Abstract
Thiopurines, including mercaptopurine (MP), 6-thioguanine (SG) and azathioprine, are widely used for the treatment of many human diseases including acute lymphoblastic leukemia (ALL). To exert their cytotoxic effect, these prodrugs need to be metabolically activated to SG nucleotides and incorporated into nucleic acids. SG in DNA can be methylated spontaneously to S6-methylthioguanine (S6mG) in the presence of S-adenosyl-L-methionine. It was proposed that S6mG, owing to its high miscoding potential (pairing preferentially with thymine), may induce cell death by triggering the post-replicative mismatch repair pathway. Understanding the implications of this pathway in the cytotoxic effect of thiopurine drugs necessitates an accurate measurement of the level of S6-methylthio-2′-deoxyguanosine (S6mdG) in DNA of cells treated with thiopurine drugs. Here we developed a sensitive HPLC coupled with tandem mass spectrometry (LC-MS/MS) method and measured the level of 6-thio-2′-deoxyguanosine (SdG) and S6mdG in genomic DNA of four human leukemia cell lines and one human colorectal carcinoma cell line. Our results revealed that, upon treatment with 3 μM SG for 24 hours, approximately 10, 7.4, 7 and 3% of guanine was replaced with SG in Jurkat T, HL-60, CCRF-CEM and K-562 cells, respectively. However, only less than 0.02% of SdG was converted to S6mdG in the above cell lines. HCT-116 cells had the lowest level (0.2%) of guanine being replaced with SG in DNA, and approximately 5 out of 104 SG was converted to its methylated counterpart. This is the first report of the simultaneous and accurate quantification of SdG and S6mdG in genomic DNA of cultured human cells treated with SG. In addition, our results suggested that DNA SG might trigger mismatch repair (MMR) pathway without being converted to S6mG.
Introduction
Thiopurines, including 6-mercaptopurine (6-MP) and 6-thioguanine (SG), have been widely used in the treatment of childhood acute lymphoblastic leukemia (ALL).1 Another thiopurine, azathioprine, which is converted to 6-MP in vivo, is a commonly prescribed immunosuppressant for the treatment of inflammatory bowel diseases, autoimmune conditions and following transplantation.2 6-MP and azathioprine were separately approved by FDA for the treatment of ALL in children in 19533 and for the prolongation of renal allograft survival in 1963.4
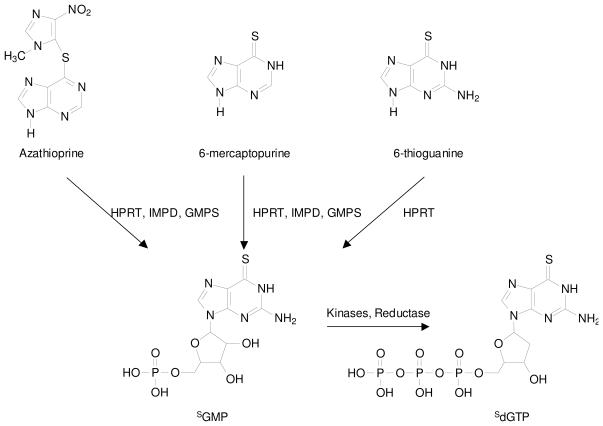
Thiopurines are inactive prodrugs and, to exert their cytotoxic effect, these prodrugs need to be metabolically activated (i.e., to thioguanine nucleotides) and incorporated into DNA (Scheme 1).2 During metabolism, SG is converted to its corresponding nucleoside monophosphate (SGMP) in a single-step reaction catalyzed by hypoxanthine phosphoribosyltransferase (HPRT), whereas three enzymatic steps are required to convert 6-MP and azathioprine to SGMP. SGMP is then phosphorylated by kinases to give SGDP and SGTP, and SGDP can be converted to SdGTP through the sequential actions of ribonucleotide reductase and kinase.5 Thiopurine methyltransferase (TPMT), which exhibits genetic polymorphism, catalyzes the S-methylation of thiopurines. Clinical studies revealed that TPMT-deficient patients accumulate excessive SG nucleotides in erythrocytes and are susceptible to develop severe hematopoietic toxicity or even death from neutropenic sepsis.6-8 In addition, S6-methylthioinosine-5′-monophosphate is a potent inhibitor for de novo purine synthesis, which may constitute an alternative mechanism for the cytotoxicity of 6-MP.9
Scheme 1.
Thiopurines and their metabolism to form 6-thioguanine nucleotides. HPRT, hypoxanthine phosphoribosyltransferase; IMPD, inosine monophosphate dehydrogenase; GMPS, guanosine monophosphate synthase; SGMP, 6-thioguanosine monophosphate; dSGTP, 6-thio-2′-deoxyguanosine triphosphate.
The incorporation of 6-thioguanine into the nucleotide pool or DNA was found to perturb various cellular pathways. In this vein, SGTP, a nucleotide metabolite of azathioprine, was shown to block the activation of Rac1, a GTP-binding protein, in human CD4+ T lymphocytes upon CD28 costimulation.10 Additionally, SG in DNA can lead to global cytosine demethylation11 and it can induce single-strand breaks, interstrand cross-links, DNA-protein cross-links, and chromatid damage.12-15
The increased chemical reactivity of SG relative to guanine may also contribute to the cytotoxic effects of thiopurine drugs. In this context, Swann et al.16 found that DNA SG can be methylated spontaneously to S6-methylthioguanine (S6mG) in the presence of S-adenosyl-L-methionine (S-AdoMet). The coding property of S6mG directs the misincorporation of thymine to give S6mG:T base pair during DNA replication, which is recognized by post-replicative mismatch repair (MMR) machinery; futile cycles of repair synthesis may give rise to cell death.16 Our recent replication study demonstrated that both 6-thio-2′-deoxyguanosine (SdG) and S6-methylthio-2′-deoxyguanosine (S6mdG) are mutagenic in Escherichia coli (E. coli) cells and could lead to G→A mutations at frequencies of 10% and 94%, respectively.17 SG:T mispair is known to be recognized more readily by MMR machinery than the S6mG:T mispair.18, 19 Thus, we hypothesized that the SG:T mispair arising from in vivo DNA replication may invoke directly the post-replicative MMR pathway without having the SG being converted to S6mG.17 In order to evaluate the relative contributions of SG:T and S6mG:T mispairs in provoking the post-replicative mismatch repair pathways, it is necessary to quantify accurately the amounts of SdG and its methylated metabolite, S6mdG, in genomic DNA of SG-treated leukemic cells.
Reverse-phase HPLC with UV or fluorescence detection was used to monitor thiopurine nucleosides and nucleotides.20-23 In addition, anion-exchange HPLC was employed to determine the mono-, di- and triphosphate forms of thiopurine nucleotides.21 Moreover, erythrocyte 6-thioguanine and 6-methylmercaptopurine nucleotides were hydrolyzed by acid to its component nucleobases and subsequently analyzed by LC-MS/MS, though stable isotope-labeled standards were not used.24 However, none of the above-described methods have been used to measure the levels of SdG and S6mdG in cellular DNA. In the latter respect, S6mdG was detected in DNA of cultured Chinese hamster ovary cells by using radiolabeling with L-[3H-methyl]methionine and it was estimated that approximately 1.6 out of 104 SG could be methylated to S6mG.16 Nevertheless, the method requires the use of radioisotopes, and it has not been applied for measuring the levels of SdG and S6mdG in DNA of SG-treated leukemic cells.
Herein, we developed an LC-MS/MS with the stable isotope dilution method for the sensitive, accurate and simultaneous quantification of SdG and S6mdG in genomic DNA of cultured human cancer cells exposed with SG.
Experimental Procedures
Materials
All chemicals and enzymes, unless otherwise specified, were from Sigma-Aldrich (St. Louis, MO). [U-15N5]-2′-deoxyguanosine ([U-15N5]-dG) and CD3I were purchased from Cambridge Isotope Laboratories (Andover, MA). Proteinase K was obtained from New England Biolabs (Ipswich, WA). Cancer cell lines, which included Jurkat T, CCRF-CEM, HL-60, K-562 and HCT-116 cells, and cell culture reagents were purchased from ATCC (Manassas, VA, USA).
Synthesis and Characterization of [U-15N5]-6-thio-2′-deoxyguanosine
The title compound was prepared at microscale following previously published method for the synthesis of the corresponding unlabeled compound.25 Briefly, [U-15N5]-dG (1.5 mg, 5.5 μmol) was dried three times by evaporation in anhydrous pyridine and suspended in 0.5 mL of anhydrous pyridine, which was cooled in an ice bath under argon atmosphere. To the solution was added trifluroacetic anhydride (12 μL, 83 μmol). After a 40-min incubation, 14 mg of NaSH in 250 μL of anhydrous DMF was added. After another 24 hrs, the reaction mixture was poured into a 1-mL vigorously stirred solution of 0.16 M ammonium bicarbonate. The solution was then concentrated to dryness. The residue was triturated with methanol and filtered. The filtrate was concentrated to dryness again and the residue was triturated with 0.1 M TEAA and filtered. The desired [U-15N5]-6-thio-2′-deoxyguanosine ([U-15N5]-SdG) was purified by HPLC on a Beckman system with pump module 125 and a UV detector (module 126). A 4.6×250 mm Apollo C18 column (5 μm in particle size and 300 Å in pore size, Grace Inc., Deerfield, IL) was used and the wavelength of the UV detector was set at 340 nm. A solution of 10 mM ammonium formate (pH 6.3, solution A) and a mixture of 10 mM ammonium formate and acetonitrile (70:30, v/v, solution B) were employed as mobile phases. The flow rate was 0.80 mL/min, and a gradient of 5 min 0-10% B, 40 min 10-35% B and 1 min 35-100% B was used. After purification, the identity of [U-15N5]-SdG was confirmed by both high-resolution ESI-QTOF MS and ion trap LC-MS/MS analyses (see Results).
Synthesis and Characterization of D3-S6mdG
This compound was synthesized at microscale following previously reported procedures for the synthesis of the corresponding unlabeled compound.26 Briefly, SdG (1.8 mg, 6.3 μmol) was dissolved in 250 mM phosphate buffer (1 mL, pH 8.5), to which solution was added 2 μL CD3I (30 μmol). The solution was stirred overnight, and the reaction mixture was concentrated to dryness. The desired D3-S6mdG was purified by using the above-described HPLC method with the wavelength of the UV detector being set at 312 nm. The structure of D3-S6mdG was confirmed by both high-resolution ESI-QTOF MS and ion trap LC-MS/MS analyses (see Results).
Cell Culture, 6-Thioguanine Treatment, and DNA Isolation
Cells were cultured in ATCC-recommended medium at 37°C under 5% CO2 atmosphere. All media were supplemented with 10% fetal bovine serum, 100 I.U./mL penicillin, and 100 μg/mL streptomycin. After growing to 70% confluence (at a density of 106 cells/mL), cells were cultured in fresh medium containing 0, 1 and 3 μM of 6-thioguanine and incubated for 24 hrs.
After treatment, the adherent HCT-116 cells (~ 2×107 cells) were first detached by trypsin-EDTA treatment. The leukemic cells (~ 2×107 cells) and the detached HCT-116 cells were harvested by centrifugation to remove the medium. The cell pellets were subsequently washed twice with 1×PBS and resuspended in a lysis buffer containing 10 mM Tris-HCl (pH 8.0), 0.1 M EDTA, and 0.5% SDS. The cell lysates were then treated with RNase A (20 μg/mL) at 37°C for 1 hr and subsequently with proteinase K (100 μg/mL) at 50°C for 3 hrs. Genomic DNA was isolated by extraction with phenol/chloroform/isoamyl alcohol (25:24:1, v/v) and desalted by ethanol precipitation. The DNA pellet was redissolved in water and its concentration was measured by UV absorbance at 260 nm.
Enzymatic Digestion
For the enzymatic digestion of DNA, nuclease P1 (4 units) was added to a mixture containing 100 μg cellular DNA, 100 fmol D3-S6mdG, 2 pmol [U-15N5]-SdG, 30 mM sodium acetate (pH 5.5) and 1 mM zinc acetate, and the mixture was incubated at 37 °C for 4 hrs. To the digestion mixture was then added 30 units of alkaline phosphatase in a 50 mM Tris-HCl buffer (pH 8.6), the digestion was continued at 37°C for 2.5 hrs, and the enzymes were subsequently removed by chloroform extraction. The aqueous DNA layer was dried and the dried residues were reconstituted in distilled water. The amount of nucleosides in the mixture was quantified by UV absorbance measurements.
HPLC Enrichment
HPLC removal of unmodified nucleosides in the digestion mixture of cellular DNA was carried out by using a 4.6×250 mm Apollo C18 column (5 μm in particle size and 300 Å in pore size, Grace Inc., Deerfield, IL). An Agilent 1100 HPLC system with a capillary pump (Agilent Technologies) and a UV detector was used, and a Peak Simple Chromatography Data System was employed for data collection (SRI Instruments Inc., Las Vegas, NV, USA). A solution of 10 mM ammonium formate (pH 4.0, solution A) and a mixture of 10 mM ammonium formate and acetonitrile (70:30, v/v, pH 4.0, solution B) were used as mobile phases. A gradient of 5 min 0-12% B, 45 min 10-35% B, 1 min 35-100% B and 15 min 100% B was employed and the flow rate was 0.80 mL/min. The fractions containing S6mdG and SdG were collected separately, dried in a Speedvac, reconstituted in distilled water and subjected to LC-MS/MS analysis. Due to high level of SdG incorporation in Jurkat T, CCRF-CEM, HL-60 and K-562 cell lines, SdG levels in DNA of these cells were directly measured by LC-MS/MS with 3 nmol of digested nucleoside mixture doped with 5 pmol [U-15N5]-SdG.
Mass Spectrometry
Electrospray ionization-mass spectrometry (ESI-MS) and tandem MS (MS/MS) experiments were carried out on an LCQ Deca XP ion-trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA). A mixture of acetonitrile and water (50:50, v/v) was used as solvent for electrospray. The spray voltage was 3.0 kV, and the temperature for the ion transport tube was maintained at 275°C. High-resolution mass spectra (HRMS) were acquired on an Agilent 6510 Q-TOF LC/MS instrument equipped with an ESI source and an Agilent HPLC-Chip Cube MS interface.
LC-MS/MS
Quantitative analysis of S6mdG and SdG in the above DNA hydrolysates was performed by online capillary HPLC-ESI-MS/MS using an Agilent 1200 capillary HPLC pump interfaced with an LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA). A 0.5×150 mm Zorbax SB-C18 column (5 μm in particle size, Agilent Technologies) was used for the separation of the DNA hydrolysis mixture and the flow rate was 6.0 μL/min. A gradient of 0-20% methanol (in 5-min) followed by 20-80% methanol (in 40-min) in water with 0.1% formic acid was employed for the detection of SdG and S6mdG. The effluent from the LC column was directed to the LTQ mass spectrometer, which was set up for MS/MS analysis, where the fragmentation of the protonated ions of unlabeled and labeled SdG and S6mdG was monitored.
Cell Viability Assay
Jurkat T, CCRF-CEM, HL-60, K-562 and HCT-116 cells were separately seeded in 6-well plates at a density of ~ 3 × 105 cells/mL. To the cultured cells was added 6-thioguanine until its final concentration was 1.0 or 3.0 μM. After 24 or 48 hrs of treatment, cells were stained with trypan blue and counted on a hemocytometer to measure cell viability.
Results and Discussion
6-MP and SG are widely used for treating acute lymphoblastic leukemia. It is generally believed that DNA SG is the ultimate metabolite for thiopurines to exert their cytotoxic effects.2 However, the cytotoxic mechanism through which DNA SG results in cell death remains elusive. Based on the observations that DNA SG can be spontaneously methylated by S-AdoMet to give S6mG and the resulting S6mG:T mispair formed from DNA replication may trigger the mismatch repair pathway, it was hypothesized that SG may confer its cytotoxic effect via futile cycles of repair synthesis triggered by DNA S6mG.16
Our recent study showed that SdG is mutagenic in E. coli cells, it can give rise to ~10% G→A mutation, whereas S6mdG is highly mutagenic, with G→A mutation occurring at a frequency of 94%.17 This result led us to suggest that SG might trigger mismatch repair pathway without being methylated to S6mG. As an important step toward understanding the relative contributions of these two pathways, we developed a sensitive LC-MS/MS coupled with stable isotope dilution method and quantified the levels of SdG and S6mdG in genomic DNA of leukemic cells treated with SG.
Synthesis and Characterization of Isotope-labeled S6mdG and SdG
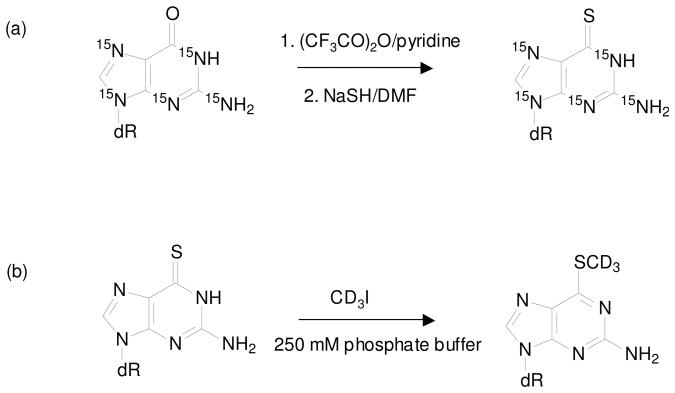
Uniformly 15N-labeled dG was reacted with trifluoroacetic anhydride in the presence of pyridine to form the putative 6-pyridyl intermediate, which was converted to [U-15N5]-SdG upon treatment of NaSH in DMF (Scheme 2a). The desired product was purified from the mixture by HPLC. High-resolution ESI MS gives m/z 289.0653 for the [M+H]+ ion of [U-15N5]- SdG, which is consistent with the calculated m/z of 289.0664 with a deviation of 3.8 ppm. Collision-induced dissociation of the ion of m/z 289.0 led to the formation of the fragment ion of m/z 173.1, which is attributed to the elimination of a 2-deoxyribose moiety (Figure S1b). Further fragmentation of the ion of m/z 173.1 gave product ions of m/z 155.1, 139.1 and 129.0, which were originated from the losses of a 15NH3, an H2S, and a C(15NH)2, respectively, from the uniformly 15N-labeled nucleobase portion (Figure S1c).
Scheme 2.
Synthesis of stable isotope-labeled 6-thio-2′-deoxyguanosine (a) and S6-methylthio-2′-deoxyguanosine (b).
D3-S6mdG was prepared from the reaction of SdG with CD3I in a phosphate buffer (Scheme 2b) and purified from the reaction mixture by HPLC. High-resolution ESI MS gives m/z 301.1145 for the [M+H]+ ion of D3-S6mdG, which is consistent with the calculated m/z of 301.1157 with a deviation of 3.9 ppm. MS/MS of the ion of m/z 300.9 (Figure S2a) shows the predominant fragment ion of m/z 185.2, which again arises from the elimination of a 2-deoxyribose moiety (Figure S2b).
Analytical Strategy for the Quantification ofSdG and S6mdG by LC-MS/MS
SG has been shown to be more efficacious than 6-MP in human ALL cell lines and in lymphoblasts from childhood ALL both in preclinical28 and clinical studies.27 In addition, SG has more direct intracellular activation pathway to form SG nucleotide than 6-MP (Scheme 1); therefore, we decided to use SG to treat human ALL cells. Viewing that the peak concentration of SG in plasma was in the range of 0.2-1.2 μM after oral SG administration (60 mg/m2) and 1.3-4.1 μM after continuous intravenous infusion (20 mg/m2/h),27 we treated human ALL cells with 1 and 3 μM of SG for 24 hrs and isolated the genomic DNA from the treated and control untreated cells. To quantify SdG and S6mdG in genomic DNA isolated from SG-treated human cancer cell lines, we first digested the DNA with nuclease P1 to yield nucleoside 5′-monophosphates, which were subsequently dephosphorylated with alkaline phosphatase. Due to the trace amount of S6mdG present in DNA, the nucleoside mixture was subsequently separated by offline HPLC, and the fractions containing these two lesions were collected and dried. After being reconstituted in H2O, SdG and S6mdG were analyzed by LC-ESI-MS/MS. Owing to the better sensitivity afforded by the positive- than the negative-ion mode, we quantified SdG and S6mdG by LC-MS/MS in the positive-ion mode, where we added 0.1% formic acid to the mobile phases to facilitate the protonation of the analytes. The quantitation of these two lesions was based on the peak area ratio between the analyte and its isotope-labeled counterpart found in the selected-ion chromatograms (SICs) and the constructed calibration curves (Figure S3).
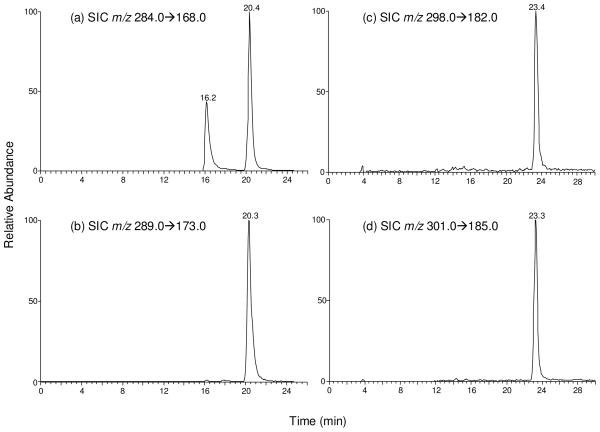
We monitored the m/z 284→168 and m/z 289→173 transitions for SdG and its uniformly 15N-labeled counterpart, respectively. The identity of the component eluting at 20.4 min in the SIC of Figure 1a was determined to be SdG based on the same retention time (Figure 1b) and similar tandem mass spectrum as those observed for [U-15N5]- SdG (Figure 2a&b). The component eluting at 16.2 min in Figure 1a was identified as 8-oxodG. In this context, 8-oxodG and SdG bear the same nominal molecular weight and these two nucleosides undergo the same fragmentation in MS/MS, i.e., with the loss of 2-deoxyribose being the dominant fragmentation pathway. However, 8-oxodG and SdG can be well resolved by capillary HPLC under the conditions we employed. Furthermore, fragmentations of the nucleobase moieties of 8-oxodG and SdG gave distinctive MS/MS/MS; the spectrum of SdG displays the loss of an NH3, C(NH)2 and H2S (Figure S1c depicts the MS/MS/MS for the uniformly 15N-labeled SdG), whereas that of 8-oxodG shows the predominant losses of H2O (m/z 150.1) and NH3 (m/z 151.0) molecules (Figure S4b).
Figure 1.
Selected-ion chromatograms (SICs) for monitoring the m/z 284→168 (a) and m/z 289→173 (b) transitions for SdG and [U-15N5]- SdG; m/z 298→182 (c) and m/z 301→185 (d) transitions for S6-mdG and D3-S6mdG, respectively, in CCRF-CEM cells treated with 3 μM 6-thioguanine for 24 hrs.
Figure 2.
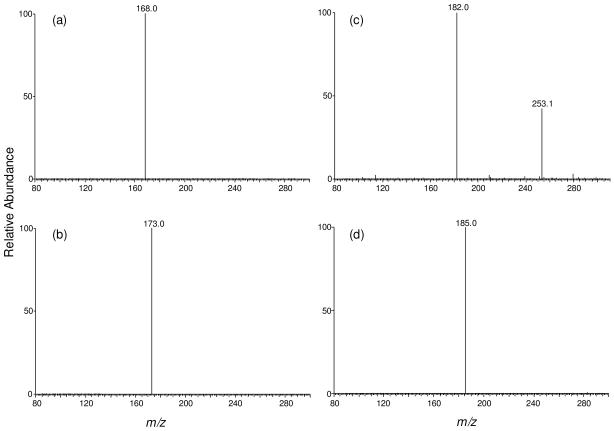
Product-ion spectra of the ions of m/z 284 (a), m/z 289 (b), m/z 298 (c), and m/z 301 (d). Panels (a) and (c) are for unlabeled, and (b) and (d) are for the [U-15N5]- SdG and D3-S6mdG, respectively. The sample was the nucleoside mixture of DNA extracted from CCRF-CEM cells treated with 3 μM 6-thioguanine for 24 hrs.
S6mdG and D3-S6mdG were monitored by the m/z 298→182 and m/z 301→185 transitions, respectively. The fraction eluting at 23.4 min in the SIC shown in Figure 1c was identified as S6mdG based on the same retention time (Figure 1d) and similar MS/MS as those of D3-S6mdG (MS/MS are shown in Figure 2c&d). In this context, aside from the most abundant ion at m/z 182.0, there was another ion of m/z 253.1 present in Figure 2c. Viewing that the corresponding fragment could not be found in the MS/MS of the internal standard (Figure 2d), we speculate that the latter fragment ion might arise from some isobaric species present in the sample. It is important to note that further cleavage of the ion of m/z 253.1 (i.e., MS3 experiment) did not give rise to the formation of the ion of m/z 182.0. Moreover, MS3 of the ion of m/z 182.0 is identical to the corresponding MS3 of the standard S6mdG (spectra not shown), suggesting that the MS/MS of the isobaric interference did not lead to the formation of the ion of m/z 182.0. Taken together, the presence of the isobaric interference should not affect the accurate the quantification of S6mdG.
Quantification of SdG and S6mdG in Leukemia Cells upon SG Treatment
LC-MS/MS with the isotope dilution method constitutes a reliable quantification method. In this study, we added isotope-labeled [U-15N5]-SdG and D3-S6mdG to the samples prior to the enzymatic hydrolysis of genomic DNA, which corrected for the potential analyte loss during various stages of sample preparation process. Limits of quantification (LOQ) for our LC-MS/MS method were assessed prior to the analysis of cellular DNA samples. It turned out that, when the pure standards were analyzed, the LOQs at an S/N of 10 were found to be 6.8 and 0.25 fmol for SdG and S6mdG, respectively. Due to the ion suppression induced by the co-eluting unmodified nucleosides and/or other species, the corresponding LOQs for SdG and S6mdG in nucleoside mixtures of genomic DNA were determined to be 62 and 0.6 fmol, respectively.
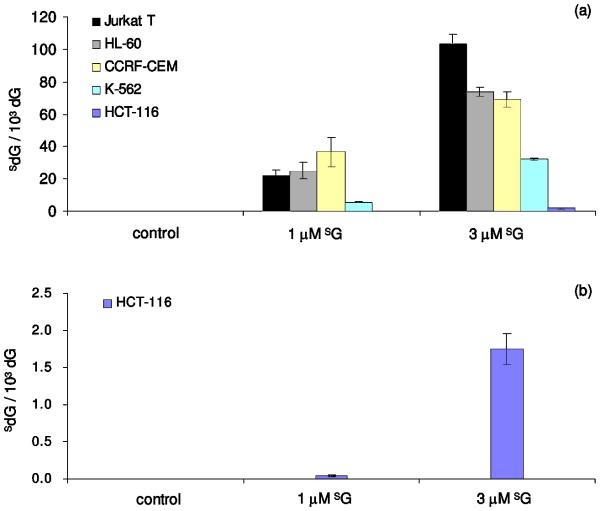
We quantified the levels of SdG and S6mdG in cellular DNA of five different cancer cell lines that were cultured for 24 hrs in a medium containing 1 or 3 μM SG. The quantification results revealed a dose-dependent incorporation of SdG in all cell lines and Jurkat T cells have the highest level of SdG incorporation in DNA; upon treatment with 1 and 3 μM of SG, genomic DNA isolated from these cells has approximately 2% and 10% of dG being replaced with SdG, respectively (Figure 3a). HL-60 and CCRF-CEM cells exhibit similar levels of SdG incorporation; a 24-h SG treatment induced 2-4% and 6-7% of dG replacement upon treatment with 1 and 3 μM of the drug, respectively. Among all studied leukemia cells, K-562 cells exhibit the lowest level of SdG incorporation, whereas less than 1% and 4% replacements were observed when the cells were treated with 1 and 3 μM of SG, respectively. HCT-116, a human colorectal carcinoma cell line, has also been investigated in this study because a previous study showed that SG replaced approximately 0.2% of DNA guanine in HCT-116 cells grown for 24 hrs in a medium containing 1 μM SG.29 We, however, observed that SG substituted approximately 0.004% and 0.2 % of DNA guanine in HCT-116 cells grown for 24 hrs in a medium containing 1 and 3 μM SG (Figure 3b).
Figure 3.
The incorporation of SdG in genomic DNA of human Jurkat T, HL-60, CCRF-CEM, K-562 and HCT-116 cells (a). In panel (b), the scale was enlarged to view better the incorporation of SdG in DNA of HCT-116 cells.
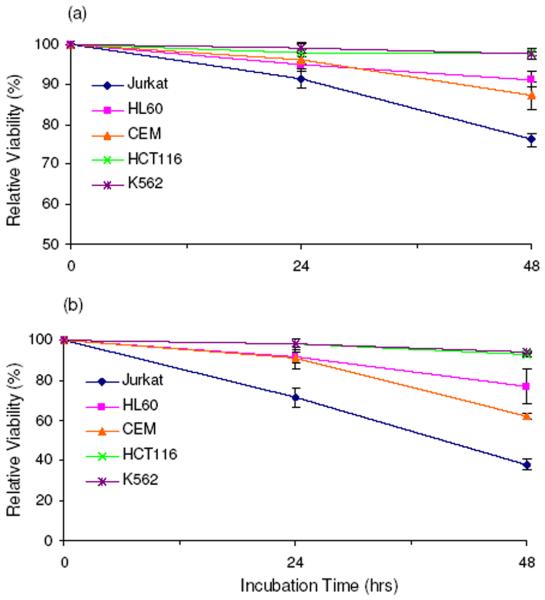
We also examined the cell viability in the above five cell lines upon SG treatment (shown in Figure 5a&b). Consistent with the incorporation rate of SG, the most pronounced cell death was found for Jurkat T cells, i.e., the cell viability dropped by 10% and 30% after a 24-hr treatment with 1 and 3 μM SG, respectively, whereas a 48-hr SG treatment resulted in 25% and 60% cell death. HL-60 and CCRF-CEM cells showed 5% and 9% cell death with the same treatment, but 9% and 30% after 48-hr incubation; in these two cell lines, the level of SdG in genomic DNA was lower than that in Jurkat T cells. Among all studied leukemia cell lines, K-562 cells displayed the maximum resistance toward SG treatment, which is in line with the lowest level of SdG incorporation being observed for this cell line (Figure 3a).
Figure 5.
The relative viabilities of human cells after 24 and 48 hrs of treatment with 1 μM (a) and 3 μM (b) 6-thioguanine compared to untreated cells.
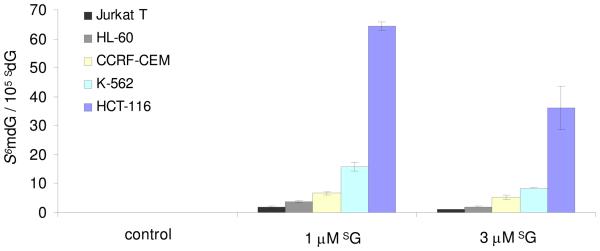
The accurate quantification of SdG and S6mdG in the above cell lines also allowed for the determination of the extent of methylation of SdG in genomic DNA from the SG-treated leukemic cells. Our results revealed that the extent of S6mdG converted from SdG was very low (Figure 4). Less than 2 out of 105 SdG in DNA was converted to S6mdG in Jurkat T cells treated with 3 μM of SG, followed by 4 and 7 per 105 SdG being replaced in HL-60 and CCRF-CEM cells, and then 16 per 105 SdG in K-562 cells. Despite the low level of SdG incorporation in HCT-116 cells, DNA isolated from this cell line was found to bear the highest level of conversion from SdG to S6mdG, i.e., 64 and 36 out of 105 SdG were substituted when the cells were treated with 1 and 3 μM SG, respectively. Considering that, in E. coli cells, SdG and S6mdG can induce G→A mutation at frequencies of 10% and 94%, respectively, and several out of 105 SdG in DNA are converted to S6mdG, we reason that the SG:T mispairs may assume the major role in triggering the MMR pathway, while the S6mG:T mispairs may be partially involved.
Figure 4.
The formation of S6mdG in genomic DNA of human Jurkat T, HL-60, CCRF-CEM, K-562 and HCT-116 cells.
Our method facilitates the accurate detection of SdG in human ALL cells induced by SG treatment from 3 nmol of digested nucleoside mixture, which is generated from ~1 μg of DNA. Furthermore, we achieved a detection limit of 0.7 fmol of S6mdG in 50 μg of digested DNA, which corresponds to 4 lesions per 109 unmodified nucleobases. This method obviates the need of 3H-labeling while maintaining good sensitivity and it also facilitates the monitoring of both SG and its methylated metabolite in DNA. Moreover, LC-MS/MS can provide structure information for the analytes, which allows for the unambiguous identification and accurate quantification of these two modified nucleosides in DNA.
Taken together, we developed a sensitive LC-MS/MS coupled with stable isotope dilution method for the simultaneous quantification of the levels of SdG and S6mdG in leukemic cells treated with SG. Our results suggest that DNA SG, instead of its methylated derivative (S6mG), may take the major role in invoking the post-replicative MMR pathway.
Supplementary Material
Acknowledgement
This work was supported by the National Institutes of Health (R56 CA96906 and R01 DK082779).
Abbreviations
- SG
6-thioguanine
- SdG
6-thio-2′-deoxyguanosine
- dG
2′-deoxyguanosine
- S6mG
S6-methylthioguanine
- S6mdG
S6-methylthio-2′-deoxyguanosine
- MMR
mismatch repair
- ESI-MS
electrospray ionization-mass spectrometry
- MS/MS
tandem MS
- ALL
acute lymphoblastic leukemia
Footnotes
Supporting Information Available
Tandem MS spectra and calibration curves. This material is available free of charge via the Internet at http://pubs.acs.org.
References:
- 1.Pui CH, Evans WE. N. Engl. J. Med. 1998;339:605–615. doi: 10.1056/NEJM199808273390907. [DOI] [PubMed] [Google Scholar]
- 2.Elion GB. Science. 1989;244:41–47. doi: 10.1126/science.2649979. [DOI] [PubMed] [Google Scholar]
- 3.Burchenal JH, Murphy ML, Ellison RR, Sykes MP, Tan TC, Leone LA, Karnofsky DA, Craver LF, Dargeon HW, Rhoads CP. Blood. 1953;8:965–999. [PubMed] [Google Scholar]
- 4.Murray JE, Harrison JH, Dammin GJ, Wilson RE, Merrill JP. N. Engl. J. Med. 1963;268:1315. doi: 10.1056/NEJM196306132682401. &. [DOI] [PubMed] [Google Scholar]
- 5.McLeod HL, Krynetski EY, Relling MV, Evans WE. Leukemia. 2000;14:567–572. doi: 10.1038/sj.leu.2401723. [DOI] [PubMed] [Google Scholar]
- 6.Schutz E, Gummert J, Mohr F, Oellerich M. Lancet. 1993;341:436–436. doi: 10.1016/0140-6736(93)93028-y. [DOI] [PubMed] [Google Scholar]
- 7.Evans WE, Horner M, Chu YQ, Kalwinsky D, Roberts WM. J. Pediatr. 1991;119:985–989. doi: 10.1016/s0022-3476(05)83063-x. [DOI] [PubMed] [Google Scholar]
- 8.McLeod HL, Miller DR, Evans WE. Lancet. 1993;341:1151–1151. doi: 10.1016/0140-6736(93)93168-z. [DOI] [PubMed] [Google Scholar]
- 9.Krynetski EY, Tai HL, Yates CR, Fessing MY, Loennechen T, Schuetz JD, Relling MV, Evans WE. Pharmacogenetics. 1996;6:279–290. doi: 10.1097/00008571-199608000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Tiede I, Fritz G, Strand S, Poppe D, Dvorsky R, Strand D, Lehr HA, Wirtz S, Becker C, Atreya R, Mudter J, Hildner K, Bartsch B, Holtmann M, Blumberg R, Walczak H, Iven H, Galle PR, Ahmadian MR, Neurath MF. J. Clin. Invest. 2003;111:1133–1145. doi: 10.1172/JCI16432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang HX, Wang YS. Biochemistry. 2009;48:2290–2299. doi: 10.1021/bi801467z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christie NT, Drake S, Meyn RE, Nelson JA. Cancer Res. 1984;44:3665–3671. [PubMed] [Google Scholar]
- 13.Pan BF, Nelson JA. Biochem. Pharmacol. 1990;40:1063–1069. doi: 10.1016/0006-2952(90)90494-6. [DOI] [PubMed] [Google Scholar]
- 14.Maybaum J, Mandel HG. Exp. Cell Res. 1981;135:465–468. doi: 10.1016/0014-4827(81)90192-0. [DOI] [PubMed] [Google Scholar]
- 15.Bodell WJ. Mutagenesis. 1991;6:175–177. doi: 10.1093/mutage/6.3.175. [DOI] [PubMed] [Google Scholar]
- 16.Swann PF, Waters TR, Moulton DC, Xu YZ, Zheng QG, Edwards M, Mace R. Science. 1996;273:1109–1111. doi: 10.1126/science.273.5278.1109. [DOI] [PubMed] [Google Scholar]
- 17.Yuan BF, Wang YS. J. Biol. Chem. 2008;283:23665–23670. doi: 10.1074/jbc.M804047200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffin S, Branch P, Xu YZ, Karran P. Biochemistry. 1994;33:4787–4793. doi: 10.1021/bi00182a006. [DOI] [PubMed] [Google Scholar]
- 19.Waters TR, Swann PF. Biochemistry. 1997;36:2501–2506. doi: 10.1021/bi9621573. [DOI] [PubMed] [Google Scholar]
- 20.Dervieux T, Chu YQ, Su Y, Pui CH, Evans WE, Relling MV. Clin. Chem. 2002;48:61–68. [PubMed] [Google Scholar]
- 21.Keuzenkampjansen CW, Deabreu RA, Bokkerink JPM, Trijbels JMF. J. Chromatogr. B. 1995;672:53–61. [Google Scholar]
- 22.Kroplin T, Weyer N, Gutsche S, Iven H. European J. Clin. Pharmacol. 1998;54:265–271. doi: 10.1007/s002280050457. [DOI] [PubMed] [Google Scholar]
- 23.Rowland K, Lennard L, Lilleyman JS. J. Chromatogr. B. 1998;705:29–37. doi: 10.1016/s0378-4347(97)00495-7. [DOI] [PubMed] [Google Scholar]
- 24.Dervieux T, Meyer G, Barham R, Matsutani M, Barry M, Boulieu R, Neri B, Seidman E. Clin. Chem. 2005;51:2074–2084. doi: 10.1373/clinchem.2005.050831. [DOI] [PubMed] [Google Scholar]
- 25.Kung PP, Jones RA. Tetrahedron Lett. 1991;32:3919–3922. [Google Scholar]
- 26.Xu YZ. Tetrahedron. 1996;52:10737–10750. [Google Scholar]
- 27.Lowe ES, Kitchen BJ, Erdmann G, Stork LC, Bostrom BC, Hutchinson R, Holcenberg J, Reaman GH, Woods W, Franklin J, Widemann BC, Balis FM, Murphy RF, Adamson PC. Cancer Chemother. Pharmacol. 2001;47:199–205. doi: 10.1007/s002800000229. [DOI] [PubMed] [Google Scholar]
- 28.Adamson PC, Poplack DG, Balis FM. Leuk. Res. 1994;18:805–810. doi: 10.1016/0145-2126(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 29.O'Donovan P, Perrett CM, Zhang XH, Montaner B, Xu YZ, Harwood CA, McGregor JM, Walker SL, Hanaoka F, Karran P. Science. 2005;309:1871–1874. doi: 10.1126/science.1114233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.