Abstract
We describe a systems biology approach for the genetic dissection of complex traits based on applying gene network theory to the results from genome-wide associations. The associations of single-nucleotide polymorphisms (SNP) that were individually associated with a primary phenotype of interest, age at puberty in our study, were explored across 22 related traits. Genomic regions were surveyed for genes harboring the selected SNP. As a result, an association weight matrix (AWM) was constructed with as many rows as genes and as many columns as traits. Each {i, j} cell value in the AWM corresponds to the z-score normalized additive effect of the ith gene (via its neighboring SNP) on the jth trait. Columnwise, the AWM recovered the genetic correlations estimated via pedigree-based restricted maximum-likelihood methods. Rowwise, a combination of hierarchical clustering, gene network, and pathway analyses identified genetic drivers that would have been missed by standard genome-wide association studies. Finally, the promoter regions of the AWM-predicted targets of three key transcription factors (TFs), estrogen-related receptor γ (ESRRG), Pal3 motif, bound by a PPAR-γ homodimer, IR3 sites (PPARG), and Prophet of Pit 1, PROP paired-like homeobox 1 (PROP1), were surveyed to identify binding sites corresponding to those TFs. Applied to our case, the AWM results recapitulate the known biology of puberty, captured experimentally validated binding sites, and identified candidate genes and gene–gene interactions for further investigation.
Keywords: bovine, complex traits, fertility, reproduction, systems biology
The analysis of genome-wide association studies (GWASs) applied to complex traits remains a challenge (1). Addressing a complex trait by a single, often binary, phenotypic measure is common practice but is limiting. It is not easy to find the right balance between applying a conservative significance threshold that gives rise to a small number of strong and hopefully biologically meaningful associations and applying a relaxed threshold yielding numerous associations, many of which are new but potentially false. In addition, an increase in sample size coupled with a denser chip results in a larger number of associations that, on average, have a much smaller effect (2). Accepting a large number of associations while simultaneously reducing the number of false positives would be ideal. It is reasonable to propose that a holistic approach applied to a relaxed significance threshold could be the solution. Such a strategy would be particularly useful when investigating the genetic basis of complex traits that, by definition, are influenced by numerous genes and pathways.
In our study, age at puberty was the complex phenotype considered. Puberty, or the progression to sexual maturity, is a developmental process with genetic drivers conserved among species (3). It is an important phenotype for the beef industry because late puberty has negative effects on reproduction rates and profitability (4). Age at puberty is moderately heritable, with estimates of heritability in cattle ranging from 0.16 to 0.57 (5, 6). In humans, ∼50% of the variation in age of puberty is genetic (7, 8). An advantage to working with cattle as a model species is that observational data on traits related to puberty are available. For example, weight and condition score are often measured on occasions throughout an animal's development. Hence, understanding genetics of cattle puberty and its biology serves two purposes: as a strategy to develop efficient livestock resources and as a model for human biology.
The focus of this work is to demonstrate a unique systems approach, which we call an association weight matrix (AWM), appropriated for GWASs of complex traits. We examined cattle puberty from a GWAS based on ∼50,000 single-nucleotide polymorphisms (SNPs) and 22 traits as an example of a typical complex phenotype. We designed the AWM with elements corresponding to the standardized additive effect of the ith SNP (in rows) on the jth trait (in columns). Rowwise, the AWM explored gene-to-gene interactions for cattle puberty across the genome; columnwise, it estimated correlations between traits influencing puberty.
Results
Commonly, GWASs are single-trait–single-SNP analyses. This analysis for our main pubertal trait, the age of occurrence of the first corpus luteum (AGECL), resulted in more associated SNPs than the number expected by chance alone: 2,799 SNPs at P < 0.05, 588 SNPs at P < 0.01, and 69 SNPs at P < 0.001. We report the results of the single-trait–single-SNP association analyses for later comparison against those from the AWM approach.
Each SNP effect, from the total set of 50,070 SNPs, was used as a data point in the calculation of all pairwise correlations between the 22 traits. As a result, AGECL correlated with weight at first corpus luteum (WTCL) (R = 0.64) and with postpartum anoestrus interval (PPAI) (R = 0.31) among other traits (Table S1). When selecting SNPs to build the AWM, we considered all 22 traits and the SNP-to-gene distance (as per Fig. S1). With increasing SNP-to-gene distance, we observed decay in the SNP significance, as measured by P value (Fig. S2). We selected 3,159 SNPs to build the AWM, where SNPs were rows and 22 related traits were columns (Fig. S3A).
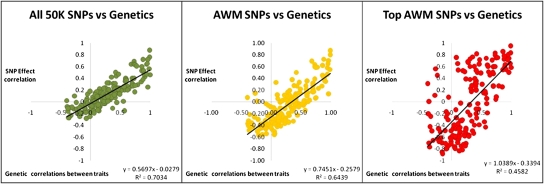
Columnwise, the AWM was used to calculate correlations between traits. This result is visualized as a hierarchical tree where AGECL clusters with WTCL and both are close to PPAI (Fig. S3B). On the hierarchical tree cluster, a strong positive correlation is displayed as proximity, whereas a strong negative correlation is displayed as a large distance. To observe negative and positive correlations equally, we developed the quantitative trait network (QTN) from AWM SNPs, which shows a high degree of interaction among all 22 traits (Fig. S3C). For visual comparison, we also present a QTN (Fig. S3D) based on published genetic correlations (5, 9). A formal comparison between these genetic correlations and SNP-based correlations showed a moderate agreement (R2 = 0.6439) between the two approaches (Fig. 1). When all SNPs were considered, the trait correlations were closer to the genetic estimates (R2 = 0.7034; Fig. 1). The same comparison using the 3,159 SNPs included in the AWM (or 6.2% of the total) captures 64% of the variation estimated by genetic correlations (Fig. 1). So AWM is equivalent to 91% (or 64/70) of the variation that was captured using the entire SNP chip. Fig. 1 also reveals a pattern of increasing regression coefficients with decreasing number of SNPs: 0.5697 (whole chip) to 0.7415 (AWM) to 1.0389 (top 71 SNPs). Hence, the more stringent we are at selecting SNPs, the more unbiased the recovery becomes (regression coefficient closer to unity). Therefore, the criteria to include SNPs in the AWM resulted in enrichment with nonredundant genetic information, implying that the AWM could be used for estimating genetic correlations, although it was not developed with this aim. Nonetheless, the higher the number of SNPs analyzed, the higher is the similarity between SNP-based and genetic correlations. This linear relationship estimates that ∼200,000 SNPs would fully recover genetic correlations between traits (Fig. S4).
Fig. 1.
Genetic correlations between traits compared with SNP-based correlations. Genetic pairwise correlations estimated for 19 traits were compared with SNP-based correlations for all 50,070 SNPs in green (R2 = 0.7034), AWM 3,159 SNPs in yellow (R2 = 0.6439), and the top 71 SNPs from the AWM in red (R2 = 0.4582).
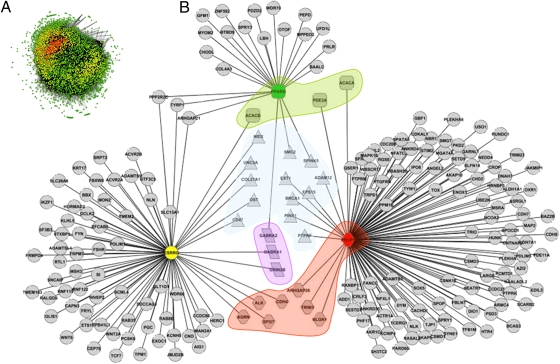
Pairwise correlations across AWM rows were used to predict gene–gene (or gene–SNP) interactions and hence build a gene network for puberty. In the network, every gene (or SNP) was a node and every significant interaction was an edge connecting two nodes. The PCIT algorithm identified 287,465 significant edges between 3,159 nodes (Fig. 2A). From this point onward, issues such as gene connectivity, annotation, and the emergence of highly connected clusters were the relevant metrics in our analysis.
Fig. 2.
Puberty network extracted from GWASs using the AWM approach. (A) Entire network. Nodes represent 3,159 genes and SNPs whereas edges represent significant correlations between nodes. The color scale corresponds to MCODE score where red nodes represent higher network density. (B) Subset of the network showing PROP1 (red node), ESRRG (yellow node), and PPARG (green node) in silico validated targets (gray nodes). Node shapes (from top) are as follows: Squares in green are genes related to lipids and fatty acid metabolism, triangles in blue are genes related to cell proliferation and apoptosis, rectangles in purple are genes related to the GABA and glutamate pathways, and hexagons in red are genes related to nervous system development.
Gene Ontology (GO) analyses showed overrepresentation for “GABA receptor activity” (P = 0.025; Fig. S5A) in our puberty network. Importantly, there were 539 genes in the AWM associated with the GO term “developmental process,” which was highly enriched (P < 1.00E-09). These genes along with those associated with “regulation of transcription” (P < 1.00E-09) would have been missed if single-trait analysis was performed (Fig. S5B). In addition, pathway analyses of the network revealed an enrichment (P < 0.001) for “calcium signaling,” “axon guidance,” and “neuroactive ligand–receptor interaction.” This last pathway includes ligands and receptors considered to be involved with pubertal signaling such as GABA receptor activity, “glutamate receptor activity,” “follicular stimulant hormone (FSH) receptor activity,” and “leptin receptor activity.” The pathway analyses also revealed enrichment for “cell growth,” “cell survival,” and “factors controlling cell cycle progression.” This last result supports a theory that implicates a role in puberty for tumor related genes (Discussion). GO term and pathway analyses applied to the AWM network identified biological processes and pathways that are relevant for cattle puberty.
To test the likelihood of AWM predictions being random we built a control gene network. As expected, the control gene network had the topology of a random network with the majority of the genes having an average number of connections (10). The distribution of number of connections per gene in the control network was different from that in the AWM network (P < 0.0001), so AWM was not random.
To test the likelihood of AWM predictions being simply a reflection of LD between SNPs from the initial GWAS, we calculated all pairwise D′ and R2. Whereas some AWM predictions were underlined by strong LD, most were not. This result was expected considering the average distance between AWM-selected SNPs (825 kb ± 1 Mb).
We selected 3 key transcription factors (TFs) from 34 available (see Methods for details and SI Text for TF list). Key TFs, Prophet of Pit 1, PROP paired-like homeobox 1 (PROP1), Pal3 motif, bound by a PPAR-γ homodimer, IR3 sites (PPARG), and estrogen-related receptor γ (ESRRG), and their AWM-predicted targets were subjected to regulatory sequence analyses. Approximately 36% of the predicted partners had at least one TF binding site (TFBS) for PROP1 and ESRRG and 18% of predicted partners had at least one TF binding site for PPARG (Table S2). We considered this high rate of identification of corresponding TFBSs to be an in silico validation of TF–target gene interactions predicted by the AWM. TFs and their in silico validated target genes are shown in Fig. 2B (for gene lists see SI Text).
A prediction of targets for PROP1, PPARG, and ESRRG was also carried out on the basis of the control network. In the control network each TF had fewer targets and a smaller (P < 0.0001) proportion of targets had corresponding TFBSs, when compared with the AWM network (Table S2). The AWM network presented more validated TF–target interactions and these interactions were not random because they contrast with the control network.
Further evidence supporting the interactions predicted by the AWM could be found for ESRRG and 19 of its targets. These targets presented a promoter model derived from published experimental data (Table S3). Thus, the AWM captured experimentally validated TFBSs.
Discussion
Our results revealed a number of appealing features of the AWM of which four are worth highlighting: (i) It identified as relevant genes that would have been missed by traditional single-trait GWASs; (ii) it predicted TF-target associations that have been experimentally validated by other authors; (iii) it captured more information than analyses exploiting LD structure; and (iv) it was more efficient than similar approaches. The following discussion is focused on demonstrating these features in the context of our GWAS for cattle puberty.
Currently, most GWASs are single-trait–single-SNP approaches that focus only on the most significant results, in terms of P values. This approach is limited for a complex phenotype because it implicates only very few genes. Previous studies used this approach to report associations of the genes CCR3, SPOCK1, LIN28B, ZNF462, TMEM38B, FKTN, FSD1L, and TAL2 with age of puberty in women (11–15). If we applied only this approach to our data, further research would consider one candidate gene, NMDAR2B, which was strongly associated (P < 0.000015) with AGECL. Despite the significance of this result and of the above-mentioned genes, it is unlikely that puberty variance could be explained by one or a handful of genes associated with a single measured trait. After all, puberty is a complex phenotype influenced by many processes, such as energy balance and brain development.
The AWM included the genes underpinning the strong associations of the single-trait–single-SNP approach but was not restricted by them. Hence, there is the potential to explain a larger proportion of the genetic variation. For example, our single-trait–single-SNP approach yielded one candidate gene, NMDAR2B, but the AWM also supported recently identified candidate genes for age of puberty from human GWASs, such as SPOCK1 (14) and ZNF462 (11). These and other results would have been missed without a systemic approach to our data. It is expected that systemic approaches that integrate the analyses of related traits (16) will identify numerous genes (many QTL), each with a small effect impacting on any complex trait (2). Indeed, puberty is likely to be affected by many interacting genes that function in a network with a high degree of redundancy to preserve the essential process of reproduction. Our approach has predicted a large and redundant gene network for puberty, in agreement with previous results (17) and consistent with the endogenous topology of a network for an essential process (18).
The exact selection criteria and thresholds proposed to include or exclude genes and SNPs from the AWM will vary according to each GWAS under investigation. Importantly, the relaxed threshold of P < 0.05 was a useful source of information for this systems approach rather than a problem. Once a SNP has been included in the AWM, its association significance no longer has a role. Instead, network theory takes command and gene connectivity, annotation, and the emergence of clusters of highly connected genes become the relevant metrics for AWM analysis.
AWM recovered the relatedness between traits using SNP effect correlations, which in our study were quite similar (R2 = 0.64) to the published genetic correlations calculated for the same population (5, 9). Ideally, one would like to have SNP effect correlations very similar to genetic correlations, but we estimated that for 100% similarity >200,000 SNPs might be required, in agreement with previous estimates of desirable SNP numbers for cattle GWASs (19). Nonetheless, the similarity we observed between genetic correlations and our SNP-based correlations show that it is possible to estimate trait correlations from GWASs.
The AWM generated a gene network, which provided a prediction of gene interactions based on SNP effect correlations. The structure of the AWM gene network was not random as it differed from the control network. Also, the number of in silico validated TF–target interactions in the AWM was superior to the number found in the control network. Hence, the AWM predicted TF–target associations that have been experimentally validated by other authors and in a frequency significantly higher than could be achieved by chance alone.
AWM gene interactions were predicted with the PCIT algorithm (10). PCIT is independent of previous knowledge and captured more information than analyses exploiting LD structure. Many predicted TF–target interactions were not underlined by linkage disequilibrium (LD). This independence from previous knowledge leads to the most important contribution of the AWM, which is to predict gene–gene interactions from GWAS data alone. These predictions either reflect known biology or are hypotheses to be tested. For example, the AWM predicted a triplet of closely associated genes: RUNDC1, BRCA1, and NBR1. BRCA1 and NBR1 are 55 kb apart and RUNDC1 is 92 kb from BRCA1 and 147 kb from NBR1 (positions of AWM SNPs). In the bovine genome, the extent of LD declines rapidly from 0 to 200 kb (20). Thus, LD between the SNPs that represent RUNDC1, BRCA1, and NBR1 in the AWM was expected: R2 = 0.97 (RUNDC1 and BRCA1; RUNDC1 and NBR1) and R2 = 0.99 (BRCA1 and NBR1). Even so, we are not alone when arguing that genomes are not random and gene proximity might also reflect gene function (21), especially when the effect of SNP over 22 traits was considered. There is previous evidence for BRCA1 and NBR1 interaction as they share the same bidirectional promoter region (22). However, the predicted interactions between RUNDC1 and BRCA1, as well as between RUNDC1 and NBR1, are yet to be tested. RUNDC1 is an inhibitor of the tumor suppressor p53 (23), a gene that embodies the link between oncogenes and reproduction (17, 24). Evidence for coexpression of p53 and BRCA1 has been only very recently published (25). Using the AWM prediction, we can hypothesize that RUNDC1 is a regulator of BRCA1, underscoring the power of the AWM for generating hypotheses.
BRCA1 has been long implicated in breast and ovarian cancer (26, 27). A risk factor for breast cancer in women is the age of puberty (28). The presence of this and other tumor-related genes in the AWM network for puberty is a confirmation and expansion of previous results (17) rather than a surprise. Previously, seven tumor-related genes were proposed as network hubs influencing puberty: OCT2, p53, MAF, CUTL1, USF2, YY1, and TTF1. The AWM network corroborates the relevance of tumor-related genes such as CUTL1, FLJ22457, SASH1, and SynCAM1 for puberty. Also, TP53BP1 that encodes a key p53 binding protein is in the AWM network, which predicts its interaction with TRIM3, a brain tumor suppressor (29). The role that each tumor-related gene may play in puberty remains unclear. As a group, they are associated with cell proliferation (oncogenes) or cell apoptosis (tumor suppressors). Genes associated with cell growth, cell survival, and factors controlling cell cycle progression were overrepresented in the AWM network. The balance between pro- and antiapoptosis signals is important for the dynamic biology of germ cells in males and females (3) and for pubertal brain development (30).
Brain remodeling, which is likely influenced by a variety of steroid hormones (31), has been shown to precede the changes in the pattern of GnRH release that trigger the onset of puberty (32). Important drivers of GnRH remodeling are the GABAergic and glutamatergic synaptic inputs (33, 34) and so genes involved in these signaling pathways might influence puberty. The strongest candidate gene from our GWAS, NMDAR2B is a glutamate receptor from the NMDA class of receptors. This class is involved in pubertal brain development (33). We expanded the list of candidates from the GABA and glutamate pathways to 19 genes using the AWM approach, which recovered this part of the known biology of puberty more effectively than the single-trait–single-SNP approach. The AWM network was enriched for the term GABA receptor activity.
We explored the promoter regions of the genes predicted to be targeted by key TFs, PROP1, ESRRG, and PPARG, to identify corresponding TFBSs. Most of the targets were not on the same chromosome as their TF. For example, the LD between SNPs for the target GABRA1 and the TF ESRRG is R2 = 0.0013. Therefore, the predicted TF–target interactions are not simply a reflection of LD. Lack of LD and simultaneous presence of TFBSs highlighted the functional potential of the interactions predicted with the AWM.
PROP1 is important for the differentiation of gonadotropes (35) and it has been associated with infertility in mice and humans (3, 35). Also, PROP1 stimulates the expression of PIT-1, which regulates growth hormone and prolactin. The PIT-1 pathway has been associated with embryonic survival rates in cattle (36). Our network predicted 320 targets for PROP1, of which 114 presented TFBSs. These included TRIM3, BRCA1, NBR1, and RUNDC1, indicating a variety of possible pathways for a known tumor-related role of PROP1 (35).
We predicted 211 targets for ESRRG, of which 76 had corresponding TFBSs, including follicle stimulating hormone receptor (FSHR), GABA receptor (GABRA1), and NMDAR2B. ESRRG targeting FSHR may reflect feedback of hormone signaling (estrogens influencing FSH release). ESRRG targeting GABRA1 and NMDAR2B indicates a link between estrogen pathways and GABA and glutamate signaling, which might influence puberty, by modifying the input on GnRH neurons.
PPARG is an important regulator of energy balance. Among the 124 AWM-predicted targets, 22 presented TFBSs for PPARG. The presence of PPARG and its 23 targets in common with ESRRG in our network was evidence for the AWM capturing known biology. There are demonstrated associations between energy balance and reproduction (37, 38). If fat deposition traits were excluded from our analysis, this PPARG and ESRRG link could have been missed. Thus, integrating traits in a systemic approach was advantageous. Three genes, ARHGAP21, PPP2R2C, and TYRP1, are targets of ESRRG and PPARG that present binding sites for both. These three targets might be components of the known metabolic link between estrogen-related receptors and PPARG (39).
Finally, the AWM was more efficient for integrating related traits and analyzing thousands of SNPs than a previous method described by Kim and Xing in 2009 (16). In that study, the authors deemed their method unfeasible to apply with >100 SNPs in a single model. In contrast, the AWM when applied to age of puberty in cattle analyzed >3,000 SNPs and thousands more could have been included.
In conclusion, the AWM approach was an appropriate method of analysis for this complex phenotype. When applied to our dataset, it predicted gene interactions that are consistent with the known biology of puberty (e.g., ESRRG and FSHR), captured known regulation binding sites, and provided candidate genes for cattle puberty (e.g., PROP1). The AWM predicted several interactions for important tumor-related genes and key TFs, indicating their potential roles in puberty and providing some promising hypotheses, which can be further investigated. Future research addressing these hypotheses and its candidate genes might contribute to better understanding of cattle puberty.
Methods
Animals, Traits, and Genotypes.
We used data from 866 cows representing 51 sire families from a tropical composite population bred in the tropical northern regions of Australia that was described elsewhere (5, 9, 40–42). In these cows, 22 traits were annotated: AGECL (days), presence or absence of corpus luteum close to the day when bulls were placed in the same paddock as the heifers (CLJOIN, score 1–0), WTCL (kg), scanned P8 site fat depth at AGECL (FATCL, mm), PPAI (days), PPAI with respect to weaning time (PW, score 1–0), live weight (WT, kg), hip height (HH, cm), serum concentration of insulin-like growth factor I (IGF-I, ng/mL), average daily weight gain (ADG, kg/d), body condition score (CS, score 1–10), scanned longissimus dorsi area (SEMA, cm2), scanned P8 site fat depth (SP8, mm) and scanned fat depth measured between the last 2 ribs (SRIB, mm). The last 8 traits were measured at two time points, T1 and T2, on average before and after AGECL. A description of the 22 traits along with summary of descriptive statistics for this herd is provided in SI Text (Table S4).
The BovineSNP50 Bead Chip (Illumina 2008) (43, 44) was used to genotype all cows. Family trios and repeat samples were included for quality assurance. SNPs with auto-calling rates <85% and SNPs with minor allele frequency <0.01 were excluded from later analyses.
LD between all possible SNP pairs was calculated using two metrics: D′ and R2 (45). SNP effects were calculated via single-trait–single-SNP association analysis. The additive effect of a SNP on each trait was calculated by regression analysis, using a mixed model (details in SI Text). Solutions to the model were estimated using ASREML (46).
The AWM.
Constructing the AWM starts with the selection of relevant SNPs from a GWAS to represent genes. The plan of the selection criteria is shown in Fig. S1. The criteria to select SNPs for the AWM include (i) significance of the allele substitution effect measured for each SNP across the 22 traits, (ii) correlations between traits, and (iii) SNP genomic position. Our selection criteria were developed to favor genes harboring SNPs with significant association across related traits. Briefly, a group of SNPs, the top 0.2%, were selected first because they were associated with ≥10 traits (P < 0.05) regardless of their distance to the nearest gene. Second, we selected SNPs that were either “close” (<2,500 bp) to or “very far” (≥1.5 Mb) from the nearest annotated gene (BTAU4.0 assembly) and were either associated (P < 0.05) with AGECL or any ≥3 traits. Definitions for close or very far SNPs are based on expectancy of LD, size of promoter region, and likelihood of cis-acting windows. Cis-acting windows could include SNPs within 100 kb, but are enriched within 250 bp of transcription end sites (47). Finally, selected SNPs were used to build the AWM with as many rows as SNPs and as many columns as traits. The rows are indexed as genes for close SNPs or as SNPs (Illumina code) otherwise. Finally, each {i, j} cell value in the AWM corresponds to the z-score normalized additive effect of the ith SNP on the jth trait. The AWM approach explores traits correlations columnwise and gene interactions rowwise.
Columnwise Pearson correlations between AGECL and the other 21 traits were calculated using the SNP effect values. First, all SNPs available from genotyping were used for the calculation, and second, the subset of SNPs selected for the AWM was used. The results of these SNP-based correlations were compared with the genetic correlations, estimated via pedigree-based restricted maximum likelihood (REML), established for the same population (5, 9). These previous genetic correlations (5, 9) and the SNP-based correlations were used to form QTNs for puberty. Rowwise AWM explores the correlations between SNP effects to predict gene interactions. We studied the predicted gene interactions using a combination of hierarchical clustering, weighted gene network, and pathway analyses to identify genetic drivers of puberty.
Visualization of the AWM and hierarchical clustering analyses were performed using PermutMatrix (48). Significant correlations between rows were identified with the PCIT algorithm (10) and reported as gene–gene or gene–SNP interactions in a network, visualized in Cytoscape (49). Overrepresented GO terms were identified using BiNGO (50) and GOrilla (51). Also, pathway mapping of genes in the network was performed using DAVID (52, 53).
Control Network.
A random matrix was built to serve as a control for our method. To build this control, we randomly shuffled each {i, j} cell value in the AWM rows, so that the values no longer corresponded with the appropriated normalized additive effect of the ith SNP on the jth trait. This random matrix was used to predict a control gene network, using the same methodology as described above. Then, we compared the connectivity distribution of the control gene network with the AWM-derived network by analyzing their structure using the Kolmogorov–Smirnov test (SAS 9.1.3).
Regulatory Sequence Analysis.
Three TFs were chosen on the basis of the following criteria: available binding site information on Genomatix (http://www.genomatix.de/), reported functional role in the context of reproduction, and position in the gene network (i.e., sufficiently separated to ensure maximum coverage of the entire network). These three were ESRRG, PPARG, and PROP1. Three lists of genes were created corresponding to AWM-predicted targets of the TFs. The promoter regions corresponding to listed genes were retrieved using the Gene2Promoter module in Genomatix (http://www.genomatix.de/). Promoter regions were systematically mined for specific TFBSs derived from the position weight matrix corresponding with the TFs using the MatInspector module (54). In addition, the target genes were explored for known promoter models (ModelInspector Module) across mammals including humans. Literature mining was also carried out using BiblioSphere (55) to obtain previous evidence for associations between TFs and TFBSs. Providing evidence for the interaction between the TF and its predicted targets via regulatory sequence analysis serves as an in silico validation for the TF–target interactions in the AWM network.
We performed regulatory sequence analysis for the control network, selecting the same TFs (ESRRG, PPARG, and PROP1) and their partners. The results of this analysis were compared with AWM results using the two-proportion z-test, under the null hypothesis of the AWM predicting fewer validated interactions than the control network.
Supplementary Material
Acknowledgments
We thank W. Barendse, J. Kijas, S. Lehnert and N. Hudson for comments on the manuscript. Collaborations with the Northern Pastoral Group, Department of Employment, Economic Development, and Innovation and Commonwealth Scientific and Industrial Research Organization Livestock Industries facilitated phenotype collection. Meat and Livestock Australia, Australian Centre for International Agricultural Research, Cooperative Research Centre for Beef Genetic Technologies, Commonwealth Scientific and Industrial Research Organization Livestock Industries, and University of Queensland gave financial support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002044107/-/DCSupplemental.
References
- 1.McCarthy MI, et al. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 2.Mackay TF, Stone EA, Ayroles JF. The genetics of quantitative traits: Challenges and prospects. Nat Rev Genet. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- 3.Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol. 2002;4(Suppl):s41–s49. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- 4.Lesmeister JL, Burfening PJ, Blackwell RL. Date of first calving in beef cows and subsequent calf production. J Anim Sci. 1973;36:1–6. [Google Scholar]
- 5.Johnston DJ, et al. Genetics of heifer puberty in two tropical beef genotypes in northern Australia and associations with heifer- and steer-production traits. Anim Prod Sci. 2009;49:399–412. [Google Scholar]
- 6.Martínez-Velázquez G, Gregory KE, Bennett GL, Van Vleck LD. Genetic relationships between scrotal circumference and female reproductive traits. J Anim Sci. 2003;81:395–401. doi: 10.2527/2003.812395x. [DOI] [PubMed] [Google Scholar]
- 7.Anderson CA, et al. A genome-wide linkage scan for age at menarche in three populations of European descent. J Clin Endocrinol Metab. 2008;93:3965–3970. doi: 10.1210/jc.2007-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson CA, Duffy DL, Martin NG, Visscher PM. Estimation of variance components for age at menarche in twin families. Behav Genet. 2007;37:668–677. doi: 10.1007/s10519-007-9163-2. [DOI] [PubMed] [Google Scholar]
- 9.Barwick SA, et al. Genetics of heifer performance in ‘wet’ and ‘dry’ seasons and their relationships with steer performance in two tropical beef genotypes. Anim Prod Sci. 2009;49:367–382. [Google Scholar]
- 10.Reverter A, Chan EK. Combining partial correlation and an information theory approach to the reversed engineering of gene co-expression networks. Bioinformatics. 2008;24:2491–2497. doi: 10.1093/bioinformatics/btn482. [DOI] [PubMed] [Google Scholar]
- 11.Perry JR, et al. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet. 2009;41:648–650. doi: 10.1038/ng.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ong KK, et al. Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet. 2009;41:729–733. doi: 10.1038/ng.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He C, et al. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009;41:724–728. doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu YZ, et al. Genome-wide association analyses identify SPOCK as a key novel gene underlying age at menarche. PLoS Genet. 2009;5:e1000420. doi: 10.1371/journal.pgen.1000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang F, et al. The chemokine (C-C-motif) receptor 3 (CCR3) gene is linked and associated with age at menarche in Caucasian females. Hum Genet. 2007;121:35–42. doi: 10.1007/s00439-006-0295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S, Xing EP. Statistical estimation of correlated genome associations to a quantitative trait network. PLoS Genet. 2009;5:e1000587. doi: 10.1371/journal.pgen.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth CL, et al. Expression of a tumor-related gene network increases in the mammalian hypothalamus at the time of female puberty. Endocrinology. 2007;148:5147–5161. doi: 10.1210/en.2007-0634. [DOI] [PubMed] [Google Scholar]
- 18.Luscombe NM, et al. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 19.de Roos AP, Hayes BJ, Spelman RJ, Goddard ME. Linkage disequilibrium and persistence of phase in Holstein-Friesian, Jersey and Angus cattle. Genetics. 2008;179:1503–1512. doi: 10.1534/genetics.107.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibbs RA, et al. Bovine HapMap Consortium. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science. 2009;324:528–532. doi: 10.1126/science.1167936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adachi N, Lieber MR. Bidirectional gene organization: A common architectural feature of the human genome. Cell. 2002;109:807–809. doi: 10.1016/s0092-8674(02)00758-4. [DOI] [PubMed] [Google Scholar]
- 22.Whitehouse C, Chambers J, Catteau A, Solomon E. Brca1 expression is regulated by a bidirectional promoter that is shared by the Nbr1 gene in mouse. Gene. 2004;326:87–96. doi: 10.1016/j.gene.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Llanos S, Efeyan A, Monsech J, Dominguez O, Serrano M. A high-throughput loss-of-function screening identifies novel p53 regulators. Cell Cycle. 2006;5:1880–1885. doi: 10.4161/cc.5.16.3140. [DOI] [PubMed] [Google Scholar]
- 24.Hu W, Feng Z, Atwal GS, Levine AJ. p53: A new player in reproduction. Cell Cycle. 2008;7:848–852. doi: 10.4161/cc.7.7.5658. [DOI] [PubMed] [Google Scholar]
- 25.Wang M, et al. Prepubertal physical activity up-regulates estrogen receptor beta, BRCA1 and p53 mRNA expression in the rat mammary gland. Breast Cancer Res Treat. 2009;115:213–220. doi: 10.1007/s10549-008-0062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 27.Futreal PA, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 28.Peeters PH, Verbeek AL, Krol A, Matthyssen MM, de Waard F. Age at menarche and breast cancer risk in nulliparous women. Breast Cancer Res Treat. 1995;33:55–61. doi: 10.1007/BF00666071. [DOI] [PubMed] [Google Scholar]
- 29.Boulay JL, et al. Loss of heterozygosity of TRIM3 in malignant gliomas. BMC Cancer. 2009;9:71. doi: 10.1186/1471-2407-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markham JA, Morris JR, Juraska JM. Neuron number decreases in the rat ventral, but not dorsal, medial prefrontal cortex between adolescence and adulthood. Neuroscience. 2007;144:961–968. doi: 10.1016/j.neuroscience.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 31.Stuart EB, Thompson JM, Rhees RW, Lephart ED. Steroid hormone influence on brain calbindin-D(28K) in male prepubertal and ovariectomized rats. Brain Res Dev Brain Res. 2001;129:125–133. doi: 10.1016/s0165-3806(01)00191-2. [DOI] [PubMed] [Google Scholar]
- 32.Terasawa E. Postnatal remodeling of gonadotropin-releasing hormone I neurons: Toward understanding the mechanism of the onset of puberty. Endocrinology. 2006;147:3650–3651. doi: 10.1210/en.2006-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarkson J, Herbison AE. Development of GABA and glutamate signaling at the GnRH neuron in relation to puberty. Mol Cell Endocrinol. 2006;254–255:32–38. doi: 10.1016/j.mce.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 34.Terasawa E. Role of GABA in the mechanism of the onset of puberty in non-human primates. Int Rev Neurobiol. 2005;71:113–129. doi: 10.1016/s0074-7742(05)71005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cushman LJ, et al. Persistent Prop1 expression delays gonadotrope differentiation and enhances pituitary tumor susceptibility. Hum Mol Genet. 2001;10:1141–1153. doi: 10.1093/hmg/10.11.1141. [DOI] [PubMed] [Google Scholar]
- 36.Khatib H, et al. Single gene and gene interaction effects on fertilization and embryonic survival rates in cattle. J Dairy Sci. 2009;92:2238–2247. doi: 10.3168/jds.2008-1767. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez-Fernandez R, et al. Novel signals for the integration of energy balance and reproduction. Mol Cell Endocrinol. 2006;254–255:127–132. doi: 10.1016/j.mce.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 38.Gasser CL, Behlke EJ, Grum DE, Day ML. Effect of timing of feeding a high-concentrate diet on growth and attainment of puberty in early-weaned heifers. J Anim Sci. 2006;84:3118–3122. doi: 10.2527/jas.2005-676. [DOI] [PubMed] [Google Scholar]
- 39.Feige JN, Auwerx J. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007;17:292–301. doi: 10.1016/j.tcb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Prayaga KC, et al. Genetics of adaptive traits in heifers and their relationship to growth, pubertal and carcass traits in two tropical beef cattle genotypes. Anim Prod Sci. 2009;49:413–425. [Google Scholar]
- 41.Barwick SA, Wolcott ML, Johnston DJ, Burrow HM, Sullivan MT. Genetics of steer daily and residual feed intake in two tropical beef genotypes, and relationships among intake, body composition, growth and other post-weaning measures. Anim Prod Sci. 2009;49:351–366. [Google Scholar]
- 42.Burrow HM, et al. Relationships between carcass and beef quality and components of herd profitability in Northern Australia. 50 Years of DNA: Proceedings of the Fifteenth Conference, Association for the Advancement of Animal Breeding and Genetics. 2003;13:359–362. [Google Scholar]
- 43.Van Tassell CP, et al. SNP discovery and allele frequency estimation by deep sequencing of reduced representation libraries. Nat Methods. 2008;5:247–252. doi: 10.1038/nmeth.1185. [DOI] [PubMed] [Google Scholar]
- 44.Matukumalli LK, et al. Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE. 2009;4:e5350. doi: 10.1371/journal.pone.0005350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao H, Nettleton D, Soller M, Dekkers JCM. Evaluation of linkage disequilibrium measures between multi-allelic markers as predictors of linkage disequilibrium between markers and QTL. Genet Res. 2005;86:77–87. doi: 10.1017/S001667230500769X. [DOI] [PubMed] [Google Scholar]
- 46.Gilmour ARCB, Gogel BJ, Welham SJ, Thompson R. ASReml, User Guide. Release 2.0. UK: VSN International, Hemel Hempstead; 2006. [Google Scholar]
- 47.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caraux G, Pinloche S. PermutMatrix: A graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics. 2005;21:1280–1281. doi: 10.1093/bioinformatics/bti141. [DOI] [PubMed] [Google Scholar]
- 49.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 51.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dennis G, Jr, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 53.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 54.Cartharius K, et al. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 55.Frisch M, Klocke B, Haltmeier M, Frech K. LitInspector: Literature and signal transduction pathway mining in PubMed abstracts. Nucleic Acids Res. 2009;37(Web Server issue):W135–W140. doi: 10.1093/nar/gkp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.