT and S484A PNPASE IPs from yeast mitochondria were used to analyze RNA degradation properties. Lower- WT or S484A mutant PNPASE was incubated with radiolabeled RNase P RNA for 10 min at 25°C to assess degradation activity. The asterisk marks degradation products.
(C) Left- Following in vitro import of radiolabeled RNase P RNA and nuclease treatment to remove non-imported RNA, mitochondria were incubated for up to 90 min at 25°C and aliquots removed at the indicated time points. The RNA was then resolved by urea-acrylamide gel electrophoresis. Right- RNase P RNA that was not degraded was quantified using FX imager; n = 3.
To determine whether PNPASE augmented either RNA import or stabilization in mitochondria, the enzymatic properties of the WT and S484A mutant protein were examined with respect to RNA turnover in vitro and in isolated yeast mitochondria. For in vitro studies, WT and S484A PNPASE were immunoprecipitated from yeast mitochondria and tested in an in vitro degradation assay with radiolabeled RNase P RNA (Figure 5B). WT PNPASE degraded the RNase P RNA, but the S484A mutant was impaired, consistent with prior results (Portnoy et al., 2008). In addition, radiolabeled RNase P RNA was imported into mitochondria, followed by incubation at 25°C for up to 90 min (Figure 5C). The internalized RNase P RNA was separated on a urea-acyrlamide gel and quantified during this time course using a phosphorimager. The rate of degradation of RNase P RNA was similar for degradation competent WT and incompetent mutant PNPASE proteins, supporting a role for PNPASE in augmenting the import of specific RNAs into the mitochondrial matrix. This result further supports PNPASE localizing to the IMS because a greater amount of WT PNPASE imported into the matrix could cause a relative increase in the rate of turnover of matrix localized RNAse P RNA.
A Predicted Stem-Loop RNA Structure Mediates PNPASE-Dependent RNA Import
Results showing PNPASE-dependent RNA import into mitochondria (Figures 3-5) do not exclude an indirect role. To establish a direct PNPASE role, a systematic search was used to identify PNPASE-dependent RNA import sequences. Primers were designed to generate distinct segments of the 340 nucleotide (nt) RNase P RNA full length sequence. RPf1 lacked the 5’ 70 nt, RPf2 lacked the 5’ 140 nt, and RPr1 lacked the 3’ 148 nt of WT PNPT1 (Figure 6A). Import assays were performed using full length or truncated in vitro transcribed RNase P RNAs (Figures 6B, 6C). Augmented RPf1 and RPr1 import into yeast mitochondria depended upon PNPASE, as did the full length RNase P RNA. In striking contrast, RPf2 was not efficiently imported into yeast mitochondria, implicating the sequence between nt 71 and 140 in PNPASE-augmented RNA import. To further refine this import signal, RNA sequences lacking the 5’ 86 (RPf3) or 102 (RPf4) nts were generated (Figure 6A). Augmented RPf3 and RPf4 import into yeast mitochondria was PNPASE-dependent (Figure 6D), further implicating an import signal between nt 103 and 140. The most obvious, predicted secondary structure of RNase P RNA in this region was a 20 nt stem-loop (Figure 6F). Interestingly, a similarly-predicted stem-loop structure was also identified in MRP RNA. To determine whether one or both stem-loop structures could mediate mitochondrial targeting of non-imported GAPDH RNA, each 20 nt stem-loop sequence was fused to the 5’-terminus of the GAPDH RNA, which is not imported (Figure 4D, 7D). Strikingly, the RNase P and MRP stem-loop structures licensed the PNPASE-dependent import of GAPDH RNA into yeast mitochondria (Figure 6E). By contrast, a control random 20nt sequence could not mediate this import. Human mitochondrial tRNAtrp with the RNase P RNA step-loop structure, but not tRNAtrp itself, was imported into isolated mouse liver mitochondria, with the tRNAtrp-PNPASE interaction captured using UV-cross linking (Figure 6G). These results strongly implicate the structural specificity of mitochondrial RNA import (Figure 6F) and the direct involvement of PNPASE in this process.
Figure 6. A Stem-Loop Structure Mediates PNPASE-Dependent RNA Import.
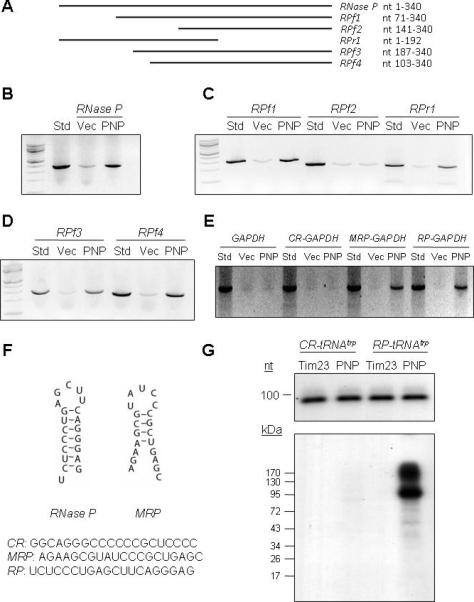
(A) Schematic depiction of human RNase P RNA and deletion fragments.
(B) Import of full length RNase P RNA into yeast mitochondria expressing PNPASE (PNP) or control (Vec) vectors, as in Figure 4A.
(C) Import of the indicated RNase P RNA fragments.
(D) Import of RNase P RNA fragments RPf3 and RPf4.
(E) Import of human GAPDH mRNA or GAPDH mRNA with control (CR), MRP RNA, or RNase P RNA 20 nt sequences fused to the 5’ end, as shown in panel F.
(F) The secondary structures and sequences of mitochondrial RNA targeting signals in RNase P (RP) and MRP (MRP) RNAs. A random sequence (CR) was used as a control.
(G) Isolated mitochondria from HEK293 cells stably expressing IMS-localized PNPASE-HisPC or TIM23-HisPC (control) dual-tagged proteins were incubated with [32P]-CTP labeled CR-tRNAtrp or RP-tRNAtrp, followed by UV-cross linking, tag-IP, separation by SDS-PAGE, and autoradiography. See also Figure S6.
Figure 7. PNPASE Augments RNA Import into Yeast and Mammalian Mitochondria In Vivo.
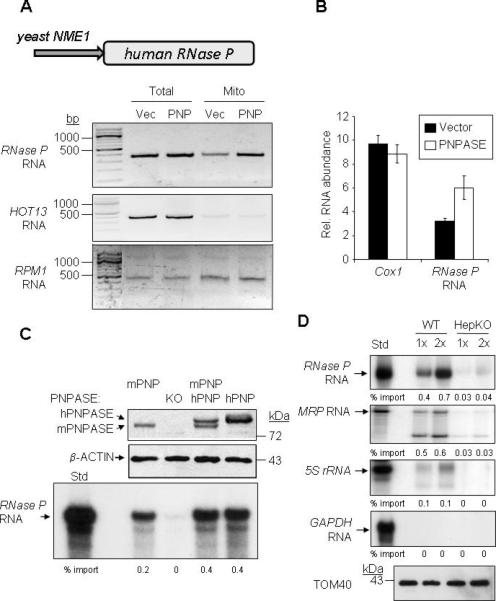
(A) Upper- Human RNase P RNA yeast expression construct is driven by the RPM1 RNA promoter, NME1. Lower-Mitochondria from yeast expressing human RNase P RNA and either PNPASE (PNP) or an empty vector (Vec) were isolated and treated with nuclease. RNA was then isolated from the total cell lysate or from nuclease-treated mitochondria (Mito) and analyzed by primer-specific RT-PCR.
(B) QPCR for Cox1 and RNase P RNAs isolated from mitochondria in panel A, normalized to the total mitochondrial RNA obtained.
(C) Radiolabeled, in vitro transcribed RNase P RNA was imported into mitochondria from MEF cell lines WT (expressing mouse PNPASE, mPNP), Pnpt1 knockout (KO), PNPT1 over-expression (expressing mPNP and hPNP), or Pnpt1 knockout plus PNPT1 over-expression (expressing hPNP). Upper panel is an immunoblot for mouse and human PNPASE expression. Middle panel is an immunoblot of β-ACTIN, a loading control. Lower panel is an autoradiogram of RNase P RNA import into isolated MEF mitochondria.
(D) Radiolabeled, in vitro transcribed RNAs were incubated with WT or HepKO liver mitochondria for 10 min at 25°C. Non-imported RNA was removed with nuclease, followed by RNA isolation and separation on a urea- acrylamide gel. Import reactions were repeated with 1X and 2X amounts of synthesized RNAs. TOM40 immunoblot provides a mitochondrial loading control. See also Figure S7.
PNPASE Augments RNA Import into Yeast and Mammalian Mitochondria in Vivo
To explore in vivo RNA import into mitochondria, a construct was generated in which the human RNase P RNA was expressed from the yeast NME1 promoter (Figure 7A). When expressed in control yeast, RNase P RNA localized to mitochondria, consistent with nuclear-encoded RNA import by a PNPASE-independent mechanism, since yeast normally lack PNPASE. By contrast, RNase P RNA import increased by ~2-fold in mitochondria from yeast expressing PNPASE compared with control cells (Figures 7A, 7B). Importantly, a RNA similar in size to RNase P RNA (340 nt), HOT13, that is translated in the cytosol and imported as a protein into mitochondria, was not localized to mitochondria. Also, mitochondrial-encoded RPM1, which codes for the yeast homologue of RNase P RNA, was sequestered in the mitochondrion at a level equivalent to control yeast mitochondria, as expected. These data indicate that PNPASE augments the import of RNase P RNA into yeast mitochondria in vivo. Finally, replacement of the human RNase P RNA stem-loop sequence with the 20nt random sequence blocked augmented RNase P RNA import into yeast mitochondria in vivo (Figure S7), confirming the role of the stem-loop in PNPASE-regulated import.
PNPASE augmented import of RNase P RNA into yeast mitochondria is non-physiologic. Therefore, we developed WT, PNPASE KO, WT expressing human PNPASE, and PNPASE KO expressing human PNPASE MEFs for import assays. PNPASE abundance in each MEF line was confirmed by immunoblot (Figure 7C). Radiolabeled RNase P RNA was not imported into mitochondria from the PNPASE KO MEFs, but was imported into mitochondria that contained mouse and/or human PNPASE. The in vitro import of RNase P, MRP, 5S rRNA, and GAPDH RNAs was also tested in liver mitochondria isolated from the HepKO mouse and WT littermates. Again, RNase P, 5S rRNA, and MRP RNAs were imported into mitochondria expressing PNPASE, whereas cytosolic GAPDH RNA was not imported (Figure 7D). As expected, more than half of the imported MRP RNA was processed into the mature ~130 nt form (Figure 7D, S7) (Chang and Clayton, 1987). By contrast, mitochondrial RNA import was severely compromised in HepKO liver mitochondria. Combined, these results strongly support PNPASE as the first RNA import factor that mediates the translocation of specific RNAs into the mammalian mitochondrial matrix.
DISCUSSION
In contrast to the protein translocation, very little is understood about the import of specific RNAs into the mitochondrion. A confounder is that the spectrum of RNAs imported and the import factors and mechanisms seem to vary greatly amongst different organisms. At an extreme is mammalian mitochondria, in which despite strong evidence for RNA import (Alfonzo and Soll, 2009; Tarassov et al., 1995), no factors have thus far been identified. Here we implicate PNPASE as the first RNA import factor for mammalian mitochondria. Our results show that PNPASE KO disrupts mitochondrial morphology and respiration in mouse liver cells, at least partially by inhibiting the import of RNAs that control the transcription and translation of the ETC proteins. Our data also suggest that a nucleic acid component of the RNase P RNA processing complex, possibly RNase P RNA (Puranam and Attardi, 2001), is imported in vivo to process linked tRNAs in long mitochondrial transcripts. PNPASE mediated RNA delivery into the mitochondrial matrix and this import was augmented over background. Strikingly, PNPASE RNA import and RNA processing functions were separable and predicted stem-loop structures were identified in two imported RNAs that could transfer PNPASE-dependent import potential to non-imported RNAs. Combined, these results open a new chapter for studies into the pathway(s) and mechanism(s) of RNA import into mammalian mitochondria.
A key question that this study presages is how PNPASE regulates the import of specific cytosolic RNAs. For this, the location of PNPASE first needs to be considered. Previously, we localized PNPASE to the IMS. Carbonate extraction studies also indicated that PNPASE was bound to the IM facing the IMS (Chen et al., 2006; Rainey et al., 2006). However, more recently, others have shown a weak interaction with the matrix-localized RNA helicase hSUV3, suggesting a matrix localization for PNPASE (Szczesny et al., 2009; Wang et al., 2009). Our sub-fractionation studies were highly reproducible and, in contrast, we favor the idea that hSUV3 bound to PNPASE after all of the mitochondrial sub-compartments were exposed during purification. The methods used to identify this interaction do not exclude this possibility and we have already shown an unanticipated interaction between PNPASE and the cytosol-localized oncoprotein TCL1 using similar limited resolution methods (French et al., 2007). Our failure to again identify a hSUV3 and PNPASE interaction using an ultra-sensitive dual-tag expression and purification system (Claypool et al., 2008) further supports this interpretation (Figure S1). An alternative explanation that we cannot exclude or confirm with current methodologies is that a small amount PNPASE could get into the matrix and interact with hSUV3.
A second interesting area opened by the current results is to determine how PNPASE controls RNA import into mitochondria. In concept, PNPASE could import RNAs from the cytosol into the IMS and then pass this RNA to another protein or complex that would assist it through the IM into the matrix (Figure 4C). Interestingly, PNPASE augments import in yeast, which does not have a PNPASE homolog, indicating a distinct RNA import mechanism that PNPASE can augment directly or independently. In mouse liver and MEF mitochondria, PNPASE KO is incomplete because cells without PNPASE are non-viable, so it is unclear whether it is absolutely essential for all RNA import or whether the amount detected in import assays is still mediated by the minimal residual amount of PNPASE present required for cell survival. Furthermore, it is not clear if this imported RNA in PNPASE KO mitochondria is stuck in the IMS and not in the matrix, which could provide further insight for the detailed function of PNPASE in RNA import.
PNPASE has two external domains (KH and S1) that bind RNA near the opening of a central processing pore in a trimeric complex (Carpousis, 2002; Symmons et al., 2000). It is not clear whether the same domains are used indiscriminately or in some distinct manner to trigger PNPASE RNA processing versus import functions. It is possible that the stem-loop structures identified in RNase P and MRP RNAs interact with PNPASE in a manner that triggers only import rather than processing. Interestingly, GAPDH RNA can be a target of PNPASE degradation activity in vitro (French et al., 2007), although when either of the identified stem-loop structures is appended to the 5’-terminus, GAPDH RNA is efficiently imported into mitochondria (Figure 6E). Indeed, RNA structural elements regulate PNPASE function in chloroplasts and prokaryotes (Lisitsky et al., 1996; Yehudai-Resheff and Schuster, 2000) and a stem-loop structure protects the RNA from degradation by PNPASE in chloroplasts (Yehudai-Resheff et al., 2001). A detailed dissection of what constitutes a trigger sequence for processing versus import activities is clearly warranted. Finally, since the overall sequence homology between the identified stem-loop structures on RNase P and MRP RNAs is not high, binding interactions between specific RNAs and PNPASE may be stronger or weaker, allowing or inhibiting detection by standard in vitro techniques such as IP, which could make the verification of additional candidate imported RNAs challenging without the functional import assay system for validation.
Studies and important applications that use RNA import in mammalian cells have been hampered by the inconsistent requirements between systems and their study in vitro versus in vivo. Although the exact mechanism for how PNPASE augments and licenses RNA import is not yet known, the identification of PNPASE represents the first receptor-like component that binds RNA in mammalian cells to mediate RNA import into the mitochondrial matrix. This finding, along with the identification of import signal sequences, should open up additional studies to determine what other pathway components are involved and what the RNA sequence or structure rules tell us about how PNPASE may decipher between processing and import.
EXPERIMENTAL PROCEDURES
Supplemental Data includes Supplemental Experimental Procedures, seven figures, and Supplemental References.
Protein and RNA Purification
Purification of the PNPASE-HisPC protein complex was performed as before (Claypool et al., 2008). For protein-RNA interactions, mitochondria (1 mg/ml) were solubilized in lysis buffer (300 mM NaCl, 10 mM imidazole, 10% glycerol, 0.25% Triton X-100, 2 mM DTT, 20 mM HEPES pH 6.6) containing protease inhibitor (Roche) and RNase inhibitor (Invitrogen). Insoluble material was removed by spinning and extracts transferred to Eppendorf tubes. 50 μl of Ni2+NTA resin (QIAGEN) was incubated in 1 ml lysis buffer with 100 μg/ml ssDNA for 1h at 4°C. The resin was then mixed with the mitochondrial lysates in the presence of 100 μg/ml ssDNA for 1h at 4°C. After incubation, the resin was washed 10X with lysis buffer containing RNase inhibitor. The protein-RNA complex was eluted with elution buffer (300 mM NaCl, 10 mM imidazole, 10% glycerol, 0.25% Triton X-100, 20 mM citrate pH 5.5) containing RNase inhibitor. RNA was isolated from the eluate using TRIzol reagent (Invitrogen).
Isolation of Mitochondrial RNA and DNA
Mitochondria (1 mg/ml) were treated with 25 μg/ml of micrococcal nuclease S7 in nuclease buffer (0.6 M Sorbitol, 20 mM MgCl2, 5 mM CaCl2, 20 mM Tris pH 8.0) for 30 min at 27°C. The reaction was stopped by addition of 20 mM EGTA. Mitochondria were collected and solubilized in SDS buffer (100 mM NaCl, 1% SDS, 20 mM Tris pH 7.4) at 65°C for 5 min. RNA was purified using TRIzol reagent, and treated with RNase-free DNase I (Roche) for 1h at 37°C. DNase I was inactivated by heating at 65°C for 10 min. Phenol-chloroform (EM Science) extractions were used for DNA purification from the mitochondrial lysates.
In Vitro Transcription
RNAs were synthesized as before (Portnoy et al., 2008). For radiolabeled RNA synthesis, [32P]-CTP (MP Biochemicals) was incorporated. The RNAs were purified using TRIzol reagent.
RNA Import Assay
Yeast mitochondria were isolated from cells grown in selection medium until stationary phase and mammalian mitochondria were isolated as previously described (Chen et al., 2006; Rainey et al., 2006). In vitro RNA import assays were performed in a 200-μl volume containing 0.5 μg of RNA, 100 μg of mitochondria, 0.6 M sorbitol, 2 mM KH2PO4, 50 mM KCl, 10 mM MgCl2, 2.5 mM EDTA, 5 mM L-methionine, 1 mg/ml BSA, 5 mM ATP, 2 mM DTT, 5 mM NADH, 50 mM HEPES, pH 7.1, at RT for 10 min. Mitochondria were spun at 11,000 × g for 5 min and washed once with wash buffer (0.6 M sorbitol, 20 mM Tris, pH 8.0). Mitochondria were spun again and resuspended in 200 μl nuclease buffer containing 25 μg/ml of micrococcal nuclease S7 and incubated for 30 min at 27°C. Mitochondria were collected and solubilized in SDS buffer at 65°C for 5 min. RNA was purified using TRIzol reagent. For import into mammalian mitochondria, 0.25 M sucrose instead of 0.6 M sorbitol, and 20 mM succinate instead of 5 mM NADH, were used. For import with radiolabeled RNA, the purified RNAs were analyzed by SDS-PAGE and autoradiography.
RNA Degradation Assay
The RNA processing activity of WT and mutant PNPASE was done as before (Portnoy et al., 2008). [32P]-RNA was incubated with the corresponding proteins in buffer E (20 mM HEPES, pH 7.9, 60 mM KCl, 12.5 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, and 17% glycerol, 0.1 mM Pi) at 25°C for 5 min. Following incubation, the RNA was isolated and analyzed by SDS-PAGE and autoradiography.
Additional Procedures
Osmotic shock was performed by incubating mitochondria for 30 min on ice in 0.03 M sorbitol and 20 mM Hepes-KOH, pH 7.4 (Claypool et al., 2006). Blue native gel electrophoresis was performed on a 6–16% linear polyacrylamide gradient using 50 μg of digitonin solubilized material (Chen et al., 2006). Northern blotting was performed as previously described (Tollervey at al., 1987). Total mtRNA was separated on a 12% agaroseformaldehyde gel and transferred to a nylon membrane. Hybridization was carried out with [32P]-dCTP (MP Biochemicals) labeled DNA probes. In organello protein synthesis assays were performed as before with minor changes (Stuart and Koehler, 2007). 100 μg mouse liver mitochondria were incubated in 100 μl translation buffer (250 mM sucrose, 100 mM KCl, 1 mM MgCl2, 10 mM Tris pH 7.4, 10 mM K2HPO4 pH 7.4, 10 mM glutamate, 10 mM malate, 5mM NADH, 1 mM ADP, 1 mg/ml BSA, 100 μg/ml emetine, 100 μg/ml cycloheximide, and 30 μM of amino acid mix without methionine) with 5 μl of L-[35S] methionine (MP Biochemicals) at 37°C for 30 min. The mitochondria were then precipitated and proteins resolved by 12% SDS PAGE.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michelle Husain for expertise in TEM. Supported by NIH grants R01GM061721 and R01GM073981 (C.M.K.), R01CA90571 (M.A.T.) and PN2EY018228 (Roadmap for Medical Research Nanomedicine Initiative) (M.A.T.), the Muscular Dystrophy Association 022398 (C.M.K.), the American Heart Association 0640076N (C.M.K.), and the California Institute of Regenerative Medicine (RS1-00313 and RB1-01397). C.M.K. is an Established Investigator of the American Heart Association and M.A.T. is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alfonzo JD, Soll D. Mitochondrial tRNA import--the challenge to understand has just begun. Biol Chem. 2009;390:717–722. doi: 10.1515/BC.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- Carpousis AJ. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem Soc Trans. 2002;30:150–155. [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Clayton DA. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987;235:1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- Chang DD, Clayton DA. Mouse RNAase MRP RNA is encoded by a nuclear gene and contains a decamer sequence complementary to a conserved region of mitochondrial RNA substrate. Cell. 1989;56:131–139. doi: 10.1016/0092-8674(89)90991-4. [DOI] [PubMed] [Google Scholar]
- Chen HW, Koehler CM, Teitell MA. Human polynucleotide phosphorylase: location matters. Trends Cell Biol. 2007;17:600–608. doi: 10.1016/j.tcb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Chen HW, Rainey RN, Balatoni CE, Dawson DW, Troke JJ, Wasiak S, Hong JS, McBride HM, Koehler CM, Teitell MA, French SW. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol Cell Biol. 2006;26:8475–8487. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J Cell Biol. 2008;182:937–950. doi: 10.1083/jcb.200801152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F, Garcia S, Hernandez D, Regev A, Rebelo A, Oca-Cossio J, Moraes CT. Pathophysiology and fate of hepatocytes in a mouse model of mitochondrial hepatopathies. Gut. 2008;57:232–242. doi: 10.1136/gut.2006.119180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doersen CJ, Guerrier-Takada C, Altman S, Attardi G. Characterization of an RNase P activity from HeLa cell mitochondria. Comparison with the cytosol RNase P activity. J Biol Chem. 1985;260:5942–5949. [PubMed] [Google Scholar]
- Duchene AM, Pujol C, Marechal-Drouard L. Import of tRNAs and aminoacyltRNA synthetases into mitochondria. Curr Genet. 2009;55:1–18. doi: 10.1007/s00294-008-0223-9. [DOI] [PubMed] [Google Scholar]
- Entelis NS, Kolesnikova OA, Martin RP, Tarassov IA. RNA delivery into mitochondria. Adv Drug Deliv Rev. 2001;49:199–215. doi: 10.1016/s0169-409x(01)00135-1. [DOI] [PubMed] [Google Scholar]
- French SW, Dawson DW, Chen HW, Rainey RN, Sievers SA, Balatoni CE, Wong L, Troke JJ, Nguyen MT, Koehler CM, Teitell MA. The TCL1 oncoprotein binds the RNase PH domains of the PNPase exoribonuclease without affecting its RNA degrading activity. Cancer Lett. 2007;248:198–210. doi: 10.1016/j.canlet.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Hollingsworth MJ, Martin NC. RNase P activity in the mitochondria of Saccharomyces cerevisiae depends on both mitochondrion and nucleus-encoded components. Mol Cell Biol. 1986;6:1058–1064. doi: 10.1128/mcb.6.4.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Koehler CM, Jarosch E, Tokatlidis K, Schmid K, Schweyen RJ, Schatz G. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science. 1998;279:369–373. doi: 10.1126/science.279.5349.369. [DOI] [PubMed] [Google Scholar]
- Kolesnikova OA, Entelis NS, Jacquin-Becker C, Goltzene F, Chrzanowska-Lightowlers ZM, Lightowlers RN, Martin RP, Tarassov I. Nuclear DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Hum Mol Genet. 2004;13:2519–2534. doi: 10.1093/hmg/ddh267. [DOI] [PubMed] [Google Scholar]
- Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, Steer CJ. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009;6:65–72. doi: 10.4161/rna.6.1.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsky I, Klaff P, Schuster G. Addition of destabilizing poly (A)-rich sequences to endonuclease cleavage sites during the degradation of chloroplast mRNA. Proc Natl Acad Sci U S A. 1996;93:13398–13403. doi: 10.1073/pnas.93.23.13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel H, Hartman C, Berkowitz D, Elpeleg ON, Manov I, Iancu TC. The hepatic mitochondrial DNA depletion syndrome: ultrastructural changes in liver biopsies. Hepatology. 2001;34:776–784. doi: 10.1053/jhep.2001.27664. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Basu S, Home P, Dhar G, Adhya S. Necessary and sufficient factors for the import of transfer RNA into the kinetoplast mitochondrion. EMBO Rep. 2007;8:589–595. doi: 10.1038/sj.embor.7400979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- Portnoy V, Palnizky G, Yehudai-Resheff S, Glaser F, Schuster G. Analysis of the human polynucleotide phosphorylase (PNPase) reveals differences in RNA binding and response to phosphate compared to its bacterial and chloroplast counterparts. Rna. 2008;14:297–309. doi: 10.1261/rna.698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Puranam RS, Attardi G. The RNase P associated with HeLa cell mitochondria contains an essential RNA component identical in sequence to that of the nuclear RNase P. Mol Cell Biol. 2001;21:548–561. doi: 10.1128/MCB.21.2.548-561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey RN, Glavin JD, Chen HW, French SW, Teitell MA, Koehler CM. A new function in translocation for the mitochondrial i-AAA protease Yme1: import of polynucleotide phosphorylase into the intermembrane space. Mol Cell Biol. 2006;26:8488–8497. doi: 10.1128/MCB.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MA, Rinehart JJ, Krett B, Duvezin-Caubet S, Reichert AS, Soll D, Alfonzo JD. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0804283105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov AV, Entelis NS, Krasheninnikov IA, Martin R, Tarassov IA. Specific features of 5S rRNA structure - its interactions with macromolecules and possible functions. Biochemistry (Mosc) 2008;73:1418–1437. doi: 10.1134/s000629790813004x. [DOI] [PubMed] [Google Scholar]
- Stuart RA, Koehler CM. In vitro analysis of yeast mitochondrial protein import. Curr Protoc Cell Biol Chapter. 2007;11:19. doi: 10.1002/0471143030.cb1119s34. Unit 11. [DOI] [PubMed] [Google Scholar]
- Symmons MF, Jones GH, Luisi BF. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure. 2000;8:1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- Symmons MF, Williams MG, Luisi BF, Jones GH, Carpousis AJ. Running rings around RNA: a superfamily of phosphate-dependent RNases. Trends Biochem Sci. 2002;27:11–18. doi: 10.1016/s0968-0004(01)01999-5. [DOI] [PubMed] [Google Scholar]
- Szczesny RJ, Borowski LS, Brzezniak LK, Dmochowska A, Gewartowski K, Bartnik E, Stepien PP. Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 2009;38:279–98. doi: 10.1093/nar/gkp903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov I, Entelis N, Martin RP. An intact protein translocating machinery is required for mitochondrial import of a yeast cytoplasmic tRNA. J Mol Biol. 1995;245:315–323. doi: 10.1006/jmbi.1994.0026. [DOI] [PubMed] [Google Scholar]
- Tollervey D. A yeast small nuclear RNA is required for normal processing of preribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SC, Engelke DR. A protein-only RNase P in human mitochondria. Cell1. 2008;35:412–414. doi: 10.1016/j.cell.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DD, Shu Z, Lieser SA, Chen PL, Lee WH. Human Mitochondrial SUV3 and Polynucleotide Phosphorylase Form a 330-kDa Heteropentamer to Cooperatively Degrade Double-stranded RNA with a 3'-to-5' Directionality. J Biol Chem. 2009;284:20812–20821. doi: 10.1074/jbc.M109.009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff S, Hirsh M, Schuster G. Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol Cell Biol. 2001;21:5408–5416. doi: 10.1128/MCB.21.16.5408-5416.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff S, Schuster G. Characterization of the E.coli poly(A) polymerase: nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucleic Acids Res. 2000;28:1139–1144. doi: 10.1093/nar/28.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


