SUMMARY
Human autoimmune regulator (AIRE) functions to control thymic expression of tissue-specific antigens via sequence-specific histone H3 recognition by its plant homeodomain (PHD) finger. Mutations in the AIRE PHD finger have been linked to autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED). Here we report the three-dimensional solution structure of the first PHD finger of human AIRE bound to a histone H3 peptide. The structure reveals a detailed network of interactions between the protein and the amino-terminal residues of histone H3, and particularly key electrostatic interactions of a conserved aspartic acid 297 in AIRE with the unmodified lysine 4 of histone H3 (H3K4). NMR binding study with H3 peptides carrying known post-translational modifications flanking H3K4 confirms that transcriptional regulation by AIRE through its interactions with histone H3 is confined to the first N-terminal eight residues in H3. Our study offers a molecular explanation for the APECED mutations and helps define a subclass of the PHD finger family proteins that recognize histone H3 in a sequence-specific manner.
INTRODUCTION
Immunological tolerance to self is essential in the prevention of autoimmune disease. Mechanisms of central tolerance are mediated in part through the expression of a wide array of otherwise tissue-specific self-antigens such as insulin and thyroglobulin in specialized medullary thymic epithelial cells (Derbinski et al., 2005; Derbinski et al., 2001; Smith et al., 1997). The thymic expression of many of these tissue-specific antigens is dependent on the autoimmune regulator (AIRE) gene (Anderson et al., 2002; Liston et al., 2003), and mutations in AIRE lead to severe, multiorgan, tissue-specific autoimmunity in both mice (Anderson et al., 2002; Ramsey et al., 2002) and humans (Nagamine et al., 1997), which is characterized by abnormal features of chronic mucocutaneous candidiasis, hypoparathyroidism, and adrenal insufficiency, a disorder termed APECED (autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy). At the cellular level, AIRE protein activity has been reported to regulate ectopic protein expression (Anderson et al., 2002). Transcriptional profiling of AIRE-positive thymic epithelia versus their null counterparts revealed that AIRE favors the expression of a subset of peripheral tissue-specific antigens, several of which are APECED autoantibody targets (Anderson et al., 2002). AIRE activity allows particular medullary thymic epithelia to generate and present the peptide fingerprints of other organs. Although some peripheral antigens are expressed in the thymus in the absence of AIRE expression, it is clear that AIRE activity generates an impressive array of peripheral tissue-specific antigens in medullary thymic epithelial cells (Ferguson et al., 2008).
Despite AIRE’s crucial role for self-tolerance, a detailed understanding of the molecular mechanism underlying its functions in cells is lacking. Given the modular architecture of proteins, the mechanism of action of the full-length protein can in many cases be inferred from the knowledge of molecular functions of constituent modules. AIRE is a protein of 545 amino acid residues consisting of several domains characteristic of many transcription regulators. These include, notably, two Zn finger-containing plant homeodomain (PHD) finger motifs and a SAND (named after Sp100, AIRE, NucP41/75, and DEAF1/suppressin) domain. Though over 60 mutations, scattered along the entire protein, have been localized on the AIRE gene of APECED patients, no structural mechanism has been determined satisfactorily to substantiate AIRE function of these mutants. A high density of these mutations localize on the first PHD finger, suggesting a crucial role of PHD1 in transcriptional regulation in thymus epithelial cells. The functional role of PHD1 in chromatin mediated transcription assembly indeed has recently been demonstrated (Koh et al., 2008; Org et al., 2008).
Recently, structural studies of two distinct classes of PHD fingers with respective cognate lignads have demonstrated its crucial role in histone/nucleosome recognition typifying it as an epigenetic “reader” module (Lan et al., 2007; Li et al., 2006; Peña et al., 2006). The first embraces trimethylated histone H3 lysine 4 (H3K4me3, regarded as an epigenetic mark for transcriptional activation) by an aromatic cage; the other, devoid of the cage, uses an amino-terminal aspartic acid to recognize nonmethylated histone H3 lysine 4 (H3K4me0, perceived as a mark for transcriptional repression). PHD1 of AIRE belongs to the second class of PHD histone reader based on its protein sequence. This is consistent with AIRE’s regulatory function in switching transcription on from a repressive state through recruiting transcriptional coactivators such as CBP to the repressive site when it is bound to H3K4me0 (Ferguson et al., 2008).
To determine the molecular basis of human AIRE PHD1 recognition of H3K4me0, we determined the high-resolution 3D solution structure of the AIRE-PHD1 in complex with its biological ligand, the unmodified amino-terminal histone H3 peptide. Guided by the complex structure, we characterized the molecular basis of AIRE PHD1 recognition of the H3Kme0 mark in the context of possible cross-talk with neighboring epigenetic modifications in histone H3 that may alter physiological behavior of AIRE.
RESULTS AND DISCUSSION
Structure Determination of AIRE PHD1 Bound to Histone H3 Peptide
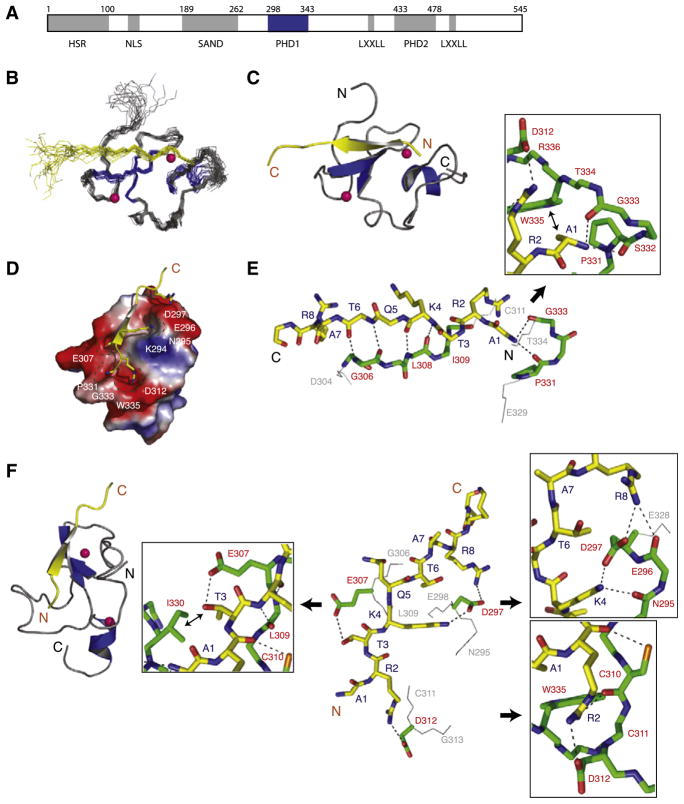
To determine the molecular basis of histone H3 recognition by the first PHD finger of human AIRE (termed PHD1; Figure 1A), we cloned, expressed, and purified the protein to homogeneity (for details, see Experimental Procedures). We solved the 3D solution structure of the complex comprising the PHD1 (residues 294–347) and the H3 peptide (residues 1–20) with a total of 935 NOE-derived distance and torsion-angle restraints (Table 1) obtained from heteronuclear multidimensional NMR spectroscopy (Clore and Gronenborn, 1994). Superimposition of an ensemble of the 20 final NMR structures of the complex is depicted in Figure 1B. Excluding the H3 residues 12–20, which show minimal contact with the protein, the complex structure is well defined. The structure of the complex shows a conserved mode of H3Kme0 recognition seen in the PHD finger of BHC80 (2puy) and PHD-like finger of Dnmt3L (2pvc) (see below for more details). The structure consists primarily of a two-strand antiparallel β sheet that coordinates two zinc atoms anchored by the Cys4-His-Cys3 motif in a cross-brace topology followed by a C-terminal helix (residues 343–347; Figure 1C). The structure of the AIRE-PHD1 bound to the H3K4me0 peptide superimposes well with the structure of the free PHD finger with a root-mean-squared deviation (rmsd) of 1.3 Å, indicating that peptide binding does not induce overall major conformational changes of the protein.
Figure 1. Three-Dimensional Solution Structure of the AIRE-PHD1/H3K4me0 Peptide Complex.
(A) Schematic representation of the functional domains in the human AIRE protein. Grey boxes represent HSR (homogenously staining region), PHD (plant homeodomain), and SAND (Sp100, AIRE-1, NucP41/P75, and Drosophila DEAF-1). PHD and SAND domain boundaries are based on Pfam HMM (Bateman et al., 1999) and remaining segments are based on Meloni et al. (2008). AIRE-PHD1 studied here is shown in blue.
(B) Backbone atoms (N, Cα, and C′) of the 20 superposed NMR structures of the AIRE-PHD1 where protein and peptide are gray and yellow, respectively (left).
(C) Ribbon representation of the complex (middle) highlights the secondary structural elements (protein, blue; peptide, yellow). Pink spheres represent Zn atoms. Only a single representation of Zn atoms of the lowest energy structure is shown in the ensemble for clarity.
(D) Electrostatic potential (isocontour value of ±70 kT/e) surface representation of the AIRE-PHD1 bound to the H3K4me0 peptide (yellow).
(E) Backbone protein-peptide interactions with inset showing the H3A1 interacting neighborhood. The peptide and protein residues are color coded by atom type with carbon atoms in yellow and green, respectively. The orientation of the peptide is the same as that in (C).
(F) Key protein-peptide side-chain interactions with insets respectively highlighting R2, K4, and T3 neighborhood and their surface grooves. The nonpolar nonbonded interacting atoms are labeled with ↔. The peptide orientation in the stick representation is depicted as in the ribbon diagram on left.
Table 1.
NMR Structural Statistics for AIRE PHD Finger/H3K4me0 Complex
| Protein | |
|---|---|
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 935 |
| Intraresidue | 442 |
| Interresidue | 493 |
| Sequential (|i − j| = 1) | 133 |
| Medium range (|i − j| < 4) | 129 |
| Long range (|i − j| > 5) | 231 |
| Protein-zinc restraints | 34 |
| Hydrogen bonds | 28 |
| Total dihedral angle restraints | |
| φ | 12 |
| ψ | 12 |
| Structure statistics | |
| Violations (mean ± SD) | |
| Distance constraints (Å) | 0.0311 ± 0.00266 |
| Dihedral angle constraints (°) | 0.238 ± 0.151 |
| Max. dihedral angle violation (°) | 0.557 |
| Max. distance constraint violation (Å) | 0.0367 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.0029 ± 0.00018 |
| Bond angles (°) | 0.487 ± 0.0214 |
| Impropers (°) | 0.401 ± 0.028 |
| Average pairwise rmsd (Å)a | |
| Heavy | 0.94 ± 0.093 |
| Backbone | 0.49 ± 0.064 |
Number of structures used in rmsd calculations is 20. Pairwise rmsd was calculated among 20 refined structures of the region with residues numbered 295–343.
Structural Basis of H3 Recognition by AIRE PHD1
The histone H3K4me0 peptide is bound in a deep and extensive binding site that engages nearly one-third of the PHD finger residues (Figure 1D; i.e., 19 of the 58 residues of the protein). The H3 peptide binding results in a major change in solvent-accessible surface, with a total of ~480 Å 2 (about 12% of the total area) of surface being buried upon peptide binding. The peptide, adopting an extended β strand-like conformation (residues 2–6), is positioned on the surface of the finger forming a third β strand antiparallel to the Gly 306-Leu 308 segment of the β sheet in the protein (Figures 1C and 1D). This mixed protein/peptide antiparallel β sheet is stabilized by hydrogen bonds between backbone amide and carbonyl groups involving peptide and protein (Figure 1E).
The amino terminus of the H3 peptide is anchored through intermolecular hydrogen bonds with the backbone carbonyl oxygen atoms of protein residues Pro 331 and Gly 333 as well as nonpolar interactions between methyl group of H3A1 and side chains of Pro 331 and Trp 335 (Figure 1E, inset). The peptide is stapled to the protein surface with electrostatic interactions formed between side-chain groups of peptide and protein residues, i.e. H3R2 and Asp 312, H3K4 and Asp297, and H3R8 and Asp 297, respectively, and by additional hydrogen bonds formed between the hydroxyl of H3T3 and the carboxylate of Glu 307 (Figure 1F). The terminal side-chain groups of R2, T3, and K4, and the amino-terminal NH3+ group of the H3 peptide are bound in tiny surface grooves, whereas that of H3R8 is completely solvent exposed. The interconnected surface grooves holding H3A1-T3 are bordered by side-chain carboxylates of Asp 312 on one end (H3R2 site), and Glu 307 (H3T3 site) on the other with backbone carbonyls of Pro 331, Gly 333, and Trp 335 forming the remaining pocket rim atoms, providing a densely electronegative environment for the amino terminus of histone H3. The H3R2 guanidinium group, pointing to the wall of the pocket, rests ~3.0 Å away from the pocket rim (Figure 1F), explaining that symmetric or asymmetric dimethylation of H3R2 sterically hinders the interactions, and phosphorylation of H3T3 also has unfavorable electrostatic effects on binding in the pocket (Koh et al., 2008).
For H3R2 and H3T3, ~75% and ~80% of their respective solvent-accessible surfaces are lost upon binding to the protein, giving a closely knit frame on the H3 amino terminus, which has reduced conformational flexibility due to highly restrained terminal NH3+ group anchored within the pocket (Figure 1E). Given the requirement for free H3 amino terminus immediately amino terminus to the ARTK residues to fit R and K in the respective groves, H4 amino-terminal sequence SGRGK, due to an extra residue, cannot be accommodated, even though RGK can replace RTK, thus confirming a recent report of binding specificity of AIRE-PHD1 for H3 over H4 (Koh et al., 2008).
Additional intermolecular interactions are observed. These are between thiol group of Cys 310 and carbonyl oxygen of H3R2 (Figure 1F, inset). As shown recently (Koh et al., 2008), mutation of C213W at the corresponding residue in the mouse AIRE-PHD1 (see alignment below) reduced binding, possibly due to the loss of this interacting thiol as well as steric constraints of a bulky Trp in the packed interface. Finally, the anchoring side chains of H3K4 and H3R8 also make hydrogen bonds with backbone carbonyl oxygen of Asn 295 and Asp 297, respectively.
Asp297 and Asp312 in AIRE-PHD1 for H3K4 and H3R2 binding, respectively, were also reported in the studies by Koh et al. (2008) and Org et al. (2008) based on a modeled structure built using BPTF-PHD2/H3K4me3 structure instead of BHC80-PHD/H3K4me0 as a modeling template. Because BPTF-PHD2 markedly differs from AIRE-PHD1 or BHC80 PHD structurally and functionally in histone H3 recognition (see below for more details), the conformation of the H3 peptide beyond K4 is significantly different between our structure and the modeled structure (Koh et al., 2008) in that the latter only partly captured AIRE-PHD1 interactions with H3 limiting to residues 1–4. For instance, the PHD1/H3 interactions that were defined in this study based on observed intermolecular NOEs but not described in the previous studies include those between H3R8 and Asp297, and H3T3 and Glu307. The latter explains the effect of H3T3 phosphoylation on steric expulsion. It is important to note that dimethylation of H3R2 has negligible effect on BPTF binding even though H3R2 is seen to form a salt bridge with Asp27 in BPTF, whereas in TAF3-PHD H3R2 is not in ionic interaction with Asp886 but still shows a dramatic drop in affinity due to H3R2me2 (van Ingen et al., 2008; Vermeulen et al., 2007). Hence, a model structure built to accommodate a salt bridge between H3R2 and an Asp/Glu in corresponding PHD finger may not predict an H3R2me2(s/a) effect. Experimentally, structural analysis is much more reliable to determine the structural and molecular basis of molecular interactions. Finally, our structure also captures the dynamic nature of the histone peptide when bound to the protein. As shown in Figure 1B, the N terminus of the peptide is highly restrained, whereas the C terminus is highly flexible.
Cross-Talk of Histone H3 Modifications
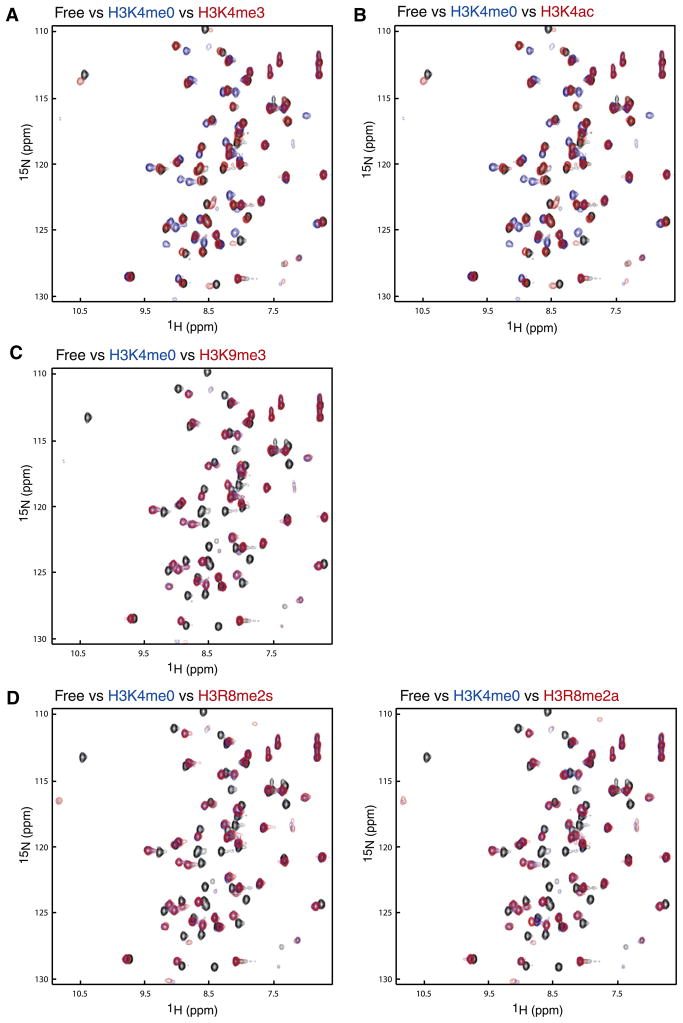
It has been suggested that binding of a reader module specific for a distinctly modified histone site may be affected positively/negatively by adjacent modification in the histone sequence (Fischle et al., 2005; Guccione et al., 2007; Ramón-Maiques et al., 2007), thereby bringing about additional level of regulation in gene transcription. For example, during cell cycle, phosphorylation of H3S10 by Aurora B in M phase disrupts the HP1 chromodomain-H3K9me3 interaction important for chromatin compaction in interphase (Fischle et al., 2005). To investigate whether such a mechanism exists for the AIRE-PHD1-H3K4me0 interaction, we carried out NMR titration of AIRE-PHD1 with various histone peptides. As shown in Figure 2A, trimethylated H3K4 peptide completely abolishes binding to AIRE-PHD1, as the 2D 1H-15N-HSQC spectrum of the protein in the presence of H3K4me3 (red) superposes almost exactly with that of the protein in the free form (black), which is a sharp contrast to that with the nonmodified H3K4me0 peptide (blue). This observation is consistent with the fact that ~85% of H3K4 solvent-accessible surface, the highest among all other H3 residues, is buried upon binding to the protein. H3K4 side-chain atoms are snuggly caged in a narrow groove with side-chain atoms of protein residues Lys 294 to Asp 297 forming the rim (Figure 1F).
Figure 2. Peptide Binding Studies.
Peptide binding studied between AIRE-PHD1 and N-terminal H3 peptides by NMR. Comparison of two-dimensional 1H-15N HSQC spectra of AIRE-PHD1 between its free form (black) and that in presence of a peptide derived from N-terminal H3 residues 1–11 with or without a known post-translational modification: (A) H3K4me0 (blue) versus H3K4me3 (red); (B) H3K4ac (red); (C) H3K9me3 (red); (D) H3R8me2a (red), left panel, and H3R8me2s (red), right panel. The concentration of the protein was 0.5 mM, and the molar ratio of the protein to peptide was kept at 1:5 for all NMR binding studies.
Recent report of H3K4ac mark in human, mouse, and Tetrahymena thermophila (Garcia et al., 2007) in a genome-wide mass spectrometry analysis prompted us to examine possible effect of this modification on AIRE regulation (Figure 2B). We observed that the loss of the positive charge on H3K4ac reduces the binding, likely due to loss of the critical ion pair with Asp 297 and the bulky nature of acetamide moiety. The loss of binding of H3K4ac contrasts that of H3K4me3, as the latter is solely due to steric expulsion. Because the abundance of H3K4me3 (Garcia et al., 2007) is much lower than that of unmodified H3K4, H3K9me3, or H3K27me3, the role of H3K4ac may be as important as H3K4me3 to impact AIRE function.
In accordance with the structural observation that side chain of H3K9 is close to the end of the recognition site (Figure 1F), trimethylation of H3K9 (blue) does not result in any change in interactions as almost no additional perturbation was observed in the 2D 1H-15N-HSQC spectrum as compared with that of non-modified H3 peptide (red) (Figure 2C). This is in agreement with earlier reports (Koh et al., 2008; Org et al., 2008). Furthermore, we investigated contribution of H3R8 in AIRE recognition, which is partially solvent exposed and forms an electrostatic interaction with carboxylate of Asp 297 (Figure 1F). Neither symmetric nor asymmetric dimethylation of H3R8 induced any significant change to the binding (Figure 2D), confirming that modification of H3R8 does not likely affect AIRE function. However, we observed small spectral changes in the 1H-15N-HSQC of AIRE-PHD1 upon addition of a H3R8A mutant peptide as compared with that of the wild-type peptide (see Figure S1A available online), suggesting that H3R8 makes limited contributions via its electrostatic interaction to the overall binding energetics. Finally, consistent with the structure, we concluded that modifications on H3 beyond K9 have no or minimal effect on AIRE binding. This conclusion is supported by the recent results of a mouse AIRE-PHD1 microarray study carried out with various histone H3 peptides (residues 1–21) that bear known modification, such as H3R17me2a/s, H3K9ac/me1/me2/me3, H3K14ac, H3K18ac, or H3K27ac (Koh et al., 2008). The strict requirement of NH3+-ARTK restricts AIRE-PHD1 to binding H3 amino terminus, not other H3, H2A, or H2B peptides (Koh et al., 2008).
APECED Disease Mutants
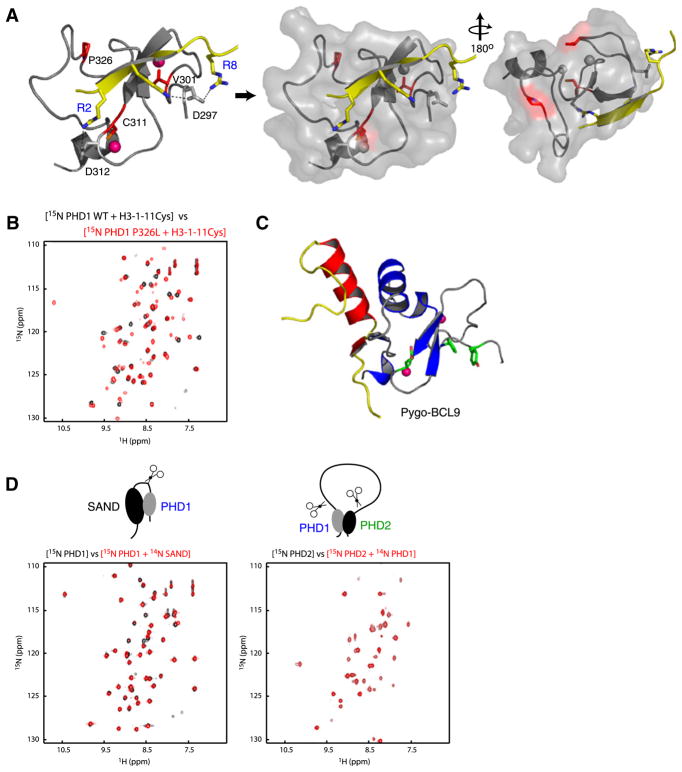
We next examined the structural insights into effects of the AIRE APECED mutants of V301M, C311Y, P326L, and P326Q on H3 binding (Figure 3A). C311Y impairs the C-terminal Zn2+ coordination and thereby destroys the fold of the PHD finger. The V301M mutation, though close to the amino-terminal Zn-coordination site, is less likely to impact the overall structure integrity of the protein or H3 binding, consistent with only a small or negligible effect observed on H3 peptide binding (Koh et al., 2008). Interestingly, although distant from the peptide recognition site, Pro 326 is the second Pro in the conserved Pro-Pro motif in the H3K4me0 binding PHD fingers, which is located in a loop positioned below the signature Trp 335 in the finger. Thus the P326L mutation may cause a partial impairment in the protein structure for the observed 2- to 3-fold reduction in binding affinity to a H3K4me0 peptide (Koh et al., 2008). However, comparison of 2D 1H-15N-HSQC spectrum of the wild-type and the mutant P326L in the presence of a H3 peptide of residues 1–11 (Figure 3B) shows negligible changes due to the mutation, indicating that this mutation does not have any significant effect on the protein structure or H3 binding. Given the relatively large solvent accessibility of Val 301 and Pro 326 in the complex (i.e., 56% and 51%, respectively), we cannot rule out a possible functional role of these two residues in AIRE interactions with other proteins in transcription regulation, as AIRE is known to associate with other transcriptional proteins (Peterson et al., 2008). Indeed, it has been recently reported that Pygo-PHD finger uses a protein surface distinct from the histone interacting site to interact with BCL9 HD1 domain (Figure 3C; Fiedler et al., 2008; van Ingen et al., 2008). Notably, Pro 326 in AIRE-PHD1 resides in a region corresponding to the nonhistone-interacting surface of Pygo and KAP-1 (see below).
Figure 3. APECED Mutants.
(A) Location of the mutant residues (red) on the protein (gray)-peptide (yellow) complex structure in ribbons (left) and transparent surface representation (middle, right). The pink Zn atoms in left panel are shown in gray in surface representation for clarity.
(B) Comparison of two-dimensional 1H-15N HSQC spectra of AIRE-PHD1 wild-type with that of P326L mutant in presence of a peptide derived from N-terminal H3 residues 1–11 (black, wild-type; red, P326L mutant).
(C) Nonhistone interacting surface of Pygo-PHD (gray) in complex with the HD1 domain of BCL9 (yellow/red) (left). The aromatic cage residues of Pygo-PHD involved in H3K4me2 are green.
(D) Two-dimensional 1H-15N HSQC spectra assessing AIRE’s interdomain interaction at 1:1 molar ratio with 0.2 mM respective labeled proteins. The 15N-labeled PHD1 and PHD2 are labeled blue and green, respectively.
The structural protein domains often appear in tandem in chromatin-associated proteins such as bromodomains in yeast Rsc4 (VanDemark et al., 2007) and human TAFII250 (Jacobson et al., 2000); tudor domains in JMJD2A (Huang et al., 2006); chromo-domains in CHD1 (Flanagan et al., 2005); PHD + GATA-like fingers in DNMT3L (Ooi et al., 2007); and PHDs in DPF3 (Lange et al., 2008), which have been shown to interact with each other in the respective functions of chromatin regulation. In AIRE, PHD2 and SAND domains (Figure 1A), also containing a number of disease-mutant sites, are recognized structural domains that are seen in many chromatin-associated proteins. We carried out NMR analysis to check the possibility of interdomain interaction in AIRE. 2D 1H-15N-HSQC spectra of AIRE-PHD1 with and without unlabeled SAND domain (Figure 3D, left) and 2D 1H-15N-HSQC spectra of AIRE-PHD2 with and without unlabeled AIRE-PHD1 (Figure 3D, right) show negligible changes in each case, indicating little if any interdomain interactions between the isolated domains. These results suggest that these structural domains of AIRE do not likely function together as combined units. Thus, the molecular basis for the APECED mutations P326L, P326Q, and V301M are not likely due to intermolecular interactions within AIRE but rather with other cellular proteins. The latter remains to be elucidated further in a separate study.
Structural Comparison with Other PHD Finger Domains
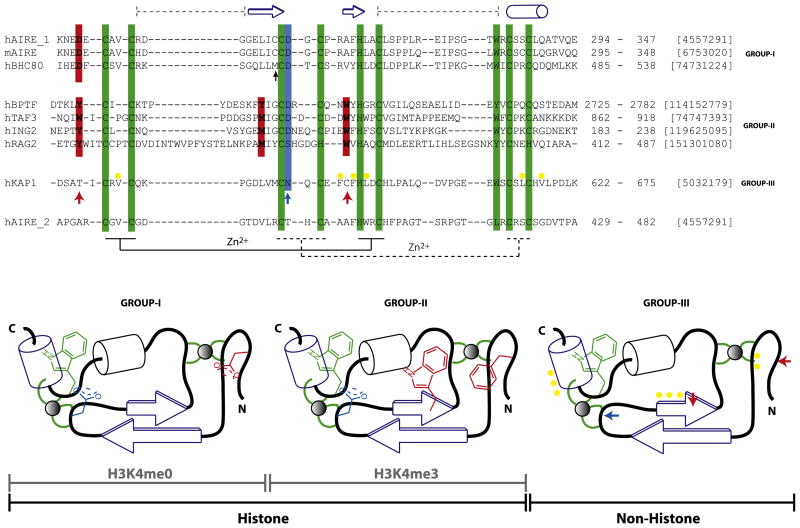
The PHD finger, a binuclear interleaved Zn-chelating domain (Aasland et al., 1995; Capili et al., 2001) of 50–80 residues, shares a core treble clef motif with other Zn-binding domains (Grishin, 2001), such as the RING finger, LIM domain, FYVE domain, and C1 domain. However, there are distinct structural features that differentiate one from the other (Aravind and Iyer, 2003). The distinctive Zn-chelating Cys4-His-Cys3 pattern with conserved characteristic aromatic residue directly amino-terminal to the distal cysteine dyad and most often another aromatic residue immediately preceding the His constituting the hydrophobic core characterizes the fold. Though certain members, such as RAG2-PHD (see below), may have altered Zn-chelating pattern, the presence of the hydrophobic core-forming residues preserves the structural fold. Due to the presence of these features, the overall fold of the AIRE-PHD1 finger is similar to other known structures of PHD fingers despite their relatively low percentage sequence identity.
Structural comparison of the AIRE-PHD1 with the PHD fingers of BHC80 (PDB, 2puy), RAG2 (2v89), BPTF (2f6j), ING2 (2g6q), KAP1 (2r01), TAF3 (2k17), and Pygo1 (2dx8) reveals that it is most closely related to the BHC80 PHD finger (rmsd of 1.42 Å) (Figure 4; Figure S2A) followed by ING2 (2.04 Å), BPTF (2.21 Å), Pygo1 (2.65 Å). KAP1 (2.73 Å), TAF3 (3.0 Å), and RAG2 (3.34 Å). The closeness of AIRE-PHD1 to BHC80 and ING2 is primarily due to the absence of the helical segment in place of the C-terminal loop corresponding to the AIRE-PHD1 residues 330–340 where the PHD fingers of RAG2, Pygo1 (not shown), and BPTF all contain an α helix in place of this loop. Even though KAP1-PHD does not contain the helix in this region, it differs from AIRE-PHD1 in the orientation of the loop seen in contact with adjacent KAP1 bromodomain (2r01; Zeng et al., 2008). The structural difference with TAF3 is also due to a difference in the orientation of the C-terminal loop. Pygo1 forms a dimer via this helix and an adjacent β strand, which is not found in the other PHD fingers (Nakamura et al., 2007), and the extended N-terminal region of TAF3 is seen to form a pair of stands that lie just outside the PHD finger domain boundary. Additionally, a large insertion in the N-terminal loop unique to RAG2 (Figure 4) leads to the large observed structural deviations. Recent structural study involving hPygo (Figure 3C; Fiedler et al., 2008), capable of binding H3K4me3 by virtue of the aromatic cage, in complex with HD1 domain of hBCL9 of the Wnt signaling pathway reveals that the interface of the complex is distinct from that of the H3 recognition site and is located in a region similar to that of the interface of PHD in the KAP1 bromodomain (Figure S2A), underscoring the functional versatility of the basic conserved PHD finger fold. Finally, the PHD2 of AIRE lacks the histone-interacting signature residues (Figure 4) such as KAP1, as well as the characteristic Trp residue adjacent to the terminal Cys dyad, explaining its inability to interact with H3 or H4 peptides (Figure S1B).
Figure 4. Classification of the PHD Finger Family.
Sequence features of structurally characterized three distinct subclasses of the PHD finger family with respect to ligand binding specificity, i.e., H3K4me0 (group I), H3K4me3 (group II), and nonhistone binding (group III). The recognition of H3K4me3 takes place by embracement of the trimethyl groups by characteristically positioned aromatic cage residues, whereas that of K4me0 is due to an ion pair formed with a distinct N-terminal Asp residue. The characteristic histone-peptide interacting positions are in red, and the Zn-chelating residues (the first and second tetrads are connected by regular and dotted lines, respectively) and conserved C-terminal aromatic residue characteristic of the entire PHD family are in green. In either of the H3 interacting PHD fingers, H3R2 often interacts with Asp/Glu (blue). These “red” and “blue” positions are absent in KAP1-PHD finger (bottom) indicated by ↑ that binds the adjacent bromodomain’s ZA helix by patch of nonpolar residues (yellow). Topology diagrams (bottom; based on Aravind et al. [2006]), not drawn to scale, highlight these features for clarity. The domain boundaries and gi numbers are indicated in the alignments. The secondary structural elements of the AIRE-PHD1 are indicated above the sequence. The black ↑ indicates position where similar interactions involving protein side chain is observed in AIRE and BHC80 PHD fingers. The sequence of AIRE-PHD2 is shown below the alignment to show its grouping with group III as non-H3 binder.
Comparison of Histone Recognition by the Different PHD Finger Domains
In general, histone H3 peptide binding mode is conserved among the majority of PHD fingers (Figure S2). The peptide, whether unmodified or modified (H3K4me0 or H3K4me3), sits in the binding sites in a similar fashion, in which the NH2 group of the amino-terminal H3A1 is hydrogen bonded to backbone carbonyls of Pro 331 and Gly 333 of AIRE-PHD1. In addition, a conserved D/E immediately following the Zn-chelating third Cys (blue; Figure 4) often forms electrostatic interactions with H3R2, as observed in the bound peptide complexes such as BPTF, ING2, and AIRE. In BHC80 and TAF3, though no interaction between H3R2 and this acidic residue is observed in the crystal and NMR structure, respectively, the negatively charged residue is expected to favor the H3R2 positioning. In the case of RAG2, this acidic residue is absent, and the methylated H3R2 is favored at this site due to the presence of adjacent aromatic residue Tyr 445 in RAG2. Though in the case of AIRE (H3K4me0) and TAF3 (H3K4me3), peptide binding is abrogated or drastically reduced with H3R2 methylation (Koh et al., 2008; van Ingen et al., 2008; Vermeulen et al., 2007), binding of cognate peptide H3K4me3 is only marginally reduced in the case of BPTF and Pygo (Fiedler et al., 2008; van Ingen et al., 2008) and enhanced with RAG2 (Ramón-Maiques et al., 2007). These results suggest that the basic PHD finger fold is capable of providing differential regulation of gene transcription by varying surface residues or conformation to interact with different forms of histone sequences.
The side-chain hydroxyl of T3 in histone H3 in AIRE-PHD1, BPTF-PHD2, and ING2-PHD is hydrogen bonded to protein residues. In general, the H3 residues 1–3 in H3K4me0 and H3K4me3 recognition are functionally similar, and major differences arise at the H3K4 and beyond (Figure 4; Figure S2). Peptide interactions beyond H3T6 are not observed in the H3K4me3 case, whereas in the H3K4me0 case the interactions extend up to H3R8. The notable differences between H3K4me3 and H3K4me0 recognition is by two distinct molecular mechanisms. H3K4me3 is embraced in an aromatic cage formed by two or more aromatic residues that anchor the positively charged H3K4me3 by cation-π interaction, whereas for the nonmodified case, K4 side-chain amine forms electrostatic interactions with the carboxylate of the conserved aspartate residue (Figure 4; Figure S2A). The positions of these residues are characteristically at fixed distance from the Zn-anchoring Cys, His residues in the protein sequence, thus enabling prediction of histone peptide-binding capability with a fair degree of certainty. An aspartic acid located at two positions N-terminal to the first Zn-chelating Cys is an indicator of H3K4me0 binder. We have confirmed a few of human PHD fingers to be H3Kme0 binder predicted based on this feature (data not shown). However, as for H3K4me3, a tryptophan located two positions N-terminal to the Zn-chelating His and an aromatic amino acid/methionine two positions N-terminal to the third Zn-chelating Cys are a strong indicator of H3K4me3 binder (Ruthenburg et al., 2007).
The presence of strong electronegative characteristics of the AIRE-PHD1 (with a theoretical pI of ~4.9) appears to complement the positively charged histone H3 sequence, resulting in a relatively strong affinity of ~5–10 μM (Org et al., 2008). A comparison of the electrostatic potential surface of AIRE-PHD1 (Figure S2B) with that of BHC80 qualitatively agrees with the observed lower affinity, ~30 μM, of BHC80 for the unmodified H3 (Lan et al., 2007), and this trend is also observed between TAF3 and BPTF PHD fingers where a higher proportion of negatively charged surface residues of the former (Figure 2B) accounts for its 5- to 10-fold higher affinity than that of the latter for H3K4me3 recognition (van Ingen et al., 2008).
DISCUSSION
Our structure reported here demonstrates that AIRE-PHD1 recognizes H3K4me0 histone sequence through an extensive network of interactions in the binding groove. Within this groove, electrostatic interactions between H3K4 and the conserved Asp 297 in AIRE stabilize the complex. The H3R2 residue is coordinated in an adjacent small pocket connected by a narrow channel, which requires a small residue at the third position in the histone amino-terminal segment. Together, these features define the specificity of the AIRE-PHD1 finger for H3K4me0. Notably, APECED mutant sites are not directly involved in peptide binding, suggesting possible yet unknown mechanism in addition to H3 recognition in chromatin. Our NMR binding study by 2D 1H-15N-HSQC spectrum-based titration suggests that there is minimal, if any, cross-talk between H3K4 and other H3 sites of modification, indicating that AIRE’s regulatory function on gene transcription is mainly and negatively dependent on lysine 4 methylation of hisotone H3. Given that H3 residues R2–K4 are buried in protein surface grooves, AIRE-PHD1 when bound to H3 may restrict the action of methylases and kinases that target to modify amino-terminal residues in histone H3, thereby providing a fine level of epigenetic regulation of gene transcription. Taken together, our study reported here on the new structure of AIRE-PHD1/H3K4me0 complex and NMR structure-guided analysis provides a perspective to understand sequence-specific histone recognition for the entire PHD family.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The PHD finger (residues 293–346) of human AIRE (gi-4557291; AIRE-PHD1) was subcloned into pGEX4T3 vector (GE Healthcare, Piscataway, NJ) using BamHI and XhoI restriction sites. The GST fusion PHD finger was expressed in E. coli BL21(DE3) cells induced with 0.25 mM isopropyl-β-D-thiogalactopyr-anoside at 15°C overnight supplemented with 0.1 mM ZnCl2 in the medium. The fusion protein was purified on glutathione-Sepharose beads (GE Healthcare). Following reduced glutathione (20 mM) elution, GST was cleaved off by thrombin (Haematologic Technologies, Inc., Essex Junction, VT) at 4°C, leaving two amino acids (GlySer) fused to the hAIRE-PHD1. The protein was further purified to homogeneity by Superdex-200 (GE Healthcare) and confirmed by mass spectrometry. NMR spectra of the purified hAIRE-PHD1 were acquired to ensure proper protein folding. Uniformly 15N- and 15N/13C-labeled proteins were prepared from cells grown in the minimal medium containing 15NH4Cl, with or without 13C6-glucose in H2O. The AIRE-PHD1 mutant was made using a QuikChange Mutagenesis Kit (Stratagene, La Jolla, CA), and the presence of appropriate mutation was confirmed by DNA sequencing and mass spectometric analysis of the purified mutant protein.
NMR Structure Determination
NMR samples contained 0.5 mM protein in 25 mM phosphate buffer (pH 6.7) with 250 mM NaCl in H2O/2H2O (9:1) or 2H2O. All NMR spectra were acquired at 30°C on 800, 600, or 500 MHz spectrometer. The protein backbone assignments were made using (1H, 13C, and 15N) triple-resonance spectra of HNCA, HN(CO)CA, HNCACB, HN(CO)CACB, and (H)C(CO)NH-TOCSY recorded with uniformly 13C and 15N-labeled protein (Clore and Gronenborn, 1994) in complex with a unlabeled H3K4me0 peptide (residues 1–20) with a molar ratio of 1:3. The side-chain assignments were obtained using HCCH-TOCSY and HCCH-COSY spectra. The H3 peptide resonance were assigned using 13C- and 15N-filtered and J-resolved NEOSY and TOCSY spectra recorded on the 13C/15N-labeled protein with unlabeled H3 peptide, and REOSY and TOCSY spectra recorded on the unlabeled free-H3 peptide. The distance restraints were obtained from 13C- and 15N-edited 3D NOESY or 2D homonuclear NOESY spectra. Slowly exchanging amide protons, identified in the 2D 15N-HSQC spectra recorded after a H2O buffer was exchanged to a 2H2O buffer, were used in the structures calculated with NOE distance restraints to generate hydrogen bond restraints for the final structure calculations. 3JHN,Hα coupling constants measured from 3D HNHA data were used to determine backbone φ-angle restraints. The intermolecular NOEs between labeled protein and unlabeled H3 peptide were obtained by 3D 13C-F−1 edited, 13C/15N-F3 filtered NOESY spectra. The structures of AIRE-PHD1/H3 peptide (residues 1–20) complex were calculated using distance geometry simulated annealing protocol with X-PLOR (Brünger, 1992). Initial protein structure calculations were performed with manually assigned NOE-derived distance restraints. Hydrogen bond distance restraints, generated from the H/D exchange data, were added at a later stage of the structure calculations for residues with characteristic NOE patterns. The converged structures were used for the iterative automated NOE assignment by ARIA (Nilges and O’Donoghue, 1998) that integrates with X-PLOR for refinement. For the final 20 lowest-energy NMR structures, no distance or torsional angle restraint was violated by more than 0.5 Å or 5°, respectively (see Table 1). Ramachandran plot analyses of the final structures with PROCHECK-NMR (Laskowski et al., 1996) showed that 83.3%, 13.2%, 2.5%, and 1.0% of the nonglycine and nonproline residues were in the most favorable, additionally allowed, generously allowed, and disallowed regions, respectively. The lowest-energy ensemble structure was used in the structural analyses. Accessible surface area was computed using NACCESS (Hubbard and Thornton, 1993). Structural superposition and alignment was carried using MODELER (Sali and Blundell, 1993) and CE (Shindyalov and Bourne, 1998). Sequence based alignments were carried out using Muscle (Edgar, 2004) and HMMR (Eddy, 1996). Only a qualitative representation of electrostatic surface potential (±70 kT/e isocontour) is represented in Figure 1 and Figure S2B using default PyMOL (www.pymol.org) contact potential, and any careful comparison will need use of advanced methods, such as adaptive Poisson-Boltzmann solver.
Supplementary Material
Acknowledgments
We thank A. Meloni of the Consiglio Nazionale delle Ricerche (CNR; Cagliari, Italy) and M.C. Rosatelli of the Universita degli Studi di Cagliari, Italy, for kindly providing the cDNA encoding the human protein. We acknowledge the use of the NMR facility at the New York Structural Biology Center for this study. We also thank Alexander Plotnikov for technical advice on protein preparation, and Rinku Jain for helpful discussion. M.-M.Z. was in part supported by funds from the Dr. Golden and Harold Lamport Chair, and grants from the National Institutes of Health (GM73207 and CA87658).
Footnotes
ACCESSION NUMBERS
The experimental constraints and coordinates for the solution structure of the human AIRE-PHD1 in complex with H3 peptide (residues 1–20) have been deposited in the RCSB Protein Data Bank with ID code rcsb101067 and the Protein Data Bank with ID code 2kft.
Supplemental Data include two figures and can be found with this article online at http://www.cell.com/structure/supplemental/S0969-2126(09)00152-X.
References
- Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- Aravind L, Iyer L. Provenance of SET-domain histone methyl-transferases through duplication of a simple structural unit. Cell Cycle. 2003;2:369–376. [PubMed] [Google Scholar]
- Aravind L, Iyer LM, Koonin EV. Comparative genomics and structural biology of the molecular innovations of eukaryotes. Curr Opin Struct Biol. 2006;16:409–419. doi: 10.1016/j.sbi.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Finn RD, Sonnhammer EL. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. X-PLOR (Version 3.1): A System for X-ray Crystallography and NMR. New Haven, CT: Yale University Press; 1992. [Google Scholar]
- Capili AD, Schultz DC, Rauscher FJ, III, Borden KL. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 2001;20:165–177. doi: 10.1093/emboj/20.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM, Gronenborn AM. Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol. 1994;239:349–363. doi: 10.1016/s0076-6879(94)39013-4. [DOI] [PubMed] [Google Scholar]
- Derbinski J, Gabler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M, Peltonen L, Walter J, Kyewski B. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2:1032–1039. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Hidden Markov models. Curr Opin Struct Biol. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BJ, Cooke A, Peterson P, Rich T. Death in the AIRE. Trends Immunol. 2008;29:306–312. doi: 10.1016/j.it.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Fiedler M, Sanchez-Barrena MJ, Nekrasov M, Mieszczanek J, Rybin V, Muller J, Evans P, Bienz M. Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Mol Cell. 2008;30:507–518. doi: 10.1016/j.molcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, Shabanowitz J, Mishra N, Strahl BD, Allis CD, Hunt DF. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- Grishin NV. Treble clef finger–a functionally diverse zinc-binding structural motif. Nucleic Acids Res. 2001;29:1703–1714. doi: 10.1093/nar/29.8.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- Hubbard SJ, Thornton JM. NACCESS (computer program) Department of Biochemistry and Molecular Biology, University College London; 1993. [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Koh AS, Kuo AJ, Park SY, Cheung P, Abramson J, Bua D, Carney D, Shoelson SE, Gozani O, Kingston RE, et al. Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc Natl Acad Sci USA. 2008;105:15878–15883. doi: 10.1073/pnas.0808470105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Collins R, De Cegli R, Alpatov R, Horton J, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange M, Kaynak B, Forster UB, Tonjes M, Fischer JJ, Grimm C, Schlesinger J, Just S, Dunkel I, Krueger T, et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008;22:2370–2384. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan E, Wysocka J, Allis C, Patel D. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Aire regulates negative selection of organ-specific T cells. Nat Immunol. 2003;4:350–354. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- Meloni A, Incani F, Corda D, Cao A, Rosatelli MC. Role of PHD fingers and COOH-terminal 30 amino acids in AIRE transactivation activity. Mol Immunol. 2008;45:805–809. doi: 10.1016/j.molimm.2007.06.156. [DOI] [PubMed] [Google Scholar]
- Nagamine K, Peterson P, Scott H, Kudoh J, Minoshima S, Heino M, Krohn K, Lalioti M, Mullis P, Antonarakis S, et al. Positional cloning of the APECED gene. Nat Genet. 1997;17:393–398. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Umehara T, Hamana H, Hayashizaki Y, Inoue M, Kigawa T, Shirouzu M, Terada T, Tanaka A, Padmanabhan B, Yokoyama S. Crystal structure analysis of the PHD domain of the transcription co-activator Pygopus. J Mol Biol. 2007;370:80–92. doi: 10.1016/j.jmb.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Nilges M, O’Donoghue S. Ambiguous NOEs and automated NOE assignment. Prog Nucl Magn Reson Spectrosc. 1998;32:107–139. [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Org T, Chignola F, Hetenyi C, Gaetani M, Rebane A, Liiv I, Maran U, Mollica L, Bottomley MJ, Musco G, Peterson P. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 2008;9:370–376. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña PV, Davrazou F, Shi X, Walter K, Verkhusha V, Gozani O, Zhao R, Kutateladze T. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson P, Org T, Rebane A. Transcriptional regulation by AIRE: molecular mechanisms of central tolerance. Nature. 2008;8:948–957. doi: 10.1038/nri2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci USA. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey C, Winqvist O, Puhakka L, Halonen M, Moro A, Kampe O, Eskelin P, Pelto-Huikko M, Peltonen L. Aire deficient mice develop multiple features of APECED phenotype and show altered immune response. Hum Mol Genet. 2002;11:397–409. doi: 10.1093/hmg/11.4.397. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell T. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- Smith KM, Olson DC, Hirose R, Hanahan D. Pancreatic gene expression in rare cells of thymic medulla: evidence for functional contribution to T cell tolerance. Int Immunol. 1997;9:1355–1365. doi: 10.1093/intimm/9.9.1355. [DOI] [PubMed] [Google Scholar]
- van Ingen H, van Schaik FM, Wienk H, Ballering J, Rehmann H, Dechesne AC, Kruijzer JA, Liskamp RM, Timmers HT, Boelens R. Structural insight into the recognition of the H3K4me3 mark by the TFIID subunit TAF3. Structure. 2008;16:1245–1256. doi: 10.1016/j.str.2008.04.015. [DOI] [PubMed] [Google Scholar]
- VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol Cell. 2007;27:817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Zeng L, Yap KL, Ivanov AV, Wang X, Mujtaba S, Plotnikova O, Rauscher FJ, III, Zhou MM. Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat Struct Mol Biol. 2008;15:626–633. doi: 10.1038/nsmb.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.