Abstract
The Golgi-associated retrograde protein (GARP) complex is a membrane-tethering complex that functions in traffic from endosomes to the trans-Golgi network. Here we present the structure of a C-terminal fragment of the Vps53 subunit, important for binding endosome-derived vesicles, at a resolution of 2.9 Å. We show that the C terminus consists of two α-helical bundles arranged in tandem, and we identify a highly conserved surface patch, which may play a role in vesicle recognition. Mutations of the surface result in defects in membrane traffic. The fold of the Vps53 C terminus is strongly reminiscent of proteins that belong to three other tethering complexes—Dsl1, conserved oligomeric Golgi, and the exocyst—thought to share a common evolutionary origin. Thus, the structure of the Vps53 C terminus suggests that GARP belongs to this family of complexes.
Keywords: endocytosis, membrane traffic
Tethering complexes are multisubunit assemblies that function in the last steps of most membrane trafficking pathways, as a transport vesicle docks to and is recognized at the target organelle (1). These complexes may physically link the arriving vesicle and acceptor membrane and may also play a role in facilitating the assembly of the SNARE complexes (2, 3), which catalyze vesicle fusion with the target organelle and cargo delivery. Distinct tethering complexes function in distinct transport pathways. They are localized to specific compartments through interactions with SNAREs, with the small activated GTPases that help to define organelle identity, and with phospholipids enriched in the membranes of target organelles (4, 5), or a combination of these interactions.
The last 5 y have seen a number of structural studies aimed at understanding the architecture and ultimately the function of tethering complexes. The TRAPPI and Dsl1 complexes, which function in traffic between the endoplasmic reticulum and the Golgi, have been particularly well studied (3, 6), and substantial effort has also been expended in investigating structures of individual subunits of the exocyst and conserved oligomeric Golgi (COG) complexes (reviewed in ref. 7). These studies have shown that subunits within each of the complexes are structurally related and, moreover, that subunits in the Dsl1, exocyst, and COG complexes (but not TRAPPI) are structurally related despite low or undetectable sequence conservation (8, 9).
In contrast, little is known regarding either the overall or subunit architecture of the Golgi-associated retrograde complex (GARP). This complex, conserved in all known eukaryotes, functions in traffic from early and late endosomes to the trans-Golgi network (TGN) (10). Mutations in GARP subunits lead to impaired retrograde traffic and, in yeast, to a vacuolar tubulation phenotype (10). In mice, mutations in the Vps54 subunit lead to embryonic lethality or motor neuron degeneration similar to amyotrophic lateral sclerosis in humans (11).
GARP consists of at least three different proteins, Vps52 (75 kDa), Vps53 (95 kDa), and Vps54 (100 kDa), which are associated in a stable 1:1:1 complex (10); and yeast GARP has an additional small subunit, Vps51 (20 kDa) (12). GARP interacts directly with small GTPases that localize to the TGN [Ypt6/Rab6 (12, 13) and Arl1 (14)], and in humans, GARP also interacts with SNARE proteins specific to the retrograde pathway (t-SNARE proteins syntaxin 6, syntaxin 16, and v-SNARE Vamp4) and may facilitate formation of the SNARE complex (2) at the TGN, the site of vesicle fusion. The SNARE interaction requires N-terminal regions in the Vps53 and Vps54 proteins (2). Interaction with transport vesicles additionally requires the C terminus of Vps53, and deletion of that region disrupts retrograde traffic even as it does not affect GARP complex assembly (2).
GARP, Dsl1, COG, and the exocyst are all multisubunit assemblies that function as membrane tethers late in membrane trafficking pathways. On the basis of weak conservation of a single amphipathic helix present at the N terminus of several subunits (including Vps53 and Vps54 in GARP), it has been speculated that GARP could be related to the Dsl1, COG, and exocyst tethering complexes in structure as well as in function (8, 9). Here we report the structure of the C-terminal portion of the Saccharomyces cerevisiae Vps53 subunit at a resolution of 2.9 Å. The fold, comprising contiguous α-helical bundles, is similar to that reported for structures of the subunits of Dsl1, COG, and the exocyst, providing strong evidence that GARP does indeed belong to this family of tethering complexes. The structural similarity is consistent with the notion that the general molecular mechanisms that underlie tethering are conserved for the complexes in this group.
Results and Discussion
Structure of the Vps53 C-Terminal Fragment.
Because we were unable to obtain crystals of the full-length Vps53 (residues 1–822), we performed limited proteolysis experiments to investigate its domain architecture. On the basis of proteolytic cleavage sites, we identified three fragments (residues 95–822, 200–822, and 554–822) that could be solubly expressed and purified from Escherichia coli. Only the C-terminal fragment (residues 554–822) crystallized. We determined its structure by single anomalous wavelength dispersion methods using data from selenomethionine substituted crystals, where residues I586 and L722 of Vps53 had been mutated to methionine to increase the anomalous signal. The final model for the Vps53 C terminus includes residues 564–730 and 742–779, 19 water molecules, and three barium ions (from barium chloride in the crystallization solution). The N and C termini of the fragment (residues 554–563 and 780–822), as well as one loop (residues 731–741), are not ordered. Data collection and refinement statistics are reported in Table 1.
Table 1.
Data collection and refinement statistics
| Parameter | Statistic |
| Data collection | |
| Space group | P6322 |
| Unit cell dimensions | a = b =127.2 Å, c = 81.7 Å |
| α = β = 90°, γ = 120° | |
| Wavelength (Å) | 0.9792 (peak) |
| Resolution (Å) | 25.0–2.90 (3.00–2.90) |
| Rsym (last shell) (%) | 8.8 (34.8) |
| I/σI | 28.8 (8.1) |
| Completeness (%) | 100.0 (100.0) |
| Redundancy | 15.4 (11.1) |
| Refinement | |
| Resolution (Å) | 24.40–2.90 |
| No. of reflections | 8,606 |
| Rwork/Rfree (%) | 18.9/22.1 |
| No. of protein atoms | 1745 |
| Average B (Å2) | 61.5 |
| Rms deviation | |
| Bond lengths (Å) | 0.017 |
| Bond angles (°) | 1.768 |
| Ramachandran plot | |
| Allowed (% residues) | 95.5 |
| Generously allowed (% residues) | 4.5 |
| Disallowed (% residues) | 0.0 |
Values in parentheses refer to the highest resolution shell.
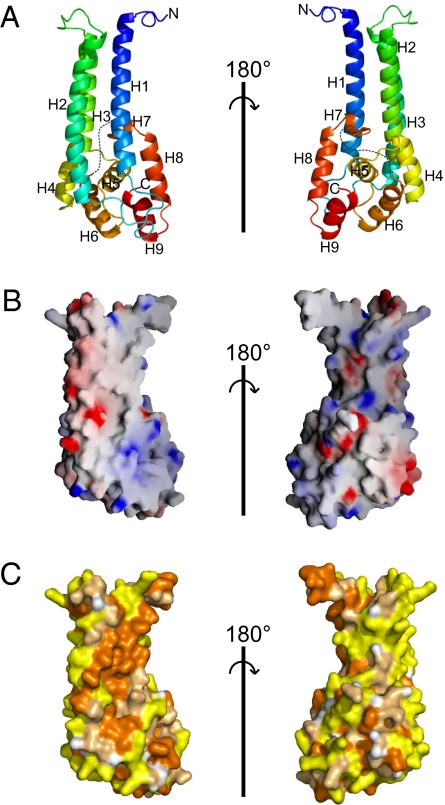
The Vps53 C terminus folds into a single domain consisting of two adjacent α-helical bundles (Fig. 1A). Three N-terminal α-helices (H1–H3) form a ≈60-Å-long stem, which adjoins a second, more globular helical bundle (H4–H9), ≈25–30 Å in diameter. An unusual feature of the surface is an extended hydrophobic region, centered on helices H1 and H2 and running along their length (Fig. 2 B and C). In the crystal lattice, this surface is buried in a crystallographic interface (Fig. S1), but this interface is not maintained in solution as the Vps53-C-terminal fragment migrates as a monomer on sizing columns.
Fig. 1.
Structure of the C-terminal fragment of Vps53 (residues 564–799). (A) Ribbon diagrams colored from blue at the N-terminal end to red at the C terminus. Sequences that are disordered in the structure are indicated by dotted lines. Two orientations, related by a 180° rotation, are shown. (B) The surface of the Vps53 C terminus (oriented as in the ribbon diagrams) is colored according to electrostatic potential. (C) The surface is colored according to hydrophobicity. Hydrophobic residues (Val, Ile, Leu, Pro, Met, Ala, Phe, and Trp) are orange, polar residues (Ser, Thr, Cys, Asn, Gln, and Tyr) are pale orange, and charged residues (Glu, Asp, Lys, Arg, and His) are yellow.
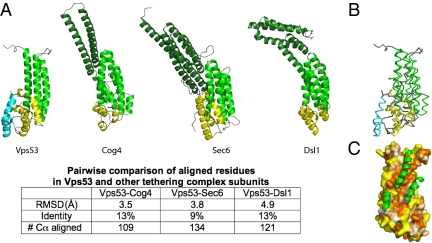
Fig. 2.
The C-terminal fragment has a similar fold to subunits in the COG, exocyst, and Dsl1 multisubunit tethering complexes. (A) Vps53, Cog4 (PDB ID 3HR0), Sec6 (PDB ID 2FJI), and Dsl1 (PDB ID 3K8P) C-terminal fragments are shown in the same orientation. Distinct helical bundles are shown in dark green, green, and yellow (at the C terminus). The C-terminal helical bundle in Vps53 has two helices not present in the other proteins; they are shown in cyan. The N-terminal-most helical bundle, or “stem,” in the Vps53 fragment lacks two or three helices present in the other proteins. A more quantitative comparison of the structures is tabulated below. (B) Superposition of the Vps53 and Cog4 C termini, showing that Cog4 lacks the two most C-terminal helices in Vps53 (cyan). Vps53 and Cog4 are colored as in A. (C) The surface of the C-terminal fragment of Vps53, with residues colored according to hydrophobicity as in Fig. 1C. The additional helices present in the Cog4 “stem” are indicated in green. They bury the Vps53 hydrophobic patch, suggesting that the full-length Vps53 protein shares these helices even as they are not present in the proteolytic fragment of Vps53 that was crystallized.
One possibility is that this surface is involved in protein–protein interactions. As discussed below, however, it is more likely that in the full-length Vps53 protein, the hydrophobic surface is buried by structural elements missing in the fragment that was crystallized.
Similarity to Other Tethering Complex Subunits.
The key finding of this work is that the structure of the Vps53 C terminus bears a strong resemblance to structures of subunits belonging to three other tethering complexes—Dsl1, the exocyst, and COG—even though sequence identity is minimal (Fig. 2). Each of these complexes has subunits that are constructed from a series of α-helical bundles arranged in tandem. The Vps53 C terminus is structurally most similar to the C terminus of Sec6p in the exocyst (15), Cog4 in the COG complex (16), and Dsl1 in the Dsl1 complex (3) (Fig. 2), with similar orientations and connectivities for the helices that comprise the two C-terminal helical bundles. The Vps53 structure differs from these other structures by the presence of two additional α-helices at the very C terminus (Fig. 2 A and B). Additionally, the “stem” of the Vps53 fragment crystallized comprises only three α-helices, whereas the equivalent bundle in the other proteins consists of five or six helices. Residues in the extended hydrophobic region on the surface of the Vps53 fragment would be buried in the presence of the additional “stem” helices (Fig. 2C), as the hydrophobic, structurally equivalent residues in Sec6, Cog4, and Dsl1 are. In these proteins, the hydrophobic residues are part of the protein core. It is likely that sequences that would form the additional helices in Vps53 were deleted in the crystallization construct, but most probably the helices are present in the full-length protein. (In the limited proteolysis experiment used to design the crystallization construct, the cleavage site would have been in the loop that connects these helices to H1 in the structure. The nature of the proteolysis experiment did not allow us to assess whether N-terminal sequences in Vps53 remain associated with the crystallization fragment in the absence of a covalent linkage, and we have not further investigated the possibility that the interaction between the hydrophobic region and these additional sequence elements might be dynamic. To our knowledge, there is no evidence that the interface is dynamic in Sec6, Cog4, or Dsl1.) The structural similarity of Vps53 in GARP to subunits in Dsl1, the exocyst, and COG suggests that Vps53 shares a common ancestor with these proteins despite low or undetectable sequence similarity.
Conserved Surface and Possible Functions.
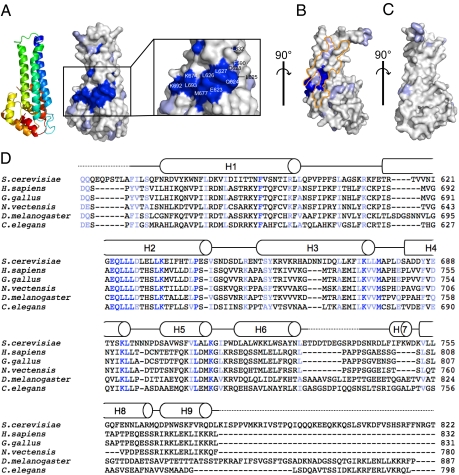
To identify functionally important surfaces, we aligned the sequences of Vps53 subunits from different organisms and mapped residues that are highly conserved onto the S. cerevisiae structure (Fig. 3). We find a cluster of conserved positions that includes residues in helices H2, H3, and H4. This surface is distinct from the elongated hydrophobic region discussed above (which likely is buried in full-length Vps53) and includes a mix of charged, polar, and hydrophobic residues (Fig. 3 A and B).
Fig. 3.
A surface of of the Vps53 C terminus is highly conserved. (A) residues on the surface of Vps53 that are most highly conserved are dark blue, and those that are conserved are light blue. The conserved patch is enlarged, and the most highly conserved residues are labeled. A ribbons diagram indicates the orientation of the molecule. (B) A second view of the Vps53 C terminus and conserved surface residues, related to that in A by a 90° rotation. Hydrophobic regions thought to be buried in the core of the Vps53 stem by additional helices not present in the crystallized fragment, as in Fig. 2C, are outlined in orange. (C) A third view, related to that in B by a 90° rotation.(D) Sequence alignments for the Vps53 C terminus. Residues that are identical or highly conserved in the alignment are indicated in dark and light blue, respectively, and were mapped in A–C. Secondary structure for the S. cerevisiae C-terminal fragment is indicated.
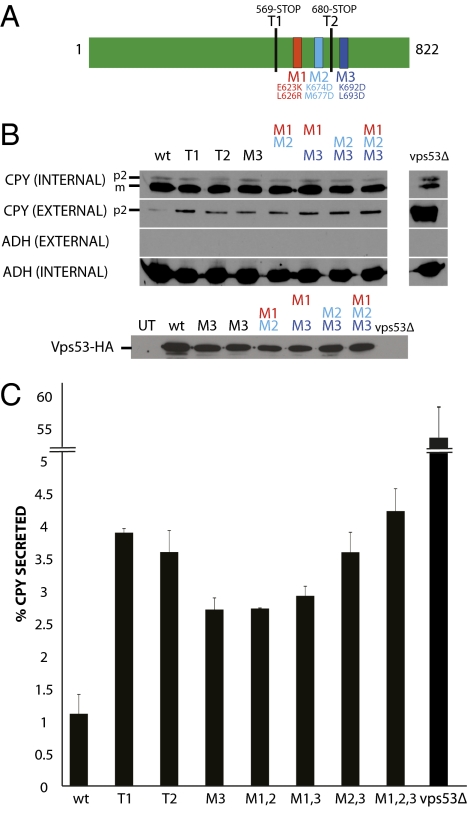
We used carboxypeptidase Y (CPY) secretion assays to test whether the conserved surface is functionally important. In these experiments, we mutated residues in the conserved surface of Vps53 and monitored the effect on CPY transport. Only ≈1% of total CPY is secreted in wild-type yeast, and levels are increased when traffic from the endosome to the Golgi is disrupted. We observe that CPY secretion is increased in strains where Vps53 bears mutations in the conserved surface (Fig. 4). The largest effect is for a hextuple mutant (M1,2,3 in Fig. 4 B and C). Secretion levels for this mutant are similar to those in which the entire Vps53 C terminus is removed (T1 in Fig. 4), suggesting that the conserved surface in the Vps53 C terminus is the functionally important part of the C terminus. The effects in all cases are appreciably smaller than effects caused by deletion of the entire Vps53 protein (Fig. 4 and ref. 10). This result would be expected given that Vps53 deletion abrogates GARP complex formation (10), whereas the Vps53 C terminus is not important for complex formation (2).
Fig. 4.
CPY secretion assays showing that the conserved surface of the Vps53 C terminus is important for membrane trafficking. (A) Schematic diagram of Vps53p and the mutations tested in the CPY secretion assay. In T1 and T2, Vps53 was truncated at residues 569 and 680, respectively. M1, E623K, L626R; M2, K674D, M677D; M3, K692D, L693D. (B) Western blots testing vps53 mutants for CPY secretion. The levels of CPY secreted into the media (external CPY) and CPY retained in the cells (internal CPY) were examined for wild-type (wt) cells and several vps53 mutants. The p2 form (Golgi-modified) and m form (mature, vacuolar) of CPY are indicated. Western blots against alcohol dehydrogenase (Adh1p) were performed to control for loading (internal ADH) and to verify that cells did not lyse upon harvesting (external ADH). Western blots against a C-terminal 3xHA-tag were used to control for levels of Vps53 constructs (except T1 and T2, which are untagged). The M3 sample was loaded twice. Levels are comparable to wild-type protein, as determined by a comparison with the ADH loading control. (C) Quantitation of the CPY secretion assays. The percentage of secreted CPY for several vps53 mutants compared with wild-type cells was quantitated from two separate trials. Error bars indicate SD (n = 2).
The defects in secretion are not explained by decreases in expression or protein instability because expression levels for the mutant proteins are comparable to that of wild-type (compare levels of Vps53 with the ADH loading control in Fig. 4B). The largest secretion effects, observed for the M2,3 and M1,2,3 mutants, do not correlate with lower protein levels. Thus, the CPY assays are consistent with the functional importance of the conserved surface of the Vps53 C terminus for membrane traffic.
Notably, the Cog4 C-terminal fragment has a conserved surface patch on an equivalent face to that in the Vps53 C terminus (16). The importance of this surface was confirmed by CPY assays similar to ours, and the effects were of similar magnitude. The finding that conserved regions of the Vps53 and Cog4 C termini are similarly located is consistent with the notion that these structurally related protein fragments may also be functionally related. The conserved surfaces in Sec6, however, are on different faces (15). Because the Vps53 C terminus is not required for the assembly of the GARP complex (2), it is unlikely that the conserved surface is involved in interactions with other GARP subunits. Instead, the Vps53 C terminus is required for interactions with the transport vesicle (2), suggesting that the conserved surface revealed by our structure may mediate this interaction.
How might the Vps53 C terminus interact with the transport vesicle? The Exo70 and Sec3 subunits in another tethering complex, the exocyst, are believed to interact with the membrane directly via extended basic surface regions (5, 17–19). However, the mixed nature of the Vps53 C terminus surface, and of the conserved surface in particular, argues against this mechanism for the Vps53 C terminus. Several tethering complexes, including Dsl1 and TRAPPI (20, 21), recognize proteins in the vesicle coat, and an intriguing possibility is that the Vps53 C terminus could interact with the retromer coat of endosome-derived vesicles, perhaps through the conserved patch.
Conclusions.
We have shown that the C terminus of Vps53 is similar in structure to C-terminal fragments of subunits in three other tethering complexes—Dsl1, the exocyst, and COG—strongly suggesting that GARP is a member of this family of tethering complexes. Most of the subunits within each of these complexes (all structurally characterized subunits except for Sec39 in Dsl1p) also seem to be related in their architecture because they are all constructed from tandem helical bundle domains. Similarly, the other GARP proteins—Vps51, Vps52, and Vps54—are predicted to be almost entirely α-helical, making it plausible that they too will feature α-helical bundles arranged in tandem. Additional structural information regarding these other subunits of GARP should elucidate whether the components of the GARP complex are also structurally related to each other.
A related question is whether certain modules—such as the structurally similar C termini of Vps53, Sec6, Cog4, and Dsl1—have evolved for related functions. Interestingly, as for Vps53, the C-terminal helical bundles of Sec6, Cog4, and Dsl1 are not required for complex formation (2, 3, 15, 16). Further, as with Vps53, the C terminus of Dsl1 is thought to interact with transport vesicles (3), and the Sec6 C terminus is thought to be important for binding to the target membrane (22). As noted above, the Vps53 and Cog4 (but not Sec6) C termini have conserved surface patches on the same face, which might serve a similar function. Thus, at this still early stage in structural and functional studies of membrane tethering complexes, it seems possible that these structurally related domains may perform related functions.
Another outstanding question is whether the higher-order organizations of the different tethering complexes have similarities, because thus far only the architecture of Dsl1 (3) is well understood. Size differences may argue against a common overall organization. Dsl1 and GARP have three subunits (or four for the yeast GARP), and COG and the exocyst each have eight (8). Dsl1 and GARP may be organizationally similar, however, and it remains possible that their architecture is repeated in forming the larger complexes. Hopefully structural and functional studies will help to answer these challenging questions in the next several years.
Methods
Cloning.
Sequences for full-length Vps53 from S. cerevisiae as well as for several C-terminal fragments thereof (residues 95–822, 200–822, and 554–822) were cloned into a modified pCOLA DUET plasmid (Novagen) between the BamHI and the NotI restriction sites. The constructs include an N-terminal hexahistidine tag followed by a tobacco etch virus (TEV) protease cleavage site. The construct used for structure determination includes residues 554–822 of the Vps53 sequence. In this construct, residues Ile586 and Leu722 were mutated to methionine using QuikChange site-directed mutagenesis (Stratagene).
Protein Expression and Purification.
E. coli BL21(DE3) cells harboring plasmids for Vps53 protein constructs were grown at 37 °C in Luria broth supplemented with 30 μg/mL of kanamycin. When the cells reached an OD600 of 0.6–0.8, the cells were shifted to 20 °C, and protein production was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside. The selenomethionine substituted protein used for structure determination was similarly expressed, except that the cells were grown on M9 media, supplemented with all amino acids except methionine. Just before induction, L-selenomethionine was added (60 mg/L) along with threonine, lysine, and phenylalanine (100 mg/L), and leucine, isoleucine, and valine (50 mg/mL) to inhibit methionine biosynthesis. Protocols for expression of selenomethionine substituted proteins are well described (23).
Cells were harvested 12–15 h after induction. They were resuspended in lysis buffer [50 mM Tris-HCl (pH 8.0), 0.5 M NaCl, 5% glycerol (wt/vol), 20 mM imidazole (pH 8.0), and 20 mM β-mercaptoethanol] supplemented with Complete Protease Inhibitor Mixture (Roche), 0.5 mg/mL lysozyme, and 0.5 mg/mL DNaseI. The cells were lysed in a cell disruptor (Avestin), and the lysate was cleared by centrifugation. Vps53 protein was isolated by batch binding to Ni-NTA resin (Qiagen). The resin was washed with 10 bed volumes of lysis buffer, followed by 10 more volumes of lysis buffer supplemented to contain 50 mM imidazole. Vps53 protein was eluted using lysis buffer supplemented to contain 250 mM imidazole. To cleave the hexahistidine tag, TEV protease was added and incubated overnight at 4 °C. The cleavage reaction was terminated by addition of Complete Protease Inhibitor Mixture (Roche), and the protein was concentrated for further purification on a MonoQ anion exchange column (GE Amersham). The MonoQ column was equilibrated with 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 5% glycerol, and 10 mM DTT, and protein was fractionated using a gradient of 0.15–0.5 M NaCl. Final purification was by gel filtration on a Superdex200 16/30 column (GE Amersham). All Vps53 constructs behaved as monomers on the gel filtration column. Protein was collected from the gel filtration column in buffer containing 20 mM Tris-HCl (pH 8.0), 0.5 M NaCl, and 10 mM DTT and concentrated to 10 mg/mL for crystallization.
Crystallization and Structure Determination.
Crystals of the C-terminal fragment of selenomethionine substituted Vps53 (residues 554–822) were grown at room temperature using the hanging drop method. The protein solution (1 μL) was mixed with an equal volume of well solution containing 50 mM Tris (pH 8.5), 10% PEG 4K, 200 mM BaCl2, and 10 mM DTT. Crystals appeared in 1 wk, growing to final dimensions of 200 × 60 × 60 μ within 2 wk. They belong to spacegroup P6322 (a = b = 127.2 Å, c = 81.7 Å). For data collection, the crystals were quick-dunked in well solution supplemented to contain 20% (vol/vol) of glycerol and flash-frozen in liquid nitrogen.
Data from the flash-frozen crystals were collected at beamline 24-IDC at the Advanced Photon Source in Chicago, IL. For structure determination, we used complete and highly redundant anomalous data from a single selenomethionine substituted crystal (Table 1). Data were integrated and scaled using HKL2000 (24). Selenium and barium positions (barium also has a large anomalous signal at the wavelength used) were located with SHELXD (25). The positions were refined and phases were calculated with the program SHARP (26). Phases were modified by solvent flipping as implemented in SHARP, and an easily interpretable electron density map was obtained at a resolution of 2.9 Å (Fig. S2). An initial model was built into the experimental density using the programs O and COOT (27, 28). There is one copy of the Vps53 C-terminal fragment in each asymmetric unit. Cycles of manual rebuilding, energy minimization, and Translation/Libration/Screw (TLS) refinement were performed using REFMAC (29, 30). The final model includes residues 564–730 and 742–779 of Vps53, 19 water molecules, and three barium ions. Final Rwork is 18.9%, and Rfree is 22.1%. Other details describing the refined structure are listed in Table 1. Coordinates and structure factors have been deposited in the Protein Data Bank (accession no. 3NS4).
CPY Secretion Assay.
Mutations were made to plasmid PLC92, an integrating plasmid bearing the Vps53 C terminus (a kind gift from Dr. T. Stevens, University of Oregon), using the Quikchange system (Stratagene). The vps53 mutations were introduced into NY1490 (NY1490: Mat a, leu2-3,112, ura3-52, trp1, his3α200, LA-) by transformation and selected on –Trp dropout medium. PCRs were performed on genomic DNA isolated from transformants to verify integration at the correct locus, and the resulting PCR products were sequenced to check that the mutations were present in the VPS53 coding sequence. To perform the CPY secretion assay, cells were grown overnight in YPD at 25 °C, and six OD600 units of cells were harvested for the assay. The cells were pelleted, and the culture supernatant was carefully removed. Proteins in the supernatant were precipitated by addition of 100% (wt/vol) trichloroacetic acid (TCA) to a final concentration of 10% TCA. After incubation on ice for 30 min, the precipitated proteins were pelleted at 20,000 × g for 15 min, washed in acetone, and resuspended in 1× SDS/PAGE loading buffer. Tris base (1 M) was added at a 1:50 dilution, when needed, to neutralize any residual TCA. The pelleted cells were washed with 1× PBS and then lysed by vortexing in the presence of glass beads and 100 μL of 1× PBS. One hundred microliters of 2× SDS/PAGE loading buffer was added to the lysate, and the samples were separated by SDS/PAGE, transferred to nitrocellulose, and probed with polyclonal antibodies against CPY. We also probed for Vps53 expression with monoclonal antibodies against C-terminal 3× HA tags present in the all Vps53 constructs except the C-terminal truncations (T1, T2).
Supplementary Material
Acknowledgments
We thank Dr. T. Stevens (University of Oregon) for his kind gift of PLC92; Dr. D. Kuemmel for help with data collection; and Dr. Kuemmel and Dr. D. W. Rodgers for discussions regarding the manuscript. Data were collected at beamline 24-IDC at the Advanced Photon Source, and we thank the Northeastern Collaborative Access Team staff for their support. Work presented here was funded by National Institutes of Health Grants GM080616 (to K.M.R.), GM035370 (to P.N.), and F30HL097628 and MSTP TG 2T32GM07205 (to N.V.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Atomic coordinates and structure factors have been deposited to the Protein Data Bank (accession no. 3NS4).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009419107/-/DCSupplemental.
References
- 1.Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Perez-Victoria JF, Bonifacino JS. Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-Golgi network. Mol Cell. 2009;29:5251–5263. doi: 10.1128/MCB.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren Y, et al. A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell. 2009;139:1119–1129. doi: 10.1016/j.cell.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, et al. Membrane association and functional regulation of Sec3 by phospholipids and Cdc42. J Cell Biol. 2008;180:145–158. doi: 10.1083/jcb.200704128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He B, Xi F, Zhang X, Zhang J, Guo W. Exo70 interacts with phospholipids and mediates the targeting of the exocyst to the plasma membrane. EMBO J. 2007;26:4053–4065. doi: 10.1038/sj.emboj.7601834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim YG, et al. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell. 2006;127:817–830. doi: 10.1016/j.cell.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 7.Hughson FM, Reinisch KM. Structure and mechanism in membrane trafficking. Curr Opin Cell Biol. 2010 doi: 10.1016/j.ceb.2010.03.011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whyte JR, Munro S. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev Cell. 2001;1:527–537. doi: 10.1016/s1534-5807(01)00063-6. [DOI] [PubMed] [Google Scholar]
- 9.Koumandou VL, Dacks JB, Coulson RM, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell. 2000;11:305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitt-John T, et al. Mutation of Vps54 causes motor neuron disease and defective spermiogenesis in the wobbler mouse. Nat Genet. 2005;37:1213–1215. doi: 10.1038/ng1661. [DOI] [PubMed] [Google Scholar]
- 12.Siniossoglou S, Pelham HR. Vps51p links the VFT complex to the SNARE Tlg1p. J Biol Chem. 2002;277:48318–48324. doi: 10.1074/jbc.M209428200. [DOI] [PubMed] [Google Scholar]
- 13.Liewen H, et al. Characterization of the human GARP (Golgi associated retrograde protein) complex. Exp Cell Res. 2005;306:24–34. doi: 10.1016/j.yexcr.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 14.Panic B, Whyte JR, Munro S. The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr Biol. 2003;13:405–410. doi: 10.1016/s0960-9822(03)00091-5. [DOI] [PubMed] [Google Scholar]
- 15.Sivaram MV, Furgason ML, Brewer DN, Munson M. The structure of the exocyst subunit Sec6p defines a conserved architecture with diverse roles. Nat Struct Mol Biol. 2006;13:555–556. doi: 10.1038/nsmb1096. [DOI] [PubMed] [Google Scholar]
- 16.Richardson BC, et al. Structural basis for a human glycosylation disorder caused by mutation of the COG4 gene. Proc Natl Acad Sci USA. 2009;106:13329–13334. doi: 10.1073/pnas.0901966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong G, Hutagalung AH, Fu C, Novick P, Reinisch KM. The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat Struct Mol Biol. 2005;12:1094–1100. doi: 10.1038/nsmb1017. [DOI] [PubMed] [Google Scholar]
- 18.Hamburger ZA, Hamburger AE, West AP, Jr, Weis WI. Crystal structure of the S.cerevisiae exocyst component Exo70p. J Mol Biol. 2006;356:9–21. doi: 10.1016/j.jmb.2005.09.099. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita M, et al. Structural basis for the Rho- and phosphoinositide- dependent localization of the exocyst subunit Sec3. Nat Struct Mol Biol. 2010;276:180–186. doi: 10.1038/nsmb.1722. [DOI] [PubMed] [Google Scholar]
- 20.Andag U, Schmitt HD. Dsl1p, an essential component of the Golgi-endoplasmic reticulum retrieval system in yeast, uses the same sequence motif to interact with different subunits of the COPI vesicle coat. J Biol Chem. 2003;278:51722–51734. doi: 10.1074/jbc.M308740200. [DOI] [PubMed] [Google Scholar]
- 21.Cai H, et al. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007;445:941–944. doi: 10.1038/nature05527. [DOI] [PubMed] [Google Scholar]
- 22.Songer JA, Munson M. Sec6p anchors the assembled exocyst complex at sites of secretion. Mol Biol Cell. 2009;20:973–982. doi: 10.1091/mbc.E08-09-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–529. [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Sheldrick G, Schneider T. SHELXL: High-resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 26.De LaFortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 27.Kleywegt GJ, Jones TA. Model building and refinement practice. Methods Enzymol. 1997;277:208–230. doi: 10.1016/s0076-6879(97)77013-7. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 31.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 32.Nicholls A, Sharp KA, Honig B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.