Abstract
Theory and empirical evidence suggest that plant–soil feedback (PSF) determines the structure of a plant community and nutrient cycling in terrestrial ecosystems. The plant community alters the nutrient pool size in soil by affecting litter decomposition processes, which in turn shapes the plant community, forming a PSF system. However, the role of microbial decomposers in PSF function is often overlooked, and it remains unclear whether decomposers reinforce or weaken litter-mediated plant control over nutrient cycling. Here, we present a theoretical model incorporating the functional diversity of both plants and microbial decomposers. Two fundamental microbial processes are included that control nutrient mineralization from plant litter: (i) assimilation of mineralized nutrient into the microbial biomass (microbial immobilization), and (ii) release of the microbial nutrients into the inorganic nutrient pool (net mineralization). With this model, we show that microbial diversity may act as a buffer that weakens plant control over the soil nutrient pool, reversing the sign of PSF from positive to negative and facilitating plant coexistence. This is explained by the decoupling of litter decomposability and nutrient pool size arising from a flexible change in the microbial community composition and decomposition processes in response to variations in plant litter decomposability. Our results suggest that the microbial community plays a central role in PSF function and the plant community structure. Furthermore, the results strongly imply that the plant-centered view of nutrient cycling should be changed to a plant–microbe–soil feedback system, by incorporating the community ecology of microbial decomposers and their functional diversity.
Keywords: buffering effect, ecological model, microbial community, nutrient cycling, plant–soil feedback
There is a long-standing view that a plant controls the soil conditions (e.g., size of inorganic nutrient pool) via litter supply in terrestrial ecosystems. On the basis of this view, a plant community and local soil conditions are understood as an outcome of the plant–soil codevelopment process. A change in the composition of a plant community leads to a change in litter quality, which alters the local nutrient cycling process and soil conditions; the changed soil conditions may in turn drive a further change in plant community composition. Those two processes taken together form a plant–soil feedback (PSF), a major driver of plant community dynamics and nutrient cycling [1, 2 (and references therein), 3].
Litter quality is a key plant trait that determines whether PSF supports or inhibits the coexistence of plant species. Both empirical (4–6) and theoretical (7–9) evidence indicate that litter-mediated PSF from the dominant species in a plant community can be positive (favoring species dominance) or negative (favoring competitor invasion), depending on the combinations of litter quality and nutrient competition strategy. If the dominant species favors nutrient-rich sites and produces a quickly decomposing litter, then the accelerated nutrient cycling maintains a competitive advantage, preventing competitor invasion and enhancing species dominance (8). Species dominance is also maintained when a species favors nutrient-poor sites and produces a slowly decomposing litter, leading to diminished nutrient cycling. With other combinations [i.e., species with a competitive advantage in nutrient-rich (or -poor) sites with a poorly (or easily) decomposing litter], plant control on nutrient cycling facilitates competitor invasion and accelerates succession (8).
Earlier studies concerning nutrient cycling PSF often neglected the roles of microbial decomposers in controlling the soil nutrient pool size, except some studies on microbial pathogens and symbionts that directly interact with plants (10–15). However, recent studies in microbial ecology have started to challenge this plant-centered view of plant–soil systems. Two specific lines of evidence suggest that microbial decomposers can modify litter-mediated PSF. First, empirical evidence indicates that direct control of the nutrient pool size by the plant litter can be weak (16–18) because nutrients such as nitrogen within the plant litter are first assimilated into a microbial biomass or soil organic matter (immobilization) (19) and then released into the pool available for plants (net mineralization). Second, recent advances in culture-independent techniques indicate that microbial communities exhibit distinct compositions (20–25) and/or functions (21–23, 25, 26), depending on the litter quality and plant species with which they are associated. It has been hypothesized that flexibility in the community composition and function of microbial decomposers either reinforces (14, 27, 28) or weakens (3, 18) plant control over nutrient mineralization, although direct evidence for either hypothesis is lacking.
Here, using a simple mechanistic model for plant–microbe–soil feedback (PMSF), we show that incorporation of the soil microbial community poses a significant effect on the plant–soil codevelopment process. More specifically, microbial diversity provides the microbial community with a functional flexibility that buffers changes in the decomposability and thus weakens the plant control over nutrient cycling. Furthermore, the microbial diversity tends to shift the sign of PMSF from positive to negative and thus may facilitate plant coexistence.
Model
Consider a plant community in a habitat comprising numerous discrete patches, each of which is either empty or is occupied by an individual of either of two plant species, a “better competitor for light” (PL) or a “better competitor for nutrient” (PN). The proportion of patches occupied by either species changes with time because of interpatch recruitment and within-patch mortality. Interpatch recruitment increases with the soil nutrient pool size (N) through increased fecundity and/or increased seedling survival rate. The plant species PL and PN are representatives of plant groups with contrasting strategies (29). A modified patch occupancy model enables us to simulate shifts in the plant community composition along a gradient of soil nutrient pool sizes (8). The better competitor for nutrient PN loses its competitive advantage and decreases its population size with increasing nutrient pool size. In contrast, the better competitor for light PL increases its population size and dominates the habitat in nutrient-rich conditions.
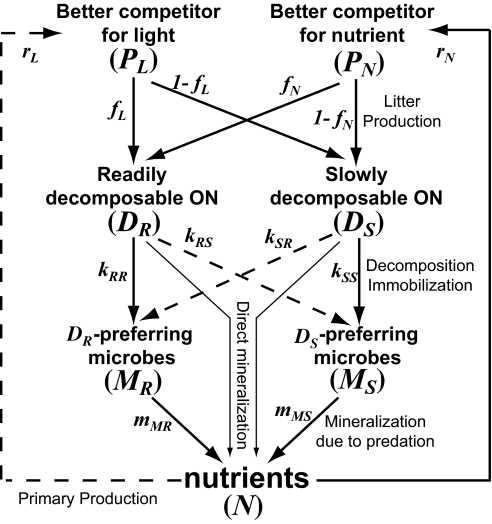
Nutrients assimilated by the plants are released to the environment as plant litter (organic matter), which is decomposed by microbes. The mineralized nutrients are released to the nutrient pool, forming a nutrient cycling process (Fig. 1). Plant litter (detritus) consists of two organic compounds: “readily decomposable organic nutrient” (DR) and “slowly decomposable organic nutrient” (DS). The fractions of DR in the litter from PL and PN are fL and fN, respectively. Litter with a higher DR fraction (i.e., higher litter decomposability) is decomposed more rapidly by microbial decomposers.
Fig. 1.
Flow diagram of the plant–microbe–soil feedback model. Flows from the inorganic nutrient pool (N) to the plant compartments (PL and PN) represent primary production processes. Flows from the plant compartments to the two litter compartments (DR and DS) represent litter production. Flows from the litter compartments to the microbial biomasses (MR and MS) represent decomposition (gross mineralization) and subsequent microbial immobilization (microbial growth) processes. Flows from the litter compartments to the inorganic nutrient pool represent direct mineralization. Flows from the microbial biomass to the inorganic nutrient pool represent net mineralization through nutrient release from microbial biomass due to predation.
It is reasonable to divide the whole microbial community into two contrasting functional groups (e.g., bacteria vs. fungi, or sugar fungi vs. cellulolytic fungi) that differ in their performance of the two organic nutrients (27, 30). “DR-preferring microbes” (MR) and “DS-preferring microbes” (MS) are better competitors for readily and slowly decomposable organic nutrient, respectively (i.e., functional complementarity) (Fig. 1). We assume that kRm > kSm represents the difference in litter quality (m = R or S) and that kRR > kRS and kSR < kSS represent the functional complementarity (31, 32), where kjm is the decomposition coefficient of Dj by Mm (j, m = R or S; Methods). The assimilation efficiency (eMj) is the fraction of mineralized organic nutrient that is assimilated into a biomass of Mj (j = R or S) (i.e., microbial immobilization), and 1 − eMj is the fraction released to the nutrient pool (i.e., net mineralization). We assume a per-capita group-specific mortality (mMj) due to predation for each group (j = R or S). Nutrients in the microbial biomass are also released to the nutrient pool via this mortality (27). Major nutrient fluxes in our model are shown in Fig. 1, the model equations are shown in Methods, and a list of parameters and their default values are found in Table S1. Although we did not explicitly include microbe predators, the following results are qualitatively robust irrespective of complexities in the microbial food webs under reasonable assumptions (Fig. S1).
Results
Plant–Soil Feedbacks Along the Nutrient Gradient.
In our simple model, the nutrient pool size N* at steady state is the key determinant of the sign of PSF (8). When the better competitor for light PL is dominant in the plant community, it prevents (or allows) the invasion of the better competitor for nutrient PN if the nutrient pool size is larger (or smaller) than 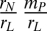 (≡ NL**), where mP represents the per-capita mortality rate of individual plants. This is defined as positive (or negative) PSF in this model. Conversely, when PN is dominant, it prevents (or allows) the invasion of PL if the nutrient pool size is smaller (or larger) than
(≡ NL**), where mP represents the per-capita mortality rate of individual plants. This is defined as positive (or negative) PSF in this model. Conversely, when PN is dominant, it prevents (or allows) the invasion of PL if the nutrient pool size is smaller (or larger) than 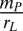 (≡ NN**), which is defined as positive (or negative) PSF. Therefore, the plant litter decomposability, which affects the nutrient recycling rate and thus the size of the soil nutrient pool, potentially affects the sign of PSF.
(≡ NN**), which is defined as positive (or negative) PSF. Therefore, the plant litter decomposability, which affects the nutrient recycling rate and thus the size of the soil nutrient pool, potentially affects the sign of PSF.
Roles of Microbial Diversity in Plant Litter Control over Nutrient Cycling.
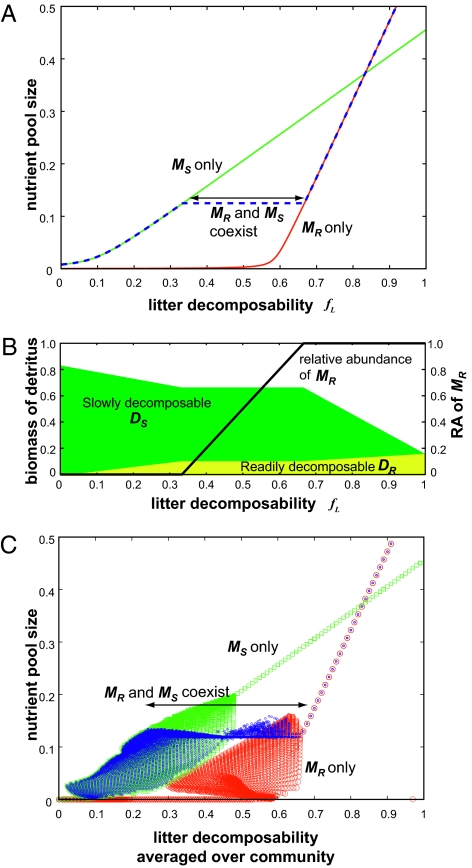
In the absence of microbial diversity, a higher litter decomposability simply leads to a larger nutrient pool size, regardless of the dominant microbial group. This is well demonstrated in a system in which a single plant species PL is dominant with a single microbial group (MR and MS for the red and green lines in Fig. 2A, respectively). This pattern agrees with the long-standing view that plant litter decomposability determines the nutrient pool size (1–3). However, this is no longer true when microbial diversity is considered; the dependence of the nutrient pool size on litter decomposability is much weaker in the presence of two microbial groups (blue broken line in Fig. 2A: 0.33 < fL < 0.66) than it is when microbial diversity is not considered (red and green lines in Fig. 2A).
Fig. 2.
Consequences of PMSF on nutrient cycling, as a function of plant litter decomposability. (A) Relationship between litter decomposability (fL) and equilibrium nutrient pool size in systems with a better competitor for light (PL) only. Red, green, and blue lines correspond to systems with only MR, only MS, and both MR and MS, respectively. In the region with coexisting MR and MS, the slope of the line is zero. (B) Relationship between litter decomposability (fL), relative abundance of DR-preferring microbes (RA of MR, dimensionless), and readily decomposable DR and slowly decomposable DS accumulations in a system with PL only. (C) Relationship between the average litter decomposability (long-term average of 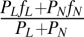 , from t = 45,000 to t = 50,000) and the average nutrient pool size (long-term average of N) in a system with two plant species. Consequences of PMSF on nutrient pool size for every combination of (fL and fN) from (0.0, 0.0) to (1.0, 1.0) with interval (ΔfL, ΔfN) = (0.01, 0.01) are plotted against the average litter decomposability in a system with microbial functional group MR only (red dots), MS only (green dots), and with two competing microbial groups (blue dots). All parameters are set as default values (Table S1).
, from t = 45,000 to t = 50,000) and the average nutrient pool size (long-term average of N) in a system with two plant species. Consequences of PMSF on nutrient pool size for every combination of (fL and fN) from (0.0, 0.0) to (1.0, 1.0) with interval (ΔfL, ΔfN) = (0.01, 0.01) are plotted against the average litter decomposability in a system with microbial functional group MR only (red dots), MS only (green dots), and with two competing microbial groups (blue dots). All parameters are set as default values (Table S1).
The decoupling of the litter decomposability and nutrient pool size is achieved by the flexible shift of the microbial community composition and mineralization process in response to variations in the plant litter decomposability. Low litter decomposability favors microbes MS, which prefer slowly decomposable DS [relative abundance (RA) of MR = MR/(MR + MS), RA of MR = 0 in Fig. 2B], whereas DR-preferring microbes MR are dominant when the litter decomposability is high (RA of MR = 1 in Fig. 2B). The two microbial groups coexist at equilibrium (0 < RA of MR < 1) at intermediate litter decomposability levels (0.33 < fL < 0.66), owing to the well-balanced supply of the two organic nutrient types (see SI Text, Section 1 and Tables S2 and S3 for the exact condition). In consequence of the competition between two microbes for two different types of organic nutrients, a higher plant litter decomposability (i.e., increased fraction of DR in plant litter) leads to a higher relative abundance of DR-preferring microbes MR (Fig. 2B). This flexibility in microbial community composition alters the accumulation pattern of readily decomposable DR and slowly decomposable DS in a way that decouples litter decomposability and nutrient pool size (Fig. 2B). When a single microbial group dominates the community with a very high or very low litter decomposability (fL < 0.33 or fL > 0.66 in Fig. 2B), the total amount of accumulated organic nutrients (DR + DS) decreases with an increasing supply of the readily decomposable fraction (DR) in the plant litter (i.e., higher litter decomposability fL), leading to a larger nutrient pool size (Fig. 2A). However, when DR- and DS-preferring microbes coexist (0.33 < fL < 0.66), the amount of accumulated organic nutrient remains constant despite increases in the litter decomposability (Fig. 2B). The increase in DR-preferring microbes (MR) allows rapid decomposition of the readily decomposable organic nutrient fraction, preventing an increase in DR accumulation. At the same time, the reduction in DS-preferring microbes (MS) causes slower decomposition of the slowly decomposable fraction, resulting in the same level of DS accumulation despite the increased litter decomposability. Therefore, the increase in litter decomposability does not increase the nutrient pool size (Fig. 2A).
Decoupling of the litter decomposability and the nutrient pool size is observed even when we assume differences in the mortalities of the microbial groups (Fig. S2A) or a more complex soil food web structure (Fig. S1). When we consider a full system comprising two plant species and two microbial groups, the link that develops between the average litter decomposability of the plant community and the realized nutrient pool size becomes weaker in the presence of microbial diversity (Fig. 2C). In the region where the two microbes coexist, the realized nutrient pool size tends to be intermediate between those realized in the two systems without microbial diversity (i.e., between that realized with the dominance of MS and that realized with the dominance of MR) (Fig. 2C).
Roles of Microbial Diversity in Determining the Sign of PSF.
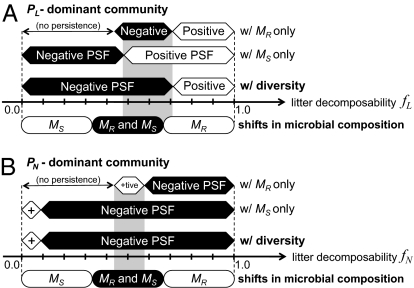
Consider a system comprising a single plant species in the absence of microbial diversity. When PL produces a litter with sufficiently high decomposability fL, it leads to a large nutrient pool size (>NL**), causing a positive PSF (Fig. 3A). The threshold litter decomposability, which separates negative and positive PSF, depends on which microbial group dominates the community. When PN is dominant, the sign of PSF is positive if its litter decomposability fN is lower than a certain threshold (which depends on the microbial community composition), leading to a small nutrient pool size (<NN**) (Fig. 3B). The buffering of the nutrient pool size achieved with functional microbial diversity (Fig. 2) alters these thresholds and shifts the sign of PSF under each combination of fL and fN (see also Tables S2 and S3).
Fig. 3.
Roles of microbial diversity in determining the sign of PSF. Dependence of the sign of PSF on litter decomposabilities fL and fN is shown in a system with PL only (“PL-dominant community”) (A) and PN only (“PN-dominant community”) (B), respectively. There are three distinct configurations for the microbial community: a system with MR only (“w/ MR only”), MS only (“w/ MS only”), and with microbial diversity (“w/ diversity”). The sign of PSF is positive (“Positive PSF”, ”Positive”, “+tive”, or “+”) or negative (“Negative PSF” or “Negative”). In a system with microbial diversity, the realized microbial composition depending on litter decomposability is shown as “Shifts in microbial composition” (MS, MR and MS, or MR). If the litter decomposability is too low, the system cannot persist (“no persistence”). Parameters are set to default values (Table S1).
Consider a system of PL with a high litter decomposability, which tends to cause a positive PSF. The high decomposability allows the invasion of MR into a system with MS (resulting in either coexistence with MS or dominance of MR) (Fig. 2B), which in turn leads to a smaller nutrient pool size (N*) than that in a system with MS alone (when 0.33 < fL < 0.82; Fig. 2A). The suppression of N* can shift the sign of PSF from positive to negative and increase the range of litter decomposabilities that cause a negative PSF (shaded ranges in Fig. 3A). Similarly, when PN has a low litter decomposability and thus tends to cause a positive PSF, the invasion of MS into a system with MR increases the nutrient pool size (when fL < 0.66; Fig. 2A) and causes a negative PSF (shaded range in Fig. 3B). Microbial diversity may shift the sign of PSF from negative to positive, decreasing the range of litter decomposabilities that cause a negative PSF (Fig. S2 B and C). Nevertheless, such a facilitation of microbial diversity on negative PSF is observed in a wide range of parameter values (Figs. S3 and S4), indicating that microbial diversity tends to facilitate negative PSF.
Roles of Microbial Diversity in Structuring the Plant Community.
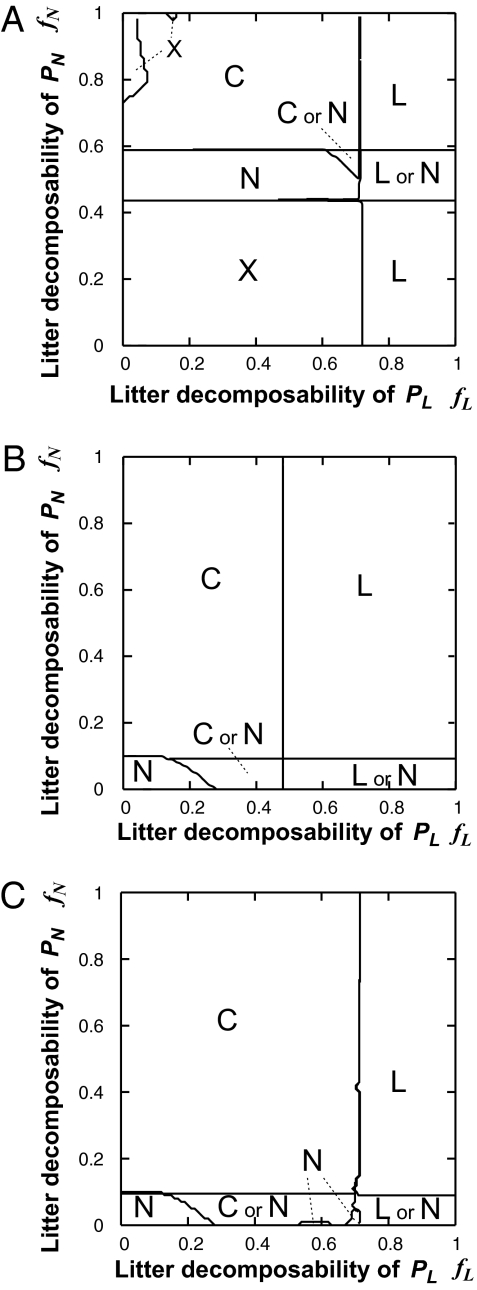
The microbial diversity that facilitates negative PSF tends to promote the coexistence of plant species. This is confirmed by comparing the dependence of plant coexistence on plant litter decomposability (0 ≤ fL, fN ≤ 1) in systems with a single microbial group (DR-preferring microbes, Fig. 4A; DS-preferring microbes, Fig. 4B) and in those with microbial functional diversity (Fig. 4C). Microbial functional diversity broadens the region where the two plant species can coexist (region C in Fig. 4C) compared with systems with a single microbial group (region C in Fig. 4 A or B) (see also Fig. S5 for temporal dynamics and Fig. S6 for the realized microbial community composition). Microbial diversity also narrows the region where both plant species cause a positive PSF and either plant species can dominate depending on the initial abundance (region L or N in Fig. 4C). However, microbial diversity may not facilitate plant coexistence. For example, when the microbial assimilation efficiency (eMj) is too high or too low, the region where the two plant species can coexist in a system with microbial diversity can be the same as that in a system with only MR or MS (Fig. S6 A and E, respectively). Yet even in such cases, microbial diversity never makes the plant coexistence region narrower than that in a system with only MR or only MS (Fig. S4).
Fig. 4.
Consequences of PMSF on plant communities, as a function of the plant litter decomposability (fL and fN). (A and B) Microbial community consisting of MR (A) or MS (B) only. (C) Microbial community consisting of MR and MS, and their realized community compositions as determined by PMSF. A better competitor for light (PL) is dominant in the plant community in region L, a better competitor for nutrient (PN) is dominant in region N, and the two plant species coexist in the region C. In region X, the ecosystem cannot persist because the plant litter decomposability of the dominant species or that of the invasible species is too low (see SI Text, Section 5). Coexistence or dominance of PN is realized in region C or N, and dominance of PL or PN is realized in region L or N, depending on the initial conditions. In a system with functional microbial diversity, dependence of the realized microbial composition (dominance of DR- or DS-preferring microbes or coexistence) on litter decomposability is not shown (Fig. S6). Although plant coexistence is realized by periodic succession with some combinations of litter decomposability, we did not distinguish this from coexistence at steady state. The presence of periodic succession may result in complex boundaries between region C or N, region N, and region L or N (see SI Text, Section 5). Parameters are set to default values (Table S1).
Discussion
Effects of Microbial Diversity on Plant Litter Control over Nutrient Cycling.
The present study is a theoretical attempt to understand the role of microbial diversity in PSF functioning. Microbial diversity with the functional complementarity in decomposing different types of plant litter weakens plant control over nutrient cycling. This is achieved by a change in the relative microbial abundance in response to plant litter decomposability. Increasing litter decomposability tends to decrease the relative abundance of microbes that favor slowly decomposable litter (Fig. 2B). This lowers the decomposition rate of, and thus enhances accumulation of, slowly decomposable litter (Fig. 2B), suppressing an increase in net mineralization rate. Thus, microbial diversity acts as a “buffer” against plant control over nutrient cycling and decouples the relationship between litter decomposability and nutrient pool size (Fig. 2 A and C). Our study presents a theoretical support for the hypothesis that microbial community weakens plant control over nutrient cycling (3).
Earlier studies, focusing on differences in the immobilization efficiencies of microbes, often argued that shifts in microbial composition reinforce plant control over nutrient cycling. This is because high (or low) litter decomposability favors bacteria (or fungi) with low (or high) immobilization efficiency and may enhance (or reduce) nutrient release rate from microbial biomass (14, 27, 28). Given the opposite prediction of the earlier hypothesis, it would be important to investigate how the immobilization efficiency and functional complementarity interact to determine the role of microbial diversity in plant control over soil. However, our model predicted that microbial diversity acts as the buffer even in the presence of differences in the immobilization efficiencies of microbes (Fig. S2A).
The decoupling of the litter decomposability and nutrient pool size predicted here can arise also from the phenotypic plasticity of the immobilization efficiency in response to changes in litter quality (18) and the buffering effect of soil arthropods that can modify microbial activities and compositions (33). These previous hypotheses and the plant litter control hypothesis can be tested against our hypothesis by manipulating the microbial community composition. The regression slope of the net mineralization rate or nutrient pool size with specific litter chemical traits (litter decomposability, e.g., C/N ratio, lignin concentration) will be smaller with increasing microbial diversity (our hypothesis), depend on the physiological flexibility of microbes (18) or the presence/absence/composition of soil arthropods (33), or be insensitive to these factors (plant litter control hypothesis).
Microbial Diversity, Negative Feedbacks, and Plant Coexistence.
Plant diversity is essential for maintaining ecosystem productivity (34). If litter-mediated plant control over soils causes a positive PSF, it may hinder plant coexistence and then negatively affect ecosystem productivity. However, it has been proposed that interactions between plants and other trophic levels (e.g., herbivores and microbes) can change the plant control over soil and may contribute to the maintenance of plant diversity (3). Recent studies have suggested that mutualistic and parasitic microbes in the rhizosphere cause negative PMSF and facilitate plant coexistence (10–15). In contrast, the diversity of free-living microbial decomposers is thought to facilitate plant coexistence by moderately enhancing nutrient recycling and plant community productivity, or by providing a diverse range of plant-available soil resources, thus contributing to niche differentiation (14, 35, 36). However, these previous studies did not explicitly consider the impact of the plant community on the community composition of microbial decomposers and thus PMSF. Our model presents a mechanism for plant coexistence through feedbacks between the plant, free-living microbial decomposers, and the soil. Microbial diversity provides the microbial community with a functional flexibility, which buffers litter-mediated plant control over nutrient cycling (Fig. 2). This buffering effect prevents the realization of an extremely large or small nutrient pool size (Fig. 2) and a positive PSF (Fig. 3). On the contrary, it leads to intermediate levels of nutrient pool size (NN** < N* < NL**), which cause a negative PSF (Fig. 3) and facilitate the coexistence of plants (Fig. 4).
Although much more difficult to detect than PMSF systems caused by symbiotic microbes in the rhizosphere over short timescales, litter-mediated PMSF systems are important determinants for the plant community over long timescales. This is particularly true in stable and closed environments where plant litter is a major nutrient source and can locally control the nutrient pool size and plant community dynamics. Although not considered in this study, microbial diversity in open environments may affect the PSF by altering the nutrient exchange rates between systems. This issue is an open one for future studies.
The classic plant-centered view of nutrient cycling may be totally changed by considering the community ecology of microbial decomposers and their functional diversity. An approach that incorporates functional diversities within plant and microbial communities into nutrient cycling can be readily applied to other ecosystems (e.g., freshwater lakes and marine ecosystems) and uncover the role of microbial diversity in structuring those ecosystems. Our study has an implication on the effect of biodiversity on ecosystem resilience (37). The presence of positive feedbacks and multiple attractors in ecosystems can cause a discontinuous shift of ecosystem states in response to small environmental changes (37). We suggest that functional diversity within a trophic level may buffer the effects of another trophic level on ecosystem processes and prevent positive feedbacks, which would contribute to high ecosystem resilience (37).
Methods
A multispecies patch occupancy model (38, 39) represents temporal changes due to interpatch recruitment and within-patch mortality in the proportion of patches occupied by PL or PN. The dynamics of PL and PN are represented by Eqs. 1 and 2, as follows:
The first term of Eqs. 1 and 2 represents new recruitment in empty patches (increasing with the inorganic nutrient level), the second term represents loss through a constant mortality rate (mP), and the last term represents the recruitment or loss resulting from individual-level competition (8). Although an individual of PL has the advantage in within-patch local competition for light and can replace the individual of PN that already occupies the patch, PN has the higher colonization ability into an empty patch (higher reproductive ability and/or higher seedling survival rate) than PL (rN > rL). Plant individuals of species i take up a nutrient (bi) from the nutrient pool (N) during each recruitment (primary production) and release it into the litter pool during each mortality (litter production). An individual plant may take up and release nutrients continuously after its establishment and until its death, at a rate of aiN (annual primary production and litter production). The default setting for parameters assumes that bL = bN > 0 and aL = aN = 0. Results with a positive value of ai and species-specific values of biomass production (bi, and ai) (40) are shown in Figs. S3 and S4.
The litter pool consists of readily and slowly decomposable organic nutrients, with biomasses (nutrient contents) DR and DS, respectively. Readily decomposable DR occupies the fraction fi of the litter from plant Pi (i = L or N). Dj (j = R or S) is decomposed by two functional groups of soil microbes, DR- and DS-preferring microbes with biomasses MR and MS, respectively, at a rate of cD(kjRMR + kjSMS)Dj. Here cD represents the decomposition coefficient that is determined by external factors such as temperature. The dynamics of DR and DL are represented by:
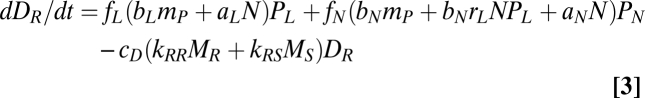 |
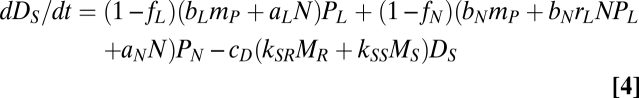 |
where the first and second terms of Eqs. 3 and 4 represent the supply from PL and PN, respectively, and the third term is the loss through microbe-mediated decomposition (i.e., gross mineralization).
Because of differences in the chemical characteristics of the organic nutrients (e.g., carbon/nutrient ratio) and in the levels of secondary metabolites (e.g., tannins and phenolics), structural carbohydrates (lignin), and nutrient partitioning (e.g., nitrogen between phytosynthetic enzymes and cell walls) (2, 41), the decomposition efficiency for DR is higher than that for DS, regardless of the microbial group (i.e., kRR > kSR and kRS > kSS). We assume a functional complementarity between two microbial functional groups for the decomposition of the two organic nutrient types, following that kRR > kRS > kSS > kSR. We can normalize kRR as 1.0 without loss of generality, because the realized decomposition rate is weighted by cD.
By separating decomposition, microbial growth (nutrient immobilization), and net mineralization, the following equations are derived for the dynamics of the microbial biomass (Mj) and the inorganic nutrient (N):
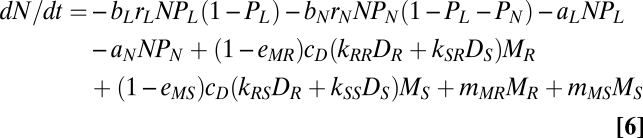 |
where the first through fourth terms of Eq. 6 represent the loss through plant uptake, and the other terms represent supply through the microbial pool (i.e., net mineralization process). The difference in the assimilation efficiency between two organic nutrients is not considered, with the assumption that microbial activity is strongly limited by nutrients and that nutrients in detritus are efficiently and maximally assimilated regardless of detritus quality. In other words, we did not consider the physiological flexibility in response to litter quality. We also did not consider complex formation between nutrient and soil minerals, because our target nutrient is nitrogen rather than phosphorus.
The ecosystem is closed, such that the total amount of nutrients (TN) is constant over time (i.e., bLPL + bNPN + DR + DS + MR+ MS + N = const ≡ TN). This allows us to focus on the effects that the plant and microbe community dynamics have on the nutrient cycling rate and distribution of nutrients (i.e., living plant, litter, microbial biomass, and inorganic nutrient) in the system. We can set TN as 1.0 without loss of generality. When either plant covers all patches, bL or bN (< TN) is allocated to the living plant biomass within the model ecosystem. We also assume that the microbial turnover rate (mortality rate) is much larger than the plant turnover rate (i.e., mMj > mP). Major nutrient fluxes in our model are shown in Fig. 1.
Supplementary Material
Acknowledgments
We thank Dr. Peter Manning and two anonymous reviewers for their valuable suggestions on the earlier versions of the manuscript. This work was supported by National Taiwan University Grant 97R0034-29 (to T.M.), a Grant-in-Aid (21-1526) for Japan Society for the Promotion of Science Fellows, and a Grant-in-Aid for the Global COE Program A06 of Kyoto University (to M.U.), a Grant-in-Aid for Young Scientists (B) (19770019), and a Grant-in-Aid for Scientific Research (B) (20370009 and 22370012) of Japan Society for the Promotion of Science (to M.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914281107/-/DCSupplemental.
References
- 1.Wedin DA, Tilman D. Species effects on nitrogen cycling: A test with perennial grasses. Oecologia. 1990;84:433–441. doi: 10.1007/BF00328157. [DOI] [PubMed] [Google Scholar]
- 2.Scott NA, Binkley D. Foliage litter quality and annual net N mineralization: comparison across North American forest sites. Oecologia. 1997;111:151–159. doi: 10.1007/s004420050219. [DOI] [PubMed] [Google Scholar]
- 3.Binkley D, Giardina C. Why do tree species affect soils? The warp and woof of tree-soil interactions. Biogeochemistry. 1998;42:89–106. [Google Scholar]
- 4.Berendse F, Oudhof H, Bol J. A comparative study on nutrient cycling in wet heathland ecosystems. I. Litter production and nutrient losses from the plant. Oecologia. 1987;74:174–184. doi: 10.1007/BF00379357. [DOI] [PubMed] [Google Scholar]
- 5.Berendse F, Bobbink R, Rouwenhorst G. A comparative study on nutrient cycling in wet heathland ecosystems. II. Litter decomposition and nutrient mineralization. Oecologia. 1989;78:338–348. doi: 10.1007/BF00379107. [DOI] [PubMed] [Google Scholar]
- 6.Frelich LE, Calcote RR, Davis MB, Pastor J. Patch formation and maintenance in an old-growth hemlock-hardwood forest. Ecology. 1993;74:513–527. [Google Scholar]
- 7.Berendse F. Litter decomposability—a neglected component of plant fitness. J Ecol. 1994;82:187–190. [Google Scholar]
- 8.Miki T, Kondoh M. Feedbacks between nutrient cycling and vegetation predict plant species coexistence and invasion. Ecol Lett. 2002;5:624–633. [Google Scholar]
- 9.Clark BR, Hartley SE, Suding KN, de Mazancourt C. The effect of recycling on plant competitive hierarchies. Am Nat. 2005;165:609–622. doi: 10.1086/430074. [DOI] [PubMed] [Google Scholar]
- 10.Bever JD, Westover KM, Antonovics J. Incorporating the soil community into plant population dynamics: The utility of the feedback approach. J Ecol. 1997;85:561–573. [Google Scholar]
- 11.Bever JD. Soil community feedback and the coexistence of competitors: Conceptual frameworks and empirical tests. New Phytol. 2003;157:465–473. doi: 10.1046/j.1469-8137.2003.00714.x. [DOI] [PubMed] [Google Scholar]
- 12.Bonanomi G, Giannino F, Mazzoleni S. Negative plant-soil feedback and species coexistence. Oikos. 2005;111:311–321. [Google Scholar]
- 13.Kardol P, Cornips NJ, van Kempen MML, Bakx-Schotman JMT, van der Putten WH. Microbe-mediated plant–soil feedback causes historical contingency effects in plant community assembly. Ecol Monogr. 2007;77:147–162. [Google Scholar]
- 14.van der Heijden MGA, Bardgett RD, van Straalen NM. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett. 2008;11:296–310. doi: 10.1111/j.1461-0248.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- 15.Kulmatiski A, Beard KH, Stevens JR, Cobbold SM. Plant-soil feedbacks: A meta-analytical review. Ecol Lett. 2008;11:980–992. doi: 10.1111/j.1461-0248.2008.01209.x. [DOI] [PubMed] [Google Scholar]
- 16.Wedin DA, Pastor J. Nitrogen mineralization dynamics in grass monocultures. Oecologia. 1993;96:186–192. doi: 10.1007/BF00317731. [DOI] [PubMed] [Google Scholar]
- 17.Nosshi MI, Butler J, Trlica MJ. Soil nitrogen mineralization not affected by grass species traits. Soil Biol Biochem. 2007;39:1031–1039. [Google Scholar]
- 18.Knops JMH, Bradley KL, Wedin DA. Mechanisms of plant species impacts on ecosystem nitrogen cycling. Ecol Lett. 2002;5:454–466. [Google Scholar]
- 19.Schimel JP, Weintraub MN. The implications of exoenzyme activity on microbial carbon and nitrogen limitation in soil: a theoretical model. Soil Biol Biochem. 2003;35:549–563. [Google Scholar]
- 20.Wardle DA, et al. Plant removals in perennial grassland: Vegetation dynamics, decomposers, soil biodiversity, and ecosystem properties. Ecol Monogr. 1999;69:535–568. [Google Scholar]
- 21.Bartelt-Ryser J, Joshi J, Schmid B, Brandl H, Balser T. Soil feedbacks of plant diversity on soil microbial communities and subsequent plant growth. Perspect Plant Ecol Evol Syst. 2005;7:27–49. [Google Scholar]
- 22.Bezemer TM, et al. Plant species and functional group effects on abiotic and microbial soil properties and plant–soil feedback responses in two grasslands. J Ecol. 2006;94:893–904. [Google Scholar]
- 23.Dornbush ME. Grasses, litter, and their interaction affect microbial biomass and soil enzyme activity. Soil Biol Biochem. 2007;39:2241–2249. [Google Scholar]
- 24.Ushio M, Wagai R, Balser TC, Kitayama K. Variations in the soil microbial community composition of a tropical montane forest ecosystem: Does tree species matter? Soil Biol Biochem. 2008;40:2699–2702. [Google Scholar]
- 25.Strickland MS, Lauber C, Fierer N, Bradford MA. Testing the functional significance of microbial community composition. Ecology. 2009;90:441–451. doi: 10.1890/08-0296.1. [DOI] [PubMed] [Google Scholar]
- 26.Bardgett RD, Shine A. Linkages between plant litter diversity, soil microbial biomass, and ecosystem function in temperate grasslands. Soil Biol Biochem. 1999;31:317–321. [Google Scholar]
- 27.Wardle DA. Community and Ecosystems: Linking the Aboveground and Belowground Components. Princeton: Princeton Univ Press; 2002. [Google Scholar]
- 28.Wardle DA, Walker LR, Bardgett RD. Ecosystem properties and forest decline in contrasting long-term chronosequences. Science. 2004;305:509–513. doi: 10.1126/science.1098778. [DOI] [PubMed] [Google Scholar]
- 29.Rees M, Condit R, Crawley M, Pacala S, Tilman D. Long-term studies of vegetation dynamics. Science. 2001;293:650–655. doi: 10.1126/science.1062586. [DOI] [PubMed] [Google Scholar]
- 30.Wardle DA, Yeates GW, Williamson W, Bonner KI. The response of a three trophic level soil food web to the identity and diversity of plant species and functional groups. Oikos. 2003;102:45–56. [Google Scholar]
- 31.Neely CL, Beare MH, Hargrove WL, Coleman DC. Relationships between fungal and bacterial substrate-induced respiration, biomass and plant residue decomposition. Soil Biol Biochem. 1991;23:947–954. [Google Scholar]
- 32.Rousk J, Bååth E. Fungal and bacterial growth in soil with plant materials of different C/N ratios. FEMS Microbiol Ecol. 2007;62:258–267. doi: 10.1111/j.1574-6941.2007.00398.x. [DOI] [PubMed] [Google Scholar]
- 33.Teuben A. Nutrient availability and interactions between soil arthropods and microorganisms during decomposition of coniferous litter: A mesocosm study. Biol Fertil Soils. 1991;10:256–266. [Google Scholar]
- 34.Loreau M, et al. Biodiversity and ecosystem functioning: current knowledge and future challenges. Science. 2001;294:804–808. doi: 10.1126/science.1064088. [DOI] [PubMed] [Google Scholar]
- 35.Loreau M. Microbial diversity, producer-decomposer interactions and ecosystem processes: A theoretical model. Proc Biol Sci. 2001;268:303–309. doi: 10.1098/rspb.2000.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reynolds HL, Packer A, Bever JD, Clay K. Grassroots ecology: Plant-microbe-soil interactions as drivers of plant community structure and dynamics. Ecology. 2003;84:2281–2291. [Google Scholar]
- 37.Folke C, et al. Regime shifts, resilience, and biodiversity in ecosystem management. Annu Rev Ecol Evol Syst. 2004;35:557–581. [Google Scholar]
- 38.Hastings A. Disturbance, coexistence, history, and competition for space. Theor Popul Biol. 1980;18:363–373. [Google Scholar]
- 39.Tilman D. Competition and biodiversity in spatially structured habitats. Ecology. 1994;75:2–16. [Google Scholar]
- 40.Adair EC, et al. Simple three-pool model accurately describes patterns of long-term litter decomposition in diverse climates. Glob Change Biol. 2008;14:2636–2660. [Google Scholar]
- 41.Takashima T, Hikosaka K, Hirose T. Photosynthesis or persistence: Nitrogen allocation in leaves of evergreen and deciduous Quercus species. Plant Cell Environ. 2004;27:1047–1054. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.