Summary
Class IIa HDACs including HDAC7 play a role in gene expression, cell differentiation, and animal development through their association with transcription factors such as myogenic enhancer factors 2 (MEF2s). In this study, we show that endogenous HDAC7 localizes to both the nucleus and the cytoplasm of C2C12 myoblasts, but is exclusively retained in the cytoplasm of myotubes after completion of differentiation process. To elucidate the role of differential distribution of HDAC7 during myogenesis, we examined the effects of stably expressed HDAC7 mutants on myogenesis. Expression of nuclear-retained HDAC7 mutants significantly inhibits myogenesis in C2C12 cells and reduces the expression of muscle-specific myosin heavy chain (MHC) and myogenin. The inhibition in myocyte differentiation can be partially relieved by introduction of a mutation disrupting HDAC7:MEF2 interaction. Since phosphorylation of HDAC7 plays an important role in its nucleocytoplasmic shuttling, we further investigated the expression and distribution of phosphorylated HDAC7. To our surprise, the phosphorylation levels of HDAC7 at S344 and S479 were slightly decreased upon differentiation, whereas the phosphorylation of S178 was unchanged. Interestingly, a significant fraction of pS344- and/or pS479-HDAC7 localizes to plasma membrane of myotubes. In addition, Ser178-phosphorylated (pS178) HDAC7 shows a predominant actin filament-like staining prior to muscle differentiation and cytoplasmic and plasma membrane staining after differentiation. Consistent with this notion, HDAC7 partially co-localizes with actin filaments; in particular, pS178-HDAC7 largely colocalizes with actin filaments as indicated by phalloidin counter staining in myocytes. Furthermore, C2C12 cells expressing nuclear-retained HDAC7 display defects in migration. Our results provide novel insight into the mechanisms that regulate myocyte differentiation and migration by controlling the subcellular distribution of HDAC7 in differentiating myoblasts.
Keywords: Nucleocytoplasmic shttling, histone deacetylase 7 (HDAC7), phosphorylation, myocyte differentiation, cell migration, myogenin
Introduction
Class IIa HDACs, HDAC4, -5, -7, and -9, are abundantly expressed in cardiac and skeletal muscle [1-3] and are mistakenly thought to function interchangeably. They contain a highly conserved catalytic domain but differ widely outside this region. All class IIa HDACs can repress transcriptional activity of myocyte enhancer factor-2 (MEF2) proteins [4-11]. MEF2s play important roles during muscle differentiation [12-17] by up-regulating many key muscle-specific genes. Class IIa HDACs are subject to regulation by covalent modification and nucleocytoplasmic trafficking [18]. As such, control of their subcellular localization may constitute an essential mechanism to regulate muscle differentiation.
Skeletal muscle differentiation is an ordered temporal process that can be recapitulated in vitro [19,20]. Studies of HDAC4 and HDAC5 in C2C12 myoblast differentiation suggest that their subcellular localization during myoblast differentiation is subjected to distinct regulation. HDAC5 is distributed in both the nucleus and the cytoplasm of myoblasts, but is localized exclusively in the cytoplasm of multinucleated myotubes and mis-localization of HDAC5 inhibits myoblast differentiation [21]. In contrast, HDAC4 is predominantly localized in the cytoplasm of myoblasts and translocates into the nucleus in differentiated myotubes [16,22]. These studies suggest that HDAC4 and HDAC5 are differentially regulated and may have distinct functions and also highlight the importance of the mechanisms underlying the subcellular localization of class IIa HDACs during myocyte differentiation.
Nucleocytoplasmic shuttling of class IIa HDACs involves dynamic interplay between nuclear import and export. In addition, we have recently shown that in response to extracellular signals, sequestration of HDAC7 to PML NBs may represent a subnuclear mechanism that prevents HDAC7 from repressing the expression of its target genes [23]. Nuclear localization of class IIa HDACs is mediated through a nuclear localization sequence (NLS) and their association with importin α [21,24-26]. Phosphorylation of class IIa HDACs at conserved serine residues by kinases such as CaMK I/IV [5], protein kinase D (PKD) [27-30], Mirk/dyr1B [31], (MARK)-Par-1 kinase [32], EMK and C-TAK1 [33] may inhibit nuclear import and/or promote nuclear export, thereby increasing their cytoplasmic accumulation [5,25]. This is likely achieved through the association of class IIa HDACs with 14-3-3 family proteins. Unphosphorylatable class IIa HDACs fail to interact with 14-3-3 proteins and are preferentially localized in the nucleus. This observation is consistent with the notion that 14-3-3 binding blocks the recognition of the nearby NLS on class IIa HDACs to inhibit their nuclear import [26].
We and others have shown that a C-terminal nuclear export sequence (NES) is absolutely essential for nuclear export of class II HDACs [5,21,26,34]. Treatment of cells with leptomycin B (LMB), a CRM1-specific inhibitor, or mutations at the NES restrict localization of class II HDACs to the nucleus [5,25]. Furthermore, overexpression of CRM1 potently promotes cytoplasmic localization of wild-type HDAC7 and the unphosphorylatable mutant, S/A, but not the NES mutant in HeLa cells [26]. These data suggest that phosphorylation at S178, S344, and S479 is not essential for cytoplasmic localization of HDAC7.
Despite the importance of the role of phosphorylated class II HDACs in myocyte differentiation, little is known in the dynamics of their subcellular distribution. In this study, we have examined the subcellular distribution of endogenous HDAC7 and phosphorylated class II HDACs during myocyte differentiation. To our surprise, phosphorylation of HDAC7 was not increased during myocyte differentiation. We further generated stable cell lines that constitutively express wild-type or HDAC7 mutants and examined their effects on myocyte differentiation. Our studies detail the mechanism underlying the subcellular distribution and establish unique requirements of HDAC7 in myoblast migration and myotube formation.
Materials and Methods
Plasmid construction
HDAC7 expression plasmids, CMX-FLAG-mHDAC7 (S/A which is S178A/S344A/S479A), CMX-FLAG-mHDAC7 (mNES), CMX-FLAG-mHDAC7 (AWA which is D692A/D694A) and CMX-FLAG-mHDAC7 (ΔNLS, in which amino acids 183-212 were removed) have been previously described [5,26,35]. Site-directed mutagenesis was carried out to generate HDAC7 (SN which is S/A + mNES), HDAC7 (SNL which is S/A + mNES + L112A), and HDAC7 (SNA which is S/A + mNES + AWA) according to manufacturer's protocol (Stratagene). The pBabe-FLAG-mHDAC7 expression plasmids and their mutant derivatives were generated from PCR reactions using CMX-FLAG-mHDAC7 as templates and subcloning into the pBabe vector according to manufacturer's protocol (Invitrogen). All constructs were verified by DNA sequencing.
Antibodies
HDAC7 antibodies and their properties have been previously described [23,26,36]. The specificity of the phospho-antibodies has been previously examined in HeLa cells [26] and is also examined in C2C12 myocytes in Supplementary Figure 3. Anti-α-tubulin antibody was purchased from Roche (2013819), while anti-FLAG (F3165) antibodies were purchased from Sigma. Anti-actin antibodies were purchased from Santa Cruz (sc-1615, sc-8432 and sc-10731) or Sigma (A5441). Rhodamin-conjugated phalloidin was purchased from Cytoskeleton (PHDR1). The myosin heavy chain (MHC) antibody (MF20) was purchased from Developmental Studies Hybridoma Bank, Iowa City, IA. Anti-Myogenin antibodies were purchased from Santa Cruz (sc-12732).
C2C12 proliferation and differentiation
Differentiation of parental and stable C2C12 myoblasts was carried out following the same protocol. Monolayers of C2C12 myoblasts and stably transfected derivatives were maintained in growth medium (GM) consisting of Dulbecco's Modified Eagle's Medium (DMEM) and 20% fetal bovine serum (FBS). Induction of C2C12 differentiation was achieved by replacing the growth medium with differentiating medium (DM) (DMEM, 2% Horse serum, and 1% Penicillin streptomycin) when the cells reached 80% confluence. The cells were fed once every two days thereafter until harvesting.
Subcellular localization of transiently transfected HDAC7 in C2C12 cells
C2C12 myoblasts were transiently transfected with YFP, YFP-HDAC7 (WT), or mutant YFP-HDAC expression plasmids followed by confocal microscopy. To evaluate the effects of CaMK and CRM1 on the subcellular distribution of HDAC7, an empty vector, CaMK, or CRM1 expression plasmid was co-transfected with wild-type or mutant HDAC7 expression plasmid.
Generation of stable C2C12 cells expressing HDAC7 mutants
C2C12 myoblasts were stably transfected with pBabe-FLAG-HDAC7 (WT) or the HDAC7 mutants described above to generate the cell lines C2C12-FLAG-HDAC7. The C2C12-pBabe (vector alone) cell line was used as a negative control in which C2C12 myoblasts were stably transfected with the empty vector (pBabe-3F vector). After transfection, stable cells were selected on puromycin (2 μg/ml) for 10 days. The stable cell lines are maintained in GM + 0.5 μg/ml puromycin. For differentiation, cells were grown to 80% confluence and switched to DM + 0.5 μg/ml puromycin for up to 5 days.
Immunofluorescence and confocal microscopy
C2C12 cells were washed in 1x PBS, fixed for 30 min with freshly prepared 3.7% paraformaldehyde in PBS. Fixed cells were washed with 1x PBS, followed by permeabilization with a 10-min treatment with permeablization buffer (10% goat serum, 1% triton-X 100, 1x PBS). After 1 hr of incubation with a blocking buffer (10% goat serum, 0.5% tween20, 1x PBS), cells were incubated for 2 hrs with the indicated antisera diluted 1:500 in blocking buffer. Samples were washed three times with 1x PBS, mounted for microscopy in mounting solution with DAPI (Vector Laboratories, Inc.) and stained with anti-FLAG, anti-HDAC7 or anti-phosphorylated HDAC7 antibodies and MHC antibodies. Following primary antibody incubation, samples were incubated with fluorochrome-coupled secondary antibodies (anti-rabbit Alexa Fluor 488 or anti-mouse Alexa Fluor 594 from Invitrogen). Epi- and confocoal microscopy were conducted according to our published protocols [36].
Preparation of whole cell lysates, immunoblotting
C2C12 myoblasts or myocytes were harvested, lysed in NETN buffer (50 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol, 0.5% NP-40, 1 mM PMSF, phosphatase inhibitor cocktail (Roche), and protease inhibitor cocktail (Roche)) for 15 min at 4°C, scraped, and centrifuged 15 min at 13,000 rpm. The cell lysates were loaded onto SDS-PAGE gels and then transferred to a nitrocellulose membrane. The proteins of interest on the membrane were detected using the indicated antibodies by Western blotting. To determine phosphorylation of HDAC7, whole cell extracts were probed with anti-HDAC7, anti-pS178, anti-pS344, or anti-pS479 antibodies.
Subcellular fractionation of myocytes and myotubes
Myocytes and myotubes were collected at Day 0 (D0) and 4 (D4) after grown in differentiation medium, respectively. Subcellular fractionation of cell lysates was performed according to a published protocol [37].
GST pulldown assays
The expression of GST-MEF2A (1-86) and GST-MEF2A (1-243) fusion proteins and GST pulldown assays were carried out according to our published protocol [38]. Immobilized GST fusion proteins were incubated with HA-HDAC7 (SN) or HDAC7 (SNL) expressed in C2C12 myocytes. The pulldown fractions were subjected to Western blotting with anti-HA antibody.
Scratch migration assays
C2C12 stable cells were grown to monolayer 100% confluence and scratched vertically with a sterile 100 ml pipette tip. Cells were then washed twice to remove cellular debris and incubated in the same growth medium. To standardize the position of the wound for photography, short lines were marked underneath the plates. The same wound fields were observed and images taken under microscope (TELAVAL31, ZEISS, Germany) equipped with SPOT LIGH camera (Model 3.2.0) at 0 and 7 hrs after wounding. The percentage of migration was then calculated by using Photoshop to measure the width of the clear zone. The percent migration at 7 hrs was calculated by subtracting the width of the scratch at 7 hrs from the total width of the scratch at 0 hrs, divided by the total width at 0 hr multiplied by 100.
Results
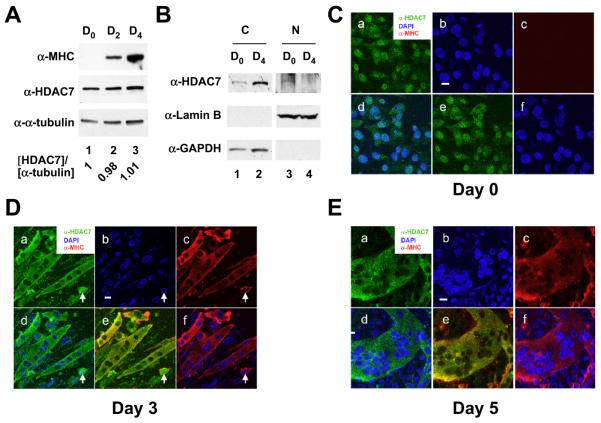
We have previously shown that mouse HDAC7 is highly expressed in cardiac and skeletal muscle [2], implying that it has a physiological role in muscle. To test this, we first examined the expression of HDAC7 during C2C12 myocyte differentiation and found that the expression levels of HDAC7 during differentiation at day 0, 2, and 4 were relatively constant (Fig. 1A). We found that HDAC7 protein level was not significantly different upon differentiation. In order to investigate whether the subcellular distribution of HDAC7 is altered during C21C2 differentiation, we performed subcelular fractionation of C2C12 myocytes and myotubes and found that no nuclear HDAC7 was detected in differentiated myotubes (Fig. 1B). We further carried out immunostaining and confocal microscopy in C2C12 myoblasts and myotubes. At day 0, prior to the induction of differentiation, HDAC7 was localized both in the nucleus and the cytoplasm (Fig. 1C, panels a-f). C2C12 cells were induced to differentiate by serum withdrawal for up to 5 days. Differentiation of myoblasts was evaluated by monitoring the expression of myosin heavy chain (MHC). Terminally differentiated myotubes were determined by their multinucleate phenotype. Upon differentiation, HDAC7 was redistributed primarily to the cytoplasm of myotubes from a nascent stage (with an average of 4 nuclei per myotube) at day 3 to a mature stage (with more than 50 nuclei per myotube) at day 5 (Fig. 1D-E). In the majority of C2C12 cells that express MHC, HDAC7 was exclusively localized in the cytoplasm. However, a few cells remained that displayed MHC staining with nuclear HDAC7 (Fig. 1D, white arrow). We also noticed that HDAC7 was localized to the cytoplasmic membrane in some myotubes (see below). Overall, these results demonstrate that muscle differentiation coincides with HDAC7 redistribution to the cytoplasm.
Fig. 1.
Subcellular localization of HDAC7 during myogenesis. (A) The expression levels of HDAC7 during C2C12 differentiation. C2C12 myoblasts were induced to differentiate and cells were harvested at the indicated times. Whole cell extracts were prepared and subjected to immunoblotting with anti-MHC, anti-HDAC7, and anti-α-tubulin antibodies. The relative expression levels of HDAC7 at day 0, 2, or 4 post differentiation are normalized by α-tubulin and are shown. (B) Subcellular fractionation of C2C12 myocytes and differentiated myotubes. Undifferentiated myocytes (D0) and differentiated myotubes (D4) were harvested and subcellular fractions prepared as described in “Materials and Methods”. Cytoplasmic and nuclear fractions of equal cell numbers were loaded followed by SDS-PAGE and Western blotting with anti-HDAC7, anti-Lamin B, or anti-GAPDH antibodies. (C) Undifferentiated C2C12 myoblasts were stained with anti-HDAC7 and anti-MHC antibodies followed by confocal microscopy. Upon induction of differentiation, cells were fixed and stained at days 3 (D) & 5 (E) with DAPI (blue) and antibodies against muscle-specific myosin heavy chain (MHC) (red) followed by confocal microscopy. The white arrow marks a myocyte that expresses MHC with nuclear HDAC7 staining. Scale bars: 20 μm. For C-E, panel a: anti-HDAC7 antibody staining; panel b: DAPI staining; panel c: anti-MHC antibody staining; panel d: merged image of DAPI and anti-HDAC7 antibody staining; panel e: merged image of anti-HDAC7 and anti-MHC antibody staining; panel f: merged image of DAPI and anti-MHC antibody staining.
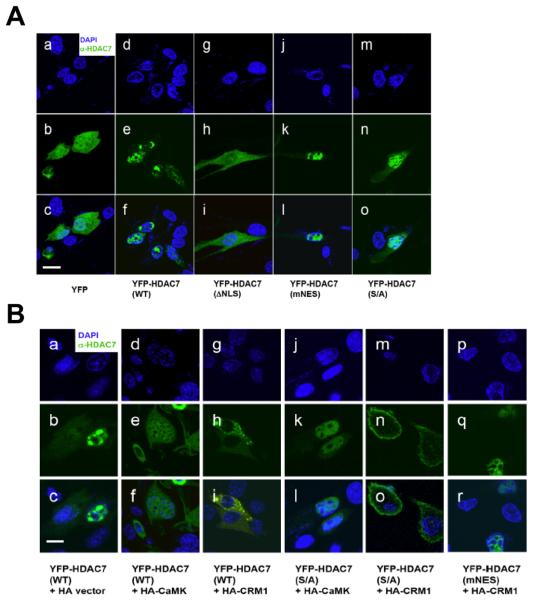
To characterize the regulation of HDAC7 subcellular distribution in C2C12 cells, YFP-HDAC7 (WT) or mutants transiently transfected into C2C12 myoblasts and their subcellular localizations were examined by confocal microscopy. Consistent with previous reports in HeLa cells [26], YFP-HDAC7 (WT) was mostly nuclear with a small fraction of the protein in the cytoplasm (Fig. 2A, panels a-f). Deletion of the NLS (ΔNLS) caused a redistribution of HDAC7 to the cytoplasm (Fig. 2A, panels g-i). In addition, mutations in the NES (mNES) resulted in accumulation of HDAC7 in the nucleus, whereas a phosphorylation site mutant (S/A) exhibited a similar staining pattern to that of the wild-type protein (Fig. 2A, panels j-o). To examine the effects of CRM1 and CaMK on the subcellular distribution of HDAC7, we co-expressed wild-type (WT) or mutant YFP-HDAC7 with constitutively active CaMK or CRM1 [26] (Fig. 2B). Overexpression of either CRM1 or CaMK promoted cytoplasmic retention of YFP-HDAC7 (WT) (panels a-i), while only CRM1 promoted nuclear export of YFP-HDAC7 (S/A) (panels j-o), but not YFP-HDAC7 (mNES) (panels p-r, Supplementary Figure 1). Taken together, these data demonstrate that CRM1 stimulates cytoplamisc retention of HDAC7 and that the C-terminal NES is essential for CRM1-mediated HDAC7 nuclear export. In addition, phosphorylation of S178, S344, or S479 enhances cytoplasmic retention of HDAC7 but is not sufficient to do so in the absence of a functional NES. These data are also consistent with our observation reported earlier in Hela cells [26].
Fig. 2.
Subcellular distribution of transfected YFP-HDAC7. (A) Wild-type (WT) and mutant HDAC7 YFP-HDAC7 expression plasmids were transiently transfected into C2C12 myoblasts followed by immunofluorescence microscopy. Panels a-c: YFP alone, panes d-f: YFP-HDAC7 (WT), panels g-i: YFP-HDAC7 (ΔNLS), panels j-l: YFP-HDAC7 (mNES), panels m-o: YFP-HDAC7 (S/A). (B) The effects of the expression of CaMK and CRM1 on subcellular distribution of YFP-HDAC7. The expression plasmids transfected into C2C12 myocytes are indicated. Scale bars: 20 μm.
A mutation at the C-terminal NES of HDAC7 inhibits myocyte differentiation
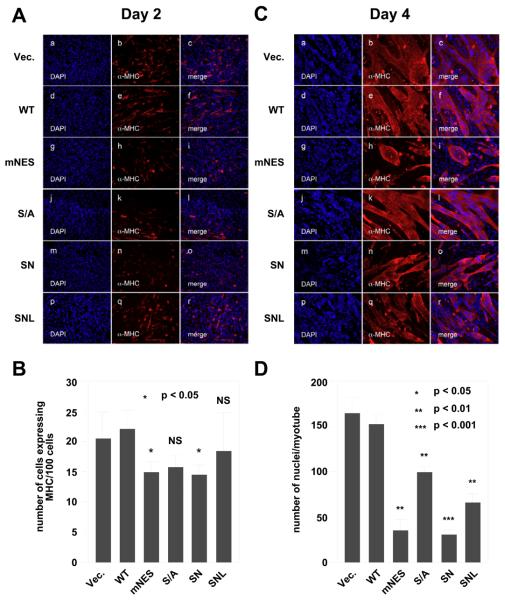
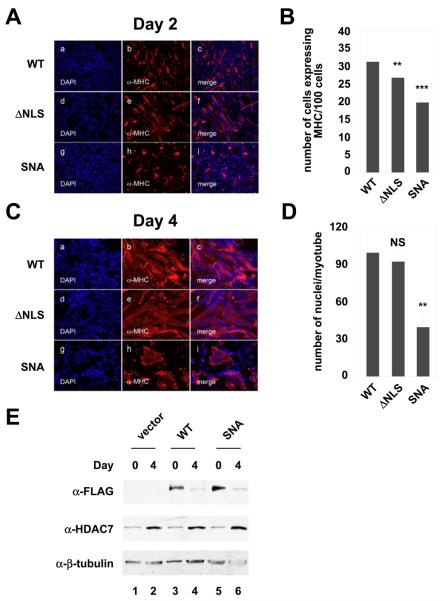
To examine whether altered cellular distribution of HDAC7 affects muscle differentiation, we tested the effects of HDAC7 mislocalization on myogenesis. C2C12 cells were stably transfected with wild-type HDAC7 (WT) or HDAC7 mutant expression constructs. The subcellular localization of the stably expressed wild-type or mutant HDAC7 in undifferentiated myocytes was examined by immunoflurescene microscopy using anti-FLAG antibodies (Supplementary Figure 2A). The subcellular localization of stably transfected HDAC7 mutants exhibited similar distributions to those of the transiently transfected proteins except for the stably expressed wild-type HDAC7 protein, which exhibited more cytoplasmic distribution than transiently transfected proteins in undifferentiated myocytes. Differentiation assays were performed to investigate the ability of these C2C12 cell lines to differentiate into myotubes. Focusing first on the 2 day samples, we noted that there were no detectable differences in differentiation between C2C12-FLAG (vector) and C2C12 FLAG-HDAC7 (WT) stable cells (Fig. 3A, panels a-f & Supplementary Figure 2B). However, the stable expression of FLAG-HDAC7 (mNES) significantly decreased the number of differentiating cells at day 2 of myogenesis (panels g-I, and 3B). In addition, the stable expression of FLAG-HDAC7 (S/A) moderately reduced the amount of differentiating myoblasts (panels j-l. and Fig. 3B). To examine whether the combined mutations (S/A and N) will affect myogenesis additively, we generated another HDAC7 mutant FLAG-HDAC7 (SN or S/A + mNES). This mutant repressed the myogenesis at day 2 to a similar extent as FLAG-HDAC7 (mNES).
Fig. 3.
The effects of HDAC7 mutations on the ability of HDAC7 to inhibit myogenesis. C2C12 cells stably expressing vector alone, HDAC7 (WT), HDAC7 (S/A), HDAC7 (mNES), HDAC7 (SN), or HDAC7 (SNL) were grown to 80% confluence and induced for differentiation as described in “Materials and Methods”. Cells were fixed and stained at days 2 (A) & 4 (C) with DAPI (blue) and antibodies against muscle-specific MHC (red) followed by epifluorescence microscopy. Representative images obtained from two independent clones of each stable cell line are shown. (B) The average number of cells expressing MHC per 100 cells at day 2. (D) The average number of nuclei per myotubes at day 4. The p values of statistical t-test comparing C2C12 cells expressing wild-type HDAC7 or mutant proteins recorded under the same conditions are shown (* p < 0.05, ** p<0.01, or *** p < 0.001). NS: not significant.
To examine whether the nuclear retained HDAC7 (SN) mutant impairs myogenesis through its transcriptional repression activity on MEF2 transcription factors, we generated a mutant HDAC7, HDAC7 (SNL) that contains an additional mutation, L112A, which abolishes its interaction with MEF2A ([38], Supplementary Figure 4). We found that the additional L112A mutation partially rescued the myogenesis deficiency in C2C12-FLAG-HDAC7 (SN) (Fig. 3A,B).
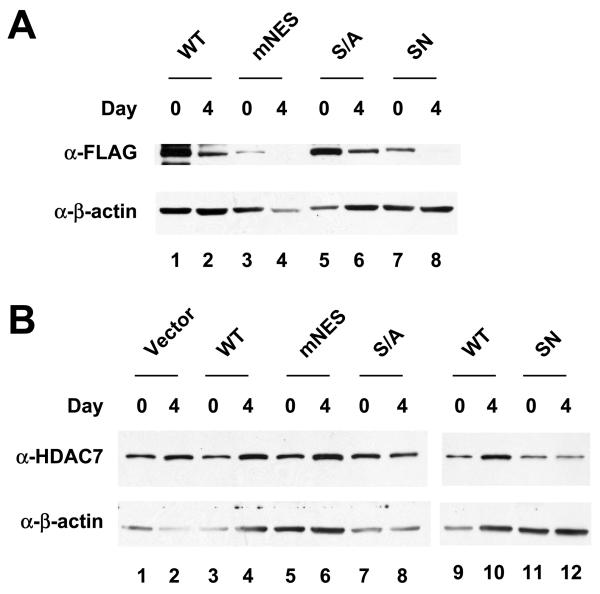
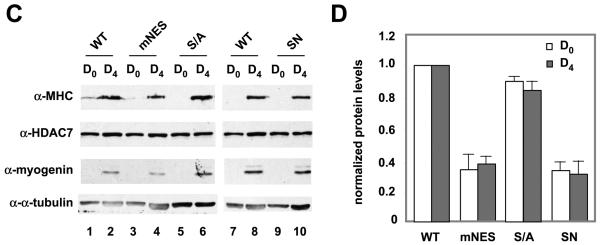
At day4, stable expression of HDAC7 (mNES), and HDAC7 (SN), but not HDAC7 (S/A), significantly reduced the size of the myotubes (Fig. 3C,D). In contrast, the C2C12 cells stably expressing HDAC7 (SNL) mutant had more myotubes than C2C12-FLAG-HDAC7 (SN) cells. Nonetheless, the number of nuclei per myotube in C2C12-FLAG-HDAC7 (SNL) was significantly lower than that in C2C12-FLAG-HDAC7 (WT). MEF2 has been shown to regulate the expression of genes involved in myogenesis including myogenin. To test whether HDAC7 mutants defective in differentiation are also impaired in MEF2 target gene expression, we first examined the expression of exogenous HDAC7 and found that the expression levels of stably transfected HDAC7 (S/A), HDAC7 (mNES), or HDAC7 (SN) were comparable to or lower than that of the wild-type HDAC7 (Fig. 4A,B). We next measured myogenin expression of wild-type and HDAC7 mutants, mNES and S/A, at day 0 and 4. Fig. 4C,D shows that HDAC7 (mNES) and HDAC7 (SN) expressing cells have lower levels of myogenin as well as MHC, a differentiation marker. In summary, these data demonstrated that stable expression of constitutively nuclear HDAC7 mutants, mNES or SN, impairs myogenic gene expression and blocks C2C12 differentiation.
Fig. 4.
Expression of MHC and myogenin in HDAC7 nuclear mutants. Whole cell extracts of HDAC7 wild-type and mutants were prepared at day 0, 2, and 4. Immunoblotting was performed to detect expression of exogenous FLAG-HDAC7 (A) or total HDAC7 (B). Whole cell extracts of HDAC7 wild-type and mutants were prepared at day 0, 2, and 4. Immunoblotting was performed to detect expression of HDAC7, MHC, and myogenin (C). The relative expression levels of MHC (white column) and myogenin (shaded column) at day 4 (D) were quantified by Photoshop software. The ratio of [MHC]/[a-tubulin] and [myogenin]/[a-tubulin] of HDAC7 (WT) at day 4 was set at 1.
The deacetylase catalytic activity of HDAC7 is not essential for the inhibition of myocyte differentiation
We further examined the effects of the NLS on C2C12 differentiation and found that stable expression of HDAC7 (ΔNLS) did not affect C2C12 differentiation (Fig. 5). We had previously shown that a D692A/D694A mutant ablates the ability of HDAC7 to deacetylate histones [Downes, 2000 #104]. To determine whether the intrinsic HDAC activity is essential for HDAC7 inhibition of C2C12 differentiation, we generated a cell line that stably expresses a nuclear export-defective, non-phophorylatable deacetylase-defective HDAC7 (SNA). We found that stable expression of HDAC7 (SNA) still inhibited C2C12 differentiation. The inhibitory effect of HDAC7 (SNA) on differentiation was not due to its higher expression (Fig. 5E). These results indicated that the HDAC catalytic activity is not required for HDAC7 to inhibit C2C12 differentiation.
Fig. 5.
The effects of HDAC7 NLS-deletion and catalytic mutation on myogenesis. C2C12 myoblasts stably expressing wild-type, mutant HDAC7 (ΔNLS), or HDAC7 (SNA) were grown to 80% confluence and induced for differentiation. Cells were fixed and stained at days 2 (A) & 4 (C) with DAPI (blue) and antibodies against muscle-specific myosin heavy chain (MHC) (red) followed by epifluorescence microscopy. Representative images from two independent clones of each stable cell lines are shown. (B) The average number of cells expressing MHC per 100 cells at day 2. (D) The average number of nuclei per myotubes at day 4. The p values of statistical t-test are shown (* p < 0.05, ** p<0.01, or *** p < 0.001). NS: not significant. (E) Expression of exogenously expressed HDAC7 (SNA) in C2C12 cells. Western blotting was performed similar to that of Figs 4A and 4B.
The phosphorylation level of HDAC7 at S178, S344, or S479 during myocyte differentiation
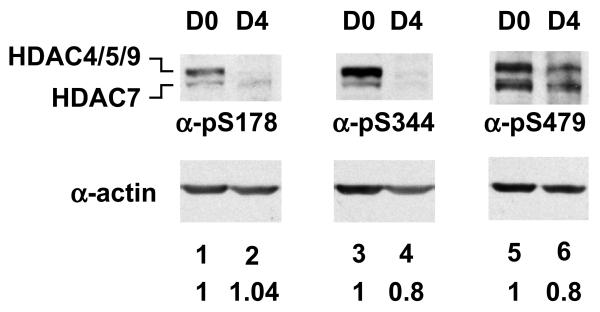
Phosphorylation of HDAC7 at S178, S344, or S479 enhances the binding between HDAC7 and 14-3-3 and promotes subsequent cytoplasmic accumulation of HDAC7 [5,27] Wild-type HDAC7 also redistributes to the cytoplasm and membranes during muscle differentiation. To investigate the role of phosphorylation of HDAC7 during muscle differentiation, we first examined the phosphorylation status of endogenous HDAC7 by carrying out immunoprecipitations followed by immunoblotting using various antibodies (anti-HDAC7, anti-pS178, anti-pS344, and anti-pS479). We found that phosphorylation level of HDAC7 at S78 in undifferentiated and differentiated C2C12 remained almost the same (Fig. 6). To our surprise, phosphorylation of HDAC7 at S344 and S479 was slightly decreased upon differentiation.
Fig. 6.
Phosphorylation of class II HDACs during C2C12 differentiation. C2C12 whole cell extracts harvested at the indicated days after the induction of differentiation were prepared. Whole cell lysates were subjected to Western blotting with anti-pS178, anti-pS344, and anti-pS479 antibodies. Relative levels ([phosphorylated HDAC7 at day 4]/[phosphrylated HDAC7 at day 0]) of phosphorylated HDAC7 at S178, S344, or S479 in differentiated myotubes and undifferentiated myocytes are normalized by actin and are shown at the bottom of each lane. The experiment was repeated 3 times. A representative image is shown. The slower migrating species likely to be other members of class IIa HDACs because phospho-antibodies cross-react with other class IIa HDACs.
Subcellular distribution of phosphorylated class II HDACs during myocyte differentiation
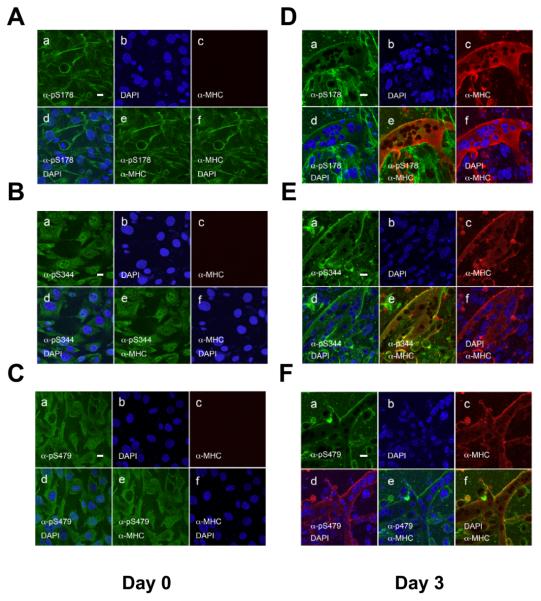
We further determined the subcellular distribution of phosphorylated HDAC7 during muscle differentiation. We have previously generated phospho-specific antibodies against phosphorylated HDAC7 at S178, S344 or and S479 and confirmed their specificity. However, due to the high sequence conservation at these phosphorylation sites, these phospho-specific antibodies crossreact with other Class IIa HDACs (HDAC4, -5 and -9) [26]. Prior to induction of differentiation, Class IIa HDACs phosphorylated on S178 or S479 were primarily localized in the cytoplasm (Fig. 7A-C). Of note, a significant fraction of pS178-phosphoryated class IIa HDACs showed an actin filament-like distribution pattern in myoblasts. In contrast, class IIa HDACs phosphorylated on S344 were distributed evenly between the nucleus and the cytoplasm of the myoblasts (Fig. 7B). Upon induction of differentiation, pS178, pS344 and pS479-phosphorylated class IIa HDACs localize in the cytoplasm and the cytoplasmic membrane of the differentiated cells (Fig. 7D,E). In summary, these data demonstrate that phosphorylation of Class IIa HDACs coincides with their cytoplasmic localization during myogenesis.
Fig. 7.
Subcellular localization of phosphorylated Class IIa HDACs in myoblasts and differentiated myotubes. Undifferentiated (A-C) and differentiated (D-F) C2C12 cells were stained with anti-pS178, anti-pS344 and anti-pS479 antibodies followed by confocal microscopy. Note that α-pS178 antibodies co-stained with actin filaments and the plasma membrane before and after muscle differentiation. Induction of myoblast differentiation was performed as described in ”Materials and Methods”. Scale bars: 20 μm. panel a: anti-phospho-HDAC7 antibody staining; panel b: DAPI staining; panel c: anti-MHC antibody staining; panel d: merged image of DAPI and anti-phospho-HDAC7 antibody staining; panel e: merged image of anti-phospho-HDAC7 and anti-MHC antibody staining; panel f: merged image of DAPI and anti-MHC antibody staining.
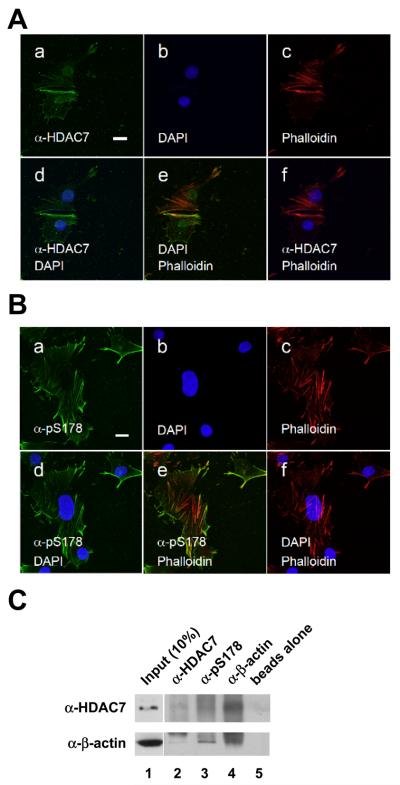
To further confirm that pS178-phosphorylated class IIa HDACs were localized to actin filaments, we performed additional immunofluorescence microscopy. C2C12 cells were co-stained with anti-HDAC7 antibody and rhodamine-conjugated phalloidin, a marker for actin filaments. We found that a significant fraction of HDAC7 colocalized with actin stress fibers (Fig. 8A). An even more pronounced staining pattern was observed when anti-pS178 antibodies were used for immunostaining. In this case, the majority of pS178-phosphorylated class IIa HDACs co-localized with actin stress fibers (Fig. 8B). We further investigated the nature of the colocalization,by coimmunoprecipitation experiments and found that HDAC7 and actin interact in C2C12 myocytes (Fig. 8C). In summary, these data indicate that HDAC7 colocalizes and associates with actin filaments.
Fig. 8.
pS178-HDAC7 colocalizes with actin filaments. (A) HDAC7 partially colocalizes with actin filaments in C2C12 cells. (B) pS178-HDAC7 colocalizes with actin filaments. Immunostaining was carried out as in Fig. 1 except that the cells were not confluent. Note that DAPI and HDAC7 staining were taken from two different sections. Scale bars: 20 μm. Panel a: anti-HDAC7 antibody staining; panel b: DAPI staining; panel c: anti-MHC antibody staining; panel d: merged image of DAPI and anti-HDAC7 antibody staining; panel e: merged image of anti-HDAC7 and anti-MHC antibody staining; panel f: merged image of DAPI and anti-MHC antibody staining. (C) HDAC7 and actin associate in C2C12 cells. Whole cell lysates prepared from C2C12 myocytes collected at day 0 prior to switching to differentiation medium were subjected to coimmunoprecipitation with beads alone or the indicated antibodies followed by Western blotting with the indicated antibodies.
HDAC7 mutants inhibit myoblast migration
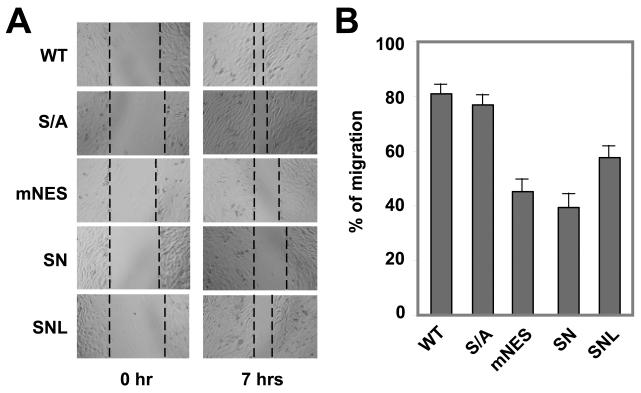
Cell migration is an important step for myoblast fusion to form myotubes. Our observation that HDAC7 is localized to the actin filaments and plasma membranes suggests that HDAC7 may play a role in cell migration. To test this, we assayed cell migration of the HDAC7 mutant stable cell lines in a migration assay. As shown in Fig. 9, C2C12 myoblasts expressing the HDAC7 (S/A) mutant did not significantly differ from the cells expressing wild-type HDAC7 protein. In contrast, cells expressing HDAC7 (mNES) or HDAC7 (SN) exhibited significant deficits in myoblast migration. Furthermore, HDAC7 (SNL) showed a better migration than that of HDAC7 (SN). These data suggest that nuclear retention of HDAC7 not only inhibits myogenesis but also impair myoblast migration most likely through its regulation of MEF2s.
Fig. 9.
The effect of HDAC7 mutations on migration of C2C12 myoblasts. (A) The HDAC7 mNES mutant inhibits migration in confluent myoblasts. Myoblasts stably integrated with wild-type or mutant HDAC7 were grown to confluence and wounds were generated and measured. Seven hours later, the width of the wound was determined. A total of six wounds were used to calculate the % migration for each stable cell line. A representative image is shown. (B) Quantitative analysis of cell migration. The data were derived from 6 independent fields and are representative of two independent experiments.
Discussion
Our present study demonstrates that endogenous HDAC7 is localized in both the nucleus and cytoplasm in undifferentiated myoblasts but becomes exclusively cytoplasmic in terminally differentiated myotubes. Furthermore, phosphorylated forms of class II HDACs, as recognized by a cross reacting phospho-specific antibody, are predominantly localized in the cytoplasm of terminally differentiated myotubes. Unexpectedly, phosphorylation of HDAC7 at S344 and S479 is slightly decreased upon induction of differentiation. We also show that a nuclear export defective mutant of HDAC7 (mNES), but not the phosphorylation-defective mutant (S/A), impairs myotube formation and cell migration of C2C12 myoblasts, Taken together, our data establish the significance of HDAC7 subcellular distribution and imply that redistribution of HDAC7 is an important event during myogenesis.
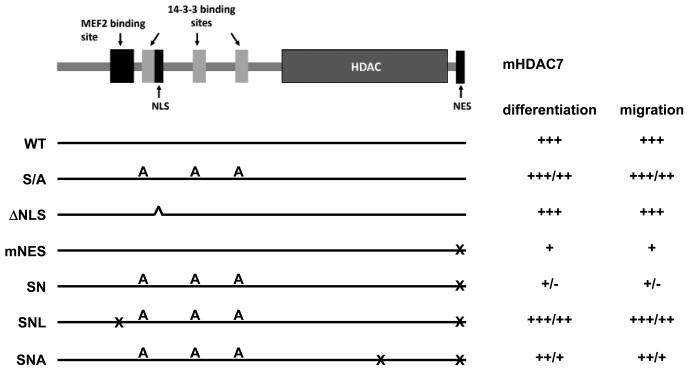
Muscle differentiation is a tightly regulated process, which is controlled by a network of proteins involved in signal transduction and transcriptional regulation [39]. MEF2 transcription factors are the central players in this network through their effects on gene expression and associations with other myogenic transcription factors. MEF2s transmit signals from membranes to downstream genes by switching binding partners from corepressors to coactivators to regulate transcription, or vice versa [40]. In this study, we show that HDAC7 is retained in the cytoplasm during myocyte differentiation. This leads to the derepression of MEF2 targeted myogenic genes such as myogenin and thus promotes the muscle differentiation process. Using a set of HDAC7 mutants that are defective in nuclear localization, nuclear export, or its ability to interact with 14-3-3 proteins (Figure 10), we have identified sequences that are critical for myocyte differentiation. We have found that C2C12 cells stably expressing HDAC7 (mNES) or HDAC7 (SN) display severe defects in myogenic gene expression and myotube formation (Fig. 3). However, a mutation (L112A) that abolished the association between HDAC7 and MEF2 [38] partially restored the normal differentiation of C2C12 cells. L112A mutant abolishes interactions between HDAC7 and MEF2 and interactions between HDAC7 and alpha actinin 4 (ACTN4) [38]. It is possible that nuclear HDAC7 inhibits myogenesis by repressing the transcriptional activity of MEF2 in the nucleus. However, we can not rule out the possibility that the interaction between HDAC7 and ACTN4 may be also involved in myogenesis. Interestingly, our data also show that the catalytically dead mutant HDAC7 (D652A/D654A) modestly affect the ability of HDAC7 (SN) to inhibit C2C12 differentiation, suggesting that deacetylase activity is dispensable for HDAC7 to inhibit myogenesis. Therefore, HDAC7 may exert its effects on myogenesis through other transcription regulatory motifs. Indeed, we have previously demonstrated that HDAC7 harbors three potent repression domains and loss of the C-terminal HDAC activity (D562A/D654A) does not completely abrogate its repression activity [Kao, 2000 #140]. Further support for this result has come from several reports demonstrating that the N-terminal region of class IIa HDACs associates with other transcriptional corepressors including CtBP1 [41] and HP1 [42]. This suggests that the N-terminal non-catalytic domain of HDAC7 can potently repress transcription, even when HDAC catalytic activity is defective.
Fig. 10.
A summary of the effect of HDAC7 mutations on C2C12 mycocyte differentiation and migration. A schematic representation of HDAC7 functional domains is shown. The HDAC7 mutants used in this study are described in the text and indicated and their effect on myocyte differentiation and migration are summarized.
We found that HDAC7 showed decreases in phosphorylation at S344 and S479 four days post differentiation, whereas phosphorylation at S178 remained constant. Immunofluorescence microscopy consistently demonstrated that the intensity of phosphorylated class II HDACs was comparable for undifferentiated and differentiated C2C12 cells. These observations suggest that the level of HDAC7 phosphorylation at these residues does not correlate with the cytoplasmic accumulation of HDAC7 during C2C12 differentiation. This is consistent with our observation that overexpression of CRM1 is capable of promoting cytoplasmic retention of the HDAC7 (S/A) mutant ([26], Figure 2). One plausible mechanism is that another factor(s) which is expressed late (becomes active) during differentiation and blocks nuclear import or promotes cytoplasmic retention of HDAC7.
The three phosphorylated class IIa HDACs appear to distribute differently prior to differentiation (Fig. 7). In undifferentiated myoblasts, pS178-HDAC predominantly co-localizes with actin filaments, whereas pS344-HDAC and pS479-HDAC localized in both the nucleus and the cytoplasm. Upon induction of differentiation, all of the phosphorylated forms of HDACs accumulated in the cytoplasm and cytoplasmic membranes. These observations raise several questions. First, why and how does pS178-HDAC7 colocalize with actin filaments? We have previously identified ACTN4, an actin-binding protein, as an HDAC7 interacting protein. It is possible that ACTN4 may anchor HDAC7 in the cytoplasm [38]. Second, does cytoplasmic or membrane-bound HDAC7 have a distinct function. It is becoming increasingly clear, like histone (protein) acetyltransferase (HATs) (or PATs), HDACs have non-histone substrates. Recently, several laboratories including ours have shown that class II HDACs such as HDAC4 and HDAC7 are capable of promoting protein sumoylation [36,43-46]. Given these observations, it is possible that class II HDACs may have distinct activity in the cytoplasm and/or cytoplasmic membranes.
Prior to fusion into multinucleated myotubes, myoblasts migrate and align with each other. The observation that pS178-HDAC7 co-localizes with the actin filaments and plasma membranes raised the possibility that HDAC7 may regulate and/or interact with the actin cytoskeleton to influence cell migration. However, our data demonstrated that C2C12 cells expressing nuclear-retained HDAC7 (mNES), but not C2C12 cells expressing phosphorylation-deficient HDAC7 (S/A), are defective in migration in wound-healing assays. These data indicate that HDAC7 regulation of myoblast migration depends on the subcellular localization of HDAC7, but not on the phosphorylation states of HDAC7. These data also suggest that nuclear-retained HDAC7 may associate with an actin binding factor that is important for the function of the actin skeleton, such as ACTN4, in the nucleus. Furthermore, the L112A mutation, which abolishes the interaction between HDAC7 with either MEF2 or ACTN4, rescued the ability of HDAC7 (SN) to inhibit cell migration. These observations indicate that the inability for HDAC7 mutant-expressing C2C12 stable cells to differentiate correlates with their defects in migration. However, the details of the mechanism will require further investigation.
Supplementary Material
Acknowledgements
We thank Drs. Samols and Reineke for their comments on the manuscript. This project is supported by NIH, DK078965, HL093269 and the Pardee Foundation to H.-Y. Kao and by the Confocal Microscopy Core Facility of the Comprehensive Cancer Center of Case Western Reserve University and University Hospitals of Cleveland (NIH P30 CA43703-12).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci U S A. 1999;96:4868–73. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kao HY, Downes M, Ordentlich P, Evans RM. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 3.Verdel A, Khochbin S. Identification of a new family of higher eukaryotic histone deacetylases. Coordinate expression of differentiation-dependent chromatin modifiers. J Biol Chem. 1999;274:2440–5. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 4.Dressel U, Bailey PJ, Wang SC, Downes M, Evans RM, Muscat GE. A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J Biol Chem. 2001;276:17007–13. doi: 10.1074/jbc.M101508200. [DOI] [PubMed] [Google Scholar]
- 5.Kao HY, Verdel A, Tsai CC, Simon C, Juguilon H, Khochbin S. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J Biol Chem. 2001;276:47496–507. doi: 10.1074/jbc.M107631200. [DOI] [PubMed] [Google Scholar]
- 6.Lemercier C, Verdel A, Galloo B, Curtet S, Brocard MP, Khochbin S. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J Biol Chem. 2000;275:15594–9. doi: 10.1074/jbc.M908437199. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, McKinsey TA, Zhang CL, Olson EN. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell. 2000;6:233–44. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 8.Miska EA, Karlsson C, Langley E, Nielsen SJ, Pines J, Kouzarides T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. Embo J. 1999;18:5099–107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang AH, et al. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol Cell Biol. 1999;19:7816–27. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Youn HD, Liu JO. Cabin1 represses MEF2-dependent Nur77 expression and T cell apoptosis by controlling association of histone deacetylases and acetylases with MEF2. Immunity. 2000;13:85–94. doi: 10.1016/s1074-7613(00)00010-8. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, McKinsey TA, Nicol RL, Olson EN. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci U S A. 2000;97:4070–5. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edmondson DG, Lyons GE, Martin JF, Olson EN. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120:1251–63. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- 13.Lilly B, Galewsky S, Firulli AB, Schulz RA, Olson EN. D-MEF2: a MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc Natl Acad Sci U S A. 1994;91:5662–6. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–93. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 15.Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science. 1999;286:785–90. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- 16.Miska EA, Langley E, Wolf D, Karlsson C, Pines J, Kouzarides T. Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res. 2001;29:3439–47. doi: 10.1093/nar/29.16.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Youn HD, Sun L, Prywes R, Liu JO. Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science. 1999;286:790–3. doi: 10.1126/science.286.5440.790. [DOI] [PubMed] [Google Scholar]
- 18.Yang XJ, Seto E. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr Opin Genet Dev. 2003;13:143–53. doi: 10.1016/s0959-437x(03)00015-7. [DOI] [PubMed] [Google Scholar]
- 19.Blau HM, Chiu CP, Webster C. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell. 1983;32:1171–80. doi: 10.1016/0092-8674(83)90300-8. [DOI] [PubMed] [Google Scholar]
- 20.Yaffe D. Retention of differentiation potentialities during prolonged cultivation of myogenic cells. Proc Natl Acad Sci U S A. 1968;61:477–83. doi: 10.1073/pnas.61.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–11. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X, Ito A, Kane CD, Liao TS, Bolger TA, Lemrow SM, Means AR, Yao TP. The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J Biol Chem. 2001;276:35042–8. doi: 10.1074/jbc.M105086200. [DOI] [PubMed] [Google Scholar]
- 23.Gao C, Cheng X, Lam M, Liu Y, Liu Q, Chang KS, Kao HY. Signal-dependent regulation of transcription by histone deacetylase 7 involves recruitment to promyelocytic leukemia protein nuclear bodies. Mol Biol Cell. 2008;19:3020–7. doi: 10.1091/mbc.E07-11-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci U S A. 2000;97:7835–40. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang AH, Yang XJ. Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol Cell Biol. 2001;21:5992–6005. doi: 10.1128/MCB.21.17.5992-6005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao C, Li X, Lam M, Liu Y, Chakraborty S, Kao HY. CRM1 mediates nuclear export of HDAC7 independently of HDAC7 phosphorylation and association with 14-3-3s. FEBS Lett. 2006;580:5096–104. doi: 10.1016/j.febslet.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 27.Dequiedt F, Van Lint J, Lecomte E, Van Duppen V, Seufferlein T, Vandenheede JR, Wattiez R, Kettmann R. Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis. J Exp Med. 2005;201:793–804. doi: 10.1084/jem.20042034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huynh QK, McKinsey TA. Protein kinase D directly phosphorylates histone deacetylase 5 via a random sequential kinetic mechanism. Arch Biochem Biophys. 2006;450:141–8. doi: 10.1016/j.abb.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Matthews SA, Liu P, Spitaler M, Olson EN, McKinsey TA, Cantrell DA, Scharenberg AM. Essential role for protein kinase D family kinases in the regulation of class II histone deacetylases in B lymphocytes. Mol Cell Biol. 2006;26:1569–77. doi: 10.1128/MCB.26.4.1569-1577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vega RB, Harrison BC, Meadows E, Roberts CR, Papst PJ, Olson EN, McKinsey TA. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol. 2004;24:8374–85. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng X, Ewton DZ, Mercer SE, Friedman E. Mirk/dyrk1B decreases the nuclear accumulation of class II histone deacetylases during skeletal muscle differentiation. J Biol Chem. 2005;280:4894–905. doi: 10.1074/jbc.M411894200. [DOI] [PubMed] [Google Scholar]
- 32.Berdeaux R, Goebel N, Banaszynski L, Takemori H, Wandless T, Shelton GD, Montminy M. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat Med. 2007;13:597–603. doi: 10.1038/nm1573. [DOI] [PubMed] [Google Scholar]
- 33.Dequiedt F, et al. New role for hPar-1 kinases EMK and C-TAK1 in regulating localization and activity of class IIa histone deacetylases. Mol Cell Biol. 2006;26:7086–102. doi: 10.1128/MCB.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang AH, Kruhlak MJ, Wu J, Bertos NR, Vezmar M, Posner BI, Bazett-Jones DP, Yang XJ. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol Cell Biol. 2000;20:6904–12. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Downes M, Ordentlich P, Kao HY, Alvarez JG, Evans RM. Identification of a nuclear domain with deacetylase activity. Proc Natl Acad Sci U S A. 2000;97:10330–5. doi: 10.1073/pnas.97.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao C, et al. Histone deacetylase 7 promotes PML sumoylation and is essential for PML nuclear body formation. Mol Cell Biol. 2008;28:5658–67. doi: 10.1128/MCB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King CC, Bouic K, Friedmann T. A fractionation method to identify qauntitative changes in protein expression mediated by IGF-1 on the proteome of murine C2C12 myoblasts. Proteome Sci. 2009;7:28–45. doi: 10.1186/1477-5956-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakraborty S, Reineke EL, Lam M, Li X, Liu Y, Gao C, Khurana S, Kao HY. Alpha-actinin 4 potentiates myocyte enhancer factor-2 transcription activity by antagonizing histone deacetylase 7. J Biol Chem. 2006;281:35070–80. doi: 10.1074/jbc.M602474200. [DOI] [PubMed] [Google Scholar]
- 39.Arnold HH, Winter B. Muscle differentiation: more complexity to the network of myogenic regulators. Curr Opin Genet Dev. 1998;8:539–44. doi: 10.1016/s0959-437x(98)80008-7. [DOI] [PubMed] [Google Scholar]
- 40.Naya FS, Olson E. MEF2: a transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr Opin Cell Biol. 1999;11:683–8. doi: 10.1016/s0955-0674(99)00036-8. [DOI] [PubMed] [Google Scholar]
- 41.Zhang CL, McKinsey TA, Lu JR, Olson EN. Association of COOH-terminal-binding protein (CtBP) and MEF2- interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J Biol Chem. 2001;276:35–9. doi: 10.1074/jbc.M007364200. [DOI] [PubMed] [Google Scholar]
- 42.Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–12. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by Histone Deacetylase 4- and SIRT1 Deacetylase-Mediated Lysine Modifications. Mol Cell Biol. 2005;25:8456–64. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gregoire S, Yang XJ. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol Cell Biol. 2005;25:2273–87. doi: 10.1128/MCB.25.6.2273-2287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghisletti S, Huang W, Ogawa S, Pascual G, Lin ME, Willson TM, Rosenfeld MG, Glass CK. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARgamma. Mol Cell. 2007;25:57–70. doi: 10.1016/j.molcel.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stankovic-Valentin N, Deltour S, Seeler J, Pinte S, Vergoten G, Guerardel C, Dejean A, Leprince D. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol Cell Biol. 2007;27:2661–75. doi: 10.1128/MCB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.