Abstract
A total of 176 genes homozygously deleted in human lung cancer were identified by DNA array-based whole genome scanning of 52 lung cancer cell lines and subsequent genomic PCR in 74 cell lines, including the 52 cell lines scanned. One or more exons of these genes were homozygously deleted in one (1%) to 20 (27%) cell lines. These genes included known tumor suppressor genes, e.g., CDKN2A/p16, RB1, and SMAD4, and candidate tumor suppressor genes whose hemizygous or homozygous deletions were reported in several types of human cancers, such as FHIT, KEAP1, and LRP1B/LRP-DIP. CDKN2A/p16 and p14ARF located in 9p21 were most frequently deleted (20/74, 27%). The PTPRD gene was most frequently deleted (8/74, 11%) among genes mapping to regions other than 9p21. Somatic mutations, including a nonsense mutation, of the PTPRD gene were detected in 8/74 (11%) of cell lines and 4/95 (4%) of surgical specimens of lung cancer. Reduced PTPRD expression was observed in the majority (>80%) of cell lines and surgical specimens of lung cancer. Therefore, PTPRD is a candidate tumor suppressor gene in lung cancer. Microarray-based expression profiling of 19 lung cancer cell lines also indicated that some of the 176 genes, such as KANK and ADAMTS1, are preferentially inactivated by epigenetic alterations. Genetic/epigenetic as well as functional studies of these 176 genes will increase our understanding of molecular mechanisms behind lung carcinogenesis.
INTRODUCTION
Lung cancer is the leading cause of cancer-related deaths in the world (Herbst et al., 2008). The majority of lung cancers are comprised of four major histological types, which are small cell lung carcinoma (SCLC) and three nonsmall cell lung carcinoma (NSCLC) types; adenocarcinoma (ADC), squamous cell carcinoma (SQC), and large cell carcinoma (LCC). Lung cancer develops through the acquisition of alterations in oncogenes, such as EGFR (10–40% of ADC) and KRAS (10–30% of ADC), and tumor suppressor genes, such as TP53 (~90% of SCLC; 50% of NSCLC), RB1 (~90% of SCLC; ~20% of NSCLC), CDKN2A/p16 (~50% of NSCLC), and LKB1/STK11 (20–30% of NSCLC) (Minna et al., 2002; Herbst et al., 2008). The EGFR, KRAS, and TP53 genes have been subjected to diagnostic and therapeutic applications (Toloza et al., 2006; Herbst et al., 2008); therefore, identification of more genes involved in lung carcinogenesis will be highly applicable to further improve the diagnosis and therapy of lung cancer. Allelic imbalance (AI) studies on lung cancer have identified several chromosome arms frequently hemizygously deleted, such as 1p, 4q, 5q, 6q, 8p, 11q, 12q, 13q, 17q, and 21q (Shiseki et al., 1996; Kawanishi et al., 1997; Virmani et al., 1998; Girard et al., 2000). Our recent comparative genome-wide AI study of noninvasive and invasive lung adenocarcinomas (ADCs) further suggested that AI on each chromosome arm has different roles in the development and progression of lung cancer (Nakanishi et al., 2009). Therefore, chromosomal deletions and inactivation of corresponding tumor suppressor genes are thought to play multiple roles in the development and/or progression of lung cancer. However, responsible tumor suppressor genes for most of these chromosomal deletions have not yet been identified.
Homozygous deletion (i.e., deletion of both alleles) is a genetic event causing inactivation of tumor suppressor genes (Minna et al., 2002; Yokota and Kohno, 2004), and has played an important role as a tool in identifying several tumor suppressor genes, such as CDKN2A/p16, PTEN, and SMAD4 (Kamb et al., 1994; Nobori et al., 1994; Hahn et al., 1996; Li et al., 1997; Steck et al., 1997). Up to the present, DNA array analyses have been performed by several groups, including ours, to find homozygously deleted regions in lung cancer genomes (Sato et al., 2005; Tonon et al., 2005; Zhao et al., 2005; Garnis et al., 2006; Imoto et al., 2006; Nagayama et al., 2007; Weir et al., 2007), and tens of genomic regions with homozygous deletions have been identified. However, only a few genes located in some of the homozygously deleted regions were focused on and investigated.
In this study, genes whose exons were removed by homozygous deletions were comprehensively searched for by a DNA array-based whole genome scanning of 52 human lung cancer cell lines followed by genomic PCR analyses. Notably, several well-known tumor suppressor genes, such as RB1, DCC, and BRCA2, have been identified from a single or a few cases of homozygous deletions detected in a large number of cancer cases analyzed (Dryja et al., 1986; Fearon et al., 1990; Wooster et al., 1995). The results indicated the significance of homozygous deletions irrespective of their frequencies for the identification of novel tumor suppressor genes. Therefore, in the present study, all genes deduced to be mapped in homozygously deleted regions were examined, even if the deletions were detected only in a single lung cancer case. Lung cancer cell lines were used for two reasons: First, the presence of a homozygous deletion can be easily validated by genomic PCR due to the lack of noncancerous cell contamination that hampers detection of homozygous deletions; second, frequencies of copy number changes in the genome were shown to be similar in cell lines and surgical specimens in our previous study (Ogiwara et al., 2008). Hence homozygous deletions detected in cell lines can be considered to have occurred mostly in vivo, and not during their establishment and cultivation in vitro. In total, 176 genes located in 45 genomic loci on 17 chromosomes were identified as genes whose exons were homozygously deleted (Supporting Information Table 1). One of the 176 genes, PTPRD, was subjected to mutation and expression analyses in surgical specimens of lung cancer as well as lung cancer cell lines to address the authenticity of this gene as a lung tumor suppressor gene.
MATERIALS AND METHODS
Human Lung Cancer Cell Lines and Surgical Specimens for Lung Cancer
Forty-three lung cancer cell lines were previously subjected to a SNP array analysis at a 100-kb resolution using an Affymetrics Mapping 100-k array (Affymetrix, Inc., Santa Clara, California), and they were 11 SCLCs, 21 ADCs, 7 SQCs, and 4 LCCs (Nagayama et al., 2007). In the present study, 27 ADC cell lines consisting of 18 lines (II-18, A549, Ma17, Ma24, H23, H322, H1395, H1437, H2009, H2087, H2122, H2347, PC3, PC7, PC9, PC14, RERF-LCMS, and VMRC-LCD) analyzed in the previous analysis (Nagayama et al., 2007) and 9 cell lines (ABC1, Ma10, Ma12, Ma26, Ma29, HCC44, HCC78, HCC193, and HCC515) prepared for this study were subjected to an array-CGH analysis at a 30-kb resolution using a Human CGH 185-k array (Agilent Technologies, Santa Clara, California). Therefore, 52 cell lines in total were scanned for homozygous deletions by using one or two DNA-array methods at 30 and 100-kb resolutions. To validate homozygous deletion, 74 cell lines consisting of 52 cell lines subjected to these array analyses and an additional 22 lung cancer cell lines consisting of 11 SCLCs (H526, H774, H1339, H1450, H1607, H1819, NCI-H1963, H2195, HCC33, Lu24, and Ms18), 3 ADCs (H2126, H1703, and RERF-LCOK), 3 SQCs (HCC95, Sq-5, and PC10), 3 LCCs (Lu99, Ma2, and Ma25), and 2 adenosquamous carcinomas (ASCs) (H596 and HCC366) were analyzed. Details of H- and HCC-series cell lines have been described elsewhere (Burbee et al., 2001). PC-, Lu-, Ma-series, and II-18 cell lines were provided by Drs. Y. Hayata (Tokyo Medical University, Tokyo, Japan), T. Terasaki (Kanagawa Institute of Technology, Kanagawa, Japan) and S. Hirohashi (National Cancer Center Research Institute, Tokyo, Japan), M. Takada (National Hospital Organization Kinki-chuo Chest Medical Center, Osaka, Japan), and K. Hagiwara (Saitama Medical University, Saitama, Japan), respectively. Cell lines were also obtained from the American Type Culture Collection (Manassas, Virginia), the Japanese Collection of Research Bioresources (Tokyo, Japan), and the RIKEN BioResource Center (Tsukuba, Japan). Genomic DNA and poly A RNA were extracted by standard protocols.
Macro-dissected and micro-dissected cancerous and noncancerous lung cells were obtained from patients who were treated at the National Cancer Center Hospital, Tokyo, Japan. Details of these materials were described previously (Matsumoto et al., 2006; Nakamura et al., 2006). Genomic DNA and total RNA were extracted by standard protocols. This study was performed under the approval of the Institutional Review Board of National Cancer Center.
Detection of Homozygous Deletions by Array CGH Analysis
Copy number changes in genomic DNAs of 27 lung cancer cell lines were assessed using a Human CGH 185-k array covering 181,988 loci and Agilent CGH Analytics Software (Version 3.3) (Agilent Technologies). Genomic DNAs from these cell lines and 10 lymphoblastoid cell lines were analyzed according to the manufacturer’s protocol using a human normal genomic DNA mix (Promega) as a reference. First, data for probes that were located in copy number variable regions deposited in the UCSC genome database and those that showed log2 ratios < −2.5 or >2.5 in 10 lymphoblastoid cell line DNAs in the present analyses were removed to mask copy number variable regions. Next, the copy number along the genome of 27 lung cancer cell lines was inferred by the Aberration Detection Method 2 (ADM2) algorithm. Autosomal regions encompassed by ≥2 consecutive probes with log2 ratios < −2.5 were defined as candidates for homozygously deleted regions. As the probes were placed at a mean interval of 15-kb, the present homozygous deletion search was undertaken at a 30-kb resolution.
Validation of Homozygous Deletions
One or more set(s) of PCR primers was designed to amplify genomic fragments that encompass an exon of genes deduced to be located in homozygously deleted regions by referring to the information in the UCSC database (http://genome.ucsc.edu/) and was subjected to multiplex PCR using the IRF1 locus as a reference (Kishimoto et al., 2005). At least one exon was examined for each gene. Primer sequences are listed in Supporting Information Table 2. PCR products were separated by electrophoresis on 3% agarose gel and visualized by staining with ethidium bromide. When no PCR product was detected, such an exon was judged as being homozygously deleted.
Expression Analysis of the 176 Genes Homozygously Deleted in Lung Cancer
Information on the expression levels for 23,583 genes of 15 lung cancer cell lines (A549, H157, H322, H1299, H1437, H1648, H2009, H2122, H2126, H2347, HCC95, HCC193, HCC366, and HCC515) was previously obtained by analysis using Affymetrix Gene Chips HG-U133A and HG-U133B (Zhou et al., 2006). Information on those of four other lung cancer cell lines (H223, H209, H841, and H2141) and cultured noncancerous lung epithelial cells (Ramirez et al., 2004) was obtained for the present study. Expression data for 160 of the 176 genes homozygously deleted in lung cancer cell lines were available, and these 160 genes were assessed by 281 probes (Supporting Information Table 1). Differences in expression levels between lung cancer cell lines and noncancerous lung epithelial cells were examined by t test, and probes with P < 0.05 were judged as significantly different. In addition, probes with P < 0.00018 were judged as significantly different after Bonferroni correction for multiple tests (i.e., 0.05/281 = 0.00018).
Mutation Analysis of the PTPRD Gene
All coding exons of the PTPRD gene were amplified from 10 ng of DNA from 74 cell lines and 95 surgical specimens of lung cancer by PCR using 44 sets of primers. PCR products from the cell lines were directly sequenced using a Big Dye Terminator Sequencing kit and an ABI Prism 3700 Genetic Analyzer (Applied Biosystems, Foster City, California, USA). PCR products from the surgical specimens were subjected to WAVE analysis according to the manufacturer’s protocol (Transgenomic, Omaha, Nebraska, USA). PCR products with different mobilities in the WAVE analysis were purified and directly sequenced.
Quantitative Real-time Reverse Transcription PCR (QRT-PCR) Analysis
Expression levels of the PTPRD gene were evaluated by QRT-PCR using ABI Prism 7900HT (Applied Biosystems). A Taqman probe (5′-AGGATCAATATCAGTTTTCCTA-3′) and a set of primers (5′-TGTTAAGAACACAACGAC CAGCTAT-3′ and 5′- TCAAAGCTGCCCAGG TACTCTAGT-3′) were used as previously described (Sato et al., 2005). PCR was performed in a single tube in duplicate. Results were expressed as the average of these two independent tests.
RESULTS AND DISCUSSION
Identification of Genes Homozygously Deleted in 52 Human Lung Cancer Cell Lines
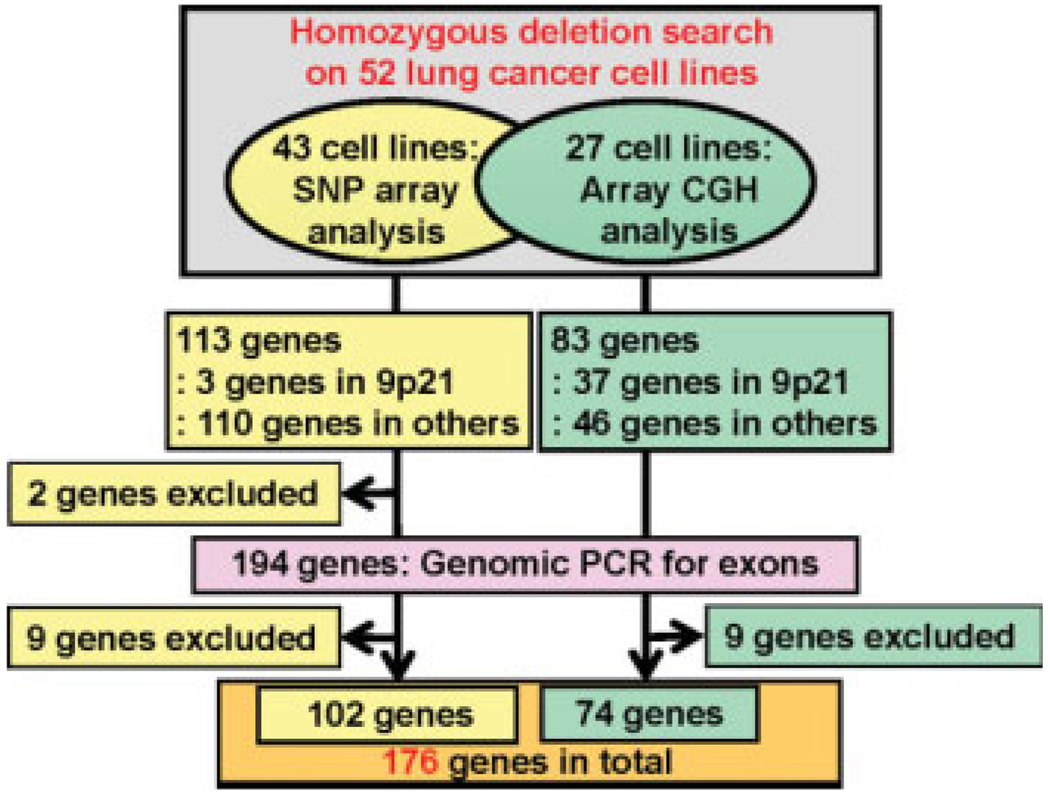
Homozygously deleted regions in 43 lung cancer cell lines were previously searched for at a 100-kb resolution by a SNP array analysis, and 113 genes were deduced to map to homozygously deleted regions in one or more cell lines (Nagayama et al., 2007). These 113 genes consisted of three genes, CDKN2A/p16, p14ARF (a gene sharing the same exons with CDKN2A/p16 but encoding a different protein) (Stone et al., 1995) and CDKN2B/p15, which are considered target tumor suppressor genes for homozygous deletions at chromosome band 9p21 (Hamada et al., 2000), and 110 genes located in regions other than 9p21 (Fig. 1).
Figure 1.
Strategy to identify genes homozygously deleted in lung cancer. A previous search on 43 lung cancer cell lines led to the identification of 113 genes deduced to be homozygously deleted (Nagayama et al., 2007). The present search on 27 lung cancer cell lines (18 cell lines overlaped) led to the identification of an additional 83 genes, and excluded two genes from the 113 genes above. Genomic PCR for exons of 194 genes led to the validation of homozygous deletions in 176 genes.
In this study, homozygous deletions were further searched for by an array CGH analysis at a 30-kb resolution in 27 lung cancer cell lines consisting of 18 cell lines previously analyzed (Nagayama et al., 2007) and 9 cell lines newly prepared (Fig. 1). Among 113 genes found to be deleted in the previous study (Nagayama et al., 2007), 111 were verified in this study. Two genes, THSD4 and C20orf133, considered to be homozygously deleted in the previous study, were not found to be deleted here. Therefore, these two genes were excluded from further analyses. In addition, 83 genes not indicated in the previous study were deduced to be homozygously deleted in the present 30-kb resolution analysis. These 83 genes consisted of 37 genes in the 9p21 region and 46 genes in other regions. Thus, in total, 194 genes (111 + 83) were considered to be homozygously deleted in 52 lung cancer cell lines by using two different DNA array analyses.
To confirm the homozygous deletions of these 194 genes, genomic PCR against DNA fragments encompassing an exon located in a homozygously deleted region was performed for all of the 194 genes (primer information in Supporting Information Table 2). One hundred seventy-six (91%) of the 194 genes showed homozygous deletions of at least one exon (Fig. 1). These 176 genes included CDKN2A/p16 and CDKN2B/p15, and the results for deletions of these two genes determined by genomic PCR were consistent with the results determined by Southern blot analysis in our previous study (Okamoto et al., 1995). On the other hand, homozygous deletions of exons were not detected in the remaining 18 genes, probably due to the fact that only intronic or intergenic sequences were deleted or the deletions deduced were spurious ones caused by experimental errors. Therefore, these 18 genes were excluded from the remaining part of this study. Homozygous deletion of one or more genes was detected in 20 (74%) of the 27 cell lines analyzed by the present array CGH analysis, but not in the remaining seven cell lines. Among a total of 52 cell lines subjected to the present and/or previous homozygous deletion scanning, homozygous deletion of one or more genes was detected in 36 (69%) cell lines.
Characteristics and Genomic Status of 176 Genes Homozygously Deleted in Lung Cancer Cell Lines
The 176 genes with verified homozygous deletions are listed in Table 1 and Supporting Information Table 1. They consisted of 171 protein-encoding genes and five miRNA genes (genes 58, 78, 167–169). These 176 genes were located in 45 regions on 17 chromosomes (Supporting Information Fig. 1). They included known tumor suppressor genes, CDKN2A/p16, p14ARF, CDKN2B/p15, RB1, and SMAD4 (genes 80–82, 131, and 155 in Table 1) (Futreal et al., 2004), as well as candidate tumor suppressor genes shown to be hemizygously or homozygously deleted in several types of human cancers, such as LRP1B/LRP-DIP, FHIT, PTPRD and KEAP1 (genes 3, 18, 47, and 157 in Table 1) (Sozzi et al., 1996; Liu et al., 2000; Sonoda et al., 2004; Sato et al., 2005; Singh et al., 2006; Stallings et al., 2006; Ohta et al., 2008).
TABLE 1.
Homozygous Deletion and Expression of 17 Genes in Cancerous and Noncancerous Cultured Lung Cells (Extracted from Supplementary Table 1)
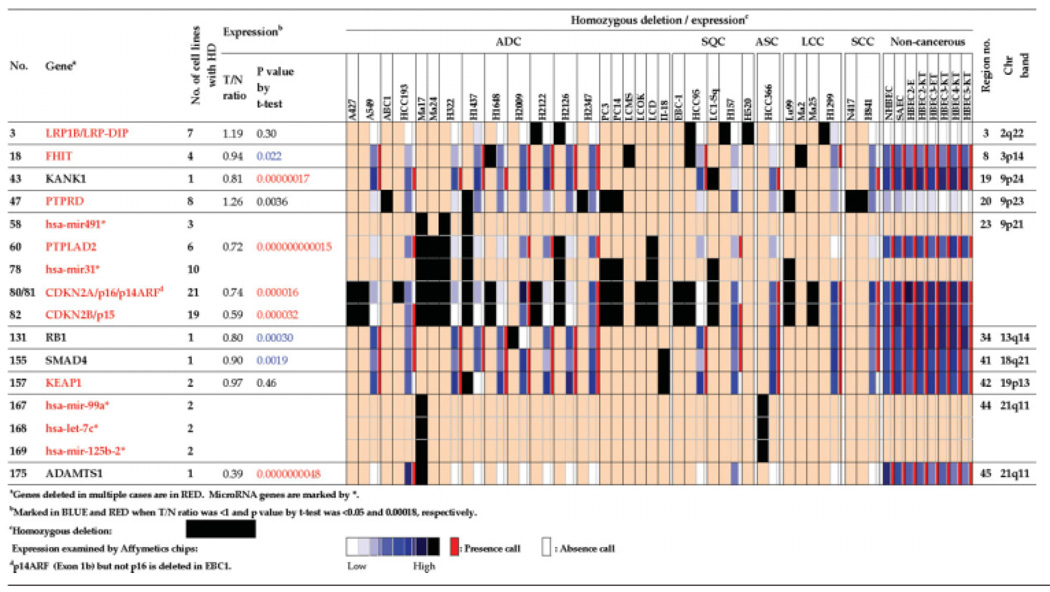 |
Frequencies of homozygous deletion for these 176 genes were examined in 74 lung cancer cell lines, consisting of 52 cell lines used for the array analyses and 22 additional cell lines, by genomic PCR using the same sets of primers as described above (Supporting Information Table 2). One to 75 of these genes were homozygously deleted in 44 of the 74 cell lines analyzed. No gene was deleted in the remaining 30 cell lines (Supporting Information Table 1). Homozygous deletion of each gene was detected in one (1%) to 20 (27%) of the 74 cell lines. Sixty-four (36%) of the 176 genes were deleted in two or more cell lines (Supporting Information Table 3), while the other 112 (64%) were deleted in a single cell line. The CDKN2A/p16 and p14ARF genes (genes 80–81) in the 9p21 region were most frequently deleted (20/74, 27%). Thirty-four genes deleted in two or more cell lines were located in regions other than 9p21 (Table 2). A candidate tumor suppressor gene, PTPRD (gene 47), was most frequently deleted (8/74, 11%) among them. Other known candidate tumor suppressors, LRP1B (gene 3), FHIT (gene 18), and KEAP1 (gene 157), were also included in these 34 genes. Therefore, other genes listed in Table 2 will be also strong candidates for lung tumor suppressors.
TABLE 2.
34 Genes Mapped to Regions Other Than 9p21 and Homozygously Deleted in Two or More Lung Cancer Cell Lines
| Chromosomal location |
Gene | Gene product | No. of cell lines with homozygous deletion |
(%) |
|---|---|---|---|---|
| 9p23 | PTPRD | Protein tyrosine phosphatase, receptor type D | 8 | (11) |
| 2q21 | LRP1B /LRP-DIP | Low density lipoprotein-related protein 1B | 7 | (9) |
| 3p14 | FHIT | Dinucleosidetriphosphatase | 4 | (5) |
| 2q24 | GRB14 | Growth factor receptor-bound protein 14 | 2 | (3) |
| 2q24 | COBLL1 | COBL-like 1 | 2 | (3) |
| 2q24 | SLC38A11 | Solute carrier family 38, member 11 | 2 | (3) |
| 2q24 | SCN3A | Sodium channel type III, alpha subunit | 2 | (3) |
| 2q24 | SCN2A | Sodium channel type II, alpha subunit | 2 | (3) |
| 2q24 | FAM130A2 | TGF-beta induced apoptosis protein 2 | 2 | (3) |
| 2q24 | GALNT3 | UDP-N-acetyl-alpha-D-galactosamine | 2 | (3) |
| 5q11 | PDE4D | Phosphodiesterase 4D | 2 | (3) |
| 5q31 | CTNNA1 | Alpha-catennin | 2 | (3) |
| 7q35 | CNTNAP2 | Contactin associated protein-like 2 | 2 | (3) |
| 10p11 | PARD3 | Par-3 partitioning defective 3 homolog | 2 | (3) |
| 18q21 | ME2 | Malate dehydrogenase 2 | 2 | (3) |
| 18q21 | ELAC1 | elaC homolog 1 | 2 | (3) |
| 19p13 | KEAP1 | Cytosolic inhibitor of Nrf2 | 2 | (3) |
| 19q13 | MZF1 | Myeloid zinc finger 1 | 2 | (3) |
| 19q13 | MGC2752 | Hypothetical LOC65996 | 2 | (3) |
| 21q11 | LIPI | Membrane-associated phospholipase A1 beta | 2 | (3) |
| 21q11 | RBM11 | RNA binding motif protein 11 | 2 | (3) |
| 21q11 | STCH | Stress 70 protein chaperone | 2 | (3) |
| 21q11 | SAMSN1 | SAM domain, SH3 domain and nuclear localization signals 1 | 2 | (3) |
| 21q11 | NRIP1 | Nuclear receptor interacting protein 1 | 2 | (3) |
| 21q11 | USP25 | Ubiquitin specific peptidase 25 | 2 | (3) |
| 21q11 | C21orf34 | Chromosome 21 open reading frame 34 | 2 | (3) |
| 21q11 | hsa-mir-99a | miRNA | 2 | (3) |
| 21q11 | hsa-let-7c | miRNA | 2 | (3) |
| 21q11 | hsa-mir-125b-2 | miRNA | 2 | (3) |
| 21q11 | CXADR | Coxsackie virus and adenovirus receptor | 2 | (3) |
| 21q11 | BTG3/ANA | BTG/Tob family protein | 2 | (3) |
| 21q11 | C21orf91 | Chromosome 21 open reading frame 91 | 2 | (3) |
| 21q11 | CHODL | Transmembrane protein MT75 | 2 | (3) |
| 21q11 | PRSS7 | Enterokinase | 2 | (3) |
Expression Status of the 176 Genes Homozygously Deleted in Lung Cancer Cell Lines
Nineteen of the 44 cell lines with homozygous deletion were available for information on expression levels of 23,583 genes obtained by microarray analysis. These cell lines included all four major histological types of lung cancers. Information on expression levels was available for 160 (91%) of the 176 genes. Most of these genes showed nonsignificant signals (i.e., absent call) in cell lines with homozygous deletion of the corresponding genes, and such genes were LRP1B (gene 3), PTPRD (gene 47), CDKN2A/p16, and p14ARF (genes 80–81 assessed by the same probes), CDKN2B (gene 82), RB1 (gene 131) and KEAP1 (gene 157) (Table 1). On the other hand, some genes with deletions of parts of genes, such as FHIT (gene 18), showed significant signals (i.e., present call) in cell lines with homozygous deletions. As for the FHIT gene, transcripts lacking exons, which are homozygously deleted, were previously shown to be expressed in lung cancer cells (Sozzi et al., 1996).
Expression data on eight cultured noncancerous lung epithelial cells were also available for the same set of genes as in lung cancer cell lines (Zhou et al., 2006). Therefore, we searched for genes whose expression was significantly lower in lung cancer cells compared to noncancerous lung epithelial cells. In 55 (31%) genes, at least one probe showed a level of expression significantly lower than that in noncancerous lung epithelial cells (T/N ratio <1 and P < 0.05 by t test, marked in blue in Table 1 and Supporting Information Table 1). Expression levels in 52 (95%) of these 55 genes remained significantly lower after removing lung cancer cases with homozygous deletion. The differences in expression of 18 genes (10%) remained significant after Bonferroni correction for multiple tests (i.e., P < 0.00018, marked in red in Table 1 and Supporting Information Table 1), and that of 13 (72%) genes remained significant after removing cases with homozygous deletion. These genes included the KANK and ADAMTS1 (genes 131 and 175) candidate tumor suppressor genes whose down-regulation by epigenetic alterations rather than genetic alterations in renal and lung cancers, respectively, were reported (Sarkar et al., 2002; Choi et al., 2008). It was noted that homozygous deletions of these three genes were detected only in one cell line, respectively. The results suggest that the present 176 genes include genes preferentially inactivated in lung cancer cells by epigenetic alterations rather than homozygous deletions.
PTPRD Alterations in Human Lung Cancer
Homozygous deletions and mutations in the PTPRD gene in human lung cancer and other cancers, as well as the ability of PTPRD protein to inhibit growth and to cause apoptosis have indicated that PTPRD is a tumor suppressor gene (Sjoblom et al., 2006; Weir et al., 2007; Ding et al., 2008; Solomon et al., 2008; Veeriah et al., 2009). Thus, we searched for mutations in the PTPRD gene in both cell lines and surgical specimens of lung cancer. Sequencing of all coding exons in 74 lung cancer cell lines revealed that eight cell lines (11%) had nonsynonymous (i.e., associated with amino acid change) nucleotide substitutions that were not deposited in the dbSNP database (Table 3). The substitution in H2171 cells was validated to be a somatic mutation, by the absence of this substitution in the corresponding lymphoblastoid cell line (Supporting Information Fig. 2A). The other seven substitutions detected in the remaining seven cell lines were also likely to be somatic mutations because these substitutions were not detected in noncancerous cells of 95 different individuals (see below), and each of them was detected in only one of the 74 lung cancer cell lines and in none of the 95 primary tumors. Among the 95 surgical specimens analyzed for PTPRD mutations, four cases (4%) were concluded as having somatic mutations because nucleotide substitutions were detected only in cancer cells and not in the corresponding noncancerous cells (Table 3). One was a nonsense mutation, two were missense mutations, and the remaining one was a mutation in an intronic sequence. By RT-PCR and sequencing, mutant alleles were shown to be expressed in all of the eight cell lines with PTPRD mutations and a surgical specimen whose RNA was available for analysis (Supporting Information Fig. 2A).
TABLE 3.
PTPRD Mutations in Lung Cancer
| Sample | Histological type | No. of exon | (nucleotide change) | Predicted effect | mRNA levela |
|---|---|---|---|---|---|
| Cell line | |||||
| Ma29 | ADC | 11 | (C1184T: Homo) | Ala395Val | 0.0015 |
| H23 | ADC | 11 | (C1201T: Homo) | Arg401Trp | 0.053 |
| Sq-5 | SQC | 20 | (C3299T: Hetero) | Thr1100Met | 0.00068 |
| H1155 | LCC | 14 | (C2057T: Hetero) | Thr686Ile | 0.035 |
| PC13 | LCC | 20 | (A3164G: Hetero) | Asp1055Gly | 0.010 |
| Lu65 | LCC | 32 | (G5258T: Homo) | Gly1753Cys | 0.00047 |
| H2171b | SCC | 5 | (G460T: Homo) | Asp154Tyr | 0.025 |
| H526 | SCC | 17 | (A2443G: Hetero) | Lys815Glu | 0.70 |
| Surgical specimen | |||||
| Na68Tb | SQC | 4 | (G235T: Hetero) | Gly79STOP | 0.10 |
| Na182Tb | SQC | 21 | (TG3472–3473AT: Hetero) | Trp1158Met | 0.27 |
| S171Tb | SCC | 15 | (G2206T: Hetero) | Val1736Leu | Not tested |
| 1662Tb | SCC | 17 | (C IVS17+16 T: Hetero) | Unknown | Not tested |
Relative expression level to noncancerous lung tissues.
Validated to be somatic mutation.
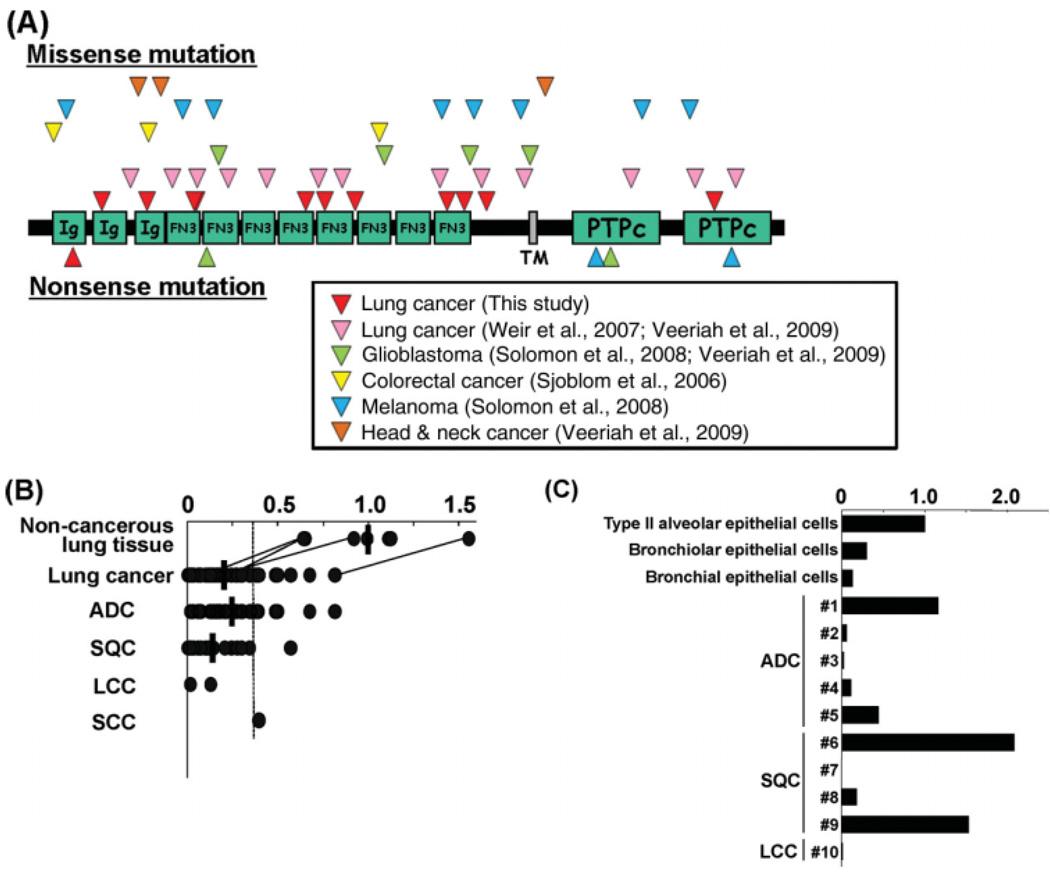
The PTPRD mutations detected in this study were dispersed through the PTPRD protein as previously observed in lung and others cancers (Fig. 2A) (Sjoblom et al., 2006; Weir et al., 2007; Solomon et al., 2008; Veeriah et al., 2009). It was noted that the same mutations were not present among the mutations detected in human cancers up to the present, and hot spots for mutations were not obvious (Supporting Information Fig. 2B and 2C). A recent study indicated that several mutant PTPRD proteins have lower abilities than the wild-type protein to inhibit growth and to cause apoptosis in cells (Solomon et al., 2008). In addition, a subset of mutations, including Gly79X detected in the present study, were nonsense mutations causing a production of truncated PTPRD proteins lacking the whole or a part of protein tyrosine phosphatase catalytic domains. These results indicate that somatic PTPRD mutations are a genetic event causing functional inactivation of the PTPRD gene in human cancer cells.
Figure 2.
PTPRD mutation and expression in lung cancer. (A) Location of missense and nonsense mutations detected in the present and previous studies. Ig, immunoglobulin-like C2-type domain; FN3, fibronectin type III domain; TM, transmembrane domain; PTPc, protein tyrosine phosphatase catalytic domain. (B) Expression in macro-dissected cancerous and noncancerous lung cells. Values for four paired noncancerous and cancerous lung tissues are connected. Expression levels are indicated after adjusting the mean for the levels of expression in seven cases of noncancerous lung tissues to 1. Mean values are indicated by horizontal bars if the group has three or more samples. The threshold level to judge as reduced expression is indicated by a dashed line. (C) Expression in micro-dissected cancerous and noncancerous lung cells. The levels of PTPRD expression relative to those of GAPDH expression are shown after adjusting the level of PTPRD expression in type II alveolar epithelial cells to 1.
We next examined the expression of the PTPRD gene in both cell lines and surgical specimens of lung cancer. We previously reported that the majority (>90%) of lung cancer cell lines, including eight cell lines with homozygous PTPRD deletions showed lower expression levels than noncancerous lung cells (Sato et al., 2005). In this study, eight cell lines with PTPRD mutations were also shown to express lower levels compared to normal lung tissue (Table 3). Sixty surgical specimens of lung tumors were also subjected to QRT-PCR expression analysis. Fifty-one (85%) specimens showed lower levels of PTPRD expression than the mean-2SD for seven noncancerous lung tissues, therefore, these samples were judged as having a reduced PTPRD expression (Fig. 2B). Two specimens with PTPRD mutations for which RNA was available also showed reduced PTPRD expression (Table 3). Differences in PTPRD levels between noncancerous lung tissues and all lung cancers, ADCs or SQCs were significant (P < 0.05 by t test). Two LCCs also showed reduced PTPRD expression. A SCC case examined also showed a lower level of PTPRD, however, was not judged as having a reduced PTPRD expression according to the criteria above.
We also examined PTPRD expression in non-cancerous lung component cells and cancerous cells which were obtained by micro-dissection of surgical specimens (Nakamura et al., 2006). PTPRD expression was detected in noncancerous lung component cells with the highest expression in type II alveolar epithelial cells, candidate precursor cells for lung ADC (Otto, 2002) (Fig. 2C). The levels of expression in five lung cancer samples were lower than those of any component cells (cases 2, 3, 4, 7, and 10 in Fig. 2C). These results suggested that reduced PTPRD expression commonly occurs in lung carcinogenesis. Recently, reduced expression of PTPRD was shown to be a frequent event in human glioblastoma, and to be caused by hypermethylation of the promoter region of the PTPRD gene (Veeriah et al., 2009). Therefore, it was strongly suggested that PTPRD is silenced by a promoter hypermethylation also in lung cancer, although methylation status of the PTPRD gene was not examined in this study.
CONCLUSION
We identified 176 genes homozygously deleted in human lung cancer. These genes included known tumor suppressor genes and candidate tumor suppressor genes, whose hemizygous or homozygous deletions as well as intragenic mutations had been reported in several types of human cancers. Furthermore, these 176 genes include genes preferentially inactivated by epigenetic alterations, such as KANK and ADAMTS1. Indeed, one of these candidates, PTPRD, was shown to be genetically and/or epigenetically altered in a considerable fraction of lung cancer. Therefore, this set of genes will be informative to identify novel lung tumor suppressor genes. The Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) suggests that genes with specific functions or involved in specific signaling pathways are not significantly enriched among these 176 genes. Thus, it was suggested that genes involved in a variety of biological processes could function as lung tumor suppressors. Further genetic/epigenetic as well as functional studies of these 176 genes will help understanding of molecular mechanism of lung carcinogenesis. In fact, the present homozygous deletion scanning was performed on a set of 52 lung cancer cell lines including all four major histological types of lung cancer. However, fractions of ADC (30/52, 58%) and SQC (7/52, 13%) were larger and smaller than those in the population of lung cancer patients (Parkin et al., 2004), respectively, therefore, scanning of other sets of lung cancer that are predominant for SQC might provide additional genes.
One to 75 genes were homozygously deleted in 44 lung cancer cell lines analyzed, while no genes was deleted in the remaining 30 cell lines. The result might imply that the intrinsic genomic stability against homozygous deletion is different among lung cancer cases. We recently reported genetic/epigenetic alteration profiles of known oncogenes and tumor suppressor genes in the cell lines used in this study (Medina et al., 2008; Blanco et al., 2009). Therefore, relationships between these alterations and homozygous deletions of those 176 genes were examined. Interestingly, numbers of genes with homozygous deletions are significantly or marginally significantly different according to alterations of tumor suppressor genes, TP53, CDKN2A/p16, LKB1, and PTEN (Supporting Information Table 4). Multivariate analysis indicated that only the CDKN2A/p16 alteration among them was independently associated with the number of genes homozygously deleted. The result suggests that CDKN2A/p16 alteration is involved in genomic instability inducing homozygous deletions as this gene is critical for the maintenance of genome integrity in human cells (McDermott et al., 2006). Interestingly, the cell lines with homozygous CDKN2A/p16 deletions carried deletions of significantly larger number of genes than those with promoter hypermethylation and mutation of the CDKN2A/p16 gene; and those without (Supporting Information Fig. 3). Therefore, homozygous CDKN2A/p16 deletion can be a marker of intrinsic instability for homozygous deletion. More detailed analysis of genetic/epigenetic interactions as well as functional interactions among genes altered in lung cancer cells will further provide insights into the molecular mechanism of lung carcinogenesis.
Supplementary Material
ACKNOWLEDGMENTS
Supported by: Grants-in-Aid from the Ministry of Health, Labor and Welfare of Japan for the 3rd-term Comprehensive 10-year Strategy for Cancer Control and for Cancer Research (16-1); from the program for promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NiBio); National Cancer Institute Lung Cancer SPORE Grant P50CA70907 and the DOD VITAL grant.
The authors thank Kaho Minoura and Yayoi Fukuoka of Agilent Technologies Japan for technical assistance in array-CGH analysis.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Blanco R, Iwakawa R, Tang M, Kohno T, Angulo B, Pio R, Montuenga LM, Minna JD, Yokota J, Sanchez-Cespedes M. A gene-alteration profile of human lung cancer cell lines. Hum Mutat. 2009;30:1199–1206. doi: 10.1002/humu.21028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S, Sekido Y, Latif F, Milchgrub S, Toyooka S, Gazdar AF, Lerman MI, Zabarovsky E, White M, Minna JD. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JE, Kim DS, Kim EJ, Chae MH, Cha SI, Kim CH, Jheon S, Jung TH, Park JY. Aberrant methylation of ADAMTS1 in non-small cell lung cancer. Cancer Genet Cytogenet. 2008;187:80–84. doi: 10.1016/j.cancergencyto.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Gonzalez-Garay ML, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, Vickery T, Sander S, Robinson J, Winckler W, Baldwin J, Chirieac LR, Dutt A, Fennell T, Hanna M, Johnson BE, Onofrio RC, Thomas RK, Tonon G, Weir BA, Zhao X, Ziaugra L, Zody MC, Giordano T, Orringer MB, Roth JA, Spitz MR, Wistuba II, Ozenberger B, Good PJ, Chang AC, Beer DG, Watson MA, Ladanyi M, Broderick S, Yoshizawa A, Travis WD, Pao W, Province MA, Weinstock GM, Varmus HE, Gabriel SB, Lander ES, Gibbs RA, Meyerson M, Wilson RK. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, Rapaport JM, Joyce JM, Petersen RA. Molecular detection of deletions involving band q14 of chromosome 13 in retinoblastomas. Proc Natl Acad Sci USA. 1986;83:7391–7394. doi: 10.1073/pnas.83.19.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, Hamilton SR, Preisinger AC, Thomas G, Kinzler KW, Vogelstein B. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science. 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnis C, Lockwood WW, Vucic E, Ge Y, Girard L, Minna JD, Gazdar AF, Lam S, Macaulay C, Lam WL. High resolution analysis of non-small cell lung cancer cell lines by whole genome tiling path array CGH. Int J Cancer. 2006;118:1556–1564. doi: 10.1002/ijc.21491. [DOI] [PubMed] [Google Scholar]
- Girard L, Zochbauer-Muller S, Virmani AK, Gazdar AF, Minna JD. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 2000;60:4894–4906. [PubMed] [Google Scholar]
- Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- Hamada K, Kohno T, Takahashi M, Yamazaki M, Tashiro H, Sugawara C, Ohwada S, Sekido Y, Minna JD, Yokota J. Two regions of homozygous deletion clusters at chromosome band 9p21 in human lung cancer. Genes Chromosomes Cancer. 2000;27:308–318. [PubMed] [Google Scholar]
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto I, Izumi H, Yokoi S, Hosoda H, Shibata T, Hosoda F, Ohki M, Hirohashi S, Inazawa J. Frequent silencing of the candidate tumor suppressor PCDH20 by epigenetic mechanism in non-small-cell lung cancers. Cancer Res. 2006;66:4617–4626. doi: 10.1158/0008-5472.CAN-05-4437. [DOI] [PubMed] [Google Scholar]
- Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, III, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- Kawanishi M, Kohno T, Otsuka T, Adachi J, Sone S, Noguchi M, Hirohashi S, Yokota J. Allelotype and replication error phenotype of small cell lung carcinoma. Carcinogenesis. 1997;18:2057–2062. doi: 10.1093/carcin/18.11.2057. [DOI] [PubMed] [Google Scholar]
- Kishimoto M, Kohno T, Okudela K, Otsuka A, Sasaki H, Tanabe C, Sakiyama T, Hirama C, Kitabayashi I, Minna JD, Takenoshita S, Yokota J. Mutations and deletions of the CBP gene in human lung cancer. Clin Cancer Res. 2005;11:512–519. [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Liu CX, Musco S, Lisitsina NM, Forgacs E, Minna JD, Lisitsyn NA. LRP-DIT, a putative endocytic receptor gene, is frequently inactivated in non-small cell lung cancer cell lines. Cancer Res. 2000;60:1961–1967. [PubMed] [Google Scholar]
- Matsumoto S, Iwakawa R, Kohno T, Suzuki K, Matsuno Y, Yamamoto S, Noguchi M, Shimizu E, Yokota J. Frequent EGFR mutations in noninvasive bronchioloalveolar carcinoma. Int J Cancer. 2006;118:2498–2504. doi: 10.1002/ijc.21670. [DOI] [PubMed] [Google Scholar]
- McDermott KM, Zhang J, Holst CR, Kozakiewicz BK, Singla V, Tlsty TD. p16(INK4a) prevents centrosome dysfunction and genomic instability in primary cells. PLoS Biol. 2006;4:e51. doi: 10.1371/journal.pbio.0040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina PP, Romero OA, Kohno T, Montuenga LM, Pio R, Yokota J, Sanchez-Cespedes M. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat. 2008;29:617–622. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1:49–52. doi: 10.1016/s1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- Nagayama K, Kohno T, Sato M, Arai Y, Minna JD, Yokota J. Homozygous deletion scanning of the lung cancer genome at a 100-kb resolution. Genes Chromosomes Cancer. 2007;46:1000–1010. doi: 10.1002/gcc.20485. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Kobayashi K, Nakamoto M, Kohno T, Sasaki H, Matsuno Y, Yokota J. Identification of tumor markers and differentiation markers for molecular diagnosis of lung adenocarcinoma. Oncogene. 2006;25:4245–4255. doi: 10.1038/sj.onc.1209442. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Matsumoto S, Iwakawa R, Kohno T, Suzuki K, Tsuta K, Matsuno Y, Noguchi M, Shimizu E, Yokota J. Whole genome comparison of allelic imbalance between noninvasive and invasive small-sized lung adenocarcinomas. Cancer Res. 2009a;69:1615–1623. doi: 10.1158/0008-5472.CAN-08-3218. [DOI] [PubMed] [Google Scholar]
- Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- Ogiwara H, Kohno T, Nakanishi H, Nagayama K, Sato M, Yokota J. Unbalanced translocation, a major chromosome alteration causing loss of heterozygosity in human lung cancer. Oncogene. 2008;27:4788–4797. doi: 10.1038/onc.2008.113. [DOI] [PubMed] [Google Scholar]
- Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, Shibata T, Yamamoto M, Hirohashi S. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- Okamoto A, Hussain SP, Hagiwara K, Spillare EA, Rusin MR, Demetrick DJ, Serrano M, Hannon GJ, Shiseki M, Zariwala M, Xiong Y, Beach DH, Yokota J, Harris CC. Mutations in the p16INK4/MTS1/CDKN2, p15INK4B/MTS2, and p18 genes in primary and metastatic lung cancer. Cancer Res. 1995;55:1448–1451. [PubMed] [Google Scholar]
- Otto WR. Lung epithelial stem cells. J Pathol. 2002;197:527–535. doi: 10.1002/path.1160. [DOI] [PubMed] [Google Scholar]
- Parkin M, Je T, Boffetta P, Samet J, Shields PG, Caporaso NE. Lung cancer epidemiology and etiology. In: Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC, editors. World Health Organization Classification of Tumors: Pathology and Genetics, Tumours of Lung, Pleura, Thymus and Heart. Lyon, France: IARC Press; 2004. pp. 31–34. [Google Scholar]
- Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, Dimaio JM, Milchgrub S, Smith AL, Souza RF, Gilbey L, Zhang X, Gandia K, Vaughan MB, Wright WE, Gazdar AF, Shay JW, Minna JD. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Roy BC, Hatano N, Aoyagi T, Gohji K, Kiyama R. A novel ankyrin repeat-containing gene (Kank) located at 9p24 is a growth suppressor of renal cell carcinoma. J Biol Chem. 2002;277:36585–36591. doi: 10.1074/jbc.M204244200. [DOI] [PubMed] [Google Scholar]
- Sato M, Takahashi K, Nagayama K, Arai Y, Ito N, Okada M, Minna JD, Yokota J, Kohno T. Identification of chromosome arm 9p as the most frequent target of homozygous deletions in lung cancer. Genes Chromosomes Cancer. 2005;44:405–414. doi: 10.1002/gcc.20253. [DOI] [PubMed] [Google Scholar]
- Shiseki M, Kohno T, Adachi J, Okazaki T, Otsuka T, Mizoguchi H, Noguchi M, Hirohashi S, Yokota J. Comparative allelotype of early and advanced stage non-small cell lung carcinomas. Genes Chromosomes Cancer. 1996;17:71–77. doi: 10.1002/(SICI)1098-2264(199610)17:2<71::AID-GCC1>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buck-haults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Solomon DA, Kim JS, Cronin JC, Sibenaller Z, Ryken T, Rosenberg SA, Ressom H, Jean W, Bigner D, Yan H, Samuels Y, Waldman T. Mutational inactivation of PTPRD in glioblastoma multiforme and malignant melanoma. Cancer Res. 2008;68:10300–10306. doi: 10.1158/0008-5472.CAN-08-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda I, Imoto I, Inoue J, Shibata T, Shimada Y, Chin K, Imamura M, Amagasa T, Gray JW, Hirohashi S, Inazawa J. Frequent silencing of low density lipoprotein receptor-related protein 1B (LRP1B) expression by genetic and epigenetic mechanisms in esophageal squamous cell carcinoma. Cancer Res. 2004;64:3741–3747. doi: 10.1158/0008-5472.CAN-04-0172. [DOI] [PubMed] [Google Scholar]
- Sozzi G, Veronese ML, Negrini M, Baffa R, Cotticelli MG, Inoue H, Tornielli S, Pilotti S, De Gregorio L, Pastorino U, Pierotti MA, Ohta M, Huebner K, Croce CM. The FHIT gene 3p14.2 is abnormal in lung cancer. Cell. 1996;85:17–26. doi: 10.1016/s0092-8674(00)81078-8. [DOI] [PubMed] [Google Scholar]
- Stallings RL, Nair P, Maris JM, Catchpoole D, McDermott M, O’Meara A, Breatnach F. High-resolution analysis of chromosomal breakpoints and genomic instability identifies PTPRD as a candidate tumor suppressor gene in neuroblastoma. Cancer Res. 2006;66:3673–3680. doi: 10.1158/0008-5472.CAN-05-4154. [DOI] [PubMed] [Google Scholar]
- Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- Stone S, Jiang P, Dayananth P, Tavtigian SV, Katcher H, Parry D, Peters G, Kamb A. Complex structure and regulation of the P16 (MTS1) locus. Cancer Res. 1995;55:2988–2994. [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toloza EM, Morse MA, Lyerly HK. Gene therapy for lung cancer. J Cell Biochem. 2006;99:1–22. doi: 10.1002/jcb.20851. [DOI] [PubMed] [Google Scholar]
- Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ, Martin ES, Yang Z, Ji H, Chin L, Depinho RA. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeriah S, Brennan C, Meng S, Singh B, Fagin JA, Solit DB, Paty PB, Rohle D, Vivanco I, Chmielecki J, Pao W, Ladanyi M, Gerald WL, Liau L, Cloughesy TC, Mischel PS, Sander C, Taylor B, Schultz N, Major J, Heguy A, Fang F, Mellinghoff IK, Chan TA. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc Natl Acad Sci USA. 2009;106:9435–9440. doi: 10.1073/pnas.0900571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virmani AK, Fong KM, Kodagoda D, McIntire D, Hung J, Tonk V, Minna JD, Gazdar AF. Allelotyping demonstrates common and distinct patterns of chromosomal loss in human lung cancer types. Genes Chromosomes Cancer. 1998;21:308–319. doi: 10.1002/(sici)1098-2264(199804)21:4<308::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, Shah K, Sato M, Thomas RK, Barletta JA, Borecki IB, Broderick S, Chang AC, Chiang DY, Chirieac LR, Cho J, Fujii Y, Gazdar AF, Giordano T, Greulich H, Hanna M, Johnson BE, Kris MG, Lash A, Lin L, Lindeman N, Mardis ER, McPherson JD, Minna JD, Morgan MB, Nadel M, Orringer MB, Osborne JR, Ozenberger B, Ramos AH, Robinson J, Roth JA, Rusch V, Sasaki H, Shepherd F, Sougnez C, Spitz MR, Tsao MS, Twomey D, Verhaak RG, Weinstock GM, Wheeler DA, Winckler W, Yoshizawa A, Yu S, Zakowski MF, Zhang Q, Beer DG, Wistuba II, Watson MA, Garraway LA, Ladanyi M, Travis WD, Pao W, Rubin MA, Gabriel SB, Gibbs RA, Varmus HE, Wilson RK, Lander ES, Meyerson M. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- Yokota J, Kohno T. Molecular footprints of human lung cancer progression. Cancer Sci. 2004;95:197–204. doi: 10.1111/j.1349-7006.2004.tb02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Weir BA, LaFramboise T, Lin M, Beroukhim R, Garraway L, Beheshti J, Lee JC, Naoki K, Richards WG, Sugar-baker D, Chen F, Rubin MA, Janne PA, Girard L, Minna J, Christiani D, Li C, Sellers WR, Meyerson M. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E, Lo Y, Baribaud F, Mikami I, Reguart N, Yang G, Li Y, Yao W, Vaddi K, Gazdar AF, Friedman SM, Jablons DM, Newton RC, Frid-man JS, Minna JD, Scherle PA. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.