Abstract
Despite its fundamental nature, bacterial chromosome segregation remains poorly understood. Viewing segregation as a single process caused multiple proposed mechanisms to appear in conflict and failed to explain how asymmetrically dividing bacteria break symmetry to move only one of their chromosomes. Here, we demonstrate that the ParA ATPase extends from one cell pole and pulls the chromosome by retracting upon association with the ParB DNA-binding protein. Surprisingly, ParA disruption has a specific effect on chromosome segregation that only perturbs the latter stages of this process. Using quantitative high-resolution imaging, we demonstrate that this specificity results from the multistep nature of chromosome translocation. We propose that Caulobacter chromosome segregation follows an ordered pathway of events with distinct functions and mechanisms. Initiation releases polar tethering of the origin of replication, distinction spatially differentiates the two chromosomes, and commitment irreversibly translocates the distal centromeric locus. Thus, much as eukaryotic mitosis involves a sequence of distinct subprocesses, Caulobacter cells also segregate their chromosomes through an orchestrated series of steps. We discuss how the multistep view of bacterial chromosome segregation can help to explain and reconcile outstanding puzzles and frame future investigation.
Keywords: Par system, quantitative image analysis, bacterial cell biology
The process of cellular division requires faithful partitioning of chromosomes into two daughter cells. In most cells, cytokinesis and its accompanying chromosome segregation are symmetric events. However, some cells use asymmetric mechanisms for completing chromosome segregation. These segregation mechanisms must break the symmetry of the two newly replicated daughter chromosomes to differentially translocate them. Multiple mechanisms have been proposed to drive bacterial chromosome segregation (1–9), and the pervasive assumption that bacteria are simple organisms that move their chromosomes in a single process has led these mechanisms to appear in conflict. Another unresolved question concerns how the two chromosomes are distinguished during asymmetric chromosome segregation to ensure that only one of the chromosomes is translocated across the cell.
The polarized cell cycle of Caulobacter crescentus makes it well-suited for studying asymmetric chromosome segregation in bacteria. Caulobacter's origin of replication (ori) localizes to a specific cell pole before replication begins (10), and upon being duplicated, one and only one of the two oris translocates from the proximal, stalked pole to the distal, nascent swarmer pole (11). The ori and its neighboring parS loci are the first chromosomal sequences to be moved and the rest of the chromosome follows, such that ori-region translocation is the guiding event of chromosome segregation (8, 11). Previous efforts to image the dynamics of Caulobacter ori translocation demonstrated that in an averaged population, ori motion is faster than the rate of cell growth and largely directional, suggesting that it is driven by an active transport process (11). One candidate mechanism for mediating this active translocation involves the Par system, which consists of two proteins, ParA and ParB, and the origin-proximal parS cis-acting DNA sequence (6, 12). ParA is an ATPase that binds to ParB, which in turn binds parS. The Par system has been generally implicated in Caulobacter chromosome segregation (6, 8, 13), but its specific role is not understood. Here, we quantitatively analyze individual cells at unprecedented spatial and temporal resolution to characterize the dynamics of ori translocation in Caulobacter and define the specific function of the Par system. We find that Caulobacter chromosome segregation is a multistep process involving distinct initiation, distinction, and commitment steps that together ensure its robust execution. We propose that ParA retraction serves as an irreversible mechanism for specifically committing one and only chromosome to the process of translocation.
Results and Discussion
Caulobacter ParA Forms a Polarized Gradient Whose Retraction Mediates the Latter Part of ParB Translocation.
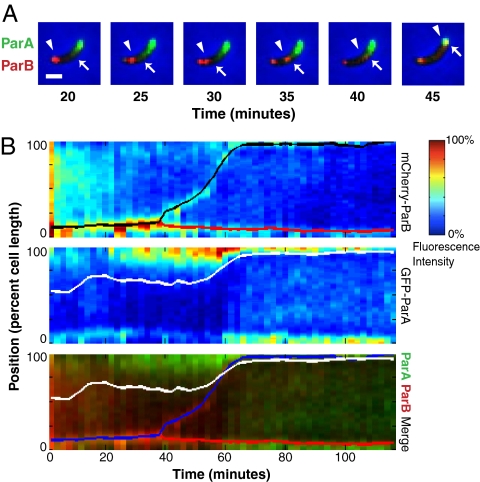
To investigate the mechanisms underlying Caulobacter chromosome segregation, we initially focused on the Par system. To examine the potential functions of ParA and ParB, we determined their relative spatial and temporal localization patterns (Fig. 1 and Fig. S1). At the beginning of the cell cycle, GFP-ParA was primarily localized to the distal pole, and showed a graded, “comet tail-like” localization that extended from the distal cell pole. Meanwhile, mCherry-ParB initially localized to a polar focus at the pole with the lowest density of ParA. As the cell cycle began, the ParB focus duplicated into two foci and one of the two foci advanced into the vicinity of the ParA comet tail. Once the ParB focus moved into the edge of the ParA comet tail, the ParA comet tail began to retract. By projecting the fluorescence intensities of both GFP-ParA and mCherry-ParB along the long axis of the cell, it became clear that ParA retraction always began only upon association with ParB, and that the translocating ParB focus moved with the retreating edge of the ParA gradient (Fig. 1B and Fig. S1). This concurrent delocalization of the retreating edge of the ParA structure with the translocating ParB continued for the duration of late translocation, until the ParB focus reached the distal pole. Once the ori reaches the distal pole, it may be anchored there by PopZ (14, 15) to ensure that this translocation is maintained. These dynamics were highly reproducible from cell to cell in over 50 cells examined, and four representative cells are illustrated. To help visualize ParA and ParB, we reduced the localization dynamics of each fusion to a kymograph and labeled the peak of the mCherry-ParB fluorescence and a normalized threshold of GFP-ParA intensity (Fig. 1B and Fig. S1). The colocalization patterns suggest that the translocation of ParB results from ParB inducing ParA retraction, which in turn pulls ParB across the cell. This model supports the conclusions of previous studies of ParA homologs from other systems (2, 6, 8, 16).
Fig. 1.
ParA retracts upon ParB association and appears to pull ParB to the distal pole. (A) Overlaid images from a time-lapse of a cell coexpressing mCherry-ParB (red) and GFP-ParA (green). Arrowhead indicates the translocating mCherry-ParB focus and the arrow indicates the edge of the GFP-ParA comet tail. (Scale bar, 1 μm.) (B) Kymographs of a cell coexpressing mCherry-ParB and GFP-ParA. The kymographs are a composite of the cell's fluorescence intensities at 2-min intervals over a 2-h time-course for mCherry-ParB (Top), GFP-ParA (Middle), and mCherry-ParB (red) and GFP-ParA (green) merge (Bottom). The red and black curves (Top) and the red and blue curves (Bottom) display the positions of the proximal (red) and distal (black/blue) mCherry-ParB labeled oris, respectively. The white line (Middle and Bottom) indicates a normalized threshold of GFP-ParA intensity (see Materials and Methods for details).
Because ParA had previously been implicated as a general mediator of DNA translocation (2, 6, 8, 16), we were surprised to note that much of ParB's motion early in the process of chromosome segregation occurred in a region of the cell that is largely devoid of ParA. To distinguish whether ParA simply cannot be detected in this region or whether it does not function in this region, we examined the effect of perturbing ParA function with a dominant-negative ParA mutant. ParAK20R-mCherry has been previously shown to inhibit both ParA function and the overall process of segregation in Caulobacter (8), but its specific effect on ParB motion had not been described. We found that cells expressing ParAK20R-mCherry still duplicated their GFP-ParB foci and moved one of the two ParB foci toward the opposite cell pole. In contrast to ParB foci in unperturbed cells, these ParB foci generally failed to progress all of the way across the cell and on average arrested at 40.6 ± 0.8% of cell length. This position corresponded well with the end of the comet tail of ParA, supporting the hypothesis that the early dynamics of ParB are independent of ParA. Together, these results suggest that ParA is necessary for Caulobacter chromosome segregation, but only mediates the latter stage of this process.
Quantitative Analysis of ParB Trajectories Identifies Distinct Phases of Chromosome Segregation.
The specific role of ParA in the latter part of ParB motion suggested that more than one type of process might govern ParB dynamics. Previous efforts to image the segregation dynamics of the origin and its neighboring loci have only identified a single type of motion (11), but these studies were performed at relatively low temporal resolution and averaged the trajectories of multiple cells, potentially obscuring distinct phases of motion in individual cells. We imaged the dynamics of ParB in living cells at a temporal resolution of 15 s, producing hundreds of images per cell cycle. To circumvent complications introduced by exogenous chromosome labeling methods, we used a strain in which the endogenous parB gene was replaced with a fully functional gfp-parB fusion that binds to and labels native parS sites in close proximity to the ori (17). To quantitatively analyze the immense amount of data produced by each of these experiments, we used our recently developed PSICIC software package (18), which takes advantage of advanced subpixel-resolution analysis methods (19). A synchronized movie of a representative cell including the raw image, 2D PSICIC tracking, and 1D representation illustrates the analysis pipeline (Movie S1). All motions were calculated as 2D projections and subsequently reduced to 1D along the cell length for ease of visualization and interpretation.
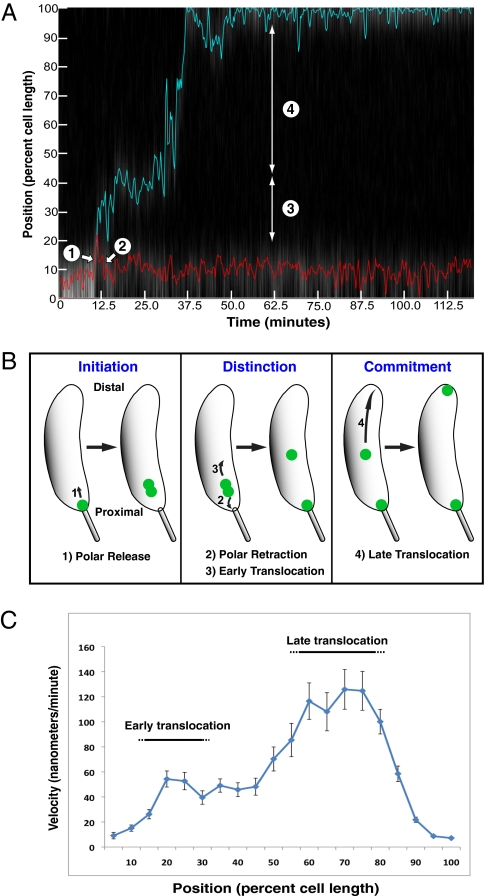
Unexpectedly, we observed that ori translocation does not proceed via a single continuous motion, but rather is a complex process with four definable steps (Fig. 2). First, the single polar ori moves away from the proximal stalked pole (we term this motion “polar release”) (14, 20). After polar release, the single GFP-ParB focus separates into two foci. One of these newly duplicated foci then returns to the proximal pole (“polar retraction”). Meanwhile, the other newly duplicated ori begins a relatively slow motion toward midcell (“early translocation”). After early translocation, the now-distal ori undergoes a final rapid burst of motion that carries it to the distal cell pole (“late translocation”). Although individual cells show variability in the timing, duration, and velocity of the above motions, these four distinct steps are seen in most cells. To illustrate these steps, five representative examples of individual cells are shown in Fig. 2A and Fig. S2, and aggregate data for over 100 cells are shown in the plots in Fig. 2C and Fig. S3.
Fig. 2.
Caulobacter chromosome segregation is a complex motion comprised of multiple steps. (A) Kymograph showing GFP-ParB labeled oris translocating within a single cell. The kymograph is a composite of fluorescence intensity measured along the cell length at 15-s intervals over a 2-h time-course. PSICIC image analysis was used to detect and track the proximal (red) and distal (blue) GFP-ParB labeled oris (see Materials and Methods for details). The steps of ori translocation are labeled: 1, polar release; 2, polar retraction; 3, early translocation; and 4, late translocation. (B) A schematic summarizing the steps of ori translocation proposed here. (C) Plot of average GFP-ParB instantaneous velocity as a function of relative cell position in bins of 5% of cell length (error bars = SEM). Velocities are based on mean squared displacement (values are positive when oriented toward the distal pole and negative when oriented toward the proximal pole).
To better characterize the four types of motion, we calculated the instantaneous velocity of the ParB foci using mean squared displacement, and plotted these velocities as a function of cellular position (Fig. 2C). Considering velocity with respect to position helped to account for variability in timing between cells. Motions oriented toward the distal pole were assigned positive values and motions oriented toward the proximal pole were assigned negative values. When all of the trajectories were combined, two velocity peaks could be observed (Fig. 2C). These two peaks corresponded to the early and late translocation steps of the distal ParB focus, as confirmed by plotting only the velocities of the distal focus (Fig. S3A). The early translocation occurred in the proximal third of the cell and peaked at 83 ± 10 nm/min, and the late translocation occurred in the distal half of the cell and peaked at 126 ± 16 nm/min (Fig. S3A). Because the polar release and polar retraction steps covered the same cellular region, they largely obscured each other when all velocities were included (Fig. 2C). These motions could be visualized by separately plotting the velocities of ParB before the focus resolved into two foci (Fig. S3B), and plotting the velocities of the most proximal ParB foci after splitting (Fig. S3C). Polar release has a relatively constant positive velocity of roughly 6 nm/min before ParB separation (Fig. S3B), but polar retraction proceeds via a brief burst of negative velocity that returns the proximal focus to the cell pole (Fig. S3C). These results demonstrate that ParB dynamics are mediated by distinct phases of motion with distinct characteristics.
To confirm that the ParB dynamics do not result from the specific fusion imaged, we repeated our tracking analysis on a strain in which multiple LacO sequences have been integrated near the origin of replication, allowing for their visualization with a LacI-CFP fusion (11). This strain photobleached more rapidly than the GFP-ParB strain, such that we had to image it at 3-fold lower temporal resolution, taking an image every 45 s. Nevertheless, the results from the two strains were largely similar. In particular, the distinct stages of polar release, polar retraction, early translocation, and late translocation were apparent in most cells (Fig. S4). The transient steps of polar release and polar retraction were observed in fewer cells than in the GFP-ParB data, likely reflecting the lower temporal resolution of these data. Thus, the multistep nature of Caulobacter origin segregation is independent of the labeling method used to track its dynamics.
ParA Mediates an Irreversible Commitment Step.
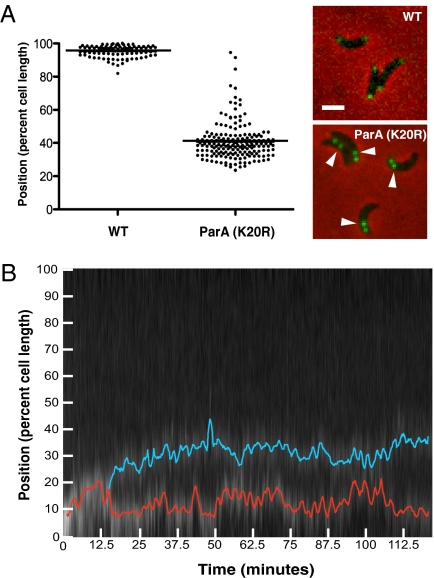
The localization of ParA at the distal pole and the incomplete segregation phenotype of ParAK20R-mCherry suggested that ParA could mediate the late translocation step. Consistently, we found that the position in the cell where ParB arrested in the presence of the dominant negative ParAK20R-mCherry (40.6 ± 0.8%) (Fig. 3A) corresponded extremely well to the position where the late translocation phase began in unperturbed cells (41.3 ± 1.3%). Furthermore, we applied the same high-resolution quantitative imaging approach to the strains expressing the dominant-negative ParAK20R-mCherry and observed that the late translocation motion was generally abolished (Fig. 3B and Fig. S5).
Fig. 3.
Perturbing ParA specifically disrupts the late translocation step of ori motion. (A) Scatter plot showing the position of the distal GFP-ParB focus 90 minutes after synchronization in WT (Left) and in cells expressing ParAK20R-mCherry (Right). Horizontal line indicates the mean. Images of representative cells are shown to the right. Arrowheads denote incompletely translocated GFP-ParB foci 90 minutes after synchronization. (Scale bar, 1 μm.) (B) A representative kymograph tracking the GFP-ParB labeled ori in a cell expressing ParAK20R-mCherry. The kymograph is a composite of the cell's fluorescent intensity at each 15-s time-point over the 2-h time-course. PSICIC image analysis was used to detect and track the proximal (red) and distal (blue) GFP-ParB foci.
An important feature of the ParB-triggered ParA retraction model is that the ParA machinery gets disassembled during ParB movement, ensuring that once ParA moves the distal ori (the first ori that it encounters) it cannot later move the second ori. Thus, Par-mediated translocation represents an irreversible commitment step after which no further oris can be segregated. ParA homologs have been shown to form cytoskeletal filaments. Consequently, the mechanism by which ParA mediates ParB translocation could be based on ParB stimulation of ParA depolymerization, which would in turn cause ParB to follow the retracting ParA structure via a Brownian ratchet type of process. Such ParB-mediated depolymerization of ParA could serve two simultaneous functions, both translocating the distal ParB focus and preventing the translocation of the proximal ParB focus.
Early Phases of Motion May Initiate Segregation and Distinguish the Proximal and Distal Chromosomes.
ParA retraction appears to translocate ParB, but how does the cell ensure that only one of the two chromosomes is translocated by ParA? The cellular solution to this problem appears to use polar retraction and early translocation to distinguish the two oris as distal and proximal loci before ParA engagement. Immediately after ori replication, the duplicated oris are equivalent and in close proximity. If ParA encountered the oris in this state, it would be incapable of distinguishing the two oris and could inadvertently move both oris to the distal pole. Consistent with this scenario, we find that when DNA replication is blocked with novobiocin (21), the single ori is occasionally translocated across the cell, resulting in a cell with an inverted ParB polarity (Fig. S6A). This aberrant translocation depends upon ParA, as it was never observed when cells expressing the ParAK20R-mCherry dominant-negative mutant were treated with novobiocin (n > 100) (Fig. S6B). To prevent such catastrophic mis-segregation by ParA, the two newly-replicated oris must consequently be spatially separated before interaction with the Par machinery. The motions of early translocation and polar retraction may thus serve as a distinction step, breaking symmetry to generate unique proximal and distal oris.
The mechanism by which this distinction occurs remains mysterious. DNA replication, RNA transcription, DNA condensation, membrane protein transertion, and entropic unmixing have all been proposed as mechanisms by which the bulk segregation of DNA could occur (1, 3–5, 7, 9) (hereafter collectively refered to as “bulk segregation mechanisms”). These mechanisms have been considered problematic as candidate drivers of ori motion because they appear incapable of generating the directionality and possibly the force required to complete the entire process of asymmetric ori translocation. However, our results suggest that the middle steps only need to spatially separate, and thereby distinguish, the two oris. As these motions do not need to be long, fast, or directional, it is therefore conceivable that they might be carried out by any combination of the bulk segregation mechanisms. Because these bulk mechanisms are essential, pleiotropic, and difficult to specifically perturb, future work will be needed to detail the precise mechanisms that mediate ori distinction.
If late translocation represents a commitment step and early translocation and polar retraction are part of a distinction step, then what might be the function of the first step of translocation, polar release? Previous studies demonstrated that ParB interacts with the polar protein, PopZ, to tether parS and the neighboring ori loci to the cell pole (14, 15). PopZ remains at the pole throughout segregation (14, 15), yet GFP-ParB moves away from the pole (14), indicating that segregation involves the release of ParB from PopZ. During this process of polar release, only one GFP-ParB focus can be observed, but this single focus could represent either one unreplicated focus or two superimposed foci. To distinguish whether polar release occurs before, during, or after the initiation of DNA replication, we imaged ParB dynamics in the presence of the DNA replication inhibitor novobiocin. As expected, novobiocin treatment inhibited ori duplication such that cells exhibited only one GFP-ParB focus throughout the time-lapse. However, these single oris still showed polar release in the form of movement of the GFP-ParB focus away from the proximal cell pole (Fig S7). On average, the focus in these cells was initially located 4.5 ± 0.4% of the cell length away from the proximal pole and moved to 10.6 ± 1.2% of the cell length during the course of the experiment. The end point of this motion corresponds well with the end point of polar release in wild-type cells (10.7 ± 0.9%). Novobiocin inhibits the activity of DNA gyrase, and a low level of DNA replication can proceed in its presence (21). Although the novobiocin result thus does not rule out a minor role for DNA replication in polar release, a recent report that used other methods for perturbing DNA replication provides yet further support that the initial phases of chromosome segregation do not require replication (20).
If polar release is independent of DNA replication, one would predict that the DNA replication machinery (replisome) should assemble at a position displaced from the cell pole. We thus examined the localization of an mCherry fusion to DnaN (CC0156), the β subunit of DNA polymerase III (22). In synchronized cells expressing mCherry-DnaN, the cellular position where DnaN initially localized was 13.8 ± 1.7% of the cell length from the proximal pole. This position corresponded well with both the end point of replication-independent motion (10.6 ± 1.2%) and the end point of unperturbed polar release (10.7 ± 0.9%). Together, these results suggest that DNA replication is not required for releasing ParB from its polar tether, and that polar release may be an initiation step for the pathway of chromosome translocation.
Conclusion
Complex cellular processes are often mediated by an ordered series of discrete steps that together ensure the overall fidelity of the process. For example, as early as 1880, Walther Flemming imaged eukaryotic mitosis and described it as proceeding through a series of steps with the distinct phases of prophase, metaphase, anaphase, and telophase (23). Because bacteria are both small and couple chromosome segregation to replication, similar analysis of bacterial chromosome dynamics only recently became feasible. Here we use high spatial- and temporal-resolution imaging to track the dynamics of the Caulobacter origin and its neighboring centromeric loci in individual cells. We find that Caulobacter chromosome segregation is also a complex multistep process that proceeds through a stereotyped pathway of distinct events. Thus, the overall process of segregating one and only one chromosome to the opposite cell pole is executed by an ordered progression: first, the ori is released from its polar tethering to initiate segregation; second, the ori is duplicated and the two daughter oris are spatially differentiated, distinguishing them as proximal and distal oris; and finally, the distal ori is committed to moving across the cell by an irreversible ParA-mediated translocation mechanism. It is worth noting that our study focuses on the initial chromosomal regions to be segregated, such that it remains unclear whether the remaining loci passively follow the origin or are transported by yet additional mechanisms.
In the past, multiple molecular mechanisms have been implicated in bacterial chromosome segregation. The view that a single mechanism mediates the entire segregation process caused these observations to appear in conflict. However, viewing chromosome segregation as a stepwise pathway of events allows a synthesis and reconciliation of much of the previous data. For example, ParA and a series of bulk segregation mechanisms have all been implicated in chromosome dynamics. Here we propose that all of these mechanisms may contribute, but to different steps: ParA mediates commitment, but the bulk segregation mechanisms might play an important role in distinguishing the two oris. The multistep nature of segregation may also help to explain why some factors, such as the MreB actin homolog, appear to influence the overall process of segregation in some contexts but not others (see ref. 13 and references contained therein). In the future, it will prove interesting to determine whether the segregation steps we define here are conserved in other bacterial species or are influenced by different growth conditions. The understanding of chromosome dynamics has been advanced by the appreciation of distinct steps in chromosome maturation such as DNA replication, decatenation, condensation, and cohesion (reviewed in ref. 24). Understanding that bacterial chromosome translocation is also a multistep developmental process rather than a single continuous event will thus similarly advance our understanding of chromosome segregation.
Materials and Methods
Media and Growth Conditions.
Cells for all experiments were grown in M2G minimal media at 30 °C (25). Novobiocin was diluted in water and administered at 5 μg/mL. Density gradient centrifugation with Percoll was used to synchronize cell populations, as previously described (26).
Microscopy.
All cells were imaged on 1% agarose pads containing M2G medium and the appropriate additives. For imaging of living cells, an asynchronous culture of OD660 0.4 was synchronized and swarmer cells were collected and transferred to a 1% agarose M2G pad. A coverslip was placed on the pad and then sealed with valap (1:1:1 parafin: lanolin: vaseline). In strains ZG192 and ZG194, expression of fluorescent proteins was induced with 0.03% xylose 3 h before and during imaging. Strain ZG195 was grown in the presence of 0.3% glucose. ParAK20R-mCherry was induced by washing the cells into media containing 0.03% xylose for 60 min before imaging. LS3833 was grown in the presence of 0.02% glucose and was induced with 0.03% xylose in the presence of 1 μM IPTG for 90 min before and during imaging. Images were collected on a Nikon 90i microscope equipped with a Nikon Plan Apo 100×/1.4 phase contrast objective, a Rolera XR cooled CCD camera, and NIS Elements software.
Strains and Strain Construction.
A strain table (Table S1) and description of strain construction is included in SI Materials and Methods.
Image Analysis.
Individual frames of time-lapse experiments were analyzed using the previously described PSICIC software (18). A binary threshold was applied to each image, and the thresholded versions of consecutive pairs of time-points were compared with to determine the shift, in pixels, between the two images. Individual cells in consecutive time-points were then matched by comparing the positions of the poles, thereby preserving the orientation of the internal coordinate system.
A 2D intensity profile was generated for each cell in every frame using bicubic interpolation at the points on the internal coordinate system generated by PSICIC. Two-dimensional peaks were found by locating positions that are brighter than one SD higher than the mean fluorescence value for that cell, and that are also local maxima in the horizontal, vertical, and diagonal dimensions. For the colocalization study of ParA and ParB and for the FROS labeling experiment, the intensity threshold was adapted to a higher value of the mean plus 1.5 SDs, which reflects the decreased amount of bleaching because of a lower imaging frequency. For peaks that are very close together, the brightest peak is chosen.
To track the path of the fluorescent foci over time, we found it useful to begin at the end of the time-course, when the foci are well separated, and move backward frame by frame. The algorithm finds peaks in each individual time-frame and connects a peak to the growing path by choosing the peak that minimizes the distance to the most recent points on the growing path. Comparing the newest point to several of the most recent points ensures that even when the two paths move close together and reseparate, the paths remain distinct. A window of 20 time-points was empirically chosen to give the best results in this data set. For the ParA-ParB colocalization and the FROS labeling experiments, the window size was reduced to three time-points. Occasionally, a fluorescent peak will not be visible in a time-frame because of noise in the data. To prevent such “missing“ peaks from interfering with the path tracking, we filtered out large “jumps,” empirically chosen to be motions within one frame of greater than 75% of the cell length (greater than 40% of the cell length in the case of the colocalization and the FROS labeling experiments). The paths were smoothed using a Savitzky–Golay smoothing filter of degree 2, with a window of five time-points.
The point of release of the old pole was defined as the latest time at which the polar focus exceeded the mean distance from the pole before separation. Separation was defined as the earliest point at which the two tracked foci do not overlap. The beginning of late translocation was measured as the latest time at which the focus crossed the midpoint of the cell. The distal polar capture, which is the end of the late translocation phase, was measured as the final time-point at which the focus was within the proximal 90% of the cell length.
For the identification of the boundary of the GFP-ParA aggregate at the distal pole (Fig. 1B and Fig. S1), we obtained the laterally averaged green fluorescent intensity as a function of the relative position along the long axis of the cell with the PSICIC analysis software (18). The boundaries of ParA were identified as the closest distance from the distal pole, at which the laterally averaged green fluorescence intensity underpasses a value of 17% of the difference between its maximum and its minimum value for each given time-point. The data-points were smoothed in time using a Savitzky–Golay smoothing filter of degree 2, with a window of five time-points.
Supplementary Material
Acknowledgments
We thank the members of the Z.G. laboratory, Ned Wingreen, Dorothy Lerit, and Coleen Murphy for helpful discussions and input on the manuscript, and Martin Thanbichler (Max Planck Institute for Terrestrial Microbiology, Marburg, Germany), Esteban Toro (Stanford University, Palo Alto, CA), and Lucy Shapiro (Stanford University, Palo Alto, CA) for materials. This work was supported in part by funds from Grant DE-FG02-05ER64136 from the US Department of Energy Office of Science, National Institutes of Health New Innovator Award 1DP2OD004389-01, and a Beckman Young Investigator Award (to Z.G.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005274107/-/DCSupplemental.
References
- 1.Dworkin J, Losick R. Does RNA polymerase help drive chromosome segregation in bacteria? Proc Natl Acad Sci USA. 2002;99:14089–14094. doi: 10.1073/pnas.182539899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fogel MA, Waldor MK. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 2006;20:3269–3282. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 4.Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: Lessons for the bacterial chromosome. Proc Natl Acad Sci USA. 2006;103:12388–12393. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemon KP, Grossman AD. Movement of replicating DNA through a stationary replisome. Mol Cell. 2000;6:1321–1330. doi: 10.1016/s1097-2765(00)00130-1. [DOI] [PubMed] [Google Scholar]
- 6.Mohl DA, Gober JW. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toro E, Hong SH, McAdams HH, Shapiro L. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc Natl Acad Sci USA. 2008;105:15435–15440. doi: 10.1073/pnas.0807448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woldringh CL. The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol Microbiol. 2002;45:17–29. doi: 10.1046/j.1365-2958.2002.02993.x. [DOI] [PubMed] [Google Scholar]
- 10.Jensen RB, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci USA. 1999;96:10661–10666. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viollier PH, et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schumacher MA. Structural biology of plasmid partition: Uncovering the molecular mechanisms of DNA segregation. Biochem J. 2008;412:1–18. doi: 10.1042/BJ20080359. [DOI] [PubMed] [Google Scholar]
- 13.Shebelut CW, Jensen RB, Gitai Z. Growth conditions regulate the requirements for Caulobacter chromosome segregation. J Bacteriol. 2009;191:1097–1100. doi: 10.1128/JB.00862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowman GR, et al. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134:945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell. 2008;134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ringgaard S, van Zon J, Howard M, Gerdes K. Movement and equipositioning of plasmids by ParA filament disassembly. Proc Natl Acad Sci USA. 2009;106:19369–19374. doi: 10.1073/pnas.0908347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 18.Guberman JM, Fay A, Dworkin J, Wingreen NS, Gitai Z. PSICIC: Noise and asymmetry in bacterial division revealed by computational image analysis at sub-pixel resolution. PLOS Comput Biol. 2008;4:e1000233. doi: 10.1371/journal.pcbi.1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reshes G, Vanounou S, Fishov I, Feingold M. Cell shape dynamics in Escherichia coli. Biophys J. 2008;94:251–264. doi: 10.1529/biophysj.107.104398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowman GR, et al. Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol Microbiol. 2010;76:173–189. doi: 10.1111/j.1365-2958.2010.07088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gellert M, O'Dea MH, Itoh T, Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci USA. 1976;73:4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collier J, Shapiro L. Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter. J Bacteriol. 2009;191:5706–5716. doi: 10.1128/JB.00525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flemming W. Contributions to the knowledge of the cell and its vital processes. J Cell Biol. 1965;25:3–69. [PMC free article] [PubMed] [Google Scholar]
- 24.Reyes-Lamothe R, Wang X, Sherratt D. Escherichia coli and its chromosome. Trends Microbiol. 2008;16:238–245. doi: 10.1016/j.tim.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 26.Alley MR. The highly conserved domain of the Caulobacter McpA chemoreceptor is required for its polar localization. Mol Microbiol. 2001;40:1335–1343. doi: 10.1046/j.1365-2958.2001.02476.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.