Abstract
Using the HPV18 genome as the backbone, we exchanged the HPV18 L2 or L1 genes with those of HPV16. The intertypical exchange of HPV18 L1 with the HPV16 L1 produced genomes that efficiently replicated and produced infectious virus. Genomes containing an intertypical exchange of HPV18 L2 for the HPV16 L2 failed to produce infectious virus in multiple independently derived cell lines. Using chimeric constructs of individual capsid proteins, we identified a type-specific domain at the N-terminus of the HPV18L1 capsid protein, which interferes with its ability to cooperate with the HPV16 L2 protein to form infectious viral particles. Deletion of this domain allows for the cooperation of the HPV18 L1 protein and HPV 16 L2 protein and production of infectious progeny. Additionally, cooperation of this N-terminal HPV18 L1 deletion mutant protein with the wild type HPV18 L2 protein efficiently replicates infectious virus but changes occur in the viral structure.
Keywords: Papillomavirus, chimera, organotypic culture, raft culture, capsid genes, virion morphogenesis
INTRODUCTION
The life cycle of human papillomaviruses (HPV) has evolved to be intimately connected to the differentiation program of host epithelial tissues (Meyers et al., 1992; Meyers, Mayer, and Ozbun, 1997; Taichman and LaPorta, 1986). The development of the organotypic (raft) epithelial culture system has allowed for the development on an in vitro culture system capable of reproducing the complete HPV life cycle, including the propagation of infectious viral particles (Alam et al., 2008; McLaughlin-Drubin, Christensen, and Meyers, 2004; McLaughlin-Drubin and Meyers, 2004; McLaughlin-Drubin et al., 2003; Meyers et al., 1992; Meyers, Mayer, and Ozbun, 1997; Sen, Alam, and Meyers, 2004). The raft culture system has been used to study in detail the steps in the HPV life cycle (Alam et al., 2008; Bedell et al., 1991; Bodily, Alam, and Meyers, 2006; Bodily and Meyers, 2005; Frattini, Lim, and Laimins, 1996; Grassmann et al., 1996; Mayer and Meyers, 1998; Ozbun and Meyers, 1996; Ozbun and Meyers, 1997; Ozbun and Meyers, 1998a; Ozbun and Meyers, 1998b; Ozbun and Meyers, 1999b; Sen, Alam, and Meyers, 2004). Additonally, two recent publications we have shown that the replication and maturation of native virus in differentiating host tissue differs in significant aspects from particles made using pseudovirus/quasivirus technologies (Conway et al., 2009a; Conway et al., 2009b).
Chimeric viruses are commonly used to compare genes from one virus with the homologous genes from a related virus to determine the similarities and differences of those genes. A chimeric virus system can be used to assign a particular viral phenotype to a specific gene or sequence. Another use of a chimeric virus system is to discover the commonalities of related viral genes. In this manuscript we used chimeric HPVs to test the hypothesis that although the HPV18 and HPV16 capsid genes have a high amount of sequence homology there are type-specific domains affecting the interaction of the two capsid proteins during virion morphogenesis.
Previously, we designed experiments to study whether the nonstructural genes from one HPV type could function with the structural genes of another HPV type. In that study we replaced the L1 and L2 capsid protein open reading frames (ORFs) from HPV type 18 (HPV18) with the L1 and L2 capsid protein ORFs from HPV16 (Meyers et al., 2002a). The resulting HPV18/16 chimeric virus was able to carry out the complete viral life cycle culminating in the production of infectious virus after introduction into keratinocytes that were allowed to terminally differentiate and stratify in raft culture. Antiserum raised against HPV16 virus-like particles (VLPs) and not HPV18 VLPs specifically neutralized the HPV18/16 chimeric virus (Meyers et al., 2002a). This study established the use of a viable chimeric virus replication system to study HPV genetics and confirmed the ability of the nonstructural genes of HPV18 to function with the structural genes of HPV16.
To extend these studies we replaced either the L1 capsid ORF of HPV18 with that of HPV16 L1 or the L2 capsid ORF of HPV18 with that of HPV16 L2. Chimeric HPV18 containing the HPV16 L1 ORF were able to behave similar to wild type HPV18 in that they completed the viral life cycle in terminally differentiating raft tissue with the production of infectious chimeric viral particles. However, while chimeric HPV18 containing the HPV16 L2 ORF immortalized primary keratinocytes, was maintained in an episomal state, induced a transformed phenotype in raft culture, amplified viral DNA (vDNA), and HPV18 L1 and the HPV16 L2 capsid gene expression appeared normal in raft cultures, we were unable to detect the production of infectious virus from this chimera. In an effort to begin to narrow down the region of the HPV18 L1 or HPV16 L2 capsid gene responsible for the interference of production of infectious particles we analyzed chimeric exchanges of only half of the L1 or L2 capsid gene ORF of HPV18 with that of HPV16. These studies identified the N-terminus of the L1 capsid protein of HPV18 to be inhibitory to the production of infectious chimeric virus containing the HPV16 L2 capsid gene.
MATERIALS AND METHODS
Plasmid Construction
HPV18 DNA, a generous gift from Harold zur Hausen, was cloned into pBluescript SK(+) [pBSSK(+)] (Stratagene, La Jolla, CA) at the unique HPV18 EcoRI site within the E1 ORF and named pBSHPV18. pBSHPV18 was used as a template to separately amplify the L2 and L1 ORFs using primers 18a5’, 18L2gHin, 18L1hHin, and 18hBgl (Table 1). The HPV16 L2/L1 ORFs were BglII digested out of the HPV18/16 chimeric genome (Meyers et al., 2002a) and ligated into the BglII site of the pGL2 Basic Vector (Promega, Madison, WI) to make pGL216L2/L1. The electroporation protocol requires the release of the HPV genomic DNA from the vector DNA sequences. Since the HPV18 genomic DNA was cloned into the vector DNA at the EcoRI site, it was necessary to mutate the EcoRI site contained within the HPV16 L1 ORF at 6819 (Meyers et al., 2002a). Using pGL216L2/L1 as a template the HPV16 L2 and L1 ORFs were separately amplified using primers 16c5’, 16L2cHin, 16L1fHin, and 16L1fBgl (Table 1). Each amplified ORF was cloned individually into the pCR2.1 vector (Invitrogen, Eugene, OR) using the TOPO TA cloning procedure according to the manufacturer’s instructions (Invitrogen). Each capsid ORF was then individually cloned into pBSSK+ vectors. The plasmid pBSHPV18ΔL2/L1 contains a HPV18 genome with its L2 and L1 ORFs removed and replaced with a BglII site in a pBSSK(+) vector (Meyers et al., 2002a). pBSHPV18ΔL2/L1 was linearized by digesting with BglII, the HPV18 and HPV16 capsid ORFs were separated from the pBSSK+ vector following digestion with BglII and HindIII, and each combination of L2 and L1 ORFs were cloned into the linearized pBSHPV18ΔL2/L1 plasmid as shown in Figure S1. All mutant genomes were analyzed by restriction digestion and sequenced to ensure they were correct.
TABLE 1.
| Oligo Name | Direction | Sequence 5’→ 3’ |
|---|---|---|
| 18a5’ | 5’ | GCTAGCAGATCTATGGTATCCCACCGTGCCGC |
| 18L2gHin | 3’ | GCTAGCAAGCTTCTAGGCCGCCACAAAGCC |
| 18L1hHin | 5’ | GCTAGAAAGCTTATGTGCCTGTATACACGG |
| 18L1hBgl | 3’ | GCTAGCAGATCTTTACTTTCCTGGCACGTAC |
| 16c5’ | 5’ | GCTAGCAGATCTATGCGACACAAACGTTCTGC |
| 16L2eHin | 3’ | GCTAGCAAGCTTCTAGGCAGCCAAGAA |
| 16L1fHin | 5’ | GCTAGCAAGCTTTTACAGCTTACGTTTTTTGCG |
| 16L1fBgl | 3’ | GCTAGCAGATCTATGCAGGTGACTTTTATTTAC |
| CM B1 | 5’ | GCTAGCGCTCTTCATGACAACCCGGCCTTTGAGCCTGTG |
| CM B2 | 3’ | GCTAGCAAGCTTCTAGGCCGCCACAAAGCC |
| CM B3 | 5’ | GCTAGCAGATCTATGCGACACAAACGTTCT |
| CM B4 | 3’ | GCTAGCGCTCTTCATCATATGTAATAAGTTTAGTGGGAGT |
| CM C1 | 5’ | GCTAGCAAGCTTATGTGCCTGTATACACGG |
| CM C2 | 3’ | GCTAGCGCTCTTCCTCACGCCGTAAGCAAAAAAACATGGA |
| CM C3 | 5’ | GCTAGCGCTCTTCGTGAACAAATGTTTGTTAGACATTTA |
| CM C4 | 3’ | GCTAGCAGATCTTTACAGCTTACGTTTTTT |
| CM E1 | 5’ | GCTAGCGCTCTTCATGATAATCCTGCATATGAAGGTATA |
| CM E2 | 3’ | GCTAGCAAGCTTCTAGGCAGCCAAAGAGAC |
| CM E3 | 5’ | GCTAGCAGATCTATGGTATCCCACCGTGCC |
| CM E4 | 3’ | GCTAGCGCTCTTCGTCATATGTAATTAAAGAGGATGGACG |
| CM F1 | 5’ | GCTAGCAAGCTTATGCAGGTGACTTTTATT |
| CM F2 | 3’ | GCTAGCGCTCTTCCCTTCGTAAATAAAAAAATAAGCT |
| CM F3 | 5’ | GCTAGCGCTCTTCGAAGGCAGCTTTTTGCTAGGCATTTT |
| CM F4 | 3’ | GCTAGCAGATCTTTACTTCCTGGCACGTAC |
| CM N-35 | 5’ | GCTAGCAAGCTTATGGTACACATTATTATTTGTGGC |
Each half (N-terminal and C-terminal) of the HPV18 and HPV16 L2 and L1 ORFs were PCR amplified using primers B1-4, C1-4, E1-4, and F1-4 (Table 1) as shown in Figure S2. This created N-terminal and C-terminal halves with the N-termini of the L2 ORFs and the C-termini of the L1 ORFs containing a BglII (BII) restriction site, and the C-termini of the L2 ORFs and the N-termini of the L1 ORFs containing a HindIII (HIII) restriction site. The other end of each amplified sequence contained a SapI (SI) restriction site. Each amplified sequence was then digested with BglII and SapI or HindIII and SapI as appropriate and isolated. To create chimeric L2 or L1 ORFs, the N-termini and C-termini of HPV18 L2 and L1 ORFs were ligated to the N-termini and C-termini of HPV16 L2 and L1 ORFs, respectively, and with the pGL2 vector that was previously linearized by double digestion with BglII and HindIII (Fig. S2). Restriction digests and sequencing were performed to verify the chimeric capsid ORFs. Following digestion with BglII and HindIII the chimeric capsid ORFs were separated and isolated from the pGL2 vector. Wild type HPV18 and HPV16 L2 and L1 ORFs were separated and isolated from the HPV18-L2(18)L1(18) and HPV18-L2(16)L1(16). A chimeric L2 or L1 ORF was then ligated with a wild type L1 or L2 ORF along with pHPV18 Δ L2/L1 that had been previously digested with BglII and HindIII (Fig. S2). This produced eight chimeric virus genomes as shown in Figure S2. We designated the resulting chimeric plasmids as HPV18-L2(18)L1(16/18), HPV18-L2(18)L1(18/16), HPV18-L2(16)L1(16/18), HPV18-L2(16)L1(18/16), HPV18-L2(16/18)L1(18), HPV18-L2(16/18)L1(16), HPV18-L2(18/16)L1(18) and HPV18-L2(18/16)L1(16) (Fig. S2). All mutant genomes were analyzed by restriction digestion and sequenced to ensure they were correct.
The HPV18/HPV16 L2 chimeras form a junction in the homologous sequence, LITYDNPA, which corresponds to amino acids 249–256 in HPV16 and amino acids 248–255 in HPV18. The HPV18/HPV16 L1 chimeras form a junction in the homologous sequence, LRREQ, which corresponds to amino acids 276–280 in HPV16 and amino acids 311–315 in HPV18. The amino acid difference in numbering for the L1 and L2 proteins is due to gaps inserted when the HPV18 and HPV16 sequences were lined up according to their homologies. The homologous sequences that form the junction site for the L2 and L1 chimeras are approximately in the middle of each ORF.
To create viral genomes containing a HPV18 L1 with the N-terminal first 35 amino acids deleted, primers N-35 and F4 (Table 1) were used to PCR amplify the ORF sequence lacking the first 105 nt (Fig. S3). Primer N-35 maintained the start sequence ATG and added a BamHII site on the 5’ end and the F4 primer added a BglII site on the 3’ end. This amplimer was then digested with BamHIII and BglII and was ligated into the pHPV18L1/L2Δ plasmid that was previously digested with BamHIII and BglII along with one of the following wild-type or chimeric L2 ORFs, 18L2, 16L2, 18/16L2, or 16/18L2 (Fig. S3). This create four mutant viral genomes, HPV18-L2(18)L1(Δ18), HPV18-L2(16)L1(Δ18), HPV18-L2(18/16)L1(Δ18) and HPV18-L2(16/18)L1(Δ18) (Fig. S3). All mutant genomes were analyzed by restriction digestion and sequenced to ensure they were correct.
Western Blot Analysis
One hundred μl aliquots of protein extracts were transferred into a 1.5 ml microfuge tube, followed by adding 100 μl Buffer A (50 mM NaPO4 (pH7.2), 5 mM EDTA, 50 mM NaF, 0.5 μg/ml leupeptin, 0.5 μg/ml pepstatin, 20 μg/ml aprotinin, 0.2 mM Na3VO4, 1 mM DTT, 1 mM PMSF). To this suspension, 200 μl Buffer B was added (identical to Buffer A but supplemented with 2% w/v SDS), and boiled in a water bath for 8 min. While boiling, the samples were vortexed for 2 min. To each sample 125 μl 5X sample-loading buffer was added (100 mM Tris, pH 8.0, 5 mM EDTA, 10 % (w/v) sucrose, 2 % (w/v) SDS, 33 mM DTT and 0.1 mg/ml Pyronin Y). Total protein concentrations were measured using the Peterson protein assay (Lowry method) as previously described (Meyers et al., 2001). To determine capsid protein expression, 90 and 120 μg of the extracts were loaded onto 7.5 % SDS-PAGE gels. Proteins were transferred to a nitrocellulose membrane (Protran; Schleicher & Schuell). Membranes were incubated overnight with primary monoclonal antibodies (kindly provided by Dr. Neil Christensen, Penn State College of Medicine), H18.E20 (L1-reactive) (Rizk et al., 2008), H18.L9 (L1-reactive) (Bishop et al., 2007b) and H16.4B4 (L2-reactive) (Embers et al., 2004). All three monoclonals cross-react with their respective capsid proteins from HPV18 and HPV16, but not with CRPV. After being washed, the blots were incubated with anti-mouse horseradish peroxidase linked secondary antibody (Amersham Life Science) as per the manufacturer’s instructions. The proteins were detected using the enhanced chemiluminescence method (Perkin-Elmer) as per the manufacturer’s instructions.
Electroporation and maintenance of keratinocyte
Primary human foreskin keratinocyte (HFK) cultures were derived from newborn foreskin via trypsin digestion at 37°C as previously described (Meyers, 1996). HFKs were maintained in monolayer cultures without feeder cells, with 154 medium (Cascade Biologics, Portland, OR), supplemented with antibiotics (Cascade Biologics) and human keratinocyte growth supplement (Cascade Biologics). Multiple batches of primary human foreskin keratinocytes were electroporated with each mutant genome as previously described (Meyers et al., 2002b).
Organotypic raft cultures and histochemical analyses
Organotypic raft cultures were grown as previously described (Meyers et al., 2002b). Once at the air liquid interface the raft cultures were fed by diffusion from below every other day with E medium lacking EGF supplemented with 10 μM 1,2-dioctanoyl-sn-glycerol (C8; Sigma Chemical Co.). Raft cultures were allowed to stratify and differentiate for 12 days, as viral gene expression has been shown to peak between 10 and 12 days in the raft system (Ozbun and Meyers, 1999a; Ozbun and Meyers, 1997; Ozbun and Meyers, 1998a; Ozbun and Meyers, 1998b; Ozbun and Meyers, 1999b). For histochemical analyses, harvested raft tissues were fixed in 10% neutral buffered formalin, and embedded in paraffin. Four-micrometer sections were cut and stained with hematoxylin and eosin.
Southern blot hybridization
Viral DNA was isolated as previously described (Meyers et al., 2002b). Five μg of total cellular DNA from each sample was digested with either EcoRI, to linearize the viral genomes, or BglII, to separate the structural genes from nonstructural genes, or left undigested and then electrophoresed in a 0.8% agarose gel and transferred onto a GeneScreen Plus membrane (New England Nuclear Research Products, Boston, MA) as previously described (Meyers et al., 2002b). Southern blot hybridization was performed as previously described (Meyers et al., 2002b). The HPV DNA probes were prepared by gel purification of the 8 kb HPV and labeled with the Random Primed DNA labeling kit (Roche Molecular Biochemicals, Indianapolis, IN). Labeled probe was purified with a Quick Spin Column for radiolabeled DNA purification (Roche Molecular Biochemicals). Blots were probed first with a complete HPV18 genomic probe and then were stripped and reprobed with a complete HPV16 genomic probe. Stripping of a membrane was accomplished by placing the blots in 0.1x SSC (1x SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-1% sodium dodecyl sulfate and boiling for 1 h.
Preparation of virus stocks
Virus stocks were prepared by peeling the epithelial tissue away from the collagen of three organotypic rafts for each HPV chimera. The peeled epithelial layers were homogenized in 0.6 ml of ice-cold 1 M NaCl/0.05 M Na-Phosphate Buffer with a 7.5 ml homogenizer. The homogenizer was washed twice with 200 μl 1 M NaCl/0.05 M Na-Phosphate Buffer, pH 8. The homogenized viral solution was centrifuged at 10.5k for 10 min at 4 °C and the supernatant was transferred to a 1.8 ml Nalgene cryovial. The virus stocks were stored at −20 °C.
HPV infection and Neutralization Assays
The infectivity measurement of chimeric HPV used the Limited Dilution RT-PCR titering assay as described previously (Alam et al., 2008; Conway et al., 2009a; Conway et al., 2009b; McLaughlin-Drubin, Christensen, and Meyers, 2004; McLaughlin-Drubin and Meyers, 2004; McLaughlin-Drubin and Meyers, 2005; McLaughlin-Drubin et al., 2003; Meyers et al., 2002b; Meyers, Mayer, and Ozbun, 1997) using HaCaT cells, an immortalized human keratinocyte cell line (kindly provided by Norbert Fusenig). The virus titer was determined to be the last dilution at which the spliced transcript could be detected.
Infectivity neutralization assays were performed by infecting HaCaT cells with a 1:20 diluted viral sample that had been preincubated with type-specific monoclonal antibodies (MAb) diluted 1:20 in HaCaT culture medium as previously described (Conway et al., 2009a; Conway et al., 2009b; McLaughlin-Drubin, Christensen, and Meyers, 2004; McLaughlin-Drubin and Meyers, 2005; McLaughlin-Drubin et al., 2003). The type-specific antibodies included L1 reactive conformation-dependent monoclonal antibodies, H18.J4 and H18.K2. Both of these antibodies were previously characterized (Bishop et al., 2007a). Following preincubation, HaCaT cells were infected with the virus-antibody mixture and incubated for 2 days as described.
ELISA
Five μl of HPV stocks were bound per well to 96-well plates overnight at 4 C in neutral PBS buffer, washed and blocked for 2 h with 5% non-fat milk in PBS. Conformation-dependent HPV18 L1-reactive monoclonal antibodies H18.J4 and H18.K2 (Bishop et al., 2007a) were used in ELISA studies. Wells containing HPV mutants were incubated with MAbs diluted 1:100 in blocking buffer for 1 h, followed by incubation with rabbit anti-mouse-AP (Pierce) at 1:1000 and development with 1 mg/ml p-nitrophenyl phosphate substrate (Sigma). Wells probed with a negative control MAb H11.B2 (a HPV11 L1 conformation-dependent, neutralizing MAb) were read and used as background values for comparison. Absorbance at 405 nm (A405) was measured via a microplate reader (ThermoLabsystems). The readings from wells probed with capsid-specific MAb minus the readings from wells probed with the negative control MAb was defined as the specific binding to each mutant HPV.
RESULTS
L2 and L1 chimeric viruses
Previously, in order to investigate whether the nonstructural genes of one HPV type could cooperate with the structural genes of a second HPV type during the complete viral life cycle, we constructed a recombinant plasmid consisting of the URR and the nonstructural early gene ORFs of HPV18 and the structural late gene ORFs of HPV16 (Meyers et al., 2002a). This chimeric virus behaved similar to wild type HPV18 or HPV16 maintaining 50–100 episomal genomes copies in infected cells; upon differentiation in the organotypic raft culture system late viral functions, including capsid gene expression and infectious virus propagation were observed (Meyers et al., 2002a). These data demonstrated that the nonstructural genes of HPV18 function with the structural genes of HPV16, allowing the complete HPV life cycle to occur. In the present study we hypothesized that the structural genes of one viral type contain domains that may affect intertype interactions during the viral life cycle and virion morphogenesis.
To test this hypothesis it was necessary to create chimeric mutant viruses exchanging either the L2 or L1 ORF of HPV18 for the L2 or L1 ORF of HPV16. An initial problem with creating these chimeric mutants is that the L2 and L1 ORFs of HPV18 and HPV16 overlap, therefore direct exchanges of one of the ORF would affect the other ORF (Fig. S1). To overcome this situation we first created mutant viruses using PCR technology to individually amplify each ORF, introducing appropriate restriction enzyme sites and cloning the amplimers into the pHPV18L1/L2Δ plasmid that lacks the L2 and L1 ORFs (Meyers et al., 2002a) (Fig. S1). This created two mutant viral genomes; one wild type for HPV18 except that the L2 and L1 ORFs do not overlap but are now separated by an unique HindIII site, the second is similar to our previous reported HPV18/16 chimera (Meyers et al., 2002a), except that the HPV16 L2 and L1 ORFs do not overlap but are now separated by an unique HindIII site (Fig. S1). We named these mutants, HPV18-L2(18)L1(18) and HPV18-L2(16)L1(16). Our main goal was to determine if capsid proteins from two different HPVs could cooperate to produce infectious virus and if not then what domain(s) of the protein is responsible. However, in creating these mutant genomes there was the possibility that the tandem duplication of the putative L1 splice acceptor would interfere with expression of the capsid protein and therefore the ability of these mutants to produce infectious virus. Therefore, we first analyzed the mutants for their ability to correctly express the capsid proteins and produce infectious virus.
Continuously infected cell lines were made using our standard electroporation protocol previously reported (Conway et al., 2009a; Conway et al., 2009b; McLaughlin-Drubin, Christensen, and Meyers, 2004; McLaughlin-Drubin and Meyers, 2004; McLaughlin-Drubin and Meyers, 2005; McLaughlin-Drubin et al., 2003; Meyers et al., 2002a; Meyers, Mayer, and Ozbun, 1997). Upon differentiation in raft culture these cell lines were tested for late viral functions of viral genome amplification by Southern blotting, capsid gene expression by Western blotting, and propagation of infectious progeny by a Limited Dilution RT-PCR titering assay. Wild type and mutant viruses had a similar phenotype including viral titers when late viral functions were analyzed (Fig. 1). This suggested that the physical separation of the L2 and L1 ORFs had no significant effect on the viral life cycles. We concluded that construction of mutant viruses having their capsid gene ORFs separated did not introduce any adverse effects in the capsid protein expression and the production of infectious virus. All mutant genomes for this section and throughout this report were analyzed by restriction digestion and sequenced to ensure they were correct. Here and throughout the manuscript three or more independently derived virus infected cell lines were tested and the results were always found to be similar.
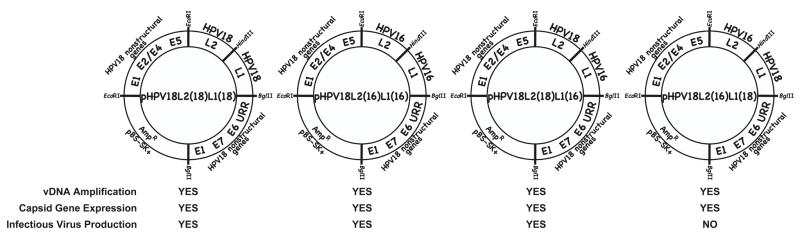
FIGURE 1.
Late functions of viral mutants with nonoverlapping L2 and L1 ORFs. Late viral life cycle functions were analyzed for the four mutant HPV18 viruses containing nonoverlapping L2 and L1 ORFs; HPV18-L2(18)L1(18), HPV18-L2(16)L1(16), HPV18-L2(18)L1(16), and HPV18-L2(16)L1(18).
We then proceeded to create two chimeric mutants, by exchanging the HPV18 L2 or L1 ORF for its counterpart from HPV16 (Fig. S1). We named these mutants, HPV18-L2(18)L1(16) and HPV18-L2(16)L1(18). Cell lines continuously infected with each mutant were made as described (Conway et al., 2009a; Conway et al., 2009b; McLaughlin-Drubin, Bromberg-White, and Meyers, 2005; McLaughlin-Drubin, Christensen, and Meyers, 2004; McLaughlin-Drubin and Meyers, 2004; McLaughlin-Drubin et al., 2003; Meyers et al., 2002a; Meyers, Mayer, and Ozbun, 1997). Upon differentiation in raft culture these cell lines were tested for late viral functions of vDNA amplification by Southern blotting, capsid gene expression by Western blotting, and propagation of infectious progeny by Limited Dilution RT-PCR titering assay. Both mutant viruses were similar to wild type in their ability to amplify their genomes and express their capsid genes, however, when infectivity was measured by Limited Dilution RT-PCR titering, HPV18-L2(18)L1(16) was capable of infectious virus synthesis similar to wild type, but the chimera HPV18-L2(16)L1(18) was not (Fig. 1). Three individual continuously infected cell lines using three separate batches of primary foreskin keratinocytes were originally made for each of the mutants. To rule out the possibility that the particular genetic background of the three batches of primary foreskin keratinocytes were responsible for the lack of infectious virus production, we created 17 more HPV18-L2(16)L1(18) continuously infected cell lines for a total of 20 cell lines. Infectious virus was undetectable in 19 of the 20 cell lines after growth in raft culture (Table 3). Following growth in raft culture we were able to detect infectious virus with only one of the twenty cell lines, but detection was at the lowest limit of our ability to detect infectious virus having a titer of 20 (Table 3). This shows that the chimeric HPV, HPV18-L2(16)L1(18), was extremely inefficient at propagating infectious virus. We concluded that the low levels of infectious virus was a consequence of cooperation between the HPV18 L1 protein and the HPV16 L2 protein.
TABLE 3.
Mutant viruses containing the HPV16 L2 C-terminus and the HPV18 L1 N-terminus.
| VIRUS | Independently immortalized lines (line/attemptsa) | Infectious virus producing lines (lines producing/total linesb) | TITERS | Independent Measurementsc | Titer Range | Ave/sdd |
|---|---|---|---|---|---|---|
| HPV18-L2(16)L1(18/16) | 4/4 | 0/4 | <20 | 4 | 0 | 0/0 |
| HPV18-L2(18/16)L1(18) | 5/5 | 1/5 | 100 | 5 | 0–100 | 20/44.7 |
| HPV18-L2(16)L1(18) | 20/20 | 1/20 | 20 | 20 | 0–20 | 1/4.5 |
The number of established continuous immortalized cell lines created with the particular HPV chimera over the number of attempts to derive such a cell line.
The number of cell lines able to produce measurable infectious virus over the number of establishe20/44.7d cell lines.
Number of independent measurements made for each HPV construct.
Titer average/standard deviation
The separation of the L2 and L1 ORFs in these two chimeric mutants could potentially affect the splicing patterns of the mRNAs encoding the late proteins. Therefore, the inability of HPV18-L2(16)L1(18) to propagate infectious virus could be due to a lack or decrease in capsid protein expression. We therefore compared capsid protein expression of the mutant viruses to wild-type virus. For these analyses we included three separate isolates of the replication deficient mutant, HPV18-L2(16)L1(18) (Figs. 2–4). Two different HPV18 L1-specifc MAbs H18.E20 and H18.L9 (Figs. 2–3) that cross-react with the HPV16 L1 protein and one HPV16 L2 MAb that cross-reacts with the HPV18 L2 protein H16.4B4 (Fig. 4) were used. A replication competent chimeric HPV18/CRPV virus (HPV18 with the capsid genes of CRPV) was used as a negative control. All HPV18/HPV16 L2/L1 chimeric viruses, including the three separate isolates of the replication incompetent virus HPV18-L2(16)L1(18), expressed similar levels of L1 (Figs. 2–3) and L2 (Fig. 4) as compared to wild-type HPV18. The physical separation of the two overlapping structural ORFs did not interfere with capsid protein expression and therefore the lack of infectivity observed with the HPV18-L2(16)L1(18) mutant virus is not due to reduced capsid protein expression. At least three separate batches were used for the Western analyses and the results were the same for each batch. Differentiating epithelium infected with HPV18-L2(16)L1(18) produced assembled virion-like particles with benzonase protected encapsidated viral genomes at levels similar to wild-type levels (data not shown). Therefore the lack of infectivity was not due to the inability to form stable viral particles.
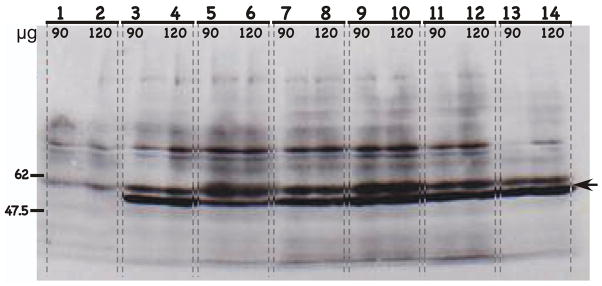
FIGURE 2.
Western blot of HPV L1 capsid protein expression with the L1-specific MAb H18.E20. H18.E20 monoclonal antibody was used to detect L1 protein expression from chimeric HPV infected raft tissues. H18.E20 was raised against HPV18 L1 protein but cross-reacts with HPV16 L1 protein. Protein isolated from chimeric HPV18/CRPV virus infected raft tissues, expressing the CRPV L2 and L1 capsid proteins was used as a negative control. Protein isolated from wild-type HPV18 infected raft tissues was used as a positive control. Protein isolated from virus infected raft tissues were loaded, 90 or 120 μg to each well as labeled. Lanes 1–2: HPV18/CRPV infected tissues; Lanes 3–4: wild-type HPV18 infected tissues; Lanes 5–6: HPV18-L2(18)L1(18) infected tissues; Lanes 7–8: HPV18-L2(18)L1(16) infected tissues; Lanes 9–10: HPV18-L2(16)L1(18) infected tissues (cell line #1); Lanes 11–12: HPV18-L2(16)L1(18) infected tissues (cell line #2); and Lanes 13–14: HPV18-L2(16)L1(18) infected tissues (cell line #3). NOTE, that Lanes 9–14 represent three individually derived cell lines of the same mutant, HPV18-L2(16)L1(18). Molecular weight markers are on the left. The arrow indicates the position of the L1 protein.
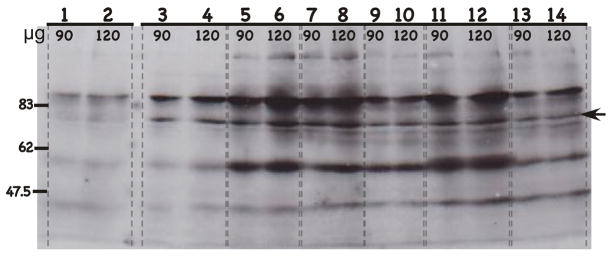
FIGURE 4.
Western blot of HPV L2 capsid protein expression with the L2-specific MAb H16.4B4. H16.4B4 monoclonal antibody was used to detect L2 protein expression from chimeric HPV infected raft tissues. H16.4B4 was raised against HPV18 L2 protein but cross-reacts with HPV16 L2 protein. Protein isolated from chimeric HPV18/CRPV virus infected raft tissues, expressing the CRPV L2 and L1 capsid proteins was used as a negative control. Protein isolated from wild-type HPV18 infected raft tissues was used as a positive control. Protein isolated from virus infected raft tissues were loaded, 90 or 120 μg to each well as labeled. Lanes 1–2: HPV18/CRPV infected tissues; Lanes 3–4: wild-type HPV18 infected tissues; Lanes 5–6: HPV18-L2(18)L1(18) infected tissues; Lanes 7–8: HPV18-L2(18)L1(16) infected tissues; Lanes 9–10: HPV18-L2(16)L1(18) infected tissues (cell line #1); Lanes 11–12: HPV18-L2(16)L1(18) infected tissues (cell line #2); and Lanes 13–14: HPV18-L2(16)L1(18) infected tissues (cell line #3). NOTE, that Lanes 9–14 represent three individually derived cell lines of the same mutant, HPV18-L2(16)L1(18). Molecular weight markers are on the left. The arrow indicates the position of the L2 protein.
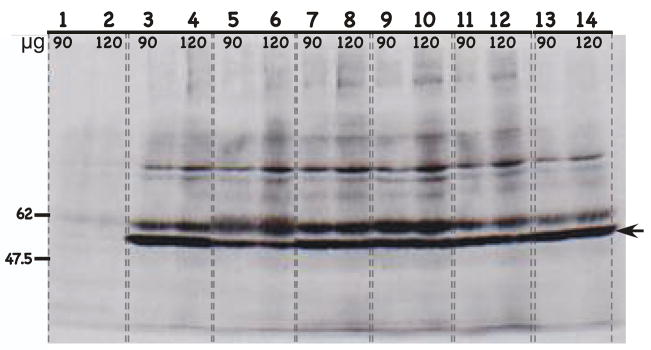
FIGURE 3.
Western blot of HPV L1 capsid protein expression with the L1-specific MAb H18.L9. H18.L9 monoclonal antibody was used to detect L1 protein expression from chimeric HPV infected raft tissues. H18.L9 was raised against HPV18 L1 protein but cross-reacts with HPV16 L1 protein. Protein isolated from chimeric HPV18/CRPV virus infected raft tissues, expressing the CRPV L2 and L1 capsid proteins was used as a negative control. Protein isolated from wild-type HPV18 infected raft tissues was used as a positive control. Protein isolated from virus infected raft tissues were loaded, 90 or 120 μg to each well as labeled. Lanes 1–2: HPV18/CRPV infected tissues; Lanes 3–4: wild-type HPV18 infected tissues; Lanes 5–6: HPV18-L2(18)L1(18) infected tissues; Lanes 7–8: HPV18-L2(18)L1(16) infected tissues; Lanes 9–10: HPV18-L2(16)L1(18) infected tissues (cell line #1); Lanes 11–12: HPV18-L2(16)L1(18) infected tissues (cell line #2); and Lanes 13–14: HPV18-L2(16)L1(18) infected tissues (cell line #3). NOTE, that Lanes 9–14 represent three individually derived cell lines of the same mutant, HPV18-L2(16)L1(18). Molecular weight markers are on the left. The arrow indicates the position of the L1 protein.
We concluded from these studies that part or all of the HPV18 L1 and/or the HPV16 L2 proteins were unable to mutually cooperate to efficiently produce infectious virus. This led us to begin to characterize the region(s) of these two proteins responsible for this inability to cooperate.
L2 and L1 half and half chimeric viruses
Many secondary structures and functional domains have been mapped in the L1 and L2 proteins (Fig. 5). It was not readily apparent which structural or functional domain may account for the inability of L1 and L2 to cooperate in the chimeric mutant HPV18-L2(16)L1(18). Consequently, we decided to characterize potential inhibitory domains by making HPV18 and HPV16 L2 or L1 chimeras containing the N-terminal half of a protein from one viral type and the C-terminal half of a protein of the other viral type. We chose to use a homologous sequence in the middle of the L2 and L1 proteins of HPV18 and HPV16 for the chimeric junction, allowing for the maintenance of sequence and structure around the junction sites (see Materials and Methods, Fig. S2, Fig. 5). In total, at least three independently derived cell lines for each of the 8 chimeric mutant HPV genomes were created (Fig. S2).
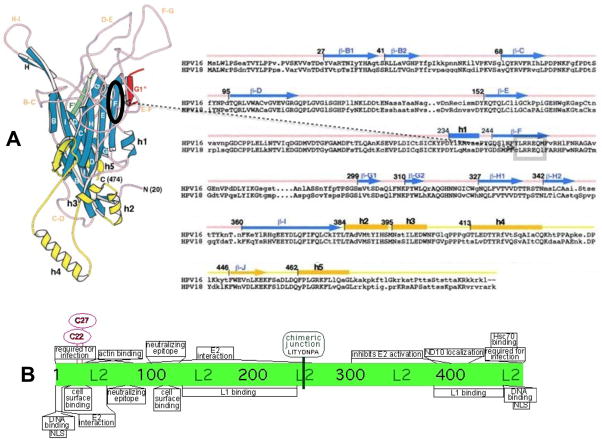
FIGURE 5.
Capsid proteins L1 and L2 chimeric junctions. Shown are the approximate positions of the cloning junctions used for the L1 and L2 HPV18 and HPV16 chimeras. (A) Shown are the three-dimensional monomeric structure of L1 (left) and the amino acid sequences of HPV16 and HPV18 L1 proteins beginning at the consensus methionine (right) as described by Chen et al. (Chen et al., 2000). On the three-dimensional structure an oval shows the region on the F ß-sheet strand containing the chimeric junction. The position of the chimeric junction can also be seen of the two-dimensional sequence inside of the box. The positions of the ß-sheets (capital letter only for the three-dimensional structure and ß-capital letter for the sequence) and the helixes (h) are shown. (B) Shown are described functional regions of papillomavirus L2 proteins (Bossis et al., 2005; Fay et al., 2004; Florin et al., 2006; Florin et al., 2002; Gornemann et al., 2002; Heino, Zhou, and Lambert, 2000; Holmgren et al., 2005; Kamper et al., 2006; Kawana et al., 1999; Laniosz, Nguyen, and Meneses, 2007; Okun et al., 2001; Roden et al., 2001; Roden et al., 1994b; Yang et al., 2003a; Yang et al., 2003b; Zhou et al., 1994). The chimeric junction is shown at its position in the center of the L2 protein.
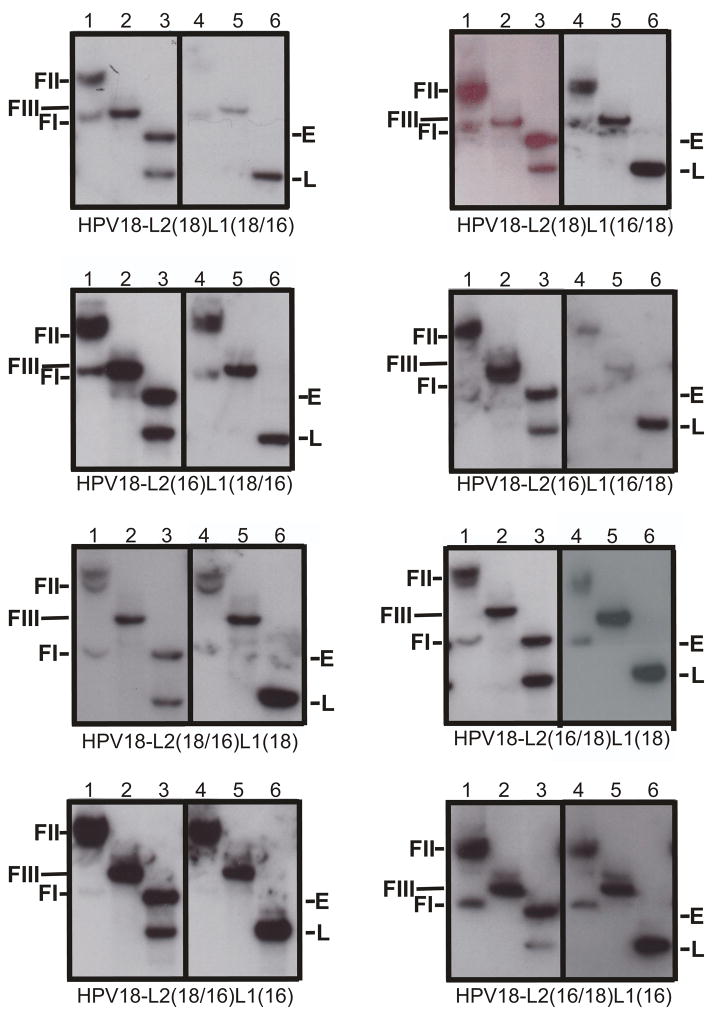
Multiple continuously infected cell lines were made with each of the eight mutant genomes and their abilities to transit the viral life cycle and replicate infectious viral stocks were studied. When grown in raft culture all cell lines were able to differentiate and their morphologies were within the parameters we have observed for tissues infected with wild type HPV18 and HPV16 (Fig. S3) (McLaughlin-Drubin, Christensen, and Meyers, 2004; McLaughlin-Drubin and Meyers, 2004; Meyers et al., 2002a; Meyers, Mayer, and Ozbun, 1997). Southern blots of whole tissue DNA were first probed with HPV18-specific radiolabeled probes then stripped and reprobed with HPV16-specific radiolabeled probes to demonstrate the presence of both HPV18 and HPV16 DNA (Fig. 6). Undigested DNA showed supercoiled (FI) and nicked (FII) genomic DNAs when probed with either HPV18 or HPV16. Viral genomes digested with EcoRI and probed with HPV18 or HPV16 demonstrated a linear (FIII) band consistent with a length of about 8 kb, the size of the HPV genomic DNA. Viral genomes digested with BglII and probed with HPV18 showed a band consistent with a length of approximately 4,900 nt, the size of the URR and early genes (E) of HPV18. Viral genomes digested with BglII and probed with HPV18 or HPV16 showed a band consistent with a length of around 3 kb, representing the L2 and L1 ORFs (L) of the chimeric HPV (Fig. 6). Southern blots of independently derived chimeric HPV infected cell lines showed similar results.
FIGURE 6.
Southern (DNA) blot hybridization of chimeric HPV DNA-electroporated HFK cell lines grown in monolayer cultures. Cell lines were analyzed for the episomal maintenance, copy number, and integrity of the chimeric genomic DNA. Left Panels: The blots were probed with an HPV18-specific probe. Right Panels: The blots were stripped and reprobed with an HPV16-specific probe. Samples in lanes 1 and 4 were undigested, Samples in lanes 2 and 5 were digested with EcoRI, a single cutter of the chimeric genomes. Samples in lanes 3 and 6 were digested with BglII to separate the capsid genes (late ORFs) from the rest of the viral genome (early ORFs). Indicated are form I DNA (FI), form II DNA (FII), form II DNA (FIII), early ORFs (E), and late ORFs (L).
Titers of L2 and L1 half and half mutants
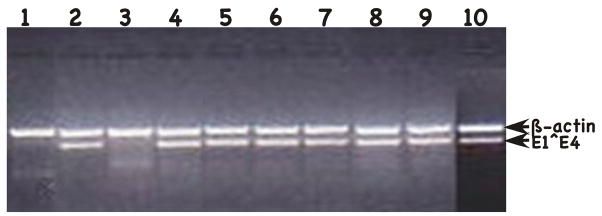
We next tested for infectious virus production. Putative viral stocks were prepared as previously described (Alam et al., 2008; Conway et al., 2009a; Conway et al., 2009b; McLaughlin-Drubin, Bromberg-White, and Meyers, 2005; McLaughlin-Drubin, Christensen, and Meyers, 2004; McLaughlin-Drubin and Meyers, 2004; McLaughlin-Drubin et al., 2003; Meyers et al., 2002a; Meyers, Mayer, and Ozbun, 1997). To measure titers HaCaT cells were infected with 1:20, 1:100, 1:1,000, 1:5,000, 1:7,500 and 1:10,000 dilutions of each viral preparation for two days, total RNA was then harvested, and nested RT-PCR to specifically amplify the HPV18 early spliced viral mRNA E1^E4 transcript and an internal cellular control mRNA β-actin. Nested RT-PCR produces a 269-bp HPV18 E1^E4 and a 429-bp β-actin product. At the lowest dilution of virus, all of the half and half chimeric HPV genomes except HPV18-L2(16)L1(18/16) were able to produce infectious virus in our infectivity assay (Fig. 7 and Table 1). While HPV18-L2(18)L1(18/16) was infectious at a titer of 5,000, stocks of HPV18-L2(16)L1(16/18) were somewhat less infectious having a titer of 1,000 (Table 2). HPV18-L2(16/18)L1(18), HPV18-L2(16/18)L1(16), HPV18-L2(18/16)L1(18) and HPV18-L2(18/16)L1(16) all had a lower titer of 100 (Table 2). Finally, HPV18-L2(18)L1(16/18) only had a titer of 20 (Table 2). Multiple cell lines and viral stocks prepared from an infected cell line were tested for titers with all giving similar results for each particular mutant. This demonstrates that while the capsid gene chimeras did not appear to affect many aspects of the viral life cycle, there was a significant effect on titers for some of the mutants. At least three independently derived chimeric HPV infected cell lines were tested for their production of infectious titers after growing in raft cultures. Titers in each case were found to be similar for each cell line infected with the same chimeric virus.
FIGURE 7.
Infectivity assay of chimeric HPVs. Shown is a 2% agarose gel of nested RT-PCR-amplified HPV18 E1^E4 and ß-actin. All assays were done with use 1:20 dilution of the viral stock. Lane 1: negative control mock infected; Lane 2: positive control wildtype HPV18; Lane 3: HPV18-L2(16)L1(18/16); Lane 4: HPV18-L2(18)L1(18/16); Lane 5: HPV18-L2(18/16)L1(18); Lane 6: HPV18-L2(18)L1(16/18); Lane 7: HPV18-L2(16)L1(16/18); Lane 8: HPV18-L2(16/18)L1(18); Lane 9: HPV18-L2(16/18)L1(16); and Lane 10: HPV18-L2(18/16)L1(16). Positions of ß-actin and E1^E4 amplified sequences are shown on the right.
TABLE 2.
Virus titers of HPV18/16 L2/L1 chimeras.
| Mutant | Titers | Independent Measurementsa | Titer Range | Ave/sdb |
|---|---|---|---|---|
| Wt HPV18 | 7,500 | 8 | 7,500–10,000 | 8,125/1157 |
| HPV18-L2(18)L1(18/16) | 5,000 | 3 | 5,000 | 5,000/0 |
| HPV18-L2(18)L1(16/18) | 20 | 3 | 20 | 20/0 |
| HPV18-L2(16)L1(18/16) | <20 | 4 | 0 | 0/0 |
| HPV18-L2(16)L1(16/18) | 1,000 | 3 | 1,000 | 1,000/0 |
| HPV18-L2(18/16)L1(18) | 100 | 5 | 0–100 | 20/44.7 |
| HPV18-L2(18/16)L1(16) | 100 | 3 | 20–100 | 73/46.2 |
| HPV18-L2(16/18)L1(18) | 100 | 3 | 100 | 100/0 |
| HPV18-L2(16/18)L1(16) | 100 | 3 | 100 | 100/0 |
Number of independent measurements made for each HPV construct.
Titer average/standard deviation
Identification of a type-specific domain in the N-terminus of the HPV18 L1 gene
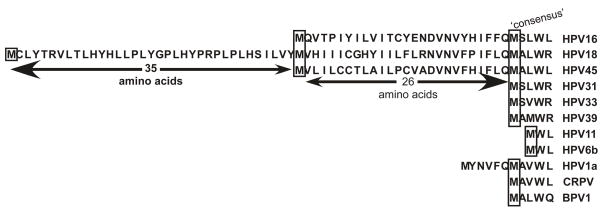
We looked for patterns to try to explain the differences in efficiency of chimeric mutant infectious virus production. One pattern we noticed (although unlikely to be the only pattern) was that chimeric viruses HPV18-L2(18/16)L1(18), HPV18-L2(16)L1(18/16) and HPV18-L2(16)L1(18) were all relatively inefficient for virus production, all had the C-terminus of HPV16 L2 and the N-terminus of HPV18 L1 (Table 3). These results suggested that at least one type-specific domain might reside in these regions affecting virion morphogenesis. We compared the amino acid sequence of the L1s of HPV16 and HPV18 and found that HPV18 L1 has an extra 35 amino acids in its N-terminus compared to HPV16 (Fig. 8). To determine if this extra 35 amino acids of HPV18 L1 affects the interaction of HPV16 L2 and HPV18 L1 during virion morphogenesis or infectivity resulting in poor viral titers, we constructed mutants deleting the first 35 amino acids of HPV18 L1 designated HPV18-L2(18)L1(Δ18), HPV18-L2(16)L1(Δ18), HPV18-L2(18/16)L1(Δ18) and HPV18-L2(16/18)L1(Δ18) (Fig. S4). At least three independently derived cell lines were created for each mutant virus. The loss of the 35 amino acids from the N-terminus had no effect of the stability of the particles.
FIGURE 8.
Papillomavirus L1 N-terminus amino acid alignment. Potential start sites for high-risk HPVs and the ‘consensus’ methionine are boxed.
All Δ18L1 mutants were able to maintain their genomes episomally. The efficiency of the Δ18L1 mutants for developing cell lines capable of stable episomal maintenance was 100%, the same as wild-type HPV18 (data not shown). All the tissues harboring HPV18 L1 N-terminal deletions were able to stratify and differentiate, with a similar morphology to that of wild-type HPV18. When we checked HPV mutant genome amplification by Southern analyses, all Δ18L1 mutants, HPV18-L2(18)L1(Δ18), HPV18-L2(16)L1(Δ18), HPV18-L2(18/16)L1(Δ18) and HPV18-L2(16/18)L1(Δ18), were able to amplify their genomes when grown in raft cultures (data not shown). All independently derived lines infected with the same mutant virus showed similar results.
Performing our standard infectivity assay we measured titers of the Δ18L1. Wild-type HPV18 had an infectious titer of 7,500 and HPV18-L2(18)L1(Δ18) had an infectious titer of 10,000 (Table 4). This result shows that deletion of the HPV18 L1 35 amino acid N-terminus did not greatly affect virus titers. However, when we compared the Δ18L1 chimeric mutants to their full-length counterparts we observed a 125-fold increase in viral titer between HPV18-L2(16)L1(Δ18) (titer = 2,500) and HPV18-L2(16)L1(18) (titer = 20) (Table 4). This result demonstrates that the first 35 amino acids in HPV18 L1 N-terminus, through a presently unknown mechanism, affects a necessary interaction of the HPV18 L1 protein and the HPV16 L2 protein resulting in lower viral titers. HPV18-L2(16)L1(18) was very inefficient in virus production, with only one out of twenty cell lines able to produce infectious virus. The only cell line able to produce infectious virus did so at the limit of the assay’s ability to detect infection. After deleting the first 35 amino acids in HPV18 L1 N-terminus, the virus titer was increased 75-fold when comparing HPV18-L2(18/16)L1(Δ18) (titer = 7,500) and HPV18-L2(18/16)L1(18) (titer = 100) (Table 4). Similarly, a 25-fold increase in viral titer was observed between HPV18-L2(16/18)L1(Δ18) (titer = 2,500) and HPV18-L2(16/18)L1(18) (titer = 100). Together these results suggest that the extra 35 amino acids of the HPV18 L1 N-terminus may cause a structural distortion in a HPV16 L2 capsid structure, which was relieved by deleting the HPV18 L1 N-terminus. These studies have identified one type-specific sequence affecting with the interaction between HPV16 L2 and HPV18 L1, resulting in lower levels of infectious virus production. At least three independently derived chimeric HPV infected cell lines were tested for their production of infectious titers after growing in raft cultures. Titers in each case were found to be similar for each cell line infected with the same chimeric virus. Additionally, mutants containing the Δ18L1 produced benzonase protected genome encapsidated viral particles in numbers similar to wild-type (data not shown). We realize that others have produced VLPs, etc., using the consensus Met and splicing data suggest this extra 35 amino acids would not contribute to the structure of the virus. However, our genetic analysis suggests the N-terminal 35 amino acids do play a role in capsid structure.
TABLE 4.
Virus titers of HPV18 N-terminal 35 aa deletion mutants.
| Mutant | Titers | Independent Measurementsa | Titer Range | Ave/sdb |
|---|---|---|---|---|
| Wt HPV18 | 7,500 | 8 | 7,500–10,000 | 8,125/1157 |
| HPV18-L2(18)L1(Δ18) | 10,000 | 3 | 10,000 | 10,000/0 |
| HPV18-L2(16)L1(18) | 20 | 20 | 0–20 | 1/4.5 |
| HPV18-L2(16)L1(Δ18) | 2,500 | 3 | 2,500 | 2,500/0 |
| HPV18-L2(18/16)L1(18) | 100 | 5 | 0–100 | 20/44.7 |
| HPV18-L2(18/16)L1(Δ18) | 7,500 | 3 | 7,500 | 7,500/0 |
| HPV18-L2(16/18)L1(18) | 100 | 3 | 100 | 100/0 |
| HPV18-L2(16/18)L1(Δ18) | 2,500 | 3 | 2,500 | 2,500/0 |
Neutralization analyses of Δ18L1 chimeric virus with L1 conformation-dependent antibodies
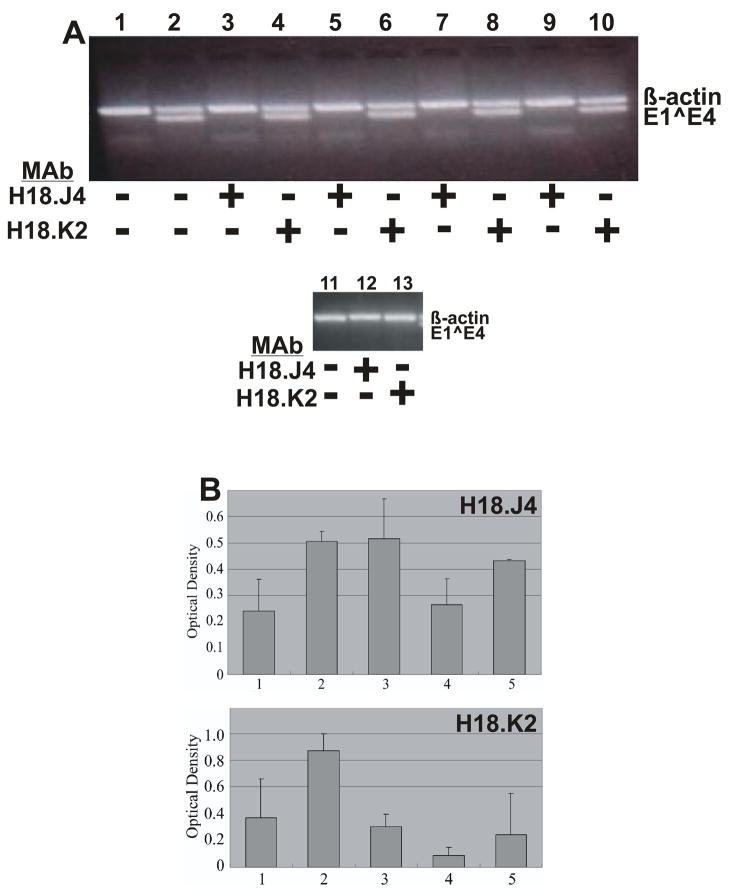
We hypothesized that changes in viral titers seen with the Δ18L1 mutants (Table 4) were due to structural changes in the virus evoked by the mutation. In order to provide preliminary evidence that this may be the case, we used conformation dependent L1 monoclonal antibodies (MAb) to measure changes in the infectivity neutralization associated with the mutations. To test whether the capsid structure had been affected, HPV18 L1 conformation-dependent antibodies H18.J4 and H18.K2 were tested for their ability to neutralize infection by HPV18-L2(18)L1(Δ18), HPV18-L2(16)L1(Δ18), HPV18-L2(18/16)L1(Δ18) and HPV18-L2(16/18)L1(Δ18). Although the epitopes for these monoclonals have not yet been mapped we know that the deleted N-terminal 35 amino acid sequence cannot contain the epitope for H18.J4 or H18.K2. This is because MAbs H18.J4 and H18.K2 were raised against HPV18 L1 VLPs lacking the first 61 amino acids (Fig. 8) (Bishop et al., 2007a). Infection by wild-type HPV18, as expected, was completely neutralized by both H18.J4 and H18.K2 (Fig. 9A). Interestingly, all four mutants were neutralized by H18.J4, but not by H18.K2 (Fig. 9A), suggesting that subtle but significant changes were affected by the removal of the N-terminal 35 amino acids from the HPV18 L1 protein. Studies were repeatable with virus from independently derived cell lines.
FIGURE 9.
(A) Neutralization of chimeric HPVs. Shown is a 2% agarose gel of nested RT-PCR-amplified HPV18 E1^E4 and ß-actin. Chimeric viruses were preincubated with the indicated MAb, H18.J4 or H18.K2. Lane 1: negative control, mock infected; Lane 2: positive control, wildtype HPV18; Lanes 3 and 4: HPV18-L2(18)L1(Δ18); Lanes 5 and 6: HPV18-L2(16)L1(Δ18); Lanes 7 and 8: HPV18-L2(18/16)L1(Δ18); Lanes 9 and 10: HPV18-L2(16/18)L1(Δ18); Lane 11: negative control, mock infected; Lanes 12 and 13: positive control, wildtype HPV18. Positions of ß-actin and E1^E4 amplified sequences are shown on the right. (B) Relative binding of conformation-dependent HPV18 L1-specific MAbs H18.J4 and H18.K2 to chimeric HPVs by ELISA assay. Relative binding for each chimeric HPV was defined as the difference between wells probed with (A) H18.J4 or (B) H18.K2 and wells probed with an irrelevant MAb. Values shown are means ± standard errors of the means. Lane 1: wildtype HPV18; Lane 2: HPV18-L2(18)L1(Δ18); Lane 3: HPV18-L2(16)L1(Δ18); Lane 4: HPV18-L2(18/16)L1(Δ18); and Lane 5: HPV18-L2(16/18)L1(Δ18).
MAb binding to Δ18L1 mutants
We reasoned that the lack of neutralization by the MAb H18.K2 was due to an inability to bind to the mutant particle or that the H18.K2 may still be able to bind but unable to block infection due to a change in conformation caused by the loss of the N-terminus 35 amino acids. To test whether the failure of neutralization correlated to the loss of antibody binding by the conformation-dependent antibodies, ELISAs were performed. To perform ELISA analysis, each chimeric HPV was used as antigen with wild-type HPV18 serving as the positive controls. Wells probed with an irrelevant MAb were included and used to subtract background signal. The ELISA measurement for each chimeric HPV was defined as the difference between wells probed with a L1 type-specific and a type-irrelevant antibodies. We first measured binding using the MAb H18.J4, which was capable of neutralizing all of the Δ18L1 mutants. As seen in Figure 9B, H18.J4 was able to bind to each of the Δ18L1 mutants as well or better than wild type HPV18. We next measured the binding of H18.K2, which was unable to neutralize the Δ18L1 mutants (Fig. 9A). With the exception of HPV18-L2(18/16)L1(Δ18), H18.K2 was still able to bind to the other three Δ18L1 mutants similar or better than its binding to wild type (Fig. 9B). Although the binding of H18.K2 was more variable than what was seen with H18.J4. These results suggest that the change in capsid structure evoked by the deletion of the N-terminus 35 aa of the HPV18 L1 protein blocked the neutralizing capacity of MAb H18.K2 but not necessarily its ability to bind to the capsid. While not quantitative the ELISA results demonstrated that at least part of the chimeric capsid proteins were correctly folded and could be recognized by conformation-dependent antibodies. These results collectively demonstrate that the extra 35 aa on the HPV18 L1 N-terminus interferes with the ability of the HPV18 L1 and the HPV16 L2 to cooperate and properly assemble into infectious viral particles. Studies were repeatable with virus from independently derived cell lines.
DISCUSSION
For these studies we used the organotypic raft culture system to test the chimeric mutants instead of co-expressing the viral structural proteins in a simple and less demanding system such as VLP or pseudovirus technology for several reasons. First is that we wanted to test the mutants in a system best capable of using the process of naturally differentiating human epithelium by which HPV replicates itself in vivo. Second we have extensive experience with raft cultures in studying the HPV life cycle and replication of infectious virus. Third, in two recent publications we have shown that the replication and maturation of native virus in differentiating host tissue differs in significant aspects from particles made using pseudovirus/quasivirus technologies (Conway et al., 2009a; Conway et al., 2009b). When the two conserved L2 cysteines were mutated in pseudo- or quasiviruses the particles produced were non infectious (Campos and Ozbun, 2009; Gambhira et al., 2009). When we tested the same mutations in native particles, not only were the mutant viruses infectious but titers were enhanced significantly (Conway et al., 2009a). This showed that viral morphogenesis can differ from particles formed in undifferentiating monolayer culture versus native particles formed in differentiating host epithelium. Finally, we have numerous other studies in the lab underway, including studies on mechanisms of infectivity, neutralization and cross-neutralization epitopes, disinfectant sensitivity, and microbicide sensitivity, all demonstrating differences between particles made in monolayer culture and native virus made in host epithelium.
Our laboratory has previously shown that both structural genes of HPV16 could function with the nonstructural genes of HPV18 for virus production (Meyers et al., 2002b). The present study sought to determine if the structural proteins, L2 and L1, could cooperate to produce infectious viral particles. We replaced either the L2 ORF or the L1 ORF of HPV18 with the counterpart from HPV16. All aspects of the viral life cycle examined including expression of the capsid proteins and particle assembly were found to be similar to wild-type, however while the HPV18 L2 protein could function with the HPV16 L1 protein to propagate infectious particles, the HPV16 L2 protein was not able to cooperate with the HPV18 L1 protein for infectious particle production.
In an effort to identify the regions of either the HPV18 L1 and/or the HPV16 L2 protein involved in the inability to propagate infectious virus, we made chimeras where only half of either the HPV18 L2 or L1 ORFs were exchanged for the complementary region of HPV16. After testing the eight resulting viral chimeras it appeared that whenever the N-terminal of the HPV18 L1 was present in the chimera with the C-terminal half of the HPV16 L2 protein, production of infectious virus synthesis was compromised. After comparing the L1 protein N-terminal sequences of HPV18 and HPV16 it was noticed that HPV18 had 35 unique amino acids on its L1 protein N-terminus. Additionally, when the L1 protein N-termini of HPV18 and HPV16 were aligned with the N-termini of other papillomaviruses, both had additional in-frame sequences on their N-termini when compared to the other papillomaviruses. Specifically, HPV18 had an additional 61 amino acids and HPV16 had an additional 26 amino acids. In studies where virus-like particles (VLP), pseudovirus, or quasivirus are used these extra sequences are deleted from the expression vectors used and all L1 expression begins at the ‘consensus’ ATG (Browne et al., 1988; Carter et al., 1991; Cason et al., 1994; Heino, Dillner, and Schwartz, 1995; Kirnbauer et al., 1992; Kirnbauer et al., 1993; Nardelli-Haefliger et al., 1997; Rossi et al., 2000; Xi and Banks, 1991; Zhou et al., 1990; Zhou et al., 1991a; Zhou et al., 1991b). While maps of HPV18 late gene mRNAs have yet to be published, splicing studies of the HPV16 capsid ORFs have mapped a splice acceptor at nt 5637 3’ to the first inframe ATG and 5’ to the second ATG of the L1 ORF (Doorbar et al., 1990; Milligan et al., 2007). Use of this splice acceptor would remove the extra 26 amino acids from the N-terminus of the HPV16 L1 protein. Evidence for the potential expression of L1 proteins initiated from the first ATG comes from a recent publication from our laboratory (Conway et al., 2009b). In this study, we demonstrated that native viral particles consisted of two L1 proteins of different sizes. Importantly, the slower migrating second L1 protein is not present in pseudo/quasiviruses (Conway et al., 2009b). While a post-translational modification is possible, the sizes of the two proteins are consistent with the larger being 26 amino acids more than the smaller. It is possible that another splice acceptor 5’ of the first ATG was missed in previous studies or it is also possible that the larger L1 protein expressed is translated from one of the several transcripts containing both the L2 and L1 ORFs (Milligan et al., 2007). In eukaryotic cells initiation of translation can occur by a mechanism of internal initiation where the ribosome is recruited to an internal start site that can be a considerable distance downstream of the 5’ terminus of the RNA (Charnay et al., 2009; Jackson, 2000; Lin, Li, and Shih, 2009; Stoneley and Willis, 2004). Using the Kozak rules for translational initiation the upstream ATG of HPV18 is either equivalent or superior to that for the downstream ATG (Kozak, 1989).
When the extra 35 amino acids (as compared to the L1 protein from HPV16) in the N-terminus of the HPV18 L1 protein was deleted, chimeric virus containing the L1 protein of HPV18 and the L2 protein of HPV16 produce viral titers equivalent to wild type. Therefore, these results demonstrate that the first 35 amino acids of HPV18 L1 N-terminus acts as a type-specific sequence affecting the proper interactions between HPV16 L2 and HPV18 L1.
HPV11 L1 has been shown to cooperate and form complexes with L2 proteins of HPV5, 6b, 12, 16, 33 and canine oral PV type 1 (COPV1), but not the L2s of either HPV1a or BPV1 (Finnen et al., 2003) demonstrating type specificity in the interaction of L2 and L1. Another study suggested that viral type-restricted domains exist affecting the cooperation between L1 and L2 of different PV types (Okun et al., 2001).
The L1 protein has been shown to be involved in viral DNA binding, cell surface binding and nuclear localization (Day and Schiller, 2006; Graham, 2006). All these known activities are concentrated in the L1 C-terminus. So far, there are no known activities reported for the L1 N-terminus. In contrast to L1, the minor capsid protein L2 has been shown to be multifunctional. For example, L2 is required to cooperate with L1 and induce the localization of viral L1 proteins to nuclear subdomain ND10 (Becker et al., 2003; Florin et al., 2002; Okun et al., 2001). In addition, the L2 protein consists of interactive sequences for the viral E2 protein (Finnen et al., 2003; Heino, Zhou, and Lambert, 2000; Okun et al., 2001), Hsc70 chaperones (Florin et al., 2004), β-actins (Yang et al., 2003b), t-SNARE (Bossis et al., 2005) and cell surface receptors (Da Silva et al., 2001; Giroglou et al., 2001; McMillan et al., 1999; Roden et al., 1994a). Incorporation of L2 into the viral capsid also enhances the VLP and pseudovirus assembly and infection (Holmgren et al., 2005).
In addition to biological activities, structural analysis of the HPV16 L1 VLP capsid has been characterized (Chen et al., 2000). Because the HPV16 L1 VLP could be crystallized for analysis only after the first 36 amino acids were deleted (the 26 that are all deleted in VLPs plus 9 more), the extreme L1 N-terminal fragment is missing and the structure is unknown. By comparison to the structurally related major capsid protein of SV40, the PV L1 N-terminus is proposed to project into the groove between the capsomeres to help stabilize the structure. The extra piece in HPV18 L1 N-terminus may be located close to the capsid surface and form extra contacts with the minor capsid protein to further stabilize the particle (Chen et al., 2000; Modis, Trus, and Harrison, 2002). Notably, 19 out of the 35 extra amino acids are hydrophobic. It is likely that this N-terminal sequence is buried inside the capsid or deep in the groove between the capsomeres. HPV18 L1 conformation dependent monoclonal antibodies H18.J4 and H18.K2 were both highly efficient at neutralizing infection of wild type HPV18 and HPV18-L2(18)L1(18) virus. All four chimeras with the HPV18 L1 N-terminal 35 amino acids deleted were neutralized by H18.J4 but not by H18.K2. The epitope recognized by H18.K2 cannot be contained within the deleted 35 amino acids because this sequence was not part of the VLP used to produce H18.K2. These results suggest that the epitope recognized by H18.K2 is sensitive to structural changes resulting from deletion. Additionally, the ELISA results suggest that the structural change does not necessarily inhibit binding of H18.K2 to the viral particle but only prevents its ability to neutralize infection. Using the in vitro raft culture system and mutating one methionine at a time may help us to identify the start codon(s) used by the native virus. Although the L1 structure has been studied, the L2 structure is largely unknown.
In this study, we used a genetic approach to study HPV morphogenesis and showed that the N-terminus of HPV18 L1 interfered with the cooperation between HPV18 L1 and HPV16 L2 during virion morphogenesis. After deleting the 35 amino acids in HPV18 L1 N-terminus, infectious virus production was greatly improved. Since HPV18 virus can be efficiently produced with or without the first 35 amino acids, its potential role in viral life cycle is intriguing. In addition, because most available conformation-dependent antibodies were raised against PV L1 VLPs, atomic structure analysis of VLPs combined with immunological studies using our chimeric viruses may also help us to better understand the similarities and differences between VLPs and authentic virions.
Supplementary Material
(A) Genomic organization of HPV18 displaying how the L2 and L1 ORFs overlap. The approximate positions of the ORFs are shown. The HPV18 major early promoter, p105, is indicated. PCR strategy was used to physically separate the two structural gene ORFs placing a HindIII (HIII) restriction sequence between the two ORFs. Primers used are listed in Table 1. (B) Cloning process to create HPV capsid mutants that have their L2 and L1 ORFs physically separated. Step 1 pHPV18L1/L2Δ was digested with BglII (BII) and HIII. pHPV18L1/L2Δ has the L2 and L1 ORFs deleted and replaced with a BII restriction sequence (41). Step 2 The amplified HPV18 and 16 L2 and L1 ORFs were digested with BII and HIII sets of one L2 ORF amplimer and one L1 ORF amplimer were ligated into the BII digested pHPV18L1/L2Δ in all four possible combinations. Step 3 The four mutant viral genomes were isolated and characterized by restriction digestion and sequencing for correctness.
L2 and L1 half and half chimeric mutants. Step 1 Each half (N-terminal and C-terminal) of the HPV18 and HPV16 L2 and L1 ORFs were PCR amplified using primers B1-4, C1-4, E1-4, and F1-4 as shown. This created N-terminal and C-terminal halves with the N-termini of the L2 ORFs and the C-termini of the L1 ORFs containing a BglII (BII) restriction site, and the C-termini of the L2 ORFs and the N-termini of the L1 ORFs containing a HindIII (HIII) restriction site. The other end of each amplified sequence contained a SapI (SI) restriction site. Step 2 Purified each L2 and L1 ORF amplimer. Step 3 Each amplimer was then digested with BII and SI or HIII and SI as appropriate. Step 4 The pGL2 vector was digested with BII and HIII. Step 5 Paired amplimers of capsid ORFs were ligated to the pGL2 vector as shown. Each recombinant plasmid containing a chimeric capsid ORF was isolated and analyzed by restriction digest. Step 6 The chimeric capsid ORFs were released from the recombinant plasmids by digestion with BII and HIII and purified. Step 7 The recombinant plasmid pHPV18L1/L2Δ was digested with BII and HIII. pHPV18L1/L2Δ has the L2 and L1 ORFs deleted and replaced with a BII restriction sequence. Step 8 Recombinant plasmids HPV18-L2(18)L1(18) and HPV18-L2(16)L1(16) (see Figs 1 and 2) were digested with BII and HII to release the wildtype HPV18 and HPV16 L2 and L1 ORFs. The wildtype ORFs were then isolated and purified. Step 9 BII and HIII digested pHPV18L1/L2Δ was ligated with sets of capsid ORFs containing one wildtype ORF (L2 or L1) and one chimeric ORF (L2 or L1) as shown. Step 10 Eight chimeric viral genomes were isolated and purified. Each chimeric genome was checked by restriction digest and sequencing for correctness.
Chimeric HPV-infected, fully stratified and differentiated raft culture tissues were stained with hematoxylin and eosin for histological analysis.
HPV18 L1 N-terminal 35 amino acid deletion mutants. Step 1 Using primers N-35 and F4 as shown, the HPV18 L1 ORF was PCR amplified excluding the first 105 nucleotides. The PCR amplification introduced a HIII restriction site on the 5’ end and a BII restriction site on the 3’ end as shown. Step 2 The amplimer was isolated and purified. Step 3 The amplimer was digested with HIII and BII. Step 4 Digested pHPV18L1/L2Δ with BII and HIII. pHPV18L1/L2Δ has the L2 and L1 ORFs deleted and replaced with a BII restriction sequence. Step 5 The BII and HIII digested pHPV18L1/L2Δ was ligated with the N-terminal deleted HPV18 L1 ORF and either a wildtype L2 or chimeric L2 ORF as shown. Recombinant plasmids containing the mutant viral genomes were isolated and purified. Each mutant was analyzed by restriction digest and sequenced for correctness.
Acknowledgments
We thank Lynn Budgeon for histological expertise; and the members of the Meyers’ laboratory for their critical reading of the manuscript and many helpful suggestions. This study was supported by a PHS grant from the National Institute of Allergy and Infectious Disease (R01AI57988).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alam S, Conway MJ, Chen HS, Meyers C. The cigarette smoke carcinogen benzo[a]pyrene enhances human papillomavirus synthesis. J Virol. 2008;82(2):1053–8. doi: 10.1128/JVI.01813-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KA, Florin L, Sapp C, Sapp M. Dissection of human papillomavirus type 33 L2 domains involved in nuclear domains (ND) 10 homing and reorganization. Virology. 2003;314(1):161–7. doi: 10.1016/s0042-6822(03)00447-1. [DOI] [PubMed] [Google Scholar]
- Bedell MA, Hudson JB, Golub TR, Turyk ME, Hosken M, Wilbanks GD, Laimins LA. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J Virol. 1991;65(5):2254–60. doi: 10.1128/jvi.65.5.2254-2260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop B, Dasgupta J, Klein M, Garcea R, Christensen N, Zhao R, Chen X. Crystal structures of four types of human papillomavirus L1 capsid proteins: understanding the specificity of neutralizing monoclonal antibodies. Journal of Biological Chemistry. 2007a;282(43):31803–31811. doi: 10.1074/jbc.M706380200. [DOI] [PubMed] [Google Scholar]
- Bishop B, Dasgupta J, Klein M, Garcea RL, Christensen ND, Zhao R, Chen XS. Crystal structures of four types of human papillomavirus L1 capsid proteins: understanding the specificity of neutralizing monoclonal antibodies. J Biol Chem. 2007b;282(43):31803–11. doi: 10.1074/jbc.M706380200. [DOI] [PubMed] [Google Scholar]
- Bodily JM, Alam S, Meyers C. Regulation of human papillomavirus type 31 late promoter activation and genome amplification by protein kinase C. Virology. 2006 doi: 10.1016/j.virol.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Bodily JM, Meyers C. Genetic analysis of the human papillomavirus type 31 differentiation-dependent late promoter. J Virol. 2005;79(6):3309–21. doi: 10.1128/JVI.79.6.3309-3321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossis I, Roden RB, Gambhira R, Yang R, Tagaya M, Howley PM, Meneses PI. Interaction of tSNARE syntaxin 18 with the papillomavirus minor capsid protein mediates infection. J Virol. 2005;79(11):6723–31. doi: 10.1128/JVI.79.11.6723-6731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne HM, Churcher MJ, Stanley MA, Smith GL, Minson AC. Analysis of the L1 gene product of human papillomavirus type 16 by expression in a vaccinia virus recombinant. J Gen Virol. 1988;69 ( Pt 6):1263–73. doi: 10.1099/0022-1317-69-6-1263. [DOI] [PubMed] [Google Scholar]
- Campos SK, Ozbun MA. Two highly conserved cysteine residues in HPV16 L2 form an intramolecular disulfide bond and are critical for infectivity in human keratinocytes. PLoS One. 2009;4(2):e4463. doi: 10.1371/journal.pone.0004463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter JJ, Yaegashi N, Jenison SA, Galloway DA. Expression of human papillomavirus proteins in yeast Saccharomyces cerevisiae. Virology. 1991;182(2):513–21. doi: 10.1016/0042-6822(91)90592-y. [DOI] [PubMed] [Google Scholar]
- Cason J, Kambo PK, Manse C, Jewers RJ, Best JM. Expression of human papillomavirus type 16 (HPV-16) major (L1) and minor (L2) capsid proteins in insect cells as polyhistidine fusion proteins. Biochem Soc Trans. 1994;22(3):336S. doi: 10.1042/bst022336s. [DOI] [PubMed] [Google Scholar]
- Charnay N, Ivanyi-Nagy R, Soto-Rifo R, Ohlmann T, Lopez-Lastra M, Darlix JL. Mechanism of HIV-1 Tat RNA translation and its activation by the Tat protein. Retrovirology. 2009;6:74. doi: 10.1186/1742-4690-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XS, Garcea RL, Goldberg I, Casini G, Harrison SC. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol Cell. 2000;5(3):557–67. doi: 10.1016/s1097-2765(00)80449-9. [DOI] [PubMed] [Google Scholar]
- Conway MJ, Alam S, Christensen ND, Meyers C. Overlapping and independent structural roles for human papillomavirus type 16 L2 conserved cysteines. Virology. 2009a;393(2):295–303. doi: 10.1016/j.virol.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway MJ, Alam S, Ryndock EJ, Cruz L, Christensen ND, Roden RB, Meyers C. Tissue-spanning redox gradient-dependent assembly of native human papillomavirus type 16 virions. J Virol. 2009b;83(20):10515–26. doi: 10.1128/JVI.00731-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva DM, Velders MP, Nieland JD, Schiller JT, Nickoloff BJ, Kast WM. Physical interaction of human papillomavirus virus-like particles with immune cells. Int Immunol. 2001;13(5):633–41. doi: 10.1093/intimm/13.5.633. [DOI] [PubMed] [Google Scholar]
- Day PM, Schiller JT. Early events in the papillomaviral life cycle. In: Campo MS, editor. PAPILLOMAVIRUS RESEARCH: Frm natural history to vaccines and beyond. Caister Academic Press; Norfolk: 2006. [Google Scholar]
- Doorbar J, Parton A, Hartley K, Banks L, Crook T, Stanley M, Crawford L. Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology. 1990;178(1):254–62. doi: 10.1016/0042-6822(90)90401-c. [DOI] [PubMed] [Google Scholar]
- Embers ME, Budgeon LR, Culp TD, Reed CA, Pickel MD, Christensen ND. Differential antibody responses to a distinct region of human papillomavirus minor capsid proteins. Vaccine. 2004;22(5–6):670–80. doi: 10.1016/j.vaccine.2003.08.037. [DOI] [PubMed] [Google Scholar]
- Fay A, Yutzy WHt, Roden RB, Moroianu J. The positively charged termini of L2 minor capsid protein required for bovine papillomavirus infection function separately in nuclear import and DNA binding. J Virol. 2004;78(24):13447–54. doi: 10.1128/JVI.78.24.13447-13454.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnen RL, Erickson KD, Chen XS, Garcea RL. Interactions between papillomavirus L1 and L2 capsid proteins. J Virol. 2003;77(8):4818–26. doi: 10.1128/JVI.77.8.4818-4826.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin L, Becker KA, Lambert C, Nowak T, Sapp C, Strand D, Streeck RE, Sapp M. Identification of a dynein interacting domain in the papillomavirus minor capsid protein l2. J Virol. 2006;80(13):6691–6. doi: 10.1128/JVI.00057-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin L, Becker KA, Sapp C, Lambert C, Sirma H, Muller M, Streeck RE, Sapp M. Nuclear translocation of papillomavirus minor capsid protein L2 requires Hsc70. J Virol. 2004;78(11):5546–53. doi: 10.1128/JVI.78.11.5546-5553.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin L, Schafer F, Sotlar K, Streeck RE, Sapp M. Reorganization of nuclear domain 10 induced by papillomavirus capsid protein l2. Virology. 2002;295(1):97–107. doi: 10.1006/viro.2002.1360. [DOI] [PubMed] [Google Scholar]
- Frattini MG, Lim HB, Laimins LA. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc Natl Acad Sci U S A. 1996;93(7):3062–7. doi: 10.1073/pnas.93.7.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambhira R, Jagu S, Karanam B, Day PM, Roden R. Role of L2 cysteines in papillomavirus infection and neutralization. Virol J. 2009;6:176. doi: 10.1186/1743-422X-6-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroglou T, Florin L, Schafer F, Streeck RE, Sapp M. Human papillomavirus infection requires cell surface heparan sulfate. J Virol. 2001;75(3):1565–70. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornemann J, Hofmann TG, Will H, Muller M. Interaction of human papillomavirus type 16 L2 with cellular proteins: identification of novel nuclear body-associated proteins. Virology. 2002;303(1):69–78. doi: 10.1006/viro.2002.1670. [DOI] [PubMed] [Google Scholar]
- Graham SV. Late events in the life cycle of human papillomaviruses. In: Campo MS, editor. PAPILLOMAVIRUS RESEARCH: From natural history to vaccines and beyond. Caister Academic Press; Norfolk: 2006. [Google Scholar]
- Grassmann K, Rapp B, Maschek H, Petry KU, Iftner T. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV–16 DNA. J Virol. 1996;70(4):2339–49. doi: 10.1128/jvi.70.4.2339-2349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heino P, Dillner J, Schwartz S. Human papillomavirus type 16 capsid proteins produced from recombinant Semliki Forest virus assemble into virus-like particles. Virology. 1995;214(2):349–59. doi: 10.1006/viro.1995.0044. [DOI] [PubMed] [Google Scholar]
- Heino P, Zhou J, Lambert PF. Interaction of the papillomavirus transcription/replication factor, E2, and the viral capsid protein, L2. Virology. 2000;276(2):304–14. doi: 10.1006/viro.2000.0342. [DOI] [PubMed] [Google Scholar]
- Holmgren SC, Patterson NA, Ozbun MA, Lambert PF. The minor capsid protein L2 contributes to two steps in the human papillomavirus type 31 life cycle. J Virol. 2005;79(7):3938–48. doi: 10.1128/JVI.79.7.3938-3948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ. In: Translational Control of Gene Expression. Sonenberg N, Hershey JWB, Matthews MB, editors. Cold Spring Harbour Press; Cold Spring Harbour: 2000. pp. 127–184. [Google Scholar]
- Kamper N, Day PM, Nowak T, Selinka HC, Florin L, Bolscher J, Hilbig L, Schiller JT, Sapp M. A membrane-destabilizing peptide in capsid protein L2 is required for egress of papillomavirus genomes from endosomes. J Virol. 2006;80(2):759–68. doi: 10.1128/JVI.80.2.759-768.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawana K, Yoshikawa H, Taketani Y, Yoshiike K, Kanda T. Common neutralization epitope in minor capsid protein L2 of human papillomavirus types 16 and 6. J Virol. 1999;73(7):6188–90. doi: 10.1128/jvi.73.7.6188-6190.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirnbauer R, Booy F, Cheng N, Lowy DR, Schiller JT. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci U S A. 1992;89(24):12180–4. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirnbauer R, Taub J, Greenstone H, Roden R, Durst M, Gissmann L, Lowy DR, Schiller JT. Efficient self-assembly of human papillomavirus type 16 L1 and L1–L2 into virus-like particles. J Virol. 1993;67(12):6929–36. doi: 10.1128/jvi.67.12.6929-6936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108(2):229–41. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laniosz V, Nguyen KC, Meneses PI. Bovine papillomavirus type 1 infection is mediated by SNARE syntaxin 18. J Virol. 2007;81(14):7435–48. doi: 10.1128/JVI.00571-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JY, Li ML, Shih SR. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res. 2009;37(1):47–59. doi: 10.1093/nar/gkn901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer TJ, Meyers C. Temporal and spatial expression of the E5a protein during the differentiation-dependent life cycle of human papillomavirus type 31b. Virology. 1998;248(2):208–17. doi: 10.1006/viro.1998.9262. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Bromberg-White JL, Meyers C. The role of the human papillomavirus type 18 E7 oncoprotein during the complete viral life cycle. Virology. 2005 doi: 10.1016/j.virol.2005.04.036. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Christensen ND, Meyers C. Propagation, infection, and neutralization of authentic HPV16 virus. Virology. 2004;322(2):213–9. doi: 10.1016/j.virol.2004.02.011. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Meyers C. Evidence for the coexistence of two genital HPV types within the same host cell in vitro. Virology. 2004;321(2):173–80. doi: 10.1016/j.virol.2003.12.019. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Meyers C. Propagation of infectious, high-risk HPV in organotypic “raft” culture. Methods Mol Med. 2005;119:171–86. doi: 10.1385/1-59259-982-6:171. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Wilson S, Mullikin B, Suzich J, Meyers C. Human papillomavirus type 45 propagation, infection, and neutralization. Virology. 2003;312(1):1–7. doi: 10.1016/s0042-6822(03)00312-x. [DOI] [PubMed] [Google Scholar]
- McMillan NA, Payne E, Frazer IH, Evander M. Expression of the alpha6 integrin confers papillomavirus binding upon receptor-negative B-cells. Virology. 1999;261(2):271–9. doi: 10.1006/viro.1999.9825. [DOI] [PubMed] [Google Scholar]
- Meyers C. Organotypic (raft) epithelial tissue culture system for the differentiation-dependent replication of papillomavirus. Methods in Cell Science. 1996;18:1–10. [Google Scholar]
- Meyers C, Alam S, Mane M, Hermonat PL. Altered biology of adeno-associated virus type 2 and human papillomavirus during dual infection of natural host tissue. Virology. 2001;287(1):30–9. doi: 10.1006/viro.2001.0968. [DOI] [PubMed] [Google Scholar]
- Meyers C, Bromberg-White JL, Zhang J, Kaupas ME, Bryan JT, Lowe RS, Jansen KU. Infectious virions produced from a human papillomavirus type 18/16 genomic DNA chimera. J Virol. 2002a;76(10):4723–33. doi: 10.1128/JVI.76.10.4723-4733.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers C, Bromberg-White JL, Zhang J, Kaupas ME, Bryan JT, Lowe RS, Jansen KU. Infectious virions produced from a human papillomavirus type 18/16 genomic DNA chimera. J Virol. 2002b;76(10):4723–33. doi: 10.1128/JVI.76.10.4723-4733.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers C, Frattini MG, Hudson JB, Laimins LA. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science. 1992;257(5072):971–3. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- Meyers C, Mayer TJ, Ozbun MA. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J Virol. 1997;71(10):7381–6. doi: 10.1128/jvi.71.10.7381-7386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan SG, Veerapraditsin T, Ahamet B, Mole S, Graham SV. Analysis of novel human papillomavirus type 16 late mRNAs in differentiated W12 cervical epithelial cells. Virology. 2007;360(1):172–81. doi: 10.1016/j.virol.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Trus BL, Harrison SC. Atomic model of the papillomavirus capsid. Embo J. 2002;21(18):4754–62. doi: 10.1093/emboj/cdf494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardelli-Haefliger D, Roden RB, Benyacoub J, Sahli R, Kraehenbuhl JP, Schiller JT, Lachat P, Potts A, De Grandi P. Human papillomavirus type 16 virus-like particles expressed in attenuated Salmonella typhimurium elicit mucosal and systemic neutralizing antibodies in mice. Infect Immun. 1997;65(8):3328–36. doi: 10.1128/iai.65.8.3328-3336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okun MM, Day PM, Greenstone HL, Booy FP, Lowy DR, Schiller JT, Roden RB. L1 interaction domains of papillomavirus l2 necessary for viral genome encapsidation. J Virol. 2001;75(9):4332–42. doi: 10.1128/JVI.75.9.4332-4342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbun M, Meyers C. Human papillomavirus type 31b transcription during the differentiation-dependent viral life cycle. Current Topics in Virology. 1999a;1:203–217. [Google Scholar]
- Ozbun MA, Meyers C. Transforming growth factor beta1 induces differentiation in human papillomavirus-positive keratinocytes. J Virol. 1996;70(8):5437–46. doi: 10.1128/jvi.70.8.5437-5446.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbun MA, Meyers C. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J Virol. 1997;71(7):5161–72. doi: 10.1128/jvi.71.7.5161-5172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbun MA, Meyers C. Human papillomavirus type 31b E1 and E2 transcript expression correlates with vegetative viral genome amplification. Virology. 1998a;248(2):218–30. doi: 10.1006/viro.1998.9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbun MA, Meyers C. Temporal usage of multiple promoters during the life cycle of human papillomavirus type 31b. J Virol. 1998b;72(4):2715–22. doi: 10.1128/jvi.72.4.2715-2722.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbun MA, Meyers C. Two novel promoters in the upstream regulatory region of human papillomavirus type 31b are negatively regulated by epithelial differentiation. J Virol. 1999b;73(4):3505–10. doi: 10.1128/jvi.73.4.3505-3510.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk RZ, Christensen ND, Michael KM, Muller M, Sehr P, Waterboer T, Pawlita M. Reactivity pattern of 92 monoclonal antibodies with 15 human papillomavirus types. J Gen Virol. 2008;89(Pt 1):117–29. doi: 10.1099/vir.0.83145-0. [DOI] [PubMed] [Google Scholar]
- Roden RB, Day PM, Bronzo BK, Yutzy WHt, Yang Y, Lowy DR, Schiller JT. Positively charged termini of the L2 minor capsid protein are necessary for papillomavirus infection. J Virol. 2001;75(21):10493–7. doi: 10.1128/JVI.75.21.10493-10497.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden RB, Kirnbauer R, Jenson AB, Lowy DR, Schiller JT. Interaction of papillomaviruses with the cell surface. J Virol. 1994a;68(11):7260–6. doi: 10.1128/jvi.68.11.7260-7266.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden RB, Weissinger EM, Henderson DW, Booy F, Kirnbauer R, Mushinski JF, Lowy DR, Schiller JT. Neutralization of bovine papillomavirus by antibodies to L1 and L2 capsid proteins. J Virol. 1994b;68(11):7570–4. doi: 10.1128/jvi.68.11.7570-7574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi JL, Gissmann L, Jansen K, Muller M. Assembly of human papillomavirus type 16 pseudovirions in Saccharomyces cerevisiae. Hum Gene Ther. 2000;11(8):1165–76. doi: 10.1089/10430340050015211. [DOI] [PubMed] [Google Scholar]
- Sen E, Alam S, Meyers C. Genetic and biochemical analysis of cis regulatory elements within the keratinocyte enhancer region of the human papillomavirus type 31 upstream regulatory region during different stages of the viral life cycle. J Virol. 2004;78(2):612–29. doi: 10.1128/JVI.78.2.612-629.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23(18):3200–7. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- Taichman LP, LaPorta RF. The expression of papillomaviruses in epithelial cells. In: Salzman N, Howley PM, editors. The Papillomaviruses. Vol. 2. Plenum Press; New York: 1986. pp. 109–139. [Google Scholar]
- Xi SZ, Banks LM. Baculovirus expression of the human papillomavirus type 16 capsid proteins: detection of L1–L2 protein complexes. J Gen Virol. 1991;72(Pt 12):2981–8. doi: 10.1099/0022-1317-72-12-2981. [DOI] [PubMed] [Google Scholar]
- Yang R, Day PM, Yutzy WHt, Lin KY, Hung CF, Roden RB. Cell surface-binding motifs of L2 that facilitate papillomavirus infection. J Virol. 2003a;77(6):3531–41. doi: 10.1128/JVI.77.6.3531-3541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Yutzy WHt, Viscidi RP, Roden RB. Interaction of L2 with beta-actin directs intracellular transport of papillomavirus and infection. J Biol Chem. 2003b;278(14):12546–53. doi: 10.1074/jbc.M208691200. [DOI] [PubMed] [Google Scholar]
- Zhou J, Crawford L, McLean L, Sun XY, Stanley M, Almond N, Smith GL. Increased antibody responses to human papillomavirus type 16 L1 protein expressed by recombinant vaccinia virus lacking serine protease inhibitor genes. J Gen Virol. 1990;71 ( Pt 9):2185–90. doi: 10.1099/0022-1317-71-9-2185. [DOI] [PubMed] [Google Scholar]
- Zhou J, Sun XY, Louis K, Frazer IH. Interaction of human papillomavirus (HPV) type 16 capsid proteins with HPV DNA requires an intact L2 N-terminal sequence. J Virol. 1994;68(2):619–25. doi: 10.1128/jvi.68.2.619-625.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Sun XY, Stenzel DJ, Frazer IH. Expression of vaccinia recombinant HPV 16 L1 and L2 ORF proteins in epithelial cells is sufficient for assembly of HPV virion-like particles. Virology. 1991a;185(1):251–7. doi: 10.1016/0042-6822(91)90772-4. [DOI] [PubMed] [Google Scholar]
- Zhou JA, McIndoe A, Davies H, Sun XY, Crawford L. The induction of cytotoxic T-lymphocyte precursor cells by recombinant vaccinia virus expressing human papillomavirus type 16 L1. Virology. 1991b;181(1):203–10. doi: 10.1016/0042-6822(91)90485-t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Genomic organization of HPV18 displaying how the L2 and L1 ORFs overlap. The approximate positions of the ORFs are shown. The HPV18 major early promoter, p105, is indicated. PCR strategy was used to physically separate the two structural gene ORFs placing a HindIII (HIII) restriction sequence between the two ORFs. Primers used are listed in Table 1. (B) Cloning process to create HPV capsid mutants that have their L2 and L1 ORFs physically separated. Step 1 pHPV18L1/L2Δ was digested with BglII (BII) and HIII. pHPV18L1/L2Δ has the L2 and L1 ORFs deleted and replaced with a BII restriction sequence (41). Step 2 The amplified HPV18 and 16 L2 and L1 ORFs were digested with BII and HIII sets of one L2 ORF amplimer and one L1 ORF amplimer were ligated into the BII digested pHPV18L1/L2Δ in all four possible combinations. Step 3 The four mutant viral genomes were isolated and characterized by restriction digestion and sequencing for correctness.
L2 and L1 half and half chimeric mutants. Step 1 Each half (N-terminal and C-terminal) of the HPV18 and HPV16 L2 and L1 ORFs were PCR amplified using primers B1-4, C1-4, E1-4, and F1-4 as shown. This created N-terminal and C-terminal halves with the N-termini of the L2 ORFs and the C-termini of the L1 ORFs containing a BglII (BII) restriction site, and the C-termini of the L2 ORFs and the N-termini of the L1 ORFs containing a HindIII (HIII) restriction site. The other end of each amplified sequence contained a SapI (SI) restriction site. Step 2 Purified each L2 and L1 ORF amplimer. Step 3 Each amplimer was then digested with BII and SI or HIII and SI as appropriate. Step 4 The pGL2 vector was digested with BII and HIII. Step 5 Paired amplimers of capsid ORFs were ligated to the pGL2 vector as shown. Each recombinant plasmid containing a chimeric capsid ORF was isolated and analyzed by restriction digest. Step 6 The chimeric capsid ORFs were released from the recombinant plasmids by digestion with BII and HIII and purified. Step 7 The recombinant plasmid pHPV18L1/L2Δ was digested with BII and HIII. pHPV18L1/L2Δ has the L2 and L1 ORFs deleted and replaced with a BII restriction sequence. Step 8 Recombinant plasmids HPV18-L2(18)L1(18) and HPV18-L2(16)L1(16) (see Figs 1 and 2) were digested with BII and HII to release the wildtype HPV18 and HPV16 L2 and L1 ORFs. The wildtype ORFs were then isolated and purified. Step 9 BII and HIII digested pHPV18L1/L2Δ was ligated with sets of capsid ORFs containing one wildtype ORF (L2 or L1) and one chimeric ORF (L2 or L1) as shown. Step 10 Eight chimeric viral genomes were isolated and purified. Each chimeric genome was checked by restriction digest and sequencing for correctness.
Chimeric HPV-infected, fully stratified and differentiated raft culture tissues were stained with hematoxylin and eosin for histological analysis.
HPV18 L1 N-terminal 35 amino acid deletion mutants. Step 1 Using primers N-35 and F4 as shown, the HPV18 L1 ORF was PCR amplified excluding the first 105 nucleotides. The PCR amplification introduced a HIII restriction site on the 5’ end and a BII restriction site on the 3’ end as shown. Step 2 The amplimer was isolated and purified. Step 3 The amplimer was digested with HIII and BII. Step 4 Digested pHPV18L1/L2Δ with BII and HIII. pHPV18L1/L2Δ has the L2 and L1 ORFs deleted and replaced with a BII restriction sequence. Step 5 The BII and HIII digested pHPV18L1/L2Δ was ligated with the N-terminal deleted HPV18 L1 ORF and either a wildtype L2 or chimeric L2 ORF as shown. Recombinant plasmids containing the mutant viral genomes were isolated and purified. Each mutant was analyzed by restriction digest and sequenced for correctness.