Abstract
The envelope spike of HIV is one of the most highly N-glycosylated structures found in nature. However, despite extensive research revealing essential functional roles in infection and immune evasion, the chemical structures of the glycans on the native viral envelope glycoprotein gp120—as opposed to recombinantly generated gp120—have not been described. Here, we report on the identity of the N-linked glycans from primary isolates of HIV-1 (clades A, B, and C) and from the simian immunodeficiency virus. MS analysis reveals a remarkably simple and highly conserved virus-specific glycan profile almost entirely devoid of medial Golgi-mediated processing. In stark contrast to recombinant gp120, which shows extensive exposure to cellular glycosylation enzymes (>70% complex type glycans), the native envelope shows barely detectable processing beyond the biosynthetic intermediate Man5GlcNAc2 (<2% complex type glycans). This oligomannose (Man5–9GlcNAc2) profile is conserved across primary isolates and geographically divergent clades but is not reflected in the current generation of gp120 antigens used for vaccine trials. In the context of vaccine design, we also note that Manα1→2Man-terminating glycans (Man6–9GlcNAc2) of the type recognized by the broadly neutralizing anti-HIV antibody 2G12 are 3-fold more abundant on the native envelope than on the recombinant monomer and are also found on isolates not neutralized by 2G12. The Manα1→2Man residues of gp120 therefore provide a vaccine target that is physically larger and antigenically more conserved than the 2G12 epitope itself. This study revises and extends our understanding of the glycan shield of HIV with implications for AIDS vaccine design.
Keywords: HIV, 2G12, gp120, glycosylation, vaccine
The surface antigen of HIV, gp120, is covered by an extensive array of N-linked glycans (1, 2). These host-derived carbohydrate structures comprise half the mass of gp120 and shield much of the underlying protein surface (3, 4). Although progress has been made in the structural elucidation of the protein component of gp120 (5, 6), the identity of the glycans that form the immunological shield on the native virus has not been established. A gp120 antigen is likely to be a component of any successful AIDS vaccine (2, 7–9), and this glycoprotein has formed the basis of extensive phase III clinical trials (10). However, half of this recombinant antigen is of unknown similarity to the native virus. To improve our picture of the antigenic structure of HIV, we therefore sought to determine the composition of the glycan shield of gp120 as expressed on virions.
The position of N-linked glycans is directly encoded within the viral genome (within the consensus sequence Asn-X-Ser/Thr-X, X ≠ Pro), whereas the type(s) of glycans found at a given Asn is determined by the biosynthetic processes of the endoplasmic reticulum (ER) and Golgi apparatus of the infected cell (11, 12) (SI Appendix, Fig. S1). The transition from oligomannose type (Man5–9GlcNAc2) to hybrid or complex type glycans arises from the action of UDP-N-acetyl-d-glucosamine:α-3-d-mannoside β1→2-N-acetylglucosaminyltransferase I (GnT I). This medial Golgi-resident enzyme catalyzes the addition of a β1→2-linked GlcNAc residue to form GlcNAcβ1→2Man5GlcNAc2, the obligate biosynthetic intermediate for mature complex glycosylation (11–13).
In contrast to this classic model of cell-directed glycosylation, a small number of glycoproteins exhibit the phenomenon of protein-directed glycosylation. In these cases, glycan processing may be enhanced by the recruitment of glycosyltransferases (14) or may be limited by the 3D structure (15) or subcellular trafficking (16) of the protein, thus preventing exposure of glycans to the full range of glycosylation enzymes. One such glycoprotein is gp120, which contains a number of unprocessed oligomannose glycans on its heavily glycosylated outer domain (1). This cluster of ectopically expressed ER glycans provides a window for immunological recognition and forms the epitope for one of the few known broadly neutralizing antibodies against HIV, IgG 2G12 (17–20). We have hypothesized that divergence from host glycan processing arises because of the limited accessibility of the ER and Golgi α1→2 mannosidases to the unusually dense arrangement of Man6–9GlcNAc2 carbohydrates on gp120 (1, 4). Here, we confirm that a small population of Man9GlcNAc2 glycans on gp120 is entirely resistant to recombinant ER α-mannosidase I.
The glycans around the variable loops and the receptor binding site are fully processed into cell type-specific complex glycans on recombinant gp120 (1, 21, 22). In this study, we report the chemical structures of the glycans of the envelope spike from infectious viral particles. We show that the N-linked glycans of native HIVJRCSF envelope, derived from both peripheral blood mononuclear cells (PBMCs) and cultured human cells, are composed of oligomannose glycans. We find that the abundance of Man6–9GlcNAc2 glycans is dramatically increased compared with recombinant, monomeric gp120. Moreover, >98% of the glycans show no evidence of further Golgi-resident glycan processing at or beyond the GnT I stage and are exclusively Man5-9GlcNAc2. The carbohydrate profile of HIV is therefore entirely divergent to normal glycosylation of the cell from which it derives. This uniquely conserved HIV-specific glycosylation pattern exists across all isolates of HIV-1 tested here (clades A, B, and C) as well as on the gp120 of simian immunodeficiency virus (SIV).
Results
Native HIV Glycans Are Oligomannose.
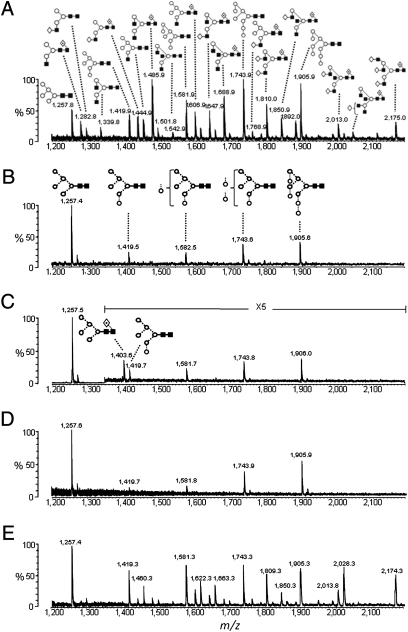
Common to all analyses of recombinant gp120 is the observation of both oligomannose and complex type glycans, which are proposed to exist in discrete domains on the protein surface (1, 21, 22). Here, we expressed recombinant monomeric HIV-1 gp120JRCSF, a primary clade B isolate, from human embryonic kidney (HEK) 293T cells and analyzed the isolated N-linked glycan component. MALDI-TOF MS analysis showed extensive chemical diversity, with glycan structures characteristic for this cell line (23) (Fig. 1A and SI Appendix, Table S1). The spectrum is composed of both oligomannose type glycans (24% of total N-linked carbohydrates) and a large array of complex type glycans (76%), and is therefore consistent with previous analyses of gp120 glycosylation (1, 21, 22).
Fig. 1.
MALDI-TOF MS analyses of the N-linked glycans released from gp120JRCSF. Spectra of released N-linked glycans ([M + Na]+ ions) from desialylated recombinant gp120 expressed in HEK 293T cells (A), native HIV envelope derived from pseudovirions from HEK 293T cells (B), recombinant gp120 expressed in GnT I-deficient HEK 293S cells (C), native HIV envelope derived from pseudovirions from GnT I-deficient HEK 293S cells (D), and desialylated envelope isolated from HEK 293T culture supernatant after removal of virions (E) are illustrated. Representative glycan structures are shown for masses that may contain multiple isobaric structures. The full assignment of A and E is presented in SI Appendix, Table S1. Symbols used for the structural formulae in this and subsequent figures are as follows: ◇, Gal; ■, GlcNAc; ○, Man;  , Fuc; ⋆, NeuNAc (50). The linkage position is shown by the angle of the lines linking the sugar residues (vertical line, two-link; forward slash, three-link; horizontal line, four-link; back slash, six-link). Anomericity is indicated by full lines for β-bonds and by broken lines for α-bonds (50).
, Fuc; ⋆, NeuNAc (50). The linkage position is shown by the angle of the lines linking the sugar residues (vertical line, two-link; forward slash, three-link; horizontal line, four-link; back slash, six-link). Anomericity is indicated by full lines for β-bonds and by broken lines for α-bonds (50).
We subsequently purified the same glycoprotein from functional JRSCF virus produced in the same HEK 293T cell line. In this case, MALDI MS analysis of isolated viral envelope glycans revealed an exclusively oligomannose population with peaks at m/z 1,257.4, 1,419.5, 1,582.5, 1,743.6, and 1,905.6 ([M + Na]+ ions) corresponding to the mannose series Man5–9GlcNAc2 (Fig. 1B). This dramatic diminution of the amount of heterogeneous complex type glycans is matched by changes in the amounts of the oligomannose glycans. Most notably, the Man5GlcNAc2 glycan, which is only found as a minor (5%) species on the monomeric envelope, is observed as the single most abundant (37%) glycan on the envelope trimer. To determine the residual amount of complex type glycans present on viral envelope, HIVJRFL virions were produced in HEK 293T cells in the presence of swainsonine (SI Appendix, Fig. S2), a Golgi α-mannosidase II inhibitor. As a result, the heterogeneity of processed glycans is reduced, allowing a GlcNAcβ1→2Man5GlcNAc2 intermediate to be observed as a trace population (SI Appendix, Fig. S2; peak at m/z 1,460.6, [M + Na]+, <2% of total N-glycans).
When interpreted in the context of the biosynthetic pathway (SI Appendix, Fig. S1), the virus, unlike recombinant monomeric gp120, exhibits a lack of processing by GnT I or any of the subsequent Golgi-resident glycan processing enzymes. To demonstrate that evasion of GnT I activity is sufficient to explain the divergence in glycosylation patterns, we used a GnT I-deficient HEK 293S cell line (13) to express recombinant gp120JRCSF (Fig. 1C). The glycan types obtained from this cell line reproduce those observed from native virus glycoproteins from GnT I-competent cells (Fig. 1B); in both cases, glycan processing appears stalled at the Man5GlcNAc2 biosynthetic intermediate. Consideration of the biosynthetic pathway indicates that the abundance of Man5GlcNAc2 can be interpreted as representing both the naturally occurring Man5GlcNAc2 population for gp120 (5%) and a population of glycans that would usually be complex glycans (76%), which are trapped in this GnT I-deficient cell line at the Man5GlcNAc2 biosynthetic intermediate. The predicted level of Man5GlcNAc2 abundance based on this addition (81%) correlates well with that observed here (83%) by MS analysis (Fig. 1C).
For comparison, we analyzed the N-linked glycans from the envelope of HIVJRCSF virus produced from this GnT I-deficient cell line, which, as for the virus expressed in the GnT I-competent HEK 293T cell line, revealed a major Man5GlcNAc2 peak and an additional range of Man6–9GlcNAc2 glycans (Fig. 1D). Thus, the major differences between glycosylation profiles observed for recombinant gp120 from GnT I-competent and -deficient cells (Fig. 1 A and C, respectively) were not found in the glycosylation profiles of native viral envelope obtained from these cell lines (Fig. 1 B and D). We conclude that the evasion of GnT I processing is sufficient to result in the purely oligomannose range of glycans observed for the native virus. However, GnT I activity is fully functional in virus-producing cells, as evidenced by the presence of complex type glycosylation on soluble nonvirion-associated envelope glycoprotein isolated from culture supernatant (Fig. 1E).
Oligomerization Prevents Golgi-Resident Fucosylation.
A crucial feature of recombinant gp120 derived from the GnT I-deficient cells is the presence of the fucosylated structure, Man5GlcNAc2Fuc1, at m/z 1,403.6 (Fig. 1C). This glycan is not observed in the corresponding analysis of the native viral glycans from the same cell line (Fig. 1D) and represents the only qualitative difference evident between the two spectra. Fucosylation of the reducing-terminal GlcNAc in the α1→6 position is performed by the Golgi-resident α1→6-fucosyltransferase. We have previously shown that such fucosylation can occur in human cells in a GnT I-independent manner to produce Man5GlcNAc2Fuc1 (24) and that this structure is evident on gp120BaL expressed in HEK 293S GnT I-deficient cells (19). This core fucosylation is also observed here for recombinant gp120JRCSF (Fig. 1C). In contrast, we note that recombinant trimers, made from artificial disulfide-stabilized envelope expressed in HEK 293S GnT I-deficient cells, exhibit no detectable core fucosylation (25). Therefore, as for GnT I activity itself, the absence of GnT I-independent fucosylation on the native envelope (Fig. 1 B and D) shows that the HIV envelope avoids medial Golgi-resident glycan processing and that this evasion appears to be driven, at least in part, by the steric consequences of oligomerization.
Cross-Clade Conservation of HIV Glycosylation.
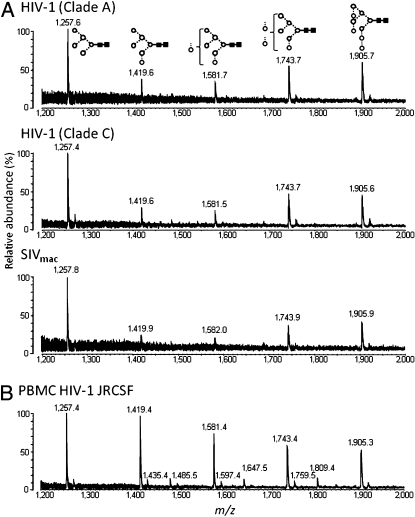
We next determined whether the oligomannose type glycosylation obtained for HIV-1JRCSF was a general feature displayed by different viral isolates. Analysis of viral envelope N-linked glycans from clade A (gp12092RW020) and clade C (gp120DU422) by MALDI-TOF MS (Fig. 2A) showed almost indistinguishable profiles of structures compared with those seen for the clade B isolate (gp120JRCSF; Fig. 1B). This demonstrates that in marked contrast to the underlying protein epitopes, there is a high degree of conservation of the surface-exposed glycan structures. Furthermore, glycans from SIVmac239 envelope were similarly lacking in complex type glycans (Fig. 2A). In all cases, Man5GlcNAc2 was by far the most abundant glycan structure detected, indicating that evasion of Golgi-resident GnT I is a conserved feature for all these immunodeficiency viruses.
Fig. 2.
MALDI-TOF MS analysis of released N-linked glycans from native HIV envelope. 92RW020 (clade A), DU422 (clade C), and SIVmac239 isolates derived from pseudovirions from HEK 293T cells (A) and JRCSF isolate derived from pseudovirions from PBMCs (B) are shown.
Native Glycosylation of HIV from PBMCs.
HEK 293T cells are routinely used to produce pseudoviral particles in research laboratories; however, these cells are not naturally infected by HIV-1. Virions thus derived do not therefore necessarily reflect the glycosylation of the native envelope. To address this concern, we infected PBMCs with HIV-1JRCSF and determined the composition of the N-linked glycosylation of the envelope. MALDI MS analysis of isolated glycans from infectious PBMC-derived virus again showed the same profile of oligomannose structures observed for virus from HEK 293T cells: Man5–9GlcNAc2 (Fig. 2B). As for the HEK 293T-derived virus, the absence of complex glycans correlates with an increase in the amount of oligomannose glycans, with Man5GlcNAc2 again observed as the single most abundant glycan structure. A trace population (1.8%) of fucosylated biantennary complex type glycans in the spectrum of the envelope glycans from PBMCs derives from the capture antibodies used to isolate viral particles [their identity as IgG Fc glycans is indicated by the highly distinctive fucosylated and partially galactosylated biantennary structures (26) and independently confirmed by MALDI MS analysis of the N-linked glycans of the capture antibodies themselves; SI Appendix, Figs. S3 and S4].
Mannosidase Processing Is Limited on the Native Trimer.
The viral envelope spike shows a resistance to glycan processing by GnT I or by subsequent Golgi-resident enzymes (Fig. 1B). However, there are also differences between virus envelope and recombinant monomer glycosylation that indicate differential processing by enzymes upstream of GnT I. Specifically, there is an increase of Manα1→2Man terminating glycans on the viral envelope when compared with those from sequence-matched monomeric recombinant gp120 from the same cell line. The amount of Man6–9GlcNAc2 on the functional virus envelope (63%; Fig. 1B) is over 3-fold greater than the amount seen on the recombinant mononer (19%; Fig. 1A). Moreover, the same trimeric HIVJRCSF envelope derived from PBMCs shows an even more extended array of these Manα1→2Man terminating glycans (Fig. 2B). The differential between the monomeric and trimeric Man6–9GlcNAc2 abundances is perhaps most clearly illustrated in the simplified spectrum provided by the GnT I-deficient system (Fig. 1 C and D), which shows a similar 3-fold increase relative to that observed for the GnT I-competent cell line. Thus, α-mannosidase processing during the early steps of viral glycoprotein assembly and secretion is limited to a greater extent on the native virus than on monomeric gp120. This would be compatible with a model in which envelope oligomerization occurs in compartments with active α1→2 mannosidases (SI Appendix, Fig. S1).
Mannosidase Kinetics Explain Divergence of HIV and Host Glycosylation.
There are clear differences in the glycosylation of recombinant monomeric gp120 and virus-associated envelope (Fig. 1). However, the initial divergence of viral from typical host glycosylation is visible even with the monomeric envelope subunit (Fig. 1A). The persistence of the 2G12 epitope and a core population of Man6–9GlcNAc2 glycans in both recombinant monomeric gp120 and native trimeric envelope suggests that this patch is intrinsic to the monomer and is formed by a process independent of that which prevents viral glycan processing beyond Man5GlcNAc2.
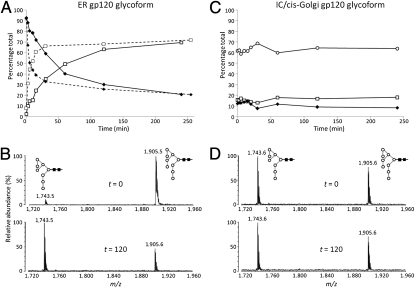
The first enzyme to process Man9GlcNAc2 is the ER-resident α-mannosidase I, which removes a single d-mannose residue from the D2 arm of Man9GlcNAc2 to generate D1,D3-Man8GlcNAc2 (SI Appendix, Fig. S1). The persistence of Man9GlcNAc2 on mature glycoproteins is therefore a marker for reduced ER glycan processing. We generated a glycoform of gp120 bearing exclusively Man9GlcNAc2, which replicates the immature early ER form of this glycoprotein (19, 27). A kinetic analysis of the hydrolysis of Man9GlcNAc2 to D1,D3-Man8GlcNAc2 on this gp120 by recombinant ER α-mannosidase I revealed dramatically differential rates of processing: 50% of the Man9GlcNAc2 was rapidly trimmed to D1,D3-Man8GlcNAc2 (Fig. 3 A and B), whereas the remaining Man9GlcNAc2 was processed at a much (≈100-fold) slower rate, with a significant population of Man9GlcNAc2 (30%) left unprocessed even after exhaustive digestion. This residual Man9GlcNAc2 population is also evident in gp120 from the GnT I-deficient cell line, which effectively represents the intermediate compartment (IC)/cis-Golgi glycoform of gp120 (SI Appendix, Fig. S1). The Man9GlcNAc2 glycans remaining on this glycoprotein are entirely resistant to ER α-mannosidase I (Fig. 3 C and D). The observed kinetics provide direct support for a model in which the rate(s) of α-mannosidase hydrolysis are responsible for the bifurcation in glycan processing, that this oligomannose patch is intrinsic to the monomer, and that this patch is generated independent of the secondary population Man6–9GlcNAc2 glycans found on trimeric envelope.
Fig. 3.
Kinetics of ER α-mannosidase I processing of gp120. Two glycoforms were used: an ER gp120 glycoform bearing predominantly the Man9GlcNAc2 glycan (expressed in the presence of kifunensine) and an IC/cis-Golgi glycoform exhibiting predominantly Man5GlcNAc2 glycans, with a residual Man6–9GlcNAc2 population (expressed in GnT I-deficient cells). The rates of mannose hydrolysis were determined by MALDI-TOF MS analysis of released N-linked glycans ([M + Na]+ ions): Abundances of Man9GlcNAc2 (◆), Man8GlcNAc2 (□), and Man5GlcNAc2 (○) were recorded. (A) Hydrolysis of the ER gp120 glycoform was monitored in the presence of 1 μg (solid line) or 5 μg (dashed line) of ER α-mannosidase I. (B) MALDI-TOF MS for the released N-linked glycans for initial (t = 0) and processed sample (t = 120 min) abundances of Man9GlcNAc2 (m/z = 1,905.5) and Man8GlcNAc2 (m/z = 1,743.5) is shown for this ER gp120 glycoform. (C) Hydrolysis of the IC/cis-Golgi gp120 was monitored in the presence of 1 μg of ER α-mannosidase I. (D) MALDI-TOF MS for the released N-linked glycans for initial (t = 0) and processed sample (t = 120 min) abundances of Man9GlcNAc2 (m/z = 1,905.6) and Man8GlcNAc2 (m/z = 1,743.6) is shown for this IC/cis-Golgi gp120 glycoform.
The epitope recognized by the broadly neutralizing antibody, 2G12, is formed by Manα1→2Manα1→2Man residues within the D1 arm of Man7–9GlcNAc2 glycans on gp120 (17, 19). 2G12 binds to both monomeric and trimeric gp120, suggesting that most of the Man7–9GlcNAc2 glycans found on the native envelope are unrelated to the 2G12 epitope. To test this hypothesis, we generated mutant viruses with glycosylation site deletions that disrupt the 2G12 epitope: N295A and N332A, either of which abrogates 2G12 neutralization of HIVJRCSF (17). Analysis of the N-linked glycans from these envelopes shows a modest decrease in abundance of Man8–9GlcNAc2 glycans, consistent with the loss of the single glycan required for productive 2G12 recognition, but the overall profile remains unperturbed (SI Appendix, Fig. S5). This confirms the exquisite sensitivity of 2G12 for a small subset of Manα1→2Man terminating glycans, with precisely defined geometries of presentation that exist within a wider array of Man6–9GlcNAc2 glycans on the native virus. These results also suggest that the overall composition of the glycan shield is not much affected by sequence variation, consistent with the abundance of Man6–9GlcNAc2 glycans on clade C envelope, which is also not recognized by 2G12 (Fig. 2A) but is recognized by Manα1→2Man-specific antibodies (19, 28). The non–self-glycosylation of HIV extends beyond the 2G12 epitope.
Discussion
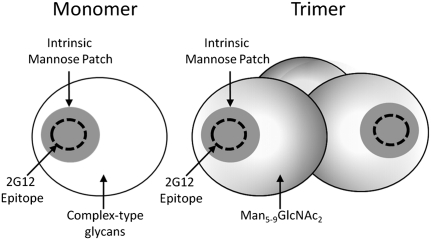
Mammalian N-linked glycosylation is strictly compartmentalized and follows a unidirectional and highly ordered processing pathway (11, 12) (SI Appendix, Fig. S1). Therefore, the presence or absence of a specific glycan structure can serve as a helpful marker of exposure to enzymatic activity restricted to intracellular processing compartment(s). In the present study, the chemical composition of the native trimeric HIV envelope glycans is shown to exhibit markedly constrained protein-directed glycan processing when compared with that of the host cell. We integrated the findings from this study into a revised model for the glycan shield of HIV (Fig. 4). The 2G12 epitope is contained within a conserved Man5–9GlcNAc2 patch that is intrinsic to gp120 and is present in both monomeric and trimeric envelopes; the complex type glycans found on recombinant monomeric gp120 are largely absent on the viral envelope trimer and are replaced by incompletely processed Man5–9GlcNAc2 glycans.
Fig. 4.
Mannose patches of recombinant monomeric (Monomer) and virion-associated trimeric (Trimer) gp120. The glycosylation of monomeric gp120 reveals an unprocessed intrinsic patch of Man5–9GlcNAc2 glycans (Fig. 1A), which appears to have resisted α-mannosidase activity, consistent with the results from the kinetic study (Fig. 3). Within this patch, in some isolates of HIV, the precise arrangement of Manα1→2Man-terminating oligomannose glycans supports binding of 2G12. The remaining glycans in the monomer are a cell-specific mixture of complex type glycans. The trimer contains the original mannose patch, as evidenced by the neutralization of functional virus by 2G12. The array of complex glycans is absent in the native trimer, however, and appears to be replaced by a further range of Man5–9GlcNAc2 glycans (Figs. 1C and 2 A and B).
Two possible explanations exist for incomplete glycan processing: (i) steric constraints prevent substrate recognition by glycosidases/glycosyltransferases, or (ii) the viral envelope is physically sequestered away from compartments containing these enzymes.
Protein-directed glycosylation via steric hindrance is perhaps most dramatically illustrated when the structure of a glycoprotein renders its conjugate glycans resistant to the enzymatic activities of the initial stages of the glycan processing pathway (15). We show here that the resistance to α-mannosidase activity offers an explanation for the core population of Man5–9GlcNAc2 glycans and provides direct support for the model of Zhu et al. (1), who argue for a clustered subdomain of oligosaccharides on gp120, which appears to exclude α-mannosidase processing. Within this intrinsic patch, a specific subset of glycans provides the Manα1→2Man termini that form the epitope recognized by 2G12 (Fig. 4).
The oligomannose type glycans found on recombinant monomeric gp120 therefore constitute a population that is independent of envelope trimerization. However, we nonetheless observe a much higher level of these Manα1→2Man terminating glycans (Man6–9GlcNAc2) on the virus envelope trimer, indicating that these additional structures derive from a mechanism separate from the one leading to formation of the monomer intrinsic mannose patch. We suggest, given the extensive opportunities for glycan-glycan and glycan-protein interactions at the trimer interface, that these structural constraints alter HIV glycan processing.
The fundamental divergence of host and viral envelope glycosylation occurs at the biosynthetic transition from precursor oligomannose to mature complex type glycans (Figs. 1 and 2). The obligate committed step in the biosynthesis of complex type glycosylation is the addition, by GnT I, of a single β1→2-linked GlcNAc residue to Man5GlcNAc2. Given the dominance of this substrate on the virus envelope trimer (Figs. 1B and 2B), the lack of processing by GnT I is remarkable. However, in contrast to the evidence that the Man6–9GlcNAc2 glycans arise as a consequence of steric occlusion, it is less obvious that the more exposed Man5GlcNAc2 glycans of gp120 are similarly protected. For example, the terminal mannose residue, which we show here does not participate in the formation of the β1→2 linkage catalyzed by GnT I, does participate in the glycosidic linkage accessible to and hydrolyzed by the α1→2 mannosidases that produce Man5GlcNAc2. Furthermore, the same Man5GlcNAc2 glycans, on the same strain of gp120, are readily accessible to GnT I and to subsequent processing enzymes when this protein is expressed as a soluble monomer (Fig. 1A). However, in the context of the functional virus, these abundantly conserved Man5GlcNAc2 glycans are entirely unprocessed. Furthermore, lack of GnT I activity cannot be attributed to infection per se, because complex glycans can still be obtained on soluble nonvirus-associated envelope glycoprotein from infected culture supernatant (Fig. 1E), which presumably has been processed in a monomeric form, and thus resembles the diversity of glycosylation observed for the recombinant monomer. An additional contribution to this glycan spectrum may also derive from material dissociated from mature virions, which would add to the observed abundances of oligomannose glycoforms. Although trimerization may induce a global ordering of glycans that limits processing of the envelope, the absence of complex type glycosylation is nonetheless surprising and raises the possibility that other factors, beyond steric constraints, may operate to exclude GnT I activity.
Regardless of its genesis, the Man5–9GlcNAc2 array of the HIV envelope represents a major component of the viral surface antigen, which is likely to affect many aspects of its structure and antigenicity. The presence of the trimer-induced mannose population may indicate that there are interactions or contacts between the carbohydrate and the neighboring subunit that are not present in the recombinant monomer. Artificially assembled, recombinant trimers may not exhibit native glycosylation, and may thus fail to mimic native spike conformation. The data presented here should therefore assist in the development of envelope trimers for structural or antigenic studies (6). We suggest that GnT I-deficient cell lines provide a helpful approximation of the native glycosylation of HIV and should be considered for use in such cases.
From the perspective of viral transmission and pathogenesis, it is proposed that oligomannose glycans are required for the interaction with dendritic cell-specific intercellular adhesion molecule-3–grabbing nonintegrin (DC-SIGN) on peripheral dendritic cells, which promotes the capture and dissemination of HIV in the early stages of infection (29). The broad conservation of the oligomannose array reported here, especially the Manα1→2Man-terminating glycans that are optimal for DC-SIGN binding (30), indicates that such interactions are highly efficient for any isolate or clade of HIV.
Another well-characterized ligand for the oligomannose glycans of HIV is the broadly neutralizing antibody 2G12. The identification of its epitope as a cluster of Manα1→2Man termini within the intrinsic mannose patch of gp120 (17) has encouraged the development of vaccines based around biological (19, 27, 31–33) and synthetic Manα1→2Man clusters (34–38). A major limitation of any vaccine based on 2G12 recognition is the absence of its cognate epitope on the epidemiologically important clade C. The specific Asn residues required for 2G12 neutralization are absent in clade C isolates, explaining their resistance to 2G12. However, the Manα1→2Man cluster(s) extend beyond the recognition motif of 2G12 (SI Appendix, Fig. S2) and are found even in resistant viruses, including those from clade C (Fig. 2A). Moreover, the prevalence of Man5GlcNAc2 compared with recombinant envelope may explain the surprising antiviral efficacy of lectins to Manα1→3 and Manα1→6 terminating glycans (39). Furthermore, as the most conserved and abundant glycan structure on the viral envelope, Man5GlcNAc2 should itself be evaluated as a target for vaccine design.
Carbohydrates modulate the antigenicity of the gp120 protein core and can have profound effects on antibody binding to gp120 (40–43). Recombinant gp120 is used extensively in animal studies and human vaccine trials. However, there are radical differences between recombinant (Fig. 1A) and viral (Figs. 1B and 2B) envelope glycosylation. Consideration should therefore be given to the use of glycoprotein immunogens with an antigenic surface resembling that of naturally circulating virus.
Materials and Methods
PBMC Virus.
Human PBMCs were isolated and stimulated as previously described (44). HIV-1JRCSF virus stocks were grown and titered on CD8+-depleted PBMCs (45). Virus production was monitored by p24 ELISA (Aalto Bioreagents).
Pseudovirus Preparation.
Pseudovirus was generated in HEK 293T or GnT I-deficient HEK 293S cells, as described (46). Briefly, cells were transfected with plasmids (pSVIII) carrying the reporter gene expressing the virus backbone PSG-3 and the functional envelope clone at a ratio of 2:1 using Fugene (Roche) according to the manufacturer's instructions. Virus was harvested after 3 d. Swainsonine was used at a concentration of 20 μM.
Envelope Isolation.
Virus particles were pelleted by ultracentrifugation (22,000 rpm for 1 h using Optima L-90K preparative ultracentrifuge, SW32 Ti Rotor [Beckman Coulter, Sunnyvale, CA]). Virus pellets were lysed with Nonidet P-40 (1% in PBS with protease inhibitors, 20 min at 4 °C). The debris was removed by centrifugation, and the envelope protein was immunoprecipitated with HIV envelope-specific monoclonal antibodies (D7324, b12, b6, F425-b4e8, 5B11, 8C7, or 7D3 depending on virus isolate) or serum from SIV-infected animals. Protein A and G beads were added and incubated overnight at 4 °C. The protein was eluted by heating in loading buffer (containing DTT) for 10 min at 100 °C and resolved by SDS/PAGE. The envelope band was confirmed by Western blot and cut to use directly in glycan analysis. Where discrete bands for gp160 and gp120 were detected, each was analyzed separately to confirm similar glycan profiles. Culture supernatant is defined as the supernatant after the virus particles have been pelleted.
Protein Expression and Purification.
Recombinant human ER α-mannosidase I and gp120BaL were cloned, expressed, and purified, as previously described (19). Recombinant gp120JRCSF (corresponding to residues 1–507, numbering based on alignment with the HxB2 reference strain) was similarly cloned and transiently expressed using the pHLSec expression vector (47). Expression of glycoproteins was determined in either HEK 293T (no. CRL-1573; American Type Culture Collection), with or without 20 μM kifunensine (Cayman Europe), or in HEK 293S cells deficient in GnT I (13).
Mannosidase Kinetic Assay.
Recombinant gp120BaL (20 μg) and ER α-mannosidase I (1 or 5 μg) were separately prewarmed at 37 °C for 5 min in a reaction buffer [80 mM piperazine-N,N′-bis(2-ethanesulfonic acid), 1μg/μL BSA, 4 mM CaCl2, 0.0016% NaN3 (pH 6.5)]. Samples were then mixed to a final volume of 250 μL, aliquots were taken at different time points, and gp120 glycans were analyzed by MS.
MALDI-TOF MS.
Oligosaccharides were released from gp120 by in-gel peptide-N-glycosidase F digestion (New England Biolabs) following the method of Küster et al. (48). Glycan samples were cleaned using a Nafion membrane (Sigma Aldrich), and mass spectra were recorded using a Shimazu AXIMA TOF2 MALDI TOF/TOF mass spectrometer (Kratos Analytical), as previously described (49).
Supplementary Material
Acknowledgments
We thank Michael Huber, Pascal Poignard, Mark Wormald, and Nicole Zitzmann for helpful discussions. This work was supported by the International AIDS Vaccine Initiative, the Oxford Glycobiology Endowment, National Institute of Allergy and Infectious Diseases Grant AI33292 (to D.R.B.), and the Ragon Institute. We acknowledge an equipment grant from the International AIDS Vaccine Initiative to purchase the Shimazu AXIMA TOF2 MALDI TOF/TOF mass spectrometer.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006498107/-/DCSupplemental.
References
- 1.Zhu X, Borchers C, Bienstock RJ, Tomer KB. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry. 2000;39:11194–11204. doi: 10.1021/bi000432m. [DOI] [PubMed] [Google Scholar]
- 2.Pantophlet R, Burton DR. GP120: Target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 3.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 4.Scanlan CN, Offer J, Zitzmann N, Dwek RA. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature. 2007;446:1038–1045. doi: 10.1038/nature05818. [DOI] [PubMed] [Google Scholar]
- 5.Pancera M. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci USA. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montefiori DC, Mascola JR. Neutralizing antibodies against HIV-1: Can we elicit them with vaccines and how much do we need? Current Opinion in HIV and AIDS. 2009;4:347–351. doi: 10.1097/COH.0b013e32832f4a4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMichael AJ. HIV vaccines. Annu Rev Immunol. 2006;24:227–255. doi: 10.1146/annurev.immunol.24.021605.090605. [DOI] [PubMed] [Google Scholar]
- 9.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: Good news for an HIV-1 vaccine? Nat Med. 2009;15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 10.Rerks-Ngarm S, et al. MOPH-TAVEG Investigators. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 11.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 12.Varki A, et al. Essentials of Glycobiology. 2nd Ed. Plainview, NY: Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 13.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanisch FG, Breloy I. Protein-specific glycosylation: Signal patches and cis-controlling peptidic elements. Biol Chem. 2009;390:619–626. doi: 10.1515/BC.2009.043. [DOI] [PubMed] [Google Scholar]
- 15.Crispin MD, et al. Monoglucosylated glycans in the secreted human complement component C3: Implications for protein biosynthesis and structure. FEBS Lett. 2004;566:270–274. doi: 10.1016/j.febslet.2004.04.045. [DOI] [PubMed] [Google Scholar]
- 16.Iacob RE, Perdivara I, Przybylski M, Tomer KB. Mass spectrometric characterization of glycosylation of hepatitis C virus E2 envelope glycoprotein reveals extended microheterogeneity of N-glycans. J Am Soc Mass Spectrom. 2008;19:428–444. doi: 10.1016/j.jasms.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scanlan CN, et al. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1→2 mannose residues on the outer face of gp120. J Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders RW, et al. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol. 2002;76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunlop DC, et al. Polysaccharide mimicry of the epitope of the broadly neutralizing anti-HIV antibody, 2G12, induces enhanced antibody responses to self oligomannose glycans. Glycobiology. 2010;20:812–823. doi: 10.1093/glycob/cwq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Go EP, et al. Glycosylation site-specific analysis of clade C HIV-1 envelope proteins. J Proteome Res. 2009;8:4231–4242. doi: 10.1021/pr9002728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizuochi T, et al. Carbohydrate structures of the human-immunodeficiency-virus (HIV) recombinant envelope glycoprotein gp120 produced in Chinese-hamster ovary cells. Biochem J. 1988;254:599–603. doi: 10.1042/bj2540599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crispin M, et al. A human embryonic kidney 293T cell line mutated at the Golgi α-mannosidase II locus. J Biol Chem. 2009;284:21684–21695. doi: 10.1074/jbc.M109.006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crispin M, et al. Inhibition of hybrid- and complex-type glycosylation reveals the presence of the GlcNAc transferase I-independent fucosylation pathway. Glycobiology. 2006;16:748–756. doi: 10.1093/glycob/cwj119. [DOI] [PubMed] [Google Scholar]
- 25.Eggink D, et al. Lack of complex N-glycans on HIV-1 envelope glycoproteins preserves protein conformation and entry function. Virology. 2010;401:236–247. doi: 10.1016/j.virol.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu Rev Immunol. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 27.Scanlan CN, et al. Inhibition of mammalian glycan biosynthesis produces non-self antigens for a broadly neutralising, HIV-1 specific antibody. J Mol Biol. 2007;372:16–22. doi: 10.1016/j.jmb.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 28.Luallen RJ, et al. Antibodies against Manalpha1,2-Manalpha1,2-Man oligosaccharide structures recognize envelope glycoproteins from HIV-1 and SIV strains. Glycobiology. 2010;20:280–286. doi: 10.1093/glycob/cwp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geijtenbeek TB, et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 30.van Liempt E, et al. Specificity of DC-SIGN for mannose- and fucose-containing glycans. FEBS Lett. 2006;580:6123–6131. doi: 10.1016/j.febslet.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 31.Dunlop DC, et al. Antigenic mimicry of the HIV envelope by AIDS-associated pathogens. AIDS. 2008;22:2214–2217. doi: 10.1097/QAD.0b013e328314b5df. [DOI] [PubMed] [Google Scholar]
- 32.Luallen RJ, et al. A yeast glycoprotein shows high-affinity binding to the broadly neutralizing human immunodeficiency virus antibody 2G12 and inhibits gp120 interactions with 2G12 and DC-SIGN. J Virol. 2009;83:4861–4870. doi: 10.1128/JVI.02537-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luallen RJ, et al. An engineered Saccharomyces cerevisiae strain binds the broadly neutralizing human immunodeficiency virus type 1 antibody 2G12 and elicits mannose-specific gp120-binding antibodies. J Virol. 2008;82:6447–6457. doi: 10.1128/JVI.00412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HK, et al. Reactivity-based one-pot synthesis of oligomannoses: Defining antigens recognized by 2G12, a broadly neutralizing anti-HIV-1 antibody. Angew Chem Int Ed Engl. 2004;43:1000–1003. doi: 10.1002/anie.200353105. [DOI] [PubMed] [Google Scholar]
- 35.Wang LX, Ni J, Singh S, Li H. Binding of high-mannose-type oligosaccharides and synthetic oligomannose clusters to human antibody 2G12: Implications for HIV-1 vaccine design. Chem Biol. 2004;11:127–134. doi: 10.1016/j.chembiol.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 36.Wang SK, et al. Targeting the carbohydrates on HIV-1: Interaction of oligomannose dendrons with human monoclonal antibody 2G12 and DC-SIGN. Proc Natl Acad Sci USA. 2008;105:3690–3695. doi: 10.1073/pnas.0712326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krauss IJ, et al. Fully synthetic carbohydrate HIV antigens designed on the logic of the 2G12 antibody. J Am Chem Soc. 2007;129:11042–11044. doi: 10.1021/ja074804r. [DOI] [PubMed] [Google Scholar]
- 38.Dudkin VY, et al. Toward fully synthetic carbohydrate-based HIV antigen design: On the critical role of bivalency. J Am Chem Soc. 2004;126:9560–9562. doi: 10.1021/ja047720g. [DOI] [PubMed] [Google Scholar]
- 39.Balzarini J, et al. Alpha-(1-3)- and alpha-(1-6)-D-mannose-specific plant lectins are markedly inhibitory to human immunodeficiency virus and cytomegalovirus infections in vitro. Antimicrob Agents Chemother. 1991;35:410–416. doi: 10.1128/aac.35.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer PB, Karlsson GB, Butters TD, Dwek RA, Platt FM. N-butyldeoxynojirimycin-mediated inhibition of human immunodeficiency virus entry correlates with changes in antibody recognition of the V1/V2 region of gp120. J Virol. 1996;70:7143–7152. doi: 10.1128/jvi.70.10.7143-7152.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duenas-Decamp MJ, Peters P, Burton D, Clapham PR. Natural resistance of human immunodeficiency virus type 1 to the CD4bs antibody b12 conferred by a glycan and an arginine residue close to the CD4 binding loop. J Virol. 2008;82:5807–5814. doi: 10.1128/JVI.02585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, et al. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J Virol. 2008;82:638–651. doi: 10.1128/JVI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolk T, Schreiber M. N-Glycans in the gp120 V1/V2 domain of the HIV-1 strain NL4-3 are indispensable for viral infectivity and resistance against antibody neutralization. Med Microbiol Immunol (Berl) 2006;195:165–172. doi: 10.1007/s00430-006-0016-z. [DOI] [PubMed] [Google Scholar]
- 44.Mann AM, et al. HIV sensitivity to neutralization is determined by target and virus producer cell properties. AIDS. 2009;23:1659–1667. doi: 10.1097/QAD.0b013e32832e9408. [DOI] [PubMed] [Google Scholar]
- 45.Rusert P, et al. Quantification of infectious HIV-1 plasma viral load using a boosted in vitro infection protocol. Virology. 2004;326:113–129. doi: 10.1016/j.virol.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 46.Li M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 48.Küster B, Wheeler SF, Hunter AP, Dwek RA, Harvey DJ. Sequencing of N-linked oligosaccharides directly from protein gels: In-gel deglycosylation followed by matrix-assisted laser desorption/ionization mass spectrometry and normal-phase high-performance liquid chromatography. Anal Biochem. 1997;250:82–101. doi: 10.1006/abio.1997.2199. [DOI] [PubMed] [Google Scholar]
- 49.Harvey DJ, Royle L, Radcliffe CM, Rudd PM, Dwek RA. Structural and quantitative analysis of N-linked glycans by matrix-assisted laser desorption ionization and negative ion nanospray mass spectrometry. Anal Biochem. 2008;376:44–60. doi: 10.1016/j.ab.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 50.Harvey DJ, et al. Proposal for a standard system for drawing structural diagrams of N- and O-linked carbohydrates and related compounds. Proteomics. 2009;9:3796–3801. doi: 10.1002/pmic.200900096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.