Abstract
Ubiquitously found in the extracellular matrix and attached to the surface of most cells, glycosaminoglycans (GAGs) mediate many intercellular interactions. Originally described in 1889 as the primary carbohydrate in cartilage and then in 1916 as a coagulation inhibitor from liver, various GAGs have since been identified as key regulators of normal physiology. GAGs are critical mediators of differentiation, migration, tissue morphogenesis, and organogenesis during embryonic development. While GAGs are simple polysaccharide chains, many GAGs acquire a considerable degree of complexity by extensive modifications involving sulfation and epimerization. Embryos that lack specific GAG modifying enzymes have distinct developmental defects, illuminating the importance of GAG complexity. Revealing how these complex molecules specifically function in the embryo has often required additional approaches, the results of which suggest that GAG modifications might instructively mediate embryonic development.
1. Introduction
GAGs are long, linear polysaccharides composed of an amino sugar and uronic acid repeating as a disaccharide unit. Variations in disaccharide identity distinguish four classes of GAGs: heparan sulfate/heparin (HS), chondroitin sulfate/dermatan sulfate (CS/DS), keratan sulfate (KS), and hyaluronan (HA, Fig 1A). Additional differences in disaccharide linkage and modification result in each GAG class assuming a unique macrostructure [1]. While HA is synthesized at the plasma membrane where it is not modified or covalently attached to a core protein, HS, CS/DS, and KS are covalently linked to core proteins and modified by sulfation and epimerization during synthesis in the golgi. Together known as proteoglycans, sulfated GAGs and their attached core proteins are presented on the cell surface, stored in secretory granules, or secreted into the extracellular matrix.
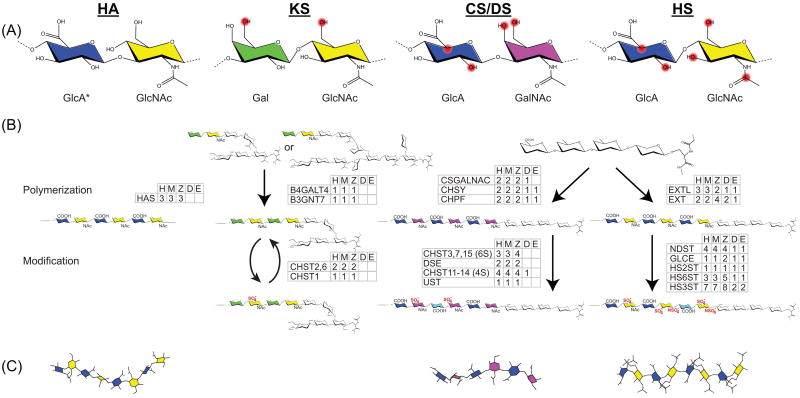
Figure 1. Interrelated biosynthesis of GAG classes yields similar yet distinct structures.
(A) GAGs are classified based on their disaccharide building blocks. Potentially modified positions are highlighted in red. (B) Numerous enzymes catalyze GAG elongation and modification. Families of enzymes involved in each step are listed with the number of homologs from humans-H compared to four models organisms: mouse-M, zebrafish-Z, Drosophila -D, and C elegans-E. Saccharides that link KS to core proteins are quite diverse and simplified here for clarity. Descriptions of each gene family along with specific gene IDs are in the Supplemental. (C) GAGs adopt distinct helical conformations. Illustrations were generated using Chem3D Pro with the pdb files 2BVK (HA), 2KQO (CS), and 1HPN (Heparin).
*Saccharides and modifications are abbreviated using the following nomenclature: GluA, β-D-glucuronic acid; GlcNAc, α/β-D-glucosamine; GalNAc, β-D-galactosamine; Gal, β-D-galactose IdoA, α-L-iduronic acid; NAc, N-acetylation; SO4−, sulfate. Specific disaccharides are condensed. For example, the disaccharide containing glucuronic acid linked to a 4-O and 6-O sulfated galactosamine is represented by GlcA-GalNAc4S, 6S.
Structurally diverse GAG chains bind and regulate the activity of a wide range of protein ligands. In a well-characterized model, HS binds various cell-cell signaling ligands, regulating both how the cell-cell signaling ligands interact with their receptors and how they form concentration gradients [2]. Recent evidence suggests that CS/DS can employ similar roles as well [3, 4]. Furthermore, distinct classes of GAGs can regulate how ligands function in different ways. For instance, all sulfated GAGs interact with collagens and mediate matrix organization: KS increases collagen spacing to aid in light transmission through the cornea [5], CS/DS decreases collagen fibril size to increase the tensile strength of skin [6], and HS remodels collagen matrices to facilitate migration [7]. Modifications on particular GAG chains add an additional level of control as embryos that lack specific GAG modifying enzymes often closely resemble others in which specific developmental signaling pathways have been blocked [8]. While these observations suggest that distinct GAG modifications might provide an extracellular code that helps direct development [8], demonstrating clear structure–function relationships has proven elusive.
A paradigm for GAG binding specificity is the interaction between antithrombin and a distinct HS pentasaccharide, a target for most heparin-related drugs [9]. Among other modifications, a specific 3-O sulfation dramatically increases the affinity of HS/heparin for antithrombin yet can be dispensable for antithrombin activation [10]. The connection between antithrombin-GAG binding and function is important clinically as a number of patient deaths were associated with heparin doses found to be contaminated with chemically-oversulfated CS [11], a GAG that can bind antithrombin but cannot stimulate antithrombin-mediated coagulation [12]. Similarly, multiple HS modifications can mediate FGF binding to HS, but specific HS modifications appear to be required to form a functional signaling complex [13]. Thus, GAG modifications that mediate ligand binding can be separated from those that mediate ligand activity. To identify GAG modifications that mediate ligand activity, multiple labs are analyzing the developing embryo which, as Viktor Hamburger suggested, may be “the only teacher who is always right” [14]. This review examines four systems where genetic loss of multiple modifying enzymes results in overlapping phenotypes. A recurring theme emerges: Distinct phenotypes are observed when the phenotype can be analyzed as several traits, suggesting that GAG classes and even specific GAG modifications do not appear to be functionally redundant but complement each other.
2. GAG biosynthesis of diverse structures
2.1. Searching for a relationship between GAG fine structures and ligand binding
GAG biosynthesis consists of several polymerization and modification steps (see Supplemental for overview of biosynthesis with enzyme descriptions). The structural variability of sulfated GAG chains is generated by multiple modifying enzymes that expanded in number during early vertebrate evolution (Fig 1B). Sulfated GAGs are thus endowed with an impressive diversity of possible disaccharides: 4 KS disaccharides, 16 CS/DS and 48 HS. However, the observed diversity is less than what is theoretically possible as only two-thirds of the HS disaccharides have been identified and homopolymeric repeats tend to predominate, particularly in KS and CS/DS. Synthesis is not template-driven, enzyme reactions do not go to completion, and substrate specificity is generally imprecise, creating heterogeneous chains with unique sulfation patterns that are challenging to characterize. The most common GAG analysis approach is to depolymerize the GAG chain and quantitate the disaccharides, akin to reading a sentence from a list of letters. This technique has revealed that disaccharide content differs between model organisms, between organs, and during different stages of development [15–18]. Curiously, no new disaccharides were observed in vertebrates that might be predicted with the significant increase in the number of modifying enzymes [16]. Since ligand binding sites typically range from a trisaccharide to a dodecasaccharide [19], disaccharide analysis might mask important patterns of sulfation. Commonly referred to as GAG fine structure, antibodies and comparable approaches have revealed that disaccharides in KS, CS\DS, and HS GAG chains are organized into domains, that domains have distinct ligand-binding sites, and that these epitopes vary between cell types [20–22].
Little is known about the mechanisms regulating the biosynthesis of GAG fine structure. There is evidence that substrate availability can have a role [23, 24], but GAG classes appear to be affected differently [25, 26]. In early Drosophila development, protein translation of HS biosynthetic enzymes is temporally controlled through internal ribosome entry sites, a mechanism that appears to be conserved in vertebrates and possibly CS biosynthesis as well [27–29]. Esko and Selleck suggested in a 2002 review that biosynthetic enzymes might form physical complexes or GAGosomes, whereby different golgi complexes would synthesize distinct GAG domains [30]. Physical association between enzymes has been demonstrated between Gpce and Hs2st1 [31] as well as between Ext2 and Ndst1 [32]. The later work also demonstrated that the level of Ext1 and Ext2 expression affected the amount of active Ndst1 in the cell, which consequently influenced HS structure. Further support for the GAGosome model also came from Kuberan and colleagues who synthesized an array of xylosides and found that the xylosides could initiate synthesis of GAG chains with distinct fine structures [33]. Defining how these xylosides affect developmental pathways might better define whether GAGosomes and GAG fine structure are functionally important.
The majority of GAG-binding proteins interact with single or multiple sulfated domains, and protein binding to HS appears to fall into two types: specific interactions with rare modifications and less-specific interactions with common modifications [34, 35]. The rare HS 3-O-sulfation is specifically required by antithrombin in coagulation, herpes simplex gD glycoprotein in viral invasion, and Notch in Drosophila neurogenesis [36, 37]. Similarly, two rare DS disaccharides, IdoA2S-GalNAc4S and IdoA-GalNAc4S,6S, can mediate binding and activation of heparin cofactor II in coagulation [38–40]. These examples of specific binding are more the exception than the rule: hepatocyte growth factor, most FGF ligands, and numerous other proteins bind both HS and CS/DS [41] [35, 42]. Recent proteomic analysis suggests that KS can bind many of these same factors [43]. Hence, biochemical and histochemical analyses suggest that fine structures exist, but ligand binding assays indicate that it is generally not utilized.
So why have a fine structure? Analysis of transcription factor binding to DNA might lend some insight as a model for GAG binding, since evidence indicates that low affinity transcription factor binding sites are functionally important [44]. Using comprehensive protein binding microarray data with genomic analysis, 41 of the 104 transcription factors examined have distinct secondary DNA binding site motifs which were just as evolutionarily conserved as the primary binding sites [45]. Moreover, dramatic changes in binding affinity of six transcription factors between Drosophila species only weakly correlated with changes in function [46]. These results demonstrate that the spatial proximity of transcription factor binding sites to each other and the transcription start site are more critical than actual affinity. An analogy might be made to GAGs: Highly sulfate domains in HS are often longer at the non-reducing end and may be important in FGF signaling [47, 48]. Additionally, binding sites for a ligand and its receptor might need to be in close proximity on a single chain [49], or the GAG might need to be expressed on a particular core protein [50]. An alternative model is that GAGs are not merely a binding scaffold, as the catalytic role of HS in fibronectin fibrillogenesis suggests [51, 52]. Further detailed analysis of mutant embryos should help clarify how GAGs are functioning.
2.2. Diverse core proteins aid in mediating function
The core proteins to which sulfated GAGs are attached dictate GAG location and, in part, their function. At least 11 KS, 30 CS/DS, and 15 HS proteoglycans have been identified in vertebrates. Multiple GAG classes can be simultaneously attached to some core proteins, such as HS, CS and KS can attach to Perlecan [53]. Other core proteins support distinct GAG chains depending on the tissue in which it is expressed. For example, Serglycin is extensively modified by the oversulfated HS heparin in mast cells from connective tissue while mast cells from mucosa and lung have CS attached [54]. The amino acid diversity of core proteins has made it challenging to identify homologs in lower vertebrates and invertebrates; but a proteomics approach using C. elegans identified 9 secreted CS proteoglycans that have no homology to vertebrate CS proteoglycans, suggesting that core proteins have evolved to serve roles that are distinct to the organism [29].
Some core protein functions are restricted to a specific core protein, such as the ability to directly bind the same ligand as the GAG or bind a receptor that might mediate the function of the ligand [55]. More generally, core proteins serve to concentrate and localize the GAG chains. Multiple GAG attachment sites are clustered as serine-glycine repeats on CS and HS core proteins [56], and some core proteins can support a significant number of GAGs (Aggrecan contains about 100 CS and 60 KS attachment sites). Multivalent GAGs usually serve to enhance the action of a single chain, but there are examples where multiple sites are required for function, as in the ability of syndecan-4 to regulate cell migration [57]. Similarly, the Drosophila Glypican Dally-like protein (Dlp) can stimulate Wnt signaling at low levels while inhibiting it at higher levels, emphasizing that the amount of the core protein can be critical to how it functions [58].
Location appears to be a critical element for how the Glypicans Dlp and Dally regulate Hedgehog signaling [59]. Dlp and Dally are glycophosphatidylinositol (GPI)-linked to the apical surface of the wing imaginal disc epithelium, but their paths diverge from there. Dlp is rapidly endocytosed with Hedgehog and its receptor Patched, cell-autonomously mediating Hedgehog signal transduction [60]. In contrast, Dally binds Hedgehog at the apical surface but is shed by the GPI-hydrolyase Notum, mediating long-range hedgehog signaling [61]. The mechanism by which these closely-related core proteins are recognized for discrete trafficking remains to be determined. Nevertheless, both Dlp and Dally can be blocked by transmembrane versions of these GPI proteins, raising the possibility that trafficking is governed by interactions that occur within lipid rafts. In mice, Glypican-1 also localizes to lipid rafts during myogenesis, sequestering fibroblast growth factor 2 (FGF2) away from the transmembrane proteoglycans [62]. FGF-signaling is thus decreased and myoblast differentiation is promoted, further emphasizing that subcellular localization is a recurring mechanism for mediating proteoglycan function. Hence, most core proteins have distinct functions during development.
3. GAGs have complementary roles in embryonic development and homeostasis
C. elegans and mice that lack CS die in early development as they fail to undergo cytokinesis [63, 64]. HS-deficient C. elegans and mice arrest during gastrulation [65, 66]. Has2(−/−) mice lack HA and die during mid-gestation with severe cardiac and vascular defects [67]. Clearly, GAGs are required for normal embryogenesis. To begin to assess how specific GAG modifications mediate development, examples are reviewed where several modifying enzymes appear to have overlapping roles in four systems. When detailed phenotypes of each of these systems is carefully dissected, specific GAG modifications repeatedly have distinct, complementary roles.
3.1 Branching morphogenesis
FGFs require HS for high affinity binding to their receptors and subsequent signaling, a finding that established a basis for understanding how GAGs function [68, 69]. These initial cell culture observations have since been supported by results from HS-deficient mice, zebrafish, and Drosophila [7, 70, 71]. FGF-mediated branching of epithelia in different tissues is particularly sensitive to the loss of HS. In the budding Drosophila trachea, HS is not essential for the secretion and distribution of the FGF ligand, but does regulate FGF signaling in the receiving cells as a co-receptor [72]. To elucidate how specific HS modifications might regulate FGF signaling, trachea were analyzed in Hs2st or Hs6st mutant embryos, but no phenotype was observed [73]. Biochemical analysis of the mutant embryos revealed that 2-O sulfation was increased in Hs6st mutants and 6-O sulfation was increased in Hs2st mutants, suggesting that increasing sulfation at one position could compensate for the loss at a different position. In Hs2st- Hs6st double mutants, tracheal branching morphogenesis was severely disrupted, thus supporting a model that overall charge density is sufficient to mediate FGF signal reception in some developmental contexts.
FGF10 is required for branching morphogenesis in multiple vertebrate tissues [74]. Using chemically-defined derivates of HS, Hoffman and colleagues reported that FGF10-induced elongation of submandibular explants required 6-O-sulfation [75]. In contrast, end bud branching of the explants required 2-O sulfation together with either an N-or 6-O-sulfate. The results suggest that different HS modifications mediate two distinct aspects of branching morphogenesis, a model that is supported by experiments with Hs2st and Ndst1 mutant mice [76, 77]. In explants from Hs2st mutant mice, branching of the uteric bud occurred normally, but kidney morphogenesis failed as the epithelia could not respond to terminal differentiation cues [76]. Similarly, Ndst1-null mammary epithelia undergo initial branching morphogenesis, but milk production failed due to defects in lobuloalveolar expansion [77]. Both of these phenotypes arise from cell-autonomous defects in HS, supporting results from Drosophila work suggesting that HS mediates FGF signal reception at the epithelial cell surface. To better understand how FGF10 functions, the HS-binding site in FGF10 was mutated to resemble that of FGF7, a ligand that binds poorly to HS [78]. Lacrimal and submandibular explants treated with the FGF10-HS-binding mutant only branched and failed to elongate, similar to FGF7-treated explants. In contrast, when the FGF receptor binding site in FGF10 was mutated, there was a reduction in the extent of explant elongation, but not in the type of response. Notably, the profile of genes expressed in submandibular explants that were induced by the FGF10-HS-binding mutant resembled that of FGF7. Taken together, these data suggest that interactions with HS mediate how FGF10 forms a morphogen gradient, independently from its interaction with the FGF receptor. Knock-in replacement of the endogenous genes with FGF10 mutant version will help further clarify the mechanism.
3.2 Stem cells
Sonic Hedgehog (Shh) has two general activities during development; it stimulates stem cell proliferation and patterns multiple tissues, including the limbs and central nervous system. To characterize how vertebrate Hedgehog signaling is mediated by its interaction with GAGs, shh was replaced in mice with a mutant that has minimal interactions with GAGs but normal receptor binding [79]. Unexpectedly, limb and central nervous system patterning was normal in mutant mice while shh-induced proliferation was defective. Analysis of stem cells from neuronal mitogenic niches indicated that gli2 signaling was defective, leading to a selective increase in the expression of genes associated with proliferation. Similarly, the Glypicans Dally and Dlp are required for maintaining the female and male germline stem cell niche, respectively [80, 81]. Which GAGs regulate stem cell proliferation is unclear, but HS and CS both appear to have roles in mediating stem cell differentiation. Embryonic stem cells (ESCs) deficient in HS from either Ext1(−/−) or Ndst1/2(−/−) null mice fail to differentiate [82, 83]. Two sets of experiments suggest that distinct GAG modifications regulate stem cell differentiation. ESCs expressing a specific HS epitope have increased potential to differentiate into hematopoietic cells [84]. Likewise, a unique CS epitope defines a population of multipotent neural progenitors in developing and adult mice [85]. Further analysis of ESCs from various mutant mice should help elucidate how the loss of specific modifying enzymes affects the developmental potential of ESCs.
3.3 Axon pathfinding
Work from several labs has demonstrated that distinct GAG modifications stimulate or inhibit neuron migration while yet other GAG modifications can actively direct axon pathfinding. Hsieh-Wilson and colleagues synthesized a series of homogeneous CS tetrasaccharides that differ in their sulfation positions [86]. Tetrasaccharides bearing a distinct GlcA-GalNAc4S6S repeat (CSE) were found to stimulate the outgrowth of various neuron types in cell culture while other closely related tetrasaccharides like GlcA-GalNAc4S that are found prominently in the brain had no effect. The CSE activity was directly associated with the binding of growth factors. Inhibiting growth factor-CSE interaction blocks the stimulatory effect on neurite outgrowth. These observations correlate well with experiments in mouse embryos: Migration of cortical neurons is disrupted when the enzyme responsible for synthesizing CSE, Chst15, is reduced [87]. When another CS-modifying enzyme, Chst14, is targeted, a comparable disruption is not observed. Knocking-down a different CS-modifying enzyme, Chst11, disrupts motor axon migration during zebrafish development [88].
Paradoxically, CS can also inhibit neuronal migration. After injury to the adult central nervous system, glial scar tissue rich in CS accumulates around the wound site, forming a barrier to axonal regeneration [89]. Enzymatically digesting the CS/DS chains in the lesion promotes the regeneration of neurons in the spinal cord [90], and CS/DS has since been the focus of several clinical approaches [91]. Nonetheless, Chst2(−/−) mice that are defective in KS in the central nervous system showed improved recovery of motor function following spinal cord injury [92]. Chst2(−/−) mice also had reduced glial scars and improved axon growth, and in vitro explants suggested that CS and KS can functionally overlap in this general activity.
HS can also mediate migration of vertebrate neurons, despite the increased cellular complexity and redundancy of HS-modifying enzymes. Migration defects in retinal ganglion cells (RGCs) and longitudinal neurons are found in three distinct zebrafish mutants that are all deficient in HS biosynthesis [93] [94]. Ext1(−/−) mice have markedly disorganized neurons throughout the brain [95], while mice that are mutant for two HS modifying enzymes have distinct learning and motor defects [96]. Analysis of RGCs in Hs2st(−/−) and Hs6st1(−/−) mice revealed that they have different axon pathfinding defects: Hs2st(−/−) RGCs are disorganized around the optic chiasm while Hs6st1(−/−) RGCs cross the chiasm normally but have abnormal project to the eye [97]. Furthermore, only axons from Hs6st1(−/−) mice had a decreased sensitivity to Slit2, suggesting that different HS modifications mediate how vertebrate RGCs respond to distinct migratory cues.
CS and HS can cooperatively, yet distinctly, regulate axon guidance [98]. In Drosophila, CS attached to the transmembrane proteoglycan Syndecan is required for axon migration across the ventral midline by specifically regulating Slit signal reception in the axon. In contrast, the Glypican Dlp to which HS is attached mediates axon guidance cell non-autonomously, possibly regulating the transport of Slit from the secreting ventral midline cells to the receiving axons. Bulow and colleagues used genetic approaches in C. elegans to demonstrate that specific neuronal subtypes require different combinations of HS-modifying enzymes: Some neurons require Glce, Hs2st, and Hs6st for proper axon guidance, other neurons require only the Glce and Hs2st, while still others do not require any HS modifying enzymes [99]. By combining mutants with targeted misexpression, specific neuronal subtypes were redirected, demonstrating that specific HS modifications are both necessary and sufficient to direct axon migration. Because of their detailed perturbations and analyses, Bulow and colleagues concluded that distinct combinations of HS modifications can instruct axon guidance. This important distinction between permissive and instructive implies that GAGs may have a more active role in directing development and should be rigorously defined in other systems.
3.4 Cardiovascular system
During heart development, endocardial cells normally migrate and differentiate into a functional valve, a process that is under the control of multiple GAGs. HA, CS/DS, and HS are all likely to be affected in the zebrafish mutant jekyll, which is defective of the enzyme responsible for UDP-GlcA synthesis [100]. Has2(−/−) mice have a strikingly similar phenotype, and transformation of endothelial cells into migratory mesenchymal cells was rescued in Has2(−/−) explants by exogenous HA [101]. HS appears to have a somewhat distinct role through its high affinity for HB-EGF, as replacing HB-EGF in mice with a mutant that has minimal interactions with GAGs but normal receptor binding develop enlarged cardiac valves [102]. CS also mediates atrioventricular valve formation as knockdown of CHSY1 or a small molecule inhibitor of CS synthesis in zebrafish results in similar defects [103]. The role of CS in heart development is supported as atrial septal and ventricular septal defects are associated with humans that carry mutations in Chst14 and Chst3, respectively [104, 105]. Microinjecting HS derivatives into the chick heart field suggests that N- and 6-O-sulfation are distinctly involved in heart looping [106], but these studies have not been corroborated by genetic analyses. These results taken together suggest that GAG classes have complementary roles in heart development.
Studies into how GAGs regulate angiogenesis and vasculogenesis tend to focus on the ability of HS to regulate the bioactivity of VEGF165, a critical angiogenic factor. Angiogenesis is blocked by peptides derived from the HS binding site in VEGF165 and small molecules that inhibit VEGF165-binding to HS [107, 108]. In vitro binding studies demonstrated that 6-O sulfation appeared to be particularly important [109]. Blocking the expression of Hs6st-2 but not Hs6st-1 in zebrafish embryos decreased vascular branching; Hs6st-1(−/−) mice have vascular defects and die at birth [110, 111]. Vasculogenesis was also impaired in NDST1-deficient zebrafish, and VEGF165 fails to induce vessel formation in Ndst1/2(−/−) embryoid bodies [112, 113]. While loss of Ndst1 disrupts both N- and O-sulfation, the removal of 6-O-sulfation by the sulfatase Sulf2 abolishes the ability of HS to bind VEGF165, supporting a model where 6-O-sulfation of HS is a key mediator of VEGF165 function and, consequently, vascular development [114, 115].
4. Conclusions and challenges
Robust phenotypes are readily observed when an entire class of GAGs is blocked, but studying changes in fine structure requires analysis with fine resolution. Detailed dissection of phenotypes from GAG modifying enzymes frequently reveals that developmental decisions require specific GAG modifications. Yet examples still arise when several GAG structures functionally overlap. At present, the developing embryo appears to use both redundant and specific GAG modifications, the preponderance of which still needs to be determined.
Further characterizing how specific GAG modifications mediate development will be aided by finer perturbations and finer analyses. Mutating GAG binding sites in ligands will continue to help define how their interactions with GAGs mediate their function. So too will substrate-specific inhibitors and/or mutations in modifying enzymes that limit or change substrate specificity. Sulfotransferase mutants with distinct substrate specificity are certainly of value in synthesizing unique GAGs using microfluidic chips or related technologies [116, 117], but replacing endogenous sulfotransferases with substrate-restricted mutants also might help unravel the role of specific HS epitopes. A systems approach will be better realized once a better understanding of what the GAG fine structure looks like and how it is generated. Arrays of defined GAGs synthesized from modular building blocks or naturally diverse libraries will certainly help refine the understanding of GAG-binding specificity [118, 119], but careful in vivo analysis will still be required [120]. An array of specific antibodies to complement the GAG array might seem farfetched, but so did the initial 60+step synthesis of the antithrombin-binding pentasaccharide that changed perspectives on what is required for specific GAG binding [121]. Thus, it is likely through combined advances in numerous disciplines that that we might better understand GAGs complexity.
Supplementary Material
Acknowledgments
I appreciated the constructive comments from Keji Zhao, Matt Hoffman, Charley Tharp, and Ashok Srinivasan. Research in my laboratory is funded by the intramural program of NHLBI, NIH. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sasisekharan R, Raman R, Prabhakar V. Glycomics approach to structure-function relationships of glycosaminoglycans. Annual Review of Biomedical Engineering. 2006;8:181–231. doi: 10.1146/annurev.bioeng.8.061505.095745. [DOI] [PubMed] [Google Scholar]
- 2.Yan D, Lin X. Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol. 2009;1:a002493. doi: 10.1101/cshperspect.a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sirko S, Holst Av, Weber A, Wizenmann A, Theocharidis U, Götz M, et al. Chondroitin Sulfates Are Required for Fibroblast Growth Factor-2-Dependent Proliferation and Maintenance in Neural Stem Cells and for Epidermal Growth Factor-Dependent Migration of Their Progeny. Stem Cells. 2010;28:775–87. doi: 10.1002/stem.309. [DOI] [PubMed] [Google Scholar]
- 4.Gualeni B, Facchini M, De Leonardis F, Tenni R, Cetta G, Viola M, et al. Defective proteoglycan sulfation of the growth plate zones causes reduced chondrocyte proliferation via an altered Indian hedgehog signalling. Matrix Biol. 2010:11. doi: 10.1016/j.matbio.2010.05.001. In Press. [DOI] [PubMed] [Google Scholar]
- 5.Hayashida Y, Akama TO, Beecher N, Lewis P, Young RD, Meek KM, et al. Matrix morphogenesis in cornea is mediated by the modification of keratan sulfate by GlcNAc 6-O-sulfotransferase. Proc Natl Acad Sci U S A. 2006;103:13333–8. doi: 10.1073/pnas.0605441103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maccarana M, Kalamajski S, Kongsgaard M, Magnusson SP, Oldberg A, Malmstrom A. Dermatan sulfate epimerase 1-deficient mice have reduced content and changed distribution of iduronic acids in dermatan sulfate and an altered collagen structure in skin. Mol Cell Biol. 2009;29:5517–28. doi: 10.1128/MCB.00430-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osterholm C, Barczyk MM, Busse M, Gronning M, Reed RK, Kusche-Gullberg M. Mutation in the heparan sulfate biosynthesis enzyme EXT1 influences growth factor signaling and fibroblast interactions with the extracellular matrix. J Biol Chem. 2009;284:34935–43. doi: 10.1074/jbc.M109.005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bulow HE, Hobert O. The molecular diversity of glycosaminoglycans shapes animal development. Annu Rev Cell Dev Biol. 2006;22:375–407. doi: 10.1146/annurev.cellbio.22.010605.093433. [DOI] [PubMed] [Google Scholar]
- 9.Petitou M, Nancy-Portebois V, Dubreucq G, Motte V, Meuleman D, De Kort M, et al. From heparin to EP217609: The long way to a new pentasaccharide-based neutralisable anticoagulant with an unprecedented pharmacological profile. Thrombosis and Haemostasis. 2009;102:804–10. doi: 10.1160/TH09-01-0063. [DOI] [PubMed] [Google Scholar]
- 10.Richard B, Swanson R, Olson ST. The Signature 3-O-Sulfo Group of the Anticoagulant Heparin Sequence Is Critical for Heparin Binding to Antithrombin but Is Not Required for Allosteric Activation. Journal of Biological Chemistry. 2009;284:27054–64. doi: 10.1074/jbc.M109.029892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerrini M, Beccati D, Shriver Z, Naggi A, Viswanathan K, Bisio A, et al. Oversulfated chondroitin sulfate is a contaminant in heparin associated with adverse clinical events. Nature Biotechnology. 2008;26:669–75. doi: 10.1038/nbt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B, Suwan J, Martin JG, Zhang F, Zhang Z, Hoppensteadt D, et al. Oversulfated chondroitin sulfate interaction with heparin-binding proteins: New insights into adverse reactions from contaminated heparins. Biochemical Pharmacology. 2009;78:292–300. doi: 10.1016/j.bcp.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashikari-Hada S, Habuchi H, Sugaya N, Kobayashi T, Kimata K. Specific inhibition of FGF-2 signaling with 2-O-sulfated octasaccharides of heparan sulfate. Glycobiology. 2009;19:644–54. doi: 10.1093/glycob/cwp031. [DOI] [PubMed] [Google Scholar]
- 14.Noden DM. Viktor Hamburger (1900–2001) Trends in Neurosciences. 2001;24:673–4. doi: 10.1016/s0166-2236(00)01961-5. [DOI] [PubMed] [Google Scholar]
- 15.Ledin J, Staatz W, Li J-P, Gotte M, Selleck S, Kjellen L, et al. Heparan Sulfate Structure in Mice with Genetically Modified Heparan Sulfate Production. Journal of Biological Chemistry. 2004;279:42732–41. doi: 10.1074/jbc.M405382200. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence R, Olson SK, Steele RE, Wang L, Warrior R, Cummings RD, et al. Evolutionary differences in glycosaminoglycan fine structure detected by quantitative glycan reductive isotope labeling. J Biol Chem. 2008;283:33674–84. doi: 10.1074/jbc.M804288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi X, Zaia J. Organ-specific heparan sulfate structural phenotypes. J Biol Chem. 2009;284:11806–14. doi: 10.1074/jbc.M809637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada S, Onishi M, Fujinawa R, Tadokoro Y, Okabayashi K, Asashima M, et al. Structural and functional changes of sulfated glycosaminoglycans in Xenopus laevis during embryogenesis. Glycobiology. 2009;19:488–98. doi: 10.1093/glycob/cwp005. [DOI] [PubMed] [Google Scholar]
- 19.Gandhi NS, Mancera RL. The structure of glycosaminoglycans and their interactions with proteins. Chemical Biology & Drug Design. 2008;72:455–82. doi: 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 20.Gallagher JT. Multiprotein signalling complexes: regional assembly on heparan sulphate. Biochem Soc Trans. 2006;34:438–41. doi: 10.1042/BST0340438. [DOI] [PubMed] [Google Scholar]
- 21.Fosang AJ, Last K, Poon CJ, Plaas AH. Keratan sulphate in the interglobular domain has a microstructure that is distinct from keratan sulphate elsewhere on pig aggrecan. Matrix Biol. 2009;28:53–61. doi: 10.1016/j.matbio.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Pacheco B, Maccarana M, Malmstrom A. Dermatan 4-O-sulfotransferase 1 is pivotal in the formation of iduronic acid blocks in dermatan sulfate. Glycobiology. 2009;19:1197–203. doi: 10.1093/glycob/cwp110. [DOI] [PubMed] [Google Scholar]
- 23.Carlsson P, Presto J, Spillmann D, Lindahl U, Kjellen L. Heparin/Heparan Sulfate Biosynthesis. Journal of Biological Chemistry. 2008;283:20008–14. doi: 10.1074/jbc.M801652200. [DOI] [PubMed] [Google Scholar]
- 24.Woloszynek JC, Kovacs A, Ohlemiller KK, Roberts M, Sands MS. Metabolic Adaptations to Interrupted Glycosaminoglycan Recycling. Journal of Biological Chemistry. 2009;284:29684–91. doi: 10.1074/jbc.M109.020818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sainio A, Jokela T, Tammi MI, Jarvelainen H. Hyperglycemic conditions modulate connective tissue reorganization by human vascular smooth muscle cells through stimulation of hyaluronan synthesis. Glycobiology. 2010:20. doi: 10.1093/glycob/cwq076. In Press. [DOI] [PubMed] [Google Scholar]
- 26.Bishop JR, Foley E, Lawrence R, Esko JD. Insulin-dependent diabetes mellitus in mice does not alter liver heparan sulfate. Journal of Biological Chemistry. 2010;285:14658–62. doi: 10.1074/jbc.M110.112391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bornemann DJ, Park S, Phin S, Warrior R. A translational block to HSPG synthesis permits BMP signaling in the early Drosophila embryo. Development. 2008;135:1039–47. doi: 10.1242/dev.017061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grobe K, Esko JD. Regulated translation of heparan sulfate N-acetylglucosamine N-deacetylase/n-sulfotransferase isozymes by structured 5′-untranslated regions and internal ribosome entry sites. J Biol Chem. 2002;277:30699–706. doi: 10.1074/jbc.M111904200. [DOI] [PubMed] [Google Scholar]
- 29.Olson SK, Bishop JR, Yates JR, Oegema K, Esko JD. Identification of novel chondroitin proteoglycans in Caenorhabditis elegans: embryonic cell division depends on CPG-1 and CPG-2. J Cell Biol. 2006;173:985–94. doi: 10.1083/jcb.200603003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esko JD, Selleck SB. Order out of chaos: Assembly of Ligand Binding Sites in Heparan Sulfate1. Annual Review of Biochemistry. 2002;71:435–71. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 31.Pinhal MA, Smith B, Olson S, Aikawa J, Kimata K, Esko JD. Enzyme interactions in heparan sulfate biosynthesis: uronosyl 5-epimerase and 2-O-sulfotransferase interact in vivo. Proc Natl Acad Sci U S A. 2001;98:12984–9. doi: 10.1073/pnas.241175798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Presto J, Thuveson M, Carlsson P, Busse M, Wilen M, Eriksson I, et al. Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proceedings of the National Academy of Sciences. 2008;105:4751–6. doi: 10.1073/pnas.0705807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Victor XV, Nguyen TK, Ethirajan M, Tran VM, Nguyen KV, Kuberan B. Investigating the elusive mechanism of glycosaminoglycan biosynthesis. J Biol Chem. 2009;284:25842–53. doi: 10.1074/jbc.M109.043208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindahl U, Li J, Kwang WJ. International Review of Cell and Molecular Biology. Academic Press; 2009. Interactions Between Heparan Sulfate and Proteins--Design and Functional Implications; pp. 105–59. [DOI] [PubMed] [Google Scholar]
- 35.Prabhakar V, Sasisekharan R. The biosynthesis and catabolism of galactosaminoglycans. Adv Pharmacol. 2006;53:69–115. doi: 10.1016/S1054-3589(05)53005-9. [DOI] [PubMed] [Google Scholar]
- 36.Kamimura K, Rhodes JM, Ueda R, McNeely M, Shukla D, Kimata K, et al. Regulation of Notch signaling by Drosophila heparan sulfate 3-O sulfotransferase. The Journal of Cell Biology. 2004;166:1069–79. doi: 10.1083/jcb.200403077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Donnell CD, Kovacs M, Akhtar J, Valyi-Nagy T, Shukla D. Expanding the role of 3-O sulfated heparan sulfate in herpes simplex virus type-1 entry. Virology. 2010;397:389–98. doi: 10.1016/j.virol.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maimone MM, Tollefsen DM. Structure of a dermatan sulfate hexasaccharide that binds to heparin cofactor II with high affinity. Journal of Biological Chemistry. 1990;265:18263–71. [PubMed] [Google Scholar]
- 39.Halldorsdottir AM, Zhang L, Tollefsen DM. N-Acetylgalactosamine 4,6-O-sulfate residues mediate binding and activation of heparin cofactor II by porcine mucosal dermatan sulfate. Glycobiology. 2006;16:693–701. doi: 10.1093/glycob/cwj117. [DOI] [PubMed] [Google Scholar]
- 40.He L, Giri TK, Vicente CP, Tollefsen DM. Vascular dermatan sulfate regulates the antithrombotic activity of heparin cofactor II. Blood. 2008;111:4118–25. doi: 10.1182/blood-2007-12-127928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asada M, Shinomiya M, Suzuki M, Honda E, Sugimoto R, Ikekita M, et al. Glycosaminoglycan affinity of the complete fibroblast growth factor family. Biochimica et Biophysica Acta (BBA) - General Subjects. 2009;1790:40–8. doi: 10.1016/j.bbagen.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 42.Deakin JA, Blaum BS, Gallagher JT, Uhrin D, Lyon M. The binding properties of minimal oligosaccharides reveal a common heparan sulfate/dermatan sulfate-binding site in hepatocyte growth factor/scatter factor that can accommodate a wide variety of sulfation patterns. J Biol Chem. 2009;284:6311–21. doi: 10.1074/jbc.M807671200. [DOI] [PubMed] [Google Scholar]
- 43.Conrad AH, Zhang Y, Tasheva ES, Conrad GW. Proteomic Analysis of Potential Keratan Sulfate, Chondroitin Sulfate A, and Hyaluronic Acid Molecular Interactions. Invest Ophthalmol Vis Sci. 2010 doi: 10.1167/iovs.09-4914. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EEM. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
- 45.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–3. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bradley RK, Li X-Y, Trapnell C, Davidson S, Pachter L, Chu HC, et al. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related drosophila species. PLoS Biol. 2010;8:e1000343. doi: 10.1371/journal.pbio.1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ibrahimi OA, Yeh BK, Eliseenkova AV, Zhang F, Olsen SK, Igarashi M, et al. Analysis of mutations in fibroblast growth factor (FGF) and a pathogenic mutation in FGF receptor (FGFR) provides direct evidence for the symmetric two-end model for FGFR dimerization. Mol Cell Biol. 2005;25:671–84. doi: 10.1128/MCB.25.2.671-684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staples GO, Shi X, Zaia J. Extended NS domains reside at the non-reducing end of heparan sulfate chains. J Biol Chem. 2010 doi: 10.1074/jbc.M110.101592. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang F, Zhang Z, Lin X, Beenken A, Eliseenkova AV, Mohammadi M, et al. Compositional analysis of heparin/heparan sulfate interacting with fibroblast growth factor. fibroblast growth factor receptor complexes. Biochemistry. 2009;48:8379–86. doi: 10.1021/bi9006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan D, Wu Y, Feng Y, Lin S-C, Lin X. The core protein of glypican dally-Like determines its biphasic activity in wingless morphogen signaling. Developmental Cell. 2009;17:470–81. doi: 10.1016/j.devcel.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mitsi M, Forsten-Williams K, Gopalakrishnan M, Nugent MA. A catalytic role of heparin within the extracellular matrix. Journal of Biological Chemistry. 2008;283:34796–807. doi: 10.1074/jbc.M806692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arrington CB, Yost HJ. Extra-embryonic syndecan 2 regulates organ primordia migration and fibrillogenesis throughout the zebrafish embryo. Development. 2009;136:3143–52. doi: 10.1242/dev.031492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chuang CY, Lord MS, Melrose J, Rees MD, Knox SM, Freeman C, et al. Heparan sulfate dependent signaling of fibroblast growth factor (FGF) 18 by chondrocyte-derived perlecan. Biochemistry. 2010 doi: 10.1021/bi1005199. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kolset S, Tveit H. Serglycin – Structure and biology. Cellular and Molecular Life Sciences. 2008;65:1073–85. doi: 10.1007/s00018-007-7455-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kramer KL, Yost HJ. Heparan sulfate core proteins in cell-cell signaling. Annu Rev Genet. 2003;37:461–84. doi: 10.1146/annurev.genet.37.061103.090226. [DOI] [PubMed] [Google Scholar]
- 56.Roch C, Kuhn J, Kleesiek K, Götting C. Differences in gene expression of human xylosyltransferases and determination of acceptor specificities for various proteoglycans. Biochemical and Biophysical Research Communications. 2010;391:685–91. doi: 10.1016/j.bbrc.2009.11.121. [DOI] [PubMed] [Google Scholar]
- 57.Gopal S, Bober A, Whiteford JR, Multhaupt HA, Yoneda A, Couchman JR. Heparan sulfate chain valency controls syndecan-4 function in cell adhesion. J Biol Chem. 2010 doi: 10.1074/jbc.M109.056945. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan D, Wu Y, Feng Y, Lin SC, Lin X. The core protein of glypican Dally-like determines its biphasic activity in wingless morphogen signaling. Dev Cell. 2009;17:470–81. doi: 10.1016/j.devcel.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams EH, Pappano WN, Saunders AM, Kim MS, Leahy DJ, Beachy PA. Dally-like core protein and its mammalian homologues mediate stimulatory and inhibitory effects on Hedgehog signal response. Proc Natl Acad Sci U S A. 2010;107:5869–74. doi: 10.1073/pnas.1001777107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gallet A, Staccini-Lavenant L, Thérond PP. Cellular trafficking of the glypican dally-like is required for full-strength hedgehog signaling and wingless transcytosis. Developmental Cell. 2008;14:712–25. doi: 10.1016/j.devcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 61.Ayers KL, Gallet A, Staccini-Lavenant L, Thérond PP. The Long-Range Activity of Hedgehog Is Regulated in the Apical Extracellular Space by the Glypican Dally and the Hydrolase Notum. Developmental Cell. 2010;18:605–20. doi: 10.1016/j.devcel.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 62.Gutierrez J, Brandan E. A novel mechanism of sequestering fibroblast growth factor 2 by glypican in lipid rafts, allowing skeletal muscle differentiation. Mol Cell Biol. 2010;30:1634–49. doi: 10.1128/MCB.01164-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mizuguchi S, Uyama T, Kitagawa H, Nomura KH, Dejima K, Gengyo-Ando K, et al. Chondroitin proteoglycans are involved in cell division of Caenorhabditis elegans. Nature. 2003;423:443–8. doi: 10.1038/nature01635. [DOI] [PubMed] [Google Scholar]
- 64.Izumikawa T, Kanagawa N, Watamoto Y, Okada M, Saeki M, Sakano M, et al. Impairment of embryonic cell division and glycosaminoglycan biosynthesis in glucuronyltransferase-I-deficient mice. J Biol Chem. 2010;285:12190–6. doi: 10.1074/jbc.M110.100941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morio H, Honda Y, Toyoda H, Nakajima M, Kurosawa H, Shirasawa T. EXT gene family member rib-2 is essential for embryonic development and heparan sulfate biosynthesis in Caenorhabditis elegans. Biochemical and Biophysical Research Communications. 2003;301:317–23. doi: 10.1016/s0006-291x(02)03031-0. [DOI] [PubMed] [Google Scholar]
- 66.Stickens D, Zak BM, Rougier N, Esko JD, Werb Z. Mice deficient in Ext2 lack heparan sulfate and develop exostoses. Development. 2005;132:5055–68. doi: 10.1242/dev.02088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine ML, Calabro A, et al. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. The Journal of Clinical Investigation. 2000;106:349–60. doi: 10.1172/JCI10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–8. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- 69.Rapraeger AC, Krufka A, Olwin BB. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science. 1991;252:1705–8. doi: 10.1126/science.1646484. [DOI] [PubMed] [Google Scholar]
- 70.Norton WHJ, Ledin J, Grandel H, Neumann CJ. HSPG synthesis by zebrafish Ext2 and Extl3 is required for Fgf10 signalling during limb development. Development. 2005;132:4963–73. doi: 10.1242/dev.02084. [DOI] [PubMed] [Google Scholar]
- 71.Lin X, Buff EM, Perrimon N, Michelson AM. Heparan sulfate proteoglycans are essential for FGF receptor signaling during Drosophila embryonic development. Development. 1999;126:3715–23. doi: 10.1242/dev.126.17.3715. [DOI] [PubMed] [Google Scholar]
- 72.Yan D, Lin X. Drosophila glypican Dally-like acts in FGF-receiving cells to modulate FGF signaling during tracheal morphogenesis. Dev Biol. 2007;312:203–16. doi: 10.1016/j.ydbio.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kamimura K, Koyama T, Habuchi H, Ueda R, Masu M, Kimata K, et al. Specific and flexible roles of heparan sulfate modifications in Drosophila FGF signaling. The Journal of Cell Biology. 2006;174:773–8. doi: 10.1083/jcb.200603129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, et al. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes & Development. 1998;12:3156–61. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patel VN, Likar KM, Zisman-Rozen S, Cowherd SN, Lassiter KS, Sher I, et al. Specific heparan sulfate structures modulate FGF10-mediated submandibular gland epithelial morphogenesis and differentiation. Journal of Biological Chemistry. 2008;283:9308–17. doi: 10.1074/jbc.M709995200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shah MM, Sakurai H, Sweeney DE, Gallegos TF, Bush KT, Esko JD, et al. Hs2st mediated kidney mesenchyme induction regulates early ureteric bud branching. Dev Biol. 2010;339:354–65. doi: 10.1016/j.ydbio.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crawford BE, Garner OB, Bishop JR, Zhang DY, Bush KT, Nigam SK, et al. Loss of the heparan sulfate sulfotransferase, ndst1, in mammary epithelial cells selectively blocks lobuloalveolar development in mice. PLoS One. 2010;5:e10691. doi: 10.1371/journal.pone.0010691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Makarenkova HP, Hoffman MP, Beenken A, Eliseenkova AV, Meech R, Tsau C, et al. Differential interactions of FGFs with heparan sulfate control gradient formation and branching morphogenesis. Sci Signal. 2009;2:ra55. doi: 10.1126/scisignal.2000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chan JA, Balasubramanian S, Witt RM, Nazemi KJ, Choi Y, Pazyra-Murphy MF, et al. Proteoglycan interactions with Sonic Hedgehog specify mitogenic responses. Nat Neurosci. 2009;12:409–17. doi: 10.1038/nn.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hayashi Y, Kobayashi S, Nakato H. Drosophila glypicans regulate the germline stem cell niche. J Cell Biol. 2009;187:473–80. doi: 10.1083/jcb.200904118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo Z, Wang Z. The glypican Dally is required in the niche for the maintenance of germline stem cells and short-range BMP signaling in the Drosophila ovary. Development. 2009;136:3627–35. doi: 10.1242/dev.036939. [DOI] [PubMed] [Google Scholar]
- 82.Kraushaar DC, Yamaguchi Y, Wang L. Heparan sulfate is required for embryonic stem cells to exit from self-renewal. J Biol Chem. 2010;285:5907–16. doi: 10.1074/jbc.M109.066837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lanner F, Lee KL, Sohl M, Holmborn K, Yang H, Wilbertz J, et al. Heparan sulfation-dependent fibroblast growth factor signaling maintains embryonic stem cells primed for differentiation in a heterogeneous state. Stem Cells. 2010;28:191–200. doi: 10.1002/stem.265. [DOI] [PubMed] [Google Scholar]
- 84.Baldwin RJ, Dam GBt, Kuppevelt THv, Lacaud G, Gallagher JT, Kouskoff V, et al. A developmentally regulated heparan sulfate epitope defines a subpopulation with increased blood potential during mesodermal differentiation. Stem Cells. 2008;26:3108–18. doi: 10.1634/stemcells.2008-0311. [DOI] [PubMed] [Google Scholar]
- 85.von Holst A, Sirko S, Faissner A. The Unique 473HD-chondroitinsulfate epitope is expressed by radial glia and involved in neural precursor cell proliferation. J Neurosci. 2006;26:4082–94. doi: 10.1523/JNEUROSCI.0422-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gama CI, Tully SE, Sotogaku N, Clark PM, Rawat M, Vaidehi N, et al. Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat Chem Biol. 2006;2:467–73. doi: 10.1038/nchembio810. [DOI] [PubMed] [Google Scholar]
- 87.Ishii M, Maeda N. Oversulfated Chondroitin Sulfate Plays Critical Roles in the Neuronal Migration in the Cerebral Cortex. Journal of Biological Chemistry. 2008;283:32610–20. doi: 10.1074/jbc.M806331200. [DOI] [PubMed] [Google Scholar]
- 88.Mizumoto S, Mikami T, Yasunaga D, Kobayashi N, Yamauchi H, Miyake A, et al. Chondroitin 4-O-sulfotransferase-1 is required for somitic muscle development and motor axon guidance in zebrafish. Biochem J. 2009;419:387–99. doi: 10.1042/BJ20081639. [DOI] [PubMed] [Google Scholar]
- 89.Fawcett JW, Asher RA. The glial scar and central nervous system repair. Brain Research Bulletin. 1999;49:377–91. doi: 10.1016/s0361-9230(99)00072-6. [DOI] [PubMed] [Google Scholar]
- 90.Bradbury EJ, Moon LDF, Popat RJ, King VR, Bennett GS, Patel PN, et al. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature. 2002;416:636–40. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- 91.Sato Y, Oohira A. Chondroitin Sulfate, a Major Niche Substance of Neural Stem Cells, and Cell Transplantation Therapy of Neurodegeneration Combined with Niche Modification. Current Stem Cell Research & Therapy. 2009;4:200–9. doi: 10.2174/157488809789057419. [DOI] [PubMed] [Google Scholar]
- 92.Ito Z, Sakamoto K, Imagama S, Matsuyama Y, Zhang H, Hirano K, et al. N-acetylglucosamine 6-o-sulfotransferase-1-deficient mice show better functional recovery after spinal cord injury. J Neurosci. 2010;30:5937–47. doi: 10.1523/JNEUROSCI.2570-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee J-S, von der Hardt S, Rusch MA, Stringer SE, Stickney HL, Talbot WS, et al. Axon Sorting in the Optic Tract Requires HSPG Synthesis by ext2 (dackel) and extl3 (boxer) Neuron. 2004;44:947–60. doi: 10.1016/j.neuron.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 94.Kastenhuber E, Kern U, Bonkowsky JL, Chien CB, Driever W, Schweitzer J. Netrin-DCC, Robo-Slit, and heparan sulfate proteoglycans coordinate lateral positioning of longitudinal dopaminergic diencephalospinal axons. J Neurosci. 2009;29:8914–26. doi: 10.1523/JNEUROSCI.0568-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 2003;302:1044–6. doi: 10.1126/science.1090497. [DOI] [PubMed] [Google Scholar]
- 96.Kalus I, Salmen B, Viebahn C, Figura Kv, Schmitz D, D’Hooge R, et al. Differential involvement of the extracellular 6-O-endosulfatases Sulf1 and Sulf2 in brain development and neuronal and behavioural plasticity. Journal of Cellular and Molecular Medicine. 2009;13:4505–21. doi: 10.1111/j.1582-4934.2008.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pratt T, Conway CD, Tian NMM-L, Price DJ, Mason JO. Heparan sulphation patterns generated by specific heparan sulfotransferase enzymes direct distinct aspects of retinal axon guidance at the optic chiasm. J Neurosci. 2006;26:6911–23. doi: 10.1523/JNEUROSCI.0505-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chanana B, Steigemann P, Jackle H, Vorbraggen G. Reception of Slit requires only the chondroitin-sulphate-modified extracellular domain of Syndecan at the target cell surface. Proceedings of the National Academy of Sciences. 2009;106:11984–8. doi: 10.1073/pnas.0901148106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bülow HE, Tjoe N, Townley RA, Didiano D, van Kuppevelt TH, Hobert O. Extracellular sugar modifications provide instructive and cell-specific information for axon-guidance choices. Current Biology. 2008;18:1978–85. doi: 10.1016/j.cub.2008.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Walsh EC, Stainier DYR. UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science. 2001;293:1670–3. doi: 10.1126/science.293.5535.1670. [DOI] [PubMed] [Google Scholar]
- 101.Camenisch TD, Schroeder JA, Bradley J, Klewer SE, McDonald JA. Heart-valve mesenchyme formation is dependent on hyaluronan-augmented activation of ErbB2-ErbB3 receptors. Nat Med. 2002;8:850–5. doi: 10.1038/nm742. [DOI] [PubMed] [Google Scholar]
- 102.Iwamoto R, Mine N, Kawaguchi T, Minami S, Saeki K, Mekada E. HB-EGF function in cardiac valve development requires interaction with heparan sulfate proteoglycans. Development. 2010;137:2205–14. doi: 10.1242/dev.048926. [DOI] [PubMed] [Google Scholar]
- 103.Peal DS, Burns CG, Macrae CA, Milan D. Chondroitin sulfate expression is required for cardiac atrioventricular canal formation. Dev Dyn. 2009;238:3103–10. doi: 10.1002/dvdy.22154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dündar M, Müller T, Zhang Q, Pan J, Steinmann B, Vodopiutz J, et al. Loss of dermatan-4-sulfotransferase 1 function results in adducted thumb-clubfoot syndrome. The American Journal of Human Genetics. 2009;85:873–82. doi: 10.1016/j.ajhg.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tuysuz B, Mizumoto S, Sugahara K, Çelebi A, Mundlos S, Turkmen S. Omani-type spondyloepiphyseal dysplasia with cardiac involvement caused by a missense mutation in CHST3. Clinical Genetics. 2009;75:375–83. doi: 10.1111/j.1399-0004.2009.01167.x. [DOI] [PubMed] [Google Scholar]
- 106.Yue X, Schultheiss TM, McKenzie EA, Rosenberg RD. Role of heparan sulfate in dextral heart looping in chick. Glycobiology. 2004;14:745–55. doi: 10.1093/glycob/cwh083. [DOI] [PubMed] [Google Scholar]
- 107.Schuksz M, Fuster MM, Brown JR, Crawford BE, Ditto DP, Lawrence R, et al. Surfen, a small molecule antagonist of heparan sulfate. Proceedings of the National Academy of Sciences. 2008;105:13075–80. doi: 10.1073/pnas.0805862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee TY, Folkman J, Javaherian K. HSPG-binding peptide corresponding to the exon 6a-encoded domain of VEGF inhibits tumor growth by blocking angiogenesis in murine model. PLoS One. 2010;5:e9945. doi: 10.1371/journal.pone.0009945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Robinson CJ, Mulloy B, Gallagher JT, Stringer SE. VEGF165-binding sites within heparan sulfate encompass two highly sulfated domains and can be liberated by k5 lyase. Journal of Biological Chemistry. 2006;281:1731–40. doi: 10.1074/jbc.M510760200. [DOI] [PubMed] [Google Scholar]
- 110.Chen E, Stringer SE, Rusch MA, Selleck SB, Ekker SC. A unique role for 6-O sulfation modification in zebrafish vascular development. Developmental Biology. 2005;284:364–76. doi: 10.1016/j.ydbio.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 111.Habuchi H, Nagai N, Sugaya N, Atsumi F, Stevens RL, Kimata K. Mice deficient in heparan sulfate 6-o-sulfotransferase-1 exhibit defective heparan sulfate biosynthesis, abnormal placentation, and late embryonic lethality. Journal of Biological Chemistry. 2007;282:15578–88. doi: 10.1074/jbc.M607434200. [DOI] [PubMed] [Google Scholar]
- 112.Harfouche R, Hentschel DM, Piecewicz S, Basu S, Print C, Eavarone D, et al. Glycome and transcriptome regulation of vasculogenesis. Circulation. 2009;120:1883–92. doi: 10.1161/CIRCULATIONAHA.108.837724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jakobsson L, Kreuger J, Holmborn K, Lundin L, Eriksson I, Kjellén L, et al. Heparan sulfate in trans potentiates VEGFR-mediated angiogenesis. Developmental Cell. 2006;10:625–34. doi: 10.1016/j.devcel.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 114.Fan G, Xiao L, Cheng L, Wang X, Sun B, Hu G. Targeted disruption of NDST-1 gene leads to pulmonary hypoplasia and neonatal respiratory distress in mice. FEBS Letters. 2000;467:7–11. doi: 10.1016/s0014-5793(00)01111-x. [DOI] [PubMed] [Google Scholar]
- 115.Uchimura K, Morimoto-Tomita M, Bistrup A, Li J, Lyon M, Gallagher J, et al. HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: effects on VEGF, FGF-1, and SDF-1. BMC Biochemistry. 2006;7:2. doi: 10.1186/1471-2091-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xu D, Moon AF, Song D, Pedersen LC, Liu J. Engineering sulfotransferases to modify heparan sulfate. Nat Chem Biol. 2008;4:200–2. doi: 10.1038/nchembio.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bethea HN, Xu D, Liu J, Pedersen LC. Redirecting the substrate specificity of heparan sulfate 2-O-sulfotransferase by structurally guided mutagenesis. Proc Natl Acad Sci U S A. 2008;105:18724–9. doi: 10.1073/pnas.0806975105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arungundram S, Al-Mafraji K, Asong J, Leach FE, 3rd, Amster IJ, Venot A, et al. Modular synthesis of heparan sulfate oligosaccharides for structure-activity relationship studies. J Am Chem Soc. 2009;131:17394–405. doi: 10.1021/ja907358k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Powell AK, Ahmed YA, Yates EA, Turnbull JE. Generating heparan sulfate saccharide libraries for glycomics applications. Nat Protoc. 2010;5:821–33. doi: 10.1038/nprot.2010.17. [DOI] [PubMed] [Google Scholar]
- 120.Thompson SM, Fernig DG, Jesudason EC, Losty PD, van de Westerlo EM, van Kuppevelt TH, et al. Heparan sulfate phage display antibodies identify distinct epitopes with complex binding characteristics: insights into protein binding specificities. J Biol Chem. 2009;284:35621–31. doi: 10.1074/jbc.M109.009712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Petitou M, Casu B, Lindahl U. 1976–1983, a critical period in the history of heparin: the discovery of the antithrombin binding site. Biochimie. 2003;85:83–9. doi: 10.1016/s0300-9084(03)00078-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.