Abstract
In both plants and animals, nucleotide-binding (NB) domain and leucine-rich repeat (LRR)-containing proteins (NLR) function as sensors of pathogen-derived molecules and trigger immune responses. Although NLR resistance (R) proteins were first reported as plant immune receptors more than 15 years ago, how these proteins activate downstream defense responses is still unclear. Here we report that the Toll-like/interleukin-1 receptor (TIR)-NB-LRR R protein, suppressor of npr1-1, constitutive 1 (SNC1) functions through its associated protein, Topless-related 1 (TPR1). Knocking out TPR1 and its close homologs compromises immunity mediated by SNC1 and several other TIR-NB-LRR–type R proteins, whereas overexpression of TPR1 constitutively activates SNC1-mediated immune responses. TPR1 functions as a transcriptional corepressor and associates with histone deacetylase 19 in vivo. Among the target genes of TPR1 are Defense no Death 1 (DND1) and Defense no Death 2 (DND2), two known negative regulators of immunity that are repressed during pathogen infection, suggesting that TPR1 activates R protein-mediated immune responses through repression of negative regulators.
Keywords: histone deacetylase 19, plant immunity, Topless, Topless-related 1
Plant resistance (R) proteins play important roles in defense against pathogens. The majority of R proteins contain either a Toll-like/interleukin 1 receptor (TIR) or a coiled coil (CC) domain at their N terminus domain, a central nucleotide-binding (NB) domain, and C-terminal leucine-rich repeats (LRRs). Downstream components for TIR- and CC-NB-LRR R proteins appear to be different. Mutations in enhanced disease susceptibility 1 (EDS1), phytoalexin deficient 4 (PAD4), and senescence-associated gene101 (SAG101) affect the resistance specified by TIR-NB-LRR but not by CC-NB-LRR R proteins (1–3). On the other hand, mutations in non-race-specific disease resistance 1 (NDR1) compromise resistance mediated by CC-NB-LRR but not by TIR-NB-LRR R proteins (1, 4). EDS1, PAD4, and SAG101 encode three related proteins with homology to acyl hydrolases (3, 5, 6). How these proteins regulate R protein signaling is not clear.
Increasing evidence suggests that certain R proteins accumulate in the nucleus and that the nuclear pools of these R proteins are important for the activation of defense responses (7–10). Multiple TIR-NB-LRR R proteins, including nicotiana glutinosa virus resistance protein (N) in tobacco and resistance to Pseudomonas syringae 4 (RPS4) and suppressor of npr1-1, constitutive 1 (SNC1) in Arabidopsis, have been shown to localize to the nucleus, and reduction of the nuclear R protein pool attenuates the activation of downstream defense responses (7–10). These findings are consistent with that the nucleocytoplasmic trafficking machinery is required for R protein-mediated immunity (9, 11, 12). However, the function of these R proteins in the nucleus and whether they participate directly or indirectly in transcriptional regulation of defense genes is unclear.
Despite tremendous progress has been made in explaining how R proteins recognize the cognate antivirulence (Avr) proteins (13), how R proteins trigger the activation of downstream signaling pathways after the recognition of pathogens remains unknown. Here we show that the TIR-NB-LRR R protein SNC1 functions through association with a transcriptional corepressor, Topless-related 1 (TPR1), and its homologs, which also are required for resistance mediated by other TIR-NB-LRR R proteins.
Results
Knockout of TPR1 Partially Suppresses snc1 Mutant Phenotypes.
Arabidopsis SNC1 encodes a TIR-NB-LRR-type R protein. A point mutation in the snc1 mutant leads to auto-activation of the R protein and enhanced disease resistance (14). An snc1 suppressor screen was carried out previously using fast neutron-treated mutant populations to identify components downstream of R proteins (11). The phenotypes of some identified suppressors were relatively weak, and it was difficult to map those mutations. To resolve this problem, we generated a transfer DNA (T-DNA) insertional mutant population in the snc1 mutant background and screened for mutants that suppress the defense-associated dwarfism of snc1. One of the mutants, modifier of snc1, 10 (mos10), partially suppresses snc1 mutant phenotypes. The snc1-mos10 double mutant is bigger than snc1 but smaller than wild type (Fig. S1A). Levels of salicylic acid (SA) in snc1-mos10 are about half those in snc1 (Fig. S1B). Also, resistance to a virulent isolate of the oomycete pathogen Hyaloperonospora arabidopsidis (H. a.) Noco2 in snc1 is partially blocked in the double mutant (Fig. S1C). The expression level of snc1 in the double mutant is comparable to that in snc1, suggesting that mos10 does not affect snc1 expression (Fig. S1D).
Inverse PCR followed by sequencing revealed that the T-DNA in mos10 resulted in a deletion affecting two genes, At1g80480 and At1g80490 (Fig. S1E). Transforming a genomic clone of At1g80490, but not At1g80480, into snc1-mos10 reverted the mutant morphology to snc1-like (Fig. 1A), suggesting that At1g80490 is MOS10. MOS10 encodes a protein with a Lissencephaly type-1-like homology (LisH) domain at the N terminus, a C-terminal to LisH (CTLH) domain, and 12 WD (tryptophan-aspartic acid)-40 repeats at the C terminus (Fig. 1B). It is closely related to Topless (TPL), which mediates auxin-dependent transcriptional repression during embryogenesis (15, 16). To be consistent with previous literature, the mos10 mutant was renamed topless-related 1 (tpr1).
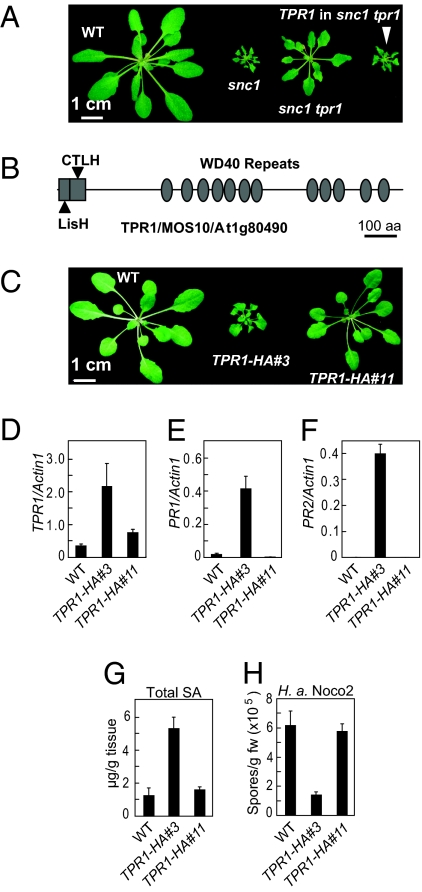
Fig. 1.
TPR1 complements the snc1-tpr1 mutant morphology, and overexpression of TPR1 leads to constitutive activation of defense responses. (A) Complementation of snc1-tpr1 mutant morphology by TPR1 (At1g80490). (B) Structure of TPR1 (MOS10) protein. (C) Morphology of wild-type (WT) and two TPR1-HA transgenic lines. (D) Real-time RT-PCR analysis of TPR1 expression in two TPR1-HA lines. (E and F) Expression of PR1 (E) and PR2 (F) in wild-type and TPR1-HA lines. (G) Total SA levels in wild-type and TPR1-HA transgenic lines. (H) Growth of H. a. Noco2 on wild-type and TPR1-HA lines.
Overexpression of TPR1 Leads to Constitutive Activation of Defense Responses.
When constructs expressing TPR1 with C-terminal HA or GFP tags under the control of its native promoter were transformed into Columbia (Col-0) plants, about one third of the transgenic lines displayed a dwarf phenotype similar to snc1. Similar dwarf plants also were obtained when a genomic clone of At1g80490 without a tag was used for transformation. TPR1 expression was found to be elevated in these plants. We analyzed two representative TPR1-HA transgenic lines in more detail. As shown in Fig. 1C, line #3 is dwarf, whereas line #11 displays wild-type morphology. TPR1 transcript levels in line #3 and line #11 are six and two times the levels of TPR1 in wild-type plants, respectively (Fig. 1D). Analysis of TPR1-HA protein expression using an anti-HA antibody showed that the TPR1-HA level also is considerably higher in line #3 (Fig. S2A). Real-time RT-PCR showed that both pathogenesis-related 1 (PR1) and pathogenesis-related 2 (PR2) are constitutively expressed in line #3 but not in line #11 (Fig. 1 E and F). The total SA level also is much higher in line #3 (Fig. 1G). Moreover, line #3 displays enhanced resistance to H. a. Noco2 (Fig. 1H), suggesting that overexpression of TPR1 leads to activation of defense responses.
Constitutive Defense Responses in TPR1-HA Line #3 Are EDS1- and PAD4-Dependent.
Mutations in EDS1 or PAD4 can suppress the mutant phenotypes of snc1 (14, 17). To test whether activation of defense responses in TPR1-HA line #3 requires EDS1 and PAD4, the eds1-2 and pad4-1 mutations were crossed into TPR1-HA line #3. eds1-2 and pad4-1 largely suppress the dwarfism (Fig. S2B), constitutive PR gene expression (Fig. S2 C and D) and resistance to H. a. Noco2 (Fig. S2E) in TPR1-HA line #3, suggesting that the overexpression effect of TPR1 requires functional EDS1 and PAD4.
TPR1 and TPL Function Redundantly in Regulating snc1-Mediated Resistance Responses.
Arabidopsis TPR1 and TPL share a remarkable 92% identity and 95% similarity at the amino acid level. To test whether TPR1 and TPL function redundantly in defense signaling, we constructed snc1-tpl double-mutant and snc1-tpr1-tpl triple-mutant lines. Although tpl has only a moderate effect on snc1-morphology dwarfism, the tpr1 and tpl mutations combined lead to almost complete suppression of the snc1 dwarfism (Fig. 2A). In addition, the expression levels of PR1 and PR2 and susceptibility to H. a. Noco2 in snc1-tpr1-tpl mutants are comparable to those in wild type (Fig. 2 B–D), suggesting that TPR1 and TPL function redundantly in regulating snc1-mediated resistance.
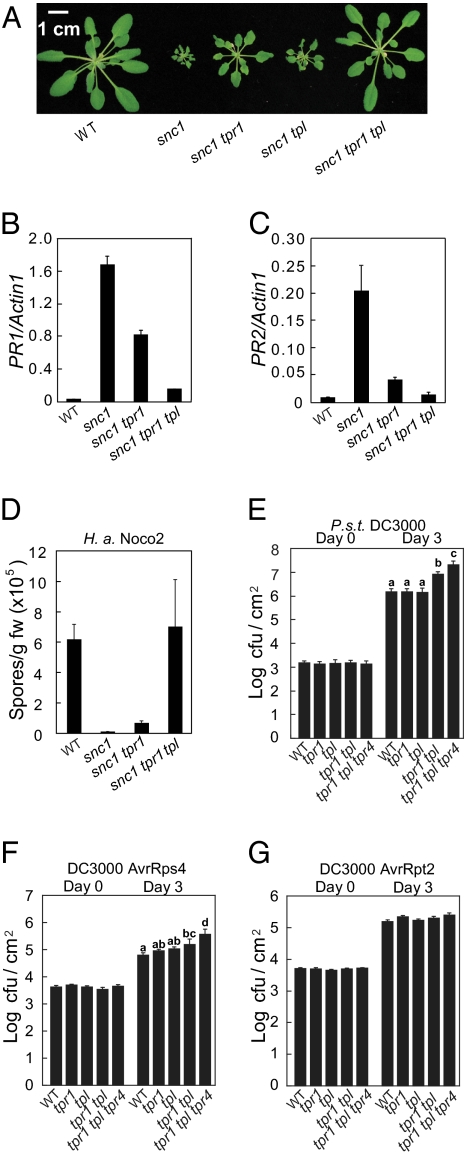
Fig. 2.
TPR1 and its close homologs function redundantly. (A) Suppression of snc1 mutant morphology by the tpr1 and tpl (SALK_097230) mutants. (B and C) Expression of PR1 (B) and PR2 (C) in the indicated genotypes. (D) Growth of H. a. Noco2 on the indicated genotypes. (E–G) Growth of P.s.t. DC3000 (E), P.s.t. DC3000 AvrRps4 (F), and P.s.t. DC3000 AvrRpt2 (G) on the indicated genotypes. tpr4, SALK_150008. Leaves of 5-wk-old plants were infiltrated with P.s.t. DC3000 (OD600 = 0.0002) or P.s.t. DC3000 carrying AvrRps4 or AvrRpt2 (OD600 = 0.001). The bacterial numbers presented are averages of six replicates ± SD. Statistical analyses of the bacterial growth at day 3 were carried out with one-way ANOVA by StatsDirect statistical software (StatsDirect Ltd.). Statistical differences among the samples were labeled with different letters in E and F (P < 0.05). No statistically significant difference was detected for bacterial growth at day 3 for P.s.t. DC3000 AvrRpt2 (G).
TPR1 and Its Close Homologs Are Required for Basal and R Protein-Mediated Resistance.
TPR1 and TPL belong to a protein family with five members. They previously were shown to function redundantly in the regulation of apical fate during embryogenesis (15). The closest homolog of TPR1 and TPL is Topless-related 4 (TPR4), which has 69% identity and 81% similarity to TPL at the amino acid level. To test whether mutations in TPR1, TPL, and TPR4 have an additive effect on pathogen resistance, tpr1-tpl double mutants and tpr1-tpl-tpr4 triple mutants were constructed. When they were infected with Pseudomonas syringae pv. tomato (P.s.t.) DC3000, the double and triple mutants supported higher bacterial growth than wild-type or single-mutant plants, with the triple mutant allowing most bacterial growth (Fig. 2E).
In addition, growth of P.s.t. DC3000 expressing the effector AvrRps4 (recognized by the TIR-NB-LRR R protein RPS4) but not P.s.t. DC3000 expressing AvrRpt2 [recognized by the CC-type NB-LRR R protein resistance to Pseudomonas syringae 2 (RPS2)] was enhanced in the tpr1-tpl double mutant and tpr1-tpl-tpr4 triple mutant (Fig. 2 F and G). We also inoculated H. a. Cala2 on the mutant plants to test whether resistance to this oomycete pathogen strain mediated by the TIR-NB-LRR R protein resistance to Peronospora parasitica 2 (RPP2) is affected. Although wild-type plants produced discrete hypersensitive-response lesions at the pathogen infection sites, trailing necrosis of plant cells was observed on inoculated tpr1 leaves (Fig. S3). In the tpr1-tpl and tpr1-tpl-tpr4 mutant plants, growth of pathogen hyphae beyond the sites of trailing necrosis was observed, suggesting that RPP2-mediated resistance is partially compromised in these mutants (Fig. S3). Taken together, these results show that TPR1, TPL, and TPR4 function redundantly in regulating basal defense and resistance mediated by several TIR-NB-LRR–type R proteins.
SNC1 Is Required for the Constitutive Activation of Defense Responses in the Transgenic Plants Overexpressing TPR1.
To identify proteins that function together with TPR1, we mutagenized seeds from the TPR1-HA line #3 (in Col-0 background) with ethyl methanesulfonate (EMS) and screened for mutants suppressing the dwarf phenotype of the transgenic plants. About 40 suppressor mutants were identified from the screen. In attempts to map two of the mutants using F2 progeny from the crosses between the mutants and Landsberg (Ler) ecotype, we found that there is a natural modifier of the overexpression phenotype of TPR1 from Ler that is closely linked to the SNC1 locus. This observation prompted us to test whether SNC1 is required for the overexpression phenotype of TPR1. Sequencing analysis of eight suppressor mutants showed that three of them contain mutations in SNC1 (Fig. S4 A and B). In these mutants, constitutive PR gene expression also was suppressed (Fig. S4 C and D).
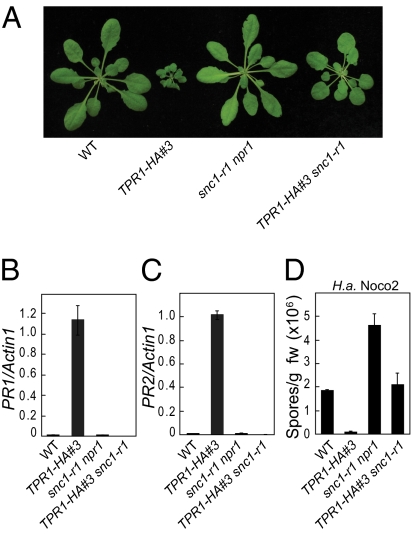
To confirm that SNC1 is required for the constitutive defense responses in TPR1-HA line #3, snc1-r1-npr1 (a loss-of-function deletion allele of SNC1) was crossed with TPR1-HA #3 to generate the TPR1-HA #3 snc1-r1 double mutant. As shown in Fig. 3A, the dwarf phenotype of the transgenic line is largely suppressed by the snc1-r1 mutation. In addition, constitutive PR gene expression (Fig. 3 B and C) and enhanced resistance to H.a. Noco2 (Fig. 3D) in TPR1-HA line #3 are completely suppressed by snc1-r1, suggesting that the overexpression effect of TPR1 requires a functional SNC1.
Fig. 3.
SNC1 is required for activation of defense responses caused by TPR1 overexpression. (A) Suppression of the dwarfism of TPR1-HA line #3 by snc1-r1. (B and C) Suppression of constitutive PR1 (B) and PR2 (C) expression in TPR1-HA line #3 by snc1-r1. (D) Suppression of enhanced resistance to H. a. Noco2 in TPR1-HA line #3 by snc1-r1.
SNC1 Associates with TPR1 in Vivo.
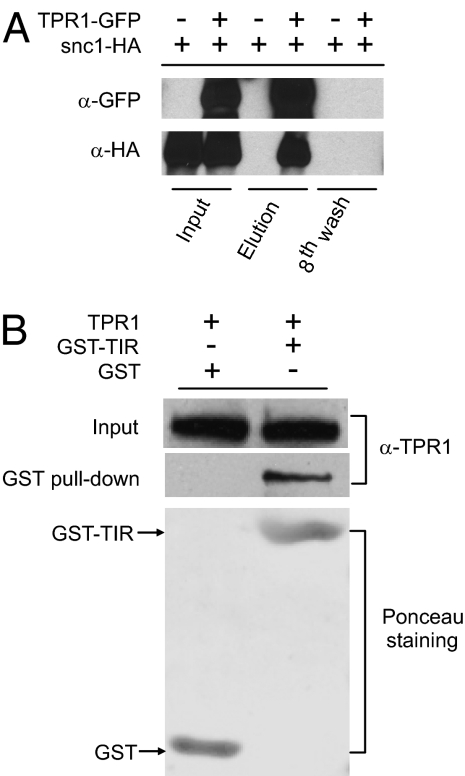
The genetic interaction between SNC1 and TPR1 prompted us to investigate the biochemical interactions between the two proteins. To test whether TPR1 associates with the snc1 mutant protein in vivo, we crossed a snc1-HA transgenic line with a TPR1-GFP transgenic line. Both snc1-HA and TPR1-GFP were expressed under their native promoters. Then coimmunoprecipitation analysis was performed using the F2 plants that carry both transgenes. When total protein extracts of these plants were incubated with anti-GFP magnetic beads to immunoprecipitate the GFP-tagged TPR1 selectively, snc1-HA coimmunoprecipitated with the GFP-tagged TPR1, whereas snc1-HA was not immunoprecipitated from the protein extract of snc1-HA transgenic plants using the same anti-GFP magnetic beads (Fig. 4A). Similar results were obtained when the nuclear protein extracts were used for the coimmunoprecipitation analysis (Fig. S4E). These results suggest that TPR1 and snc1 associate with each other in vivo.
Fig. 4.
SNC1 interacts with TPR1. (A) Coimmunoprecipitation of snc1-HA with TPR1-GFP in protein extracts of TPR1-GFP and snc1-HA double-tagged transgenic plants. Total protein extracts were subjected to immunoprecipitation with anti-GFP magnetic beads as previously described (32). Crude lysates (Input) and immunoprecipitated proteins (elution) were detected with anti-GFP or anti-HA antibodies. (B) In vitro analysis of the interaction between TPR1 and the TIR domain (amino acid 1–182) of SNC1. Crude lysates of E. coli expressing GST-tagged TIR domain of SNC1 (GST-TIR) or GST were mixed with crude lysates of E. coli expressing TPR1 before GST pull-downs. Aliquots of the mixtures (input) and proteins pulled down by GST were subjected to anti-TPR1 immunoblot analysis. GST-TIR and GST were detected by Ponceau staining. The polyclonal anti-TPR1 antibody was generated in rabbit using an N-terminal fragment (amino acid 1–356) of TPR1 expressed in E. coli.
The TIR Domain of SNC1 Interacts with TPR1.
Recently it was reported that the TIR domains of several TIR-NB-LRR R proteins are sufficient to induce cell death (18). To test whether the TIR domain of SNC1 interacts with TPR1, we expressed the TIR domain of SNC1 with a GST tag and full-length TPR1 with a 6×His tag in Escherichia coli. Although the GST-tagged TIR domain expressed well, and we were able to obtain large quantity of the protein, the amount of TPR1 expressed in E. coli is very small. We subsequently performed in vitro GST pull-down assays using the E. coli-expressed proteins. As shown in Fig. 4B, TPR1 copurified with the GST-tagged TIR domain but not with GST, suggesting that the TIR domain of SNC1 interacts with TPR1.
TPR1 Functions as a Transcriptional Corepressor and Associates with Histone Deacetylase 19 in Vivo.
When constructs expressing the TPR1-GFP or TPR1-HA fusion proteins under its native promoter were transformed into snc1-tpr1-tpl, most transgenic plants displayed snc1 morphology, suggesting that the fusion proteins are functional (Fig. S5 A and B). Fluorescence microscopy revealed that the fusion protein was localized to the nucleus (Fig. S5 C and D).
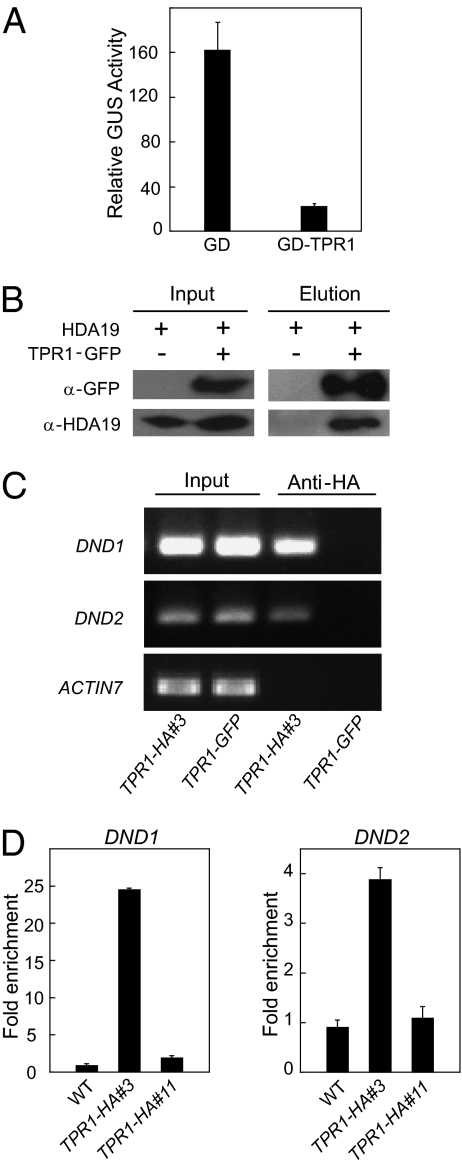
Previously, TPL was shown to function as a transcriptional corepressor (16). To test whether TPR1 also serves as a transcriptional corepressor, we used a protoplast transient assay established for studying the transcriptional repression by Aux/IAA (19). In this assay, the β-glucuronidase (GUS) reporter gene is driven by the –46 35S promoter containing 2×LexA and 2×Gal4 DNA-binding sites. Activation of the GUS reporter gene is achieved by cotransformation with a 35S-driven transactivator gene encoding a chimeric protein consisting of the LexA DNA-binding domain and the HSV VP16 activation domain (LD-VP16). Cotransformation of TPR1 fused to the Gal4 DNA-binding domain together with the reporter constructs resulted in repression of GUS expression (Fig. 5A), indicating that TPR1 functions as a transcriptional corepressor.
Fig. 5.
TPR1 functions as a transcriptional corepressor and is targeted to the promoters of DND1 and DND2. (A) Relative GUS activities in Arabidopsis mesophyll protoplasts cotransfected with a GUS reporter gene, LD-VP16, and constructs expressing GAL4 DNA-binding domain (GD) or a GD-TPR1 fusion were shown. A 35S-driven luciferase reporter was included in the assays as internal transfection controls. Diagrams of the constructs used in the assays are shown in Fig. S6. (B) Coimmunoprecipitation of HDA19 with TPR1-GFP in nuclear extracts of TPR1-GFP transgenic plants. Nuclear protein extracts were subjected to immunoprecipitation with anti-GFP magnetic beads. Crude lysates (Left, Input) and immunoprecipitated proteins (Right) were detected with anti-GFP or anti-HDA19 antibodies. (C) Semiquantitative PCR analysis of promoter fragments of DND1, DND2, and Actin7 after ChIP. ChIP was performed on TPR1-HA line #3 using anti-HA antibody. TPR1-GFP transgenic plants were used as negative controls. (D) ChIP analysis of recruitment of TPR1 to the promoter of DND1 (Left) and DND2 (Right) in wild-type plants and two TPR1-HA transgenic lines. TPR1-HA was overexpressed in line #3 but not in line #11. ChIP was performed using anti-HA antibody. The amount of DND1 and DND2 promoter DNA from ChIP was determined by real-time PCR. The fold of enrichment was obtained by dividing the amount of DNA from ChIP with anti-HA antibody by that from control ChIP with no antibody added.
Arabidopsis histone deacetylase 19 (HDA19) was suggested to function together with TPL in regulating plant developmental processes because some hda19 phenotypes also are observed in the tpl mutant (15). Knocking out HDA19 also leads to compromised pathogen resistance (20). These observations prompted us to test whether TPR1 associates with HDA19. To test that possibility, the TPR1-GFP fusion protein was immunoprecipitated from nuclear extracts of TPR1-GFP transgenic plants using a GFP antibody as described above. Endogenous HDA19 was found to be coimmunoprecipitated with the TPR1-GFP protein (Fig. 5B), suggesting that TPR1 associates with HDA19 in vivo.
Identification of TPR1 Target Genes in Defense Responses.
Because TPR1 is a transcriptional corepressor, and loss of TPR1 function results in compromised immunity, its target genes probably are negative regulators that are repressed during immune responses. The repression of these target genes is most likely EDS1- and PAD4-dependent. To identify defense-related target genes of TPR1, we first analyzed data from microarray experiments used to identify genes with EDS1- and PAD4-dependent expression changes after bacterial pathogen infections (21). Inoculation of P.s.t. DC3000 AvrRps4 resulted in EDS1-dependent repression of a set of genes. We then tested whether TPR1 is targeted to the promoters of these genes with ChIP using TPR1-HA transgenic plants. Real-time PCR was used to determine whether promoter fragments of selected genes were enriched by ChIP with an HA antibody. TPR1 was recruited to the promoters of 12 of the 48 genes tested (Fig. S7), suggesting that these 12 genes probably are direct targets of TPR1.
Among the 12 genes identified, Defense no Death 1 (DND1) and Defense no Death 2 (DND2) are known negative regulators of plant innate immunity (22–24). ChIP followed by semiquantitative PCR also showed that TPR1 was recruited to the promoters of DND1 and DND2 (Fig. 5C). To test whether the expression level of TPR1 affects its recruitment to the target promoters, we performed additional ChIP analysis on two different TPR1-HA transgenic lines. TPR1-HA was expressed to a higher level in line #3 than in line #11 (Fig. S2A). As shown in Fig. 5D, binding of TPR1 to the promoters of DND1 and DND2 was observed clearly in line #3, but not in line #11, suggesting that overexpression of TPR1 leads to increased association of the protein with the target promoters.
The rapid repression of DND1 and DND2 after inoculation with P.s.t. DC3000 AvrRps4 was confirmed further by real-time RT-PCR analysis. As shown in Fig. S8 A and B, this repression relies on functional EDS1. In snc1 and TPR1-HA line #3, repression of DND1 and DND2 is not as clear as the repression by infection with P.s.t. DC3000 AvrRps4 (Fig. S8 C–F), suggesting that repression of DND1 and DND2 during defense responses may be transient.
Discussion
Our study showed that the transcriptional corepressor TPR1 and its close homologs function as critical regulators of TIR-NB-LRR R protein-mediated resistance. Knocking out TPR1 and its close homolog TPL suppresses the constitutive activation of immune responses in the auto-activated R gene mutant snc1 and compromises resistance mediated by several other TIR-NB-LRR R proteins but not by the CC-NB-LRR R protein RPS2. In addition, overexpression of TPR1 constitutively activates immune responses that are fully dependent on EDS1 and PAD4, further indicating that TPR1 is a regulator of TIR-NB-LRR but not of CC-NB-LRR R protein-mediated immunity.
TPR1 is structurally related to Transducin beta-like protein 1 (TBL1) and its receptor TBLR1, which also contain the N-terminal LisH domain and C-terminal WD-40 repeats. TBL1 and TBLR1 are part of large protein complexes containing the nuclear receptor corepressor (N-CoR), the silencing mediator for retinoic and thyroid receptors (SMRT), and histone deacetylase 3 (HDAC3) that function as corepressors for nuclear receptors such as thyroid hormone receptors and retinoic acid receptors (25, 26). Our data show that the plant TPR1 complex also contains HDA19. Like TBL1, TPR1 serves as a transcriptional corepressor when it is targeted to the promoter of a reporter gene. Similarly, TPL, the close homolog of TPR1, also functions as a transcriptional corepressor in auxin-dependent transcriptional repression during embryogenesis in Arabidopsis (16).
From ChIP analysis, we identified DND1, DND2, and several other EDS1-regulated genes as target genes of TPR1. Loss-of-function mutations in either of these genes led to constitutive activation of resistance responses similar to that observed in snc1 or other deregulated R gene mutants (22–24, 27), suggesting that transcriptional repression of DND1, DND2, and possibly other negative regulators by TPR1 is a mechanism of activating R protein-mediated immune responses (Fig. 6).
Fig. 6.
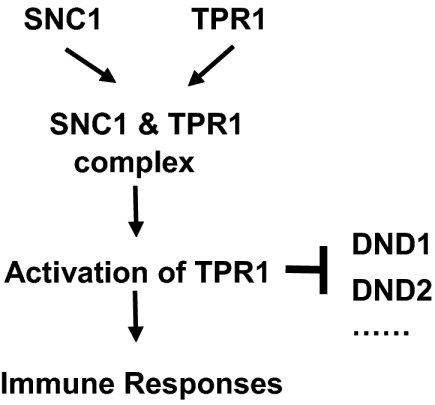
A model for SNC1-mediated defense activation through repression of negative regulators of defense. When plants are not under pathogen attack, R protein-mediated immune responses are repressed by DND1, DND2, and other negative regulators. The mutation in snc1 or overexpression of TPR1 leads to association of SNC1 and TPR1 in a protein complex. The association of SNC1 and TPR1 results in activation of the TPR1 corepressor. Activated TPR1 represses the expression of negative regulators of defense responses, which leads to activation of immune responses.
Both genetic and biochemical evidence suggests that TPR1 and SNC1 function together in a protein complex in the regulation of defense responses. Not only do snc1-mediated defense responses require TPR1 and its homolog TPL, but activation of defense responses in transgenic plants overexpressing TPR1 also requires functional SNC1. A remaining question is how the mutation in snc1 activates TPR1-dependent defense responses. One possibility is that the mutation located in the NL linker in snc1 changes the conformation of the protein and makes its TIR domain more accessible to the binding of TPR1. Increased formation of the TPR1 and SNC1 complex subsequently leads to activation of TPR1 and downstream signaling. Constitutive defense responses observed in transgenic plants overexpressing SNC1 or TPR1 probably also are caused by increased association of TPR1 and SNC1 because of elevated SNC1 or TPR1 protein levels.
The interaction of SNC1 and TPR1 suggests a model (Fig. 6) in which SNC1 activates downstream defense responses by modulating the transcriptional repression activity of TPR1, which targets negative regulators of immune responses. Our data suggest that TIR-NB-LRR R proteins participate directly in transcriptional reprogramming of downstream defense genes. Suppression of negative regulators, rather than direct activation of positive regulators, may be the driving force for the initiation of TIR-NB-LRR R protein-mediated immunity.
Methods
Mutant Isolation.
The snc1-mos10 mutant was isolated from a T-DNA–mutagenized population consisting of about 60,000 independent T1 transgenic lines generated by transforming snc1 with pSKi015. About 1.2 million T2 plants were analyzed for suppression of snc1 morphology. The tpl and tpr4 mutants were obtained from the Arabidopsis Biological Resource Center. The mos10 (later renamed tpr1) single mutant was obtained by backcrossing snc1-mos10 to Col-0 wild-type plants. The snc1-tpl, snc1-tpr1-tpl, and tpr1-tpl mutant plants were obtained by crossing tpl with snc1-tpr1. The tpr1-tpl -tpr4 triple mutant was obtained by crossing tpr4 with tpr1-tpl. The suppressor mutants of the TPR1 overexpression line were isolated from an EMS-mutagenized population.
Complementation of the snc1-mos10 Double Mutant.
An 8.6-kb genomic fragment containing At1g80490 was amplified with primers 5′-cggggtaccgaccataatttagttcaggcg-3′and 5′-gaagcaacaagtgacccatc-3′ by PCR from wild-type genomic DNA and cloned into the binary vector pCAMBIA1300 to create pCAMBIA1300-MOS10g. A similar genomic DNA fragment without the stop codon and 3′ UTR was cloned into modified pCAMBIA1305 vectors to obtain pCAMBIA1305-MOS10-HA and pCAMBIA1305-MOS10-GFP for expressing the TPR1 fusion proteins under the control of its own promoter. The plasmids were electroporated into Agrobacterium and subsequently transformed into the snc1-mos10 double mutant by floral dipping (28).
Expression Analysis and Pathogen Infections.
For gene expression analysis, RNA was extracted from 2-wk-old seedlings grown on MS medium at 22 °C under 16-h/8-h light/dark cycles using Takara RNAiso reagent. Reverse transcription was carried out using the M-MLV RTase cDNA synthesis kit from Takara. Real-time PCR was performed using the SYBR Premix Ex (Takara). SA was extracted and measured by HPLC using a previously described procedure (29). Infection of plants with various strains of P.s.t. DC3000 was carried out by infiltrating bacterial suspensions into leaves of 5-wk-old plants grown at 22 °C under 12-h/12-h light/dark cycles. Infection of H. a. Noco2 was performed by spraying 2-wk-old seedlings grown at 22 °C under 16-h/8-h light/dark cycles with a H. a. Noco2 spore suspension at a concentration of 5 × 104 spores/mL water and scored as previously described (30).
ChIP Analysis.
About 2 g of 2-wk-old MS plate-grown seedlings were harvested and used for ChIP analysis with HA antibody (11867423001; Roche). ChIP was performed as described previously (31). The immunoprecipitated DNA was resuspended in TE buffer (10 mM Tris-HCl, pH 7.5; 1 mM EDTA, pH 8.0) and subjected to real-time PCR analysis. Primers used to amplify the promoters of the target genes are listed in Table S1.
Supplementary Material
Acknowledgments
We thank Dr. Jane Parker (Max Plank Institute for Plant Breeding Research, Koln, Germany) for providing seeds of eds1-2 in Col and thoughtful comments on the manuscript and Dr. Shucai Wang (University of British Columbia, Vancouver, BC, Canada) for providing plasmid vectors for transcriptional repression analysis. Y. Z. received financial support from the Chinese Ministry of Science and Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002828107/-/DCSupplemental.
References
- 1.Aarts N, et al. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feys BJ, Moisan LJ, Newman MA, Parker JE. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001;20:5400–5411. doi: 10.1093/emboj/20.19.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feys BJ, et al. Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell. 2005;17:2601–2613. doi: 10.1105/tpc.105.033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Century KS, Holub EB, Staskawicz BJ. NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc Natl Acad Sci USA. 1995;92:6597–6601. doi: 10.1073/pnas.92.14.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jirage D, et al. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA. 1999;96:13583–13588. doi: 10.1073/pnas.96.23.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falk A, et al. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA. 1999;96:3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burch-Smith TM, et al. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 2007;5:e68. doi: 10.1371/journal.pbio.0050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wirthmueller L, Zhang Y, Jones JD, Parker JE. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol. 2007;17:2023–2029. doi: 10.1016/j.cub.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 9.Cheng YT, et al. Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell. 2009;21:2503–2516. doi: 10.1105/tpc.108.064519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen QH, et al. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315:1098–1103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Li X. A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell. 2005;17:1306–1316. doi: 10.1105/tpc.104.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palma K, Zhang Y, Li X. An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr Biol. 2005;15:1129–1135. doi: 10.1016/j.cub.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 13.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Goritschnig S, Dong X, Li X. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell. 2003;15:2636–2646. doi: 10.1105/tpc.015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006;312:1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- 16.Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Clarke JD, Zhang Y, Dong X. Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact. 2001;14:1131–1139. doi: 10.1094/MPMI.2001.14.10.1131. [DOI] [PubMed] [Google Scholar]
- 18.Swiderski MR, Birker D, Jones JD. The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol Plant Microbe Interact. 2009;22:157–165. doi: 10.1094/MPMI-22-2-0157. [DOI] [PubMed] [Google Scholar]
- 19.Tiwari S, Wang S, Hagen G, Guilfoyle TJ. Transfection assays with protoplasts containing integrated reporter genes. Methods Mol Biol. 2006;323:237–244. doi: 10.1385/1-59745-003-0:237. [DOI] [PubMed] [Google Scholar]
- 20.Kim KC, Lai Z, Fan B, Chen Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 2008;20:2357–2371. doi: 10.1105/tpc.107.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartsch M, et al. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu IC, Parker J, Bent AF. Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci USA. 1998;95:7819–7824. doi: 10.1073/pnas.95.13.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balagué C, et al. HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell. 2003;15:365–379. doi: 10.1105/tpc.006999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurkowski GI, et al. Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol Plant Microbe Interact. 2004;17:511–520. doi: 10.1094/MPMI.2004.17.5.511. [DOI] [PubMed] [Google Scholar]
- 25.Li J, et al. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 27.Clough SJ, et al. The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci USA. 2000;97:9323–9328. doi: 10.1073/pnas.150005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Zhang Y, Clarke JD, Li Y, Dong X. Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell. 1999;98:329–339. doi: 10.1016/s0092-8674(00)81962-5. [DOI] [PubMed] [Google Scholar]
- 30.Bi D, Cheng YT, Li X, Zhang Y. Activation of plant immune responses by a gain-of-function mutation in an atypical receptor-like kinase. Plant Physio. 2010 doi: 10.1104/pp.110.158501. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palma K, et al. Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 2007;21:1484–1493. doi: 10.1101/gad.1559607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.