Turnover of endosomal PI(3)P by mtm maintains endolysosomal homeostasis and cortical remodeling in Drosophila hemocytes during migration.
Abstract
Reversible phosphoinositide phosphorylation provides a dynamic membrane code that balances opposing cell functions. However, in vivo regulatory relationships between specific kinases, phosphatases, and phosphoinositide subpools are not clear. We identified myotubularin (mtm), a Drosophila melanogaster MTM1/MTMR2 phosphoinositide phosphatase, as necessary and sufficient for immune cell protrusion formation and recruitment to wounds. Mtm-mediated turnover of endosomal phosphatidylinositol 3-phosphate (PI(3)P) pools generated by both class II and III phosphatidylinositol 3-kinases (Pi3K68D and Vps34, respectively) is needed to down-regulate membrane influx, promote efflux, and maintain endolysosomal homeostasis. Endocytosis, but not endolysosomal size, contributes to cortical remodeling by mtm function. We propose that Mtm-dependent regulation of an endosomal PI(3)P pool has separable consequences for endolysosomal homeostasis and cortical remodeling. Pi3K68D depletion (but not Vps34) rescues protrusion and distribution defects in mtm-deficient immune cells and restores functions in other tissues essential for viability. The broad interactions between mtm and class II Pi3K68D suggest a novel strategy for rebalancing PI(3)P-mediated cell functions in MTM-related human disease.
Introduction
Different phosphoinositide phosphates (PIPs) under the control of kinase/phosphatase regulation are enriched in distinct membranes and direct localized cell functions (Di Paolo and De Camilli, 2006). A confounding issue in deciphering the relationship between PIP regulation and function is that different members of kinase/phosphatase families exhibit the same PIP selectivity in vitro, yet when mutated in vivo, are associated with specific diseases (Vicinanza et al., 2008). Myotubularins (MTMs) encode conserved phosphoinositide 3-phosphate phosphatases selective for phosphatidylinositol 3-phosphate (PI(3)P) and PI(3,5)P2 (Taylor et al., 2000; Berger et al., 2002; Kim et al., 2002) found as large gene families in metazoans (15 human, 7 fly, and 1 yeast; Laporte et al., 1998; Robinson and Dixon, 2006; Tosch et al., 2006). MTM1 is associated with human myotubular myopathy (Laporte et al., 1996), whereas the closely related MTMR2 is associated with Charcot-Marie-Tooth neuropathy (Bolino et al., 2000), both characterized by distinct morphological defects. In cell cultures, MTM1 and MTMR2 were detected on endosomes and isolated in complexes with the class III phosphatidylinositol 3-kinase (PI3-kinase) Vps34 (Tsujita et al., 2004; Cao et al., 2007, 2008), which is consistent with MTM in vitro substrates and suggesting coordinated regulation of localized PIP pools. However, the identity of the PIPs that require MTM1/MTMR2 in cells and the relationship to requirements in animals remain largely unexplored.
Membrane influx from distinct PI(3)P pools converges at late endosomes (Simonsen et al., 2001; Lindmo and Stenmark, 2006). PI(3)P is highly enriched on early endosomes (Gillooly et al., 2000) with well-described roles defined by conserved Vps34 functions (Schu et al., 1993). The recruitment of proteins with PI(3)P-binding domains mediate protein sorting, membrane transport, homotypic and heterotypic fusion with lysosomes, and endolysosomal maturation (Wurmser and Emr, 1998; Lindmo and Stenmark, 2006; Mima and Wickner, 2009; Saftig and Klumperman, 2009). PI(3)P is also considered central to autophagy, as demonstrated by an essential role for Vps34 in autophagosome formation and delivery of cytoplasmic content to endolysosomes for degradation (Kihara et al., 2001; Juhász et al., 2008; Simonsen and Tooze, 2009). Additionally, PI(3)P pools generated by class II PI3-kinase isoforms restricted to metazoans (MacDougall et al., 1995) are implicated in functions at the plasma membrane (MacDougall et al., 2004; Maffucci et al., 2005; Falasca and Maffucci, 2007; Falasca et al., 2007; Wen et al., 2008; Srivastava et al., 2009), although with unknown endosomal roles. Homeostasis of late endosomes depends on a balance between membrane influx and efflux that leads to diverging retrograde and recycling routes (Johannes and Popoff, 2008; Grant and Donaldson, 2009). Specificity in endocytic trafficking is emerging as a key site of regulation for many cellular and disease states (Gonzalez-Gaitan, 2008; Mosesson et al., 2008). Thus, specific kinases and phosphatases may be dedicated to the synthesis and turnover of PI(3)P subpools that intersect at endosomes.
Drosophila melanogaster presents an ideal system to elucidate metazoan PIP regulation and the functional nexus between specific PIP regulators, substrate identities, and their cell developmental roles (Hafen, 2004). We demonstrate an important functional relationship between the single Drosophila MTM1/MTMR2 orthologue Mtm and the class II PI3-kinase Pi3K68D. We show that Mtm and Pi3K68D coregulate PI(3)P to mediate endolysosomal flux and cortical dynamics in hemocytes, insect immune cells, and essential roles in animals.
Results
Identification of mtm function that promotes cell remodeling
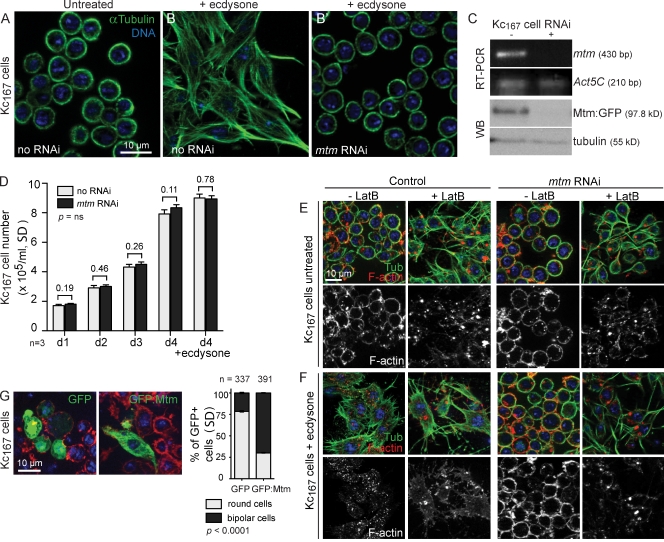
Remodeling of the cell cortex, an integrated result of cytoskeletal and membrane reorganization, is important for diverse cell functions. To identify genes required for cellular remodeling, we performed RNAi of Drosophila kinases and phosphatases for functions that affect a hormone-induced cell shape change in 20-hydroxyecdysone (ecdysone)–responsive hemocyte-derived Kc167 cells (Fig. 1, A and B; and Fig. S1; Echalier, 1997). Depletion of mtm inhibited remodeling from a round to an elongated cell shape, and instead, cells remained round without microtubule bipolar protrusions (Fig. 1, B′ and C). In the absence of ecdysone, the mtm-depleted cells exhibited normal morphology, growth, and survival (Fig. 1, D and E; and Fig. S2), as well as normal cell survival and hallmarks of hormone reception upon ecdysone addition, including increased cell size and up-regulated levels of ecdysone receptor (Fig. S2). The ecdysone-induced mtm RNAi cells inappropriately retained a uniform band of cortical filamentous actin (F-actin), which when destabilized, uncovered the ability for microtubule polymerization (Fig. 1 F). Overexpression of wild-type (WT) mtm cDNA drove the elongation of bipolar Kc167 cells in the absence of ecdysone addition (70.3% GFP:Mtm bipolar cells; Fig. 1 G). These results suggest an mtm function separable from ecdysone reception that promotes F-actin reorganization and cortical remodeling.
Figure 1.
mtm promotes remodeling of cell shape. (A and B) Microtubules in Drosophila Kc167 cells reveal normal round (A) and bipolar (B)-shaped cells 1 d after ecdysone. (B’) mtm RNAi inhibited ecdysone-induced cell elongation. (C) RNAi depletion of mtm transcript and Mtm-GFP (anti-GFP). WB, Western blot. (D) Cell numbers 1–4 d after RNAi. (E) Normal morphology of mtm-depleted cells in absence of ecdysone. (E and F) Latrunculin B (+LatB) destabilized F-actin (red) and permitted microtubule protrusions (green) in untreated (E) and ecdysone-treated (F) mtm RNAi cells. (G, right) GFP:Mtm induced cell elongation in absence of ecdysone. (left) GFP cells remained round. Percentage of transfected round or bipolar-shaped cells. Error bars indicate mean ± SD.
Essential tissue-specific roles for conserved mtm phosphoinositide phosphatase
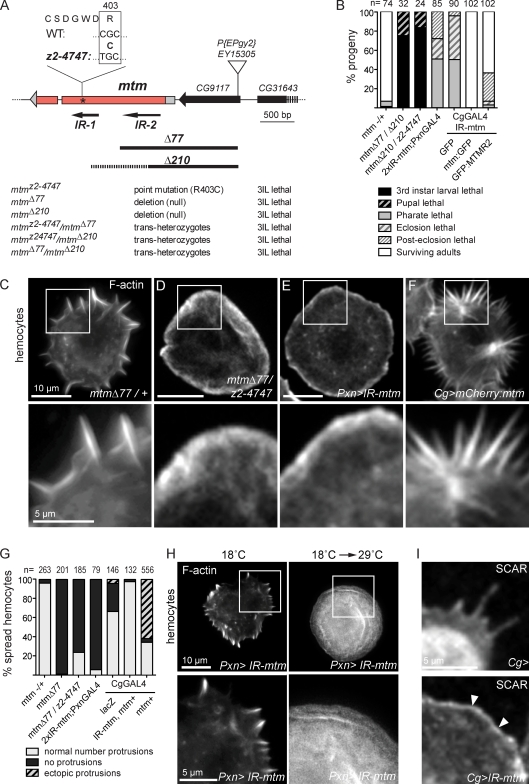
To investigate the significance of an mtm function for cell remodeling in animal development, we generated Drosophila mutant alleles (Fig. 2 A). A point mutation within the catalytic CX5R motif (mtmz2-4747) and null excision alleles (mtmΔ77 and mtmΔ210) exhibited larval lethality, indicating an essential requirement for mtm phosphatase activity (Fig. 2, A and B). Targeted expression of mtm RNAi hairpins uncovered requirements in specific tissues, including independent roles likely to act in muscle and hemocytes (Table S1). Coexpression of either fly mtm or human MTMR2 cDNAs rescued RNAi-induced animal lethality, demonstrating mtm knockdown specificity and functional conservation with the human gene (Fig. 2 B and Table S1).
Figure 2.
mtm promotes hemocyte protrusions and essential roles in Drosophila. (A) Larval lethal mtm mutant alleles: z2-4747 mutation in CX5R catalytic motif, Δ77 and Δ210 deletions, and UAS-IRmtm RNAi hairpins. (B) Percentage of lethal progeny for mtm alleles or hemocyte-targeted RNAi (PxnGAL4; CgGAL4) rescued to viability with mtm or human MTMR2 cDNA. (C–F) F-actin in primary hemocytes (top) and zoom of cell edges (bottom). (C) Normal radial protrusions in mtmΔ77/+. (D and E) Lack of protrusions in mtmΔ77/mtmz2-4747 and hemocyte mtm RNAi. (F) Increased number of protrusions in mtm overexpression. (G) Percentage of hemocytes with normal, no, or ectopic protrusions. (H, right) Lack of hemocyte protrusions with 1-d induced mtm RNAi (18 to 29°C shift). IR-mtm/+; Pxn-GAL4/+. (I) SCAR protein in control (top) and enriched along cell cortex in mtm-depleted hemocytes (bottom, arrowheads; Cg-GAL4/IRmtm). Boxed areas are shown at magnified views in the panels below.
Mtm is necessary and sufficient for immune cell protrusion formation
An mtm-dependent hemocyte function suggested unexplored roles for MTMs in immune cells (Fig. 2 B). As predicted from Kc167 cells, mtm function impaired cortical remodeling in hemocytes. In postembryonic stages, hemocytes exist as round cells distributed in the body cavity and as spread cells in sessile populations adherent to the body wall (Lanot et al., 2001). Upon dissection from larvae, primary hemocytes undergo remodeling from a round to spread cell shape, exhibiting F-actin–rich radial protrusions that dynamically extend and retract perpendicular to the periphery (Fig. 2, C and G; and Video 1). In contrast, both mtm mutants and hemocyte-autonomous depletion resulted in spread cells that completely lacked radial protrusions (control, 3.8%; mtm mutant cells, 76.8–99.5%; Fig. 2, D, E, and G), even with knockdown induced in larvae (Fig. 2 H). Mutant cells exhibited smooth or ruffled edges, often a larger cell footprint, and a broad band of circumferential F-actin and SCAR protein (Fig. 2 I; Zallen et al., 2002). The mtm-depleted cells exhibited a dynamic cell cortex, and although short protrusions appeared, they abnormally moved radially along the periphery (Video 1). Strikingly, mtm overexpression in WT hemocytes led to an opposite phenotype of excessive radial protrusions (control, 4.1%; mCherry:mtm cells, 62.2%; Fig. 2, F and G).
Mtm function in vivo is important for immune cell distribution and recruitment to wounds
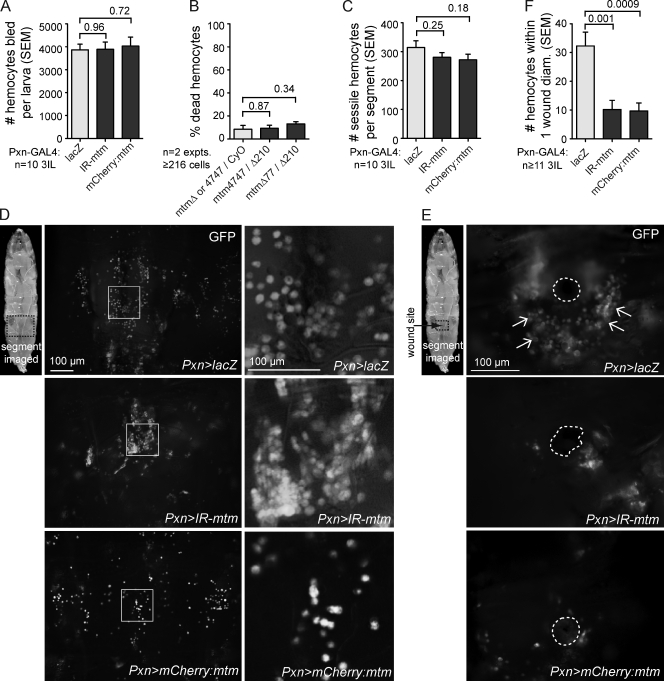
Consistent with a specific role for mtm affecting cell shape, larval hemocyte number, cell viability, and differentiation were unaffected in mtm mutants (Fig. 3, A–D). Although similar numbers of sessile hemocytes were observed for both, either mtm knockdown or WT mtm cDNA expression led to aberrantly clumped or dispersed hemocyte distribution, respectively (Fig. 3, C and D). Defects in cell morphogenesis and dispersion could disrupt hemocyte recruitment abilities needed in immune and wounding responses (Williams, 2007; Babcock et al., 2008). Puncture wounds in WT larvae led to the recruitment of 32.3 ± 4.9 hemocytes at the clot 6 h after wounding (Fig. 3, E and F). In contrast, either mtm knockdown or overexpression in hemocytes, corresponding to lack of or ectopic cell protrusions, respectively (Fig. 2, E and F), disrupted hemocyte numbers at wounds (10.2 ± 3.2; 9.7 ± 2.7 cells; Fig. 3, E and F). Altogether, mtm function is both necessary and sufficient for the formation of hemocyte protrusions, with in vivo consequences on hemocyte dispersion and efficient recruitment to wounds.
Figure 3.
mtm disrupts hemocyte distribution and recruitment to larval wounds. (A–C) mtm function did not affect the number of bled hemocytes (A), percentage of dead or dying hemocytes (B; see Materials and methods; genotypes as shown), or number of sessile hemocytes per segment of third instar larva (C). (D) GFP+ hemocytes imaged in dorsal segment: control (top), mtm RNAi (middle), and mtm expression (bottom). Boxes represent the higher magnified views at right. (E) GFP+ hemocytes (arrows) recruited 6 h after wounding to puncture sites in dorsal segments. (F) Either mtm depletion or expression reduced the number of hemocytes within one-wound diameter. (A and C–F) IR-mtm/+ or mCherry:mtm/+ with Pxn-GAL4, UAS-GFP/+. Error bars indicate SEM.
Mtm is necessary and sufficient for homeostasis of endolysosomes
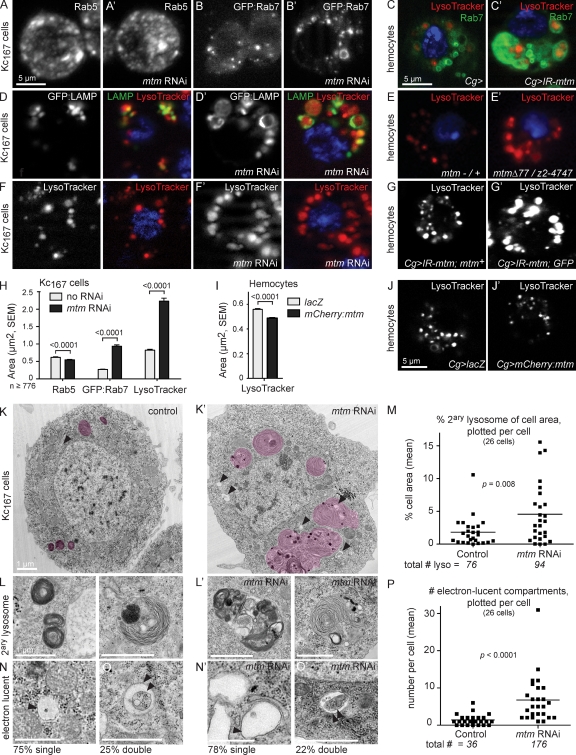
To pinpoint mtm cellular mechanisms, we investigated the identity of the Mtm functional PIP substrate by addressing effects on specific PIP-containing membranes, regulation of PIP levels, and distributions and genetic requirement for specific PIP pools. We first tested whether mtm function affected specific endosomal or autophagic membrane identities. Rab5-containing early endosomes, known sites of abundant PI(3)P synthesis (Wucherpfennig et al., 2003), were unaltered in morphology and number, although with a slight decrease in size detected in mtm RNAi cells (Fig. 4, A, A′, and H). In contrast, enlarged GFP:Rab7 and GFP:LAMP compartments suggested an mtm-dependent function for late endosome–lysosomes (endolysosomes; Saftig and Klumperman, 2009), which are known sites of transition from PI(3)P to PI(3,5)P2 synthesis (Fig. 4, B–D′ and H; Di Paolo and De Camilli, 2006). Giant compartments detected with acidic pH-sensitive LysoTracker were associated with late endolysosomal proteins (Fig. 4, D′–G′) and more than threefold enlarged (Fig. 4 H), indicating normal traffic and function of lysosomal H+-ATPases despite increased organelle size. The LysoTracker compartment in mtm-depleted cells further increased to nearly fourfold in size upon ecdysone addition, indicating an mtm function separable from ecdysone reception that may impact membrane flux (Fig. S2 G). The size of LysoTracker-positive organelles was restored with mtm cDNA expression in mtm-depleted hemocytes (Fig. 4 G), whereas the size further decreased with expression in WT hemocytes (Fig. 4, I–J′). Ultrastructural analysis in mtm-depleted Kc167 cells confirmed an increased cell area of secondary lysosomes (lamellar bodies and autophagolysosomes; Sunio et al., 1999). Secondary lysosomes were evident both in small clusters and as single, giant membrane–delimited structures (Fig. 4, K–M), which is consistent with transport, tethering, and fusion of late endosomes with lysosomes. The ultrastructure also revealed an increase in size and frequency of both single and double membrane-delimited electron-lucent compartments (Fig. 4, K′ and N–P). Double-membrane structures were characteristic of autophagosomes (Fig. 4, O and O′), organelles involved in the PI(3)P-dependent process of autophagy (Juhász et al., 2008; Simonsen and Tooze, 2009). The ultrastructure confirmed formation of intraluminal vesicles of multivesicular bodies (unpublished data). These combined results indicate that mtm function affects endolysosomal homeostasis and is both necessary and sufficient to restrict endolysosomal size.
Figure 4.
mtm controls endolysosome homeostasis. (A’–B’, C’, D’, E’, F’, and G’) mtm is necessary and sufficient to restrict size of acidified endolysosomes in Kc167 cells (A’–B’, D’, and F’) and primary hemocytes (C’, E’, and G’). (A’ and B’–C’) Normal early endosomes (A’; anti-Rab5) and enlarged and convoluted late endosomes (B’–C’; GFP:Rab7; green) with mtm RNAi are shown. (D’–G’) Both mtm RNAi and null alleles resulted in enlarged acidified endolysosomes (LysoTracker, red; GFP-LAMP, green) that reverted to normal size with mtm:GFP cDNA (G). (H) Area of individual Rab5, GFP:Rab7, and LysoTracker-stained organelles in Kc167 cells. (I–J’) Area of LysoTracker-positive organelles decreased in mCherry:mtm-expressing hemocytes. (K and K’) Ultrastructure of mtm RNAi Kc167 cells revealed an increase in secondary lysosomes (shaded pink) and electron-lucent membrane compartments (arrowheads). (L and L’) Examples of secondary lysosomes. (M) Secondary lysosome area shown as the percentage of cell area (means: control, 1.8%; mtm RNAi cell area, 5.0%). (N–O′) Examples of electron-lucent membrane structures. Single membrane bound (N and N’) and double membrane bound, typical of autophagosomes (O and O’). (P) Number of individual electron-lucent compartments (means: WT, 1.4; mtm RNAi cell, 6.8). Proportion of single (75 and 78%) and double (25 and 22%) membrane-bound electron-lucent structures remained unaffected. Error bars indicate SEM.
Endocytosis contributes to separable mtm effects on endolysosomes and cell remodeling
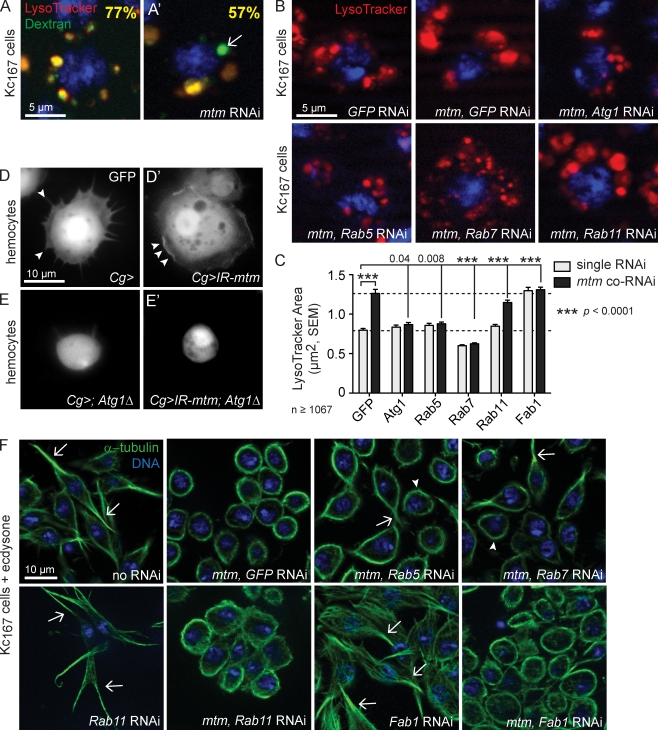
Endolysosome homeostasis reflects a balance between membrane influx and efflux. We performed kinetic assays to test an mtm impact on trafficking to endolysosomes. The mtm-depleted cells exhibited normal early steps in phagocytosis, fluid phase, and cargo-mediated endocytosis. Escherichia coli, F-dextran, or BSA colocalization with the enlarged LysoTracker-positive compartments indicated successful traffic to endolysosomes (Fig. 5 A′; and Fig. S3, A and B), although the late steps of F-dextran delivery were delayed (Fig. 5 A′). To address the balance of specific membrane influx at late endosomes, we tested whether codepletion of mtm and effectors of endocytic or autophagic trafficking could suppress the enlarged endolysosomes (Fig. 5 B; and Fig. S3, C and D). Codepletion of mtm with either Atg1, a kinase involved in initiation of autophagy (Scott et al., 2004; Xie and Klionsky, 2007), or the endosomal GTPases Rab5 or Rab7 each restored or further decreased the size of LysoTracker organelles (Fig. 5, B and C). These results indicate that autophagic and endocytic routes contribute to the mtm endolysosomal defect and suggest that Mtm may negatively regulate influx of both routes either independently or at a single site of membrane convergence.
Figure 5.
mtm interacts with both endocytic and autophagic effectors, with separable effects on endolysosomes and cell remodeling. (A and A’) Arrow indicates delay in late F-dextran (green) trafficking to LysoTracker-positive organelles (red) in mtm RNAi Kc167 cells, shown as the percent of colocalization 20 min after uptake. (B and C) Screen of endolysosome size in single and mtm coRNAi–treated Kc167 cells, identified suppressors of mtm-enlarged endolysosomes. (B) LysoTracker (red) with coRNAi as shown. (C) Quantification of mean endolysosomal area upon single RNAi (gray) or mtm coRNAi (black). Bottom dashed line, mean area GFP RNAi; top dashed line, mean area of mtm, GFP coRNAi. (D) GFP-labeled Atg1Δ3D/+ hemocytes with extended protrusions (arrowheads). (D’) Lack of cell protrusions (ruffles, arrowheads) in spread mtm-depleted hemocytes. (E) Round Atg1Δ3D−/− hemocytes failed to spread. (E’) Round cells with Atg1Δ3D−/− in combination with mtm depletion. (F) Microtubules (green) in Kc167 cells 1 d after ecdysone with coRNAi as shown. With mtm and Rab5 or Rab7 codepletion, mixed examples of cells that partially reverted cell extensions (arrows) and cells that remained round (arrowheads). No effect of Rab11 or Fab1 RNAi alone or on mtm RNAi defect. Error bars indicate SEM.
The two distinct defects in cortical remodeling and endolysosomal homeostasis raised the question of whether both mtm roles are related either as primary and secondary consequences or through a shared mtm function. If related, suppressors of the mtm-dependent enlarged endolysosomes could also revert the lack of cell protrusions. The codepletion of mtm with Atg1 modified the phenotype but did not rescue lack of protrusion formation in either Kc167 cells or hemocytes (Fig. 5 E′). Instead, Atg1 single- and double-mutant cells exhibited novel mutant phenotypes with failure of cell elongation or spreading (Fig. 5, E and E′), respectively. This suggests an mtm role antagonistic to autophagy that mediates endolysosomal homeostasis separable or downstream from protrusion formation and signifies an additional role for autophagy in cellular remodeling (Kadandale et al., 2010). We found that although neither Rab5 nor Rab7 was required for the induced cell shape change, codepletion of either endosomal effector with mtm partially restored ecdysone-induced Kc167 cellular elongation (Fig. 5 F). Together, these results suggest that endocytosis, but not endolysosomal size, impacts cortical remodeling and point to an mtm endocytic function with divergent effects on endolysosomal flux and cell protrusion formation.
Conversely, mtm may positively regulate the competing process of membrane efflux. Rab11 RNAi on its own had little effect on endolysosomal size or Kc167 cell elongation (Fig. 5, B, C, and F). This suggests that disrupted efflux of Rab11-recycling endosomes does not account for mtm mutant phenotypes without excluding a possible role for other membrane retrieval routes. Cell protrusions still formed upon knockdown of Fab1 phosphoinositide kinase (Fig. 5 F and Fig. S3 E), which is shown to result in enlarged lysosomes (Fig. 5 C and Fig. S3 C) and disruption of PI(3,5)P2 (Rusten et al., 2006), further dissociating endolysosomal size from cortical remodeling. In addition, Fab1 coRNAi failed to rescue mtm RNAi phenotypes (Fig. 5, C and F), indicating that Mtm turnover of PI(3,5)P2 alone also is unlikely to account for an mtm function in endolysosome homeostasis or cellular remodeling.
Mtm down-regulates PI(3)P for endolysosomal membrane identity and dynamics
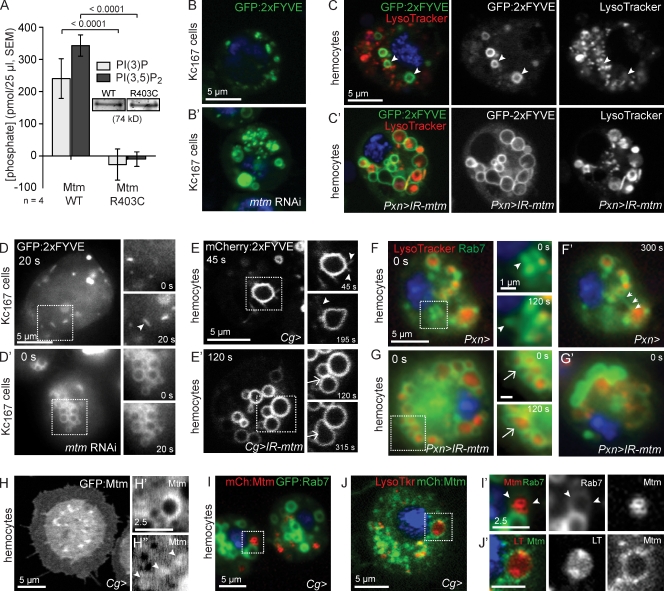
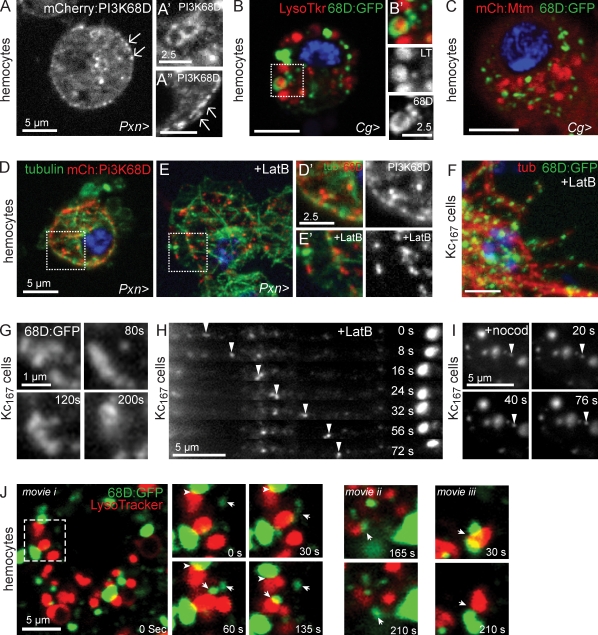
Common defects observed with null and catalytic mutant alleles suggest that mtm roles are phosphoinositide dependent (Figs. 2 and 3). As predicted, WT Mtm protein exhibited in vitro PI(3)P and PI(3,5)P2 phosphatase activity, whereas the MtmR403C form, mutated within the catalytic motif, was inactive (Fig. 6 A). If mtm function is mediated through its role in PI(3)P turnover, then mtm depletion or expression may alter PI(3)P accumulation. In WT Kc167 cells and hemocytes, the GFP:2xFYVE PI(3)P biosensor (Gillooly et al., 2000; Wucherpfennig et al., 2003) localized primarily to puncta (Fig. 6 B) and rings (Fig. 6 C). Upon mtm depletion, we detected an expanded distribution of GFP:2xFYVE both on more numerous puncta and on enlarged rings (Fig. 6, B′ and C′). In contrast, overexpression of WT mtm cDNA disrupted GFP:2xFYVE localization to a diffuse cytoplasmic pattern (Fig. S4 D). Although GFP:2xFYVE showed extensive Rab5 colocalization and minimal overlap with LysoTracker-positive compartments in WT cells (Fig. 6 C, and not shown), GFP:2xFYVE surrounded large LysoTracker-positive organelles in mtm-depleted hemocytes (Fig. 6 C′), indicating a shift in the site of highest PI(3)P detected by the biosensor from early endosomes to endolysosomes.
Figure 6.
mtm controls PI(3)P turnover, membrane compartment identity, and flux. (A) Mtm-dephosphorylated PI(3)P and PI(3,5)P2. Mutant Mtm-R403C was inactive for both substrates (released phosphate/reaction). (B–E’) PI(3)P distribution detected by GFP:2xFYVE. PI(3)P expanded distribution upon mtm RNAi in Kc167 cells (B’) and abnormal association with LysoTracker-positive organelles upon mtm depletion in hemocytes (C’). (D) PI(3)P pools detected within small dots with rapid, directional movement (arrowhead; motile particle imaged as line) by time-lapse microscopy in control Kc167 cells. (D’) PI(3)P pools detected on expanded, tethered rings and lack of particle motility upon mtm RNAi. (E) Time-lapse microscopy of PI(3)P pools detected by mCherry:2xFYVE on rings with obvious tubulation (arrowheads) in hemocytes. (E’) PI(3)P pools detected on expanded, tethered rings that underwent fusion (arrows) without detectable tubulation in mtm-depleted hemocytes; still images from Video 2 are shown. (F and F’) GFP:Rab7 motility (F, arrowheads) and dynamic extension and retraction of tubules (F’, arrows) in control hemocytes (Video 3). LysoTracker, red; DAPI, blue. (G and G’) Tethered motion and fusion (arrows) between enlarged GFP:Rab7 compartments in mtm-depleted hemocytes. (H–H″) GFP:Mtm localized to dots (H) and big (H’) and small rings (H”, arrowheads). (I and I’) mCherry:Mtm on small rings detected within GFP:Rab7 compartments (GFP:Rab7 ring, arrowheads). (J and J’) Mtm at periphery of acidified endolysosomes. Red, LysoTracker. Boxed areas are shown in higher magnification on the right. Error bars indicate SEM.
Endosomal PI(3)P recruits effectors that promote membrane transport, tethering, and fusion (Lindmo and Stenmark, 2006). We observed extensive dispersion of GFP:2xFYVE upon Rab5 but not Rab7 RNAi (Fig. S4, A and A’), suggesting that genetic interactions between mtm and Rab5 may be at the level of PI(3)P modulation, whereas interactions with Rab7 may be at the level of PI(3)P effectors. Consistent with contribution of inappropriate or excessive membrane-tethering fusion, endolysosomal size was reverted with codepletion of mtm and genes encoding machinery of the HOPS complex or SNAREs (Fig. S3, C and D; Banta et al., 1988; Seals et al., 2000; Jahn et al., 2003).
PI(3)P-containing compartments in WT cells exhibited highly dynamic behavior with directional movement (Fig. 6 D) and tubulation indicative of exiting membrane (Fig. 6 E and Video 2). In contrast, mtm-depleted cells lacked motile PI(3)P-containing particles. Instead, 2xFYVE was detected on rings with restricted, conjoined motion and little obvious tubulation (Fig. 6, D′–E′; and Video 2), which is consistent with inappropriate PI(3)P accumulation disrupting membrane efflux and promoting excessive tethering and fusion between compartments (Fig. 6 E′). Reflecting the PI(3)P membrane dynamics in WT hemocytes, we similarly observed motile GFP:Rab7- or LysoTracker-positive particles (Fig. 6 F) and dynamic GFP:Rab7 tubules suggestive of efflux from endolysosomes (Fig. 6 F′ and Video 3). In contrast, mtm-depleted cells contained adjacent LysoTracker-positive GFP:Rab7 rings that exhibited localized movements and infrequent tubulation (Fig. 6, G and G′; and Video 3). In both WT and mtm-depleted conditions, we observed GFP:Rab7 fusion events (Fig. 6 G). These results suggest that Mtm is important, perhaps through or in addition to a role in promoting efflux, to down-regulate PI(3)P-mediated tethering and fusion of Rab7-containing late endosomes.
Mtm associates with internal membranes, including endolysosomes
Given its multiple cellular functions, we asked where Mtm protein localized. When expressed as different tagged forms, Mtm was diffuse throughout the cell and localized to puncta and rings (Fig. 6, H–H″), including faint, small rings of uniform size (Fig. 6 H″, arrowheads), which are indicative of membrane structures. Mtm did not colocalize with Rab5 or GFP:Rab7 endosomes. However, small rings of mCherry:Mtm could be found inside a subset of GFP:Rab7 compartments (Fig. 6, I and I′), which is suggestive of intraluminal vesicles or autophagosomes (Xie and Klionsky, 2007). A small fraction of Mtm was detected at acidified compartments as puncta or completely surrounding LysoTracker-positive organelles (Fig. 6, J and J′), which is consistent with roles in endolysosomal homeostasis.
Both class II and class III PI(3)-kinases contribute to PI(3)P pool antagonized by mtm
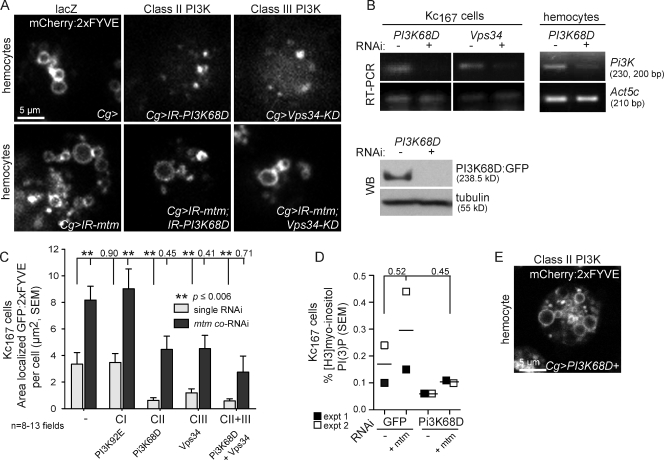
If Mtm dephosphorylates a distinct PI(3)P pool, mtm function could antagonize the PI(3)P production by a specific PI3-kinase. As in mammals, Drosophila encodes three classes of PI3-kinases, with one member per class capable of PI(3)P synthesis in vitro (class I, Pi3K92E; class II, Pi3K68D; and class III, Vps34). Vps34 was a likely candidate for production of an Mtm-functional substrate given known roles for PI(3)P synthesis on early endosomes and for autophagy (Lindmo and Stenmark, 2006). In testing all three PI3-kinases, we found that knockdown or expression of kinase-dead form of Vps34 and, surprisingly to an even greater extent, knockdown of Pi3K68D each individually resulted in dispersion of localized 2xFYVE biosensors (Fig. 7, A–C; and Fig. S4, B and C), demonstrating that both class II and III PI3-kinases are required for significant PI(3)P pools in immune cells. A recovery of the 2xFYVE-localized distribution was obtained from codisruption of mtm with Pi3K68D or Vps34 (Fig. 7, A and C; and Fig. S4 C), indicating that interference with either PI3-kinase was sufficient to restore the mtm-dependent PI(3)P imbalance. Given that Pi3K68D contribution to PI(3)P synthesis has not been characterized in vivo, we further investigated its role directly. We confirmed the effects on PI(3)P total cellular levels from myo-inositol radiolabeled Kc167 cell lysates (Fig. 7 D). Altered levels of PI(3)P were observed upon knockdown of mtm phosphatase (1.7-fold increase) and Pi3K68D kinase (2.8-fold decrease), respectively, that returned nearer to normal levels upon their codepletion (1.6-fold decrease; Fig. 7 D), mirroring the genetic interaction seen with 2xFYVE distribution. Overexpression of Pi3K68D cDNA phenocopied mtm depletion effects of expanded 2xFYVE distribution (Fig. 7 E and Fig. S4 E), which is consistent with Pi3K68D synthesis of PI(3)P.
Figure 7.
mtm interacts with both class II Pi3K68D and class III Vps34 for PI(3)P homeostasis. (A) PI(3)P distribution, as detected by mCherry:2xFYVE in single hemocyte for genotypes as shown, was expanded (IR-mtm), depleted (IR-Pi3K68D or Vps34-KD), or restored (bottom; IRmtm with IR-Pi3K68D or Vps34-KD). (B) Depletion of Pi3K68D or Vps34 transcripts in RNAi-treated Kc167 cells (left) and purified hemocytes (right; Pxn-GAL4/UAS-IR-Pi3K68D). Pi3K68D:GFP (bottom; anti-GFP) in RNAi-treated Kc167 cells. WB, Western blot. (C) GFP:2xFYVE area in RNAi-treated Kc167 cells (Fig. S4 C) normalized per cell number. Single-kinase RNAi (gray) or mtm coRNAi (black). (D) Percent PI(3)P of total myo-inositol from RNAi-treated Kc167 cells. Similar trends were found across two experiments. (E) Pi3K68D:GFP expression expanded PI(3)P detected by mCherry:2xFYVE. Error bars indicate SEM.
Class II Pi3K68D and mtm have opposing functions for homeostasis of endolysosome size
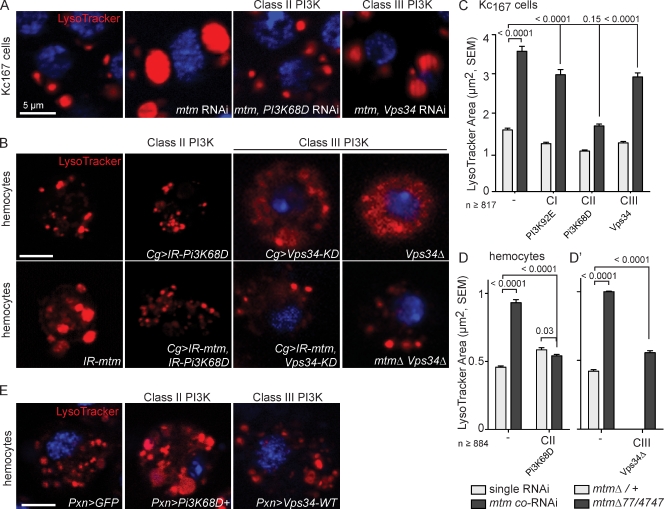
If Mtm roles are mediated through down-regulation of a distinct PI(3)P pool, then mtm could antagonize the function of a specific PI3-kinase. As observed for PI(3)P, codepletion of Pi3K68D with mtm suppressed the giant endolysosome size (Fig. 8, A–D). Conversely, we found that overexpression of Pi3K68D cDNA in WT hemocytes resulted in greatly enlarged LysoTracker-positive organelles (Fig. 8 E). This condition phenocopyied mtm depletion and is consistent with Pi3K68D coregulation of a PI(3)P pool important for endolysosomal size. Although both Vps34 and Pi3K68D disruption reduced 2xFYVE-detected PI(3)P distribution to a similar degree (Fig. 7, A and C), knockdown of Vps34 alone or in combination with mtm exhibited minor effects on LysoTracker-positive organelles in Kc167 cells (Fig. 8, A and C). Disruption of Vps34 with either a null mutant allele or targeted expression of a kinase-dead form in hemocytes, however, resulted in diffuse LysoTracker staining throughout the cells (Fig. 8 B), suggesting disruption of lysosomal H+-ATPase trafficking or of the integrity or size of acidified organelles. Codepletion or double mutants of Vps34 and mtm suppressed both individual endolysosomal defects in hemocytes. Unlike Pi3K68D, overexpression of Vps34 WT cDNA in hemocytes had little to no effect on LysoTracker-positive organelles (Fig. 8 E). These results suggest that mtm is antagonistic to both Pi3K68D and Vps34 functions but that each kinase exhibits distinct roles for normal acidified endolysosomes and differential requirements in Kc167 cells and hemocytes.
Figure 8.
mtm and class II Pi3K68D have opposing roles for endolysosome homeostasis, distinct from class III Vps34. LysoTracker, red; DAPI, blue; shown for single cell. (A) Rescue of endolysosomal size upon mtm, Pi3K68D coRNAi in Kc167 cells. (B, bottom) In hemocytes, class II PI3K depletion (IRPi3K68D) rescued mtm-dependent endolysosome size. Class III PI3K disruption alone (Vps34-KD, kinase dead, or vps34Δm22) resulted in diffuse LysoTracker staining (top) rescued by mtm disruption. (C–D’) Size of individual LysoTracker-positive organelles upon single RNAi (gray) and mtm coRNAi (black) in Kc167 cells or hemocytes (D and D’; Vps34Δ unquantifiable). (E) Pi3K68D:GFP expression (Pxn-GAL4) led to enlarged endolysosomes, whereas Vps34-WT had little effect. Error bars indicate SEM.
Pi3K68D localizes at endolysosomes and cortex with microtubule-dependent motility
Similar to Mtm, multiple tagged forms of Pi3K68D protein localized to internal puncta and rings (Fig. 9, A and A′) and failed to colocalize with Rab5 or GFP:Rab7 but partially associated with acidified organelles (Fig. 9, B and B′). Mtm and Pi3K68D did not colocalize nor obviously deviate in their patterns when coexpressed (Fig. 9 C), suggesting either non- or only transiently overlapping domains. Strikingly, Pi3K68D also aligned along the hemocyte periphery (Fig. 9 A). The cortical Pi3K68D often appeared filamentous (Fig. 9 A″, arrows) and colocalized with microtubules (Fig. 9, D and D′) independent of F-actin (Fig. 9, E–F). The distinct localization patterns raised the questions of how Mtm and Pi3K68D might coregulate a common PI(3)P pool and whether Pi3K68D localization was dynamic. Time-lapse microscopy revealed Pi3K68D:GFP oscillations (Fig. 9 G) and directed motility as small particles, including movement to and away from LysoTracker-positive organelles (Fig. 9 J and Video 4). Pi3K68D motility was coincident (Fig. 9 H) and dependent on microtubules (Fig. 9 I), which is consistent with known mechanisms of vesicular transport and endosomal dynamics (Soldati and Schliwa, 2006). Thus, conserved class II Pi3K microtubule-dependent dynamics (PI3KC2α; Zhao et al., 2007) may mediate Pi3K68D roles at cortical and internal sites.
Figure 9.
Pi3K68D localizes at endolysosomes and cell periphery with microtubule-dependent motility. (A–A”) mCherry:Pi3K68D localized to dots (A and A’), rings (A and A”), and cell periphery (arrows) in hemocytes. (B and B’) Pi3K68D:GFP surrounding acidified endolysosomes. Red, LysoTracker. (C) Mtm (red) and Pi3K68D (green) did not colocalize when coexpressed. (D–I) Pi3K68D microtubule-dependent motility and localization. (D and D’) mCherry:Pi3K68D (red) along microtubules (green). (E and E’) Localization persisted with latrunculin B (+LatB). (F) Pi3K68D:GFP colocalized with microtubules (red) in latrunculin B–treated Kc167 cells. (G–I) Pi3K68D:GFP time-lapse imaging in live Kc167 cells. (G) Pi3K68D on oscillating puncta. (H) Disruption of F-actin (+LatB) revealed extensive Pi3K68D:GFP particle motility (arrowheads). (I) Disruption of microtubules (+nocod) arrested all Pi3K68D:GFP motility. Arrowheads, examples of stationary Pi3K68D:GFP particles. (J) Pi3K68D:GFP dynamic colocalization relating to LysoTracker (red) compartments in single z sections from time-lapse microscopy of hemocytes (the following show frames from Video 4: PI3K68D:GFP movement toward [i] and away [ii and iii] from endolysosomes). Examples of persistent (arrowheads) and transient colocalization between motile Pi3K68D:GFP (arrows) and Lysotracker. Blue, DAPI. Boxed areas are shown in higher magnification on the right.
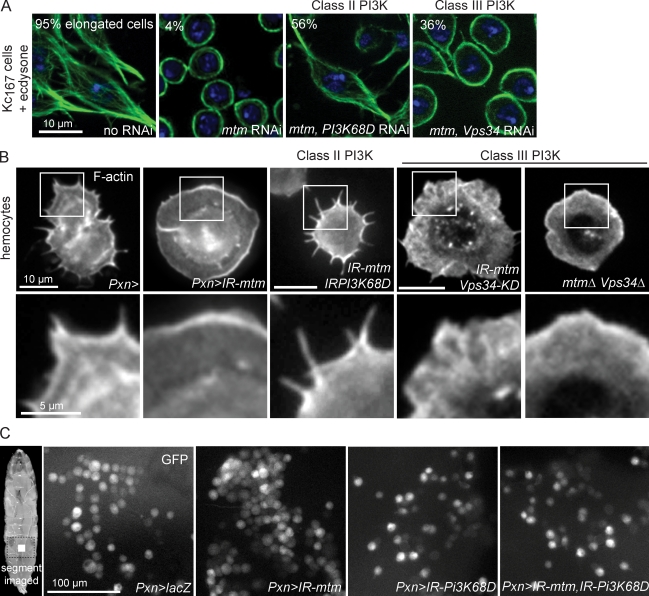
Class II Pi3K68D loss of function suppresses mtm requirement for cortical remodeling
The mtm roles for endolysosomes and in cortical remodeling could be related to regulation of a single PI(3)P pool or independent regulation of different spatial pools or PIP forms. Neither disruption nor overexpression of either Pi3K68D or Vps34 alone had a noticeable effect on hemocyte spreading or F-actin protrusions (Fig. S4, F and G). We found that Pi3K68D strongly suppressed the mtm RNAi block in Kc167 cell elongation (Fig. 10 A) and completely rescued hemocyte protrusion formation (Fig. 10 B). In contrast, Vps34 knockdown partially modified mtm RNAi inhibition of Kc167 cell elongation (Fig. 10 A) and showed infrequent and only mild modification of hemocyte protrusions in mtm double-mutant combinations (Fig. 10 B). Importantly, the effect of mtm RNAi on abnormally clumped hemocyte distribution in larvae was also reverted by Pi3K68D codepletion (Fig. 10 C). These results point to a functional interaction between Pi3K68D and mtm that modulates cortical dynamics and suggests that their direct coregulation of a distinct PI(3)P pool plays an important role in the balance of hemocyte protrusion formation and distribution in vivo.
Figure 10.
mtm and class II Pi3K68D interact for normal hemocyte protrusions and distribution. (A) Microtubules (green) are shown. 95% Kc167 cells elongated 1 d after ecdysone. mtm RNAi blocked elongation (4%) and was reverted upon Pi3K68D coRNAi (56%) and less so by Vps34 coRNAi (36%). (B) F-actin in hemocytes (top) and zoom of cell edges (bottom). Pi3K68D, mtm coknockdown rescued lack of hemocyte protrusions (UAS-IRPi3K68D16240, UAS-IRmtm3.5/Pxn-GAL4). Weakly modified or persistent lack of protrusions with coexpression of mtm RNAi and Vps34-KD (UAS-Vps34-KDm8, UAS-IRmtm3.1/+; Pxn-GAL4/+) or in double mutants (mtmΔ77, Vps34Δm22/mtmz2-4747, Vps34Δm22). (C) GFP-positive hemocytes (Pxn-GAL4) in intact larvae. mtm and Pi3K68D coRNAi reverted clumped to normal hemocyte distribution. Boxed areas are shown in higher magnification on the bottom.
Mtm and class II Pi3K68D collaborate broadly for animal viability
Unlike mtm, class II or III PI3-kinase functions in hemocytes were not required for animal viability. However, animal lethality resulted from Pi3K68D cDNA expression in tissues that also exhibited essential mtm functions (Cg-GAL4 and DMef2-GAL4; Table S1), consistent with overproduction of an Mtm PI(3)P substrate pool. Furthermore, Pi3K68D and mtm codisruption in other tissues, including in muscle, fully rescued (GAL4 drivers 24B, DMef2, 69B, and Ptc) or suppressed (GAL4 drivers Act5c, Cg, and Pxn) viability or visible phenotypes (Table S1). This indicates disruption of class II PI3-kinase as a potent and broad suppressor of mtm-dependent functions in vivo.
Discussion
We identified a class II Pi3K68D-dependent PI(3)P pool as a functional and likely direct Mtm substrate. We demonstrated that Pi3K68D and mtm played major roles in the coregulation of a hemocyte PI(3)P pool and that both were necessary and sufficient for PI(3)P-mediated endolysosomal homeostasis. Alternatively, mtm and Pi3K68D could interact through interconverted PIP pools, e.g., if class II PI3K synthesis of PI(3,4)P2 (MacDougall et al., 1995) led to inositol polyphosphate 4-phosphatase generation of endosomal PI(3)P (Norris and Majerus, 1994).
Importantly, Pi3K68D loss of function suppressed multiple mtm-dependent hemocyte functions and essential roles in multiple tissues. Our results suggest that a conserved pathway linking MTM1/MTMR2 and class II PI3-kinases could also be important for similar roles in mammals. Expression of human MTMR2 in flies rescued the lethality associated with mtm depletion in different tissues, highlighting potential significance from use of the fly to better understand MTMR2-related human disease. Recent studies in T cells identified roles for PI3KC2α and MTMR6 in PI(3)P-mediated regulation of a calcium-activated K+ channel (Srivastava et al., 2005, 2009), indicating that class II PI3-kinases may play broad and dedicated roles in conjunction with different MTM family members.
We found that a subset of mtm functions also shared interactions with class III PI3-kinase, Vps34. The genetic interactions observed between mtm and Vps34 in PI(3)P and endolysosomal homeostasis, but not cell remodeling or essential functions in different tissues, suggest several possibilities. Vps34 function may regulate a Pi3K68D function or be partially redundant with Pi3K68D for certain mtm functions; Vps34 may indirectly interact with mtm through converging PI(3)P membrane pools, and/or there may be additional essential consequences of Vps34 functions, e.g., that antagonize different MTM family member functions. Similar partial and redundant interactions have been observed in Caenorhabditis elegans, where reduction of mtm-1 rescued endocytosis defects but not lethality of vps-34 mutants (Xue et al., 2003), and increased apoptotic cell corpse engulfment upon mtm-1 depletion was found dependent on both vps-34 and the class II PIKI-1 functions (Zou et al., 2009).
We found that mtm was not only required for but could also promote cortical remodeling, specifically modulating cell protrusion formation. MTMs have not previously been ascribed specific roles in cellular remodeling, although MTM1/MTMR2 and PI3KC2 isoforms have been associated with the cortex, and MTM1 overexpression led to cell protrusions (Kim et al., 2002). Cortical F-actin organization and dynamics are under control of competing Rho GTPase activities, namely roles for Rac, Rho, and Cdc42 in lamellipodia versus protrusion formation (Ridley, 2006). A mutant form of MTM1 was detected at the plasma membrane upon constitutive Rac1 GTPase activation (Laporte et al., 2002), mtm-1 was identified as a negative regulator of Rac-mediated engulfment (Zou et al., 2009), and Rho1 pathway hyperactivation resulted from combined essential function of ymr1 (MTM), sjl2, and sjl3 lipid phosphatases in yeast PI(3)P regulation (Parrish et al., 2005). However, PI3KC2β-expressing cell lysates exhibited increased levels of activated Cdc42 (Domin et al., 2005), and PI3KC2α depletion interfered with Rho-mediated smooth muscle contraction (Yoshioka et al., 2007). Interestingly, endocytic trafficking of Rac was shown important for its spatially regulated activity (Palamidessi et al., 2008). Thus, one consequence of opposing Pi3K68D/mtm functions in hemocytes may be in the cortical balance of specific Rho GTPases, either through PI(3)P-mediated membrane trafficking or recruitment of PI(3)P-binding regulatory proteins to discrete membrane domains.
Our experiments show that Mtm has a role in policing traffic at the late endosome, which is consistent with a normal function to down-regulate membrane influx and promote efflux. mtm was important to maintain the balance, but not ability, for membrane influx from endocytic and autophagic routes. We found through genetic interactions, marker analysis, and time-lapse microscopy that mtm function antagonizes PI(3)P-mediated membrane flux consistent with known roles in transport, tethering, and fusion of endosomes with lysosomes and of autophagosomes with late endosomes (Simonsen and Tooze, 2009). Importantly, mtm-dependent functions for endolysosomal size and cortical remodeling were separable, as indicated by mtm interactions with Atg1 or Vps34 that rescued endolysosome size but not hemocyte protrusions. Live cell imaging also revealed lack of dynamic tubulation, indicative of exiting membrane, in mtm-depleted hemocytes, suggesting that mtm function promotes undetermined routes of membrane efflux from PI(3)P-containing compartments. In addition, several results point to a role for mtm in autophagy: the increased number of double-membrane–bound structures and autophagolysosomes in mtm-depleted cells, reversion of enlarged endolysosomal size with mtm and Atg1 codepletion, and Mtm localization to small rings associated with LysoTracker-positive organelles and within Rab7 compartments, suggestive of autophagosomes. Given the PI(3)P dependence and intersection with endolysosomes, there are likely roles for MTM phosphatase regulation in autophagy (Vergne et al., 2009).
Collectively, we favor a model that a PI(3)P pool directly coregulated by Pi3K68D-mediated synthesis and Mtm-mediated turnover is involved in membrane delivery and exit, respectively, at an endosomal compartment that maintains homeostasis of both cortical dynamics and endolysosome size. Pi3K68D localization and motility suggest interaction at the level of dynamic PI(3)P pools synthesized at the cortex or on internal membranes. The lack of cell protrusions upon mtm disruption could result from elevated Pi3K68D-dependent PI(3)P that inhibits membrane efflux to undefined recycling endosomes and, thus, blocks redelivery of a cortical regulator that promotes cell protrusions. Pi3K68D overexpression did not phenocopy the lack of protrusions, which may indicate that Pi3K68D requires a limiting cofactor or scaffold protein or that levels of Mtm are sufficient to override ectopic activity. Conversely, ectopic cell protrusions that form upon Mtm overexpression could result from inappropriate depletion of a Pi3K68D-synthesized PI(3)P pool that leads to excessive efflux, and, thus, persistent recycling of the same cortical regulator. Consistent with this, an endosomal-tethered form of MTM1 was able to induce membrane tubulation (Fili et al., 2006). In turn, mtm function down-regulates PI(3)P-mediated endosome transport, tethering, and fusion, restricting endolysosome size.
Our genetic analysis uncovered critical requirements for mtm- and phosphoinositide-dependent muscle and immune cell functions in Drosophila. Defects in remodeling cell shape upon either knockdown or overexpression of mtm both corresponded with defects in hemocyte dispersion and recruitment to wound sites. These results indicate the significance of mtm-dependent cellular regulation to immune cell behaviors in the animal, analogous to those performed by mammalian macrophages in response to wounding and infection. The identification of Pi3K68D-generated PI(3)P pools as a likely in vivo substrate of Mtm, and the specific cellular roles modulated by the balance of this pool in animals, has significance in better understanding roles for conserved MTM1/MTMR2 and PI3KC2 in mammals. Our results highlight the potential that class II PI3K-activating mutations could underlie unassigned MTM-related human diseases. Furthermore, class II PI3K could serve as a therapeutic target to oppose deleterious effects of MTM mutations associated with human disease.
Materials and methods
Drosophila mtm mutations and genetics
Excision alleles mtmΔ77 and mtmΔ210 were isolated after Δ2-3 mobilization of viable P(EPgy2)CG9117 for lethality over Df(2L)ED338 and molecularly determined by sequencing of genomic DNA amplicons spanning the excisions from homozygous mutant larvae. Point mutation allele mtmz2-4747 was identified with help from the Fly-TILL service (Cooper et al., 2008). The mtm genomic alleles were balanced over CyO, P{w[+mC]=ActGFP}JMR1, and presence or absence of GFP was used for determination of lethal phase and selection of homozygous and trans-heterozygous larvae raised at 25°C.
RNAi hairpin constructs made from 395- (201–202) or 565-bp (203–204) mtm genomic amplicons with primers (201F) 5′-GTACTCTAGATATTGACAACACACTGTGGCTAAA-3′, (202R) 5′-GTACTCTAGAAGTACAGATGATCCACGATGGTTA-3′, (203F) 5′-GTACTCTAGACTTACCATAACCAACTACCGTCTG-3′, and (204R) 5′-GTACTCTAGAACCTTGTAATGGTCGCTTGAGT-3′ were cloned with XbaI restriction sites into upstream activating sequence (UAS)–containing pWIZ vector sequentially at NheI and AvrII sites (Lee and Carthew, 2003), and orientation was confirmed by sequencing. Fly injections (Best Gene) generated viable transformant lines, including UAS-IR:mtm3.1 (II) and UAS-IR:mtm3.5 (III) with 201–202 fragment in “heads-in” orientation used in this study. When 2xIRmtm is noted, UAS-IR:mtm3.1; UAS-IR:mtm3.5 stock was used. Crosses using RNAi hairpins were raised at 29°C to maximize GAL4-driven expression.
Genotypes used in this study included (1) w; UAS-GFP:Rab7/CyO (Entchev et al., 2000) and w; UAS-GFP:Rab7, UAS-IRmtm3.1/CyO, (2) w; UAS-GFP:myc:2xFYVE/CyO (Wucherpfennig et al., 2003) and w; UAS-GFP:myc:2xFYVE, UAS-IRmtm3.1/CyO, (3) w; UAS-mCherry:2XFYVE2, (4) w; UAS-mtm:eGFP7 and w; UAS-IRmtm3.1; UAS-mtm:eGFP7, (5) w; UAS-eGFP:MTMR25 and w; UAS-IRmtm3.1, UAS-eGFP:MTMR25, (6) w; UAS-mCherry:mtm7 and w; UAS-eGFP:mtm4, (7) w[1118]; UAS-IRPi3K68D transformant lines 16239 and 16240, and w[1118]; UAS-IRFab127591 (Dietzl et al., 2007) and w; UAS-IRPi3K68D16240, UAS-IRmtm3.5, (8) w; UAS-mCherry:Pi3K68D2.1/CyO, w; UAS-mCherry:Pi3K68D2.2/TM6 Hu Tb and w; UAS-Pi3K68D:eGFP, (9) yw, hs-FLP; UAS-Vps34-KDm8 (Juhász et al., 2008) and w; UAS-Vps34-KDm8, UAS-IRmtm3.1, (10) yw, hs-FLP; UAS-Vps34-WTm7 (Juhász et al., 2008), (11) w; Vps34Δm22/CyO-GFP-B (Juhász et al., 2008), w; mtmΔ77, Vps34Δm22/CyO,ActGFP, and w; mtmz2-4747, Vps34Δm22/CyO,ActGFP, (12) w[1118]; P{w[+mC]=Cg-GAL4.A}2 (Asha et al., 2003), (13) w; Cg-GAL4, P{w[+mC]=UAS-2xEGFP}AH2, (14) w; Cg-GAL4, UAS-mCherry:myc:2xFYVE2, (15) w; Cg-GAL4, UAS-mCherry:mtm7, (16) w; Pxn-GAL48.1.1, (17) w; Pxn-GAL48.1.1, UAS-GFP (Stramer et al., 2005), (18) w[1118]; P{w[+mC]=UAS-lacZ.B}Bg4-1-2, (19) Cg-GAL4; Atg1Δ3D/TM6C Sb Tb (derived from Atg1Δ3D; Scott et al., 2004), and w; UAS-IRmtm3.1; Atg1Δ3D/TM6C Sb Tb.
Determination of animal viability and lethal phase
For trans-heterozygous mtm mutants, first instar larvae were collected on grape juice agar plates 1 d after egg laying. Larvae were transferred to fresh plates supplemented with yeast paste and allowed to develop at 25°C. Viable larvae were counted each day up to 12 d after embryo laying. For RNAi hairpin–expressing flies, crosses were incubated at 25°C for 12 h to allow for sufficient egg laying, parents were removed, and progeny were allowed to develop in the vials at 29°C. The number of animals reaching pupal, pharate, and adult stages were counted for each WT and mutant genotype.
Characterization of hemocytes
To test developmental timing for mtm requirement in hemocytes, experiments using the temperature sensitivity of GAL4 were performed on Pxn-GAL4/UAS-lacZ and UAS-IRmtm3.1/+; Pxn-GAL4/+ staged larvae reared either continuously at 18 or 29°C or raised at 18°C for 3 d after embryo collection then shifted to 29°C for 1 d. For hemocyte imaging (detailed in the following paragraph), hemocytes were bled from three to four wandering third instar larvae 4 d after egg laying into 100 µl complete medium (for live cell imaging) or PBS and were allowed to attach to a glass coverslip for 1 h at 25°C. Using bled hemocytes, a hemocytometer was used to determine cell numbers, and cell viability was determined using the Live/Dead Assay for Cell Viability kit (Invitrogen). To assess efficiency of RNAi knockdown in hemocytes, RT-PCR was performed on a population of primary hemocytes isolated to near purity from third instar larvae. For hemocyte purification, third instar larvae were washed with 70% ethanol then PBS. Approximately 150 mg wet WT larvae was used per preparation. The larvae were added to a cell strainer with a 70-µm nylon mesh placed in a shallow collecting dish and crushed with the back of an Eppendorf tube. The released hemocytes within the crude preparation were filtered through the 70-µm filter on the cell strainer into a 50-ml tube, and the crushed larvae were washed three times with PBS, pH 7.4. The hemocytes were centrifuged at 200 rpm for 5 min, and the supernatant containing the hemocytes collected centrifuged at 2,000 rpm for 10 min. The supernatant was discarded, and the collected hemocytes were used for mRNA extraction and RT-PCR as described for molecular analyses.
Hemocyte wound recruitment assay and imaging of hemocytes in larvae
Wandering third instar larvae raised at 29°C were washed in PBS, briefly cleaned in 100% ethanol, and rinsed with PBS. The larvae were placed on a piece of paper to dry and mounted dorsal side up on a strip of double-sided tape fastened on a glass slide. Larvae were wounded in abdominal segment 5 or 6 using a Straight Stab pin (0.03-mm tip diameter; Fine Science Tools). Immediately after wounding, a drop of PBS was placed on the larvae to release them from the tape. The larvae recovered on standard grape agar plates for 6 h at 25°C, were briefly immobilized on ice, then remounted dorsal side up on double-sided tape. A glass coverslip (#1.5; 22-mm square) was placed on the larvae and attached with tape on two edges to minimize larval body wall contractions during imaging. Care was taken not to squash the larvae. The larvae were imaged with a 20× 0.5 NA objective (HC PL FLUOTAR; Leica) on a fluorescence microscope (DM1600; Leica). To estimate sessile hemocyte populations, wandering third instar larvae were mounted on double-sided tape and imaged. For each larva, images of GFP-positive hemocytes throughout abdominal segments 5 or 6 were collected and quantified.
Cell staining and microscopy
For visualization of lysosomes, live cells were stained for 5 min with LysoTracker red DND-99 (1:7,500; Invitrogen) and Hoechst 33342 (1:1,000; Invitrogen) in complete medium, then washed and kept in complete medium for fluorescence microscopy at room temperature. Digital images of Kc167 cells were taken on a microscope (Axiovert 200M; Carl Zeiss, Inc.) using a 63× 1.4 NA Plan Apo objective, transmission grid (ApoTome; Carl Zeiss, Inc.), and a camera (ORCA-ER; Hamamatsu Photonics). AxioVision software (Carl Zeiss, Inc.) was used for image acquisition and 3D reconstruction. Digital imaging of hemocytes was performed on a point-scanning microscope (FV1000; Olympus) controlled by the FluoView program (Olympus) using a 60× 1.2 NA Plan Apo N objective with 2× zoom. ImageJ (National Institutes of Health) was used for exporting tagged image file formats (TIFFs) and 3D reconstruction. For fluid phase uptake and trafficking, cells were given a 5-min pulse with dextran Alexa Fluor 488 (molecular mass, 10,000; 1 mg/ml; Invitrogen) together with LysoTracker red (1:10,000) and Hoechst (1:1,000) in complete Schneider’s medium at room temperature. Cells were washed five times in PBS and chased in complete Schneider’s medium for 20 min. Cells were imaged live with a microscope (Axiovert 200M). Similar experiments were performed using Alexa Fluor 488–conjugated BSA (Invitrogen). For assessment of bacterial engulfment and trafficking, hemocytes of genotypes Pxn-GAL4/UAS-lacZ and UAS-IRmtm3.1/+; UAS-IRmtm3.5/Pxn-GAL4 dissected in Schneider’s complete medium and allowed to spread for 30 min were given a pulse of fluorescein-labeled E. coli (Vybrant phagocytosis assay; Invitrogen) in PBS for 5 min, washed with PBS, and incubated for 1.5 h. Fluorescence of extracellular bacteria was quenched with Trypan blue for 5 min, washed in PBS for 5 min, and LysoTracker red and Hoechst were added before imaging with a microscope (Axiovert 200M) using a 63× objective and transmission grid (ApoTome). Photoshop (Adobe) was used to adjust the levels and curves and to crop and resize images.
For F-actin visualization and immunohistochemistry, hemocytes or Kc167 cells were fixed for 10 min in 3.7% formaldehyde 1x PBS and washed twice for 5 min. PBST (PBS with 0.1% Triton X-100) was blocked for 10 min. PBSTB (PBST with 3% BSA) was stained overnight at 4°C with Alexa Fluor 546 phalloidin (1:100; Invitrogen), mouse anti–α-tubulin (1:500; Sigma-Aldrich), rabbit anti-Rab5 (1:50; Wucherpfennig et al., 2003), mouse anti-Golgi (1:200; EMD), or guinea pig anti-SCAR (1:100; Zallen et al., 2002) in PBSTB and for 1 h with Alexa Fluor secondary antibodies as needed (1:1,000; Invitrogen) in PBSTB, washed in PBS for 10 min, stained with DAPI for 10 min, and washed twice with PBS for 10 min. Cells were kept in PBS and visualized using fluorescence microscopy. Digital images were taken at room temperature either on an epifluorescence microscope (Axiovert 200M) or a point-scanning confocal microscope (FV1000) or using a 60x objective with a spinning-disk fluorescence microscope (DSU; Olympus) using a 60x 1.2 NA Plan Apo N objective and SlideBook software (Intelligent Imaging Innovations). Images were exported as 16-bit TIFFs using ImageJ. Photoshop was used to adjust the levels and curves and to crop and resize images.
Time-lapse microscopy
For time-lapse imaging of cell morphology and dynamics, GFP-expressing hemocytes were imaged every 10 s for 5 min using a 60x 1.2 NA Plan Apo N objective on a spinning-disk fluorescence microscope (DSU). Time-lapse imaging of mCherry:2xFYVE or Pi3K68D:GFP and LysoTracker in hemocytes were obtained every 15 s for 5 min with three z sections spaced 0.3-µm apart at each time point on a point-scanning confocal microscope (FV1000) using a 60x 1.4 NA objective and 488- or 543-nm laser. Acquisition was performed with FluoView software, and image analysis was performed with ImageJ software. Time-lapse imaging of Pxn>GFP:Rab7 and LysoTracker red in hemocytes was captured every 10 s for 60 frames on a spinning-disk fluorescence microscope (DSU). TIFF files were exported as a video interleave file at three frames per second. Time-lapse imaging of Pi3K68D:GFP in Kc167 cells was collected every 6 s (or 4 s with drug treatments) for 60 frames with an epifluorescence microscope (Axiovert 200M). TIFF files were exported as QuickTime files (Apple) at three frames per second. Photoshop and ImageJ were used for digital editing and production of time-lapse videos. Time-lapse microscopy imaging was performed at room temperature with cells in complete Schneider’s media.
Fixation, embedding, and transmission electron microscopy imaging
RNAi was performed in 100-mm Petri dishes. After 5 d, cells were harvested by centrifugation for fixation. Samples were immersed in modified Karnovsky’s fixative (1.5% glutaraldehyde, 3% paraformaldehyde, and 2% sucrose in 0.1 M cacodylate buffer, pH 7.4) for 8 h, postfixed in 1% osmium tetroxide in 0.1 M cacodylate buffer for 1 h, and stained en bloc in 1% uranyl acetate for 1 h. Samples were dehydrated in ethanol, embedded in epoxy resin, sectioned at 60–70 nm, and picked up on Formvar- and carbon-coated copper grids. Grids were stained with uranyl acetate and lead nitrate, viewed using a transmission electron microscope (Philips CM-10; FEI), and photographed using a digital camera (Gatan).
Generation of DNA constructs
Monomeric mCherry cDNA (Shaner et al., 2004) was inserted into pTW1129 tagless destination vector (provided by T. Murphy, Carnegie Institution, Baltimore, MD; Drosophila Gateway Vector Collection) upstream of the Gateway cassette to create pUAST-mCherry-ccdB. Full-length mtm cDNA was PCR amplified (5′-CACCATGGATCCCTCCGCG-3′ and 5′-GTGCATAGGCGAGGCCAGA-3′), cloned into the pENTR/D-TOPO entry vector (Invitrogen), subcloned by an LR recombination reaction into Gateway destination vectors pTGW-1075 (UAS-eGFP:mtm) and pUAST-mCherry-ccdB (UAS-mCherry:mtm), and confirmed by sequencing and protein expression. EcoRI restriction digest of PCR-amplified full-length mtm cDNA (clone LD28822; Drosophila Genomics Resource Center [DGRC]) was subcloned to make UAST-mtm:eGFP. UAS-GFP:mtm and UAS-FLAG:mtm were generated from insertion of PCR-amplified full-length mtm cDNA into Gateway pENTR/D-TOPO and recombination into tag-containing destination vectors. The mtm cDNA mutation of codon 403 (CGC to TGC) was made using site-directed mutagenesis (QuickChange; Agilent Technologies). NotI restriction digest of human GFP:MTMR2 (provided by J. Dixon, University of California, San Diego, La Jolla, CA) was subcloned into pUAST for UAS-GFP:MTMR2. Full-length Pi3K68D splice form A (clone LD28067; DGRC) was subcloned into pENTR1A vector (Invitrogen), excluding the STOP codon, to generate pENTR1A-Pi3K68D RA. LR recombination was performed with destination vectors pUAST-ccdB-GFP (Drosophila Gateway Vector Collection) and pUAST-mCherry-ccdB to obtain C- and N-terminally tagged Pi3K68D:GFP and mCherry:Pi3K68D, respectively. myc:2XFYVE was amplified from pUAST-GFP:myc:2XFYVE (provided by M. González-Gaitán, University of Geneva, Geneva, Switzerland) and inserted into pUAST-mCherry to generate pUAST-mCherry:myc:2XFYVE.
Kc cell culture, DNA transfection, RNAi, and molecular analyses
Drosophila Kc167 cells (Echalier and Ohanessian, 1969) were cultured in Schneider’s Drosophila medium (Invitrogen) containing 10% FBS and 1% penicillin/streptomyocin (Sigma-Aldrich) at 24°C. Effectene transfection reagent (QIAGEN) was used for DNA delivery of metallothionein-GAL4 along with the UAS constructs of interest, UAS-GFP:Rab7, UAS-GFP:myc:2xFYVE (provided by M. González-Gaitán), UAS-GFP:LAMP (provided by H. Krämer, University of Texas Southwestern, Dallas, TX), UAS-mtm:GFP, UAS-GFP:mtm, or UAS-Pi3K68D:GFP. To induce the metallothionein-GAL4, transfected cells were incubated with 0.75 mM CuSO4 in complete media for 6–8 h before imaging. To test effects of cytoskeletal disruption on mtm RNAi phenotype or Pi3K68D localization and motility, cells were treated with 50 µM latrunculin B (EMD) for 2 h in complete medium or 15 µM nocodazole (Sigma-Aldrich) in PBS for 2 h and imaged live.
Double-stranded RNAs were generated from genomic DNA sequences, and RNAi for 3–5 d was performed as described previously (Kiger et al., 2003). To induce cellular elongation, cells were incubated 3 d after RNAi with a final concentration of 1 µM ecdysone (Sigma-Aldrich) in complete medium for 36 h before fixation. RNAi efficiency was determined from mRNA isolated from Trizol cell extracts and RT-PCR of cDNA (Invitrogen). Cell extracts in RIPA lysis buffer with protease inhibitor cocktail (Sigma-Aldrich) were used for Western blots with rabbit anti-GFP (1:2,000; Santa Cruz Biotechnology, Inc.), mouse anti–α-tubulin (1:5,000; Sigma-Aldrich), or mouse Ag10.2 anti-EcR (Developmental Studies Hybridoma Bank) followed by anti–rabbit HRP or anti–mouse HRP (1:10,000; Invitrogen).
To determine Kc167 cell numbers after RNAi, control or mtm RNAi cells in poly-d-lysine–coated glass-bottom 96-well plates were incubated at 24°C for 4 d. After 3 d, some wells were treated with 1 µM ecdysone (Sigma-Aldrich) in complete medium for 24 h. Cells from each condition were imaged then scraped, resuspended in media, and counted by hemocytometer on d 1–4 after plating. To assess cell viability and relative cell size by flow cytometry, control and mtm RNAi-treated Kc167 cells 4 d after plating with or without addition of 1 µM ecdysone for 18 h were scraped, filtered, and resuspended in PBS. Flow cytometric analysis of 20,000 ungated cells for forward and side scatter properties were collected from replicate samples from three experiments. A uniform gate drawn for forward and side scatter plots was applied to all samples collected in the same experiment and used to quantify the percentage of live and dead cells.
Phosphatase activity assay
Kc167 cells (107 cells/two 100-mm plates) transfected with UAS-FLAG:mtm or UAS-FLAGmtmR403C and metallothionein-GAL4 were induced 48 h later with 1 mM CuSO4 overnight. Plates were washed twice with ice-cold PBS and lysed in 500 µl lysis buffer (25 mM Tris-HCl, 1 mM EDTA, 0.1 mM EGTA, 5 mM MgCl2, 150 mM NaCl, 10% glycerol, 1% NP-40, 1 mM DTT, and 1x protease inhibitor [Sigma-Aldrich]). Lysates were spun at 13,000 rpm for 10 min, the supernatant was collected, and equal volumes of cell lysates were immunoprecipitated using anti-Flag M2 agarose beads (Sigma-Aldrich) for 2 h at 4°C. Beads were washed three times with lysis buffer, once with lysis buffer without NP-40, twice in reaction buffer (20 mM Tris-HCl, pH 7.4, 5 mM MgCl2, and 0.5 mM EDTA), and then split into four tubes (two used for PI(3)P and two used for PI(3,5)P2). Phosphatase activity was measured using the malachite green assay (Echelon Biosciences, Inc.) following the manufacturer’s instructions. Reaction mix (reaction buffer plus 50 µM PIPs) was added to each tube, incubated for 20 min at 25°C, then stopped by addition of 7 µl 0.05 mM Na3VO4 and heated at 95°C for 2 min. Free phosphate in solution was measured by plate reader. Beads were loaded onto a 4–12% SDS polyacrylamide gel (Invitrogen) to estimate the amount of immunoprecipitated protein. The gel was stained with Sypro Ruby red stain (Invitrogen), and it was found that the Mtm-WT and Mtm-R403C amounts were equal.
Radiolabeling, extraction, and quantification of PI(3)P concentrations
After 3 d of RNAi, Kc167 cells were labeled for 40 h with 90 µCi/ml myo-[2-3H]inositol (PerkinElmer) in inositol-free Schneider’s Drosophila medium (Invitrogen). Cells were scraped in PBS and collected by centrifugation. Phosphoinositides were extracted as described previously (Rudge et al., 2004), and the dried extracts were analyzed by HPLC as described previously (Baird et al., 2008). Radiolabeled phospholipids from yeast extracts were used as standards.
Statistical analysis
The area of subcellular objects (Rab5, GFP:Rab7, and LysoTracker) were scored and quantified using either AxioVision or CellProfiler software. Images were thresholded, segmented, and measured in marker channel to determine object areas and in nuclei channel to determine total cell count. Per object measurements for ≥745 compartments from ≥60 cells pooled from at least two experiments were analyzed with Prism software (GraphPad Software, Inc.) to determine object area mean, standard error, and Student’s t test, and results were converted from pixels to squared micrometers for Figs. 4 (H and I), 5 C, 8 (C–D′), S2 G, and S3 D. Colocalization of dextran and LysoTracker was scored and quantified with the MetaMorph software (MDS Analytical Technologies) colocalization module per entire field and reported as percentage of dextran versus LysoTracker overlap in Fig. 5 (A and A′). The area of localized PI(3)P per cell was determined using AxioVision software. Images were thresholded, segmented, and measured for field area of GFP:2xFYVE channel and DAPI channel to count nuclei, then used to calculate normalized GFP:2xFYVE area per cell per field. Prism software was used to calculate the mean-normalized GFP:2xFYVE area from 8–13 fields (31–88 cells) across two experiments, standard error, and Student’s t test for Fig. 7 C. Cell morphology of F-actin–labeled spread hemocytes or GFP-positive transfected Kc167 cells was assessed manually and categorized for all detectable cells within entire image fields (≥12) from three experiments, and results were plotted as the mean percentage of total scored cells for Figs. 1 G and 2 G. Prism was used to calculate SD and Student’s t test.
Transmission electron microscopy data in Fig. 4 (M and P) were quantified from 26 cells selected randomly for each condition, with the only criteria that the entire cell with nucleus cross section was represented. The cell perimeter and secondary lysosomes, as visually identified by membrane whorls, were manually outlined in Photoshop. The total area of secondary lysosomes as a percentage of the entire cell area was calculated in Excel (Microsoft), analyzed in Prism with Student’s t test, and graphed as a scatter plot with mean percent cell area. Electron-lucent membrane structures were visually identified, and the number of single-membrane–bound and double-membrane–bound structures were counted for each cell. Prism software was used to determine Student’s t test and graph results as a scatter plot with the mean number of structures per cell.
Hemocytometer counts of bled hemocytes were made independently from 10 larvae. CellProfiler software was used to estimate sessile hemocyte populations per segment from images of GFP-positive hemocytes in abdominal segments 5 or 6 from10 larvae. Images of wound sites from ≥11 larvae per genotype were analyzed in Photoshop by manually counting hemocytes within one-wound diameter. Prism software was used to calculate mean, standard error, and Student’s t test for hemocyte numbers graphed in Fig. 3 (A, C, and F). Hemocytometer counts of RNAi-treated Kc167 cells from three experiments were used to calculate mean, SD, and Student’s t test for Fig. 1 D. Percentage of live and dead cells, for 20,000 cells for each of two experiments, and forward scatter from four experiments, each of 16,000 gated cells, were used to calculate mean, standard error, and Student’s t test for Fig. S2 (C and D). Phosphatase activities were analyzed for mean, standard error, and Student’s t test from four experiments for Fig. 6 A. Mean percent radiolabeled PI(3)P and Student’s t test were calculated from two experiments.
Online supplemental material
Fig. S1 shows results from Kc cell RNAi screen. Fig. S2 demonstrates normal cell number, viability, and ecdysone reception in mtm RNAi Kc cells. Fig. S3 shows normal phagocytosis in mtm-depleted hemocytes, dependence of mtm RNAi lysosomal defects on HOPS complex, and that Fab1 RNAi shares similar lysosomal but not morphology defects. Fig. S4 shows GFP:2xFYVE-altered distribution in Kc cells and hemocytes and the affect of Pi3K functions on hemocyte protrusions. Table S1 displays adult viability and visible phenotypes detected with tissue-targeted mtm depletion and genetic interactions. Video 1 shows time-lapse microscopy of altered cortical dynamics in mtm-depleted hemocytes. Video 2 shows time-lapse microscopy of fusion between mCherry:2xFYVE compartments with lack of tubulation in mtm-depleted hemocytes. Video 3 shows time-lapse microscopy of GFP:Rab7 and LysoTracker in hemocytes. Video 4 shows time-lapse microscopy of Pi3K68D:GFP motility and association with LysoTracker compartments. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200911020/DC1.
Acknowledgments
We are grateful to T. Neufeld, M. González-Gaitán, H. Kramer, M. Galko, J. Dixon, H. Stenmark, J. Zallen, Bloomington Drosophila Stock Center, and Vienna Drosophila RNAi Center for reagents. We thank C. Garza, J. Dennis, G. Kim, K. Lau, and S. Cox for technical assistance. We thank B. Wakimoto and C. Zuker for mtmz2-4747 alleles. We thank T. Meerloo for technical assistance with electron microscopy and M. Farquhar for use of the Center for Molecular Medicine Electron Microscope Facility. We are grateful to S. Emr for the use of HPLC instrumentation. Microscopy was performed in part at the University of California San Diego Neuroscience Microscopy Shared Imaging Facility.
This work was supported by the Fonds de la Recherche en Santé du Québec (to S. Jean), the Kimmel Foundation (grant KF06-120 to A.A. Kiger), the Packard Foundation (grant 2005-29096 to A.A. Kiger), and the National Institutes of Health (grants R01 GM078176 to A.A. Kiger, RO1 DK17780 to M. Farquhar and the CMM Electron Microscope Facility, and P30 NS047101 to the University of California, San Diego Neuroscience Microscopy Shared Imaging Facility).
Footnotes
Abbreviations used in this paper:
- ecdysone
- 20-hydroxyecdysone
- F-actin
- filamentous actin
- MTM
- myotubularin
- PI3-kinase
- phosphatidylinositol 3-kinase
- PI(3)P
- phosphatidylinositol 3-phosphate
- PIP
- phosphoinositide phosphate
- TIFF
- tagged image file format
- UAS
- upstream activating sequence
- WT
- wild type
References
- Asha H., Nagy I., Kovacs G., Stetson D., Ando I., Dearolf C.R. 2003. Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics. 163:203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock D.T., Brock A.R., Fish G.S., Wang Y., Perrin L., Krasnow M.A., Galko M.J. 2008. Circulating blood cells function as a surveillance system for damaged tissue in Drosophila larvae. Proc. Natl. Acad. Sci. USA. 105:10017–10022 10.1073/pnas.0709951105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird D., Stefan C., Audhya A., Weys S., Emr S.D. 2008. Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J. Cell Biol. 183:1061–1074 10.1083/jcb.200804003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta L.M., Robinson J.S., Klionsky D.J., Emr S.D. 1988. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J. Cell Biol. 107:1369–1383 10.1083/jcb.107.4.1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger P., Bonneick S., Willi S., Wymann M., Suter U. 2002. Loss of phosphatase activity in myotubularin-related protein 2 is associated with Charcot-Marie-Tooth disease type 4B1. Hum. Mol. Genet. 11:1569–1579 10.1093/hmg/11.13.1569 [DOI] [PubMed] [Google Scholar]
- Bolino A., Muglia M., Conforti F.L., LeGuern E., Salih M.A., Georgiou D.M., Christodoulou K., Hausmanowa-Petrusewicz I., Mandich P., Schenone A., et al. 2000. Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularin-related protein-2. Nat. Genet. 25:17–19 10.1038/75542 [DOI] [PubMed] [Google Scholar]
- Cao C., Laporte J., Backer J.M., Wandinger-Ness A., Stein M.P. 2007. Myotubularin lipid phosphatase binds the hVPS15/hVPS34 lipid kinase complex on endosomes. Traffic. 8:1052–1067 10.1111/j.1600-0854.2007.00586.x [DOI] [PubMed] [Google Scholar]
- Cao C., Backer J.M., Laporte J., Bedrick E.J., Wandinger-Ness A. 2008. Sequential actions of myotubularin lipid phosphatases regulate endosomal PI(3)P and growth factor receptor trafficking. Mol. Biol. Cell. 19:3334–3346 10.1091/mbc.E08-04-0367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.L., Till B.J., Henikoff S. 2008. Fly-TILL: reverse genetics using a living point mutation resource. Fly (Austin). 2:300–302 [DOI] [PubMed] [Google Scholar]
- Di Paolo G., De Camilli P. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443:651–657 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 448:151–156 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Domin J., Harper L., Aubyn D., Wheeler M., Florey O., Haskard D., Yuan M., Zicha D. 2005. The class II phosphoinositide 3-kinase PI3K-C2beta regulates cell migration by a PtdIns3P dependent mechanism. J. Cell. Physiol. 205:452–462 10.1002/jcp.20478 [DOI] [PubMed] [Google Scholar]
- Echalier G. 1997. Drosophila Cells in Culture. Academic Press, New York: 702 pp [Google Scholar]
- Echalier G., Ohanessian A. 1969. [Isolation, in tissue culture, of Drosophila melangaster cell lines]. C. R. Acad. Sci. Hebd. Seances Acad. Sci. D. 268:1771–1773 [PubMed] [Google Scholar]
- Entchev E.V., Schwabedissen A., González-Gaitán M. 2000. Gradient formation of the TGF-beta homolog Dpp. Cell. 103:981–991 10.1016/S0092-8674(00)00200-2 [DOI] [PubMed] [Google Scholar]
- Falasca M., Maffucci T. 2007. Role of class II phosphoinositide 3-kinase in cell signalling. Biochem. Soc. Trans. 35:211–214 10.1042/BST0350229 [DOI] [PubMed] [Google Scholar]
- Falasca M., Hughes W.E., Dominguez V., Sala G., Fostira F., Fang M.Q., Cazzolli R., Shepherd P.R., James D.E., Maffucci T. 2007. The role of phosphoinositide 3-kinase C2alpha in insulin signaling. J. Biol. Chem. 282:28226–28236 10.1074/jbc.M704357200 [DOI] [PubMed] [Google Scholar]
- Fili N., Calleja V., Woscholski R., Parker P.J., Larijani B. 2006. Compartmental signal modulation: endosomal phosphatidylinositol 3-phosphate controls endosome morphology and selective cargo sorting. Proc. Natl. Acad. Sci. USA. 103:15473–15478 10.1073/pnas.0607040103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., Stenmark H. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19:4577–4588 10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gaitan M. 2008. The garden of forking paths: recycling, signaling, and degradation. Dev. Cell. 15:172–174 10.1016/j.devcel.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Grant B.D., Donaldson J.G. 2009. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 10:597–608 10.1038/nrm2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafen E. 2004. Cancer, type 2 diabetes, and ageing: news from flies and worms. Swiss Med. Wkly. 134:711–719 [DOI] [PubMed] [Google Scholar]
- Jahn R., Lang T., Südhof T.C. 2003. Membrane fusion. Cell. 112:519–533 10.1016/S0092-8674(03)00112-0 [DOI] [PubMed] [Google Scholar]
- Johannes L., Popoff V. 2008. Tracing the retrograde route in protein trafficking. Cell. 135:1175–1187 10.1016/j.cell.2008.12.009 [DOI] [PubMed] [Google Scholar]
- Juhász G., Hill J.H., Yan Y., Sass M., Baehrecke E.H., Backer J.M., Neufeld T.P. 2008. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J. Cell Biol. 181:655–666 10.1083/jcb.200712051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadandale P., Stender J.D., Glass C.K., Kiger A.A. 2010. Conserved role for autophagy in Rho1-mediated cortical remodeling and blood cell recruitment. Proc. Natl. Acad. Sci. USA. 107:10502–10507 10.1073/pnas.0914168107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger A.A., Baum B., Jones S., Jones M.R., Coulson A., Echeverri C., Perrimon N. 2003. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2:27 10.1186/1475-4924-2-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Noda T., Ishihara N., Ohsumi Y. 2001. Two distinct Vps34 phosphatidylinositol 3–kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152:519–530 10.1083/jcb.152.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.A., Taylor G.S., Torgersen K.M., Dixon J.E. 2002. Myotubularin and MTMR2, phosphatidylinositol 3-phosphatases mutated in myotubular myopathy and type 4B Charcot-Marie-Tooth disease. J. Biol. Chem. 277:4526–4531 10.1074/jbc.M111087200 [DOI] [PubMed] [Google Scholar]
- Lanot R., Zachary D., Holder F., Meister M. 2001. Postembryonic hematopoiesis in Drosophila. Dev. Biol. 230:243–257 10.1006/dbio.2000.0123 [DOI] [PubMed] [Google Scholar]
- Laporte J., Hu L.J., Kretz C., Mandel J.L., Kioschis P., Coy J.F., Klauck S.M., Poustka A., Dahl N. 1996. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat. Genet. 13:175–182 10.1038/ng0696-175 [DOI] [PubMed] [Google Scholar]
- Laporte J., Blondeau F., Buj-Bello A., Tentler D., Kretz C., Dahl N., Mandel J.L. 1998. Characterization of the myotubularin dual specificity phosphatase gene family from yeast to human. Hum. Mol. Genet. 7:1703–1712 10.1093/hmg/7.11.1703 [DOI] [PubMed] [Google Scholar]
- Laporte J., Blondeau F., Gansmuller A., Lutz Y., Vonesch J.L., Mandel J.L. 2002. The PtdIns3P phosphatase myotubularin is a cytoplasmic protein that also localizes to Rac1-inducible plasma membrane ruffles. J. Cell Sci. 115:3105–3117 [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Carthew R.W. 2003. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 30:322–329 10.1016/S1046-2023(03)00051-3 [DOI] [PubMed] [Google Scholar]
- Lindmo K., Stenmark H. 2006. Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 119:605–614 10.1242/jcs.02855 [DOI] [PubMed] [Google Scholar]
- MacDougall L.K., Domin J., Waterfield M.D. 1995. A family of phosphoinositide 3-kinases in Drosophila identifies a new mediator of signal transduction. Curr. Biol. 5:1404–1415 10.1016/S0960-9822(95)00278-8 [DOI] [PubMed] [Google Scholar]
- MacDougall L.K., Gagou M.E., Leevers S.J., Hafen E., Waterfield M.D. 2004. Targeted expression of the class II phosphoinositide 3-kinase in Drosophila melanogaster reveals lipid kinase-dependent effects on patterning and interactions with receptor signaling pathways. Mol. Cell. Biol. 24:796–808 10.1128/MCB.24.2.796-808.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffucci T., Cooke F.T., Foster F.M., Traer C.J., Fry M.J., Falasca M. 2005. Class II phosphoinositide 3-kinase defines a novel signaling pathway in cell migration. J. Cell Biol. 169:789–799 10.1083/jcb.200408005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima J., Wickner W. 2009. Phosphoinositides and SNARE chaperones synergistically assemble and remodel SNARE complexes for membrane fusion. Proc. Natl. Acad. Sci. USA. 106:16191–16196 10.1073/pnas.0908694106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosesson Y., Mills G.B., Yarden Y. 2008. Derailed endocytosis: an emerging feature of cancer. Nat. Rev. Cancer. 8:835–850 10.1038/nrc2521 [DOI] [PubMed] [Google Scholar]
- Norris F.A., Majerus P.W. 1994. Hydrolysis of phosphatidylinositol 3,4-bisphosphate by inositol polyphosphate 4-phosphatase isolated by affinity elution chromatography. J. Biol. Chem. 269:8716–8720 [PubMed] [Google Scholar]
- Palamidessi A., Frittoli E., Garré M., Faretta M., Mione M., Testa I., Diaspro A., Lanzetti L., Scita G., Di Fiore P.P. 2008. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 134:135–147 10.1016/j.cell.2008.05.034 [DOI] [PubMed] [Google Scholar]
- Parrish W.R., Stefan C.J., Emr S.D. 2005. PtdIns(3)P accumulation in triple lipid-phosphatase-deletion mutants triggers lethal hyperactivation of the Rho1p/Pkc1p cell-integrity MAP kinase pathway. J. Cell Sci. 118:5589–5601 10.1242/jcs.02649 [DOI] [PubMed] [Google Scholar]
- Ridley A.J. 2006. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 16:522–529 10.1016/j.tcb.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Robinson F.L., Dixon J.E. 2006. Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol. 16:403–412 10.1016/j.tcb.2006.06.001 [DOI] [PubMed] [Google Scholar]
- Rudge S.A., Anderson D.M., Emr S.D. 2004. Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol. Biol. Cell. 15:24–36 10.1091/mbc.E03-05-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten T.E., Rodahl L.M., Pattni K., Englund C., Samakovlis C., Dove S., Brech A., Stenmark H. 2006. Fab1 phosphatidylinositol 3-phosphate 5-kinase controls trafficking but not silencing of endocytosed receptors. Mol. Biol. Cell. 17:3989–4001 10.1091/mbc.E06-03-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftig P., Klumperman J. 2009. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10:623–635 10.1038/nrm2745 [DOI] [PubMed] [Google Scholar]
- Schu P.V., Takegawa K., Fry M.J., Stack J.H., Waterfield M.D., Emr S.D. 1993. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 260:88–91 10.1126/science.8385367 [DOI] [PubMed] [Google Scholar]
- Scott R.C., Schuldiner O., Neufeld T.P. 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 7:167–178 10.1016/j.devcel.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Seals D.F., Eitzen G., Margolis N., Wickner W.T., Price A. 2000. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA. 97:9402–9407 10.1073/pnas.97.17.9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Simonsen A., Tooze S.A. 2009. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol. 186:773–782 10.1083/jcb.200907014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Wurmser A.E., Emr S.D., Stenmark H. 2001. The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 13:485–492 10.1016/S0955-0674(00)00240-4 [DOI] [PubMed] [Google Scholar]
- Soldati T., Schliwa M. 2006. Powering membrane traffic in endocytosis and recycling. Nat. Rev. Mol. Cell Biol. 7:897–908 10.1038/nrm2060 [DOI] [PubMed] [Google Scholar]
- Srivastava S., Li Z., Lin L., Liu G., Ko K., Coetzee W.A., Skolnik E.Y. 2005. The phosphatidylinositol 3-phosphate phosphatase myotubularin- related protein 6 (MTMR6) is a negative regulator of the Ca2+-activated K+ channel KCa3.1. Mol. Cell. Biol. 25:3630–3638 10.1128/MCB.25.9.3630-3638.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., Di L., Zhdanova O., Li Z., Vardhana S., Wan Q., Yan Y., Varma R., Backer J., Wulff H., et al. 2009. The class II phosphatidylinositol 3 kinase C2beta is required for the activation of the K+ channel KCa3.1 and CD4 T-cells. Mol. Biol. Cell. 20:3783–3791 10.1091/mbc.E09-05-0390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stramer B., Wood W., Galko M.J., Redd M.J., Jacinto A., Parkhurst S.M., Martin P. 2005. Live imaging of wound inflammation in Drosophila embryos reveals key roles for small GTPases during in vivo cell migration. J. Cell Biol. 168:567–573 10.1083/jcb.200405120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunio A., Metcalf A.B., Krämer H. 1999. Genetic dissection of endocytic trafficking in Drosophila using a horseradish peroxidase-bride of sevenless chimera: hook is required for normal maturation of multivesicular endosomes. Mol. Biol. Cell. 10:847–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor G.S., Maehama T., Dixon J.E. 2000. Inaugural article: myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. USA. 97:8910–8915 10.1073/pnas.160255697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosch V., Rohde H.M., Tronchère H., Zanoteli E., Monroy N., Kretz C., Dondaine N., Payrastre B., Mandel J.L., Laporte J. 2006. A novel PtdIns3P and PtdIns(3,5)P2 phosphatase with an inactivating variant in centronuclear myopathy. Hum. Mol. Genet. 15:3098–3106 10.1093/hmg/ddl250 [DOI] [PubMed] [Google Scholar]
- Tsujita K., Itoh T., Ijuin T., Yamamoto A., Shisheva A., Laporte J., Takenawa T. 2004. Myotubularin regulates the function of the late endosome through the gram domain-phosphatidylinositol 3,5-bisphosphate interaction. J. Biol. Chem. 279:13817–13824 10.1074/jbc.M312294200 [DOI] [PubMed] [Google Scholar]
- Vergne I., Roberts E., Elmaoued R.A., Tosch V., Delgado M.A., Proikas-Cezanne T., Laporte J., Deretic V. 2009. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J. 28:2244–2258 10.1038/emboj.2009.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicinanza M., D’Angelo G., Di Campli A., De Matteis M.A. 2008. Function and dysfunction of the PI system in membrane trafficking. EMBO J. 27:2457–2470 10.1038/emboj.2008.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen P.J., Osborne S.L., Morrow I.C., Parton R.G., Domin J., Meunier F.A. 2008. Ca2+-regulated pool of phosphatidylinositol-3-phosphate produced by phosphatidylinositol 3-kinase C2alpha on neurosecretory vesicles. Mol. Biol. Cell. 19:5593–5603 10.1091/mbc.E08-06-0595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M.J. 2007. Drosophila hemopoiesis and cellular immunity. J. Immunol. 178:4711–4716 [DOI] [PubMed] [Google Scholar]
- Wucherpfennig T., Wilsch-Bräuninger M., González-Gaitán M. 2003. Role of Drosophila Rab5 during endosomal trafficking at the synapse and evoked neurotransmitter release. J. Cell Biol. 161:609–624 10.1083/jcb.200211087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser A.E., Emr S.D. 1998. Phosphoinositide signaling and turnover: PtdIns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J. 17:4930–4942 10.1093/emboj/17.17.4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D.J. 2007. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9:1102–1109 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- Xue Y., Fares H., Grant B., Li Z., Rose A.M., Clark S.G., Skolnik E.Y. 2003. Genetic analysis of the myotubularin family of phosphatases in Caenorhabditis elegans. J. Biol. Chem. 278:34380–34386 10.1074/jbc.M303259200 [DOI] [PubMed] [Google Scholar]
- Yoshioka K., Sugimoto N., Takuwa N., Takuwa Y. 2007. Essential role for class II phosphoinositide 3-kinase alpha-isoform in Ca2+-induced, Rho- and Rho kinase-dependent regulation of myosin phosphatase and contraction in isolated vascular smooth muscle cells. Mol. Pharmacol. 71:912–920 10.1124/mol.106.032599 [DOI] [PubMed] [Google Scholar]
- Zallen J.A., Cohen Y., Hudson A.M., Cooley L., Wieschaus E., Schejter E.D. 2002. SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J. Cell Biol. 156:689–701 10.1083/jcb.200109057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Gaidarov I., Keen J.H. 2007. Phosphoinositide 3-kinase C2alpha links clathrin to microtubule-dependent movement. J. Biol. Chem. 282:1249–1256 10.1074/jbc.M606998200 [DOI] [PubMed] [Google Scholar]
- Zou W., Lu Q., Zhao D., Li W., Mapes J., Xie Y., Wang X. 2009. Caenorhabditis elegans myotubularin MTM-1 negatively regulates the engulfment of apoptotic cells. PLoS Genet. 5:e1000679 10.1371/journal.pgen.1000679 [DOI] [PMC free article] [PubMed] [Google Scholar]