Abstract
This review recounts the early history of Drosophila phototransduction genetics, covering the period between approximately 1966 to 1979. Early in this period, the author felt that there was an urgent need for a new approach in phototransduction research. Through inputs from a number of colleagues, he was led to consider isolating Drosophila mutants that are defective in the electroretinogram. Thanks to the efforts of dedicated associates and technical staff, by the end of this period, he was able to accumulate a large number of such mutants. Particularly important in this effort was the use of the mutant assay protocol based on the “prolonged depolarizing afterpotential.” This collection of mutants formed the basis of the subsequent intensive investigations of the Drosophila phototransduction cascade by many investigators.
Keywords: Drosophila mutants, mutagenesis, early history, electroretinogram, prolonged depolarizing afterpotential
Only a few decades ago, the field of Drosophila phototransduction did not exist. As late as the mid-80's, Drosophila was widely considered to be one of the poorest organisms in which to study sensory mechanisms because of its small size. Today, although much still needs to be learned, most of the phototransduction steps have become elucidated in some detail (reviews: Wang & Montell, 2007; Katz & Minke, 2009; Raghu & Hardie, 2009). More importantly, insights gained from Drosophila phototransduction research have begun to impact wide ranging fields of biology and biomedicine. Although many examples exist, one of the clearest is the founding of the new TRP superfamily of ion channels by the Drosophila phototransduction channel, TRP (reviews: Minke, 2006; Minke & Parnas, 2006; Ramsey, Moran, Chong, & Clapham, 2006; Hardie, 2007; Venkatachalam & Montell, 2007; Talavera, Nilius, & Voets, 2008). These channels are conserved throughout animal phylogeny and have been implicated in a diverse range of biological functions, including sensory perception, secretion, T cell activation, regulation of smooth muscle tone, growth cone guidance, apoptosis, etc. These channels, particularly the human members of the superfamily, are currently subjects of intensive investigation by many investigators around the world.
In this review, I would like to recount, as accurately as I can recall, how the field of Drosophila phototransduction began during the period between about 1966 and 1979. The key element that made the study of Drosophila phototransduction effective was the use of mutants. Use of mutants required that a non-traditional approach be applied to phototransduction. Any discussion of new approaches to phototransduction must begin with Max Delbrück who had been championing the cause of sensory transduction since the mid-1950's. He understood its importance as a scientific discipline and the need for a new approach in its study. These sentiments can be found in their most explicit form in his Nobel lecture of 1969. He said, “Sensory physiology in a broad sense contains hidden at its kernel an as yet totally undeveloped but absolutely central science: transducer physiology.” He then went on to state that the key to understanding sensory transduction was finding the biological material “most suitable for bringing us decisive insights in this field.” I read these words many years after Delbrück's lecture was published. When I read them, however, they had a special ring for me because they seemed to capture how I felt when I started delving into the phototransduction field.
In 1964/65, I was working on the early receptor potential (ERP) of the vertebrate retina. The ERP is a light-evoked potential of very rapid time course that had been discovered by Brown and Murakami (1964) in the eye of Cynomolgus monkey. Because it had essentially no latency, there was the hope that it might reveal something about the nature of the transduction events that intervene between the photochemistry of visual pigments and physiological events. The work was exciting at the time. By the end of 1965, however, I was coming to the realization that the ERP most likely represented electrophysiological manifestations of photochemical transitions of visual pigments themselves rather than any subsequent events leading to the generation of the receptor potential. It no longer seemed likely that the ERP would lead one to any new insight into phototransduction. Although I had only a vague idea as to what needed to be done, I was becoming increasingly convinced that to make any real progress in phototransduction, we needed new approaches. Although I was not even aware of it at the time, I was essentially retreading the ground Delbrück had trod more than a decade earlier.
Delbrück had chosen the sporangiophores of the mold Phycomyces as the biological material for studying phototransduction. To learn more about Phycomyces, I took Delbrück's course on Phycomyces at Cold Spring Harbor Laboratory in August, 1965. Unfortunately, I came out of this course convinced that Phycomyces did not have the answer to phototransduction. The phototropism of the Phycomyces sporangiophores was nothing like the phototransduction of animal vision I was familiar with. Moreover, I could not find evidence of any potential breakthrough that could make this an organism of choice for studying phototransduction.
The key to phototransduction turned out to be genetics. However, in 1965 there was no one even remotely suggesting the use of genetics to study phototransduction or any other neuronal function. An enormously influential paper entitled, “Behavioral mutants of Drosophila isolated by countercurrent distribution,” by Seymour Benzer appeared in PNAS in June, 1967. This paper directly influenced the development of a number of subfields in neurogenetics. However, phototransduction was not one of them. For one thing, Benzer was very focused on “behavior.” Moreover, as to be substantiated shortly, Drosophila phototransduction research was going through its own independent development and was already beginning to generate its own mutants by the time the Benzer paper appeared. However, this development required some key inputs from several scientists who were not as well known as the above two, and I wish to formally acknowledge their inputs.
I went to Purdue as an Assistant Professor in the fall of 1965. Seymour Benzer was a Distinguished Professor at Purdue at the time, but he had taken a leave of absence to study neurobiology with Roger Sperry at Caltech before my arrival at Purdue, precluding any overlap between us. However, at the time, Purdue boasted a very strong group in microbial genetics. One of the key members of this group was Fred Neidhardt, whose research was broadly in E. coli molecular growth physiology and gene expression. He left Purdue in 1970 to go to University of Michigan from which he retired as the F.G. Novy Distinguished University Professor of Microbiology and Immunology in 2000. As is probably the case in every other science department, every fall, the Department of Biological Sciences at Purdue organized its professors in small groups to make presentations on their research to incoming graduate students. Fred Neidhardt was in the group of professors I was assigned to that year. One statement Fred made in his presentation made an indelible impression on me. He said, “The most important tools in biology are mutants.” My previous background as either a physicist or an electrophysiologist did not prepare me to consider mutants in this light. This statement more than anything else made me seriously consider the use of mutants in my search for a new approach to phototransduction. If one starts thinking about generating mutants, it is difficult not to consider Drosophila. Moreover, if one wants to generate mutants defective in vision, one of the more obvious selection schemes would be one based on phototaxis, since a body of literature already existed on Drosophila phototaxis (Hirsch & Boudreau, 1958; Hadler, 1964).
By the summer of 1966, I had come to the decision to use Drosophila to attempt to generate mutants defective in phototransduction, at least to carry out preliminary feasibility studies. Accordingly, I decided to take some concrete steps to implement this decision that fall. First of all, I wrote to Seymour Benzer. Secondly, I wrote to about 10 people, who I thought would be familiar with Drosophila genetics, to ask if they had ever encountered “blind” mutants. And thirdly I designed and constructed a simple phototaxis apparatus to start experimenting. By that time, I was hearing rumors that Benzer was using Drosophila mutants to “dissect behavior.” I wrote to Benzer to tell him about my ideas to use Drosophila to generate mutants to study phototransduction and related processes. He wrote back with a strong endorsement of my ideas. He also mentioned that he had developed a countercurrent method to isolate flies according to their phototactic ability. He gave no details of the method except to say that he would send a copy of the manuscript. The manuscript never arrived. However, his encouragement and endorsement were important to me at the time.
I decided to write to Drosophila experts because I wanted to know if anyone had ever noticed flies that behaved as though they were blind. Since no one had ever shown that it was possible to generate and isolate mutants with a block in the visual pathway, I thought that it might be a good idea to see if anyone had ever noticed “blind mutants” before going to the trouble of trying to isolate them. A few of the 10 people I wrote to in November, 1966, responded with thoughtful replies. Two of these are reproduced as Figs. 1 and 2. The first one (Fig. 1) is from Winifred Doane, who was then at Yale University. Subsequently, she moved to Arizona State (1977) and retired from there in 1998. The letter was dated Nov. 25, 1966. She basically said that she had never noticed any blind mutants. That did not mean that they did not exist. If you wanted them, however, you would have to isolate them yourself. You could do this by a phototaxis assay, but you need to be careful about the literature in this field. She had apparently consulted two of her colleagues before replying. The second one was from Irwin Oster at Bowling Green State University in Bowling Green, Ohio (Fig. 2). Although this letter was also in response to my query of November, 1966, it did not arrive until January, 1967, and he apologized for the delay to begin his letter (Fig. 2). Irwin Oster, now long deceased, was a graduate student of the Nobel laureate, Hermann Muller, at Indiana University. When Muller retired in 1964, Oster took over his stocks and basically operated a stock center at Bowling Green. He also said he did not “know of any stocks which might be termed `blind'.” However, he had some specific suggestions for mutagenesis. He suggested using X-rays as mutagen and females of an attached-X stock for X-chromosome mutagenesis. Even more importantly, he sent us an attached-X stock (see Fig. 2). This was my first introduction to the use of attached-X females for X-chromosome mutagenesis. Thus, when we began experimenting with mutagenesis, we did so by using the attached-X female stock he sent and X-rays as mutagen. It was only somewhat later we used the chemical mutagen, ethyl methanesulfonate (EMS) (Alderson, 1965; Lewis & Bacher, 1968).
Figure 1.
Letter from Dr. Winifred W. Doane, dated November 25, 1966.
This letter was in response to the author's letter, dated November 16, 1966, inquiring if she had ever come across Drosophila mutants that acted as though blind but had no obvious external morphological defects. Reproduced with permission from Dr. Winifred W. Doane.
Figure 2.
Letter from Dr. Irwin I. Oster, dated January 5, 1967.
This letter was also in response to the same letter sent to Winifred Doane (Fig. 1) inquiring about blind mutants sent on the same day, November 16, 1966.
The third thing we did in late 1966 was to design and build a simple phototaxis apparatus and to start experimenting with it. From the beginning, our objective was to isolate mutants which are defective in the electroretinogram (ERG), a light-evoked, extracellularly recorded, mass response of the eye. While ERG-defective mutants were not all expected to be impaired in phototransduction, a pool of such mutants, we thought, would be enriched in phototransduction-defective mutants. One obvious way of doing this would be to isolate behaviorally nonphototactic mutants first and test these mutants for ERG defects. Since we expected to go through a large number of flies, we sought to make the phototactic assay as simple as possible. Fig. 3 shows the device we built. It consisted of a black box of about 56 cm (22”) in length containing two 1½” od, 7¾” length test tubes placed mouth-to-mouth with a trap door in between. A flashlight bulb served as the light source. To begin the experiment, we introduced the flies into the tube on the dark side (right in Fig. 3), turned on the light, and opened the trap door for a prescribed length of time (usually one min) while manually agitating the tubes. At the end of the one-min period, the trap door was closed and the flies entrapped in each tube were examined and counted. Under the conditions we used, basically all wild-type flies went to the light-side tube.
Figure 3.
Phototaxis apparatus.
A simple phototaxis apparatus consisted of a black box of about 17 × 17 × 56 cm in dimensions. It contained two test tubes of 1½” (3.8 cm) outside diameter and 7¾” (19.7 cm) length, placed mouth to mouth with a trap door in between. A flashlight bulb served as a light source (left compartment).
When we were ready to do exploratory mutagenesis, we followed Oster's advice and began with X-ray mutagenesis of the X-chromosome. We X-rayed male flies and mated them to virgin females of the attached-X stock. The F1 offspring of the above cross were subjected to the phototactic assay using the apparatus. In this (F1) cross, any mutation induced on the X-chromosome of the male parent would be inherited only by the male offspring. If the mutation caused impairment of phototaxis, F1 male offspring carrying the mutation would fail to go toward light and remain in the dark side, while males that do not carry such mutations would go toward light normally. Accordingly, the F1 males remaining in the dark tube were selected and single male-mated to virgin attached-X females. The offspring of each male was tested for phototaxis, line by line. All male offspring, but none of the female offspring, of this cross (F2) would carry the mutations on the X-chromosome of the male parent. If the failure of F1 males to go toward light was due indeed to X-chromosome mutations that caused impairment of phototaxis, none of the F2 male offspring, but all F2 female offspring, would go toward light in the phototaxis assay. At least this was the expected scenario.
Shown in Fig. 4 are raw data obtained in phototaxis assays of F2 offspring of four different lines in June 1 and 2, 1967. In all four cases, essentially all females ended up in the light-side tube, while males remained in the dark side. For example, in the tx1 line, there were 76 females and nine males in the light-side tube, while there were 62 males and five females in the dark-side tube (Fig. 4). A similar gender-dependent fractionation of the F2 offspring was also observed in the other three lines (Fig. 4). In a similar fashion, we isolated three other presumptive mutant lines a week or two later (data not shown). Initially, these were labeled tx1…tx7, but later x1…x7, and still later P1…P7. Of these, only x7 showed a mutant ERG phenotype. This mutant lacked the on- and off-transients of the ERG and was shown later to be an allele of the well-known body color mutant, tan. To my knowledge, this was the first artificially induced mutant with an ERG phenotype ever isolated. Some of these results were published later (Pak, Grossfield, & White, 1969). Isolation of this mutant, though not a new one, provided us with the much needed encouragement that generation and isolation of ERG defective mutants were indeed possible.
Figure 4.
Raw phototaxis data obtained following X-ray mutagenesis.
The X-rayed parent males were mated to virgin females of the attached-X stock. The F1 offspring of the above cross were tested by the “phototaxis box” (Fig. 3), and the F1 males remaining in the dark side tube (right side tube in Fig. 3) were single-male mated to virgin attached-X females. The results shown are those from the F2 offspring of this second cross, obtained on June 1 and 2, 1967. As may be seen, the phototaxis test fractionated the population of flies in each line according to their gender, almost all females going toward light and almost all males remaining in the dark. The results were consistent with the supposition that mutations induced on the X-chromosome caused the impairment of phototaxis in males. In a similar fashion, three other presumptive non-phototactic lines were isolated a week or two later.
Our group received much needed infusion of genetic expertise when Joe Grossfield joined us in June, 1967. He discontinued the use of X-rays and began using EMS as a mutagen exclusively. He also switched the base wild-type stock for mutagenesis from Canton S to Oregon R after testing a number of wild-type stocks for their phototactic ability using our device. However, the basic strategy of phototactic assay for mutants remained the same as described above. Most of our X-chromosome mutants were isolated using this strategy.
By 1971, three laboratories independently reported isolating a series of artificially induced mutants with ERG phenotypes (Hotta & Benzer, 1970; Pak, Grossfield, & Arnold, 1970; Heisenberg, 1971). Pak (1975) summarized the status of mutant isolation in the three laboratories as of about 1973. Hotta and Benzer used their countercurrent apparatus (Benzer, 1967) to fractionate the mutagenized fly population according to their phototactic behavior. Heisenberg fractionated the mutagenized flies according to their ability to follow rotating stripes for study of optomotor response. We used the phototactic apparatus previously described for the purpose of isolating phototransduction-defective mutants. Although the objectives were different and methods used also differed, in the early rounds of mutagenesis, the three laboratories isolated, with some exceptions, mutants in the same five genes: norpA (no receptor potential A), tan, nonA(no on or off-transient A), rdgA (receptor degeneration A), and rdgB (receptor degeneration B). These mutants were characterized by either very small ERG amplitudes or an almost complete absence of the on- and off-transients of the ERG, indicative of a severe defect in neurotransmission at the photoreceptor synapses. Apparently, these early mutants were isolated because of their severe impairment in vision regardless of selection methods applied. Some of the genes identified by these mutants were later shown to encode important components of the phototransduction cascade. The most important of these was the norpA (no receptor potential A) gene encoding phospholipase Cβ (PLCβ) (Bloomquist et al., 1988), which catalyzes the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) to generate two potential second messengers, inositol 3,4,5-trisphosphate (IP3) and diacylglycerol. Since severe norpA mutants lack the photoreceptor potential, the above finding established that Drosophila phototransduction is based on a phosphoinositide-mediated signaling cascade. Two other genes were also found to be important, rdgA and rdgB. They were shown to encode, respectively, diacylglycerol kinase (DGK) (Masai, Okazaki, Hosoya, & Hotta, 1993) and phosphatidyl inositol transfer protein (PITP) (Vihtelic, Hyde, & O'Tousa, 1991), which are involved in the phosphoinositide regeneration cycle (e.g. reviews by Wang & Montell, 2007; Hardie & Postma, 2008; Katz & Minke, 2009).
Although the phototaxis assay tended to favor mutants with severe visual defects, mildly affected mutants were also isolated using this method. Among the mildly affected mutants was inaEP19, which was originally called x-19. Its ERG phenotype was originally unrecognized, and it was classified as a non-phototactic mutant without an ERG phenotype (Pak et al., 1970). Only many years later, after careful analysis, it was realized that this mutant did have an ERG phenotype and that it was an allele of N125, which we had received from Martin Heisenberg. These mutants eventually led to the identification of the sn-1 specific diacylglycerol lipase (DAGL) gene, which appears to play a role in the generation of excitatory signals to the TRP/TRPL channels (Leung et al., 2008).
By the fall of 1971, we decided to begin mutagenesis of the autosomes. Mike Deland, who was Arthur Chovnick's student at the University of Connecticut, joined the lab, and he designed the cross schemes for autosomal mutagenesis (described in Pak, 1979). At this point, we also decided to abandon prescreening mutants by phototaxis and screen for mutants directly by ERG recording. We had become acutely aware that the phototaxis assay is not the most efficient or effective way to isolate phototransduction-defective mutants. To begin with, this assay tended to be biased toward selection of mutants with grossly deficient vision. Many putative mutants with subtle ERG defects went unrecognized. Moreover, the severity of mutant defect assessed from phototaxis did not necessarily coincide with that determined electrophysiologically. In addition, only a fraction of presumptive non-phototactic mutants isolated had electrophysiological phenotypes. In short, the method generated too many false negatives as well as false positives. Finally, the phototaxis assay that worked reasonably well for X-chromosome mutagenesis did not work well for autosomes, at least in our hands. We had used as a baseline stock for mutagenesis a wild-type stock that had been population-selected for positive phototaxis for several generations to render the baseline flies highly phototactic. The autosomal mutagenesis schemes were a little more involved than that of the X-chromosome and required additional manipulations to enable the identification of offspring flies homozygous for the mutagenized chromosome for testing (see Pak, 1979). Apparently the additional steps introduced partially nullified the effect of selection. All these problems could be obviated by using the ERG as a mutant selection tool. The ERG recording of Drosophila eye was first reported by Hengstenberg and Götz (1967). In spite of the small size of the Drosophila eye, ERG recordings could be done relatively easily; and with optimization of some of the steps, they could be done rapidly enough to be incorporated into a mutant assay.
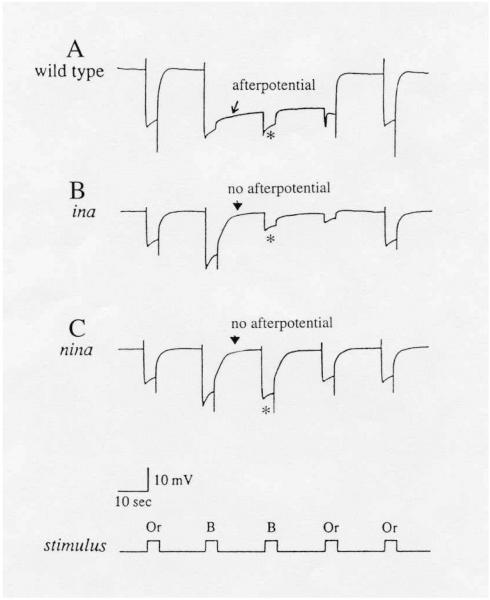
Around this time, an intriguing electrophysiological phenomenon known as the “prolonged depolarizing afterpotential (PDA)” was reported in the lateral ocellus of the barnacle (Hillman, Hochstein, & Minke, 1972) and the median ocellus of Limulus (Nolte & Brown, 1972). Normally, invertebrate photoreceptors respond to the onset of a light stimulus with a depolarization and repolarize with the cessation of the stimulus (Fig. 5). If the stimulus is bright enough to photoconvert a sufficiently large fraction (> 20%) of rhodopsin to its active state, metarhodopsin, however, the light-evoked depolarization persists even after the stimulus is turned off (Fig. 5A, arrow). The potential that persists in the dark was named the PDA (Hillman et al., 1972). We now know that repolarization of the receptor potential at the end of a stimulus occurs through inactivation of metarhodopsin by binding an inhibitory protein, arrestin, and that the PDA is generated if the stimulus photoconverts rhodopsin to metarhodopsin in molar excess of the available arrestin (~20% of rhodopsin) (Dolph et al., 1993). At the time, however, the origin of the PDA was obscure. Nevertheless, the available evidence suggested that the PDA might be an important component of the phototransduction process. We thus thought that incorporating the PDA into the mutant selection scheme might lead to isolation of novel phototransduction-defective mutants. Since we had already begun using ERG for mutant assay, we decided to devise an ERG stimulus protocol that would allow the detection of PDA as well. The major Drosophila rhodopsin, Rh1, absorbs maximally at about 485 nm and metarhodopsin at about 575 nm (Ostroy, Wilson, & Pak, 1974; Pak & Lidington, 1974). Accordingly, the stimulus protocol shown at the bottom of Fig. 5, consisting of a series of orange and blue stimuli, was devised to photoconvert the visual pigment back and forth between the rhodopsin and metarhodopsin states. With this protocol, blue stimuli would generate the PDA, and orange stimuli would cancel the potential (Fig. 5). To observe the intensity dependence of PDA, the protocol was repeated at several different intensities. Finally, we introduced a white (w) mutation in the baseline wild-type stock to eliminate the screening pigments in pigment cells in the eye. These pigments strongly absorb in the blue (Langer, 1967; Strother & Casella, 1972; Stark, 1973), severely attenuating the intensity of blue stimulus reaching rhodopsin in the rhabdomere. Consequently, in the presence of these pigments, the PDA cannot be readily observed.
Figure 5.
ERG's of PDA-defective mutants, ina (inactivation but no afterpotential) (B) and nina (neither inactivation nor afterpotential) (C), compared to that of wild type (A).
A bright blue stimulus elicits the prolonged depolarizing afterpotential (PDA) in wild type (arrow in A), but not in ina (arrowhead in B) or nina (arrowhead in C) mutants. A second bright blue stimulus delivered during the PDA elicits only a small response, originating from R7 and R8 photoreceptors, in wild type and ina (* in A & B). The R1-6 photoreceptors are inactivated and do not respond. By contrast, in nina mutants, R1-6 photoreceptors are not inactivated and respond with a full amplitude response to the second blue stimulus (* in C). All flies were marked with white (w) or brown;scarlet (bw;st) to eliminate the screening pigments in pigment cells. Or: orange; B: blue. Reproduced from Pak and Leung (2003) with permission from Taylor & Francis, Inc.
We carried out mutagenesis of the two major autosomes, the 2nd and the 3rd, separately using slightly different cross schemes (see Pak, 1979). We began mutagenizing the second chromosome in late 1971 and isolated the first second chromosome mutants the following year. Mutagenesis of the third chromosome began in 1973, and the first mutants in this chromosome appeared in 1974. The PDA protocol of ERG screening was put in place by about 1973/1974. The first PDA-defective mutant isolated was inaCP207, obtained in January, 1974. In the next five years or so, we isolated over 100 autosomal mutants representing over 30 separate genetic loci.
The first four 3rd chromosome mutants to be isolated were all trp (transient receptor potential) mutants (obtained in 1974). Ultimately, we ended up isolating 9 alleles in this gene (compiled in Lindsley & Zimm, 1990; with an additional allele described in Yoon et al., 2000). This gene is now widely known for encoding the Drosophila phototransduction channel, TRP, (Hardie & Minke, 1992) which has become the founding member of a new superfamily of TRP cation channels (e.g. reviews by Minke, 2006; Hardie, 2007; Venkatachalam & Montell, 2007). The first mutant to be reported in this gene was a spontaneously occurring one (Cosens & Manning, 1969). These authors showed that ERG responses decay prematurely in the mutant. This finding, though potentially interesting, raised a number of concerns at the time. One is that their results were based on one single spontaneously occurring mutant with no description of how it was obtained. It was difficult to know what genetic alterations this strain represented. For example, the results could have been due to additive effects of alterations in several genes mapping to the same chromosome. The isolation of multiple alleles from mutagenesis of a baseline stock of known genetic background was important in establishing that the effect observed was indeed due to mutation in a single gene. Another concern was that at the time the cellular origins of ERG components were not well established. One could not be certain how much of the effect seen in the ERG of this strain originated from photoreceptors and how much of it post-photoreceptorally. This question was settled by performing intracellular recordings from photoreceptors (Minke, Wu, & Pak, 1975). Only after determining that the defect arose from the photoreceptors, did we feel comfortable enough to name these mutants, with consent from Derek Cosens, transient receptor potential (trp) (Minke et al., 1975).
The PDA-defective mutants turned out to be particularly useful in identifying some important components of the phototransduction pathway. These mutants fell into two classes according to their ERG phenotypes, which we called nina (neither inactivation and no afterpotential) and ina (inactivation but no afterpotential), as defined in Fig. 5. During the period covered by this review, we isolated 42 nina mutants falling into eight complementation groups, ninaA, …ninaH, and eight ina mutants representing five genetic loci, inaA, … inaE. Additional nina and ina mutants, such as ninaI and J (unpublished) and inaF (Li et al., 1999), were isolated much later and were not products of the original mutagenesis. Subsequent analyses by many investigators revealed that the nina and ina series of mutants identified an array of genes encoding proteins important in the phototransduction cascade or photopigment cycle. Presented below are thumbnail descriptions of the functions mediated by the nina- and ina-encoded proteins.
ninaA encodes an eye-specific cyclophilin which serves as a chaperone for nascent opsin during its maturation and intracellular transport (Schneuwly et al., 1989; Shieh, Stamnes, Seavello, Harris, & Zuker, 1989; Colley, Baker, Stamnes, & Zuker, 1991). This was the first chaperone protein to be discovered for any G-protein coupled receptor (Brady & Limbird, 2002).
ninaB encodes a β'-carotene-15,15'-monooxygenase, which catalyzes the centric cleavage of carotenoids to form all-trans retinal in one of the first steps in the formation of the rhodopsin chromophore (von Lintig & Vogt, 2000; von Lintig, Dreher, Kiefer, Wernet, & Vogt, 2001).
ninaC encodes two protein isoforms both consisting of linked protein kinase and class IIIa myosin domains (Montell & Rubin, 1988). These are multifunctional proteins, and one of their major functions appears to be to keep calmodulin enriched in the rhabdomeres (Porter, Yu, Doberstein, Pollard, & Montell, 1993; Porter, Minke, & Montell, 1995).
ninaD encodes a class B scavenger receptor that is presumed to mediate the cellular uptake of carotenoids (Kiefer, Sumser, Wernet, & von Lintig, 2002; Minke & Parnas, 2006).
ninaE encodes the major class of opsin, Rh1, in the Drosophila eye (O'Tousa et al., 1985; Zuker, Cowman, & Rubin, 1985). It was the first invertebrate opsin to have its sequence elucidated.
ninaG encodes an oxydoreductase which is proposed to act in the biosynthesis of rhodopsin chromophore by catalyzing the conversion of (3R)-3-hydroxyretinol to the 3S enantiomer (Sarfare, Ahmed, Joyce, Boggess, & O'Tousa, 2005; Ahmed, Joyce, Boggess, & O'Tousa, 2006).
inaC encodes an eye-preferentially expressed protein kinase C (ePKC or INAC) (Schaeffer, Smith, Mardon, Quinn, & Zuker, 1989). It is a member of the INAD supramolecular signaling complex (see inaD below).
inaD encodes a PDZ domain scaffold protein that binds at least three key signaling proteins, TRP, NORPA, and INAC, to nucleate the formation of supramolecular signaling complexes (Shieh & Niemeyer, 1995; Huber et al., 1996; Chevesich, Kreuz, & Montell, 1997; Tsunoda et al., 1997). The INAD complex was the first supramolecular signaling complex to be discovered in a sensory transduction cascade and analyzed extensively (review: Huber, 2001; Minke & Parnas, 2006).
inaE encodes a diacylglycerol lipase which appears to be important in the generation of the excitatory signal to the phototransduction channels, TRP and TRPL (Leung et al., 2008).
Unfortunately, inaA and B mutants were lost before they could be subjected to analysis.
I will conclude this review by describing some of the autosomal ERG defective mutants generated by other groups. The John Merriam group at UCLA also carried out autosomal mutagenesis for the isolation of ERG-defective mutants. Koenig and Merriam (1977) reported the isolation of nine autosomal ERG-defective mutants, representing eight separate loci, five on the second chromosome and three on the third. The motivation behind this work was never described. The mutants were reported to have been isolated by phototaxis assay using the countercurrent apparatus of Benzer (1967). For some reason, this group of mutants appeared to be dominated by those that lack the on- and off-transients of the ERG, suggesting that they are defective in synaptic transmission between the major photoreceptors R1-6 and their target laminar neurons. A notable exception was the third chromosome mutant, JK84. It was initially reported that, in this mutant, the rhabdomeres of the major class of photoreceptors R1-6 do not form while the rhabdomeres of R7 and R8 are intact, and it was thus named ora (outer rhabdomeres absent) (Harris & Stark, 1977). Scavarda, O'Tousa, and Pak (1983) showed that oraJK84 fails to complement all mutations then identified in the ninaE gene, which encodes the major class of opsin, Rh1 (O'Tousa et al., 1985; Zuker et al., 1985), present in R1-6 rhabdomeres. However, oraJK84 also fails to complement mutations in another gene, ort (ora transientless) (O'Tousa, Leonard, & Pak, 1989), complicating the interpretation of oraJK84. ort encodes a histamine-gated chloride channel, which functions as the synaptic target of R1-6 photoreceptors (Geng et al., 2002). It was not until 1989 that O'Tousa et al. established conclusively that oraJK84 is a double mutant with lesions in both ninaE and ort. oraJK84 was isolated at least by August, 1973 and was brought to the Neurobiology of Drosophila course at Cold Spring Harbor Laboratory by Jane Koenig. Thus, although the ninaE gene was cloned and characterized using the ninaE mutants, isolated on the basis of their PDA phenotype (Pak, 1979; Stephenson, O'Tousa, Scavarda, Randall, & Pak, 1983; O'Tousa et al., 1985; Zuker et al., 1985), oraJK84 probably was the first mutant with a lesion in this gene to be isolated.
Subsequent to the Koenig and Merriam (1977) work, N. Orevi, R.W. Hardy, and J.R. Merriam (personal communication) continued the autosomal mutagenesis for the isolation of ERG defective mutants. However, this work was never published. In 1989, when he was cleaning up his stocks, John Merriam kindly sent us his collection of ERG-defective mutants. Unfortunately, by Merriam's own admission, “they had not been taken care of and attrition had set in” by then. The shipment contained a total of 23 mutant lines, but five of these were our mutants and one was Heisenberg's that had been sent to Merriam earlier, and two were ones Koenig and Merriam had reported earlier. Many of the remaining ones no longer had mutant phenotypes. It is difficult to know the true range of mutants isolated by these investigators or the dates of their isolation. As far as we can determine, most of them were isolated during the first half of the 80's. Because most investigators were not aware of these mutants, they were not utilized in the molecular genetic investigation of phototransduction, which began in the mid-80's. However, some of their mutants did find use in other labs. For example, many of the ort alleles O'Tousa et al. (1989) and Geng et al. (2002) used came from the Orevi, Hardy, and Merriam work. O'Tousa et al. (1989) used them to show that oraJK84 is a double mutant carrying mutations in both the ort and ninaE genes. Geng et al. (2002) used them to show that ort encodes the synaptic target of R1-6 photoreceptors.
In this review, I attempted to present an account of the early years of phototransduction genetics that is personal and yet as objective and accurate as I can make it. During this period, our preoccupation was to generate mutants that could potentially be useful in the molecular analysis of the phototransduction process. We had no idea when molecular analysis would be possible. For a small number of mutants, such as norpA, it was clear from early on that phototransduction is blocked, and these mutants were studied using the techniques that were available. However, for most mutants it was not at all obvious how the mutation affected the phototransduction process, if at all. The molecular analysis of phototransduction began in the mid 80's with cloning of the ninaE gene (O'Tousa et al., 1985; Zuker et al., 1985) followed by that of the norpA gene (Bloomquist et al., 1988). Once molecular techniques began to be applied to the phototransduction system, progress was quite rapid, with many investigators contributing. It is virtually impossible to cite them all without slighting some by omission. Nevertheless, particularly noteworthy were the contributions by Baruch Minke, Roger Hardie, Craig Montell, and Charles Zuker. While the availability of mutants was important, equally important were the contributions of all these investigators in driving the progress in this field.
A portion of this account was presented at the Erich Buchner Symposium in Würzburg, Germany on September 28, 2009.
Acknowledgements
I thank Dr. Chun-Fang Wu for asking me to write this review. I am deeply indebted to Dr. Joseph Grossfield and Dr. Michael Deland, who oversaw the mutagenesis program with technical help from: Hilary Helmrich ('66–'68), Martha Honeycutt ('67–'70), Andrea Coggeshall ('70–'72), Sally Timberlake ('70–`71), Sherry Conrad ('73–'78), and Jessica Ringgold Miller ('77–'79). Special thanks go to Michael Deland and Sherry Conrad who were responsible for the isolation of the vast majority of the mutants in the lab collection. I thank Donna Miller for her help with the manuscript and Shikoh Shino and Marion Pak for their comments on the manuscript. The mutagenesis project was supported by grants from National Science Foundation and National Eye Institute (T32 EY07008; R01 EY00033) to the author.
References
- Ahmed ST, Joyce MV, Boggess B, O'Tousa JE. The role of Drosophila ninaG oxidoreductase in visual pigment chromophore biogenesis. J. Biol. Chem. 2006;281:9205–9209. doi: 10.1074/jbc.M510293200. [DOI] [PubMed] [Google Scholar]
- Alderson T. Chemically induced delayed germinal mutation in Drosophila. Nature. 1965;207:164–167. doi: 10.1038/207164a0. [DOI] [PubMed] [Google Scholar]
- Benzer S. Behavioral mutants of Drosophila isolated by countercurrent distribution. Proc. Natl. Acad. Sci. USA. 1967;58:1112–1119. doi: 10.1073/pnas.58.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomquist BT, Shortridge RD, Schneuwly S, Perdew M, Montell C, Steller H, et al. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell. 1988;54(5):723–733. doi: 10.1016/s0092-8674(88)80017-5. [DOI] [PubMed] [Google Scholar]
- Brady AE, Limbird LE. G protein-coupled receptor interacting proteins: emerging roles in localization and signal transduction. Cell Signal. 2002;14:297–309. doi: 10.1016/s0898-6568(01)00239-x. [DOI] [PubMed] [Google Scholar]
- Brown KT, Murakami M. A new receptor potential of the monkey retina with no detectable latency. Nature. 1964;201:626–628. doi: 10.1038/201626a0. [DOI] [PubMed] [Google Scholar]
- Chevesich J, Kreuz AJ, Montell C. Requirement for the PDZ domain protein, INAD, for localization of the TRP store-operated channel to a signaling complex. Neuron. 1997;18:95–105. doi: 10.1016/s0896-6273(01)80049-0. [DOI] [PubMed] [Google Scholar]
- Colley NJ, Baker EK, Stamnes MA, Zuker CS. The cyclophilin homolog ninaA is required in the secretory pathway. Cell. 1991;67:255–263. doi: 10.1016/0092-8674(91)90177-z. [DOI] [PubMed] [Google Scholar]
- Cosens DJ, Manning A. Abnormal electroretinogram from a Drosophila mutant. Nature. 1969;224:285–287. doi: 10.1038/224285a0. [DOI] [PubMed] [Google Scholar]
- Dolph PJ, Ranganathan R, Colley NJ, Hardy RW, Socolich M, Zuker CS. Arrestin function in inactivation of G protein-coupled receptor rhodopsin in vivo. Science. 1993;260:1910–1916. doi: 10.1126/science.8316831. [DOI] [PubMed] [Google Scholar]
- Geng C, Leung H-T, Skingsley DR, Iovchev MI, Yin Z, Semenov EP, et al. Target of Drosophila photoreceptor synaptic transmission is a histamine-gated chloride channel encoded by ort (hclA) J. Biol. Chem. 2002;277:42113–42120. doi: 10.1074/jbc.M207133200. [DOI] [PubMed] [Google Scholar]
- Hadler NM. Heritability and phototaxis in Drosophila melanogaster. Genetics. 1964;50:1269–1277. doi: 10.1093/genetics/50.6.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie RC. TRP channels and lipids: from Drosophila to mammalian physiology. J. Physiol. 2007;578(1):9–24. doi: 10.1113/jphysiol.2006.118372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie RC, Minke B. The trp gene is essential for a light-activated Ca2+ channel in Drosophila photoreceptors. Neuron. 1992;8:643–651. doi: 10.1016/0896-6273(92)90086-s. [DOI] [PubMed] [Google Scholar]
- Hardie RC, Postma M. Phototransduction in microvillar photoreceptors of Drosophila and other invertebrates. In: Basbaum AI, Kaneko A, Shepherd GM, Westheimer G, editors. The Senses: A Comprehensive Reference, Volume 1, Vision 1. Academic Press; San Diego, CA: 2008. pp. 77–130. [Google Scholar]
- Harris WA, Stark WS. Hereditary retinal degeneration in Drosophila melanogaster: a mutant defect associated with the phototransduction process. J. Gen. Physiol. 1977;69:261–291. doi: 10.1085/jgp.69.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M. Isolation of mutants lacking the optomotor response. DrosophilaInformation Service. 1971;46:68. [Google Scholar]
- Hengstenberg R, Götz KG. Effect of facet-separating pigments on the perception of light and contrast in eye mutants of Drosophila (in German) Kybernetik. 1967;3(6):276–285. doi: 10.1007/BF00271510. [DOI] [PubMed] [Google Scholar]
- Hillman P, Hochstein S, Minke B. A visual pigment with two physiologically active stable states. Science. 1972;175:1486–1488. doi: 10.1126/science.175.4029.1486. [DOI] [PubMed] [Google Scholar]
- Hirsch J, Boudreau JC. Studies in experimental behavior genetics. I. The heritability of phototaxis in a population of Drosophila melanogaster. J. Comp. Physiol. Psychol. 1958;51:647–651. doi: 10.1037/h0039498. [DOI] [PubMed] [Google Scholar]
- Hotta Y, Benzer S. Genetic dissection of the Drosophila nervous system by means of mosaics. Proc. Natl. Acad. Sci. USA. 1970;67:1156–1163. doi: 10.1073/pnas.67.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A. Scaffolding proteins organize multimolecular protein complexes for sensory signal transduction. Eur. J. Neurosci. 2001;14:769–776. doi: 10.1046/j.0953-816x.2001.01704.x. [DOI] [PubMed] [Google Scholar]
- Huber A, Sander P, Gobert A, Bähner M, Hermann R, Paulsen R. The transient receptor potential protein (Trp), a putative store-operated Ca2+ channel essential for phosphoinositide-mediated photoreception, forms a signaling complex with NorpA, InaC and InaD. EMBO J. 1996;15:7036–7045. [PMC free article] [PubMed] [Google Scholar]
- Katz B, Minke B. Drosophila photoreceptors and signaling mechanisms. Front. Cell. Neurosci. 2009;3(2):1–18. doi: 10.3389/neuro.03.002.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer C, Sumser E, Wernet MF, von Lintig J. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc. Natl. Acad. Sci. USA. 2002;99:10581–10586. doi: 10.1073/pnas.162182899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig J, Merriam JR. Autosomal ERG mutants. Drosophila Information Service. 1977;52:50–51. [Google Scholar]
- Langer H. Über die pigmentgranula im facettenauge von Calliphora erythrocephala. Z. Vergl. Physiol. 1967;55:354–377. [Google Scholar]
- Leung H-T, Tseng-Crank J, Kim E, Mahapatra C, Shino S, Zhou Y, et al. DAG lipase activity is necessary for TRP channel regulation in Drosophila photoreceptors. Neuron. 2008;58:884–896. doi: 10.1016/j.neuron.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB, Bacher F. Method of feeding ethyl methane sulfonate (EMS) to Drosophila males. Drosophila Information Service. 1968;43:193. [Google Scholar]
- Li C, Geng C, Leung H-T, Hong YS, Strong LLR, Schneuwly S, Pak WL. INAF, a protein required for transient receptor potential Ca2+ channel function. Proc. Natl. Acad. Sci. USA. 1999;96:13474–13479. doi: 10.1073/pnas.96.23.13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley DL, Zimm G. The genome of Drosophila melanogaster, Part 4: Genes L-Z. Drosophila Information Service. 1990;68:287. [Google Scholar]
- Masai I, Okazaki A, Hosoya T, Hotta Y. Drosophila retinal degeneration A gene encodes an eye-specific diacylglycerol kinase with cysteine-rich zinc-finger motifs and ankyrin repeats. Proc. Natl. Acad. Sci. USA. 1993;90:11157–11161. doi: 10.1073/pnas.90.23.11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minke B. TRP channels and Ca2+ signaling. Cell Calcium. 2006;40(3):261–275. doi: 10.1016/j.ceca.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minke B, Parnas M. Insights on TRP channels from in vivo studies in Drosophila. Ann. Rev. Physiol. 2006;68:649–684. doi: 10.1146/annurev.physiol.68.040204.100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minke B, Wu C-F, Pak WL. Induction of photoreceptor voltage noise in the dark in Drosophila mutant. Nature. 1975;258:84–87. doi: 10.1038/258084a0. [DOI] [PubMed] [Google Scholar]
- Montell C, Rubin GM. The Drosophila ninaC locus encodes two photoreceptor cell specific proteins with domains homologous to protein kinases and the myosin heavy chain head. Cell. 1988;52:757–772. doi: 10.1016/0092-8674(88)90413-8. [DOI] [PubMed] [Google Scholar]
- Nolte J, Brown JE. Electrophysiological properties of cells in the median ocellus of Limulus. J. Gen. Physiol. 1972;59:167–185. doi: 10.1085/jgp.59.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroy SE, Wilson M, Pak WL. Drosophila rhodopsin: Photochemistry, extraction and differences in the norpAP12 phototransduction mutant. Biochem. Biophys. Res. Commun. 1974;59:960–966. doi: 10.1016/s0006-291x(74)80073-2. [DOI] [PubMed] [Google Scholar]
- O'Tousa JE, Baehr W, Martin RL, Hirsh J, Pak WL, Applebury ML. The Drosophila ninaE gene encodes an opsin. Cell. 1985;40(4):839–850. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- O'Tousa JE, Leonard DS, Pak WL. Morphological defects in oraJK84 photoreceptors caused by mutation in R1-6 opsin gene of Drosophila. J. Neurogenet. 1989;6:41–52. doi: 10.3109/01677068909107099. [DOI] [PubMed] [Google Scholar]
- Pak WL. Mutations affecting the vision of Drosophila melanogaster. In: King RC, editor. Handbook of Genetics. Vol. 3. Plenum; New York, NY: 1975. pp. 703–733. [Google Scholar]
- Pak WL. Study of photoreceptor function using Drosophila mutants. In: Breakefield X, editor. Neurogenetics: Genetic Approaches to the Nervous System. Elsevier North-Holland; New York, NY: 1979. pp. 67–99. [Google Scholar]
- Pak WL, Grossfield J, Arnold KS. Mutants of the visual pathway of Drosophila melanogaster. Nature. 1970;227(5257):518–520. doi: 10.1038/227518b0. [DOI] [PubMed] [Google Scholar]
- Pak WL, Grossfield J, White NV. Nonphototactic mutants in a study of vision of Drosophila. Nature. 1969;222:351–354. doi: 10.1038/222351a0. [DOI] [PubMed] [Google Scholar]
- Pak WL, Leung H-T. Genetic approaches to visual transduction in Drosophila melanogaster. In: Chidiac P, editor. Emerging aspects of heterotrimeric G protein-mediated signaling. Receptors Channels. Vol. 9. 2003. pp. 149–167. [PubMed] [Google Scholar]
- Pak WL, Lidington KJ. Fast electrical potential from a long-lived, long-wavelength photoproduct of fly visual pigment. J. Gen. Physiol. 1974;63:740–756. doi: 10.1085/jgp.63.6.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JA, Minke B, Montell C. Calmodulin binding to Drosophila NinaC required for termination of phototransduction. EMBO J. 1995;18:4450–4459. doi: 10.1002/j.1460-2075.1995.tb00124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JA, Yu M, Doberstein SK, Pollard TS, Montell C. Dependence of calmodulin localization in the retina on the ninaC unconventional myosin. Science. 1993;262:1038–1042. doi: 10.1126/science.8235618. [DOI] [PubMed] [Google Scholar]
- Raghu P, Hardie RC. Regulation of Drosophila TRPC channels by lipid messengers. Cell Calcium. 2009;45:566–573. doi: 10.1016/j.ceca.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Ramsey IS, Moran MM, Chong JA, Clapham DE. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440:1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarfare S, Ahmed ST, Joyce MV, Boggess B, O'Tousa JE. The Drosophila ninaG oxidoreductase acts in visual pigment chromophore production. J. Biol. Chem. 2005;280:11895–11901. doi: 10.1074/jbc.M412236200. [DOI] [PubMed] [Google Scholar]
- Scavarda NJ, O'Tousa J, Pak WL. Drosophila locus with gene-dosage effects on rhodopsin. Proc. Natl. Acad. Sci. USA. 1983;80:4441–4445. doi: 10.1073/pnas.80.14.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer E, Smith D, Mardon G, Quinn W, Zuker C. Isolation and characterization of two new Drosophila protein kinase C genes, including one specifically expressed in photoreceptor cells. Cell. 1989;57:403–412. doi: 10.1016/0092-8674(89)90915-x. [DOI] [PubMed] [Google Scholar]
- Schneuwly S, Shortridge RD, Larrivee DC, Ono T, Ozaki M, Pak WL. Drosophila ninaA gene encodes an eye-specific cyclophilin (cyclosporine A binding protein) Proc. Natl. Acad. Sci. USA. 1989;86:5390–5394. doi: 10.1073/pnas.86.14.5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh B-H, Niemeyer B. A novel protein encoded by the InaD gene regulates recovery of visual transduction in Drosophila. Neuron. 1995;14:201–210. doi: 10.1016/0896-6273(95)90255-4. [DOI] [PubMed] [Google Scholar]
- Shieh B-H, Stamnes MA, Seavello S, Harris GL, Zuker CS. The ninaA gene required for visual transduction in Drosophila encodes a homologue of cyclosporine A-binding protein. Nature. 1989;338:67–70. doi: 10.1038/338067a0. [DOI] [PubMed] [Google Scholar]
- Stark WS. Effect of eye color pigments on the action spectrum of Drosophila. J. Insect Physiol. 1973;19:999–1006. doi: 10.1016/0022-1910(73)90026-7. [DOI] [PubMed] [Google Scholar]
- Stephenson RS, O'Tousa J, Scavarda NJ, Randall LL, Pak WL. Drosophila mutants with reduced rhodopsin content. In: Cosens D, Vince-Price D, editors. The Biology of Photoreception. Cambridge University Press; Cambridge: 1983. pp. 477–501. [PubMed] [Google Scholar]
- Strother GK, Casella AJ. Microspectrophotometry of arthropod visual screening pigments. J. Gen. Physiol. 1972;59:616–636. doi: 10.1085/jgp.59.5.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera K, Nilius B, Voets T. Neuronal TRP channels: thermometers, pathfinders and life-savers. Trends Neurosci. 2008;31(6):287–295. doi: 10.1016/j.tins.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Tsunoda S, Sierralta J, Sun Y, Bodner R, Suzuki E, Becker A, et al. A multivalent PDZ-domain protein assembles signalling complexes in a G-protein-coupled cascade. Nature. 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- Venkatachalam K, Montell C. TRP channels. Annu. Rev. Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR, O'Tousa JE. Isolation and characterization of the Drosophila retinal degeneration B (rdgB) gene. Genetics. 1991;127:761–768. doi: 10.1093/genetics/127.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lintig J, Dreher A, Kiefer C, Wernet MF, Vogt K. Analysis of the blind Drosophila mutant ninaB identifies the gene encoding the key enzyme for vitamin A formation in vivo. Proc. Natl. Acad. Sci. USA. 2001;98:1130–1135. doi: 10.1073/pnas.031576398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lintig J, Vogt K. Filling the gap in vitamin A research: Molecular identification of an enzyme cleaving beta-carotene to retinal. J. Biol. Chem. 2000;275:11915–11920. doi: 10.1074/jbc.275.16.11915. [DOI] [PubMed] [Google Scholar]
- Wang T, Montell C. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 2007;454(5):821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- Yoon J, Ben-Ami HC, Hong YS, Park S, Strong LLR, Bowman J, et al. Novel mechanism of massive photoreceptor degeneration caused by mutations in the trp gene of Drosophila. J. Neurosci. 2000;20(2):649–659. doi: 10.1523/JNEUROSCI.20-02-00649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker CS, Cowman AF, Rubin GM. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 1985;40:851–858. doi: 10.1016/0092-8674(85)90344-7. [DOI] [PubMed] [Google Scholar]