Abstract
Established techniques for global gene expression profiling, such as microarrays, face fundamental sensitivity constraints. Due to greatly increasing interest in examining minute samples from micro-dissected tissues, including single cells, unorthodox approaches, including molecular nanotechnologies, are being explored in this application. Here, we examine the use of single molecule, ordered restriction mapping, combined with AFM, to measure gene transcription levels from very low abundance samples. We frame the problem mathematically, using coding theory, and present an analysis of the critical error sources that may serve as a guide to designing future studies. We follow with experiments detailing the construction of high density, single molecule, ordered restriction maps from plasmids and from cDNA molecules, using two different enzymes, a result not previously reported. We discuss these results in the context of our calculations.
1. Introduction
1.1. Quantifying gene expression from very small samples
Global gene expression analysis is the quantification of gene transcription, across all genes, from a cell or tissue at the time of sampling [1, 2]. Detection of differences and modulation in global expression patterns has yielded a deeper appreciation for the interconnected circuitries of normal and diseased tissues, and is now commonplace in biomedical research and drug discovery; global gene expression profiling is also beginning to be used in the clinical setting, to aid in disease prediction, diagnostics and treatment [3–7]. Importantly, there is increasing demand for expression profiling of small samples, as large amounts of material can be difficult, if not impossible, to obtain in clinical and experimental settings [8–19]. Recent methodological advances make possible the global expression profiling of minute samples from micro-dissected tissues, including single cells, thereby avoiding the confounding biological effects of tissue heterogeneity. Fine needle aspirates and fine needle core biopsies offer practical clinical sampling procedures of limited material. Technologies that facilitate the isolation of individual, specialized cells, such as by laser capture micro-dissection, yield homogenous material for analysis [3, 12, 18, 19].
Each cell contains approximately 300 000 mRNA molecules, representing more than 3 × 104 different species, while each low abundance species may be present in only a few copies per cell [18, 20, 21]. Genes transcribed at low levels, such as regulatory proteins, exert large biological effects from small changes in expression level [13, 18, 22]. Considerable work with enzymatic amplification, including PCR-based [23–33], multiple displacement amplification-based (MDA) [24, 34–55], and RNA polymerase-based [23, 56–63] protocols, has enabled the use of hybridization microarrays and sequence tag methods to characterize low abundance mRNA samples [64–72]. This includes several recent reports of message profiling of single cells [73–82]. Unfortunately, serious methodological drawbacks remain.
1.2. Critical limitations of enzymatic amplification
Recent studies of mRNA amplification protocols have found a significant decrease in correlation coefficients between low copy number species, pre- and post-amplification. Single cell transcript profiling studies have found that only large magnitude changes in expression can be quantified for moderate- and low-abundance transcripts [24, 27, 28, 30, 35, 41, 46, 57, 62, 63, 67, 83–88]. In several papers, Nygarrd et al [85, 88] have argued strongly that fundamental, stochastic effects prevent reliable enzymatic amplification of all species from minute samples; they conclude that high and medium abundance species can be quantitatively amplified, but low copy number species will always be amplified unevenly. This may prove to be an enormous limitation, as the majority of transcripts in a cell can be ‘low abundance’, defined as having 1–5 copies per cell.
1.3. Non-amplified, single molecule technologies
Non-amplified single molecule approaches provide the most direct solution to the above limitations, as they offer theoretically unlimited, unbiased sensitivity. They also require inherently fewer processing steps, reagent use is limited, and with parallelization, reasonably low amortized instrument costs are incurred [89–126]. Unfortunately, non-amplified, single molecule sequencing methods, such as nanopores or in situ synthesis-based chemistries, also face extremely challenging signal detection and sequencing chemistry hurdles, and despite extensive ongoing research, remain in the earliest stages of development; the same is true for single molecule high density oligonucleotide probe hybridization approaches [127–154].
In contrast, single molecule, high density ordered restriction mapping presents an interesting, but largely unexplored alternative in this application. Type II restriction enzymes, the most common variety, are unparalleled in their robustness and simplicity as detection systems for specific, short DNA sequences (usually less than eight bp). They bind and cleave their recognition sequences up to 106 times more specifically than similar, non-cognate sequences [155]. Over 3500 different Type II restriction enzymes exist and hundreds are available commercially. Single molecule ordered restriction mapping has been used extensively in small sample genome mapping studies using optical detection [156–165], and in a few cases using AFM [166–168]. For a combination of reasons, which we discuss in detail below, none of these studies demonstrated high resolution mapping of short DNA molecules (<2 kb) as would be required for identifying individual message transcripts. The current study examines high density, ordered restriction mapping using AFM as a method for gene expression profiling. We begin by defining the problem mathematically, using coding theory, to determine the relationship between molecule size, AFM sizing accuracy and site labelling (binding or cleavage) efficiency. We detail the construction of highly accurate, dense single molecule ordered restriction maps of actual cDNA molecules and short plasmids, using AFM and the in situ cleavage approach, a result not previously reported in the literature.
2. Materials and methods
2.1. AFM
AFM images were acquired with both a Digital Instruments Bioscope AFM and a Dimension 3000 AFM, in tapping mode, using manufacturer-supplied TESP diving board cantilevers. Imaging was conducted at 22°C and ∼30% relative humidity. DNA was processed and imaged on freshly cleaved mica derivatized with 3-aminopropyl triethoxysilane to provide a positive charge for DNA retention, as previously described [162]. Nanoscope image processing software was used to flatten and plane-fit all AFM images and NIH Image was used to manually measure DNA backbone length profiles. We have found that the DNA molecule itself serves as a very good reference for scan quality and tip condition. The true width of DNA is ∼2 nm and generally appears 8–15 nm wide in our images. Tip quality and scan parameters were assessed using the apparent DNA width.
2.2. DNA samples and sizing
DNA sizing studies used six linear fragments from pEYFP-C1 (Clontech), prepared by cleavage in solution, deposited as described above, skipping the surface cleavage and washing steps. The fragment sizes (nm/bp) were 191/579, 230/760, 447/1355, 589/1785, 788/2388, and 1561/4731. A constant 0.33 nm/bp derived from the calculated pitch of B-DNA [169] was used as a nm-to-bp conversion factor. A truncated splice variant of CD44 in the pOTB7 plasmid vector was obtained from ATCC. The CD44 plus pOTB7 sample was produced by double cleavage with XhoI and EcoRI to release the cDNA insert from the pOTB7 vector. The CD44v cDNA sequence corresponds to Genbank accession number BC052287.1. Stretching DNA on surfaces has been researched extensively [170–181]. We used fluid flow to stretch cDNAs, a standard technique. Orientation and spacing of cDNAs on the substrate could be controlled by the direction of fluid flow and sample concentration during application, as observed elsewhere [157, 162, 163, 179].
2.3. DNA cleavage and mapping
Linear DNA molecules were elongated and deposited onto derivatized mica surfaces using capillary fluid flow as described previously [162]. Surface bound molecules were exposed to aqueous salt buffer containing enzyme RsaI or PstI for 15–30 min at room temperature. Processed samples were washed with ultra pure water and dried under a stream of nitrogen gas. AFM images were taken from dried samples directly.
Breaks are scored simply from the local topography of the DNA backbone. Double-stranded DNA consistently appears between 0.5 and 1.0 nm tall and 8 and 15 nm wide in our AFM images. The height contrast between the backbone and the surrounding surface is more than sufficient to identify breaks via local threshold, i.e. the breaks appear to have height equal to the surrounding surface. Molecules with gaps larger than 100 nm are rejected. It is very unlikely that two different molecules will appear co-aligned on the surface with their ends 100 nm or less apart. However, to virtually eliminate such molecules from consideration, we only include a molecule if its fragments sum up to the known length of molecules in the sample. In a ‘real world’ sample one can easily label the ends of a molecule with a moiety detectable via AFM to disambiguate closely spaced ends. We make no attempt to automate gap finding or otherwise use more complicated criteria. This is an appropriate subject for further study, but our present criteria are sufficient given the scope of this work.
Both pOTB7 and CD44 are relatively short (594 nm/1800 bp) and loss or displacement of cleaved fragments during sample processing reduces the yield of measurable molecules. Molecules were deemed measurable if the ends were distinct, they contained one clear break, the fragments summed to full length, and the molecule was sufficiently elongated to manually follow the backbone contour. The cleavage rate (% cleavage/total sites) varied from image to image, generally with a range of 30–40%, on average. The rate of false positives (false cuts) is largely a function of image quality, sample age, and similar variables. Other in situ restriction mapping studies suggest that false cuts are more likely to be non-specific breaks than the enzyme cutting in the wrong place [162, 163].
2.4. Mathematical analysis
We calculated the probability of uniquely distinguishing cDNA molecules present in a sample containing many similar species using AFM-determined ordered restriction maps. In this analysis, we treat each map as a unique ‘molecular signature.’ The first step in determining this probability is to calculate the Hamming distance between molecular signatures, HamDist, assuming a total number of ‘good signatures’, S. Each signature is randomly selected from the set of all possible binary vectors, with a probability π. The computation of this probability proceeds as follows: start with a selected signature f0 from the set S, and compute all the possible signatures whose Hamming distances from f0 range between 1 and HamDist; there are:
| (1) |
such signatures, and with high probability, they do not contain even a single signature from the set S (probability > (1 − 10−12) > (1 − π)vol). We compute the uniqueness of the identification probability, given a fixed sizing accuracy, α, enzyme recognition site frequency, pc, and cleavage rate, pd: we compute this probability as follows:
| (2) |
That is, we sum the probabilities that starting with a signature with (a + b) unit bits, exactly b unit bits are lost from the mapped signature as a consequence of incomplete cleavage. These calculations were performed with Mathematica and the code is available upon request.
3. Results and discussion
3.1. Mathematical analysis
We examined the case of uniquely determining the identity of a population of mRNAs obtained from a limited sample, such as a single cell, using single molecule ordered restriction mapping. Our analysis takes into account molecule size, and the two primary sources of error: inaccuracy in locating the ordered restriction sites, and missing real sites (false negatives) due to noise, problems with the enzyme, and similar suboptimal situations. Mammalian mRNAs have an approximately log-normal length distribution, with the median length ∼1.5 kb [182–185]. Roughly 80% of the species are within the size range 1–8 kb, while only a few per cent are shorter than 0.5 kb. mRNAs are converted to a double-stranded DNA form, cDNA, so that they become templates for restriction enzymes (a common technique). A single cell contains approximately 300 000 mRNA molecules, representing more than 3 × 104 different species. The median size of the mRNAs and the number of distinct species dictates that whichever restriction enzyme is used, each of the molecules should contain several sites. The most practical choice is to use an enzyme with a 4 bp recognition sequence, called a 4-cutter. The 4-cutter has a recognition sequence every 256 bp on average, so the average cDNA molecule would contain seven or eight sites. The average spacing of sites on a linear molecule would be physically quite small, ∼85 nm, which in a practical sense eliminates far-field optical detection methods. This is discussed further in the next section. The required site density is also one reason that oligonucleotide probes are inferior to restriction enzymes for this application. A short probe (4- or 6 bp), even using modified nucleotides such as PNA, would not bind strongly enough to achieve the required labelling efficiency.
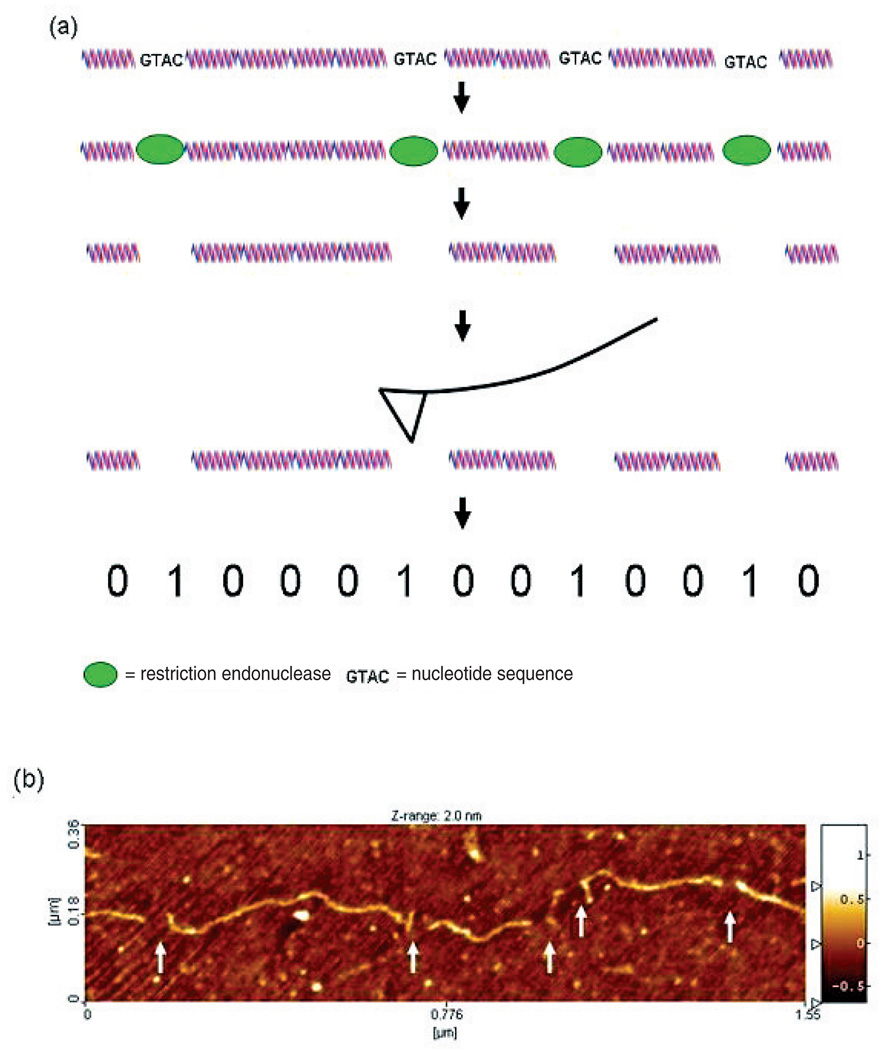
There are generally two experimental schemes for detecting the ordered restriction sites in a single molecule, and both require fixing the molecule to a flat surface for imaging. In the first method, which is the subject of this paper, the molecule is digested while fixed in situ and then the cleavage points are detected by AFM imaging (figure 1). In the second method, the enzyme is made to bind in place but not cut, and the whole molecule, with enzymes in place, is imaged. This amounts to ‘labelling’ in either case, and we will use the term ‘labelling efficiency’ to denote percentage of actual sites detected. Our mathematical analysis applies to both methods, and the relative merits of each approach will be discussed in the next section.
Figure 1.
Experimental scheme for generating single molecule, ordered restriction maps by in situ enzyme cleavage. (a) Enzyme proteins bind oligomeric nucleotide recognition sequences within a surface fixed, duplex DNA molecule. The enzymes cleave the DNA strand at the recognition sites in situ, leaving small gaps (generally <50 nm) visible in the AFM image. Because the molecule remains fixed to the surface during the entire process the order and distance between the cleavage sites is retained. This serves as a partial nucleotide sequence fingerprint that can be used to identify the molecule. (b) An AFM image of a 4700 bp DNA plasmid molecule (pEYFPC1) in situ digested with enzyme RsaI. Five cleavage sites are visible (white arrows) as breaks in the molecule backbone that correspond to the locations of the RsaI recognition sequence 5′GATC.
Several studies have examined the problem of sizing DNA molecules by AFM using backbone contour length as a metric, determined automatically in some cases and by hand in others. In spite of its simplicity, backbone contour length appears remarkably accurate for the molecule sizes tested (∼300–20 000 bp) [167, 168, 186–189]. While the conditions varied among studies, single measurement sizing accuracy (defined here as population CV) better than ±2–5% was reported for most cases for distances larger than 1000 bp. Only three studies presented data for shorter distances and only Fang et al [189] have reported analysis of fragments shorter than 500 bp. For these smaller fragments, the sizing accuracy appears to be between ±7% and ±10%. One reason for the lower accuracy of sizing small fragments is likely to be tip convolution effects, which have not been corrected for in the published studies. Our own sizing experiments, discussed below, agree with the data from Fang et al. Therefore, it is reasonable to assume that 7% accuracy is achievable with AFM, with some optimization. In the case of our analysis, ±7% error on the average 256 bp spacing of 4-cutter restriction sites equates to 36 bp overall measurement accuracy.
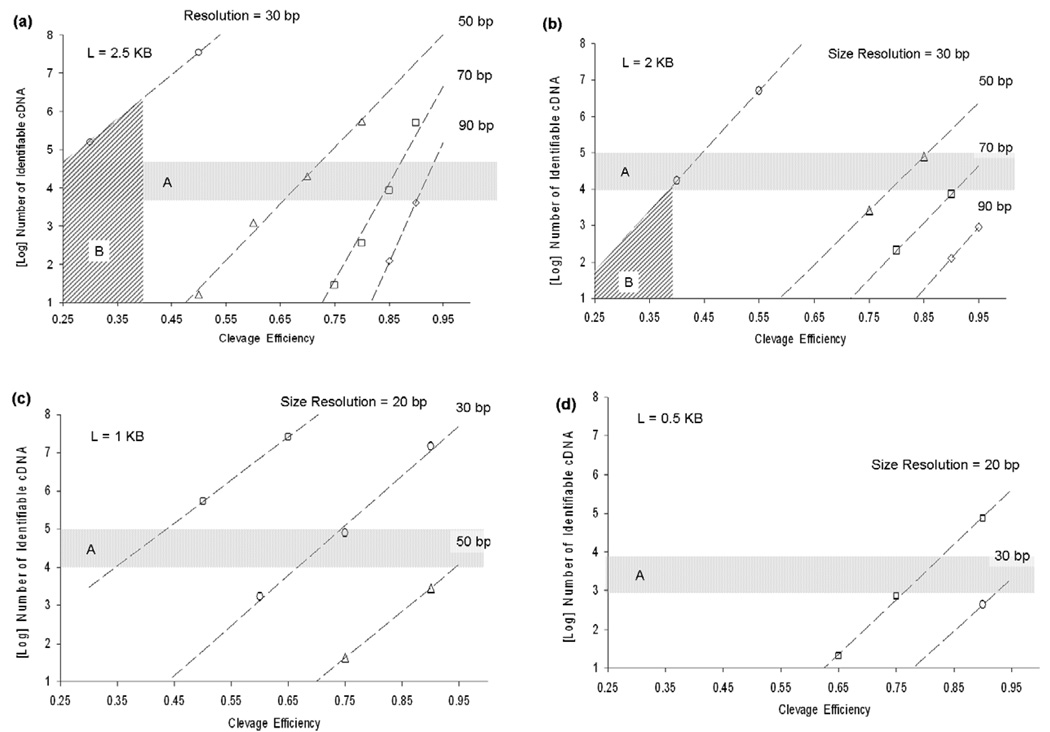
The cDNA molecule is represented as a digital binary signature, (e.g., 00100110), in which each detected site is noted by a non-zero bit, and the distance between two neighbouring detected sites by the number of intervening consecutive zero-bits. In this analysis, the physical length represented by each bit is an integer number, and is determined by the precision with which one can measure the molecular fragments. This distance is a function of the AFM imaging resolution and the conversion factor used to calculate length in bp from molecular dimensions. Here we use the conversion of 0.33 nm/bp, derived from the known bp pitch of B-DNA (see section 2). As the molecule becomes shorter, or sizing resolution worsens, the signatures contain fewer coding bits, and as the digestion rate drops, the corrupted molecular signature deviates from the true signature. In each case, our ability to disambiguate pairs of cDNAs belonging to different species becomes progressively impaired. We assume a 4-cutter enzyme is used, which cleaves at any site in a random cDNA sequence with a probability pc = 4−4 = 1/256; thus a 2 kb molecule/signature would have about eight non-zero bits (cleavages) on average. To illustrate, consider a sample calculation that assumes a resolution α = 10 bp. The 2 kb molecule is then divided up into 200 bins of width 10 bp; therefore, the signatures are of length M = 200 bits. At this value of M, there are an enormous number of possible signatures: 2M ≈ 1.61 × 1060. In actuality, a mammalian cDNA sample would contain a very small subset of these possibilities. Following this logic, we can calculate the probability with which one could uniquely distinguish cDNA molecules present in a sample containing many similar species using AFM-determined restriction maps. Figures 2(a)–(d) shows the number of unambiguously identifiable 2.5, 2, 1 and 0.5 kb cDNAs (>95% probability), for a given bp sizing accuracy, as a function of labelling efficiency. The horizontal band, region A, indicates the approximate number of cDNA species of a specific size that might be expected per cell [182, 183, 185]. For cDNA of length 2.5 kb, as sizing resolution degrades from 50 to 90 bp, difficult-to-achieve labelling efficiency (>80%) is needed to distinguish many species (>104). Conversely, as sizing resolution approaches 30 bp, 105−107 species can be detected, even at low labelling efficiencies (30%–50%).
Figure 2.
Computations of the number of unambiguously identifiable cDNA species (>95% probability) for a given bp sizing accuracy as a function of cleavage efficiency and cDNA size: 2.5 kb, (b) 2 kb, (c) 1 kb and (d) 0.5 kb. For cDNA length 2kb, as sizing resolution degrades from 50 to 90 bp, difficult-to-achieve cleavage efficiency (>80%) is needed to distinguish many species (>104). As sizing resolution approaches 30 bp, 104 to 106 species can be detected, even at very low cleavage rates (30%–50%). Region B indicates the parametric space accessible given the resolution (∼30 bp) and cleavage efficiency demonstrated (∼40%) in our experiments.
3.2. Experimental systems for high density ordered restriction mapping
As discussed above, published reports of single molecule ordered restriction mapping have used two different schemes, in situ digestion with wild type enzymes or stable binding of the enzyme to the restriction site, using modified enzymes or buffer conditions. Using the latter method, Allison et al reported in 1996 [166] and 1997 [167] accurate, AFM-based EcoRI maps of large molecules, plasmids ranging from 3200 to 6800 bp, a cosmid vector (35 000 bp) and the lambda phage genome (48 000 bp). Importantly, they used a special mutant version of EcoRI, obtained from Modrich [190], that binds with reasonably high affinity to its recognition sites, but does not cut. While interesting, their method is an unlikely candidate for cDNA restriction mapping for two reasons: first, EcoRI recognition sites occur too infrequently, on average every 4096 bp; and second, mutagenesis techniques that efficiently separate specific binding from cleavage, if applied to more frequent cutting restriction enzymes, are likely to prove to be very difficult; we refer the reader to several good works on the subject [155, 191, 192].
A more promising approach is to use wild type enzymes but eliminate the Mg++ cofactor that is required for cleavage. This has been demonstrated by Oana et al using fluorescently labelled wild type EcoRI to map restriction sites on single molecules of the lambda genome DNA [165]. Here, the binding efficiency, while not thoroughly characterized, seemed to be too poor (∼10%) for cDNA profiling, based on our above calculations The role of divalent cations in restriction enzymology is currently an active area of research [191–194]. Recent work has shown that divalent cations, while being absolutely required for cleavage activity, also play a critical role in increasing enzyme binding avidity and their ability to distinguish cognate sites from similar sequences [193]. Therefore, it remains unclear whether or not removal or replacement of Mg++ will prove to be a robust strategy for single molecule restriction mapping. Both Oana and Allison observed some non-specific binding, but the level was not well quantified.
The alternative approach of in situ digestion using wild type restriction enzymes has been studied extensively [156–164, 179, 195, 196]. In this method, genomic DNA molecules are elongated and fixed to a glass substrate, followed by in situ digestion and imaging. No sequence specific reporter is used—restriction cleavage sites are photographed directly on fluorescently stained DNA molecules. This technology has proven to be quite robust, enabling Schwartz and coworkers to map 6-cutter restriction sites across whole genomes of several microbes [159, 160]. It requires no amplification and the biochemistry is a single step and highly parallelizable. Only common, unmodified restriction enzymes are required, and most of those actually tested cleave with high efficiency [156, 159, 160, 162–164, 195]. Because the restriction sites are detected optically, this method, as reported, has difficulty resolving sites spaced closer than 2–5 kb apart. Unfortunately, most gene transcripts are shorter than 2 kb in length, and optical techniques can resolve at best one or two restriction sites on such short molecules [195], which is insufficient to discriminate more than a few species. AFM techniques can overcome this limitation; however, risk lies in the uncertainty that existing single molecule ordered restriction mapping methods can be adapted to work with much shorter molecules than used previously (<2 kb versus >30 kb), while accommodating the stringent sample preparation requirements of AFM.
Recent advances in AFM technology suggest that AFM can be used in high throughput applications, under certain circumstances. A recent report actually captured restriction enzyme cleavage of DNA in real time at a rate of 6 frames s−1 [197], though not in conditions compatible with ordered restriction mapping. Most high resolution studies of DNA by AFM use scanning speeds of ∼3–5 µm s−1. Multipurpose AFMs are not constructed for high-speed scanning, since they have to fulfill conflicting requirements, such as large vertical motion range and various modes of operation. AFMs designed for high-speed scanning can image with molecular resolution at speeds up to ∼60–75 µm s−1; some emerging designs may be able to image at a cm s−1 rate [198–216]. High-speed AFMs have emerged over the past 5–7 years, and incorporate more compact scanner designs, smaller and piezo-actuated cantilevers, and improved feedback electronics. Viani et al imaged DNA on mica to high resolution, in liquid, at rates up to 1.7 s/image, and also recorded fast protein binding dynamics [217, 218]. Ando used a high frequency piezo scanner and tip (250 kHz+) to record 100 × 100 pixel images in 80 ms. Tip speeds in their study reached 600 µm s−1 at 2 nm pixel resolution [207, 219]. Manalis has developed high-speed techniques where the cantilever itself is piezo-actuated [220]. Rogers et al used actuated tips to image E. coli and mica steps at speeds up to 75 µm s−1 [221]. Hobbs et al have developed VideoAFM, a design which replaces the cantilever with a high frequency tuning fork (micro-resonator) and circumvents the feedback control speed by using a passive technique. They have recorded 256 × 256 pixel images at rates exceeding 1 cm s−1, though it is unclear if this technique can provide the resolution required to observe restriction cleavage sites in DNA [205, 213].
3.3. Profiling cDNA molecules with AFM
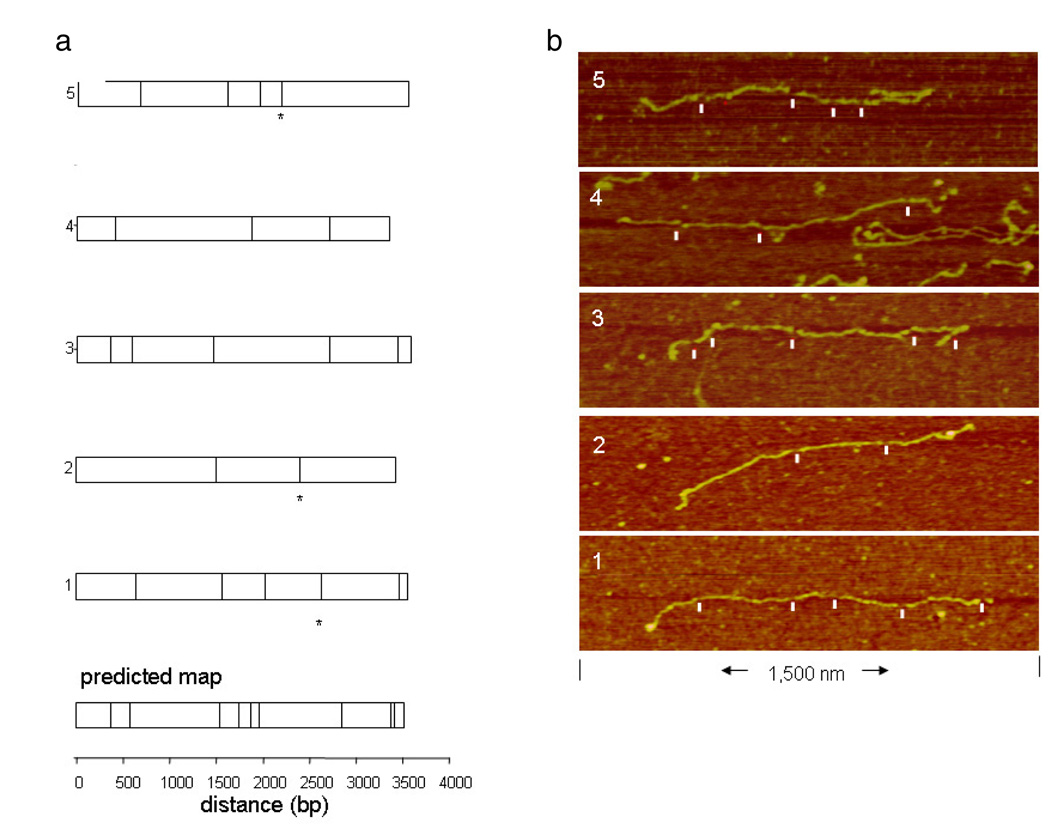
We conducted two series of experiments using the in situ cleavage approach, combined with AFM, to construct fine restriction maps of a short plasmid and actual cDNAs. In the first series of experiments, the recognition sequences of RsaI, a 4-cutter, were mapped to high resolution on a 3.5 kb linearized plasmid (see section 2). This plasmid contained nine 5′GCAT3′ sites, and produced ten fragments when fully digested. The shortest spacing between sites was 34 bp, and the largest 950 bp. Six partially digested molecules were imaged to high resolution, using a square pixel size of 1 nm and a linear scan rate of 1.5–3.0 µm s−1. When the pattern of observed breaks was compared to the predicted RsaI map for the plasmid, five molecules aligned very well (figure 3). The one molecule that did not align appeared to have six spurious breaks, which we will discuss below. To align each molecule, the observed, ordered fragments were compared to the corresponding predicted fragment by size, based on the known sequence. The width of the breaks in the molecules ranged from 8 to 42 nm, with an average value of 17 nm. In two of the molecules, the small end fragments of 101 and 34 bp were missing and may have desorbed. Otherwise, the individual fragments remained stable in situ throughout processing. Of the twenty fragments total, the median sizing error of 2% was quite good. Eighteen total cleavage sites were observed, out of a predicted 45; however, three sites appeared to be non-specific breaks, based on the map alignment. This indicates a cleavage efficiency of 33%. However, the two closely spaced restriction sites, 34 bp apart, produce an 11 nm long fragment that may be easily, and undetectably, desorbed. Correcting for this, the true cleavage efficiency would approach 40%. More non-specific breaks were observed than expected, and we speculate that this is a function of the high fluid shear required to fully elongate these molecules, rather than spurious cleavage by RsaI, which is not known to produce non-specific cleavages when the proper buffer conditions are used. Our previous optical mapping work has shown that ‘false cuts’ are much more likely to be non-specific breaks than the enzyme cutting in the wrong place (so-called ‘star activity’) [162, 163], which can be eliminated by using the correct digestion buffer conditions. Non-specific breaks in the DNA backbone, caused by excessive strain during deposition, chemical degradation, and other strain processes degrade our ability to profile cDNAs. Image artefacts also can produce apparent ‘false cuts’. We have found, both here and in previous optical mapping work [156, 159, 160, 162, 163], that the rate of ‘false cuts’ is largely a function of sample handling, sample age, and image quality.
Figure 3.
(a) Five observed molecules aligned with the predicted RsaI map for the plasmid. To align each molecule, the observed, ordered fragments were compared to the corresponding predicted fragment by size, based on the known sequence. Breaks in the molecules that did not align with the predicted map are indicated with an asterisk. (b) AFM images of the molecules with scored cleavage sites marked by white ticks.
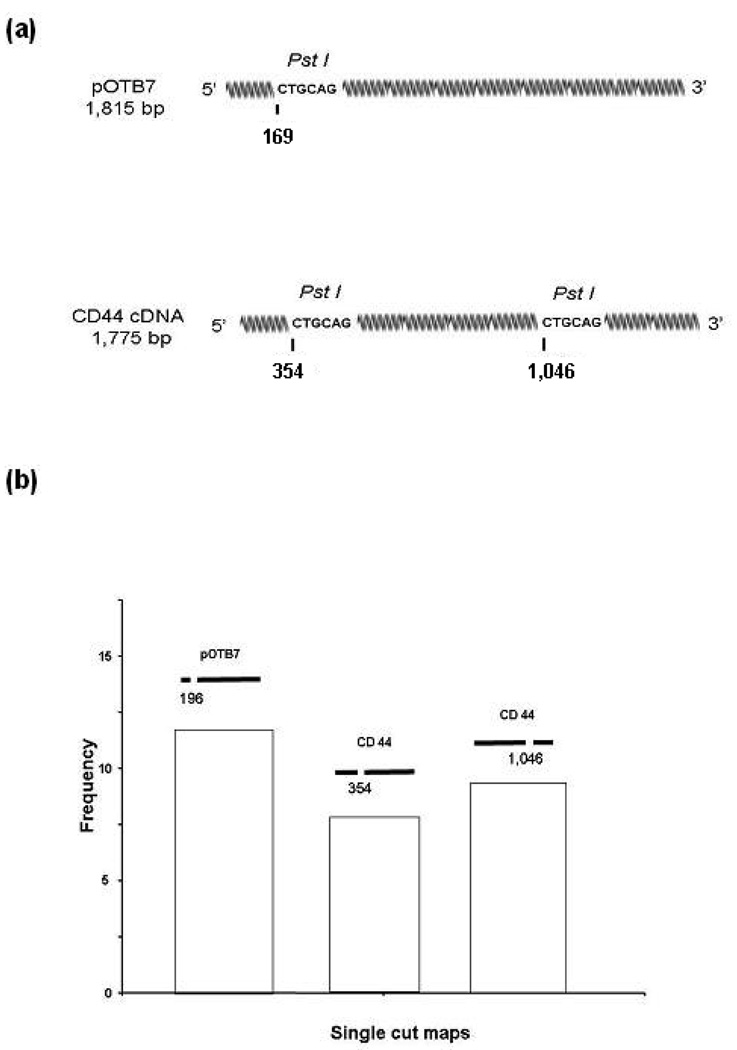
We also used AFM profiling to measure the components of a mixture containing one part DNA from the cancer-related human CD44 gene [222, 223] and one part linearized DNA plasmid pOTB7 with no insert. Both CD44 and pOTB7 molecules are approximately 1800 bp (594 nm) in length. While the enzyme used, PstI, is a 6-cutter with recognition sequence 5′CTGCAG3′, it produces smaller than average fragments in the two test molecules: pOTB7 contains a PstI recognition sequence 354 bp (117 nm) from its 5′ end, and CD44 contains a PstI site 169 bp (65 nm) for its 5′ end, and has an additional PstI site 1046 bp (345 nm) from its 5′ end (figure 4(a)). Molecules cleaved once with PstI rather than twice were chosen for measurement to increase yield. Images were collected using square 3 nm pixels and a linear scan rate of 2–4 µm s−1. The frequency of 1-cut molecules determined from a collection of fifty 1 µm × 1 µm AFM images was determined (figure 4(b)). In the sample, molecules with a PstI site ∼169 bp ±10% from one end, indicative of pOTB7, were approximately as prevalent as those with a site either 354 bp ±10% or 1046 bp ±10% from an end, indicative of CD44 (figure 4(b)). As the sample contained a pure mixture of two species, this distribution of 1-cut molecules is statistically significant and provides the expected frequency from a 1:1 mixture of the two molecules.
Figure 4.
(a) Using the single molecule profiling technique, the relative abundance of pOTB7 and CD44 cDNA was measured in a sample containing less than 500 molecules. Undigested, the two species appear identical in an AFM image. By in situ cleavage with enzyme PstI, each molecule was identified by the pattern of breaks in its backbone corresponding to the enzyme recognition sequence. (b) Frequency of molecules versus PstI cleavage pattern determined from a 1:1 mixture of pOTB7and CD44v plasmids. Molecules with the pattern corresponding to pOTB7 were equally prevalent as those with each of the two patterns corresponding to CD44v.
Returning to our mathematical calculations, in figure 2, region B indicates the parametric space accessible given the resolution and labelling efficiency inferred from published studies and from our experiments. For cDNAs 2.5 kb in length or longer, high density restriction mapping can distinguish >106 different species in the best case (40% cleavage efficiency, 30 bp resolution). This decreases to <104 species for cDNAs 2 kb in size (figure 2(b)). For 1 kb cDNAs (figure 2(c)), either an increase in cleavage efficiency, to 65%, or an increase in resolution, to 20 bp, is required to distinguish a minimum of 104 species uniquely. For cDNAs 0.5 kb in length, both an increase in resolution to 20 bp and high cleavage efficiency (>75%) is required to distinguish at least 103 species uniquely (figure 2(d)).
3.4. AFM sizing experiments on a population of small DNA fragments
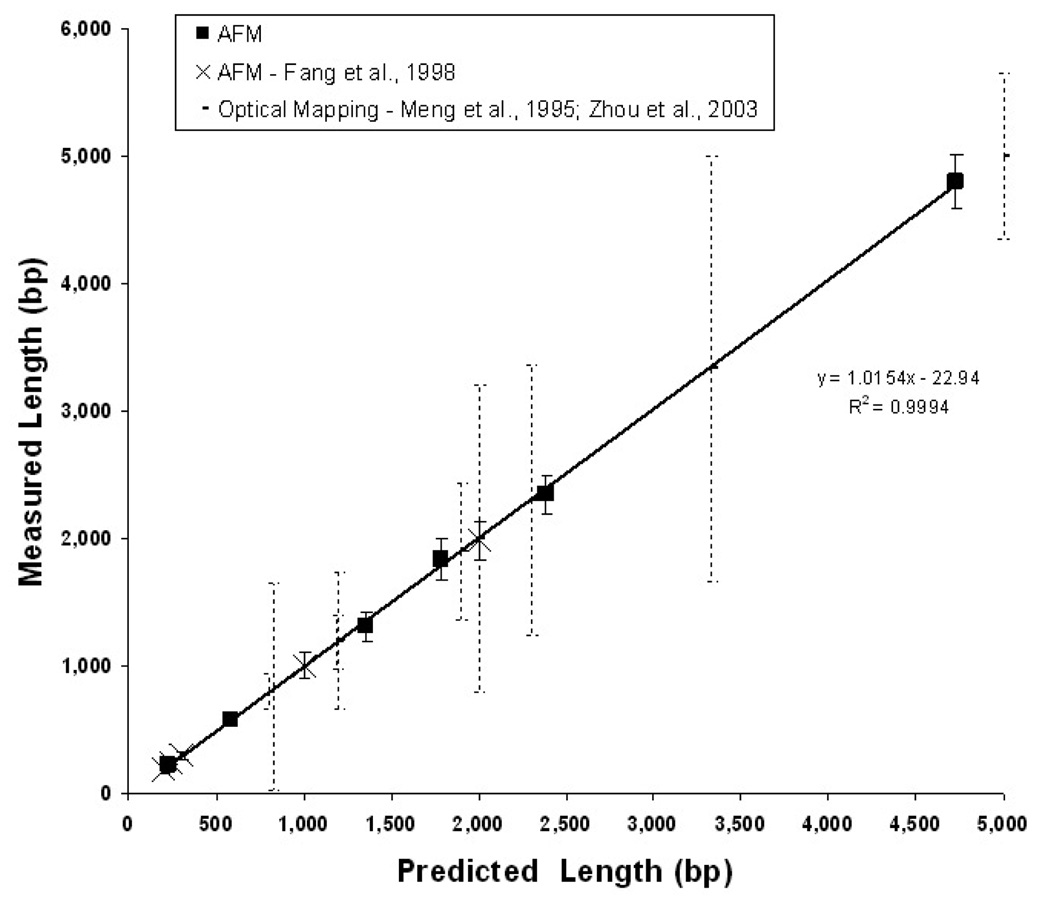
One key difference between the study by Feng et al [189] and the current one is that in their work individual molecules were deposited from solution rather than being generated as in situ fragments from larger DNA molecules. In terms of surface chemistry, the current method differs in that it includes increased surface fixation avidity, which enables the use of higher ionic strength washing solutions. These differences in technique have been reported to alter the observed chain length of DNA, which is an anionic polymer [189]. To better characterize measurement accuracy in our system, a series of six DNA fragments, 230 to 4731 bp, were measured using the AFM to determine backbone contour length. Assuming a conversion of 0.33 nm bp−1, our results duplicated the reported data in sizing accuracy, defined as population CV [189]. The linear regression slope coefficient for these data was 1.0154 with an R2 of 0.9994 (figure 5). As a reference, the data also compared favourably with single DNA molecule sizing based on fluorescence [159], which is the closest comparable surface-biochemical system. The clear advantage of AFM in single fragment sizing is apparent in the lower sizing dispersion and the ability to accurately size very small molecules (<300 bp). The coefficient of variation was 8–10% for small fragments (<600 bp) and as low as 5% for the largest fragment (4700 bp) using AFM as compared to >16% using fluorescence. Also, fluorescence methods require internal references in each image to convert fluorescence intensity into molecular length, while AFM can directly convert backbone contour length accurately to bp.
Figure 5.
AFM sizing of surface fixed, double-stranded DNA. The mean length in bp for pools of six different linear DNA molecules are plotted as dark squares (n = 10–40 molecules). Backbone contour length in nanometres is converted to bp using the nominal pitch of duplex beta DNA (0.33 nm/bp) [189]. Fragments range in size from 230 to 4731 bp (90–1561 nm). Error bars represent sample standard deviation. The x-axis is the length predicted from the sequence and the y-axis is the contour length as measured with AFM. A linear regression of the measurements is displayed. These measurements are consistent with those of Fang et al [189] who used a similar AFM sizing method (white circles). To illustrate the relative precision of this technique, we include data from a distinct optical single molecule sizing method (broken vertical lines; [156, 159, 162, 163].
3.5. Sample preparation issues
The substrates used in this study have to fulfill three conflicting criteria: (1) maximum smoothness, (2) stringent molecular adhesion, and (3) permit normal activity of the restriction enzyme. First, for AFM imaging the surfaces must be smooth on a roughness scale that is less than the diameter of the DNA molecule (∼2 nm). Second, surfaces used for in situ restriction digestion here must bind and retain small fragments (<1000 bp) in moderate ionic strength buffer. APTES silanization generates roughness that is generally proportional to the amount of silane molecules deposited on the surface [224, 225]. More adhesive surfaces that hold small fragments require more aminosilane to be adsorbed, which in turn creates greater roughness. Third, the surface must not bind the molecules so stringently so as to sterically hinder the enzyme, which could cause incomplete digestion and/or nonspecific cleavage.
To address theses requirements, we developed an APTES application protocol to produce an AFM-compatible surface that retains enough positive charge to bind and hold small DNA fragments. The contour profile of one of these substrates (figure 6) shows surface irregularities <1 nm in height and an RMS roughness of ∼0.4 nm, which is smooth enough to resolve DNA molecules using AFM. Silane hydrolysis and surface adsorption kinetics indicate that polymerized aggregates of multifunctional silanes accumulate in solution rapidly after 10 min in aqueous solvent, and these adsorbed aggregates increase roughness on silanized silica substrates [225–227]. We therefore chose short, less than 1 h, derivatization reaction times.
Figure 6.
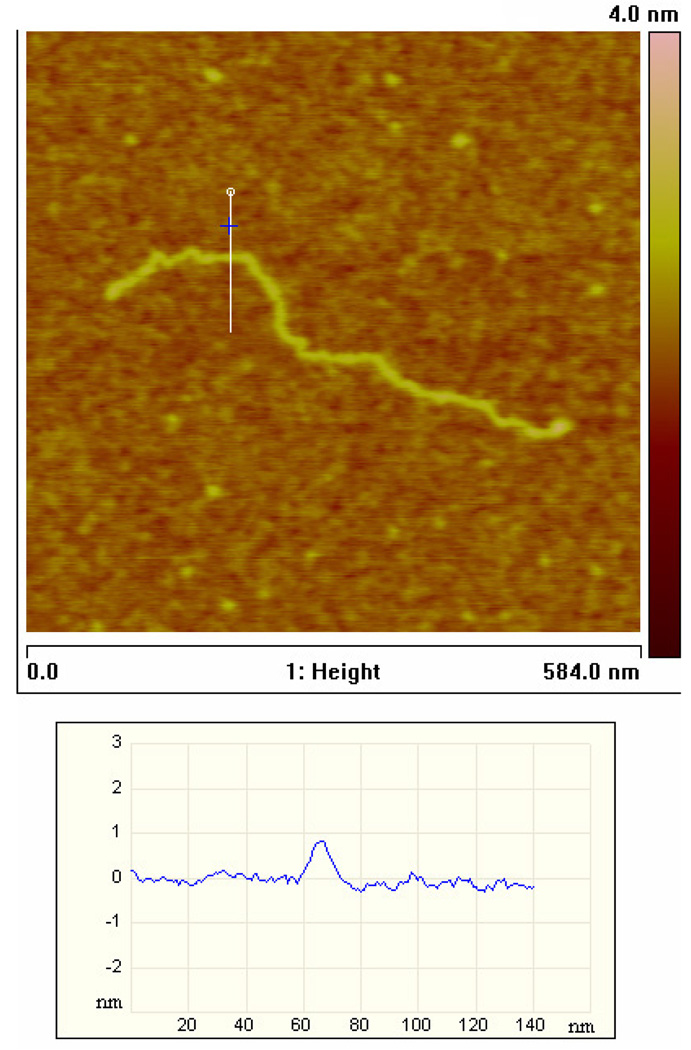
AFM image of a CD44 cDNA molecule, 1800 bp, bound to APTES surface, imaged under typical conditions.
We determined that in situ DNA digestion increases surface roughness, resulting in reduced contrast AFM images, although this reduction was not sufficient to preclude sharp AFM imaging. One source of roughness is the restriction enzyme, which adheres to the positively charged surface, though less avidly than negatively charged DNA. Even without enzyme treatment, the contrast in AFM images was reduced after treatment with enzyme digestion buffer; we suspect that adsorption of salt from the restriction enzyme buffer or rearrangement of the APTES layer itself when exposed to aqueous solution may be responsible. After some trial and error, we were able to repeatedly produce molecularly smooth samples, using low silane concentrations, which were compatible with DNA stretching and enzyme digestion. The large body of organosilane research suggests ways to improve the performance of these surfaces (durability, hydrolytic stability) in numerous ways, including using spin-coating, oven curing, applying mixtures of silanes, and potentially multi-layer coatings [160, 228–238].
In particular, it is possible that appropriate surface chemistry ‘tuning’ will improve the enzymatic digestion rate substantially. Based on our experience with this particular surface/biochemical system, we believe that it is possible to approach the 70–80% digest rates we have previously achieved in optical mapping studies [156, 159, 160, 162, 163]. The enzyme digestion rate will also improve as the incubation time is increased. The incubation time used in these studies was constrained because the APTES monolayer begins to degrade after 45 min−1 h in aqueous buffer, we believe, due to partial hydrolysis [227, 238]. We have determined that simply increasing the thickness of the APTES layer greatly improves resistance to hydrolysis, but results in unacceptable surface roughness for AFM imaging. Reports suggest the use of pre-cross linked, or bis-silanes, among other options, will increase hydrolysis resistance substantially, while retaining monolayer smoothness. Benkoski et al produced a molecularly smooth bis-silane film of thickness 1–10 nm, roughly equivalent to the thickness of our APTES layers [234]. The RMS roughness of these bis-silane layers varied from 0.15 to 0.4 nm for the roughest samples.
4. Conclusions
Our mathematical analysis of single molecule, ordered restriction mapping of cDNAs yielded benchmarks for sizing accuracy and labelling/cleavage efficiency that can guide future studies. The sizing accuracy and labelling efficiencies observed in our experiments, and inferred from the cited literature, suggests AFM profiling of cDNA with restriction maps allows, in theory, unique identification of up to ∼104 individual, 2 kb long, species, and greater than 106 individual species 2.5 kb or longer. Given that roughly 40% of mammalian cDNAs are 2 kb or longer [182, 183, 185], this approach could, in principle, quantify a sizable fraction of the message transcripts within a single cell to single molecule precision. In reference to our experiment, achievable improvements in size resolution (20%) or cleavage efficiency (30%) would permit complete quantification of 1 kb or longer cDNAs, or roughly 80% of all transcripts [182, 183, 185]. Species ∼0.5 kb are more difficult. A sufficient number of species in this category can be resolved with simultaneous improvements in cleavage efficiency (2×) and size resolution (1.5×).
Improvements in sizing accuracy are likely to come from tip deconvolution techniques, which are especially relevant to molecules smaller than 1 kb. More sophisticated metrological methods, which take into account information beyond molecule contour length, such as apparent volume, or frictional information, may also improve sizing accuracy. It is harder to speculate on specific ways to improve labelling efficiency, because inter-molecular binding can be modified with so many chemistries.
Both the in situ cleavage approach and the bind-but-not-cut approach to single molecule, ordered restriction mapping appear viable; however, the former is much better characterized. AFM throughput was not a focus of this work, but it is clearly a critical issue. The most commonly used AFMs require careful operator attention to produce high quality data and prevent damaging the tip or the sample. This is particularly true with many biological samples, such as cells, because they have highly variable shapes and material properties. In contrast, our samples used here are very smooth and the structures imaged, DNA strands, are all essentially identical. Because the samples are so smooth and regular, industrial automation techniques can be implemented. Automated AFM is used widely in the semiconductor fabrication industry for chip inspection, and is favoured for its ability to inspect nanometre features while not damaging the sample. Industrial AFMs run unassisted for extremely long periods at high duty cycles, and have the capability to replace tips automatically. We have found that the DNA molecule itself serves as a very good reference for scan quality and tip condition.
Acknowledgments
Part of this work was supported by NIH grants R21GM074509 (JG, JR and MT), R21HG003714-01 (JG, JR and BM), R01CA74929 (MT), R01CA107300 (MT), PN2EY018228 (MT), the Margaret E. Early Medical Research Trust (MT), and CMISE (JG and MT), a NASA URETI Institute (NCC 2-1364). MT is a Scholar of the Leukemia and Lymphoma Society. BM was also supported by a USAMRMC grant (W81XWH-05-1-0026) and a NIST grant (no. 60NANB5D1199).
Footnotes
Based on invited talk at the International Conference on Nanoscience and Technology 2006.
References
- 1.Ruan YJ, Le Ber P, Ng HH, Liu ET. Interrogating the transcriptome. Trends Biotechnol. 2004;22:23–30. doi: 10.1016/j.tibtech.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Bashiardes S, Lovett M. cDNA detection and analysis. Curr. Opin. Chem. Biol. 2001;5:15–20. doi: 10.1016/s1367-5931(00)00161-7. [DOI] [PubMed] [Google Scholar]
- 3.Hoheisel JD. Microarray technology: beyond transcript profiling and genotype analysis. Nat. Rev. Gen. 2006;7:200–210. doi: 10.1038/nrg1809. [DOI] [PubMed] [Google Scholar]
- 4.Shyamsundar R, Kim YH, Higgins JP, Montgomery K, Jorden M, Sethuraman A, van de Rijn M, Botstein D, Brown PO, Pollack JR. A DNA microarray survey of gene expression in normal human tissues. Genome Biol. 2005;6(9) doi: 10.1186/gb-2005-6-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shih IM, Wang TL. Apply innovative technologies to explore cancer genome. Curr. Opin. Oncol. 2005;17:33–38. doi: 10.1097/01.cco.0000147382.97085.e4. [DOI] [PubMed] [Google Scholar]
- 6.Ewis AA, Zhelev Z, Bakalova R, Fukuoka S, Shinohara Y, Ishikawa M, Baba Y. A history of microarrays in biomedicine. Expert Rev. Mol. Diagn. 2005;5:315–328. doi: 10.1586/14737159.5.3.315. [DOI] [PubMed] [Google Scholar]
- 7.Clarke PA, te Poele R, Workman P. Gene expression microarray technologies in the development of new therapeutic agents. Eur. J. Cancer. 2004;40:2560–2591. doi: 10.1016/j.ejca.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Adjaye J, Bolton V, Monk M. Developmental expression of specific genes detected in high-quality cDNA libraries from single human preimplantation embryos. Gene. 1999;237:373–383. doi: 10.1016/s0378-1119(99)00329-7. [DOI] [PubMed] [Google Scholar]
- 9.Bengtsson M, Stahlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15:1388–1392. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galvin JE. Neurodegenerative diseases: Pathology and the advantage of single-cell profiling. Neurochem. Res. 2004;29:1041–1051. doi: 10.1023/b:nere.0000023592.53769.4d. [DOI] [PubMed] [Google Scholar]
- 11.Glanzer JG, Eberwine JH. Expression profiling of small cellular samples in cancer: less is more. Br. J. Cancer. 2004;90:1111–1114. doi: 10.1038/sj.bjc.6601668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinmoller E, Schlake G, Renke B, Liu Q, Hill KA, Sommer SS, Ruschoff J. Microdissection and molecular analysis of single cells or small cell clusters in pathology and diagnosis—significance and challenges. Anal. Cell. Pathol. 2002;24:125–134. doi: 10.1155/2002/972732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawasaki ES. Applications of Bioinformatics in Cancer Detection. New York: New York Acad Sciences; 2004. Microarrays and the gene expression profile of a single cell; pp. 92–100. [DOI] [PubMed] [Google Scholar]
- 14.Nygaard V, Hovig E. Options available for profiling small samples: a review of sample amplification technology when combined with microarray profiling. Nucleic Acids Res. 2006;34:996–1014. doi: 10.1093/nar/gkj499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peixoto A, Monteiro M, Rocha B, Veiga-Fernandes H. Quantification of multiple gene expression in individual cells. Genome Res. 2004;14:1938–1947. doi: 10.1101/gr.2890204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz DJ, Goaillard JM, Marder E. Variable channel expression in identified single and electrically coupled neurons in different animals. Nat. Neurosci. 2006;9:356–362. doi: 10.1038/nn1639. [DOI] [PubMed] [Google Scholar]
- 17.Smirnov DA, Foulk BW, Doyle GV, Connelly MC, Terstappen L, Lara SM. Global gene expression profiling of circulating endothelial cells in patients with metastatic carcinomas. Cancer Res. 2006;66:2918–2922. doi: 10.1158/0008-5472.CAN-05-4003. [DOI] [PubMed] [Google Scholar]
- 18.Todd R, Margolin DH. Challenges of single-cell diagnostics: analysis of gene expression. Trends in Mol. Med. 2002;8:254–257. doi: 10.1016/s1471-4914(02)02350-x. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 20.Soller M. Pre-messenger RNA processing and its regulation: a genomic perspective. Cell. Mol. Life Sci. 2006;63:796–819. doi: 10.1007/s00018-005-5391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C, Roy M. Analysis of alternative splicing with microarrays: successes and challenges. Genome Biol. 2004;5(7) doi: 10.1186/gb-2004-5-7-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans SJ, Watson SJ, Akil H. Evaluation of sensitivity, performance and reproducibility of microarray technology in neuronal tissue. Integrative Comparative Biol. 2003;43:780–785. doi: 10.1093/icb/43.6.780. [DOI] [PubMed] [Google Scholar]
- 23.Zhu BM, Xu F, Baba Y. An evaluation of linear RNA amplification in cDNA microarray gene expression analysis. Mol. Gen. Metabol. 2006;87:71–79. doi: 10.1016/j.ymgme.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Subkhankulova T, Livesey FJ. Comparative evaluation of linear and exponential amplification techniques for expression profiling at the single-cell level. Gen. Biol. 2006;7(3) doi: 10.1186/gb-2006-7-3-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pike BL, Groshen S, Hsu Y, Shai RM, Wang XM, Holtan N, Futscher BW, Hacia JG. Comparisons of PCR based genome amplification systems using CpG island microarrays. Hum. Mutat. 2006;27:589–596. doi: 10.1002/humu.20329. [DOI] [PubMed] [Google Scholar]
- 26.Feher LZ, Balazs M, Kelemen JZ, Zvara A, Nemeth I, Varga-Orvos Z, Puskas LG. Improved DOP-PCR-based representational whole-genome amplification using quantitative real-time PCR. Diagn. Mol. Pathol. 2006;15:43–48. doi: 10.1097/00019606-200603000-00007. [DOI] [PubMed] [Google Scholar]
- 27.Zhou WL, Abruzzese RV, Polejaeva I, Davis S, Ji W. Amplification of nanogram amounts of total RNA by the SMART-based PCR method for high-density oligonucleotide microarrays. Clin. Chem. 2005;51:2354–2356. doi: 10.1373/clinchem.2005.056721. [DOI] [PubMed] [Google Scholar]
- 28.Suslov O, Steindler DA. PCR inhibition by reverse transcriptase leads to an overestimation of amplification efficiency. Nucleic Acids Res. 2005;33(20) doi: 10.1093/nar/gni176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy ZB, Kelemen JZ, Feher LZ, Zvara A, Juhasz K, Puskas LG. Real-time polymerase chain reaction-based exponential sample amplification for microarray gene expression profiling. Anal. Biochem. 2005;337:76–83. doi: 10.1016/j.ab.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 30.Wang G, Brennan C, Rook M, Wolfe JL, Leo C, Chin L, Pan H, Liu WH, Price B, Makrigiorgos GM. Balanced-PCR amplification allows unbiased identification of genomic copy changes in minute cell and tissue samples. Nuclic. Acids Res. 2004;32(9) doi: 10.1093/nar/gnh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi JX, Liu Q, Nguyen VQ, Sommer SS. Elimination of locus-specific inter-individual variation in quantitative PCR. Biotechniques. 2004;37:934–938. doi: 10.2144/04376ST01. [DOI] [PubMed] [Google Scholar]
- 32.Goff LA, Bowers J, Schwalm J, Howerton K, Getts RC, Hart RP. Evaluation of sense-strand mRNA amplification by comparative quantitative PCR. Bmc Genomics. 2004;5:76. doi: 10.1186/1471-2164-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohuchi S, Nakano H, Yamane T. In vitro method for the generation of protein libraries using PCR amplification of a single DNA molecule and coupled transcription/translation 10.1093/nar/26.19.4339. Nucl. Acids Res. 1998;26:4339–4346. doi: 10.1093/nar/26.19.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spits C, Le Caignec C, De Rycke M, Van Haute L, Van Steirteghem A, Liebaers I, Sermon K. Optimization and evaluation of single-cell whole, genome multiple displacement amplification. Hum. Mutat. 2006;27:496–503. doi: 10.1002/humu.20324. [DOI] [PubMed] [Google Scholar]
- 35.Peano C, Severgnini M, Cifola I, De Bellis G, Battaglia C. Transcriptome amplification methods in gene expression profiling. Expert Rev. Mol. Diagn. 2006;6:465–480. doi: 10.1586/14737159.6.3.465. [DOI] [PubMed] [Google Scholar]
- 36.Lovmar L, Syvanen AC. Multiple displacement amplification to create a long-lasting source of DNA for genetic studies. Hum. Mutat. 2006;27:603–614. doi: 10.1002/humu.20341. [DOI] [PubMed] [Google Scholar]
- 37.Inoue J, Shigemori Y, Mikawa T. Improvements of rolling circle amplification (RCA) efficiency and accuracy using Thermus thermophilus SSB mutant protein. Nucleic Acids Res. 2006;34(9) doi: 10.1093/nar/gkl350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evanko D. Cloning keeps on rolling. Nat. Methods. 2006;3:8–9. [Google Scholar]
- 39.Brukner I, Labuda D, Krajinovic M. Phi29-based amplification of small genomes. Anal. Biochem. 2006;354:154–156. doi: 10.1016/j.ab.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 40.Paul P, Apgar J. Single-molecule dilution and multiple displacement amplification for molecular haplotyping. Biotechniques. 2005;38:553. doi: 10.2144/05384ST01. [DOI] [PubMed] [Google Scholar]
- 41.Panelli S, Damiani G, Espen L, Sgaramella V. Ligation overcomes terminal underrepresentation in multiple displacement amplification of linear DNA. Biotechniques. 2005;39:174. doi: 10.2144/05392BM03. [DOI] [PubMed] [Google Scholar]
- 42.Kurn N, Chen PC, Heath JD, Kopf-Sill A, Stephens KM, Wang SL. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clin. Chem. 2005;51:1973–1981. doi: 10.1373/clinchem.2005.053694. [DOI] [PubMed] [Google Scholar]
- 43.Jiang ZW, Zhang XQ, Deka R, Jin L. Genome amplification of single sperm using multiple displacement amplification. Nucl. Acids Res. 2005;33(10) doi: 10.1093/nar/gni089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hellani A, Coskun S, Tbakhi A, Al-Hassan S. Clinical application of multiple displacement amplification in preimplantation genetic diagnosis. Reprod. Biomed. Online. 2005;10:376–380. doi: 10.1016/s1472-6483(10)61799-3. [DOI] [PubMed] [Google Scholar]
- 45.Bergen AW, Haque KA, Qi Y, Beerman MB, Garcia-Closas M, Rothman N, Chanock SJ. Comparison of yield and genotyping performance of multiple displacement OmniPlex (TM) whole genome amplified DNA generated from multiple DNA sources. Hum. Mutat. 2005;26:262–270. doi: 10.1002/humu.20213. [DOI] [PubMed] [Google Scholar]
- 46.Bergen AW, Qi Y, Haque KA, Welch RA, Chanock SJ. Effects of DNA mass on multiple displacement whole genome amplification and genotyping performance. Bmc Biotechnol. 2005;5:24. doi: 10.1186/1472-6750-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paez JG, et al. Genome coverage and sequence fidelity of phi 29 polymerase-based multiple strand displacement whole genome amplification. Nucleic Acids Res. 2004;32(9) doi: 10.1093/nar/gnh069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hellani A, Coskun S, Benkhalifa M, Tbakhi A, Sakati N, Al-Odaib A, Ozand P. Multiple displacement amplification on single cell and possible PGD applications. Mol. Hum. Reprod. 2004;10:847–852. doi: 10.1093/molehr/gah114. [DOI] [PubMed] [Google Scholar]
- 49.Handyside AH, Robinson MD, Simpson RJ, Omar MB, Shaw MA, Grudzinskas JG, Rutherford A. Isothermal whole genome amplification from single and small numbers of cells: a new era for preimplantation genetic diagnosis of inherited disease. Mol. Hum. Reprod. 2004;10:767–772. doi: 10.1093/molehr/gah101. [DOI] [PubMed] [Google Scholar]
- 50.Luthra R, Medeiros LJ. Isothermal multiple displacement amplification—a highly reliable approach for generating unlimited high molecular weight genomic DNA from clinical specimens. J. Mol. Diagn. 2003;6:236–242. doi: 10.1016/S1525-1578(10)60516-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nelson JR. Phi29 DNA polymerase-based methods for genomics applications. J. Clin. Ligand Assay. 2002;25:276–279. [Google Scholar]
- 52.Dean FB, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dean FB, et al. Comprehensive human genome amplification using multiple displacement amplification 10.1073/pnas.082089499. Proc. Natl Acad. Sci. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lizardi PM, Huang XH, Zhu ZR, Bray-Ward P, Thomas DC, Ward DC. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 55.Zhong XB, Lizardi PM, Huang XH, Bray-Ward PL, Ward DC. Visualization of oligonucleotide probes and point mutations in interphase nuclei and DNA fibers using rolling circle DNA amplification. Proc. Natl Acad. Sci. USA. 2001;98:3940–3945. doi: 10.1073/pnas.061026198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilhelm J, Muyal JP, Best J, Kwapiszewska G, Stein MM, Seeger W, Bohle RM, Fink L. Systematic comparison of the T7-IVT and SMART-based RNA preamplification techniques for DNA microarray experiments. Clin. Chem. 2006;52:1161–1167. doi: 10.1373/clinchem.2005.062406. [DOI] [PubMed] [Google Scholar]
- 57.Cope L, Hartman SM, Gohlmann HWH, Tiesman JP, Irizarry RA. Analysis of Affymetrix GeneChip (R) data using amplified RNA. Biotechniques. 2006;40:165. doi: 10.2144/000112057. [DOI] [PubMed] [Google Scholar]
- 58.Wadenback J, Clapham DH, Craig D, Sederoff R, Peter GF, von Arnold S, Egertsdotter U. Comparison of standard exponential and linear techniques to amplify small cDNA samples for microarrays. Bmc Genomics. 2005;6 doi: 10.1186/1471-2164-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pahl A. Gene expression profiling using RNA extracted from whole blood: technologies and clinical applications. Expert Rev. Mol. Diagn. 2005;5:43–52. doi: 10.1586/14737159.5.1.43. [DOI] [PubMed] [Google Scholar]
- 60.Schneider J, Buness A, Huber W, Volz J, Kioschis P, Hafner M, Poustka A, Sultmann H. Systematic analysis of T7 RNA polymerase based in vitro linear RNA amplification for use in microarray experiments. Bmc Genomics. 2004;5:29. doi: 10.1186/1471-2164-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moll PR, Duschl J, Richter K. Optimized RNA amplification using T7-RNA-polymerase based in vitro transcription. Anal. Biochem. 2004;334:164–174. doi: 10.1016/j.ab.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 62.Li Y, Li T, Liu SZ, Qiu MY, Han ZY, Jiang ZL, Li RY, Ying K, Xie Y, Mao YM. Systematic comparison of the fidelity of aRNA, mRNA and T-RNA on gene expression profiling using cDNA microarray. J. Biotechnol. 2004;107:19–28. doi: 10.1016/j.jbiotec.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 63.Zhao HJ, Hastie T, Whitfield ML, Borresen-Dale AL, Jeffrey SS. Optimization and evaluation of T7 based RNA linear amplification protocols for cDNA microarray analysis. Bmc Genomics. 2002;3:31. doi: 10.1186/1471-2164-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mizuarai S, Takahashi K, Kobayashi T, Kotani H. Advances in isolation and characterization of homogeneous cell populations using laser microdissection. Histol. Histopathol. 2005;20:139–146. doi: 10.14670/HH-20.139. [DOI] [PubMed] [Google Scholar]
- 65.Taylor TB, Nambiar PR, Raja R, Cheung E, Rosenberg DW, Anderegg B. Microgenomics: identification of new expression profiles via small and single-cell sample analyses. Cytometry A. 2004;59A:254–261. doi: 10.1002/cyto.a.20051. [DOI] [PubMed] [Google Scholar]
- 66.Sun M, Zhou GL, Lee S, Chen JJ, Shi RZ, Wang SM. SAGE is far more sensitive than EST for detecting low-abundance transcripts. Bmc Genomics. 2004;5:1. doi: 10.1186/1471-2164-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kenzelmann M, Klaren R, Hergenhahn M, Bonrouhi M, Grone HJ, Schmid W, Schutz G. High-accuracy amplification of nanogram total RNA amounts for gene profiling. Genomics. 2004;83:550–558. doi: 10.1016/j.ygeno.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 68.Heidenblut AM, et al. aRNA-longSAGE: a new approach to generate SAGE libraries from microdissected cells. Nucl. Acids Res. 2004;32(16) doi: 10.1093/nar/gnh130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davis JE, Eberwine JH, Hinkle DA, Marciano PG, Meaney DF, McIntosh TK. Methodological considerations regarding single-cell gene expression profiling for brain injury. Neurochem. Res. 2004;29:1113–1121. doi: 10.1023/b:nere.0000023598.04937.83. [DOI] [PubMed] [Google Scholar]
- 70.Vilain C, Libert F, Venet D, Costagliola S, Vassart G. Small amplified RNA-SAGE: an alternative approach to study transcriptome from limiting amount of mRNA. Nucl. Acids Res. 2003;31(6) doi: 10.1093/nar/gng024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Evans SJ, Datson NA, Kabbaj M, Thompson RC, Vreugdenhil E, De Kloet ER, Watson SJ, Akil H. Evaluation of affymetrix gene chip sensitivity in rat hippocampal tissue using SAGE analysis. Eur. J. Neurosci. 2002;16:409–413. doi: 10.1046/j.1460-9568.2002.02097.x. [DOI] [PubMed] [Google Scholar]
- 72.Datson NA, van der Perk J, de Kloet ER, Vreugdenhil E. Expression profile of 30,000 genes in rat hippocampus using SAGE. Hippocampus. 2001;11:430–444. doi: 10.1002/hipo.1058. [DOI] [PubMed] [Google Scholar]
- 73.Yao FY, Yu F, Gong LJ, Taube D, Rao DD, MacKenzie RG. Microarray analysis of fluoro-gold labeled rat doparnine neurons harvested by laser capture microdissection. J. Neurosci. Meth. 2005;143:95–106. doi: 10.1016/j.jneumeth.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 74.Jiang YM, et al. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann. Neurol. 2005;57:236–251. doi: 10.1002/ana.20379. [DOI] [PubMed] [Google Scholar]
- 75.Jen IT, Rihel J, Dulac CG. Single-cell transcriptional profiles and spatial patterning of the mammalian olfactory epithelium. Int. J. Dev. Biol. 2005;49:201–207. doi: 10.1387/ijdb.041939it. [DOI] [PubMed] [Google Scholar]
- 76.Fassunke J, Majores M, Ullmann C, Elger CE, Schramm J, Wiestler OD, Becker AJ. In situ-RT and immunolaser microdissection for mRNA analysis of individual cells isolated from epilepsy-associated glioneuronal tumors. Lab. Invest. 2004;84:1520–1525. doi: 10.1038/labinvest.3700165. [DOI] [PubMed] [Google Scholar]
- 77.Seshi B, Kumar S, King D. Multilineage gene expression in human bone marrow stromal cells as evidenced by single-cell microarray analysis. Blood Cells Mol. Diseases. 2003;31:268–285. doi: 10.1016/s1079-9796(03)00150-5. [DOI] [PubMed] [Google Scholar]
- 78.Oda R, Yaoi T, Okajima S, Kobashi H, Kubo T, Fushiki S. A novel marker for terminal Schwann cells, homocysteine-responsive ER-resident protein, as isolated by a single cell PCR-differential display. Biochem. Biophys. Res. Commun. 2003;308:872–877. doi: 10.1016/s0006-291x(03)01499-2. [DOI] [PubMed] [Google Scholar]
- 79.Kamme F, et al. Single-cell microarray analysis in hippocampus CA1: Demonstration and validation of cellular heterogeneity. J. Neurosci. 2003;23:3607–3615. doi: 10.1523/JNEUROSCI.23-09-03607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mufson EJ, Counts SE, Ginsberg SD. Gene expression profiles of cholinergic nucleus basalis neurons in Alzheimer’s disease. Neurochem. Res. 2002;27:1035–1048. doi: 10.1023/a:1020952704398. [DOI] [PubMed] [Google Scholar]
- 81.Liss B. Improved quantitative real-time RT-PCR for expression profiling of individual cells. Nucleic Acids Res. 2002;30(17) doi: 10.1093/nar/gnf088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brandt S, Kloska S, Altmann T, Kehr J. Using array hybridization to monitor gene expression at the single cell level. J. Exp. Botany. 2002;53:2315–2323. doi: 10.1093/jxb/erf093. [DOI] [PubMed] [Google Scholar]
- 83.Guillaud-Bataille M, et al. Detecting single DNA copy number variations in complex genomes using one nanogram of starting DNA and BAC-array CGH. Nucleic Acids Res. 2004;32(13) doi: 10.1093/nar/gnh108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schindler H, Wiese A, Auer J, Burtscher H. CRNA target preparation for microarrays: Comparison of gene expression profiles generated with different amplification procedures. Anal. Biochem. 2005;344:92–101. doi: 10.1016/j.ab.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 85.Nygaard V, Holden M, Loland A, Langaas M, Myklebost O, Hovig E. Limitations of mRNA amplification from small-size cell samples. Bmc Genomics. 2005;6:147. doi: 10.1186/1471-2164-6-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ginsberg SD. RNA amplification strategies for small sample populations. Methods. 2005;37:229–237. doi: 10.1016/j.ymeth.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 87.Petalidis L, Bhattacharyya S, Morris GA, Collins VP, Freeman TC, Lyons PA. Global amplification of mRNA by template-switching PCR: linearity and application to microarray analysis. Nucleic Acids Res. 2003;31(22) doi: 10.1093/nar/gng142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nygaard V, Loland A, Holden M, Langaas M, Rue H, Liu F, Myklebost O, Fodstad O, Hovig E, Smith-Sorensen B. Effects of mRNA amplification on gene expression ratios in cDNA experiments estimated by analysis of variance. Bmc Genomics. 2003;4:11. doi: 10.1186/1471-2164-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stutz JAR, Richert C. Tuning the reaction site for enzyme-free primer-extension reactions through small molecule substituents. Chem. Eur. J. 2006;12:2472–2481. doi: 10.1002/chem.200501008. [DOI] [PubMed] [Google Scholar]
- 90.Sakata T, Miyahara Y. DNA sequencing based on intrinsic molecular charges. Angew. Chem.-Int. Edn. 2006;45:2225–2228. doi: 10.1002/anie.200503154. [DOI] [PubMed] [Google Scholar]
- 91.Pourmand N, Karhanek M, Persson HHJ, Webb CD, Lee TH, Zahradnikova A, Davis RW. Direct electrical detection of DNA synthesis. Proc. Natl Acad. Sci. USA. 2006;103:6466–6470. doi: 10.1073/pnas.0601184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin L, Wang HD, Liu Y, Yan H, Lindsay S. Recognition imaging with a DNA aptamer. Biophys. J. 2006;90:4236–4238. doi: 10.1529/biophysj.105.079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Flagella M, et al. A multiplex branched DNA assay for parallel quantitative gene expression profiling. Anal. Biochem. 2006;352:50–60. doi: 10.1016/j.ab.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 94.Dorfman A, Kumar N, Hahm JI. Highly sensitive biomolecular fluorescence detection using nanoscale ZnO platforms. Langmuir. 2006;22:4890–4895. doi: 10.1021/la053270+. [DOI] [PubMed] [Google Scholar]
- 95.Burghardt TP, Ajtai K, Borejdo J. In situ single-molecule imaging with attoliter detection using objective total internal reflection confocal microscopy. Biochemistry. 2006;45:4058–4068. doi: 10.1021/bi052097d. [DOI] [PubMed] [Google Scholar]
- 96.Sauer S, Lange BMH, Gobom J, Nyarsik L, Seitz H, Lehrach H. Miniaturization in functional genomics and proteomics. Nat. Rev. Genet. 2005;6:465–476. doi: 10.1038/nrg1618. [DOI] [PubMed] [Google Scholar]
- 97.Mulder BA, Anaya S, Yu PL, Lee KW, Nguyen A, Murphy J, Willson R, Briggs JM, Gao XL, Hardin SH. Nucleotide modification at the gamma-phosphate leads to the improved fidelity of HIV-1 reverse transcriptase. Nucleic Acids Res. 2005;33:4865–4873. doi: 10.1093/nar/gki779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McCullough RM, Cantor CR, Ding CM. High-throughput alternative splicing quantification by primer extension and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Nucl. Acids Res. 2005;33(11) doi: 10.1093/nar/gni098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Illangkoon HI, Benner SA. Scaffolds for the development of nucleosides with the ability to form four hydrogen bonds. Abstracts of Papers Am. Chem. Soc. 2005;229:U561–U561. [Google Scholar]
- 100.Edwards JR, Ruparel H, Ju JY. Mass-spectrometry DNA sequencing. Mutat. Res.-Fundamental and Molecular Mechanisms of Mutagenesis. 2005;573:3–12. doi: 10.1016/j.mrfmmm.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 101.Costanzo PJ, Liang EZ, Patten TE, Collins SD, Smith RL. Biomolecule detection via target mediated nanoparticle aggregation and dielectrophoretic impedance measurement. Lab on a Chip. 2005;5:606–610. doi: 10.1039/b417535b. [DOI] [PubMed] [Google Scholar]
- 102.Burbulis I, Yamaguchi K, Gordon A, Carlson R, Brent R. Using protein-DNA chimeras to detect and count small numbers of molecules. Nat. Methods. 2005;2:31–37. doi: 10.1038/nmeth729. [DOI] [PubMed] [Google Scholar]
- 103.Bennett ST, Barnes C, Cox A, Davies L, Brown C. Toward the $1000 human genome. Pharmacogenomics. 2005;6:373–382. doi: 10.1517/14622416.6.4.373. [DOI] [PubMed] [Google Scholar]
- 104.Winter H, Korn K, Rigler R. Direct gene expression analysis. Curr. Pharmaceut. Biotechnol. 2004;5:191–197. doi: 10.2174/1389201043376995. [DOI] [PubMed] [Google Scholar]
- 105.Twist CR, Winson MK, Rowland JJ, Kell DB. Single-nucleotide polymorphism detection using nanomolar nucleotides and single-molecule fluorescence. Anal. Biochem. 2004;327:35–44. doi: 10.1016/j.ab.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 106.Takeishi S, et al. Observation of electrostatically released DNA from gold electrodes with controlled threshold voltages. J. Chem. Phys. 2004;120:5501–5504. doi: 10.1063/1.1643729. [DOI] [PubMed] [Google Scholar]
- 107.Stroh C, Wang H, Bash R, Ashcroft B, Nelson J, Gruber H, Lohr D, Lindsay SM, Hinterdorfer P. Single-molecule recognition imaging-microscopy. Proc. Natl Acad. Sci. USA. 2004;101:12503–12507. doi: 10.1073/pnas.0403538101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Krieg A, Laib S, Ruckstuhl T, Seeger S. Fast detection of single nucleotide polymorphisms (SNPs) by primer elongation with monitoring of supercritical-angle fluorescence. ChemBioChem. 2004;5:1680–1685. doi: 10.1002/cbic.200400044. [DOI] [PubMed] [Google Scholar]
- 109.Hart JR, Johnson MD, Barton JK. Single-nucleotide polymorphism discovery by targeted DNA photocleavage. Proc. Natl Acad. Sci. USA. 2004;101:14040–14044. doi: 10.1073/pnas.0406169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alivisatos P. The use of nanocrystals in biological detection. Nat. Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 111.Reddy MS, Hardin SH. Features in short guanine-rich sequences that stimulate DNA polymerization in vitro. Biochemistry. 2003;42:350–362. doi: 10.1021/bi020380w. [DOI] [PubMed] [Google Scholar]
- 112.Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 113.Kourentzi KD, Fox GE, Willson RC. Hybridization-responsive fluorescent DNA probes containing the adenine analog 2-aminopurine. Anal. Biochem. 2003;322:124–126. doi: 10.1016/S0003-2697(03)00429-9. [DOI] [PubMed] [Google Scholar]
- 114.Korlach J, Levene M, Foquet M, Turner SW, Craighead HG, Webb WW. Single molecule DNA sequence profiling in zero-mode waveguides using gamma-phosphate linked nucleotide analogs. Biophys. J. 2003;84:141A–141A. [Google Scholar]
- 115.Ding CM, Cantor CR. A high-throughput gene expression analysis technique using competitive PCR and matrix-assisted laser desorption ionization time-of-flight MS. Proc. Natl Acad. Sci. USA. 2003;100:3059–3064. doi: 10.1073/pnas.0630494100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Braslavsky I, Hebert B, Kartalov E, Quake SR. Sequence information can be obtained from single DNA molecules. Proc. Natl Acad. Sci. USA. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tong AK, Li ZM, Ju JY. Combinatorial fluorescence energy transfer tags: New molecular tools for genomics applications. IEEE J. Quantum Electron. 2002;38:110–121. [Google Scholar]
- 118.Korlach J, Levene M, Turner SW, Craighead HG, Webb WW. Single molecule analysis of DNA polymerase activity using zero-mode waveguides. Biophysical J. 2002;82:507A–507A. [Google Scholar]
- 119.Tong AK, Li ZM, Jones GS, Russo JJ, Ju JY. Combinatorial fluorescence energy transfer tags for multiplex biological assays. Nat. Biotechnol. 2001;19:756–759. doi: 10.1038/90810. [DOI] [PubMed] [Google Scholar]
- 120.Korlach J, Levene M, Turner SW, Larson DR, Foquet M, Craighead HG, Webb WW. A new strategy for sequencing individual molecules of DNA. Biophys. J. 2001;80:147A–147A. [Google Scholar]
- 121.Heikal AA, Korlach J, Webb WW. Time-resolved fluorescence and anisotropy of free and DNA-bound fluorescently labeled nucleotides. Biophys. J. 2001;80:8A–8A. [Google Scholar]
- 122.Cui XD, Primak A, Zarate X, Tomfohr J, Sankey OF, Moore AL, Moore TA, Gust D, Harris G, Lindsay SM. Reproducible measurement of single-molecule conductivity. Science. 2001;294:571–574. doi: 10.1126/science.1064354. [DOI] [PubMed] [Google Scholar]
- 123.Welch MB, Martinez CI, Zhang AJ, Jin S, Gibbs R, Burgess K. Syntheses of nucleosides designed for combinatorial DNA sequencing. Chem. Eur. J. 1999;5:951–960. [Google Scholar]
- 124.Lutz MJ, Horlacher J, Benner SA. Recognition of a non-standard base pair by thermostable DNA polymerases. Bioorg. Med. Chem. Lett. 1998;8:1149–1152. doi: 10.1016/s0960-894x(98)00177-2. [DOI] [PubMed] [Google Scholar]
- 125.Cheng J, Waters LC, Fortina P, Hvichia G, Jacobson SC, Ramsey JM, Kricka LJ, Wilding P. Degenerate oligonucleotide primed polymerase chain reaction and capillary electrophoretic analysis of human DNA on microchip-based devices. Anal. Biochem. 1998;257:101–106. doi: 10.1006/abio.1997.2531. [DOI] [PubMed] [Google Scholar]
- 126.Ju JY, Ruan CC, Fuller CW, Glazer AN, Mathies RA. Fluorescence energy-transfer dye-labeled primers for DNA-sequencing and analysis. Proc. Natl Acad. Sci. USA. 1995;92:4347–4351. doi: 10.1073/pnas.92.10.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aksimentiev A, Schulten K, Heng J, Ho C, Timp G. Molecular dynamics simulations of a nanopore device for DNA sequencing. Biophys. J. 2004;86:480A–480A. [Google Scholar]
- 128.Aksimentiev A, Heng JB, Timp G, Schulten K. Microscopic kinetics of DNA translocation through synthetic nanopores. Biophys. J. 2004;87:2086–2097. doi: 10.1529/biophysj.104.042960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aksimentiev A, Heng JB, Cruz-Chu ER, Timp G, Schulten K. Microscopic kinetics of DNA translocation through synthetic nanopores. Biophys. J. 2005;88:352A–352A. doi: 10.1529/biophysj.104.042960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ashkenasy N, Sanchez-Quesada J, Bayley H, Ghadiri MR. Single nucleobase sensitivity of alpha-hemolysin (alpha-HL) transmembrane protein pore: toward single DNA sequencing. Abstracts of Papers Am. Chem. Soc. 2005;229:U336–U336. [Google Scholar]
- 131.Ashkenasy N, Sanchez-Quesada J, Bayley H, Ghadiri MR. Recognizing a single base in an individual DNA strand: a step toward DNA sequencing in nanopores. Angew. Chem.-Int. Edn. 2005;44:1401–1404. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Astier Y, Braha O, Bayley H. Toward single molecule DNA sequencing: Direct identification of ribonucleoside and deoxyribonucleoside 5′-monophosphates by using an engineered protein nanopore equipped with a molecular adapter. J. Am. Chem. Soc. 2006;128:1705–1710. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 133.Bai XP, Edwards J, Ju JY. Molecular engineering approaches for DNA sequencing and analysis. Expert Rev. Mol. Diagn. 2005;5:797–808. doi: 10.1586/14737159.5.5.797. [DOI] [PubMed] [Google Scholar]
- 134.Butler TZ, Gundlach JH, Troll MA. Determination of RNA orientation during translocation through a biological nanopore. Biophys. J. 2006;90:190–199. doi: 10.1529/biophysj.105.068957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fologea D, Gershow M, Ledden B, McNabb DS, Golovchenko JA, Li JL. Detecting single stranded DNA with a solid state nanopore. Nano Lett. 2005;5:1905–1909. doi: 10.1021/nl051199m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fologea D, Uplinger J, Thomas B, McNabb DS, Li JL. Slowing DNA translocation in a solid-state nanopore. Nano Lett. 2005;5:1734–1737. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ghadiri MR, Granja JR, Buehler LK. Artificial transmembrane ion channels from self-assembling peptide nanotubes. Nature. 1994;369:301–304. doi: 10.1038/369301a0. [DOI] [PubMed] [Google Scholar]
- 138.Gracheva ME, Xiong AL, Aksimentiev A, Schulten K, Timp G, Leburton JP. Simulation of the electric response of DNA translocation through a semiconductor nanopore-capacitor. Nanotechnology. 2006;17:622–633. [Google Scholar]
- 139.Gracheva ME, Aksimentiev A, Leburton JP. Electrical signatures of single-stranded DNA with single base mutations in a nanopore capacitor. Nanotechnology. 2006;17:3160–3165. [Google Scholar]
- 140.Heng JB, Ho C, Kim T, Timp R, Aksimentiev A, Grinkova YV, Sligar S, Schulten K, Timp G. Sizing DNA using a nanometer-diameter pore. Biophys. J. 2004;87:2905–2911. doi: 10.1529/biophysj.104.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Heng JB, Aksimentiev A, Ho C, Marks P, Grinkova YV, Sligar S, Schulten K, Timp G. Stretching DNA using the electric field in a synthetic nanopore. Nano Lett. 2005;5:1883–1888. doi: 10.1021/nl0510816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Heng JB, Aksimentiev A, Dimitrov V, Grinkova Y, Ho C, Marks P, Schulten K, Sligar S, Timp G. Stretching DNA using an artificial nanopore. Biophys. J. 2005;88:659A–659A. [Google Scholar]
- 143.Heng JB, Aksimentiev A, Ho C, Marks P, Grinkova YV, Sligar S, Schulten K, Timp G. The electromechanics of DNA in a synthetic nanopore. Biophys. J. 2006;90:1098–1106. doi: 10.1529/biophysj.105.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Howorka S, Cheley S, Bayley H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nat. Biotechnol. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 145.Karhanek M, Kemp JT, Pourmand N, Davis RW, Webb CD. Single DNA molecule detection using nanopipettes and nanoparticles. Nano Lett. 2005;5:403–407. doi: 10.1021/nl0480464. [DOI] [PubMed] [Google Scholar]
- 146.Lagerqvist J, Zwolak M, Di Ventra M. Fast DNA sequencing via transverse electronic transport. Nano Lett. 2006;6:779–782. doi: 10.1021/nl0601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Ion-beam sculpting at nanometre length scales. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 148.Li JL, Gershow M, Stein D, Brandin E, Golovchenko JA. DNA molecules and configurations in a solid-state nanopore microscope. Nat. Mater. 2003;2:611–615. doi: 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- 149.Li JL, Stein D, Qun C, Brandin E, Huang A, Wang H, Branton D, Golovchenko J. Solid state nanopore as a single DNA molecule detector. Biophys. J. 2003;84:134A–135A. [Google Scholar]
- 150.Mannion JT, Reccius CH, Cross JD, Craighead HG. Conformational analysis of single DNA molecules undergoing entropically induced motion in nanochannels. Biophys. J. 2006;90:4538–4545. doi: 10.1529/biophysj.105.074732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl Acad. Sci. USA. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nakane JJ, Akeson M, Marziali A. Nanopore sensors for nucleic acid analysis. J. Phys.: Condens. Matter. 2003;15:R1365–R1393. [Google Scholar]
- 153.Storm AJ, Storm C, Chen JH, Zandbergen H, Joanny JF, Dekker C. Fast DNA translocation through a solid-state nanopore. Nano Lett. 2005;5:1193–1197. doi: 10.1021/nl048030d. [DOI] [PubMed] [Google Scholar]
- 154.Wang GL, Zhang B, Wayment JR, Harris JM, White HS. Electrostatic-gated transport in chemically modified glass nanopore electrodes. J. Am. Chem. Soc. 2006;128:7679–7686. doi: 10.1021/ja061357r. [DOI] [PubMed] [Google Scholar]
- 155.Jeltsch A, Wenz C, Wende W, Selent U, Pingoud A. Engineering novel restriction endonucleases: principles and applications. Trends Biotechnol. 1996;14:235–238. doi: 10.1016/0167-7799(96)10030-5. [DOI] [PubMed] [Google Scholar]
- 156.Meng X, Benson K, Chada K, Huff EJ, Schwartz DC. Optical mapping of bacteriophage-lambda clones using restriction endonucleases. Nat. Genet. 1995;9:432–438. doi: 10.1038/ng0495-432. [DOI] [PubMed] [Google Scholar]
- 157.Dimalanta ET, et al. A microfluidic system for large DNA molecule arrays. Anal. Chem. 2004;76:5293–5301. doi: 10.1021/ac0496401. [DOI] [PubMed] [Google Scholar]
- 158.Casey W, Mishra B. A nearly linear-time general algorithm for genome-wide bi-allele haplotype phasing. Hipc 2003: High Perform. Comput. 2003:204–215. [Google Scholar]
- 159.Zhou SG, et al. A whole-genome shotgun optical map of Yersinia pestis strain KIM. Appl. Environ. Microbiol. 2002;68:6321–6331. doi: 10.1128/AEM.68.12.6321-6331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lim A, et al. Shotgun optical maps of the whole Escherichia coli O157: H7 genome. Genome Res. 2001;11:1584–1593. doi: 10.1101/gr.172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Parida L, Mishra B. Partitioning single-molecule maps into multiple populations: algorithms and probabilistic analysis. Discr. Appl. Math. 2000;104:203–227. [Google Scholar]
- 162.Reed J, Singer E, Kresbach G, Schwartz DC. A quantitative study of optical mapping surfaces by atomic force microscopy and restriction endonuclease digestion assays. Anal. Biochem. 1998;259:80–88. doi: 10.1006/abio.1998.2640. [DOI] [PubMed] [Google Scholar]
- 163.Jing JP, et al. Automated high resolution optical mapping using arrayed, fluid-fixed DNA molecules. Proc. Natl Acad. Sci. USA. 1998;95:8046–8051. doi: 10.1073/pnas.95.14.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Anantharaman TS, Mishra B, Schwartz DC. Genomics via optical mapping.2. Ordered restriction maps. J. Comput. Biol. 1997;4:91–118. doi: 10.1089/cmb.1997.4.91. [DOI] [PubMed] [Google Scholar]
- 165.Oana H, Ueda M, Washizu M. Visualization of a specific sequence on a single large DNA molecule using fluorescence microscopy based on a new DNA-stretching method. Biochem. Biophys. Res. Commun. 1999;265:140–143. doi: 10.1006/bbrc.1999.1614. [DOI] [PubMed] [Google Scholar]
- 166.Allison DP, Kerper PS, Doktycz MJ, Spain JA, Modrich P, Larimer FW, Thundat T, Warmack RJ. Direct atomic force microscope imaging of EcoRI endonuclease site specifically bound to plasmid DNA molecules. Proc. Natl Acad. Sci. USA. 1996;93:8826–8829. doi: 10.1073/pnas.93.17.8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Allison DP, Kerper PS, Doktycz MJ, Thundat T, Modrich P, Larimer FW, Johnson DK, Hoyt PR, Mucenski ML, Warmack RJ. Mapping individual cosmid DNAs by direct AFM imaging. Genomics. 1997;41:379–384. doi: 10.1006/geno.1997.4686. [DOI] [PubMed] [Google Scholar]
- 168.Nakamura T, Maeda Y, Oka T, Tabata H, Futai M, Kawai T. Atomic force microscope observation of plasmid deoxyribose nucleic acid with restriction enzyme. J. Vacuum Sci. Technol. B. 1999;17:288–293. [Google Scholar]
- 169.Lewin B. Genes VII. Oxford: Oxford University Press; 1999. [Google Scholar]
- 170.Michalet X, et al. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science. 1997;211:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- 171.Dessinges MN, Maier B, Zhang Y, Peliti M, Bensimon D, Croquette V. Stretching single stranded DNA, a model polyelectrolyte. Phys. Rev. Lett. 2002;89(24) doi: 10.1103/PhysRevLett.89.248102. [DOI] [PubMed] [Google Scholar]
- 172.Allemand JF, Bensimon D, Croquette V. Stretching DNA and RNA to probe their interactions with proteins. Curr. Opin. Struct. Biol. 2003;13:26674. doi: 10.1016/s0959-440x(03)00067-8. [DOI] [PubMed] [Google Scholar]
- 173.Morii N, Kido G, Suzuki H, Nimori S, Morii H. Molecular chain orientation of DNA films induced by both the magnetic field and the interfacial effect. Biomacromolecules. 2004;5:2297–2307. doi: 10.1021/bm0496460. [DOI] [PubMed] [Google Scholar]
- 174.Gu Q, Cheng CD, Haynie DT. Cobalt metallization of DNA: toward magnetic nanowires. Nanotechnology. 2005;16:1358–1363. [Google Scholar]
- 175.Kawakami T, Taniguchi T, Hamamoto T, Kitagawa Y, Okumura M, Yamaguchi K. Possibilities of molecule-based spintoronics of DNA wires, sheets, and related materials. Int. J. Quantum Chem. 2005;105:655–671. [Google Scholar]
- 176.Lin HY, Tsai LC, Chi PY, Chen CD. Positioning of extended individual DNA molecules on electrodes by non-uniform AC electric fields. Nanotechnology. 2005;16:2738–2742. [Google Scholar]
- 177.Zhang JM, Ma YF, Stachura S, He HX. Assembly of highly aligned DNA strands onto Si chips. Langmuir. 2005;21:4180–4184. doi: 10.1021/la050129s. [DOI] [PubMed] [Google Scholar]
- 178.Gu Q, Cheng CD, Gonela R, Suryanarayanan S, Anabathula S, Dai K, Haynie DT. DNA nanowire fabrication. Nanotechnology. 2006;17:R14–R25. [Google Scholar]
- 179.Randall GC, Schultz KM, Doyle PS. Methods to electrophoretically stretch DNA: microcontractions, gels, and hybrid gel-microcontraction devices. Lab on a Chip. 2006;6:516–525. doi: 10.1039/b515326c. [DOI] [PubMed] [Google Scholar]
- 180.Shin M, Kim T, Kwon C, Kim SK, Park JB, Lee H. Alignment of A-DNA on organic monolayer surface patterned by scanning probe lithography. Japan. J. Appl. Phys. 1. 2006;45:2076–2081. [Google Scholar]
- 181.Terao K, Kabata H, Washizu M. Extending chromosomal DNA in microstructures using electroosmotic flow. J. Phys.: Condens. Matter. 2006;18:S653–S663. [Google Scholar]
- 182.Lewin B. Gene Expression. 2nd edn. NY: Wiley; 1980. [Google Scholar]
- 183.Sommer SS, Cohen JE. The size distributions of proteins, messenger-RNA and nuclear-RNA. J. Mol. Evol. 1980;15:37–57. doi: 10.1007/BF01732582. [DOI] [PubMed] [Google Scholar]
- 184.Draper MP, August PR, Connolly T, Packard B, Call KM. Efficient cloning of full-length cDNAs based on cDNA size fractionation. Genomics. 2002;79:603–607. doi: 10.1006/geno.2002.6738. [DOI] [PubMed] [Google Scholar]
- 185.Kuschel M. Analysis of messenger RNA using the Agilent 2100 Bioanalyzer and the RNA 6000 LabChip kit. Application Note. 2000 Agilent Technologies. [Google Scholar]
- 186.Marek J, Demjenova E, Tomori Z, Janacek J, Zolotova I, Valle F, Favre M, Dietler G. Interactive measurement and characterization of DNA molecules by analysis of AFM images. Cytometry A. 2005;63A:87–93. doi: 10.1002/cyto.a.20105. [DOI] [PubMed] [Google Scholar]
- 187.Ficarra E, Benini L, Macii E, Zuccheri G. Automated DNA fragments recognition and sizing through AFM image processing. IEEE Trans. Inform. Technol. Biomed. 2005;9:508–517. doi: 10.1109/titb.2005.855546. [DOI] [PubMed] [Google Scholar]
- 188.Woolley AT, Guillemette C, Cheung CL, Housman DE, Lieber CM. Direct haplotyping of kilobase-size DNA using carbon nanotube probes. Nat. Biotechnol. 2000;18:760–763. doi: 10.1038/77760. [DOI] [PubMed] [Google Scholar]
- 189.Fang Y, Spisz TS, Wiltshire T, D’Costa NP, Bankman IN, Reeves RH, Hoh JH. Solid-state DNA sizing by atomic force microscopy. Anal. Chem. 1998;70:2123–2129. doi: 10.1021/ac971187o. [DOI] [PubMed] [Google Scholar]
- 190.Wright DJ, King K, Modrich P. The negative charge of Glu-111 is required to activate the cleavage centre of ecori endonuclease. J. Biol. Chem. 1989;264:11816–11821. [PubMed] [Google Scholar]
- 191.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 192.Nastri HG, Evans PD, Walker IH, Riggs PD. Catalytic and DNA binding properties of P upsilon ull restriction endonuclease mutants. J. Biol. Chem. 1997;272:25761–25767. doi: 10.1074/jbc.272.41.25761. [DOI] [PubMed] [Google Scholar]
- 193.Bowen LM, Dupureur CM. Investigation of restriction enzyme cofactor requirements: a relationship between metal ion properties and sequence specificity. Biochemistry. 2003;42:12643–12653. doi: 10.1021/bi035240g. [DOI] [PubMed] [Google Scholar]
- 194.Martin AM, Horton NC, Lusetti S, Reich NO, Perona JJ. Divalent metal dependence of site-specific DNA binding by EcoRV endonuclease. Biochemistry. 1999;38:8430–8439. doi: 10.1021/bi9905359. [DOI] [PubMed] [Google Scholar]
- 195.Skiadas J, Aston C, Samad A, Anantharaman TS, Mishra B, Schwartz DC. Optical PCR: genomic analysis by long-range PCR and optical mapping. Mamm. Genome. 1999;10:1005–1009. doi: 10.1007/s003359901148. [DOI] [PubMed] [Google Scholar]
- 196.Phillips KM, Larson JW, Yantz GR, D’Antoni CM, Gallo MV, Gillis KA, Goncalves NM, Neely LA, Gullans SR, Gilmanshin R. Application of single molecule technology to rapidly map long DNA and study the conformation of stretched DNA. Nucleic. Acids Res. 2005;33:5829–5837. doi: 10.1093/nar/gki895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yokokawa M, Yoshimura SH, Naito Y, Ando T, Yagi A, Sakai N, Takeyasu K. Fast-scanning atomic force microscopy reveals the molecular mechanism of DNA cleavage by ApaI endonuclease. IEE Proc.-Nanobiotechnol. 2006;153:60–66. doi: 10.1049/ip-nbt:20050018. [DOI] [PubMed] [Google Scholar]
- 198.Uchihashi T, Kodera N, Itoh H, Yamashita H, Ando T. Feed-forward compensation for high-speed atomic force microscopy imaging of biomolecules. Japan. J. Appl. Phys. 1. 2006;45:1904–1908. [Google Scholar]
- 199.Onaran AG, Balantekin M, Lee W, Hughes WL, Buchine BA, Guldiken RO, Parlak Z, Quate CF, Degertekin FL. A new atomic force microscope probe with force sensing integrated readout and active tip. Rev. Sci. Instrum. 2006;77(2) [Google Scholar]
- 200.Kokavecz J, Marti O, Heszler P, Mechler A. Imaging bandwidth of the tapping mode atomic force microscope probe. Phys. Rev. B. 2006;73(15) [Google Scholar]
- 201.Kawai S, Kawakatsu H. Atomically resolved dynamic force microscopy operating at 4.7 MHz. Appl. Phys. Lett. 2006;88(13) [Google Scholar]
- 202.Kawai S, Kawakatsu H. Atomically resolved amplitude modulation dynamic force microscopy with a high-frequency and high-quality factor cantilever. Appl. Phys. Lett. 2006;89(1) [Google Scholar]
- 203.Jeong Y, Jayanth GR, Jhiang SM, Menq CH. Direct tip-sample interaction force control for the dynamic mode atomic force microscopy. Appl. Phys. Lett. 2006;88(20) [Google Scholar]
- 204.Jayanth GR, Jeong Y, Menq CH. Direct tip-position control using magnetic actuation for achieving fast scanning in tapping mode atomic force microscopy. Rev. Sci. Instrum. 2006;77(5) [Google Scholar]
- 205.Hobbs JK, Vasilev C, Humphris ADL. VideoAFM-a new tool for high speed surface analysis. Analyst. 2006;131:251–256. doi: 10.1039/b511330j. [DOI] [PubMed] [Google Scholar]
- 206.Beyder A, Spagnoli C, Sachs F. Reducing probe dependent drift in atomic force microscope with symmetrically supported torsion levers. Rev. Sci. Instrum. 2006;77(5) [Google Scholar]
- 207.Ando T, Uchihashi T, Kodera N, Miyagi A, Nakakita R, Yamashita H, Sakashita M. High-speed atomic force microscopy for studying the dynamic behaviour of protein molecules at work. Japan. J. Appl. Phys. 1. 2006;45:1897–1903. [Google Scholar]
- 208.Takahashi T, Ono S. Sample-and-hold atomic force microscopy for fast operation. Ultramicroscopy. 2005;105:42–50. [Google Scholar]
- 209.Stemmer A, Schitter G, Rieber JM, Allgower F. Control strategies towards faster quantitative imaging in atomic force microscopy. Eur. J. Control. 2005;11:384–395. [Google Scholar]
- 210.Salapaka S, De T, Sebastian A. Sample-profile estimate for fast atomic force microscopy. Appl. Phys. Lett. 2005;87(5) [Google Scholar]
- 211.McMaster TJ, Brayshaw D, Miles MJ, Walsby AE, Dunton P. A new ultra high speed AFM technique for biophysics: 3-dimensional imaging of surfaces, molecules and processes with true millisecond resolution. Biophys. J. 2005;88:541A–541A. [Google Scholar]
- 212.Kodera N, Yamashita H, Ando T. Active damping of the scanner for high-speed atomic force microscopy. Rev. Sci. Instrum. 2005;76(5) [Google Scholar]
- 213.Hobbs JK, Vasilev C, Humphris ADL. Real time observation of crystallization in polyethylene oxide with video rate atomic force microscopy. Polymer. 2005;46:10226–10236. [Google Scholar]
- 214.Schitter G, Allgower F, Stemmer A. A new control strategy for high-speed atomic force miciroscopy. Nanotechnology. 2004;15:108–114. [Google Scholar]
- 215.Schitter G, Stemmer A. Identification and open-loop tracking control of a piezoelectric tube, scanner for high-speed scanning-probe microscopy. IEEE Trans. Control Syst. Technol. 2004;12:449–454. [Google Scholar]
- 216.Schitter G, Stark RW, Stemmer A. Fast contact-mode atomic force microscopy on biological specimen by model-based control. Ultramicroscopy. 2004;100:253–257. doi: 10.1016/j.ultramic.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 217.Viani MB, Pietrasanta LI, Thompson JB, Chand A, Gebeshuber IC, Kindt JH, Richter M, Hansma HG, Hansma PK. Probing protein-protein interactions in real time. Nat. Struct. Biol. 2000;7:644–647. doi: 10.1038/77936. [DOI] [PubMed] [Google Scholar]
- 218.Viani MB, et al. Fast imaging and fast force spectroscopy of single biopolymers with a new atomic force microscope designed for small cantilevers. Rev. Sci. Instrum. 1999;70:4300–4303. [Google Scholar]
- 219.Ando T, Kodera N, Takai E, Maruyama D, Saito K, Toda A. A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl Acad. Sci. USA. 2001;98:12468–12472. doi: 10.1073/pnas.211400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Manalis SR, Minne SC, Quate CF. Atomic force microscopy for high speed imaging using cantilevers with an integrated actuator and sensor. Appl. Phys. Lett. 1996;68:871–873. [Google Scholar]
- 221.Rogers B, et al. High speed tapping mode atomic force microscopy in liquid using an insulated piezoelectric cantilever. Rev. Sci. Instrum. 2003;74:4683–4686. [Google Scholar]
- 222.Jackson DG, Screaton GR, Bell MV, Bell JI. Cd44 and cancer. Lancet. 1993;341:252. doi: 10.1016/0140-6736(93)90126-2. [DOI] [PubMed] [Google Scholar]
- 223.Matsumura Y, Tarin D. Significance of Cd44 gene-products for cancer-diagnosis and disease evaluation. Lancet. 1992;340:1053–1058. doi: 10.1016/0140-6736(92)93077-z. [DOI] [PubMed] [Google Scholar]
- 224.Vandenberg ET, Bertilsson L, Liedberg B, Uvdal K, Erlandsson R, Elwing H, Lundstrom I. Structure of 3-aminopropyl triethoxy silane on silicon-oxide. J. Colloid Interface Sci. 1991;147:103–118. [Google Scholar]
- 225.Bunker BC, Carpick RW, Assink RA, Thomas ML, Hankins MG, Voigt JA, Sipola D, de Boer MP, Gulley GL. The impact of solution agglomeration on the deposition of self-assembled monolayers. Langmuir. 2000;16:7742–7751. [Google Scholar]
- 226.Schwartz DK. Mechanisms and kinetics of self-assembled monolayer formation. Annu. Rev. Phys. Chem. 2001;52:10737. doi: 10.1146/annurev.physchem.52.1.107. [DOI] [PubMed] [Google Scholar]
- 227.Zhang FX, Srinivasan MP. Self-assembled molecular films of aminosilanes and their immobilization capacities. Langmuir. 2004;20:2309–2314. doi: 10.1021/la0354638. [DOI] [PubMed] [Google Scholar]
- 228.Nishiyama N, Ishizaki T, Horie K, Tomari M, Someya M. Novel polyfunctional silanes for improved hydrolytic stability at the polymer silica interface. J. Biomed. Mater. Res. 1991;25:213–221. doi: 10.1002/jbm.820250208. [DOI] [PubMed] [Google Scholar]
- 229.Parikh AN, Allara DL, Azouz IB, Rondelez F. An intrinsic relationship between molecular-structure in self-assembled N-alkylsiloxane monolayers and deposition temperature. J. Phys. Chem. 1994;98:7577–7590. [Google Scholar]
- 230.Mohsen NM, Craig RG. Hydrolytic stability of silanated zirconia–silica-urethane dimethacrylate composites. J. Oral Rehab. 1995;22:213–220. doi: 10.1111/j.1365-2842.1995.tb01566.x. [DOI] [PubMed] [Google Scholar]
- 231.Craig RG, Dootz ER. Effect of mixed silanes on the hydrolytic stability of composites. J. Oral Rehab. 1996;23:751–756. doi: 10.1046/j.1365-2842.1996.d01-194.x. [DOI] [PubMed] [Google Scholar]
- 232.Yim H, Kent MS, Hall JS, Benkoski JJ, Kramer EJ. Probing the structure of organosilane films by solvent swelling and neutron and x-ray reflection. J. Phys. Chem. B. 2002;106:2474–2481. [Google Scholar]
- 233.Pan GR, Yim H, Kent MS, Majewski J, Schaefer DW. Neutron reflectivity investigation of bis-amino silane films. J. Adhes. Sci. Technol. 2003;17:2175–2189. [Google Scholar]
- 234.Benkoski JJ, Kramer EJ, Yim h, kent MS, Hall J. The effects of network structure on the resistance of silane coupling agent layers to water-assisted crack growth. Langmuir. 2004;20:3246–3258. doi: 10.1021/la035920k. [DOI] [PubMed] [Google Scholar]
- 235.Marcinko S, Fadeev AY. Hydrolytic stability of organic monolayers supported on TiO2 and ZrO2. Langmuir. 2004;20:2270–2273. doi: 10.1021/la034914l. [DOI] [PubMed] [Google Scholar]
- 236.Zhu DQ, van Ooij WJ. Enhanced corrosion resistance of AA 2024-T3 and hot-dip galvanized steel using a mixture of bis-triethoxysilylpropyl tetrasulfide and bis-trimethoxysilylpropyl amine. Electrochim. Acta. 2004;49:1113–1125. [Google Scholar]
- 237.Yim H, Kent MS, Tallant DR, Garcia MJ, Majewski J. Hygrothermal degradation of (3-glycidoxypropyl)trimethoxysilane films studied by neutron and x-ray reflectivity and attenuated total reflection infrared spectroscopy. Langmuir. 2005;21:4382–4392. doi: 10.1021/la0474870. [DOI] [PubMed] [Google Scholar]
- 238.Pan GR, Schaefer DW. Morphology and water-barrier properties of silane films on aluminum and silicon. Thin Solid Films. 2006;503:259–267. [Google Scholar]