Abstract
Dominant mutations in two functionally related DNA/RNA-binding proteins, trans-activating response region (TAR) DNA-binding protein with a molecular mass of 43 KDa (TDP-43) and fused in sarcoma/translocation in liposarcoma (FUS/TLS), cause an inherited form of ALS that is accompanied by nuclear and cytoplasmic aggregates containing TDP-43 or FUS/TLS. Using isogenic cell lines expressing wild-type or ALS-linked TDP-43 mutants and fibroblasts from a human patient, pulse-chase radiolabeling of newly synthesized proteins is used to determine, surprisingly, that ALS-linked TDP-43 mutant polypeptides are more stable than wild-type TDP-43. Tandem-affinity purification and quantitative mass spectrometry are used to identify TDP-43 complexes not only with heterogeneous nuclear ribonucleoproteins family proteins, as expected, but also with components of Drosha microprocessor complexes, consistent with roles for TDP-43 in both mRNA processing and microRNA biogenesis. A fraction of TDP-43 is shown to be complexed with FUS/TLS, an interaction substantially enhanced by TDP-43 mutants. Taken together, abnormal stability of mutant TDP-43 and its enhanced binding to normal FUS/TLS imply a convergence of pathogenic pathways from mutant TDP-43 and FUS/TLS in ALS.
Keywords: mass spectrometry, protein stability, amyotrophic lateral sclerosis, microRNA, ribonucleoproteins
Pathological protein aggregation is one of the hallmarks of neurodegenerative diseases (1). In 2006, trans-activating response region (TAR) DNA-binding protein with a molecular mass of 43 KDa (TDP-43) was identified as a major component of ubiquinated inclusions found in frontotemporal lobar degeneration with ubiquitin aggregates (FTLD-U) and ALS patients (2, 3). Since then, intracellular TDP-43–positive inclusions have been found in an array of neurodegenerative diseases, including Alzheimer's disease (AD), Pick's disease, various forms of Parkinson's diseases (PD), and others (4).
Starting in 2008, multiple studies identified over 30 dominant mutations in TDP-43 in both sporadic and familial ALS patients but not in other neurodegenerative diseases including AD or PD, indicating that these mutations are specific to ALS pathogenesis (4–7). This evidence has shaped an emerging TDP-43 proteinopathy hypothesis in which sequestration of nuclear TDP-43 into pathological inclusions is proposed to contribute to disease pathogenesis (8). Additionally, mutations in a second functionally related gene, fused in sarcoma/translocation in liposarcoma (FUS/TLS), were found to be linked with ALS (9, 10).
In a normal context, both TDP-43 and FUS/TLS are involved in RNA transcription and splicing regulation (4). However, how ALS-linked mutations in TDP-43 and FUS/TLS contribute to cellular toxicity is not understood. TDP-43 is thought to be predominantly a nuclear protein found in nuclear bodies (TDP bodies) that are distinct from other known nuclear structures (11). The normal functions of TDP-43 are not established, although it has been proposed that TDP-43 is involved in transcription repression (12, 13) and splicing regulation (14–16). TDP-43 was first identified as a transacting factor binding to the TAR DNA promoter region of HIV to repress the expression of the TAR gene (13). Similarly, repression of the mouse SP-10 gene during spermatogenesis by TDP-43 was also proposed (12). Reduction of TDP-43 in cell culture leads to down-regulation of cyclin-dependent kinase 6 and histone deacetylase 6, further supporting TDP-43’s role in regulating gene expression (17, 18). There is, however, growing evidence indicating that TDP-43 functions in RNA processing. In particular, presence of TDP-43 affects the exon usage of the cystic fibrosis transmembrane regulator (CFTR), apolipoprotein A-II, and survival of motor neuron (SMN) transcripts (14–16). TDP-43 interacts with heterogeneous nuclear ribonucleoproteins (hnRNP) A2/B1 and hnRNP C in vitro, and these interactions may be required for splicing site selection (19). In addition to a nuclear function, TDP-43 also colocalizes with fragile X mental-retardation protein (FMRP) and Staufen proteins in the neurites of primary neurons, which suggests a role in RNA transport and localization (20).
Whether nuclear or cytoplasmic, the RNA targets and protein interactors of TDP-43 have not yet been systematically identified, and it is not known how ALS-linked mutations in TDP-43 affect its normal function(s). In disease conditions, affected neurons typically lose nuclear TDP-43 staining, possibly before the formation of intracellular aggregates (21). In addition, TDP-43 seems to be ubiquitinylated, phosphorylated, and fragmented in pathological conditions (3). Transient expression of TDP-43 fragments in mammalian cell lines and yeast have led to a widely held view that the C-terminal fragment of TDP-43 is extremely toxic (22, 23), although it remains unresolved how C-terminal fragments are generated under physiological or pathological conditions (24, 25).
Two key questions for understanding the TDP-43 proteinopathies are (i) what are the normal functions of TPD-43 and (ii) what are the acquired toxicities (gain of function) and/or perturbed normal functions (loss of function) of TDP-43 in disease conditions. Here, we address these questions by the use of site-directed recombination to produce a series of isogenic cell lines expressing a single copy gene encoding ALS-linked mutations. With these isogenic cell lines as well as fibroblasts from a human patient, we show that the mutant TDP-43 proteins are more stable than the wild-type protein. Additionally, with isotope labeling to produce quantitative mass spectrometry, we define core TDP-43 protein complexes to contain hnRNP family proteins and components of Drosha microprocessor complexes, establishing a direct link of TDP-43 to microRNA biogenesis. Furthermore, a fraction of TDP-43 interacts with FUS/TLS. Most intriguingly, FUS/TLS interacts more prominently with mutant TDP-43, even in the absence of TDP-43 nuclear aggregates. Our results suggest that mutations in TDP-43 perturb normal FUS/TLS function, which may be an early event before any mislocalization and aggregation, and possible convergence of pathogenic pathways in ALS by TDP-43 and FUS/TLS.
Results
ALS-Linked TDP-43 Mutations Exhibit Longer Protein Half-Lives.
A hallmark of TDP-43 protein pathology is intracellular inclusions of aggregated TDP-43. Neither the stability of TDP-43 nor its ALS-linked mutations have been determined. Although several prior reports using transient transfection to express abnormally high levels of TDP-43 (or portions of it) have lead to the proposal that C-terminal TDP-43 fragments are toxic (22, 26), we sought to establish the half-lives of wild-type and ALS-linked mutations in TDP-43 at physiologically relevant endogenous levels. We first determined the normal abundance of TDP-43. Using immunoblotting of known amounts of recombinant full-length human TDP-43 (Fig. S1) in parallel with total protein extracts from a known number of cells, endogenous TDP-43 was identified to comprise 0.04% of total cell protein (8.3 ng per 3 × 104 cells containing 20 μg of total cell protein). Thus, endogenous TDP-43 corresponds to ∼4 × 106 molecules per cell (Fig. 1).
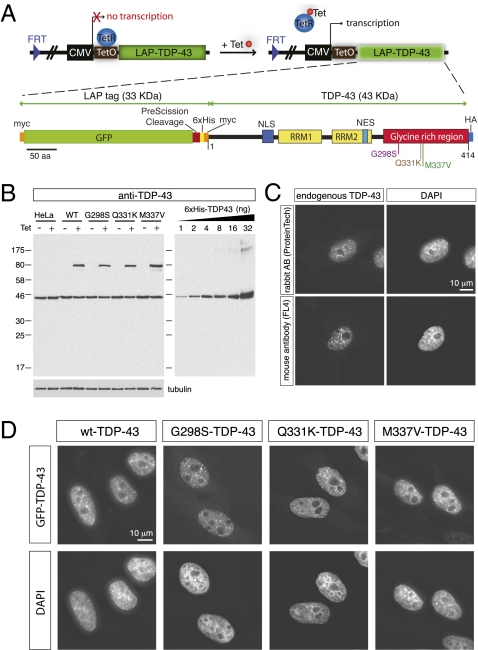
Fig. 1.
Characterization of isogenic cell lines expressing a single copy of wild-type and ALS-linked TDP-43 mutations. (A) Schematic representation of site-directed recombinase-based system to generate isogenic stable cell lines, in which CMV promoter was used to drive the expression of tetracycline (Tet)-inducible wild-type and mutant genes that were integrated at a common locus [Flp Recognition Target (FRT) site]. Normally, Tet repressor (TetR) binds to Tet operator (TetO), repressing transcription. On addition, binding of Tet to TetR induces a conformation change and releases TetR from TetO, allowing transcription to start. Lower shows the LAP tag of TDP-43. The LAP tag is composed of GFP followed by PreScission protease cleavage sequences and 6× histidine tag. TDP-43 is tagged at the N terminus with myc peptide (EQKLISSEEDL) and at the C terminus with HA peptide (YPYDVPDYA). Three different mutations, G298S, Q331K, and M337V, were used in this study. (B) Expression of transgene in isogenic stable cell lines. The transgenes are under TetR control. The transgenes express on incubating with tetracycline (−, without tetracycline; +, with tetracycline); 20 μg total cell extract was loaded for each lane. Recombinant TDP-43 of known amount is loaded for the quantification, and tubulin is used as loading control. Exposure shown was taken on the same blot for quantification. (C) Immunofluorescence images of TDP-43 in HeLa cells. Both rabbit polyclonal antibody (ProteinTech) and mouse monoclonal antibody (FL4) showed similar staining pattern for nuclear TDP bodies. (D) Fluorescent images of isogenic cell lines. Upper is GFP signal, and Lower is DAPI-stained to mark the nucleus. All LAP-tagged TDP-43 form nuclear speckles that are similar to immunofluorescence images of endogenous TDP-43 (C). (Scale bar, 10 μm.)
To achieve similar levels of expression from wild-type or mutant-encoding transgenes, we initially used a site-directed (Flp) recombinase-based system to generate isogenic cell lines (27) in which a cytomegalovirus (CMV) promoter was used to drive the expression of tetracycline-inducible wild-type and mutant genes that were integrated at a common locus (28). Each single copy transgene of TDP-43 was multiply tagged, including a localization-affinity purification (LAP) tag (comprised of GFP and hexa-histidine tags) useful for visualization and purification (29) and additional amino and carboxyl-terminal epitope tags [myc (EQKLISSEEDL) and HA (YPYDVPDYA), respectively] (Fig. 1A). Using this approach, isogenic cell lines expressing wild-type or each of three ALS-linked mutations (G298S, Q331K, and M337V) in TDP-43 were obtained (Fig. 1).
On tetracycline addition, the full-length transgene-encoded polypeptide accumulated to a level similar to endogenous TDP-43 (Fig. 1B). Furthermore, no smaller fragments were detected for wild-type TDP-43 or any of the mutants with either myc or HA antibodies (Fig. S2A), showing that none of these ALS-linked mutations generated fragments that accumulate in these cells. Moreover, except for a slight, apparent elevation in the cytoplasmic pool in some cells, LAP-tagged TDP-43 (Fig. 1D and Fig. S2) localized indistinguishably from the endogenous protein (Fig. 1C), suggesting that the LAP tag did not interfere with TDP-43 function or cause protein misfolding. At steady state, both endogenous (Fig. 1C) and LAP-tagged TDP-43 (Fig. 1D and Fig. S2) appeared as nuclear proteins and formed similar nuclear foci.
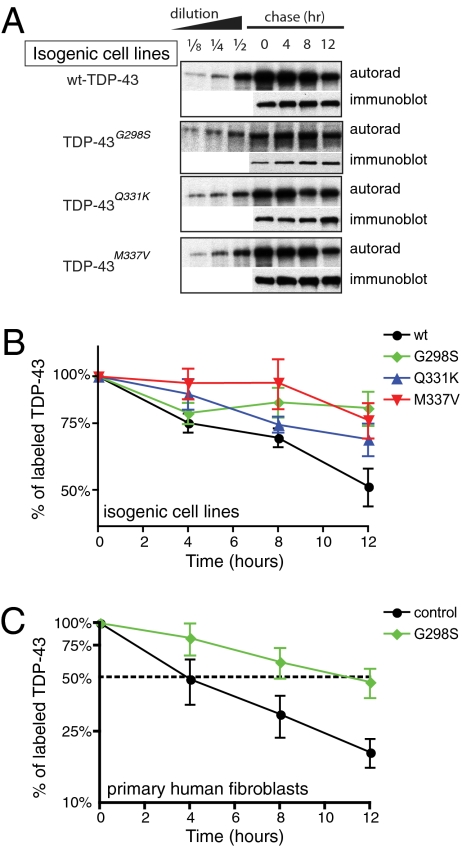
Half-lives of wild-type and TDP-43 mutations were measured in randomly cycling cells through use of short-term incubation with [35S]methionine/cysteine to radiolabel newly synthesized proteins, and the stability of the labeled proteins followed with time (30). This analysis revealed, surprisingly, that, in this in vivo context, the TDP-43 mutations were degraded two (for TDP-43Q331K) to four (for TDP-43G298S and TDP-43M337V) times more slowly than was wild-type TDP-43, yielding estimated half-lives for the mutants of ∼24–48 h versus 12 h for wild-type TDP-43 (Fig. 2B).
Fig. 2.
ALS-linked TDP-43 mutations exhibit longer half-lives. (A) Representative autoradiogram and immunoblots of the pulse-chase assay using isogenic cell lines expressing wild type (wt), G298S, Q331K, and M337V mutations in TDP-43. LAP-tagged wt–TDP-43 and its mutants were immunoprecipitated and run on SDS-PAGE for autoradiography and immunoblotting. Serial dilutions of 0-h point (start of chase) were used to monitor the linearity of the autoradiograph signal. The autoradiograph signals were normalized to the immunoblotting signals and plotted using 0 h as 100%. (B) Half-lives of LAP-tagged TDP-43 and its ALS-linked mutations (n = 5). Error bar represents SEM. (C) Half-lives of TDP-43 in primary human fibroblasts. Wild-type TDP-43 exhibits 4-h half-life, whereas TDP-43 in heterozygote harboring a copy of G298S mutant exhibits 11-h half-life (n = 5). Error bar represents SEM.
To extend this test to a more disease-relevant setting for TDP-43 half-life, we used primary fibroblasts collected from a human patient containing a dominant G298S mutation in TDP-43 in which one copy of TDP-43 carries a G to A substitution, which, in turn, leads to glycine to serine substitution (31). Analysis of pulse radiolabeling of these cells revealed that TDP-43 in wild-type fibroblasts exhibited a 4-h half-life, whereas, in cells heterozygous for one copy of the G298S mutation in TDP-43, the half-life of TDP-43 was 11 h (Fig. 2C), showing a 2.8-fold slower turnover rate for wild-type plus mutant TDP-43 in these cells.
TDP-43 Associates with hnRNP Complexes and microRNA Processing Machinery.
It has been shown that TDP-43 interacts with hnRNP A2/B1 and hnRNP C in vitro using a blot-overlay assay (19). In addition, using yeast two-hybrid screening, TDP-43 was identified as a putative direct interactor with the Xrn2 (5′ → 3′ exonuclease) involved in RNA degradation (32). Additionally, 261 putative TDP-43 interacting proteins have been proposed from an approach using a single-step immunoprecipitation from transiently transfected cells (33), albeit many of these are probably abundant, contaminant proteins rather than actual interactors. Indeed, eukaryotic translation elongation and initiation factors and ribosomal proteins, factors known to be common contaminants for their binding to affinity matrices (34), were among these proposed TDP-43 interactors (33).
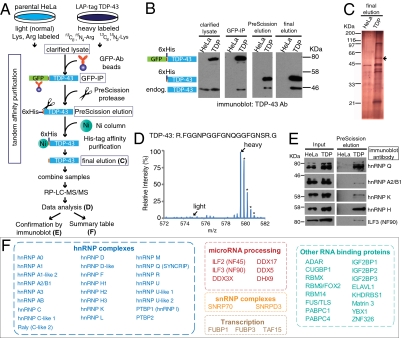
To identify specific interactors, even those in low abundance, while eliminating abundant contaminant proteins, we combined (i) tandem-affinity purification (TAP) with two sequential affinity purification and elution schemes (29, 35) and (ii) quantitative mass-spectrometry analysis using stable isotope labeling by amino acids in cell culture (SILAC) (36). A typical SILAC–TAP experiment is outlined in Fig. 3A. Cell lines stably expressing the LAP double-affinity tag containing wild-type TDP-43 were grown in isotopically heavy medium containing 13C6,15N4-arginine, and 13C6,15N2-lysine, whereas the parental line (i.e., no transgene) was grown in light medium containing normal arginine and lysine. Immunoblotting with an antibody recognizing both endogenous and transgene-encoded TDP-43 revealed that tagged TDP-43 was associated with endogenous TDP-43 throughout the purification (Fig. 3B). From the ratio of 1.6:1 molecules of transgene-encoded and endogenous TDP-43 in the initial extract, the first GFP immunoprecipitation step produced a 4:1 ratio. A comparable ratio carried through the first elution and subsequent second affinity steps (Fig. 3B), showing that (i) tagged TDP-43 forms a co-complex with native TDP-43 and (ii) endogenous TDP-43 is present in complexes with more than one TDP-43 molecule.
Fig. 3.
Quantitative proteomic analysis of TDP-43 using SILAC coupled with TAP. (A) Schematic representation of TAP purification and quantitative analysis using SILAC. (B) Samples from key steps (clarified lysate, immunoprecipitation for GFP (GFP-IP), PreScission elution, and final elution) in purification were probed with TDP-43 antibody, showing that tagged TDP-43 associates and copurifies with endogenous TDP-43. (C) Silver stain of the samples from the final elution of TAP step. Arrow indicates tagged TDP-43. (D) Representative mass spectrum for TDP-43 peptide. The corresponding light isotope-containing peptide is below detection limit, whereas the heavy Lys/Arg-containing peptide is detected. Asterisks indicate the natural occurrences of 13C/15N in the peptide, as revealed by high resolution mass spectrometry. (E) Confirmation of putative TDP-43–associated proteins by immunoblotting, including hnRNP Q, hnRNP A2/B1, hnRNP K, hnRNP H, and interleukin-enhancer binding factor 3/nuclear factor 90 KDa (ILF3/NF90). (F) Summary for putative TDP-43–associated proteins. TDP-associated proteins were identified with peptides containing only heavy Lys/Arg and by multiple different peptides from at least two independent runs. The associated proteins were grouped according to the known assigned functions for the proteins.
Silver staining revealed that the final eluates after tandem-affinity chromatography contained several polypeptides in addition to tagged TDP-43 (Fig. 3C). To identify these TDP-43–related proteins, quantitative mass spectrometry was used. The spectrum for a representative peptide is shown as Fig. 3D (see also Fig. S3). Each peptide yielded a characteristic spectrum of monoisotopic distributions of mass to charge species (m/z) as expected from the natural abundances of isotypes of 13C and 15N. Stringent selection criteria for TDP-43–associating proteins were defined as: (i) enrichment of all peptide signals to at least 8-fold or higher compared with the TAP control (purification from the parental HeLa cell line), (ii) all proteins must be identified with more than one unique peptide detected, and (iii) only proteins identified in at least two independent runs were retained.
All of the proteins whose peptides were found to be TDP-43–associated in the SILAC mass-spectrometric analyses, the TDP-43 interactome, are summarized in Fig. 3F. TDP-43 was identified to be associated with the majority of the known hnRNP proteins (A0, A1, A2/B1, C, D, F, H1, H2, H3, I, K, L, M, Q, R, and U), consistent with TDP-43 as an integral component of hnRNP complexes. Several of the protein partners found by SILAC mass spectrometry were subsequently confirmed by immunoblotting, including the hnRNP components, hnRNP A2/B1, hnRNP Q, hnRNP K, and hnRNP H. In addition, TDP-43 was associated with multiple RNA-binding proteins previously implicated in other human diseases. These included CUG-BP1 (involved in myotonic dystrophy) (37) and perhaps of highest interest, FUS/TLS, another ALS-linked DNA/RNA-binding protein (see below, Preferential ALS-Linked Mutant TDP43 Association with FUS/TLS).
Our analysis also identified a second major complex with which TDP-43 was associated: the Drosha microprocessing complex, whose action is essential for microRNA biogenesis. Drosha components identified within the TDP-43 complexes included interleukin-enhancer binding factor 2/nuclear factor 45 KDa (ILF2/NF45), interleukin-enhancer binding factor 3/nuclear factor 90 KDa (ILF3/NF90), DEAD (Asp-Glu-Ala-Asp) box polypeptide 17, also known as p72 (DDX17), and DEAD (Asp-Glu-Ala-Asp) box polypeptide 5, also known as p68 (DDX5). All of these latter components were previously found by Gregory et al. (38) after affinity purification of epitope-tagged Drosha. ILF3/NF90 was confirmed to be TDP-43–associated by immunoblotting of the PreScission elution of purified TDP-43 (Fig. 3E). Thus, our SILAC mass-spectrometric data show TDP-43 association with complexes that mediate microRNA biogenesis in addition to proposed functions in transcription repression and splicing regulation.
Preferential ALS-Linked Mutant TDP43 Association with FUS/TLS.
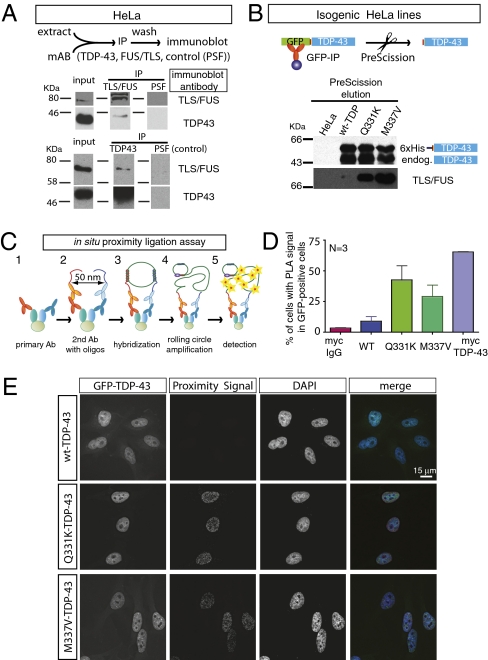
Discovery of FUS/TLS, another ALS-linked gene product, in TDP-43 complexes raised the possibility of a common link underlying disease pathogenesis. We first tested an association of endogenous FUS/TLS and TDP-43 by an additional immunoprecipitation for TDP-43. Approximately 20% of TDP-43 was successfully precipitated along with <1% of endogenous FUS (Fig. 4A). Reciprocal FUS immunoprecipitation followed by immunoblotting for TDP-43 confirmed an interaction, albeit only with a very small proportion of FUS/TLS. Ten percent of endogenous FUS/TLS was successfully precipitated, but, as judged by the input and amount of protein pull down, a much smaller percentage (<1%) of TDP-43 was coprecipitated. To test if the FUS/TLS interaction with TDP-43 was affected by ALS-linked mutation in TDP-43, we used GFP-affinity purification with PreScission elution of extracts from our isogenic cell lines expressing LAP-tagged wild-type or mutant TDP-43. Strikingly, although the distribution of FUS/TLS seemed normal in the presence of mutant TDP-43 (Fig. S4), the association of FUS/TLS with TDP-43 was sharply enhanced for both TDP43Q331K and TDP43M337V ALS-linked mutations (Fig. 4B).
Fig. 4.
ALS-linked TDP-43 mutations associate with FUS/TLS. (A) Reciprocal immunoprecipitation shows that TDP-43 interacts with FUS/TLS using HeLa cells. A monoclonal antibody against splicing factor proline/glutamine-rich or polypyrimidine tract binding protein-associated splicing factor (SFPQ/PSF) was used for control. (B) PreScission elution fractions from GFP pull-down samples showed that FUS/TLS associates more prominently with Q331K and M337V mutations in TDP-43. (C) Schematic representation of in situ proximity ligation assay. (D) Quantification of in situ proximity ligation assay (PLA) results (n = 3; cell numbers > 100 per experiment). Error bar represents SEM. A combination of anti-myc antibody and rabbit IgG molecules was used as negative control, whereas a combination of anti-myc and anti–TDP-43 antibodies was used as positive control. The monitored signal used a combination of anti-myc and anti-TLS/FUS antibody. (E) Representative results for in situ proximity ligation assay. (Scale bar, 15 μm.)
To further confirm the association between TDP-43 and FUS/TLS and to eliminate the possibility that a TDP-43 and TLS/FUS interaction was an artifact arising during cell lysis (39), we used an in vivo, in situ proximity ligation assay (Fig. 4C). In this assay, cell membranes are gently lysed, primary antibodies against wild-type or mutant TDP-43 (in this case, we used an myc antibody recognizing LAP-tagged TDP-43) and FUS/TLS were added, much as is done in conventional immunofluorescence. Instead of fluorescence-conjugated secondary antibodies, however, two different oligonucleotides were linked to the secondary antibodies (one for each of the two different primary antibodies). If the distance between the two different oligonucleotides is less than 50 nm, they can be hybridized and used as primers for rolling-circle amplification. Subsequently, fluorescent-labeled oligonucleotides were then hybridized with the amplification products, and the signals were observed with a fluorescence microscope. A combination of anti-myc antibody and rabbit IgG molecules was used as negative control, whereas a combination of anti-myc and anti–TDP-43 antibodies was used as positive control. Although FUS/TLS was seen to be associated with wild-type TDP-43 in only a small proportion (10%) of cells (consistent with the immunoprecipitation evidence above) (Fig. 4D), intranuclear proximity signals to FUS/TLS were observed in ∼40% cells expressing either of two ALS-linked TDP-43 (TDP-43Q331K and TDP-43M337V).
Discussion
A key question in understanding how ALS-linked dominant mutations in TDP-43 cause cellular toxicity is if, and if so how, these point mutations alter the normal function of TDP-43. In contrast to Cu/Zn superoxide dismutase 1 (SOD1) where ALS-linked mutations destabilize the mutant protein (30), using isogenic stable cell lines expressing a single copy of each transgene (wild type, TDP43G298S, TDP43Q331K, and TDP43M337V), we showed that all three of these ALS-linked mutations exhibit longer protein half-lives compared with wild-type protein, suggesting that abnormal stability may be a common feature for ALS-linked TDP-43 mutations. Furthermore, we showed that one of the ALS-linked TDP-43 mutants (G298S) generates higher TDP-43 stability in primary fibroblasts from a human patient (Fig. 2C), where only one copy of the TARDBP gene is mutated and under the authentic promoter. This highly unexpected discovery suggests that an inherently increased half-life may be, or at least may contribute to, the underlying mechanism for the accumulation of TDP-43 aggregations found in ALS patients.
Perhaps even more importantly, we have shown that a significantly higher proportion of endogenous, wild-type FUS/TLS is associated with both of two ALS-linked mutations tested (TDP43Q331K and TDP43M337V). This interaction is exclusively intranuclear but without apparent nuclear aggregation. Taken together, our findings imply that the increased association between mutant TDP-43 and FUS/TLS may be driven, in part, by the increasing stability of mutant TDP-43 (Fig. S5). Conceivably, this aberrant association caused by the dominant mutations in TDP-43 could lead to potential perturbations of the normal functions of both TDP-43 and FUS/TLS, suggesting a possible convergence of pathogenic pathways in ALS by TDP-43 and FUS/TLS.
Interestingly, familial PD-linked A53T substitution of α-synuclein also shows increased stability, which, in turn, probably contributes to the age-dependent accumulation of mutant α-synuclein in transgenic mice expressing mutant α-synuclein (40). Prolonged stability could give rise to at least two additional nonmutually exclusive effects on TDP-43: (i) permitting additional or aberrant posttranslational modifications, such as phosphorylation and ubiquitinylation, which were reported in disease conditions (3, 41) and/or (ii) permitting aberrant interactions with other proteins, such as with FUS/TLS reported in this study (Fig. 4). It will now be important to determine whether ALS-linked TDP-43 mutations show age-dependent accumulation and mutant-specific interactions in genetically engineered animals, such as transgenic mice (42). Additionally, because accumulation is the net balance between synthesis and degradation, it will be essential to determine how TDP-43 is degraded and whether the ALS-linked mutations cause any defect in the degradation process. Although recent evidence indicates that TDP-43 may be degraded through either autophagosome and/or proteasome pathways (43, 44), it is not clear whether one of the pathways is the major default path. Understanding the degradation process may aid the design of future therapeutic interventions, because intracellular TDP-43 inclusions are hallmarks of various neurodegeneration diseases.
The major advantage of using quantitative mass-spectrometry analysis is that the common and abundant contaminant proteins can be easily eliminated and low abundant but specific interactors can be readily identified (45). Using the SILAC–TAP approach, we identified the stable core component for TDP-43 complexes. The majority of the TDP-43–associating proteins are hnRNP family proteins, indicating that TDP-43 is an integral part of hnRNP complexes, which associate with nascent transcripts and influence their fate (46). The biochemical findings are consistent with TDP-43’s role in RNA transcription and processing and complement a recent ultrastructural study showing that TDP-43 is enriched in perichromatin fibrils, nuclear sites of transcription, and cotranscriptional splicing (47). Beside hnRNPs, several additional RNA-binding proteins were identified within TDP-43 complexes, including RBM9 (or FOX2) and CUG-BP1 (Fig. 3F). Mice overexpressing CUG-BP1 in muscles reproduce the pathological features found in myotonic dystrophy patients accompanied by disrupted normal splicing patterns (37). Similarly, RBM9/FOX2 influences splicing-site usages by positioning near the exon–intron junctions in embryonic stem cells (48). These findings are consistent with a proposed role of TDP-43 in splicing regulation.
Several TDP-43 interacting proteins were found to be components of Drosha microRNA processing complexes, including ILF2/NF45, ILF3/NF90, DDX5, DDX17, and DDX3X (38). The overlapping components between epitope-tagged Drosha (38) and TDP-43 (in this study) strongly suggest that TDP-43 is involved in microRNA biogenesis. Indeed, NF45/NF90 has been shown to complex with pre-miRNAs, and this association reduces the generation of mature miRNAs (49). In addition, adenosine deaminases (ADARs), found to bind to TDP-43 in our proteomic study, are known to influence microRNA processing through their editing activities (50). Defects in glutamine (Q) to arginine (R) substitution of glutamate AMPA receptors by ADAR-mediated RNA editing have been linked to sporadic ALS (51), suggesting yet another potential pathogenic mechanism for TDP-43. Indeed, a recent study showed reduced ADAR immunoactivity in ALS is accompanied by presence of phosphorylated (pathological) TDP-43 in ALS patients (52). Taken together, TDP-43 may be involved in both RNA transcription and processing through its interaction with hnRNP complexes and microRNA biogenesis by its association with microprocessing complexes (Fig. S5). Because gene expression is coordinated and coupled through interconnected multicomponent machineries (53), it is tempting to speculate that TDP-43 may coordinate and regulate mRNA processing with microRNA biogenesis pathway and through this linkage, regulate expression of various transcripts (54, 55).
Lastly, FUS/TLS, another ALS-linked protein (9, 10), was found associated with TDP-43. Although less than 1% of wild-type FUS/TLS interacts with wild-type TDP-43, the association is strongly enhanced by ALS-linked mutations (Fig. 4). It is tempting to speculate that dominant mutations in TDP-43 may perturb normal FUS/TLS function, thus providing possible convergence of pathogenic pathways in ALS by mutant TDP-43 and FUS/TLS. Now needed are efforts to determine whether the increasing association between mutant TDP-43 and FUS/TLS shown here affects the RNA targets for TDP-43, FUS/TLS, or both.
Materials and Methods
In Situ Proximity Ligation Assay.
In situ proximity ligation assays were done following manufacture’s protocol (Olink Bioscience, Sweden). Anti-myc (clone 4A6, BD Bioscience), which detected LAP-tag TDP-43, was paired with anti-TDP43 (ProteinTech), anti-TLS/FUS (Aviva) and purified rabbit IgG (Sigma-Aldrich) for the primary antibodies. Anti-myc and anti-TDP43 pair serves as a positive control, and anti-myc and purified rabbit IgG as a negative control. Detailed methods for plasmids and recombinant protein purification, cell culture and creations of isogenic stable cell lines, pulse-chase assay, tandem-affinity purification and quantitative mass-spectrometry analysis using SILAC, immunoprecipitation, immunofluorescence, immunoblotting, and antibodies are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Stephen Taylor (University of Manchester, Manchester, UK) for Flp-In reagents, Dr. Christopher Shaw (King’s College London, London, UK) for TDP-43 cDNA constructs, Dr. Thomas Bird (University of Washington, Seattle, WA) for primary human fibroblasts, Dr. Vladimir Gelfand (Northwestern University, Chicago, IL) for GFP binder, and Dr. Boquan Jin (Fourth Military Medical University, Xian, China) for the production of monoclonal antibodies. We are grateful for the help of Sherry Nissen, James Thompson, and John Yates for the initial protein identification, Ngoc Bui and Kristen Watanabe for their technical assistance, and Cleveland laboratory members for helpful suggestions and stimulating discussion. S.-C. L. is a recipient of a National Institutes of Health Neuroplasticity of Aging training grant (T32 AG 000216). C. L.-T. is a recipient of an Amytrophic Lateral Sclerosis Association postdoctoral fellowship. This work was supported by grants to D.W.C. from the National Institutes of Health (R37NS27036, RC1NS069144, and P30 NS0471401-08). D.W.C. receives salary support from the Ludwig Institute for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008227107/-/DCSupplemental.
References
- 1.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 2.Arai T, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 3.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 4.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: Emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kabashi E, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 6.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pesiridis GS, Lee VM, Trojanowski JQ. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:R156–R162. doi: 10.1093/hmg/ddp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forman MS, Trojanowski JQ, Lee VM. TDP-43: A novel neurodegenerative proteinopathy. Curr Opin Neurobiol. 2007;17:548–555. doi: 10.1016/j.conb.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 10.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang IF, Reddy NM, Shen CK. Higher order arrangement of the eukaryotic nuclear bodies. Proc Natl Acad Sci USA. 2002;99:13583–13588. doi: 10.1073/pnas.212483099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Acharya KK, Govind CK, Shore AN, Stoler MH, Reddi PP. cis-requirement for the maintenance of round spermatid-specific transcription. Dev Biol. 2006;295:781–790. doi: 10.1016/j.ydbio.2006.04.443. [DOI] [PubMed] [Google Scholar]
- 13.Ou SH, Wu F, Harrich D, García-Martínez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bose JK, Wang IF, Hung L, Tarn WY, Shen CK. TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J Biol Chem. 2008;283:28852–28859. doi: 10.1074/jbc.M805376200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buratti E, et al. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercado PA, Ayala YM, Romano M, Buratti E, Baralle FE. Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 2005;33:6000–6010. doi: 10.1093/nar/gki897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayala YM, Misteli T, Baralle FE. TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc Natl Acad Sci USA. 2008;105:3785–3789. doi: 10.1073/pnas.0800546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiesel FC, et al. Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 2009;29:209–221. doi: 10.1038/emboj.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buratti E, et al. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: An important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 20.Wang IF, Wu LS, Chang HY, Shen CK. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem. 2008;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- 21.Giordana MT, et al. TDP-43 redistribution is an early event in sporadic amyotrophic lateral sclerosis. Brain Pathol. 2010;20:351–360. doi: 10.1111/j.1750-3639.2009.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci USA. 2008;105:6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nonaka T, et al. Phosphorylated and ubiquitinated TDP-43 pathological inclusions in ALS and FTLD-U are recapitulated in SH-SY5Y cells. FEBS Lett. 2009;583:394–400. doi: 10.1016/j.febslet.2008.12.031. [DOI] [PubMed] [Google Scholar]
- 24.Igaz LM, et al. Enrichment of C-terminal fragments in TAR DNA-binding protein-43 cytoplasmic inclusions in brain but not in spinal cord of frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Am J Pathol. 2008;173:182–194. doi: 10.2353/ajpath.2008.080003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang YJ, et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci USA. 2009;106:7607–7612. doi: 10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nonaka T, Kametani F, Arai T, Akiyama H, Hasegawa M. Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum Mol Genet. 2009;18:3353–3364. doi: 10.1093/hmg/ddp275. [DOI] [PubMed] [Google Scholar]
- 27.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson VL, Scott MI, Holt SV, Hussein D, Taylor SS. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci. 2004;117:1577–1589. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- 29.Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE. 2005;2005:p11. doi: 10.1126/stke.2662005pl1. [DOI] [PubMed] [Google Scholar]
- 30.Borchelt DR, et al. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc Natl Acad Sci USA. 1994;91:8292–8296. doi: 10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Deerlin VM, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: A genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehner B, Sanderson CM. A protein interaction framework for human mRNA degradation. Genome Res. 2004;14:1315–1323. doi: 10.1101/gr.2122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freibaum BD, Chitta RK, High AA, Taylor JP. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trinkle-Mulcahy L, et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J Cell Biol. 2008;183:223–239. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 36.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 37.Ho TH, Bundman D, Armstrong DL, Cooper TA. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 38.Gregory RI, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 39.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W, et al. Stabilization of alpha-synuclein protein with aging and familial Parkinson's disease-linked A53T mutation. J Neurosci. 2004;24:7400–7409. doi: 10.1523/JNEUROSCI.1370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hasegawa M, et al. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann Neurol. 2008;64:60–70. doi: 10.1002/ana.21425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA. 2009;106:18809–18814. doi: 10.1073/pnas.0908767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filimonenko M, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim SH, et al. Potentiation of amyotrophic lateral sclerosis (ALS)-associated TDP-43 aggregation by the proteasome-targeting factor, ubiquilin 1. J Biol Chem. 2009;284:8083–8092. doi: 10.1074/jbc.M808064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 46.Dreyfuss G, Matunis MJ, Piñol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 47.Casafont I, Bengoechea R, Tapia O, Berciano MT, Lafarga M. TDP-43 localizes in mRNA transcription and processing sites in mammalian neurons. J Struct Biol. 2009;167:235–241. doi: 10.1016/j.jsb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakamoto S, et al. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol. 2009;29:3754–3769. doi: 10.1128/MCB.01836-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang W, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawahara Y, et al. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 52.Aizawa H, et al. TDP-43 pathology in sporadic ALS occurs in motor neurons lacking the RNA editing enzyme ADAR2. Acta Neuropathol. 2010;120:75–84. doi: 10.1007/s00401-010-0678-x. [DOI] [PubMed] [Google Scholar]
- 53.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 54.Pawlicki JM, Steitz JA. Nuclear networking fashions pre-messenger RNA and primary microRNA transcripts for function. Trends Cell Biol. 2010;20:52–61. doi: 10.1016/j.tcb.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shomron N, Levy C. MicroRNA-biogenesis and pre-mRNA splicing crosstalk. J Biomed Biotechnol. 2009;2009:594678. doi: 10.1155/2009/594678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.