Abstract
The large (about 2200 amino acids) L polymerase protein of nonsegmented negative-strand RNA viruses (order Mononegavirales) has six conserved sequence regions (“domains”) postulated to constitute the specific enzymatic activities involved in viral mRNA synthesis, 5′-end capping, cap methylation, 3′ polyadenylation, and genomic RNA replication. Previous studies with vesicular stomatitis virus identified amino acid residues within the L protein domain VI required for mRNA cap methylation. In our recent study we analyzed four amino acid residues within domain VI of the Sendai virus L protein and our data indicated that there could be differences in L protein sequence requirements for cap methylation in two different families of Mononegavirales — rhabdoviruses and paramyxoviruses. In this study, we conducted a more comprehensive mutational analysis by targeting the entire SeV L protein domain VI, creating twenty-four L mutants, and testing these mutations for their effects on viral mRNA synthesis, cap methylation, viral genome replication and virus growth kinetics. Our analysis identified several residues required for successful cap methylation and virus replication and clearly showed the importance of the K-D-K-E tetrad and glycine-rich motif in the SeV cap methylation. This study is the first extensive sequence analysis of the L protein domain VI in the family Paramyxoviridae, and it confirms structural and functional similarity of this domain across different families of the order Mononegavirales.
Keywords: Sendai virus, Paramyxovirus, mRNA cap methylation, Methyltransferase, L polymerase protein
Introduction
Viruses of the order Mononegavirales include diverse human, animal and plant pathogens that share structurally similar nonsegmented, negative-strand RNA genomes with similar strategies for viral RNA genome replication, transcription and posttranscriptional modifications of viral mRNAs (Lamb and Parks, 2007, Lyles and Rupprecht, 2007, Whelan et al., 2004). All members of this order encode the large (L) polymerase protein which has six highly conserved regions (“domains”) postulated to be responsible for the specific enzymatic activities of the viral polymerase complex which include viral genome replication, transcription, mRNA 5′ capping, cap methylation and 3′ polyadenylation. Currently, there is no structural data available for the entire L or any region of L. However, site-directed mutagenesis and computational analyses support the multifunctional nature of the L protein as targeted amino acid substitutions in the different L domains were able to inactivate individual functions of viral polymerase (Cartee et al., 2003, Cortese et al., 2000, Grdzelishvili et al., 2005, Li et al., 2005, Li et al., 2008, Ogino and Banerjee, 2010, Ogino et al., 2010, Poch et al., 1990, Schnell and Conzelmann, 1995, Sidhu et al., 1993, Sleat and Banerjee, 1993, Smallwood et al., 2002).
Similarly to eukaryotic mRNA, most of Mononegavirales synthesize mRNA containing a 5′ cap structure methylated at the guanine-N7 (G-N-7) and 2′-O-adenosine (2′-O) positions. The caps are required for mRNA stability, and cap methylation, especially at G-N-7 position, is required for efficient mRNA translation (Gingras et al., 1999, Horikami et al., 1984, Horikami and Moyer, 1982). The methyltransferase (MTase) activity was originally mapped to the L protein following the characterization of two vesicular stomatitis virus (VSV, family Rhabdoviridae) host range (hr) mutants. These mutants exhibited severe defects in cap methylation (Horikami et al., 1984, Horikami and Moyer, 1982), but this function was successfully complemented with purified wild type (wt) L protein in vitro, demonstrating that L possesses the viral mRNA MTase activities (Hercyk et al., 1988). More recently, computational analyses predicted that the L protein domain VI has a typical 2′-O MTase fold and identified a putative K-D-K-E catalytic tetrad and a glycine-rich motif (GxGxG) as the putative AdoMet binding site (Bujnicki and Rychlewski, 2002, Ferron et al., 2002, Martin and McMillan, 2002). These predictions were experimentally confirmed by several studies with the VSV L protein leading to the identification of the amino acid residues important for cap methylation within domain VI (Galloway et al., 2008, Grdzelishvili et al., 2005, Li et al., 2005, Li et al., 2006). Grdzelishvili et al. (2005) showed that one of the VSV hr mutants, hr1, had a single substitution D to V within the glycine-rich motif (GDGSG in VSV) and was completely defective in cap methylation. Further site-directed mutagenesis of the glycine-rich motif and putative K-D-K-E catalytic tetrad by Li et al., 2005, Li et al., 2006 showed that they are important for mRNA cap methylation at both the G-N-7 and 2′-O positions. Most of previous studies suggest that the L protein uses a single AdoMet binding site for both G-N-7 and 2′-O MTase activity, and that, at least in VSV, 2′-O methylation precedes G-N-7 methylation (Rahmeh et al., 2009, Testa and Banerjee, 1977).
While most of the cap methylation studies were conducted using VSV, limited studies using Sendai virus (SeV, family Paramyxoviridae) demonstrated similarities as well as differences in the cap methylation between these two distantly related viruses. SeV produces mRNA that is capped and methylated at both the G-N-7 and 2′-O positions (Takagi et al., 1995), but interestingly, purified SeV L protein or just its C-terminal portion retaining domain VI, catalyzed only G-N-7, but not the 2′-O cap methylation (Ogino et al., 2005). In both VSV and SeV, alanine substitutions of the lysine upstream of the glycine-rich motif (also the first lysine of a putative K-D-K-E catalytic tetrad) led to cap methylation defects in these two viruses (Li et al., 2006, Murphy and Grdzelishvili, 2009). However, while alanine substitutions at the first two positions of the glycine-rich motif (G1804 and G1806) had little effect on MTase activity in SeV (Murphy and Grdzelishvili, 2009), in VSV the substitutions at the homologous glycine positions G1670 and G1672 affected cap methylation at either the G-N-7 or both the 2′-O and G-N-7 positions (Li et al., 2006). Studies with VSV also identified a region upstream of domain VI important for cap methylation (Grdzelishvili et al., 2006). Interestingly, the recent study showed that VSV tolerates an insertion of GFP gene between domain VI and this upstream region, and a recombinant virus with such insertion showed a normal growth in cell culture but no virion-associated activity in vitro (Ruedas and Perrault, 2009). The upstream region has not been studied in SeV as there is no homology in this variable region between rhabdoviruses and paramyxoviruses, although the L protein of measles virus (a paramyxovirus) was also reported to tolerate GFP insertion in a region just upstream of domain VI (Duprex et al., 2002).
To dissect the L protein sequence requirements for cap methylation in SeV in more detail, we conducted a more comprehensive analysis by targeting the entire SeV L protein domain VI and created twenty-four L mutants by site-directed mutagenesis at highly conserved positions within this domain, using sequence conservation between L proteins in Mononegavirales as a guide (Bujnicki and Rychlewski, 2002). The L mutations were analyzed in the context of infectious mutant viruses for their effect on viral mRNA cap methylation and virus growth in vitro, and we found a good correlation between attenuation in cell culture and defects in MTase activity for most of SeV mutants. Our analysis experimentally confirms previous computational predictions suggesting the importance of the glycine-rich motif and K-D-K-E tetrad in cap methylation across different families of the order Mononegavirales. In addition, the majority of L mutants were tested for their ability to synthesize viral mRNA and replicate viral genomic RNA. This study is the first detailed analysis of the L protein domain VI in the family Paramyxoviridae.
Results
Site-directed mutagenesis of the SeV L protein domain and recovery of infectious SeV mutants
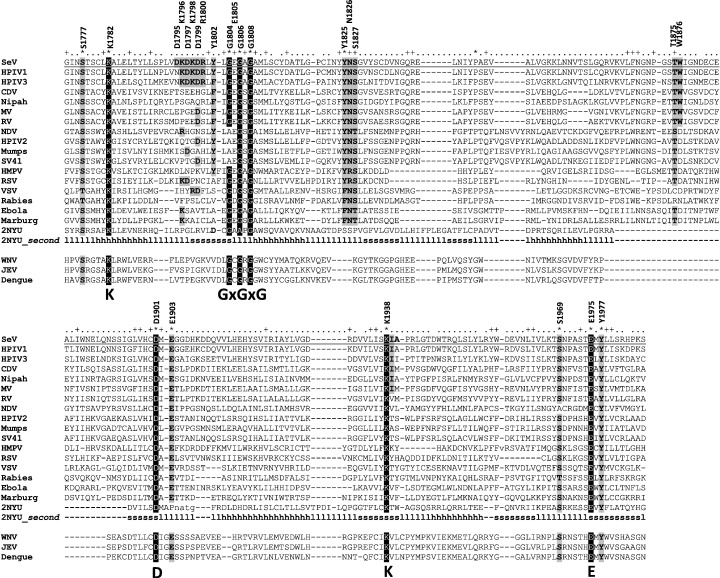
In our recent study using SeV we targeted four amino acid residues in domain VI and showed a discrepancy between VSV and SeV in their L protein sequence requirements for cap methylation (Murphy and Grdzelishvili, 2009). In this work, we analyzed the entire domain VI of SeV in more detail, targeting the residues highly conserved in most members of the order Mononegavirales (Fig. 1 ). In addition to targeting the glycine-rich motif (the putative AdoMet binding site) and the putative catalytic tetrad K-D-K-E, which also is conserved in other known 2′-O MTases including NS5 protein of flaviviruses (Fig. 1), we also targeted residues D1799 and Y1802 located just upstream of the glycine-rich motif and conserved only in some paramyxoviruses (Fig. 1). Many classes of MTases of known structure contain either a conserved aspartate or glutamate, or a tyrosine residue within the beta strand that precedes the conserved glycine-rich motif, but rarely do they contain both these residues. A polar residue in this position has been implicated in reaction mechanism as the fifth catalytic entity (Kozbial and Mushegian, 2005), and we were interested in determining whether one or both of these amino acids may play a functional role in SeV MTase. We also targeted the DKDK sequence located immediately upstream of the residue D1799. Although this DKDKD sequence is present only in some paramyxoviruses, we wanted to determine whether such high concentration of aspartates and lysines and its close proximity to the glycine-rich motif may play some role in cap methylation catalysis. In addition to single amino acid substitutions, several double and triple alanine substitutions were created, and the glycines within the glycine-rich motif were changed to leucines to address a possibility that this motif, while important for cap methylation in SeV, is more tolerant to single glycine-to-alanine substitutions as compared to VSV. All told, twenty-four L mutant genes were generated in the context of the pGEM-L vector with the L gene under control of the T7 promoter for their analysis using VVT7 system (see below). In addition, we cloned these mutations into a SeV full-length antigenomic plasmid, and we were able to generate 19 infectious SeV mutants using a reverse genetics system (Table 1 ). Infectious virus particles could not be recovered for the remaining five mutant L genes (indicated as NR in Table 1).
Fig. 1.
SeV L protein mutants generated and analyzed in this study. Multiple alignment of the L protein domain VI for members of the order Mononegavirales in comparison to the MTase domain of NS5 protein in flaviviruses. Multiple alignment was conducted using AliBee Multiple Alignment program from the GeneBee website (www.genebee.msu.su/genebee.html) (Brodsky et al., 1993). The K-D-K-E and glycine-rich motif mutated positions are highlighted in black, while gray shadows indicate other amino acid substitutions generated in this study. “SeV”: Sendai virus L protein [NCBI gene ID (gi):297180]; “HPIV1 or 3”: human parainfluenza virus 1 [gi:19718373] or 3 [gi:3510306]; “CDV”: canine distemper virus [gi:39938470]; “Nipah”: Nipah virus [gi:253559849] “MV”: measles virus [gi:1041625] “RV”: Rinderpest virus [gi:56410436] “NDV”: Newcastle disease virus [gi:11545725]; “HPIV2”: human parainfluenza virus 2 [gi:19525727]; “Mumps”: mumps virus [gi:50404170] “SV41”: Simian virus 41 [gi:55770827]; “HMPV”: human metapneumovirus [gi:46852141]; “RSV”: human respiratory syncytial virus [gi:1695266]; “VSV”: vesicular stomatitis virus [gi:336028] “Rabies”: rabies virus [gi:237688385] “Ebola”: Ebola virus [gi:10313999] “Marburg”: Marburg virus [gi:158539115]; 2NYU — ortholog of RNA MTase FtsJ [gi:119390696]; “WNV”: West Nile virus [gi:27735310] “JEV”: Japanese encephalitis virus [gi:189086643] “Dengue”: Dengue virus 2 [gi:158851624]. 2NYU_second: secondary structure elements observed in the three-dimensional structure of the human ortholog of RNA MTase FtsJ (PDB code 2NYU) are shown: h stands for alpha helix, s stands for beta strand, and l stands for loop.
Table 1.
Cap methylation analysis using SeV detergent-activated purified virions.
| Viruses | Virus stock infectivity (CIU/ml) | Txn − AdoHcy (% of rWT − AdoHcy) | Txn + AdoHcy (% of rWT − AdoHcy) | † Cap Methylation − AdoHcy (% of rWT − AdoHcy) | † Cap Methylation + AdoHcy (% of rWT − AdoHcy) |
|---|---|---|---|---|---|
| rWT | 3.8 × 108 | 100.0 | 67.0 ± 26.0 | 100.0 | 2.7 ± 0.2 |
| rS1777A | 4 × 107 | 100.0 ± 25.8 | 70.3 ± 5.2 | 45.8 ± 11.9 | 0.7 ± 0.2 |
| rK1782A* | 2 × 105 | 25.8 ± 2.6 | 32.8 ± 11.7 | 6.6 ± 1.2 | 1.4 ± 0.7 |
| rD1795A | 8 × 107 | 55.4 ± 12.4 | 51.3 ± 17.0 | 51.6 ± 16.9 | 1.7 ± 0.6 |
| rK1796A | 8 × 107 | 54.0 ± 13.5 | 41.6 ± 3.9 | 82.2 ± 13.2 | 1.7 ± 0.3 |
| rD1797A | 8 × 107 | 79.4 ± 0.5 | 64.4 ± 25.2 | 120.9 ± 20.8 | 2.7 ± 0.1 |
| rK1798A | 1 × 108 | 48.9 ± 7.1 | 24.8 ± 0.5 | 104.0 ± 0.8 | 4.2 ± 0.6 |
| rD1799A | 1.4 × 107 | 64.2 ± 12.7 | 53.1 ± 15.1 | 80.2 ± 19.8 | 2.7 ± 0.4 |
| rR1800A | 4.8 × 107 | 70.1 ± 7.3 | 70.7 ± 9.9 | 48.5 ± 17.8 | 1.5 ± 1.0 |
| rY1802A | 1.4 × 108 | 14.6 ± 5.6 | 15.4 ± 3.0 | 7.2 ± 0.5 | 1.8 ± 1.7 |
| rG1804A* | 3.4 × 107 | nd | nd | nd | nd |
| rG1804L | 1 × 106 | 27.9 ± 4.6 | 30.1 ± 5.8 | 9.9 ± 0.7 | 1.2 ± 0.5 |
| rE1805A* | 3.2 × 107 | 34.0 ± 15.8 | 50.0 ± 2.8 | 22.8 ± 5.2 | 5.3 ± 5.7 |
| rE1805L | NR | NR | NR | NR | NR |
| rG1806A* | 2.2 × 108 | nd | nd | nd | nd |
| rG1806L | < 102 | nd | nd | nd | nd |
| rG1808A | 8 × 107 | 21.8 ± 8.2 | 18.1 ± 6.3 | 31.4 ± 0.1 | 1.8 ± 1.5 |
| rG1808L | NR | NR | NR | NR | NR |
| rG1804A/G1806A* | 8 × 107 | 114.8 ± 5.9 | 86.8 ± 12.8 | 50.9 ± 1.6 | 12.5 ± 0.3 |
| rG1804A/G1806A/G1808A | 1.4 × 108 | 25.0 ± 5.0 | 17.2 ± 3.1 | 36.8 ± 5.0 | 2.1 ± 0.5 |
| rY1825A/N1826A/S1827A | NR | NR | NR | NR | NR |
| rW1876A | 1.6 × 108 | 20.3 ± 2.8 | 21.9 ± 6.2 | 46.8 ± 10.3 | 1.5 ± 0.3 |
| rT1875A/W1876A | NR | NR | NR | NR | NR |
| rD1901A | 1.4 × 107 | 37.4 ± 5.7 | 27.3 ± 4.9 | 2.1 ± 0.5 | 0.2 ± 0.3 |
| rE1903A | 4 × 105 | 26.1 ± 4.4 | 27.3 ± 3.5 | 7.3 ± 2.4 | 0.2 ± 0.5 |
| rK1938S | 1.2 × 106 | 91.0 ± 12.7 | 60.1 ± 3.1 | 8.1 ± 0.9 | 2.2 ± 0.6 |
| rK1938A/I1939L | 1.2 × 106 | 46.9 ± 23.2 | 28.4 ± 16.1 | 2.4 ± 0.5 | 1.1 ± 0.4 |
| rS1969A | 4.4 × 107 | 71.5 ± 10.1 | 67.0 ± 12.4 | 73.7 ± 20.9 | 2.1 ± 0.1 |
| rE1975A | NR | NR | NR | NR | NR |
| rY1977A | 8 × 107 | 55.5 ± 16.7 | 48.5 ± 23.2 | 28.6 ± 5.7 | 5.1 ± 3.3 |
− AdoHcy: in vitro transcription in the absence of S-adenosylhomocysteine (AdoHcy).
+ AdoHcy: in vitro transcription in the presence of 100 μM AdoHcy.
† methylation of viral mRNA (NP + P) produced in vitro is expressed as the ratio (% of rWT) of [3H]-AdoMet incorporation into viral mRNA by scintillation counting to the mRNA levels determined analyzed by Northern blot (mRNA levels are shown in the “Txn” columns).
NR = not rescued (unable to rescue after multiple attempts).
nd = not determined.
*Viruses were also analyzed in our previous study (Murphy and Grdzelishvili, 2009).
CIU = cell infectious units.
The data represent the mean ± standard deviation of two independent experiments.
Growth analysis of SeV L mutants in Vero cells
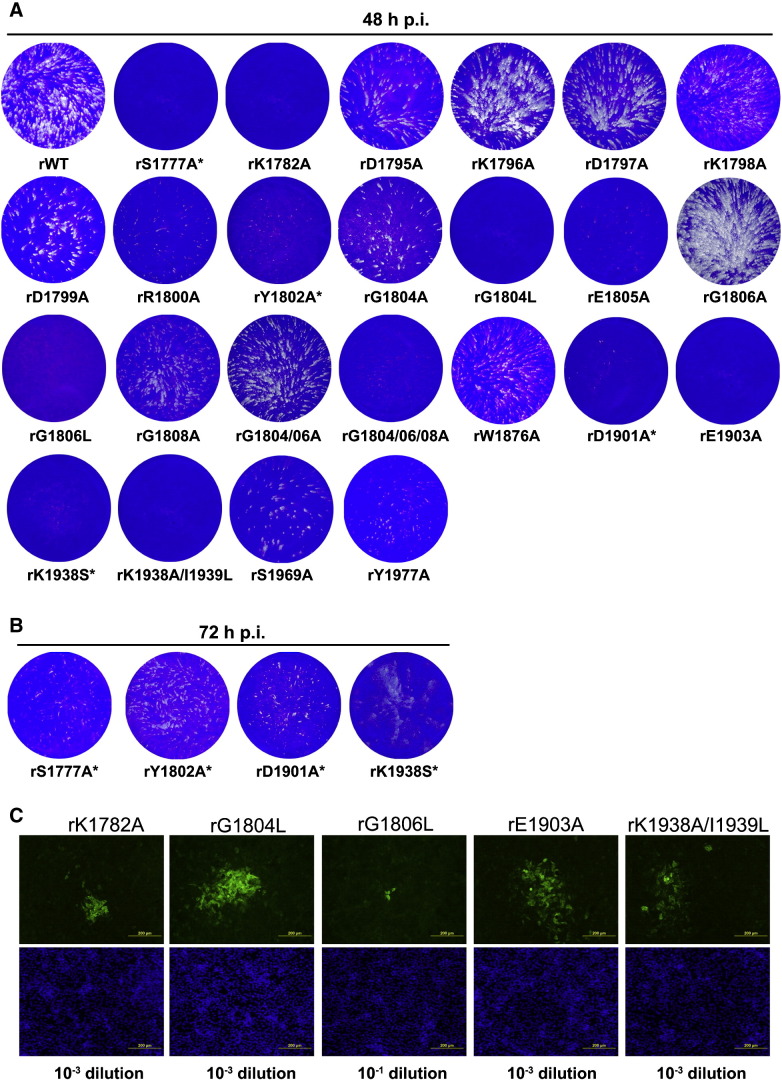
All infectious SeV mutants were initially tested for their ability to infect and produce a cytopathic effect (CPE) in Vero cells. Infectious foci were visualized by crystal violet staining or IF at 48 or 72 h p.i. Several mutants behaved similarly to rWT, while some mutants had obvious defects in growth based on their inability to form visible infectious foci (Fig. 2A). All of the mutants with the substitutions between positions 1795 and 1800 (DKDKDR) were capable of forming visible infectious foci and grew to rWT-like titers (Fig. 2A and Table 1). The rG1804A, rE1805A, rG1806A mutants and the double-mutant rG1804A/G1806A behaved as in our previous report (Murphy and Grdzelishvili, 2009), i.e., they showed similar growth to rWT. Even the rE1805A mutant, which had a slight delay in growth, still grew to high titers. Additional alanine mutants in or around glycine-rich motif behaved with minor variations: the rG1808A and especially the triple mutant rG1804A/G1806A/G1808A had smaller than rWT infectious foci, but grew to a high titer (1.4 × 108 CIU/ml), and the rY1802A mutant had small infectious foci 48 h p.i. but at 72 h p.i. had foci size and titer similar to rWT (Fig. 2B). In contrast to these mutants, leucine substitutions had dramatic effect on virus growth: E1805L and G1808L could not be recovered despite all effort; rG1804L was dramatically attenuated in Vero cells and infectious foci could only be detected by IF (Fig. 2C); and rG1806L was severely attenuated in cell culture, and even after several passages on Vero cells, titers remained extremely low (< 102 CIU/ml) (Fig. 2C and Table 1). Thus, the GxGxG motif, which in the cases of homologous MTases with the known structure is invariably located in the loop between the first beta strand and the alpha helix of the Rossmann fold, tolerates substitutions to small side chain residue such as alanine, but the bulkier aliphatic side chain of leucine appears incompatible with the structure or function of this region. The rules for E1805 were similar: negative charge turned out to be unimportant for virus viability, even though this residue is conserved in all Mononegavirales, but a bulky aliphatic leucine residue was not tolerated.
Fig. 2.
Growth analysis of recombinant SeV mutants in Vero cells. (A) Crystal violet staining of infectious foci of all rescued SeV L mutants. Plaque assays were performed on Vero cells and infectious foci from the lowest dilution displaying infectious foci were stained 48 h p.i. Wells with no visual infectious foci at 48 h p.i. were analyzed at 72 h p.i. or by IF. (B) For delayed mutants (indicated by *), plaque assays were again performed and crystal violet staining of infectious foci was done at 72 h p.i. (C) For severely attenuated mutants that had no visible infectious foci at 48 or 72 h p.i. an IF assay was performed at 72 h p.i. Upper panels represent cells stained with an anti-SeV primary antibody and an IgG secondary antibody conjugated to FITC. Lower panels represent Hoeschst staining of nuclei from the same fields shown in upper panels.
We reported previously that the SeV L mutant with an alanine substitution at the first position of the putative K-D-K-E catalytic tetrad, rK1782A, was attenuated in cell culture (Murphy and Grdzelishvili, 2009). Here we created alanine substitutions at the other positions of this motif and rescued viruses with substitutions at the D1901 and K1938 positions; however, we were not able to rescue a virus with a mutation at E1975. Interestingly, after sequencing, the K1938 position mutant had a substitution to serine and not alanine. This serine substitution apparently has been selected in vivo, as the input plasmids used for virus recovery were confirmed by sequencing to have the alanine substitution. In addition to targeting the K-D-K-E tetrad, we created alanine mutants of two other positions invariant in Mononegavirales, S1777 and E1903, and a double mutant, rK1938A/I1939L. All SeV L mutants with substitutions in and around this putative K-D-K-E catalytic tetrad were attenuated when grown on Vero cells (Fig. 2A). rK1782A, rE1903A, and rK1938A/I1939L produced infectious foci detectable only by IF (Fig. 2C).
The amino acids of the K-D-K-E tetrad are widely spaced in the sequence of virus MTase, but are brought to close distance in the homologous MTases of the known structure and in the predicted spatial structures of the L domain VI proteins (Bujnicki and Rychlewski, 2002, Galloway et al., 2008). They form a semi-circle on the outer rim of the AdoMet binding pocket and are thought to work together in transferring the methyl group from that donor to the 5′ nucleotides of virus mRNA, though exact role of each residue, as well as the details of the reaction mechanism (and indeed, the native three-dimensional structures of MTases of Mononegavirales), remain to be investigated. Residues S1777 and E1903 are predicted to be further outwards from the AdoMet binding pocket, and may be expected not to interact with the methyl donor, but rather perhaps play a role in recognition of the RNA substrate. Our results indicate that single alanine mutations in most of these residues result in attenuation of virus infection.
We also recovered several mutants with alanine substitutions at other invariant positions of domain VI, i.e., W1876A, S1969A, and Y1977A. These mutant viruses behaved similarly to rWT in Vero cells and grew to high titers (Fig. 1A and Table 1). The corresponding residues are predicted to be located outside of the ligand-binding pocket and may not be involved in any intramolecular interactions. We, however, were unable to recover virus progeny in the mutants that had multiple alanine substitutions in these patches of amino acids predicted to face outwards, i.e., a triple substitution Y1825A/N1826A/S1827A or a double substitution T1875A/W1876A. These highly conserved regions might be necessary for interactions of SeV MTase with other regions within the L protein or with other proteins.
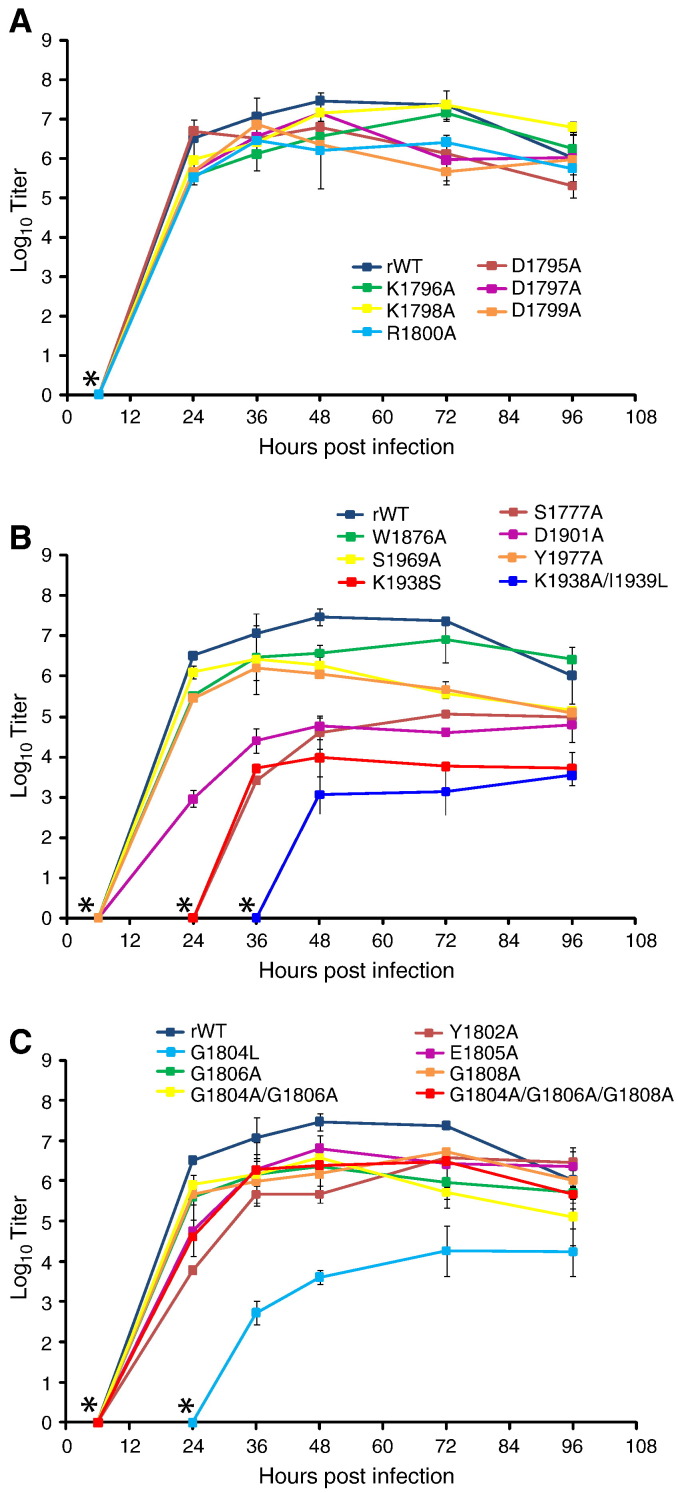
To further examine the growth characteristics of all rescued mutant viruses, a one-step growth kinetics analysis was performed as described in Materials and methods (Fig. 3 ). rWT reached its highest titer at 48 h p.i. and then declined at 72 and 96 h p.i. Mutants with substitutions within the DKDKD sequence upstream of the glycine-rich motif had slightly lower titers than rWT overall, but still reached their highest titers at 48 h p.i. (Fig. 3A). For mutants with substitutions in the glycine-rich motif, the majority behaved similarly to rWT with slightly lower titers overall and maximum titers (107–108 CIU/ml) reached at 48 h p.i. (Fig. 3C). rG1804L was severely attenuated with infectious particles detected only after 24 h p.i. and maximum titers reaching only 105 CIU/ml at 96 h p.i. L mutants with substitutions in and around the putative K-D-K-E catalytic tetrad were delayed in growth and had dramatically lower maximum titers (104–105 CIU/ml) as compared to rWT.
Fig. 3.
One-step growth kinetics of SeV mutants in Vero cells. Monolayer cultures of Vero cells were infected at an MOI of 3 CIU/cell with each mutant SeV. Vero cells were incubated with viruses for 1 h, then unabsorbed viruses were aspirated, cells were washed two times with PBS and fresh was added to each well. Supernatants were collected at 6, 24, 36, 48, 72, and 96 h p.i. and flash frozen. Plaque assays were performed on Vero cells and virus titers were determined for each time interval. * Zero titer indicates that virus titer at the indicated time point was below our detection threshold (50 CIU/ml). (A) SeV mutants with alanine substitutions in a DKDKD motif upstream of the glycine-rich motif. (B) SeV mutants that showed delayed growth in Vero cells or lower titers as compared to rWT. (C) SeV mutants with substitutions in and around the glycine-rich motif. The data represent the mean ± standard deviation of two independent experiments.
Correlation between virus attenuation and defects in cap methylation
To determine whether the L mutations affected the ability of viruses to methylate mRNA 5′ cap structures, in vitro transcription was performed using detergent-activated purified virions in the presence of [3H]-AdoMet with or without S-adenosylhomocysteine (AdoHcy), a competitive inhibitor of AdoMet-dependent MTases. RNA products were isolated and analyzed by scintillation counting for [3H]-AdoMet incorporation and by Northern blot for total mRNA levels (Table 1). Cap methylation activity of each L mutant protein was expressed as a ratio of [3H]-AdoMet incorporation into viral mRNA to the NP + P mRNA products (Table 1, columns 5 and 6). This normalization was done to account for variability in overall transcription levels between different viruses (Table 1, columns 3 and 4). This variability could be due to mutations specifically effecting viral mRNA synthesis or due to variability in the transcriptional activity of individual virion preparations (see below).
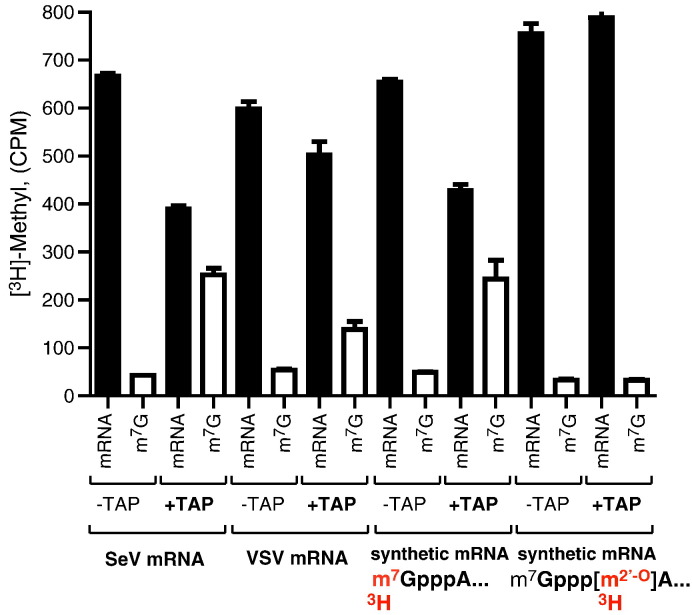
Unfortunately, a high-resolution analysis of cap structure in SeV mRNA is beyond our reach due to the more than 200-fold lower abundance of mRNA produced by SeV virion-associated L protein as compared to VSV (Murphy and Grdzelishvili, 2009). However, to confirm that [3H]-labeled SeV mRNA (Table 1) had methylated cap structure, we analyzed [3H]-AdoMet labeled mRNA generated as described above, using tobacco acid pyrophosphatase (TAP) (Fig. 4 ). TAP is commonly used to specifically remove the 5′-terminal guanosine monophosphate from the cap of mRNA, while uncapped mRNA cannot serve as a substrate for the TAP (Shinshi et al., 1976). In addition to SeV mRNA, we used three different control mRNAs (described in detail in Materials and methods): i) [3H]-AdoMet labeled VSV mRNA generated using detergent-activated wt VSV virions using in vitro transcription conditions identical to SeV; ii) synthetic SeV NP mRNA containing Cap 0 (m7GpppA...) structure labeled with [3H] at the G-N-7 position, and iii) synthetic SeV NP mRNA containing Cap 1 structure (m7Gppp[m2′-O]A...) labeled with [3H]-AdoMet only at the 2′-O position (2′-O methylated by the vaccinia virus enzyme). For TAP analysis, all these mRNAs were normalized by [3H] counts and digested by TAP (or mock-treated using the same conditions by without TAP). Fig. 4 shows [3H] counts associated with the released 5′-terminal G (“m7G”) or with mRNA after TAP treatment and separation using Sephadex G-50 columns. As expected, TAP treatment did not affect an association of [3H] with the synthetic mRNA having Cap 1 structure (m7Gppp[m2′-O]A...) but labeled only at the 2′-O position, but resulted in the release of [3H]-m7G from the synthetic Cap 0 mRNA labeled at the G-N-7 position (Fig. 4), confirming that TAP specifically hydrolyzed the phosphoric acid anhydride bonds in the triphosphate bridge of the cap structure. Importantly, TAP treatment clearly resulted in the release of [3H]-m7G from SeV mRNA produced by determent-activated virions in vitro, indicating that SeV mRNA was at least partially methylated at the G-N-7 position. A similar result was obtained for VSV virion-produced mRNA, which is consistent with the previous studies demonstrating that the fully methylated VSV mRNA has Cap 1 structure (Abraham et al., 1975, Rahmeh et al., 2009, Testa and Banerjee, 1977).
Fig. 4.
Cap methylation analysis using tobacco acid pyrophosphatase. SeV rWT (“SeV mRNA”) and VSV rWT (“VSV mRNA”) were produced in vitro transcription by detergent-activated purified virions in the presence of [3H]AdoMet. Synthetic SeV NP mRNA controls were synthesized in vitro using T7 RNA polymerase and then used to make mRNA containing Cap 0 (m7GpppA...) labeled with [3H]AdoMet at the G-N-7 position, or mRNA containing Cap 1 (m7Gppp[m2′-O]A...) labeled with [3H]AdoMet only at the 2′-O position using vaccinia virus enzymes as described in Materials and methods. For TAP analysis, all RNAs were normalized by [3H] counts and digested by TAP (“+TAP”) or mock-treated using the same reactions but without TAP (“-TAP”). mRNA was then separated from [3H]m7G using Sephadex G-50 mini-columns. [3H]Met incorporation into the G-N-7 or 2′-O cap positions was measured by scintillation counting of the entire flow through (for mRNA containing [3H-m2′-O]A) and columns (for removed [3H]m7G). The data represent the mean ± standard deviation of two independent experiments.
Although our data (Fig. 4) demonstrate that SeV methylates its mRNA at least partially at the G-N-7 position, at this point we cannot make any conclusions about cap structure of SeV mRNA produced under our experimental conditions. Fig. 4 shows that TAP released more [3H]-m7G from SeV mRNA than from VSV mRNA, and similar amounts of [3H]-m7G were released from SeV mRNA and the synthetic Cap 0 mRNA, which could indicate that SeV was methylated exclusively at the G-N-7 position. However, it would be premature to make this conclusion based solely on the TAP experiments. Thus, we did not observe a complete removal of [3H]-m7G from the synthetic Cap 0 mRNA labeled only at G-N-7 position, indicating that under our experimental conditions TAP was only about 50% effective in hydrolyzing triphosphate bridges. Furthermore, mRNA products analyzed in Fig. 4 were normalized by [3H] counts. However, it is likely that the efficiency of cap methylation is different for SeV, VSV and vaccinia virus MTases and, therefore, different molar amounts of capped mRNA were likely present in these reactions further complicating conclusions about cap structure. Further studies using alternative biochemical assays are needed to determine cap structure of SeV mRNA produced under our experimental conditions, although a dramatic improvement in our in vitro transcription conditions is needed to obtain sufficient amounts of viral mRNA for cap analysis.
Most of the SeV L mutants of the DKDKD sequence had methylation levels similar to rWT, while rD1795A and rR1800A had about a 50% decrease in cap methylation compared to rWT. Consistent with our previous results (Murphy and Grdzelishvili, 2009), the SeV L mutant rG1804A/G1806A had only slightly decreased methylation compared to rWT and the rE1805A mutant had methylation levels about 23% of rWT. The other SeV L mutants of the glycine-rich motif all had lower levels of methylation as compared to rWT ranging from about 10% for rY1802A and rG1804L to about 40% for rG1808A and rG1804A/G1806A/G1808A. Except for rS1777A, all SeV L mutants with substitutions in and around the K-D-K-E tetrad (rK1782A, rD1901A, rE1903A, rK1938S, and rK1938A/I1939L) were severely defective in cap methylation (2–8% of rWT). In general, most of the SeV mutants defective in cap methylation were attenuated in Vero cells, indicating the importance of this function in the SeV life cycle.
Interestingly, while rWT and most of our mutants showed only residual mRNA methylation in the presence of AdoHcy, this value was surprisingly high (12.5%) for rG1804A/G1806A. At this point, we do not have an explanation for this result, but it was consistently reproduced in several independent experiments. We cannot exclude an interesting possibility that this mutation improves the preference of L towards AdoMet over AdoHcy, making it less sensitive to the excess of this product. Further studies are needed to address this possibility.
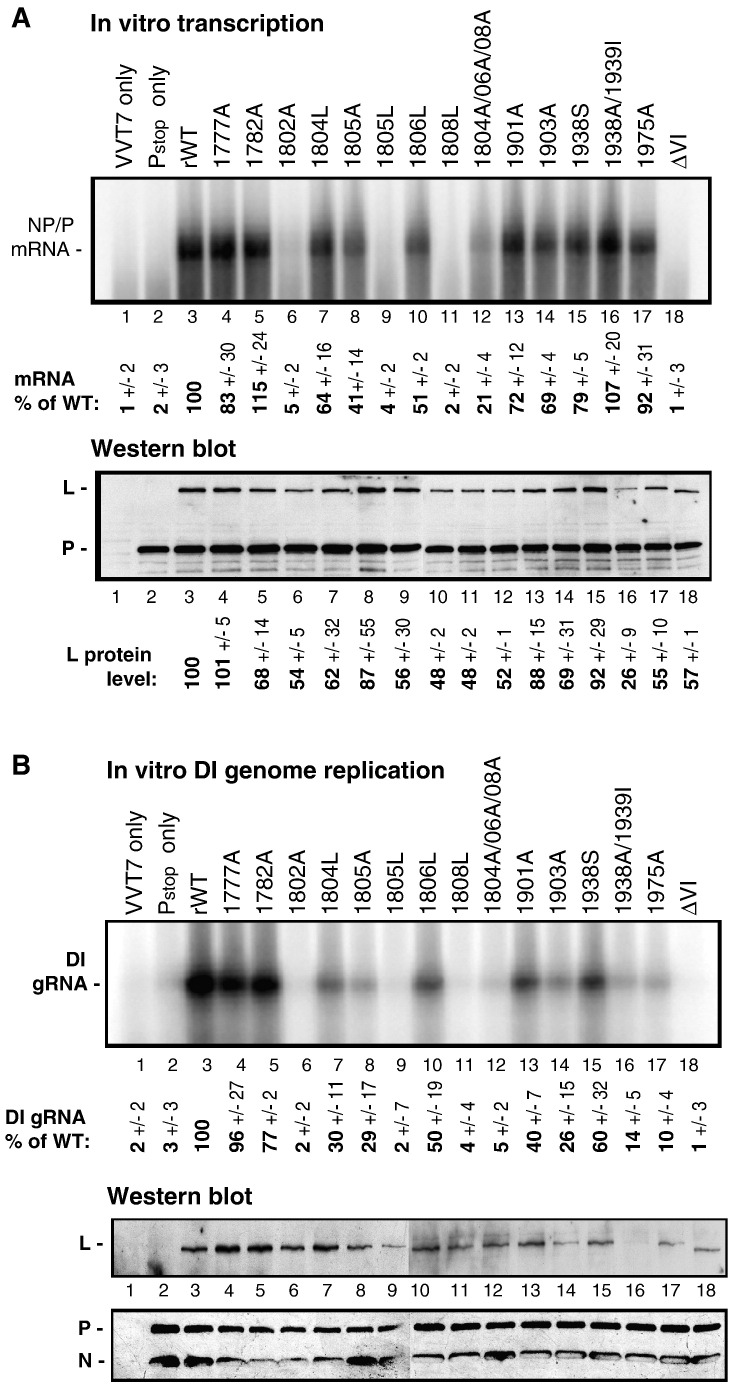
Analysis of the ability of expressed L proteins to synthesize viral mRNA and replicate genomic RNA in vitro
To examine the abilities of L mutants to synthesize viral genomic negative-stranded, as well as mRNA, we selected 15 different mutations resulting in virus attenuation in Vero cells, or low cap methylation levels, or “not rescued” phenotype (Table 1), and analyzed the effects of these mutations on L function using plasmid based expression assays, which do not depend on the viability of SeV mutant viruses. These assays are based on the exogenously provided polymerase-free wt genomic (for transcription) or DI (for replication) RNA-N template and cell extracts containing T7 RNA polymerase-expressed SeV L (wt or mutant), P and NP (for DI replication only) proteins as described in Materials and methods. Briefly, A549 cells were infected with vaccinia virus (VV) expressing T7 polymerase (VV-T7), transfected with SeV P wt and L (wt or mutant) and NP wt (only for DI replication), and incubated at 34 °C. At 18 h p.t., cytoplasmic extracts, containing P-L or NP-P-L complexes, were prepared and supplied with the exogenous wt SeV polymerase-free RNA-N template (genomic or DI) and [α32P]CTP to assay for mRNA or DI RNA synthesis.
In agreement with their putative lethal phenotype (unable to rescue mutant viruses having these mutations after about 20 independent attempts), two L mutations — E1805L and G1808L, as well as earlier described mutant ∆VI (SeV L protein with the entire domain VI deleted) (Murphy and Grdzelishvili, 2009) produced no detectable mRNA and were also defective in genomic RNA replication (< 5% of rWT) (Fig. 5 ). The E1975A mutant L protein, which could also not be rescued in vivo, had wt-like levels of transcription in the VV-mediated system, but replication levels were 10% of rWT. Several other mutant L proteins had transcription levels higher than 50% of rWT but lower levels of replication (< 40% of rWT) including D1901A, E1903A, and K1938A/I1938L (but not K1938S), all of which are members of the K-D-K-E catalytic tetrad. G1804L, E1805A, G1806L, and G1804A/G1806A/G1808A, all members of the glycine-rich motif, had similar levels of transcription and replication ranging from 30 to 60% of rWT. Intriguingly, the Y1802A mutant was less than 10% active in transcription and replication even though virions bearing this mutation were recovered and grew to similar titers as compared to rWT. This indicates that the intactness of the primary AdoMet binding site and the majority of the catalytic residues are required for multiple functions of L protein, most likely including synthesis of the negative-strand genomic RNA and its transcription into mRNAs. Only two mutant L proteins, S1777A and K1782A, had rWT levels of transcription and replication, indicating a more limited role of the predicted first helix in the Rossmann fold in SeV RNA synthesis.
Fig. 5.
In vitro mRNA synthesis and genome replication with SeV mutant L proteins using a VVT7 expression system. Plasmids expressing P and wt or mutant L proteins were transfected into VV-T7 infected A549 cells. N protein was also expressed for DI replication. Total RNA was isolated from cell lysates and in vitro transcription (A) or DI replication (B) reactions were performed in the presence of [α32P]-CTP. Western blot analyses of cell lysates used in (A) or (B) demonstrate relative expression of the SeV L, P and N (for DI replication) proteins in A549 cells. Transcription and DI replication data represent the mean ± standard deviation of two independent experiments.
Interestingly, we observed that many amino acid substitutions negatively affected L protein accumulation in the plasmid based expression assays. Some of these mutations could potentially affect L gene expression or L protein stability of the protein, which could impact viral RNA synthesis. However, we did not see any clear correlation between L protein levels and transcriptional activities for most mutant proteins (Fig. 5). Interestingly, the lowest protein accumulation (26% of wt) was shown for the K1938A/I1939L mutation; however, it had 107% transcriptional activity. At the same time, this mutant showed only 14% DI replication activity suggesting a possibility that L protein accumulation may specifically affect replication (but not transcription) activity of viral polymerase. However, other two mutations, E1903A and E1975A, which had disproportionally low DI replication activities (relative to transcription), did not show any dramatic decreases in protein accumulation (Fig. 5).
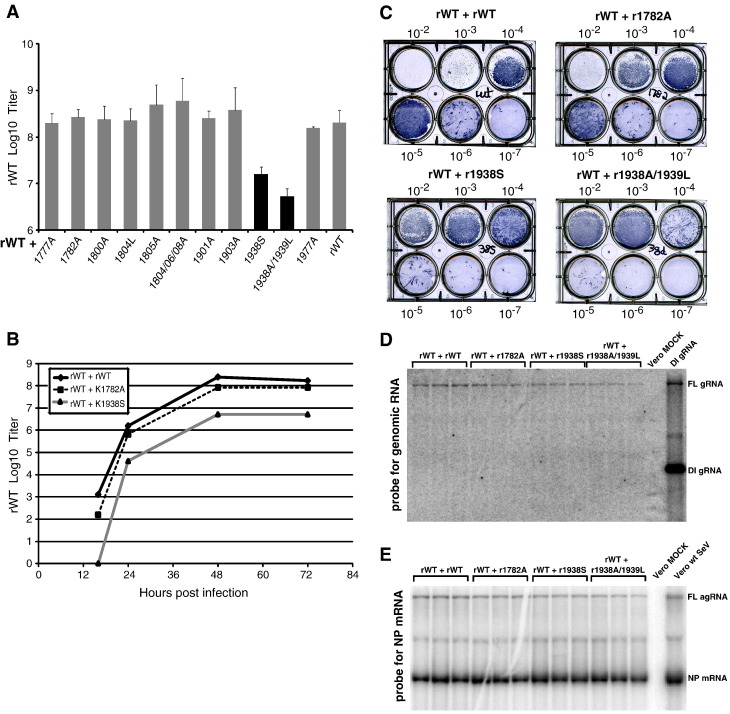
Superinfection analysis
To test whether SeV mutants with substitutions at different positions within domain VI could potentially complement each other when grown together, we infected Vero cells with various combinations of defective mutants at an MOI of 1 CIU/cell per virus (total MOI of 2). Under this conditions, most of the cells are co-infected with both viruses. In addition to viruses with the phenotypes described above, we included rWT and other mutants that behaved similarly to rWT as controls (rS1777A, rR1800A, rE1805A, rG1804A/G1806A/G1808A, and rY1977A). We did not see any increase in CPE when defective mutants were combined with other defective mutants (data not shown). Instead, when two of the defective mutants (rK1938S and rK1938A/I1939L) were combined with rWT, we observed an inhibition of CPE with less cell rounding and cells remaining attached to the plastic (data not shown), suggesting they interfered with rWT replication. To investigate it further, we repeated the experiment looking at all SeV L methylation defective mutants in combination with rWT only (Fig. 6A). All combinations grew to titers comparable to supernatant collected from Vero cells infected with rWT only with the exception of K1938S and K1938A/I1939L which had 99% lower titers for rWT (rWT foci can be easily discriminated from those generated by mutants due to their significantly larger size and earlier appearance). We then compared growth kinetics of two mutants, K1938S and K1782A in combination with rWT by infecting Vero cells and collecting supernatants at 18–72 h p.i. Similar to our titration experiments (Fig. 6A), K1782A when combined with rWT had similar growth kinetics to rWT alone while K1938S in combination with rWT had delayed growth and lower maximal titers (Fig. 6B), indicating that cap methylation defect alone is not sufficient for the interference of rK1938S and rK1938A/I1939L with rWT replication and that some additional factors are involved in the dominant-negative phenotype of these SeV mutants.
Fig. 6.
Superinfection of Vero cells with SeV mutants. (A) Vero cells were infected with combinations of SeV mutant viruses and rWT at an MOI of 1 CIU/cell (total MOI of 2). Cells were observed and CPE was visualized by light microscopy at 24, 48, and 72 h p.i. The supernatant was collected at the same time points and each sample was further analyzed by plaque assays performed on Vero cells. Data represent the mean of three independent infections ± standard deviation. (B) One-step growth kinetics for rWT grown in combination with K1782A and K1938S mutant viruses were analyzed. Vero cells were infected at MOI 1 CIU/cell (total MOI of 2), supernatants collected at 18, 24, 48, and 72 h p.i. and titered on Vero cells. (C–E) Vero cells were infected with combinations of SeV mutant viruses plus rWT at an MOI of 1 CIU/cell for each virus (total MOI of 2). 48 h p.i., cells were analyzed by Northern blot (D–E) and the medium was tittered on fresh Vero cells (C). Titration was done on 6-well plates (C) with the rWT infection foci visualized 48 h p.i. using 4 CN Peroxidase Substrate staining Kit (KPL) and anti-SeV primary antibodies. Representative plates are shown. Only large SeV rWT (but not mutant) infectious foci are visible at this time point. (D–E) Cell pellets were analyzed by Northern blot analysis for genomic RNA using a probe for full-length (FL) genomic RNA (gRNA) as well as DI genomic RNA (D) or a riboprobe for the SeV NP mRNA as well as the FL antigenomic RNA (agRNA) (E), as described in Materials and methods. “Vero Mock” sample: total RNA isolated from mock-infected Vero cells. 7 μg of total RNA was used for each sample. “Vero wt SeV” sample: total RNA isolated from Vero cells infected with SeV rWT and collected at 48 h p.i. “DI gRNA” sample: total RNA isolated from a preparation of SeV DI particles (DI-H). This preparation had some particles containing FL genomic RNA.
At this time, we cannot explain why viruses containing amino acid substitution at the L position 1938 interfere with rWT replication. One possibility is that such inhibition may simply reflect elevated levels of DI particles generated by these mutants. To test this hypothesis, we infected Vero cells again (as in Fig. 6A), collected cells and the medium at 48 h p.i. Consistent with Fig. 6A, both rK1938S and rK1938A/I1939L inhibited production of rWT as was determined by titration of the collected medium on fresh Vero cells (Fig. 6B). For analysis of RNA synthesized during superinfection, both collected supernatants and cells were analyzed by Northern blot as described in Materials and methods. To detect DI genomes, a probe complimentary to the first 54 5′-nucleotides of the SeV genomic RNA was used, which should be able to detect full-length genomic RNA of SeV as well as DI genomes independent on the mechanism of their generation. As shown in Fig. 6D, we were unable to detect any DI genomes in Vero cells under our experimental conditions. Interestingly, despite clear inhibition of SeV rWT virion production (Figs. 6A–C), we did not see any statistically significant decrease in SeV NP mRNA accumulation in the superinfected cells (Fig. 6E) using a probe against coding NP gene sequences, although a modest decrease in the full-length genomic RNA (wt plus mutant) levels was observed (Fig. 6D, FL gRNA band). A similar lack of DI genomes and a very modest reduction in full-length genomic RNA levels was observed when virus particles from the medium were pelleted by ultracentrifugation, and RNA from these particles was analyzed by Northern blot as above (data not shown). Together, out data suggest that rK1938S and rK1938A/I1939L are able to over-compete rWT during superinfection and, while the mechanism is unclear, this effect is apparently DI-independent.
Discussion
This study is a more detailed sequence–function analysis of the L protein domain VI in paramyxoviruses than earlier mutagenesis studies that we are aware of. Despite some differences between VSV and SeV presented here and in our recent study (Murphy and Grdzelishvili, 2009), our results clearly show the importance of the putative catalytic tetrad K-D-K-E and glycine-rich motif, thus confirming previous computational predictions that these regions are important for MTase function across different families of the order Mononegavirales (Bujnicki and Rychlewski, 2002, Ferron et al., 2002, Martin and McMillan, 2002).
To analyze domain VI, we conducted site-directed mutagenesis targeting the residues highly conserved among Mononegavirales, including the K-D-K-E tetrad and glycine-rich motif, also present in the known 2′-O cap MTases including NS5 protein of flaviviruses (Fig. 1) and nonstructural protein 16 of coronaviruses (Decroly et al., 2008). Similar to VSV, alanine substitutions in the putative K-D-K-E catalytic tetrad of SeV generated mutant viruses defective in cap methylation. The K-D-K-E tetrad was previously shown to catalyze a 2′-O methyl transfer in the known 2′-O MTases, including NS5 protein of flaviviruses (Bollati et al., 2009, Egloff et al., 2007, Ray et al., 2006, Selisko et al., 2010) and nonstructural protein 16 of coronaviruses (Bouvet et al., 2010, Decroly et al., 2008). In West Nile virus (WNV, a flavivirus), amino acid substitutions of the homologous residues abolished 2′-O cap methylation by NS5 protein, but had only modest effect on G-N-7 methylation (Ray et al., 2006, Zhou et al., 2007). In VSV, similar substitutions abolished both G-N-7 and 2′-O methylation (Li et al., 2005). This discrepancy between WNV and VSV can be explained by the different order of methylation events shown for flaviruses (GpppA → m7GpppA → m7Gppp[m2′-O]A) (Zhou et al., 2007) and VSV (GpppA → GpppAm → m7Gppp[m2′-O]A) (Li et al., 2006, Rahmeh et al., 2009, Testa and Banerjee, 1977). While the inactivation of 2′-O methylation did not significantly affect G-N-7 methylation in WNV, it prevented it in VSV where 2′-O methylation precedes and apparently facilitates subsequent G-N-7 methylation (Rahmeh et al., 2009). A high-resolution analysis of cap structure in SeV mRNA is still beyond our reach due to the more than 200-fold lower abundance of mRNA produced by SeV virion-associated L protein as compared to VSV (Murphy and Grdzelishvili, 2009). However, the critical importance of K-D-K-E tetrad for the overall cap methylation in SeV presented in this study may indicate that the SeV L protein catalyzes cap methylation using the same order as VSV (GpppA → GpppAm → m7Gppp[m2′-O]A).
Our data also demonstrates the importance of glycine-rich motif (putative AdoMet binding site of domain VI) in SeV cap methylation but with different effect of mutations in the glycine-rich motif in SeV as compared to VSV. For VSV, it was shown that virions with alanine substitutions at the first two glycines had decreased cap methylation, while alanine substitution at the third glycine did not affect MTase activity (Li et al., 2006). Here, we determined that the third glycine of the glycine-rich motif seems to play a more significant role in cap methylation in SeV than VSV, decreasing cap methylation by about 70% (compared to rWT). As further confirmation of the importance of this position, a triple mutant with all three glycines substituted to alanines also had low methylation levels (37% of rWT) while a double mutant with only the first two glycines substituted to alanines had rWT-like levels of growth and higher methylation. Our previous partial results showed so much tolerance to substitution in the first two glycines in the glycine-rich motif as to even doubt that this is a functional AdoMet binding site (Murphy and Grdzelishvili, 2009). Our data presented here, including the role of the third glycine in the SeV cap methylation and dramatic effect of glycine-to-leucine substitutions in this motif restore a more conventional view of the role of this conserved sequence motif.
Another mutation resulting in defective cap methylation (10% of rWT) and virus attenuation was Y1802A. This residue has never been studied before in any Mononegavirales and is highly conserved in most paramyxoviruses. It possesses a hydroxyl group and may substitute for the aspartic or glutamic acid residues frequently found in the middle of the first beta strand of the Rossmann fold. It has been proposed (Kozbial and Mushegian, 2005) that the side chain of this residue makes either direct or water molecule-mediated contact with the methionine portion of AdoMet and may be directly involved in catalysis. In several paramyxoviruses, just next to this residue there is a motif DKDKD1799 with potential importance in catalysis due to the high concentration of aspartates and lysines and its close proximity to the glycine-rich motif. We targeted these positions for substitution to alanines, but all these SeV L mutants were easily recovered, grew to high titers with similar kinetics to rWT in Vero cells and had high levels of cap methylation.
Consistent with the role of cap methylation in the translatability of mRNA, SeV mutants with decreased methylation (< 10%) were attenuated in Vero cells. The majority of these mutants had substitutions in and around the putative K-D-K-E catalytic tetrad. In agreement with our recent study (Murphy and Grdzelishvili, 2009), we observed that alanine substitutions at positions in and around the glycine-rich motif were well tolerated; however, a triple mutant with all three glycines replaced with alanines did show slight attenuation, confirming the importance of this motif for SeV L function and virus replication. We also observed that two cap methylation defective mutants, rK1938S and rK1938A/I1939L, had strong interfering effect on replication of wt SeV during superinfection. Interestingly, another cap methylation defective mutant, rK1782A, did not interfere with rWT replication indicating that cap methylation defect alone is not sufficient for this dominant-negative mutant phenotype and that some additional factors are involved. Currently, we cannot explain how K1938S mutation could inhibit the growth of rWT and further studies are warranted to explore this interesting observation.
Interestingly, a single alanine substitution at the putative catalytic K1938 position was successfully introduced into plasmids containing the SeV L gene and the full-length antigenome of SeV. However, when infectious virus particles, recovered using a reverse genetics system, were sequenced, the substitution was to serine rather than alanine. Based on the importance of this position in the catalytic tetrad, we speculate that the change to the polar serine residue led to a more favorable configuration of hydrogen bond network linking catalytic residues to AdoMet and perhaps additionally supporting proper conformation of the L protein, which may be important for its function. When tested in a VV-T7 in vitro transcription or replication systems, the K1938A mutant L had slightly lower transcription (∼ 80%) and lower replication levels (∼ 60%) as compared to rWT. Mutant virions with the K1938S substitution were defective in cap methylation (8% of rWT), but had high transcription levels (91% of rWT): apparently, a serine substitution is more favorable for the overall activity of the L protein (although not necessarily for cap methylation).
A better understanding of the biology of these SeV L mutants could lead to the rational design of live attenuated viruses and their use as vaccine, oncolytic and gene therapy vectors based on different members of Mononegavirales.
Materials and methods
Cell lines and viruses
African green monkey (Vero, ATCC# CCL-81), human lung carcinoma (A549, ATCC# CCL-185) and BSR-T7/5 cells [derived from baby hamster kidney (BHK-21) cells that constitutively express bacteriophage T7 polymerase (Buchholz et al., 1999)] were used for virus infections and plasmid transfections. For virus-driven expression of the bacteriophage T7 RNA polymerase, A549 cells were infected with T7-expressing vaccinia virus (VV-T7) (Fuerst et al., 1986). Recombinant wt (rWT) SeV (Fushimi strain) (Leyrer et al., 1998) was kindly provided by Dr. Wolfgang J. Neubert (Max-Planck-Institute of Biochemistry, Germany). SeV rWT or mutants were grown and purified as described in (Murphy and Grdzelishvili, 2009). SeV infectivity was measured by virus titration on Vero cells and expressed as cell infectious units/ml (CIU/ml). For faster growing SeV mutants, wells were stained with 0.1% Crystal Violet in 10% formalin for 10 min, washed with PBS and dried. An immunofluorescence (IF) assay was used for severely attenuated SeV mutants as described in (Murphy and Grdzelishvili, 2009). Following IF, 2 μM Hoechst reagent (Invitrogen) was added to each well for 10 min, cells were washed with PBS and viewed under a fluorescent microscope. For one-step growth kinetics, Vero cells were incubated for 1 hour (h) with SeV wt or mutants at an MOI of 3 CIU/cell in 24 well plates. At 1 h post infection (p.i.), unabsorbed viruses were aspirated, cells were washed two times with PBS and MegaVir HyQSFM4 serum-free medium (SFM) (Hyclone) with 4 μg/ml acetylated trypsin (Sigma) was added to each well. Plaque assays were performed on Vero cells using supernatants collected at different time points. For superinfection experiments, virus infection foci were detected by IF as described above and in (Murphy and Grdzelishvili, 2009) or by 4 CN Peroxidase Substrate colorimetric staining (KPL). Briefly, cells were fixed in 3% paraformaldehyde (Sigma) for 10 min and permeabilized for 2 min on ice. Cells were blocked in PBS with 5% bovine serum albumin (Sigma) for 20 min and incubated with anti-SeV primary antibodies (1:100) for 1 h. Cells were then washed, incubated with goat anti-rabbit IgG-HRP (Jackson ImmunoResearch) secondary antibodies (1:500) for 1 h. To visualize infectious foci, cells were washed and incubated with equal volumes 4 CN Peroxidase Substrate and Peroxidase Substrate Solution B (KPL).
Plasmids and mutagenesis
The pGEM plasmids containing wt genes for SeV NP, L, and Pstop (expressing P but not C due to a stop codon in the C open reading frames, and referred to here as wt P), under the control of the T7 promoter have been described previously (Curran et al., 1991). The pGEM-L plasmid was used for site-directed mutagenesis as described in (Murphy and Grdzelishvili, 2009). The SeV pTM-NP, pTM-P and pTM-L plasmids, and the pRS3Gg (Leyrer et al., 1998) full-length SeV antigenomic plasmid used for the rescue of recombinant SeV viruses were kindly provided by Dr. Wolfgang J. Neubert (Max-Planck-Institute of Biochemistry, Germany). The SeV L protein mutations were introduced into pRS3Gg as described in (Murphy and Grdzelishvili, 2009). Recombinant viruses were purified as in (Grdzelishvili et al., 2005) and all mutations were confirmed by sequence analysis for the presence of the desired mutations and absence of any spontaneous secondary mutations in the L gene. Total protein concentrations of purified virions were determined by a Bradford assay used in conjunction with viral titers to determine infectivity per total protein (CIU/μg).
In vitro transcription and DI RNA replication with T7-expressed L proteins
To produce cell lysates containing SeV wt P and wt or mutant L proteins for in vitro transcription, 60-mm dishes of A549 cells were infected with vaccinia virus (VV)-T7 at MOI of 2.5 CIU/cell for 1 h at 37 °C, washed with Opti-MEM (Gibco), transfected with 1.5 μg of SeV pGEM-Pstop and 1 μg of pGEM-L (wt L or one of the mutant L genes) plasmids using lipofectamine (Invitrogen), and incubated at 34 °C in Opti-MEM. At 18 h p.t., cytoplasmic extracts were prepared exactly as described previously (Chandrika et al., 1995, Grdzelishvili et al., 2005). To assay for SeV mRNA synthesis, 1 μg of wt SeV polymerase-free RNA-N template and 18 μCi of [α32P]CTP were added to each extract, and reactions were incubated for 2 h at 30 °C. To produce cell lysates containing SeV wt NP, wt P and wt or mutant L proteins for in vitro replication of SeV defective interfering (DI) RNA, 60-mm dishes of A549 cells were infected and transfected as above using 5 μg of SeV pGEM-Pstop, 2 μg SeV pGEM-NP and 0.5 μg of pGEM-L (wt L or one of the mutant L genes). At 18 h p.t., cytoplasmic extracts were prepared as for in vitro transcription. To assay for SeV genome synthesis, 2 μg of detergent disrupted DI-H (Carlsen et al., 1985) and 18 μCi of [α 32P]CTP were added to each extract, and reactions were incubated for 2 h at 30 °C and then treated with micrococcal nuclease to digest unpackaged RNA (mRNA). For both in vitro transcription and DI replication, total RNA was purified using Quick-RNA Miniprep (Zymo Research) and analyzed by 1.5% agarose/6 M urea gel electrophoresis. Gels were fixed in 7% acetic acid, dried, and quantitated using a Typhoon 8600 PhosphorImager and ImageQuant software (Molecular Dynamics).
Cap methylation analysis using purified SeV virions
SeV in vitro transcription by detergent-activated purified virions was conducted as described in (Murphy and Grdzelishvili, 2009). To test for viral mRNA cap methylation, RNA was synthesized in a 200 μl reaction with cold NTPs (1 mM each) and 9 μCi of [3H]AdoMet (81.5 Ci/mmol, 0.55 μM AdoMet final concentration) in the presence or absence of 100 μM of the methylation inhibitor S-adenosylhomocysteine (AdoHcy). Total RNA was purified using the Mini RNA Isolation II Kit (Zymo Research), diluted in 60 μl of diH2O, and used for measurement of [3H]AdoMet incorporation by scintillation counting (50 μl) or analyzed by Northern blot to measure viral mRNA levels (5 μl).
To generate viral mRNA for cap analysis using tobacco acid pyrophosphatase (TAP), SeV rWT and VSV rWT in vitro transcription by detergent-activated purified virions was performed as described above using [3H]AdoMet and the same reaction conditions for SeV and VSV. VSV mRNA was synthesized in a 100 μl reaction. To obtain sufficient amounts of SeV mRNA for this analysis, ten 200-μl transcription reactions were used for SeV rWT, and viral mRNA products were isolated and pooled together for TAP treatments. For preparation of synthetic mRNA controls, uncapped SeV NP mRNA was synthesized in vitro with the MAXI-Script T7 kit (Ambion) using SeV pGEM3-NP plasmid digested at the BamHI restriction site located immediately after the stop codon for the NP gene as a template. This RNA was divided to generate: i) Cap 0 containing mRNA (m7GpppA...) labeled with [3H] only at the G-N-7 position, or ii) Cap 1 containing mRNA (m7Gppp[m2′-O]A...) labeled with [3H] only at the 2′-O position. To make synthetic mRNA containing Cap 0, uncapped mRNA transcripts were capped and G-N-7 methylated in the presence of 1.75 μM [3H]AdoMet using the ScriptCap m7G Capping System (Epicentre Biotechnologies) based upon the tri-functional vaccinia virus capping enzyme, and purified using Spin-50 Sephadex G-50 mini-columns (USA Scientific). To make synthetic mRNA with Cap 1 but [3H]-labeled only at the 2′-O position, uncapped mRNA transcripts were first capped in the presence of 100 μM cold AdoMet using the ScriptCap m7G Capping System to make mRNA with the unlabeled Cap 0 structure (which is the template for 2′-O methylation by the vaccinia virus 2′-O MTase). After purification using Spin-50 Sephadex G-50 mini-columns, the unlabeled Cap 0 mRNA was 2′-O methylated in the presence of 1.75 μM [3H]AdoMet using ScriptCap 2′-O MTase (Epicentre Biotechnologies) based upon the vaccinia virus 2′-O MTase, and purified again using Spin-50 Sephadex G-50 mini-columns.
For TAP analysis, virion-produced viral mRNA (SeV or VSV) and synthetic (G-N-7 or 2′-O labeled) mRNAs were normalized by [3H] counts and digested by TAP (Epicentre Biotechnologies) in 10 μl reactions in the presence or absence of 5 U of TAP for 1 h at 37 °C. All reactions were then adjusted to 25 μl and passed through Spin-50 Sephadex G-50 mini-columns. Spin columns were then placed in new microfuge tubes and 25 μl diH2O was passed each column to retrieve the residual column-bound RNA. Separate optimization experiments demonstrated effective separation of RNA from nucleotides using this procedure (data not shown). [3H]Met incorporation into the G-N-7 or 2′-O cap position was measured by scintillation counting of the entire flow through (contained mRNA) and the Sephadex G-50 column material removed from mini-columns after separation (contained removed G).
Northern blot analysis
Northern blot analysis was conducted as described in (Murphy and Grdzelishvili, 2009) to measure mRNA levels from in vitro transcription reactions with [3H]AdoMet (described above). Briefly, mRNA products were separated on a 1.2% agarose–formaldehyde gel, transferred to a Hybond-N+ nylon membrane (GE Healthcare), and incubated with an RNA probe complementary to the SeV NP gene. The probe was synthesized by digestion of the SeV pGEM3-NP plasmid at the EcoRV restriction site and transcribed in vitro in the presence of [α-32P]-CTP using the MAXI-Script SP6 kit (Ambion). For analysis of RNA synthesized during superinfection, both supernatants and cells were collected at 48 h p.i. Virus particles in the supernatant were pelleted by centrifugation at 50,000 rpm for 1 h using a Beckman TLA 100.2 rotor and total RNA was extracted from both cells and pelleted virions using the Zymo Research Quick-RNA MiniPrep kit. 0.7 μg or 7 μg of total RNA from the supernatant or cells respectively was separated on a 1.5% agarose–formaldehyde gel and transferred to a nylon membrane. Membranes were originally probed for full length and DI genomic RNA using an oligonucleotide [5′-ACAAGAAGACAAGAAAATTTAAAAGAATAAATATCTCTTAAACTCTTGTCTGGT-3′ (Integrated DNA Technologies)] complimentary to the first 54 5′-nucleotides of the SeV genomic RNA. The primer was labeled with [γ-32P]-ATP using bacteriophage T4 polynucleotide kinase (New England Biolabs). Membranes were then reprobed for NP mRNA using the riboprobe described above. Radioactive signals were measured using a Typhoon 8600 Phosphorimager and ImageQuant software (Molecular Dynamics).
Protein analysis
To compare the amounts of the P and L proteins in cell lysates used for in vitro transcription, total protein samples from transfected cytoplasmic lysates (5 μl out of 100 μl) were separated by 7.5% SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane (PVDF, Sigma). Membranes were blocked with 5% non-fat dry milk in TBST [0.5 M NaCl, 20 mM Tris pH (7.5), 0.1% Tween 20] and antibodies were diluted in the same buffer. Blots were initially incubated with a mixture of rabbit antibodies against the SeV L protein (a-TrpE-SeV-L #5 (“1-19-90”) and a-TrpE-SeV-L #1 (“10-23-89”) (Horikami et al., 1992), and developed with a horseradish peroxidase-conjugated secondary antibody using the Enhanced Chemiluminescence Plus protein detection system (GE Healthcare) in accordance with manufacturer's instructions. Blots were then reprobed with a rabbit anti-SeV antibody (“1-4-83”) (Carlsen et al., 1985) and developed in the same manner. Protein bands were quantified using VisionWorksLS software (UVP).
Acknowledgments
We are grateful to Sue Moyer (University of Florida College of Medicine) for providing materials for this project. We thank Wolfgang J. Neubert (Max-Planck-Institute of Biochemistry, Germany) for a kind gift of SeV plasmids. This work was supported by NIH grant 1R15GM084422 (to V. Z. G.).
References
- Abraham G., Rhodes D.P., Banerjee A.K. The 5′ terminal structure of the methylated mRNA synthesized in vitro by vesicular stomatitis virus. Cell. 1975;5:51–58. doi: 10.1016/0092-8674(75)90091-4. [DOI] [PubMed] [Google Scholar]
- Bollati M., Milani M., Mastrangelo E., Ricagno S., Tedeschi G., Nonnis S., Decroly E., Selisko B., de Lamballerie X., Coutard B., Canard B., Bolognesi M. Recognition of RNA cap in the Wesselsbron virus NS5 methyltransferase domain: implications for RNA-capping mechanisms in Flavivirus. J. Mol. Biol. 2009;385:140–152. doi: 10.1016/j.jmb.2008.10.028. [DOI] [PubMed] [Google Scholar]
- Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6(4):e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky L.I., Drachev A.L., Leontovich A.M., Feranchuk S.I. A novel method of multiple alignment of biopolymer sequences. Biosystems. 1993;30:65–79. doi: 10.1016/0303-2647(93)90063-i. [DOI] [PubMed] [Google Scholar]
- Buchholz U.J., Finke S., Conzelmann K.K. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujnicki J.M., Rychlewski L. In silico identification, structure prediction and phylogenetic analysis of the 2′-O-ribose (cap 1) methyltransferase domain in the large structural protein of ssRNA negative-strand viruses. Protein Eng. 2002;15:101–108. doi: 10.1093/protein/15.2.101. [DOI] [PubMed] [Google Scholar]
- Carlsen S.R., Peluso R.W., Moyer S.A. In vitro replication of Sendai virus wild-type and defective interfering particle genome RNAs. J. Virol. 1985;54:493–500. doi: 10.1128/jvi.54.2.493-500.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartee T.L., Megaw A.G., Oomens A.G., Wertz G.W. Identification of a single amino acid change in the human respiratory syncytial virus L protein that affects transcriptional termination. J. Virol. 2003;77:7352–7360. doi: 10.1128/JVI.77.13.7352-7360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrika R., Horikami S.M., Smallwood S., Moyer S.A. Mutations in conserved domain I of the Sendai virus L polymerase protein uncouple transcription and replication. Virology. 1995;213:352–363. doi: 10.1006/viro.1995.0008. [DOI] [PubMed] [Google Scholar]
- Cortese C.K., Feller J.A., Moyer S.A. Mutations in domain V of the Sendai virus L polymerase protein uncouple transcription and replication and differentially affect replication in vitro and in vivo. Virology. 2000;277:387–396. doi: 10.1006/viro.2000.0615. [DOI] [PubMed] [Google Scholar]
- Curran J., Boeck R., Kolakofsky D. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 1991;10:3079–3085. doi: 10.1002/j.1460-2075.1991.tb07860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Imbert I., Coutard B., Bouvet M., Selisko B., Alvarez K., Gorbalenya A.E., Snijder E.J., Canard B. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J. Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprex W.P., Collins F.M., Rima B.K. Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame. J. Virol. 2002;76:7322–7328. doi: 10.1128/JVI.76.14.7322-7328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Decroly E., Malet H., Selisko B., Benarroch D., Ferron F., Canard B. Structural and functional analysis of methylation and 5′-RNA sequence requirements of short capped RNAs by the methyltransferase domain of dengue virus NS5. J. Mol. Biol. 2007;372:723–736. doi: 10.1016/j.jmb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Ferron F., Longhi S., Henrissat B., Canard B. Viral RNA-polymerases — a predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem. Sci. 2002;27:222–224. doi: 10.1016/s0968-0004(02)02091-1. [DOI] [PubMed] [Google Scholar]
- Fuerst T.R., Niles E.G., Studier F.W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway S.E., Richardson P.E., Wertz G.W. Analysis of a structural homology model of the 2′-O-ribose methyltransferase domain within the vesicular stomatitis virus L protein. Virology. 2008;382:69–82. doi: 10.1016/j.virol.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Raught B., Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Grdzelishvili V.Z., Smallwood S., Tower D., Hall R.L., Hunt D.M., Moyer S.A. A single amino acid change in the L-polymerase protein of vesicular stomatitis virus completely abolishes viral mRNA cap methylation. J. Virol. 2005;79:7327–7337. doi: 10.1128/JVI.79.12.7327-7337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grdzelishvili V.Z., Smallwood S., Tower D., Hall R.L., Hunt D.M., Moyer S.A. Identification of a new region in the vesicular stomatitis virus L polymerase protein which is essential for mRNA cap methylation. Virology. 2006;350:394–405. doi: 10.1016/j.virol.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Hercyk N., Horikami S.M., Moyer S.A. The vesicular stomatitis virus L protein possesses the mRNA methyltransferase activities. Virology. 1988;163:222–225. doi: 10.1016/0042-6822(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Horikami S.M., Moyer S.A. Host range mutants of vesicular stomatitis virus defective in in vitro RNA methylation. Proc. Natl. Acad. Sci. U. S. A. 1982;79:7694–7698. doi: 10.1073/pnas.79.24.7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikami S.M., De Ferra F., Moyer S.A. Characterization of the infections of permissive and nonpermissive cells by host range mutants of vesicular stomatitis virus defective in RNA methylation. Virology. 1984;138:1–15. doi: 10.1016/0042-6822(84)90142-9. [DOI] [PubMed] [Google Scholar]
- Horikami S.M., Curran J., Kolakofsky D., Moyer S.A. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J. Virol. 1992;66:4901–4908. doi: 10.1128/jvi.66.8.4901-4908.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozbial P.Z., Mushegian A.R. Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 2005;5:19. doi: 10.1186/1472-6807-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R.A., Parks G.D. Paramyxoviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., editors. Fields Virology. 5th edition. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1449–1496. [Google Scholar]
- Leyrer S., Neubert W.J., Sedlmeier R. Rapid and efficient recovery of Sendai virus from cDNA: factors influencing recombinant virus rescue. J. Virol. Methods. 1998;75:47–58. doi: 10.1016/s0166-0934(98)00095-0. [DOI] [PubMed] [Google Scholar]
- Li J., Fontaine-Rodriguez E.C., Whelan S.P. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J. Virol. 2005;79:13373–13384. doi: 10.1128/JVI.79.21.13373-13384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang J.T., Whelan S.P. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8493–8498. doi: 10.1073/pnas.0509821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Rahmeh A., Morelli M., Whelan S.P. A conserved motif in region v of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J. Virol. 2008;82:775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles D.S., Rupprecht C.E. Rhabdoviridae. In: Knipe D.M., Howley P.M., editors. Fields Virology. 5th edition. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1363–1408. [Google Scholar]
- Martin J.L., McMillan F.M. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- Murphy A.M., Grdzelishvili V.Z. Identification of sendai virus L protein amino acid residues affecting viral mRNA cap methylation. J. Virol. 2009;83:1669–1681. doi: 10.1128/JVI.01438-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino T., Banerjee A.K. The HR motif in the RNA-dependent RNA polymerase L protein of Chandipura virus is required for unconventional mRNA-capping activity. J. Gen. Virol. 2010;91:1311–1314. doi: 10.1099/vir.0.019307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino T., Kobayashi M., Iwama M., Mizumoto K. Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J. Biol. Chem. 2005;280:4429–4435. doi: 10.1074/jbc.M411167200. [DOI] [PubMed] [Google Scholar]
- Ogino T., Yadav S.P., Banerjee A.K. Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2010;107:3463–3468. doi: 10.1073/pnas.0913083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch O., Blumberg B.M., Bougueleret L., Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 1990;71:1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- Rahmeh A.A., L.J., Kranzusch P.J., Whelan S.P. Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J. Virol. 2009;83:11043–11050. doi: 10.1128/JVI.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D., Shah A., Tilgner M., Guo Y., Zhao Y., Dong H., Deas T.S., Zhou Y., Li H., Shi P.Y. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 2006;80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruedas J.B., Perrault J. Insertion of enhanced green fluorescent protein in a hinge region of vesicular stomatitis virus L polymerase protein creates a temperature-sensitive virus that displays no virion-associated polymerase activity in vitro. J. Virol. 2009;83:12241–12252. doi: 10.1128/JVI.01273-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell M.J., Conzelmann K.K. Polymerase activity of in vitro mutated rabies virus L protein. Virology. 1995;214:522–530. doi: 10.1006/viro.1995.0063. [DOI] [PubMed] [Google Scholar]
- Selisko B., Peyrane F.F., Canard B., Alvarez K., Decroly E. Biochemical characterization of the (nucleoside-2′O)-methyltransferase activity of dengue virus protein NS5 using purified capped RNA oligonucleotides (7Me)GpppAC(n) and GpppAC(n) J. Gen. Virol. 2010;91:112–121. doi: 10.1099/vir.0.015511-0. [DOI] [PubMed] [Google Scholar]
- Shinshi H., Miwa M., Sugimura T. Enzyme cleaving the 5′-terminal methylated blocked structure of messenger RNA. FEBS Lett. 1976;65:254–257. doi: 10.1016/0014-5793(76)80492-9. [DOI] [PubMed] [Google Scholar]
- Sidhu M.S., Menonna J.P., Cook S.D., Dowling P.C., Udem S.A. Canine distemper virus L gene: sequence and comparison with related viruses. Virology. 1993;193:50–65. doi: 10.1006/viro.1993.1102. [DOI] [PubMed] [Google Scholar]
- Sleat D.E., Banerjee A.K. Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J. Virol. 1993;67:1334–1339. doi: 10.1128/jvi.67.3.1334-1339.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood S., Hovel T., Neubert W.J., Moyer S.A. Different substitutions at conserved amino acids in domains II and III in the Sendai L RNA polymerase protein inactivate viral RNA synthesis. Virology. 2002;304:135–145. doi: 10.1006/viro.2002.1644. [DOI] [PubMed] [Google Scholar]
- Takagi T., Muroya K., Iwama M., Shioda T., Tsukamoto T., Mizumoto K. In vitro mRNA synthesis by Sendai virus: isolation and characterization of the transcription initiation complex. J. Biochem. (Tokyo) 1995;118:390–396. doi: 10.1093/oxfordjournals.jbchem.a124919. [DOI] [PubMed] [Google Scholar]
- Testa D., Banerjee A.K. Two methyltransferase activities in the purified virions of vesicular stomatitis virus. J. Virol. 1977;24:786–793. doi: 10.1128/jvi.24.3.786-793.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S.P., Barr J.N., Wertz G.W. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 2004;283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Ray D., Zhao Y., Dong H., Ren S., Li Z., Guo Y., Bernard K.A., Shi P.Y., Li H. Structure and function of flavivirus NS5 methyltransferase. J. Virol. 2007;81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]