Abstract
Ribosomal protein L10 (RPL10) proteins are ubiquitous in the plant kingdom. Arabidopsis (Arabidopsis thaliana) has three RPL10 genes encoding RPL10A to RPL10C proteins, while two genes are present in the maize (Zea mays) genome (rpl10-1 and rpl10-2). Maize and Arabidopsis RPL10s are tissue-specific and developmentally regulated, showing high levels of expression in tissues with active cell division. Coimmunoprecipitation experiments indicate that RPL10s in Arabidopsis associate with translation proteins, demonstrating that it is a component of the 80S ribosome. Previously, ultraviolet-B (UV-B) exposure was shown to increase the expression of a number of maize ribosomal protein genes, including rpl10. In this work, we demonstrate that maize rpl10 genes are induced by UV-B while Arabidopsis RPL10s are differentially regulated by this radiation: RPL10A is not UV-B regulated, RPL10B is down-regulated, while RPL10C is up-regulated by UV-B in all organs studied. Characterization of Arabidopsis T-DNA insertional mutants indicates that RPL10 genes are not functionally equivalent. rpl10A and rpl10B mutant plants show different phenotypes: knockout rpl10A mutants are lethal, rpl10A heterozygous plants are deficient in translation under UV-B conditions, and knockdown homozygous rpl10B mutants show abnormal growth. Based on the results described here, RPL10 genes are not redundant and participate in development and translation under UV-B stress.
Ribosomal protein L10 (RPL10) is a key factor in joining the 40S and 60S ribosomal subunits into a functional 80S ribosome (Eisinger et al., 1997; Loftus et al., 1997). In yeast, a mutation in Rpl10 demonstrated that it is essential for viability, as RPL10 organizes the union site to aminoacyl-tRNA; also, its incorporation into the 60S subunit is a prerequisite for the union of subunits and the initiation of translation (West et al., 2005; Hofer et al., 2007). Besides its role in translation, L10 has additional cellular roles. For example, human L10 was first identified as QM, a putative suppressor of Wilms’ tumor (Dowdy et al., 1991). QM is highly homologous to the Jun-binding protein (Jif-1), a putative tumor suppressor; it has been reported that Jif-1 forms a complex with c-Jun and represses its ability to transactivate gene expression via inhibition of binding of c-Jun to DNA (Monteclaro and Vogt, 1993). Most QM proteins are localized in the cytoplasm, and subcellular fractionation assays have shown that QM is peripherally localized to the endoplasmic reticulum (Nguyen et al., 1998). However, QM was described to be associated with presenilin 1, and it translocates from the cytoplasm to the nucleus, resulting in suppression of the complex formation of the c-Jun homodimer and acting as a transcription regulatory protein (Inada et al., 1997; Imafuku et al., 1999). In addition, it has been demonstrated that the yeast Qm homologous genes, Grc5 and Qsr1, participate in translational control of gene expression in yeast and are required for cell growth and differentiation throughout mRNA translation; deletion of Qsr1 is lethal (Tron et al., 1995; Koller et al., 1996). In animals, the expression of Qm is higher in rapidly dividing tissues than in adult and differentiated tissues, implying that QM may regulate development and differentiation (Green et al., 2000; Hwang et al., 2000). Similar expression patterns were described in the silkworm Bombyx mandarina, where Qm displays a tissue/stage-dependent expression (Hwang et al., 2000). In addition, in the plant species Nicotiana tabacum, a transcript homologous to Qm is highly expressed in young tissues and is almost undetectable in mature tissues, suggesting its contribution in cell growth beyond the role in ribosome structure (Marty et al., 1993). Furthermore, it has been recently demonstrated that QM confers protection against oxidative damage (Chen et al., 2006).
Ribosomal protein genes exist as families of multiple expressed members that could be incorporated in the cytosolic ribosome under specific situations, such as certain developmental stages, tissues, and stress conditions (Schmid et al., 2005; Byrne, 2009). In the Arabidopsis (Arabidopsis thaliana) genome, 249 genes have been identified that encode 80 families of cytosolic ribosomal proteins (Barakat et al., 2001). Identification of proteins from purified ribosomes by two-dimensional (2D) electrophoretic analyses followed by mass spectrometry (MS) demonstrated the presence of 79 families of ribosomal proteins in the 80S ribosome. Nearly half are represented by two or more protein spots on 2D gels, indicating that proteins are posttranslationally modified and/or present as different isoforms (Chang et al., 2005; Giavalisco et al., 2005; Carroll et al., 2008). It is hypothesized that this ribosomal heterogeneity fosters selective translation of specific mRNA under particular cell conditions (Barakat et al., 2001; Szick-Miranda and Bailey-Serres, 2001; Giavalisco et al., 2005; Carroll et al., 2008).
Previously, by transcriptome profiling, the expression of a number of ribosomal proteins was found to be up-regulated by UV-B light in maize (Zea mays) plants exposed under different light regimes (Casati and Walbot, 2003). In those experiments, rpl10 was found to be up-regulated by UV-B light. The reduction of the ozone layer in the stratosphere has increased the terrestrial UV-B levels, with deleterious consequences for all organisms (Ballaré et al., 2001; Searles et al., 2001; Paul and Gwynn-Jones, 2003). The increase in transcription of translation-related genes is probably the consequence of ribosomal damage by UV-B, which occurs via the formation of cross-links between RNA and specific ribosomal proteins, resulting in a 50% reduction in protein synthesis (Casati and Walbot, 2004). Cellular recovery is accompanied by selective transcription and translation of ribosomal proteins and translation factors (Casati and Walbot, 2004). To further investigate the role of ribosomal proteins in UV-B responses, in particular the participation of RPL10 under conditions of UV-B stress, and to investigate if RPL10 proteins in plants have similar roles in development as described in other species, we studied the family of maize and Arabidopsis RPL10 genes. First, tissue and developmental expression patterns were investigated in both species. Maize and Arabidopsis RPL10s are expressed with tissue and developmental stage specificity, showing high levels of expression in tissues with active cell division. Coimmunoprecipitation experiments indicate that RPL10s in Arabidopsis associate with translation proteins, demonstrating that it is a component of the 80S ribosome. In addition, Arabidopsis insertional mutants defective in particular RPL10 genes were obtained; characterization of the mutants indicates that RPL10 proteins are not functionally equivalent and participate in development and translation under UV-B stress.
RESULTS
Expression of RPL10 Transcripts in Arabidopsis and Maize
Three RPL10 genes were identified in the Arabidopsis genome as orthologs to the Homo sapiens Qm gene (Dowdy et al., 1991) and named RPL10A, RPL10B, and RPL10C, while two genes encoding RPL10 were identified in the maize genome and named rpl10-1 and rpl10-2 (version 3b.50 at maizesequence.org; Schnable et al., 2009). Arabidopsis RPL10s share between 66% and 67% (A versus B and A versus C) and 75% (B versus C) similarity at the RNA level, with a higher similarity in the coding regions (86%–88%, Supplemental Fig. S1, A and B); the predicted amino acid sequences exhibit 94% to 97% similarity (Supplemental Fig. S1C). Likewise, comparison of maize RPL10 sequences shows 95% identity at the nucleotide level for the corresponding open reading frames (ORFs) and 99% amino acid identity (Supplemental Fig. S1, A–C). In addition, the identity between RPL10 genes from Arabidopsis and maize is 52% to 62% at the transcript level (73%–74% between open reading frames) and encode proteins with 85% to 86% amino acid similarity (Supplemental Fig. S1, A–C). The plant proteins also share a high degree of primary sequence conservation with other eukaryotic orthologs, with the presence of conserved putative sites of posttranslational modifications (Supplemental Fig. S1D).
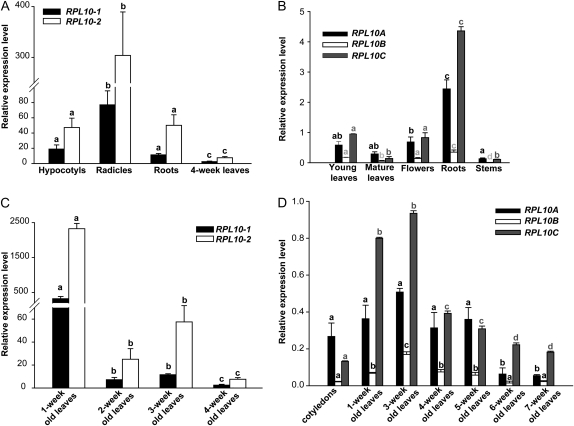
To examine the expression of RPL10, quantitative reverse transcription (qRT)-PCR was carried out using maize and Arabidopsis tissues at different developmental stages. In maize, RPL10s are expressed in all tissues analyzed, showing higher levels of expression in rapidly dividing tissues such as hypocotyls and radicles in comparison with postmitotic adult and differentiated organs such as 4-week-old leaves (Fig. 1A). In Arabidopsis, RPL10s are ubiquitously expressed in all organs tested, with the highest levels of the three transcripts measured in roots and the lowest in stems (Fig. 1B). RPL10 expression was also analyzed at different stages of leaf development; in maize, the highest level of both transcripts was in 1-week-old leaves, while lower levels of RPL10 transcripts were detected in older developmental stages (Fig. 1C). In Arabidopsis plants, the three genes show high levels of expression in tissues with active division, with transcripts showing the highest levels in 3-week-old leaves and lowest levels in senescent leaves (7 weeks old; Fig. 1D). In maize plants, rpl10-2 shows a higher level of expression (8-fold on average) than rpl10-1 in all tissues analyzed, while in Arabidopsis, RPL10A and RPL10C transcripts represent nearly 90% of the total, with the RPL10B contribution at only 10%. It is worth mentioning that the expression levels described above are consistent with AtGenExpress data from the Genevestigator microarray database (Zimmermann et al., 2004; http//www.genevestigator.ethz.ch). Although RPL10B transcript levels were consistently lower than those of RPL10A and RPL10C, this gene is highly regulated during development: its expression profile is similar to other characterized Arabidopsis ribosomal proteins, such as RPS5A and RPS5B (Weijers et al., 2001), RPS18A to RPS18C (Van Lijsebettens et al., 1994; Vanderhaeghen et al., 2006), RPL23aA and RPL23aB (Degenhardt and Bonham-Smith, 2008a, 2008b), and RPL11A and RPL11B (Williams and Sussex, 1995), where transcripts accumulate to the highest levels in mitogenic tissues and the lowest in nondividing tissues (Byrne, 2009).
Figure 1.
Tissue/stage-dependent expression of RPL10. A and B, qRT-PCR of RPL10 in different tissues of maize (A) and Arabidopsis (B) plants. C and D, Relative RPL10 transcript abundance in different stages of leaf development of maize (C) and Arabidopsis (D) plants analyzed by qRT-PCR. Each reaction was normalized using the Ct values corresponding to the ACTIN1 and POLYUBIQUITIN10 mRNAs for maize and Arabidopsis, respectively. The means of the results obtained using three independent RNAs as a template are shown; error bars indicate sd of the samples. For each RPL10 transcript analyzed, different letters indicate significant differences (P < 0.05).
Coimmunoprecipitation of RPL10 Proteins in Arabidopsis
To investigate the presence of Arabidopsis RPL10s in the ribosome, and to identify proteins that may interact with L10 in vivo, we conducted coimmunoprecipitation assays using a polyclonal antibody raised against an N-terminal peptide of human QM. Arabidopsis and human RPL10 proteins have 82% identity in this region (14 of 17 amino acids are conserved; Supplemental Fig. S1D), and the three Arabidopsis RPL10 proteins exhibit the same reactivity against this antibody as verified by western-blot analysis with the recombinant fusion RPL10 proteins (Supplemental Fig. S2A). After immunoprecipitation, proteins were separated by SDS-PAGE (Supplemental Fig. S2B), and all associated proteins were identified by tandem mass spectrometry (MS/MS) analysis. A total of 445 proteins were identified; as expected, a high percentage of these associated factors correspond to proteins involved in protein metabolism (42%), including 60S and 40S ribosomal proteins (75% of this group), and initiation and elongation factors. Thus, these results indicate that RPL10 is a constituent of the ribosome (Supplemental Fig. S2C). The identities of some proteins associated with RPL10 were verified by coimmunoprecipitation followed by western blot (Supplemental Fig. S2D). In addition, we identified other groups of proteins that are involved in photosynthesis (14%), respiration (7%), amino acid metabolism (5%), cell division, cell organization, and cell cycle (6%), transport (5%), signaling (2%), and stress response and detoxification (5%); these proteins may be contaminants or proteins that are being translated or associated with the translation machinery (Supplemental Fig. S2C). Interestingly, we also identified proteins involved in nucleic acid metabolism that regulate gene expression and proteins involved in cytoplasm-nuclear trafficking (Supplemental Table S1). Computational sorting prediction programs (sport, TargetP, iPSORT, and others) indicate that RPL10 proteins have a 50% probability of nuclear localization. Although Arabidopsis RPL10 proteins lack a nuclear localization signal, the possibility that these proteins can move to the nucleus associated with other proteins like the human QM protein cannot be ruled out.
Identification and Characterization of rpl10 Arabidopsis Insertional Mutant Lines
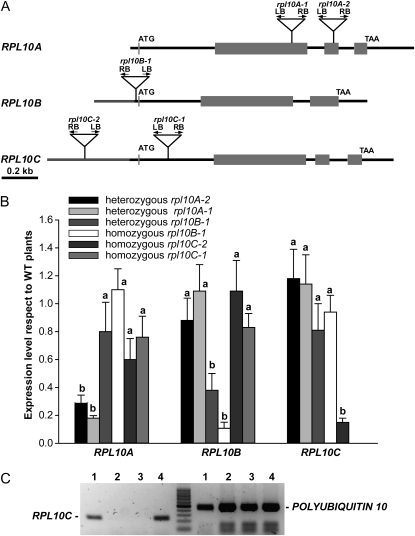
Several T-DNA mutants defective in each of the RPL10 genes were identified in the SALK and SAIL collections. For the RPL10A gene, two independent T-DNA insertional lines, SALK 010170 and SALK 106656, designated rpl10A-1 and rpl10A-2, with insertions in the second and third exons, respectively, were identified (Fig. 2A). In both lines, only heterozygous plants were viable, and they exhibited a decrease in the RPL10A expression of 5- and 3.5-fold, respectively (Fig. 2B). Both heterozygous rpl10A mutant lines showed no visible phenotype in comparison with wild-type plants under standard growth chamber conditions. No homozygous rpl10A mutant plants were found in the selfed progeny of both lines; the proportion of heterozygous to wild-type plants was 2:1, indicating that RPL10A is essential for plant viability (Supplemental Table S2).
Figure 2.
Characterization of rpl10 mutant lines. A, Structure of Arabidopsis RPL10 genes. Gray boxes indicate exons, and black and gray lines represent noncoding regions and promoter regions, respectively. The triangles show the T-DNA positions. B, RPL10 transcript abundance in T-DNA lines with respect to wild-type (WT) plants analyzed by qRT-PCR. Wild-type levels were set at 1. The means of the results obtained using three independent RNAs as a template are shown; error bars indicate sd of the samples. For each RPL10 transcript analyzed, different letters over the bars indicate statistically significant differences between wild-type and rpl10 mutant plants (P < 0.05). Each reaction was normalized using the Ct values corresponding to the POLYUBIQUITIN10 mRNA. C, RPL10C expression in rpl10C knockout line (rpl10C-1, SALK_140517) and wild-type plants analyzed by RT-PCR. Lanes 1 and 4, cDNA from leaves of wild-type plants; lanes 2 and 3, cDNA from leaves of rpl10C mutant plants. POLYUBIQUITIN10 was used as a control. The results from one of three independent experiments with similar results are shown.
For the RPL10B gene, we were able to identify heterozygous and homozygous plants with a T-DNA insertion in the 5′ untranslated region (UTR) in the only mutant available from the European Arabidopsis Stock Center seed stock (SAIL S500_01_06; Fig. 2A). Homozygous plants were found in a very low proportion compared with heterozygous plants (about 1:10). The heterozygous rpl10B plants showed a decrease of 2.6-fold in RPL10B expression compared with wild-type plants, while a decrease of 7.5-fold was observed in the homozygous mutants (Fig. 2B). The heterozygous plants were phenotypically identical to wild-type plants; however, homozygous mutants exhibited an abnormal phenotype with alterations in the vegetative and reproductive growth. The anomalous growth is according to the developmental stage-dependent expression pattern described above and suggests that RPL10B could be involved in plant development.
For RPL10C, we found two homozygous plant lines for this gene. The first line, designated rpl10C-1 (SALK_140517), with the T-DNA insertion in the first intron (Fig. 2A), showed no detectable expression of the corresponding gene as analyzed by RT-PCR (Fig. 2C). The second line analyzed has the T-DNA insertion in the promoter region (rpl10C-2 [SALK_135647]; Fig. 2A), and homozygous rpl10C mutant plants showed a 3.5-fold decrease in the transcript level compared with wild-type plants (Fig. 2B). Both homozygous rpl10C mutant lines did not exhibit any visible phenotype under standard growth chamber conditions.
For all homozygous and heterozygous rpl10 mutant plants, the expression of the other RPL10 active genes was not modified in comparison with wild-type plants, indicating that there is no compensation for the reduced or absent genes by the paralogous genes (Fig. 2B).
The RPL10B Gene Has a Role in the Development of Arabidopsis Plants
Mutations in ribosomal proteins have been reported as showing abnormalities in growth and development (Van Lijsebettens et al., 1994; Ito et al., 2000; Weijers et al., 2001; Nishimura et al., 2005; Degenhardt and Bonham-Smith, 2008a; Imai et al., 2008, Pinon et al., 2008; Yao et al., 2008; Byrne, 2009; Fujikura et al., 2009; Rosado et al. 2010). As mentioned above, homozygous rpl10B plants were significantly underrepresented. Consequently, we investigated the phenotypes of homozygous rpl10B mutant plants in more detail using the selfed progeny. Although this mutant is not a knockout line, it showed a dramatically abnormal phenotype with an overall reduced size and narrow and pointed first leaves (Fig. 3, A and B). We also observed a 77% reduction in seedling leaf size (Fig. 3, C, D, and G). In addition, mutants displayed both a reduced leaf width (58%) and a reduced leaf length (45%), resulting in a decrease of the leaf index (the ratio of leaf length to leaf width) from 1.36 ± 0.11 in the wild type to 1.03 ± 0.02 in the rpl10B mutant (P < 0.01, Student's t test). Mutants appeared to have normal vascular patterning (Fig. 3, C and D). However, root growth was affected in the rpl10B mutant. From germination, rpl10B mutant primary roots were significantly shorter than those from wild-type plants, showing an important reduction in root growth rate. For example, in 14-d-old seedlings, roots were 1.66 ± 0.31 cm long in the wild type but 0.41 ± 0.12 cm long in rpl10B (P < 0.01, Student's t test; Fig. 3H).
Figure 3.
Characterization of homozygous rpl10B mutant plants. A, Twelve-day-old plants: the wild type (left) and rpl10B mutant (right). Bar = 1 cm. B, Forty-day-old plants: the wild type (left) and rpl10B mutant (right). Bar = 5 cm. C and D, Vascular systems of wild-type (C) and rpl10B (D) first leaves. Whole-mount preparations and dark-field optics were used. Bars = 2 mm. E and F, Adaxial subepidermal palisade cells in the first leaves of wild-type (E) and rpl10B (F) plants. Bars = 20 μm. G, Palisade cell number in the adaxial subepidermal layer (left), the projected area of palisade cells (middle), and the area of the leaf blade (right) of the wild type (WT) and the rpl10B mutant. The asterisks over the bars indicate statistically significant differences between wild-type and rpl10B mutant plants (Student's t test, P < 0.05). Data show averages and sd of four seedlings and are representative of at least two independent experiments. H, Ten-day-old seedlings of rpl10B (left) and the wild type (right) grown vertically on a Murashige and Skoog agar plate. I, Open siliques from rpl10B mutant (top) and wild-type (bottom) plants. [See online article for color version of this figure.]
Before flowering, rpl10B mutant plants produced a similar number of leaves as wild-type plants, but at flowering time, rpl10B continued producing leaves with substantially increased rosette branching. Mutant plants also showed a notably shorter inflorescence shoot, clearly indicating that the RPL10B mutation has an effect on stem elongation. Furthermore, the rpl10B mutants displayed a reduction in the silique length by 48% (from 1.51 ± 0.12 cm in the wild type to 0.78 ± 0.16 in rpl10B; P < 0.01, Student's t test). Moreover, the number of seeds per silique in the rpl10B mutant was also decreased (50 ± 12 seeds in the wild-type and 11 ± 4 in rpl10B; P < 0.01, Student's t test). Taking into account that the silique length in the mutants is reduced, mutant plants produce fewer seeds per centimeter than wild-type plants (from 33 seeds in the wild type to 14 seeds in rpl10B mutants). When the seeds contained in the siliques were observed, the rpl10B mutant showed both fertilized seeds and aborted embryos; this correlates with the failure observed in seed production (Fig. 3I).
Leaves are determinate organs whose characteristic final size and shape depend on the coordination of cell proliferation and cell expansion (Piazza et al., 2005; Tsukaya, 2006). In Arabidopsis, numerous mutants have been described with altered leaf size and/or shape (Van Lijsebettens et al., 1994; Ito et al., 2000; Weijers et al., 2001; Nishimura et al., 2005; Degenhardt and Bonham-Smith, 2008a; Pinon et al., 2008; Yao et al., 2008; Fujikura et al., 2009; Rosado et al., 2010). rpl10B mutant plants displayed smaller palisade cells in the subepidermal layer (76% reduction) and also showed a moderate but significant decrease in cell number in the first leaves (32% reduction; Fig. 3, E–G). Consequently, the mutation in the RPL10B gene affects more severely postmitotic cell expansion than cell proliferation.
To validate that the abnormal phenotype in rpl10B homozygous mutant plants is due to the T-DNA insertion in the RPL10B gene, and to analyze if the rpl10B mutants could be rescued by constitutive overexpression of the wild-type RPL10B protein, we transformed the Arabidopsis rpl10B mutant plants with a plasmid expressing the Arabidopsis RPL10B wild-type ORF from the constitutive cauliflower mosaic virus 35S promoter. Kanamycin-resistant transformed plants were selected, and the presence of the transgene was examined by PCR analysis of the genomic DNA (Supplemental Fig. S3A). Expression of the transgene in the transformed seedlings was verified by qRT-PCR, and transgenic plants showed increased levels of RPL10B compared with the mutant plants (Supplemental Fig. S3B). Successful complementation of the Arabidopsis rpl10B homozygous mutant plants was demonstrated by phenotype observations of the complemented seedlings. Complemented plants showed normal development like wild-type plants, demonstrating that the wild-type phenotype was restored by overexpressing the RPL10B cDNA in the rpl10B mutant (Supplemental Fig. S3C).
Taken together, these results indicate that the RPL10B gene has an important role in plant development, both at the vegetative and reproductive stages.
Regulation of RPL10 Expression by UV-B Light
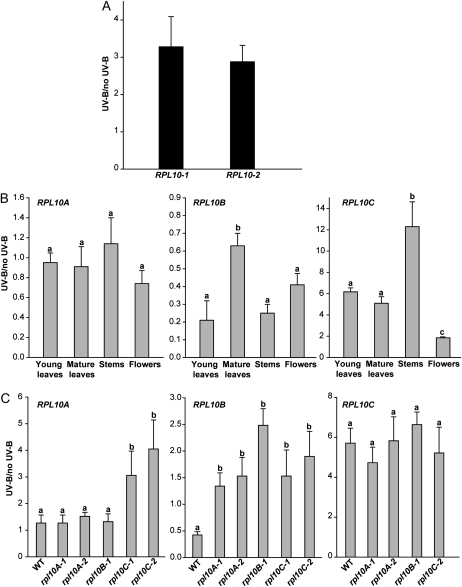
In previous experiments, by microarray analysis, we showed that a maize rpl10 gene is induced by UV-B radiation (Casati and Walbot, 2003). To validate this result, we analyzed the effect of UV-B radiation on the expression of RPL10 genes in maize and Arabidopsis plants by qRT-PCR. In maize, UV-B light increases RPL10 expression 3-fold, in agreement with our previous microarray results (Fig. 4A). In Arabidopsis, the RPL10 genes show a differential response to UV-B in all organs tested (Fig. 4B). RPL10C transcript increases after UV-B light in young and mature leaves, flowers, and stems, with the highest increase observed in stems (12-fold; Fig. 4B). RPL10A expression is not UV-B light regulated, while RPL10B shows a decrease in its expression by UV-B light in all organs.
Figure 4.
Regulation of RPL10 expression by UV-B light. A, Induction of RPL10 expression by UV-B light in leaves of maize plants analyzed by qRT-PCR. B, RPL10 expression after UV-B treatment in Arabidopsis plants analyzed by qRT-PCR. Each reaction was normalized using the Ct values corresponding to the THIOREDOXINE-LIKE and CALCIUM PROTEIN KINASE3 mRNAs for maize and Arabidopsis, respectively. The means of the results obtained using three independent RNAs as a template are shown; error bars indicate sd of the samples. C, Regulation of RPL10 expression by UV-B light in leaves of Arabidopsis insertional mutant lines. Each reaction was normalized using the Ct values corresponding to the CALCIUM PROTEIN KINASE3 mRNA. The means of the results obtained using three independent RNAs as a template are shown; error bars indicate sd of the samples. For each RPL10 transcript analyzed, different letters over the bars indicate significant differences at P < 0.05. WT, Wild type.
The regulation of RPL10 expression by UV-B light was also studied in mature leaves of Arabidopsis T-DNA insertional mutant lines. When rpl10A and rpl10B heterozygous plants were irradiated with UV-B light, RPL10A and RPL10C expression was similar to wild-type plants. On the contrary, RPL10B expression did not change by UV light exposure (rpl10A mutants); rather, a 2-fold increase was observed (rpl10B mutants), unlike in wild-type plants, where its expression was decreased (Fig. 4C). In rpl10C knockout mutants (rpl10C-1 line) exposed to UV-B treatment, the absence of RPL10C transcript affects RPL10A expression; this transcript shows an increase of 4-fold, whereas RPL10B shows an increase of 2-fold in its expression (Fig. 4C). However, it is important to emphasize that despite the increase in RPL10A expression by UV-B light, the transcript levels after the UV-B treatment are not high enough to compensate for the absence of RPL10C in the mutants. Likewise, in the rpl10C-2 line, RPL10A and RPL10B expression was similar to the rpl10C-1 line, while RPL10C transcript levels increased by UV-B light as in wild-type plants (Fig. 4C).
Participation of RPL10s in Translation under UV-B Light
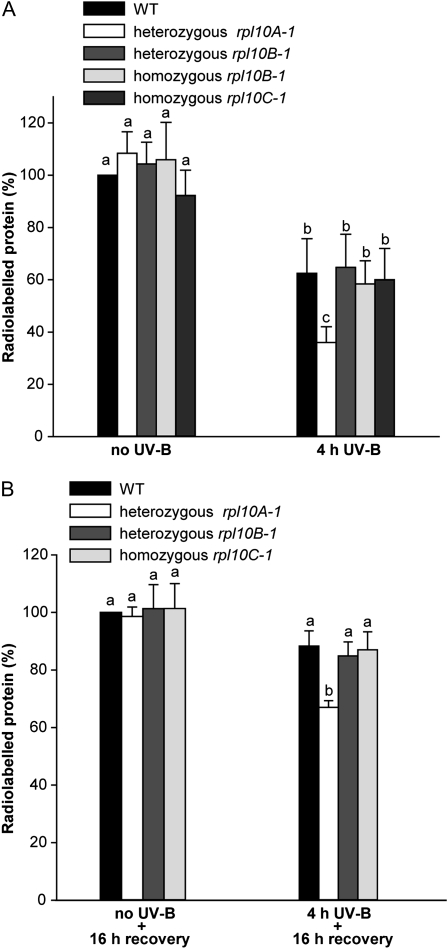
In a previous work, we found that UV-B radiation induces ribosomal damage, which occurs via formation of cross-links between RNA and specific ribosomal proteins, resulting in a 50% reduction in protein synthesis (Casati and Walbot, 2004). To study the role of RPL10 isoforms in translation, and their participation in translation under UV-B stress, we performed in vivo labeling of leaf proteins with [35S]Met in control plants (no UV-B) and after exposure under UV-B radiation for 4 h (4 h UV-B). For this experiment, we used wild-type plants to analyze the impact of UV-B radiation on translation and single rpl10A, rpl10B, and rpl10C mutants to investigate if translation under control and UV-B conditions is affected when one of the RPL10 proteins is decreased or absent. There is a significant decrease (38%) in the amount of newly synthesized protein after UV-B exposure in wild-type plants (Fig. 5A; Supplemental Fig. S4), in agreement with results obtained in experiments with maize plants exposed under UV-B radiation (Casati and Walbot, 2004). rpl10C and rpl10B mutant plants did not show significant differences in protein synthesis under control and UV-B exposure conditions compared with wild-type plants (Fig. 5A); in the heterozygous rpl10A mutant plants, while no significant difference in translation was measured under control conditions, the reduction measured in protein synthesis after the UV-B treatment was more pronounced than in the wild type and the other mutants studied (64%; Fig. 5A). After a period of 16 h of recovery in the absence of UV-B light, wild-type, rpl10C, and rpl10B plants showed restored amino acid incorporation, similar to untreated control plants (Fig. 5B). In contrast, heterozygous rpl10A mutant plants recovered protein synthesis capacity more slowly, showing only 73% of the wild-type incorporation levels 16 h after the end of the UV-B treatment (Fig. 5B; Supplemental Fig. S4). Because homozygous rpl10B mutant plants did not show any difference with respect to heterozygous rpl10B mutants in protein synthesis under control and UV-B treatment conditions, and also due to the low availability of leaf tissue from the rpl10B homozygous plants, we only used rpl10B heterozygous plants for recovery studies of protein synthesis. Similar results were obtained when precipitated counts were measured after desalting all samples, validating the data obtained by densitometric analysis.
Figure 5.
Effects of UV-B in protein synthesis in Arabidopsis wild-type (WT) plants and rpl10 mutants. In vivo labeling is shown for leaf proteins with [35S]Met in control plants (no UV-B) and after exposure under UV-B radiation for 4 h (4 h UV-B; A) and after a period of 16 h of recovery in the absence of UV-B (no UV-B + 16 h recovery) and 4 h of UV-B radiation (4 h UV-B + 16 h recovery; B). Data show averages and sd of the quantification of radioactive bands by densitometry of the autoradiographs. The percentage of labeling was corrected for loading differences per lane in SDS-PAGE. The experiments were repeated three times using four plants in each experiment. For each mutant plant and condition analyzed, different letters over the bars indicate significant differences at P < 0.05.
To rule out that the decrease in translation observed after UV-B irradiation results from cell death, physiological properties were assessed immediately after and 16 h after the 4 h of UV-B treatment. Different stress parameters (chlorophyll and flavonoid contents and maximum efficiency of PSII) were evaluated in wild-type and mutant plants under control conditions in the absence of UV-B and after the UV-B treatment. Maximum efficiency of PSII and chlorophyll a showed small but significant declines immediately after UV-B irradiation, but these parameters did not decline further and recovery was complete by the next day (Supplemental Fig. S5). No changes in the levels of chlorophyll b, flavonoids, or total proteins (Supplemental Fig. S5) were detected. Therefore, there is no evidence of UV-B-induced cell death at the wavelengths and fluence rate used in our experiments. However, we cannot rule out that differences in overall transcription and protein stability contribute to the decrease observed in newly synthesized protein after UV-B light exposure.
Proteome Analysis in the Heterozygous rpl10A Line after UV-B Irradiation
Because translation is impaired in rpl10A heterozygous plants after a 4-h UV-B treatment, we compared changes in the leaf proteome of rpl10A heterozygous plants in control conditions in the absence of UV-B and after UV-B irradiation with those in wild-type plants by 2D gel electrophoresis. The differential proteome was analyzed in two sets of comparisons: (1) proteomes from mutant plant leaves were compared with those from wild-type plants under the same radiation conditions (no UV-B and UV-B), and (2) for each plant type (wild type and mutant), we compared their proteomes under control conditions (no UV-B) and after UV-B irradiation; only proteins that were differentially UV-B regulated in the wild-type and the mutant plants were selected. From the analysis, a total of 43 protein spots showed differential levels (increased or decreased 1.5-fold or more) in at least one of the comparisons. Representative gels in Supplemental Figure S6 show the leaf proteome of rpl10A heterozygous mutant and wild-type plants after a 4-h UV-B treatment.
The 43 spots were excised from a preparative gel and subjected to protein identification by MS analysis. Interpretable MS/MS spectra were obtained for 38 of the 43 spots, and identified proteins are listed in Supplemental Table S3. For some spots, peptide sequences for more than one protein were obtained. In these cases, spot identities were assigned based on the fit of the theoretical pI and Mr of each protein to that experimentally derived from the 2D gels.
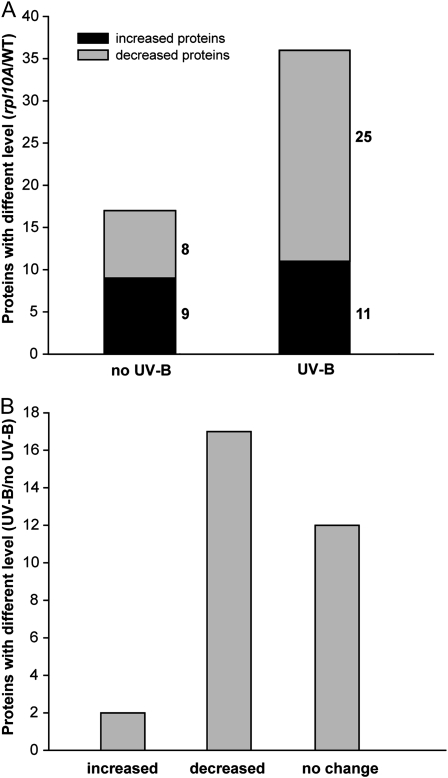
To identify coordinately regulated proteins in the rpl10A heterozygous mutants, a hierarchical clustering method was applied (Fig. 6; Supplemental Fig. S7). In control conditions in the absence of UV-B light (no UV-B), 17 protein spots were changed in rpl10A mutants compared with wild-type plants (nine increased and eight decreased); however, after a 4-h UV-B treatment, the number of proteins that changed in rpl10A mutants was increased to 36 (11 increased and 25 decreased), indicating that the rpl10A mutation more severely disrupts the protein expression pattern after a UV-B treatment than under control conditions (Fig. 6A). Similarly, when we compared the proteomes of both plants after UV-B exposure, rpl10A mutant plants exhibited a higher number of proteins that were decreased by UV-B (17 protein spots) than those increased (two protein spots; Fig. 6B). Also, some proteins that were changed by UV-B in wild-type plants did not show any difference in the mutant after the same treatment (12 protein spots; Fig. 6B). Thus, it is evident that RPL10A has an important role in translation after UV-B treatment.
Figure 6.
Proteome analysis of heterozygous rpl10A mutant plants in comparison with wild-type plants under control conditions and after a 4-h UV-B treatment by 2D gel electrophoresis. A, Protein numbers with different levels in heterozygous rpl10A mutants (at least 1.5-fold) in comparison with wild-type plants under control or UV-B conditions. B, Protein numbers with differential abundance (at least 1.5-fold) after a UV-B treatment; these proteins changed differentially in heterozygous rpl10A mutant from the levels in wild-type plants.
Finally, identified proteins were classified according to their functions (Supplemental Fig. S8). The functional groups with the largest number of proteins differentially changed in the rpl10A mutant in comparison with the wild-type plants after the UV-B treatment are involved in photosynthesis and stress response-detoxification. Other groups include proteins implicated in nucleotide metabolism, protein biosynthesis and posttranslational mechanisms, DNA metabolism (duplication and chromatin structure), RNA metabolism (transcription and regulation), and cell division and cycle. We also found proteins participating in carbohydrate, amino acid, and secondary metabolism and involved in signal transduction. Consequently, general processes for plant metabolism and growth are affected in the rpl10A mutant after a 4-h UV-B exposure, probably reflecting the deficiency in translation that this mutant shows under UV-B conditions.
DISCUSSION
RPL10 is a ubiquitous protein that participates in joining the 40S and 60S ribosomal subunits into a functional 80S ribosome; it organizes the union site to aminoacyl-tRNA; also, its incorporation into the 60S subunit is a prerequisite for the union of subunits and the initiation of translation (Eisinger et al., 1997; Loftus et al., 1997; West et al., 2005; Hofer et al., 2007). In addition, increasing evidence indicates that RPL10 protein from various organisms has multiple extraribosomal functions, besides being a constituent of the ribosome and participating in protein synthesis (Chan et al., 1996; Wool, 1996; Nika et al., 1997; Mills et al., 1999; Chávez-Ríos et al., 2003; Wen et al., 2005). Previously, by transcriptome profiling using different UV-B regimes and maize lines, we found that the functional group with the largest number of genes up-regulated by UV-B light corresponds to proteins involved in protein synthesis, including cytoplasmic ribosomal proteins, initiation and elongation factors, and poly(A)-binding proteins (Casati and Walbot, 2003). Among them, rpl10, a homolog of a human transcript encoding the QM protein, showed increased levels after the UV-B treatments (Casati and Walbot, 2003). In this work, we identified two rpl10 genes in maize, while Arabidopsis has a gene family with three members (RPL10A to RPL10C). Arabidopsis and maize RPL10s share high similarity at the transcript and protein levels (Supplemental Fig. S1). In addition, the identity and similarity between Arabidopsis and maize rpl10 genes and proteins are also high (Supplemental Fig. S1). Furthermore, the high level of similarity that maize and Arabidopsis RPL10 proteins exhibit with other eukaryotic orthologs suggests that RPL10 has been conserved during eukaryotic evolution.
The RPL10 genes studied in this work, like their homologs in other species, are regulated in a tissue- and development-dependent manner (Fig. 1), showing high levels of expression in tissues with active division, in accordance with the expression patterns of other Arabidopsis ribosomal proteins (Van Lijsebettens et al., 1994; Williams and Sussex, 1995; Weijers et al., 2001; Vanderhaeghen et al., 2006; Degenhardt and Bonham-Smith, 2008a, 2008b; Byrne, 2009). Homozygous rpl10B mutant plants exhibit an abnormal phenotype with alterations in vegetative and reproductive growth (Fig. 3). Despite the fact that, in the RPL10 family, RPL10B transcript only represents approximately 10% of the RPL10 transcript pool, we only observed an abnormal development phenotype in rpl10B mutants, which can be reverted if plants are complemented. Moreover, the Arabidopsis RPL10B gene is highly developmentally regulated. Similar results were observed for other Arabidopsis ribosomal protein mutants showing developmental defects, in which the knockout or knockdown genes represent only a small contribution to the total family transcripts (e.g. RPS18A, RPS13B [Van Lijsebettens et al., 1994; Ito et al., 2000]). Recently, it has been reported that rpl4A and rpl4D mutant plants show not only auxin-related development defects but also partial secretion of vacuole-targeted proteins. Thus, based on r-protein mutant plants that exhibit similar phenotypes, it has been proposed that aberrant phenotypes are consequences of a common regulatory mechanism for all ribosomal proteins, the auxin-regulated ribosomal biogenesis (Rosado et al., 2010). Ribosomal protein expression may be important in development because, during cell growth and proliferation, a large proportion of total cell energy is used for the synthesis of new ribosomes (Warner, 1999). Consequently, synthesis of both RNA and protein components of the ribosome is highly regulated. In vertebrates, a characteristic motif of mRNAs encoding for the 80S ribosomal proteins and some translational factors is a 5′ terminal oligopyrimidine (5′TOP) sequence that is necessary for growth-associated translational regulation (Levy et al., 1991; Avni et al., 1997; Iadevaia et al., 2008). Only the RPL10B transcript in the RPL10 family has a 5′TOP sequence (Supplemental Table S4); however, the role of the 5′TOP motif in the regulation of translation has not been demonstrated in plants. The analysis of the features in mRNA sequences responsible for translational regulation in Arabidopsis showed important determinants in the 5′ UTR, although probably other unknown mRNA sequence motifs also contribute to differential mRNA translation in plants (Kawaguchi and Bailey-Serres, 2005). Furthermore, it is well known that the amount of mRNA does not necessarily correlate with the abundance of the protein present in plant cells (Kawaguchi et al., 2004; Branco-Price et al., 2008, Mustroph et al., 2009). The translation process is primarily regulated at the initiation step involving the recruitment of the 43S preinitiation complex to the mRNA (Proud, 2007). Thus, the association of an mRNA with polysomes provides information of its translation status. It has been reported that in Arabidopsis seedlings, hypoxia causes a nearly 50% decrease in polysomes, indicating an inhibition of translational initiation that is rapidly reversed upon reoxygenation (Branco-Price et al., 2008). The comparison of total and polysomal mRNA populations showed a significant decrease in polysomal abundance without a concomitant decrease in transcript levels. Transcripts for RPL10A to RPL10C, along with other mRNAs encoding for components of the machinery for the synthesis of cytosolic proteins, show minor changes in transcript accumulation but are decreased in polysomes after hypoxia; this indicates that ribosome biogenesis is restricted under the energy crisis caused by hypoxia and also other stresses such as mild dehydration and Suc starvation (Kawaguchi et al., 2004; Kawaguchi and Bailey-Serres, 2005; Branco-Price et al., 2008). In conclusion, these observations indicate that RPL10 translation is highly regulated (Branco-Price et al., 2008). Moreover, the analysis of immunopurified mRNAs in ribosome complexes (translatome; Mustroph et al., 2009) in specific cell populations of roots and shoots shows that RPL10s are differentially expressed at cell-, region-, and organ-specific levels, with high levels of all three transcripts in leaf guard cells, the root cortex meristematic zone, the endodermis, and vasculature, while RPL10A mRNA is also specifically enriched in cotyledons, leaf epidermis, and the atrichoblast epidermis of roots. Overall, the fact that cells of different identities have distinct translatomes that are reconfigured by stress suggests that RPL10s could be involved in protein synthesis as well as other unknown processes in specific cells under different conditions.
Based on the evidence described above and our findings that RPL10B transcript expression is regulated in a tissue- and development-dependent manner, added to the abnormal phenotype observed only in rpl10B homozygous mutant plants, it is possible that the RPL10B gene could be regulated at both transcriptional and translational levels; because of this, the growth-associated translational regulation could be completely lost by the T-DNA insertion in both copies of the gene in the 5′ UTR. We conclude that the RPL10B protein has a specific and essential participation during plant growth, which cannot be replaced by the other RPL10 proteins. Moreover, the presence of the RPL10B isoform has not yet been confirmed in the Arabidopsis 80S ribosome, as no specific tryptic peptide from this protein has been identified by MS/MS in survey experiments (Chang et al., 2005; Giavalisco et al., 2005; Carroll et al., 2008). Alternatively, it cannot be ruled out that RPL10B has extraribosomal functions during development.
The absence of homozygous rpl10A mutant plants indicates that RPL10A is essential for plant viability, in agreement with Imai et al. (2008), who previously reported that this ribosomal protein is essential for female gametophyte development. In addition, a point mutation in RPL10A (named sac52-d for suppressor of acaulis) restores stem elongation in acl5 mutants (Imai et al., 2008). A possible model of action proposes that the mutation in RPL10A destabilizes the ribosome stalled on the upstream ORF of the SAC51 gene, allowing that the small ribosomal subunit efficiently reaches the start codon of the main ORF.
We also demonstrated that most of the RPL10 genes studied in this work are regulated by UV-B light. UV-B photons generate photoproducts in DNA and can also directly damage proteins, lipids, and RNA (Britt, 1996; Gerhardt et al., 1999; Casati and Walbot, 2004). Thus, plants have evolved mechanisms of protection (Stapleton and Walbot, 1994), repair (Waterworth et al., 2002; Bergo et al., 2003), and avoidance (Mazza et al., 2000; Bieza and Lois, 2001). Although UV-B photon perception and signal transduction are largely unknown (Brosche and Strid, 2003; Frohnmeyer and Staiger, 2003; Ulm and Nagy, 2005), it was recently shown that the basic domain/Leu zipper transcription factor long hypocotyl 5 (HY5), first identified as a factor for normal growth in visible light, is also an important participant in the long-wavelength (300–315 nm) UV-B light-induced signal transduction cascade in Arabidopsis (Ulm et al., 2004). Up-regulation of HY5 is mediated by UVR8, a UV-B light-specific factor with sequence similarity to the eukaryotic guanine nucleotide exchange factor RCC1 (Brown et al., 2005), in cooperation with COP1 (Oravecz et al., 2006). UVR8 and COP1 interact directly and rapidly in the nucleus in planta after UV-B light exposure. It is proposed that this very early step in UV-B light signaling coordinates responses, ensuring UV-B acclimation and protection (Favory et al., 2009). In maize plants, UV-B light exposure induced RPL10 expression, in agreement with our previous microarray results (Fig. 4A). In Arabidopsis, RPL10C shows increased levels after UV-B, while RPL10B shows a decrease in its expression in all organs studied (Fig. 4B). Regulation of the expression of some RPL10 transcripts was affected when T-DNA insertion mutants in each of the Arabidopsis RPL10 genes were irradiated with UV-B light (Fig. 4C). One possibility is that under UV-B conditions, high levels of RPL10 may be necessary for translation or for other unknown roles of this protein. If a deficiency in the expression of one isoform exists, increased levels of other isoforms may be required. However, it is important to note that a compensation effect does not occur under control conditions in the absence of UV-B light (Fig. 2B); therefore, this regulation is possibly UV-B light specific. To further investigate why RPL10 genes are differentially regulated by UV-B light and during development, we did a database survey for putative transcription factor-binding sites that could explain differences observed in the regulation of each L10 gene. Thus, we compared the RPL10 promoters (1 kb) and the 5′ UTRs; the analysis showed the presence of several common putative transcription factor-binding sites (such as a CCAAT box, a GATA box, ACE, a UV box, a GT1 motif, an I box, and MRE), whereas some putative binding sites are only found in one of the RPL10 promoters. For example, a W box is present in the RPL10A promoter, while an abscisic acid response element site is found in the RPL10B promoter (Supplemental Fig S9); the presence of these sites could explain the differential expression of RPL10 genes. However, additional studies are needed to define specific sequences involved in the regulation of the expression of RPL10 genes.
UV-B regulation of the expression of some RPL10s may be a consequence of ribosomal damage. We previously reported that UV-B radiation damages maize ribosomes by cross-linking specific ribosomal proteins to RNA; this ribosomal damage correlated with a progressive decrease in new protein production; cellular recovery was accompanied by selective transcription and translation of ribosomal proteins and translation factors (Casati and Walbot, 2004). In Arabidopsis wild-type plants, there is also a significant decrease in protein synthesis after UV-B exposure, and rpl10C and rpl10B mutant plants do not show significant differences in comparison with wild-type plants (Fig. 5). However, for the heterozygous rpl10A mutant plants, there was a significant reduction in protein synthesis only after the UV-B treatment. After a period of 16 h of recovery in the absence of UV-B, the wild type, rpl10C, and rpl10B showed restored amino acid incorporation, but heterozygous rpl10A mutants showed a much slower recovery of protein synthesis (Fig. 5). These results demonstrate that RPL10A has an important role in protein synthesis under UV-B exposure. It is striking that RPL10A is the only gene in this family that is not UV-B light regulated in Arabidopsis wild-type plants. It is possible that constitutive expression of this gene is necessary to rapidly respond to the dramatic UV-B effects on translation. Although a participation of the other two RPL10 proteins, B and C, in translation under UV-B light was not demonstrated here, it is still possible that these proteins may have a role under different UV-B light conditions or under a different developmental stage. Some evidence indicates that RPL10/QM act as coactivators/repressors of transcription in human, Entamoeba histolytica, and chicken (Monteclaro and Vogt, 1993; Stanbridge et al., 1994; Chávez-Ríos et al., 2003). QM protein has also been shown to interact with the proto-oncogene c-Yes, a Src family kinase, and it can thus participate in signal transduction pathways in different intracellular processes, including cell stability, division, proliferation, migration, and differentiation (Oh et al., 2002). Thus, RPL10B and RPL10C genes are UV-B light regulated, so if they do not participate in translation under UV-B light, it is possible that extraribosomal activities of one or both proteins may be important under UV-B conditions. A high similarity exists between the response to UV-B light and senescence at both biochemical and molecular levels (John et al., 2001). Several genes were identified that are up-regulated during leaf senescence, which are called senescence-associated genes (SAGs). In Arabidopsis, one of the SAGs is SAG24, encoding RPL10C (Ay et al., 2009). Consequently, we cannot rule out that the increase of RPL10C expression by UV-B light could be due to the similarity of both processes.
The comparison of the changes in the proteome of rpl10A heterozygous Arabidopsis mutants with that of wild-type plants in normal conditions and after a 4-h UV-B treatment showed that after the UV-B treatment, the number of proteins that changed in rpl10A mutants compared with the wild type is higher than under control conditions, suggesting that the rpl10A mutation severely affects protein synthesis after the UV-B treatment. In addition, rpl10A mutant plants exhibited a higher number of proteins decreased than those increased by UV-B, demonstrating again that RPL10A has an important role in translation after a UV-B treatment. The classification of the proteins according to their functions clearly shows that general processes for plant metabolism, growth, and development are affected in the rpl10A mutant after UV-B light exposure, probably due to the deficiency in translation that this mutant shows under UV-B conditions. Specific roles for a ribosomal protein under stress situations have previously been observed in Arabidopsis. Arabidopsis rps27 mutant plants are normal under standard conditions but show elevated sensitivity to genotoxic stress and UV irradiation by their inability for the elimination of damaged transcripts (Revenkova et al., 1999).
In conclusion, in this article we demonstrate that maize and Arabidopsis RPL10 genes are regulated during development and by UV-B radiation. RPL10 in Arabidopsis associates with proteins involved in protein biosynthesis, demonstrating that it is a component of the 80S ribosome. However, it also coimmunoprecipitates with other groups of proteins, including nuclear proteins, so at least one of the isoforms may have an extraribosomal function, such as associating with transcriptional activators or repressors, as was previously described in human and yeast. Mutants deficient in RPL10A or RPL10B have important developmental phenotypes; knockout rpl10A mutants are lethal, while knockdown rpl10B mutants show abnormal growth and plant development. Mutants in rpl10C, which is the only gene of this family that is up-regulated by UV-B light, do not have any developmental phenotype under normal growth conditions; however, they may show a particular phenotype under specific stress conditions, such as under an extreme UV-B irradiation. These were not used in the experiments described in this article. Finally, rpl10A mutants are particularly deficient in translation under UV-B light, and this is reflected by altered protein biosynthesis. Based on the results described here, RPL10 genes are not functionally equivalent, and they have important functions in development and UV-B responses.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and UV-B Treatment
Following cold treatment (72 h at 4°C in the dark), Arabidopsis (Arabidopsis thaliana) ecotype Columbia and the T-DNA insertion lines were grown in a growth chamber under light (100 μE m−2 s−1) with a 16-h-light/8-h-dark photoperiod at 22°C. The rpl10A and rpl10C mutants are from the SALK T-DNA insertion mutant collection (Alonso et al., 2003) and were obtained from the Arabidopsis Biological Resource Center. The rpl10B mutant originated in the SAIL T-DNA insertion mutant collection and was obtained from the European Arabidopsis Stock Center. The maize (Zea mays) B73 line was grown in a greenhouse with supplemental visible light (1,000 μE m−2 s−1) under a 15-h-light/9-h-dark regime without UV-B light for 28 d during the summer. Arabidopsis plants (wild-type Columbia ecotype and rpl10B mutants) were also germinated and grown in Murashige and Skoog salt-agar medium containing 3% (w/v) Suc. Arabidopsis seeds were sterilized and germinated on Murashige and Skoog medium, and growth was measured after vernalization for 3 d a 4°C.
For Arabidopsis plants, UV-B treatments were done in a growth chamber, while for maize, UV-B experiments were done in a greenhouse. Arabidopsis plants were illuminated using UV-B lamps mounted 30 cm above the plants (Bio-Rad) for 4 h, and maize plants were irradiated for 8 h using UV-B lamps mounted 30 cm above the plants (F40UVB 40 W and TL 20 W/12; Phillips) at a UV-B light intensity of 2 W m−2 and a UV-A light intensity of 0.65 W m−2 in both cases. The bulbs were covered with cellulose acetate filters (100-mm extraclear cellulose acetate plastic; Tap Plastics); the cellulose acetate sheeting does not remove any UV-B radiation from the spectrum but excludes wavelengths lower than 280 nm. Control plants without UV-B light were exposed for the same period of time under the same lamps covered with polyester filters (100-μm clear polyester plastic; Tap Plastics; 0.04 W m−2 UV-A light and 0.4 W m−2 UV-B light). This polyester filter absorbs both UV-B and wavelengths lower than 280 nm. The lamp output was recorded using a UV-B/UV-A radiometer (UV203 AB radiometer; Macam Photometrics) to ensure that both the bulbs and filters provided the designated UV light dosage in all treatments. Samples were collected immediately after irradiation and stored at −80°C. The experiments were repeated at least three times.
qRT-PCR
Tissues from three independent biological replicates were frozen in liquid nitrogen and stored at −80°C. Total RNA was isolated from 100 mg of tissue using the TRIzol reagent (Invitrogen). RNA was converted into first-strand cDNA using SuperScript II reverse transcriptase (Invitrogen) with oligo(dT) as a primer previous to treatment with DNase (Promega). The resultant cDNA was used as a template for quantitative PCR amplification in a MiniOPTICON2 apparatus (Bio-Rad) using the intercalation dye SYBR Green I (Invitrogen) as a fluorescent reporter and Platinum Taq Polymerase (Invitrogen). Primers were designed to generate unique 150- to 250-bp fragments using the PRIMER3 software (Rozen and Skaletsky, 2000). Three replicates were performed for each sample plus a negative control (reaction without reverse transcriptase). To normalize the data of UV treatments, primers for THIOREDOXIN-LIKE transcript were used for maize cDNAs and primers for CALCIUM-DEPENDENT PROTEIN KINASE3 were used for Arabidopsis cDNAs. For tissue/stage-dependent expression studies, primers for ACTIN1 and POLYUBIQUITIN10 were used for maize and Arabidopsis, respectively. All primer sequences are listed in Supplemental Table S5. Amplification conditions were as follows: 2 min of denaturation at 94°C; 40 to 45 cycles at 94°C for 15 s, 57°C for 20 s, and 72°C for 20 s; followed by 10 min at 72°C. Melting curves for each PCR were determined by measuring the decrease of fluorescence with increasing temperature (from 65°C to 98°C).To confirm the size of the PCR products and to check that they corresponded to a unique and expected product, the final products were separated on a 2% (w/v) agarose gel. The PCR products were purified from the gel and sequenced to verify their identities.
RT-PCR for RPL10C expression analyses in the knockout T-DNA line (SALK_140517) was performed under the following conditions: 1× buffer GoTaq, 3 mm MgCl2, 0.2 mm deoxyribonucleotide triphosphate, 0.25 μm of each primer, 0.625 units of GoTaq (Promega), and sterile water added to obtain a volume of 25 μL. Cycling conditions were as follows: 2 min of denaturation at 95°C; followed by 35 cycles of 15 s of denaturation at 95°C, 20 s of annealing at 57°C, and 30 s of amplification at 72°C; and finally, 7 min of amplification at 72°C. PCR products were separated on a 2% (w/v) agarose gel and stained with SYBR Green (Invitrogen).
Identification of Insertional T-DNA Mutants
The genotype of the insertion lines was determined using a PCR-based approach. Basically, genomic DNA was isolated from leaves using a modified cetyl-trimethyl-ammonium bromide method (Sambrook et al., 1989). The genotype was determined by PCR on genomic DNA using specific primers for each gene and one primer that hybridizes with the left border of the T-DNA. Three combinations of primers were used to identify homozygous, heterozygous, and wild-type plants for RPL10. Primer sequences are listed in Supplemental Table S5.
Complementation of rpl10B Mutant Plants
A full-length ORF encoding RPL10B was amplified from cDNA from leaves of Arabidopsis Columbia using the primers At RPL10B-for-3 and At RPL10B-rev-2, with KpnI and SalI restriction sites, respectively (for sequences, see Supplemental Table S5). PCR was performed with GoTaq (Promega) and Pfu Polymerase (Invitrogen; 10:1) under the following conditions: 1× GoTaq buffer, 1.5 mm MgCl2, 0.5 μm of each primer, and 0.5 mm of each deoxyribonucleotide triphosphate in a 25-μL final volume. Cycling conditions were as follows: 2 min of denaturation at 95°C; followed by 35 cycles of 30 s of denaturation at 95°C, 30 s of annealing at 58°C, and 60 s of amplification at 72°C; and finally, 7 min of amplification at 72°C. The amplified product was purified, cloned into pGEMT-Easy vector (Promega), and sequenced. The KpnI-SalI fragment was subcloned into pCHF3 binary vector, generating the construct p35S:AtRPL10B.
The p35S:AtRPL10B construct was transformed into Agrobacterium tumefaciens strain GV3101 by electroporation, and the transformation of Arabidopsis rpl10B homozygous mutant plants by the resulting bacteria was performed by the floral dip method (Clough and Bent, 1998). Transformed seedlings (T1) were identified by selection on solid Murashige and Skoog medium containing kanamycin (50 mg L−1), and then the plants were transferred to soil. The presence of the wild-type AtRPL10B in transformed plants was analyzed by PCR on the genomic DNA using the primers prom35-for and At RPL10B-rev-1 (product size of 500 bp). The expression of the wild-type AtRPL10B in transformed plants was analyzed by qRT-PCR, and each reaction was normalized using the cycle threshold (Ct) values corresponding to the UBQ10 transcript (for sequences, see Supplemental Table S5).
Immunoprecipitation Studies and MS
For immunoprecipitation analyses, Arabidopsis leaves were homogenized in a buffer containing 50 mm Tris-HCl, pH 7.5, 10% glycerol, 2 mm EDTA, 5 mm MgCl2, 150 mm NaCl, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride (PMSF), and 1× Complete protease inhibitor cocktail (Sigma). After centrifugation, 1 mL of crude extract (0.75 mg of total protein) was incubated with 15 μL (3 μg) of affinity-purified rabbit polyclonal antibody raised against an N-terminal peptide of human QM (Santa Cruz Biotechnology) for 3 h at 4°C with gentle agitation. After this, 20 μL of protein A agarose was added, and the samples were incubated at 4°C with gentle agitation for 1 h. The agarose beads were pelleted by centrifugation and washed three times with 1 mL of lysis buffer (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 150 mm NaCl, and 1% Triton X-100) for 5 min at 4°C and once with LNDET buffer (250 mm LiCl, 1% Nonidet P-40, 1% [w/v] deoxycholic acid, 1 mm EDTA, and 10 mm Tris-HCl, pH 8.0) for 5 min at 4°C. Proteins were eluted by incubation at 95°C for 5 min in 50 μL of SDS sample buffer. Samples were loaded on 10% SDS-PAGE gels. Gels were stained with Coomassie Brilliant Blue stain, and the immunoprecipitated protein-loaded lane was cut in eight strips of 0.5 mm. The strips were subjected to in-gel digestion (donatello.ucsf.edu/ingel.html) with trypsin (porcine, side chain protected; Promega). Briefly, specific excised samples were washed twice with 50% acetonitrile in 25 mm ammonium bicarbonate (NH4HCO3) and vacuum dried. The gel samples were next reduced with dithiothreitol (DTT; 10 mm in 25 mm NH4HCO3, 56°C for 1 h) and alkylated with iodoacetamide (55 mm in 25 mm NH4HCO3, room temperature for 45 min). Then, the gel pieces were vacuum dried, rehydrated in 8 μL of digestion buffer (10 ng μL−1 trypsin in 25 mm NH4HCO3), and covered with 20 μL of NH4HCO3. After overnight digestion at 37°C, peptides were extracted twice with a solution containing 50% acetonitrile and 5% formic acid. The supernatants were concentrated to 5 μL by centrifugation under vacuum. The digests were analyzed by capillary HPLC-MS/MS as described by Casati et al. (2006).
Immunoblot Analysis
For immunodetection, eluted samples were subjected to 12% SDS-PAGE, and proteins were electroblotted onto a nitrocellulose membrane for immunoblotting according to Burnette (1981). Affinity-purified rabbit polyclonal antibody raised against an N-terminal peptide of human QM was used for the detection of RPL10. Antibodies against Triticum aestivum eIF2α and eIF2β were a gift by B.A. Larkins (School of Plant Sciences, University of Arizona, Tucson, AZ). Bound antibody was visualized by linking to alkaline phosphatase- conjugated goat anti-rabbit IgG according to the manufacturer's instructions (Bio-Rad). The molecular masses of the polypeptides were estimated from a plot of the log of the molecular masses of marker standards (Bio-Rad) versus migration distance.
In Vivo Labeling and Protein Extraction
After UV-B treatments, in vivo protein labeling was done on 0.3 g of leaf segments cut into 1-mm strips perpendicular to the veins for wild-type, rpl10A heterozygous, rpl10B heterozygous, and rpl10C homozygous plants, while for rpl10B homozygous plants, 0.015 g was used. Leaf pieces were incubated with 20 μCi mL−1 [35S]Met (330 μCi g−1 fresh weight) in 0.02% Tween 40 for 1 h at 25°C. After the labeling period, the leaf pieces were rinsed extensively with water, powdered with liquid N2, and extracted with buffer containing 100 mm Tris-HCl, pH 7.5, 1 mm EDTA, 10 mm MgCl2, 15 mm 2-mercaptoethanol, 20% (v/v) glycerol, 1 mm PMSF, and Complete protease inhibitor cocktail (Sigma). Thirty micrograms of total protein determined using Bradford reagent (Bio-Rad) was loaded on a 12% (w/v) SDS-PAGE gel. Gels were stained with Coomassie Brilliant Blue stain, and radioactive proteins were visualized using a Typhoon 9200 phosphorimager screen (STORM840; GE Healthcare). Quantification was achieved by densitometry of the radioactive proteins and Coomassie Brilliant Blue-stained gels using ImageQuant software version 5.2. The amount of radiolabeled protein was normalized to equal amounts of protein on the gels after quantification; the percentage of labeling was calculated for the same amount of protein loaded on the gel. The experiment was repeated three times. Supplemental Figure S4 shows the results of one experiment; the same bands were detected in all experiments. In addition, the samples were desalted by the method of Penefsky (1977) using 3-mL columns of Sephadex G-25, and the eluates were taken for quantifying radioactive counts using a liquid scintillation counter (Wallac 1214 Rackbeta).
Protein Extraction under Denaturing Conditions, and Labeling with Alexa 610 and Alexa 532 Dyes
Approximately 0.5 g of leaves was ground in liquid nitrogen using a mortar and pestle, transferred to a tube containing 2.5 mL of extraction buffer (100 mm Tris-HCl, pH 8.8, 2% [w/v] SDS, 0.4% [v/v] 2-mercaptoethanol, 10 mm EDTA, 1 mm PMSF, and 0.9 m Suc) and 2.5 mL of phenol saturated with ice-cold Tris-HCl, pH 8.8, and then agitated at 4°C for 30 min. The aqueous phases were back extracted with extraction medium and phenol by vortexing. Tubes were centrifuged at 5,000g for 15 min at 4°C, and the phenolic phase was transferred to a new tube, leaving the interface intact. Proteins were precipitated with 5 volumes of 0.1 m cold ammonium acetate in methanol at –20°C overnight. Samples were collected by centrifugation at 20,000g at 4°C for 20 min. Next, the pellet was washed with 1.5 mL of 70% (v/v) cold ethanol. Finally, the pellet was resuspended in a buffer containing 30 mm Tris-HCl, pH 8.5, 7 m urea, 2 m thiourea, and 4% CHAPS. After pelleting the debris by centrifugation at 20,000g, proteins were labeled with succinimidyl ester derivatives of Alexa 610 or Alexa 532 after adjusting the pH to 8.5 (Invitrogen). Proteins were labeled at the ratio 75 μg of protein:60 nmol of Alexa labeling dye in dimethyl sulfoxide. After vortexing, samples were incubated for at least 2 h on ice. The reaction was quenched by 1 μL of 1 mm Lys and 20 mm DTT, then 4% (v/v) ampholyte buffers 3 to 10 were added (Bio-Rad).
2D Gel Electrophoresis
Seventy-five micrograms of an Alexa 610-labeled sample was mixed with 75 μg of Alexa 532-labeled protein. A Protean IEF Cell apparatus (Bio-Rad) was used for isoelectric focusing with precast immobilized pH gradient strips (pH 3–10; linear gradient, 17 cm [Bio-Rad]). Samples (300 μL final volume) were loaded by in-gel rehydration. The strips were subjected to isoelectric focusing for 60,000 V h−1. Focused gel strips were equilibrated in SDS equilibration buffer (50 mm Tris-Cl, pH 8.8, 30% glycerol, 2% SDS, and 6 m urea), first with buffer containing 1% (w/v) DTT for 15 min and afterward with buffer containing 4% iodoacetamide for 15 min. The strips were washed briefly with SDS-PAGE running buffer, loaded on top of a prepared SDS-PAGE Laemmli gel cast with 12.5% acrylamide, and covered with 0.5% agarose. Proteins were separated at 1 W per gel for 12 to 15 h at 15°C using a Hoefer TMSE 600 system (GE Healthcare), and the fluorescent images corresponding to Alexa 610 (excitation, 612 nm; emission peak, 628 nm) and Alexa 532 (excitation, 532 nm; emission peak, 554 nm) were acquired using an EpiChemi3 fluorescent scanner (UVP BioImaging Systems). Data were saved in TIF format. Triplicate gels of biological replicates were run; the dye label was swapped for one gel. To excise samples for MS, a preparative gel loaded with 0.6 mg of total protein was run.
Gel Image Analysis
Images were analyzed using Image Master 2D-Platinum (GE Healthcare) using the protocol described by Casati et al. (2008). Difference thresholds were applied to identify the protein spots with a statistically significant 1.5-fold difference in normalized spot volume (P < 0.05).
In-Gel Digestion, MS, and Database Searching
Before the spots were removed, the gel was stained using Coomassie Brilliant Blue stain. Gel spots of interest were manually excised from the gels and sent to CEBIQUIEM facilities (Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires) for further analyses. Spots were subjected to in-gel digestion (donatello.ucsf.edu/ingel.html) with trypsin according to Casati et al. (2006). The mass spectrometric data were obtained using a matrix-assisted laser-desorption ionization time of flight spectrometer (Ultraflex II; Bruker). The spectra obtained were submitted for National Center for Biotechnology Information database searching using MASCOT (www.matrixscience.com; Perkins et al., 1999) and analyzed as described previously (Casati et al., 2006). Protein functional classification was carried out according to literature data (Usadel et al., 2006).
Microscopic Observations
To obtain paradermal views of palisade cells, leaves were fixed with formaldehyde-acetic acid and cleared with chloral hydrate solution (200 g of chloral hydrate, 20 g of glycerol, and 50 mL of deionized water) as described (Horiguchi et al., 2005). Palisade leaf cells were observed using differential interference contrast microscopy. The density of palisade cells per unit area of this region was determined, and the area of the leaf blade was divided by this value to calculate the total number of palisade cells in the subepidermal layer. To determine the cell area, 20 palisade cells were measured in each leaf. Experiments were carried out in duplicate with four seedlings, giving similar results.
Chlorophyll and Flavonoid Extraction, and Maximum Efficiency of PSII Measurement
Total chlorophylls were determined by standard procedures (Wintermans and De Mots, 1965). Flavonoid extraction was performed as described previously (Casati and Walbot, 2003). Chlorophyll fluorescence parameters were measured using a fluorometer (Qubit Systems). The minimum chlorophyll fluorescence at an open PSII center (Fo) was determined using light (655 nm) at an intensity of 0.05 to 0.1 mE m−2 s−1. A saturation pulse of white light (2,500 mE m−2 s−1 for 0.8 s) was applied to determine the maximum chlorophyll fluorescence at closed PSII centers in the dark (Fm). Maximum efficiency of PSII (Fv/Fm) was calculated as (Fm − Fo)/Fm (Ifuku et al., 2005). The measurements were made four times in at least three different plants.
Statistical Analysis
Data presented were analyzed using one-way ANOVA. Minimum significant differences were calculated by the Bonferroni, Holm-Sidak, Dunett, and Duncan tests (α = 0.05) using the SigmaStat Package. In some cases, data were compared using Student's t test (n = 4–10 biological replicates in a single experiment; P < 0.05), and significant differences are indicated in the figures with asterisks.
Sequence data from this article can be found in the Arabidopsis Genome Initiative, the maize genome sequence (version 3b.50 at maizesequence.org), and GenBank databases under the following accession numbers: RPL10A, At1g14320; RPL10B, At1g26910; RPL10C, At1g66580; POLYUBIQUITIN10 (UBQ10), At4g05320; CALCIUM-DEPENDENT PROTEIN KINASE3 (CDPK3), At4g23650; maize RPL10-1, GRMZM2G087233; maize RPL10-2, GRMZM2G027451; maize THIOREDOXIN-LIKE, AW927774; maize ACTIN1, J01238. Protein sequences used in this work for alignments are as follows. Gallus gallus, Q08200; Mus musculus, NP_443067; Caenorhabditis elegans, Q09533; Homo sapiens, M64241; Saccharomyces cerevisiae, AAA81534; Solanum melongena, AB001891; Solanum lycopersicum, AAY97865; Pinus taeda, AAB66347; Trypanosoma brucei, AAK53755; and Entamoeba histolytica, AAL68397.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. RPL10 sequences are highly conserved between different organisms at nucleotide and amino acid levels.
Supplemental Figure S2. Coimmunoprecipitation of RPL10 proteins in Arabidopsis.
Supplemental Figure S3. Complementation of Arabidopsis homozygous rpl10B mutants with wild-type Arabidopsis RPL10B.
Supplemental Figure S4. Inhibition of protein synthesis by UV-B light in Arabidopsis wild-type and rpl10 mutant plants.
Supplemental Figure S5. UV-B treatment is not lethal to Arabidopsis plants.
Supplemental Figure S6. Examples of 2D gels of leaf proteins from heterozygous rpl10A-1 mutant and wild-type plants after a 4-h UV-B treatment.
Supplemental Figure S7. Hierarchical cluster analysis of proteins showing different levels in rpl10A mutant plants in comparison with wild-type plants under control conditions and after a 4-h UV-B treatment identified by MS.
Supplemental Figure S8. Classification of proteins showing different levels in the rpl10A mutant in comparison with wild-type plants based on their cell functions.
Supplemental Figure S9. RPL10 promoter sequences with predicted cis-elements.
Supplemental Table S1. Immunoprecipitation and identification of RPL10-associated proteins by MS/MS.
Supplemental Table S2. Segregation of RPL10A alleles.
Supplemental Table S3. Proteins showing different levels in rpl10A mutant plants in comparison with wild-type plants under control conditions and after a 4-h UV-B treatment identified by MS.
Supplemental Table S4. 5′ UTR sequences in Arabidopsis RPL10 transcripts.
Supplemental Table S5. Primer sequences used for PCR.
Supplementary Material
Acknowledgments
We thank Dr. Ramiro Rodriguez for his excellent technical assistance with analysis of the rpl10B mutant, Dr. Eduardo Rodriguez for taking the photographs, and Dr. Virginia Walbot for critical reading of the manuscript. The Arabidopsis Biological Resource Center provided SALK seed stock.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Avni D, Biderman Y, Meyuhas O. (1997) The 5′terminal oligopyrimidine tract confers translational control on TOP mRNAs in a cell type- and sequence context-dependent manner. Nucleic Acids Res 25: 995–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ay N, Irmler K, Fischer A, Uhlermann R, Reuter G, Humbeck K. (2009) Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J 58: 333–346 [DOI] [PubMed] [Google Scholar]
- Ballaré CL, Rousseau MC, Searles PS, Zaller JG, Giordano CV, Robson TM, Caldwell MM, Sala OE, Scopel AL. (2001) Impacts of solar ultraviolet-B radiation on terrestrial ecosystems of Tierra del Fuego (southern Argentina): an overview of recent progress. J Photochem Photobiol B 62: 67–77 [DOI] [PubMed] [Google Scholar]
- Barakat A, Szick-Miranda K, Chang IF, Guyot R, Blanc G, Cooke R, Delseny M, Bailey-Serres J. (2001) The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol 127: 398–415 [PMC free article] [PubMed] [Google Scholar]
- Bergo E, Segalla A, Giacometti GM, Tarantino D, Soave C, Andreucci F, Barbato R. (2003) Role of visible light in the recovery of photosystem II structure and function from ultraviolet-B stress in higher plants. J Exp Bot 54: 1665–1673 [DOI] [PubMed] [Google Scholar]
- Bieza K, Lois R. (2001) An Arabidopsis mutant tolerant to lethal ultraviolet-B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiol 126: 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJ, Larive CK, Bailey-Serres J. (2008) Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56: 743–755 [DOI] [PubMed] [Google Scholar]
- Britt AB. (1996) DNA damage and repair in plants. Annu Rev Plant Physiol Plant Mol Biol 4: 75–100 [DOI] [PubMed] [Google Scholar]
- Brosche M, Strid A. (2003) Molecular events following perception of ultraviolet-B radiation by plants. Physiol Plant 117: 1–10 [Google Scholar]
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI. (2005) A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA 102: 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette WN. (1981) Western blotting: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem 112: 195–203 [DOI] [PubMed] [Google Scholar]
- Byrne ME. (2009) A role for the ribosome in development. Trends Plant Sci 14: 512–519 [DOI] [PubMed] [Google Scholar]
- Carroll AJ, Heazlewood JL, Ito J, Millar AH. (2008) Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol Cell Proteomics 7: 347–369 [DOI] [PubMed] [Google Scholar]
- Casati P, Campi M, Chu F, Suzuki N, Maltby D, Guan S, Burlingame AL, Walbot V. (2008) Histone acetylation and chromatin remodeling are required for UV-B-dependent transcriptional activation of regulated genes in maize. Plant Cell 20: 827–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Walbot V. (2003) Gene expression profiling in response to ultraviolet radiation in Zea mays genotypes with varying flavonoid content. Plant Physiol 132: 1739–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Walbot V. (2004) Crosslinking of ribosomal proteins to RNA in maize ribosomes by UV-B and its effects on translation. Plant Physiol 136: 3319–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Zhang X, Burlingame AL, Walbot V. (2006) Analysis of leaf proteome after UV-B irradiation in maize lines differing in sensitivity. Mol Cell Proteomics 4: 1673–1685 [DOI] [PubMed] [Google Scholar]
- Chan YL, Diaz JJ, Denoroy L, Madjar JJ, Wool IG. (1996) The primary structure of rat ribosomal protein L10: relationship to a Jun-binding protein and to a putative Wilms’ tumor suppressor. Biochem Biophys Res Commun 225: 952–956 [DOI] [PubMed] [Google Scholar]
- Chang IF, Szick-Miranda K, Pan S, Bailey-Serres J. (2005) Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol 137: 848–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez-Ríos R, Arias-Romero LE, Almaraz-Barrera MJ, Vargas M. (2003) L10 ribosomal protein from Entamoeba histolytica share structural and functional homologies with QM/Jif-1: proteins with extraribosomal functions. Mol Biochem Parasitol 127: 151–160 [DOI] [PubMed] [Google Scholar]
- Chen C, Wanduragala C, Becker DF, Dickman MB. (2006) Tomato QM-like protein protects Saccharomyces cerevisiae cells against oxidative stress by regulating intracellular proline levels. Appl Environ Microbiol 72: 4001–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Degenhardt RF, Bonham-Smith PC. (2008a) Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development. Plant Physiol 147: 128–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt RF, Bonham-Smith PC. (2008b) Transcript profiling demonstrates absence of dosage compensation in Arabidopsis following loss of a single RPL23a paralog. Planta 228: 627–640 [DOI] [PubMed] [Google Scholar]
- Dowdy SF, Weissman BE, Matsui Y, Hogan BLM, Stanbridge EJ. (1991) The isolation and characterization of a novel cDNA demonstrating an altered mRNA level in nontumorigenic Wilms’ microcell hybrid cells. Nucleic Acids Res 19: 5763–5769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger DP, Dick FA, Trumpower BL. (1997) Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol Cell Biol 17: 5136–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohnmeyer H, Staiger D. (2003) Ultraviolet-B radiation mediated responses in plants: balancing damage and protection. Plant Physiol 133: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura U, Horiguchi G, Ponce MR, Micol JL, Tsukaya H. (2009) Coordination of cell proliferation and cell expansion mediated by ribosome-related processes in the leaves of Arabidopsis thaliana. Plant J 59: 499–508 [DOI] [PubMed] [Google Scholar]
- Gerhardt KE, Wilson MI, Greenberg BM. (1999) Tryptophan photolysis leads to a UVB-induced 66 kDa photoproduct of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) in vitro and in vivo. Photochem Photobiol 70: 49–56 [Google Scholar]
- Giavalisco P, Wilson D, Kreitler T, Lehrach H, Klose J, Gobom J, Fucini P. (2005) High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol Biol 57: 577–591 [DOI] [PubMed] [Google Scholar]
- Green H, Canfield AE, Hillarby MC, Grant ME, Boot-Handford RP, Freemont AJ, Wallis GA. (2000) The ribosomal protein QM is expressed differentially during vertebrate endochondral bone development. J Bone Miner Res 15: 1066–1075 [DOI] [PubMed] [Google Scholar]
- Hofer A, Bussiere C, Johnson AW. (2007) Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. J Biol Chem 282: 32630–32639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H. (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Hwang JS, Goo TW, Yun EY, Lee JH, Kang SW, Kim KY, Kwon OY. (2000) Tissue-/stage-dependent expression of a cloned Bombyx mandarina QM homologue. Biomol Eng 16: 211–215 [DOI] [PubMed] [Google Scholar]
- Iadevaia V, Caldarola S, Tino E, Amaldi F, Loreni F. (2008) All translation elongation factors and the e, f, and h subunits of translation initiation factor 3 are encoded by 5′-terminal oligopyrimidine (TOP) mRNAs. RNA 14: 1730–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifuku K, Yamamoto Y, Ono T, Ishihara S, Sato F. (2005) PsbP protein, but not PsbQ protein, is essential for the regulation and stabilization of photosystem II in higher plants. Plant Physiol 139: 1175–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imafuku I, Masaki T, Waragai M, Takeuchi S, Kawabata M, Hirai S, Ohno S, Nee LE, Lippa CF, Kanazawa I, et al. (1999) Presenilin 1 suppresses the function of c-Jun homodimers via interaction with QM/Jif-1. J Cell Biol 147: 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai A, Komura M, Kawano E, Kuwashiro Y, Takahashi T. (2008) A semi-dominant mutation in the ribosomal protein L10 gene suppresses the dwarf phenotype of the acl5 mutant in Arabidopsis thaliana. Plant J 56: 881–890 [DOI] [PubMed] [Google Scholar]
- Inada H, Mukai J, Matsushima S, Tanaka T. (1997) QM is a novel zinc-binding transcription regulatory protein: its binding to c-Jun is regulated by zinc ions and phosphorylation by protein kinase C. Biochem Biophys Res Commun 230: 331–334 [DOI] [PubMed] [Google Scholar]
- Ito T, Kim GT, Shinozaki K. (2000) Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon mediated mutagenesis causes aberrant growth and development. Plant J 22: 257–264 [DOI] [PubMed] [Google Scholar]
- John CF, Morris K, Jordan BR, Thomas B, A-H-Mackerness S. (2001) Ultraviolet-B exposure leads to up-regulation of senescence-associated genes in Arabidopsis thaliana. J Exp Bot 52: 1367–1373 [PubMed] [Google Scholar]
- Kawaguchi R, Bailey-Serres J. (2005) mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res 33: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Girke T, Bray EA, Bailey-Serres J. (2004) Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Koller HT, Klade T, Ellinger A, Breitenbach M. (1996) The yeast growth control gene GRC5 is highly homologous to the mammalian putative tumor suppressor gene QM. Yeast 12: 53–65 [DOI] [PubMed] [Google Scholar]
- Levy S, Avni D, Hariharan N, Perry RP, Mayuhas O. (1991) Oligopyrimidine tract at the 5′ end of mammalian ribosomal protein mRNAs is required for their translational control. Proc Natl Acad Sci USA 88: 3319–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus TM, Nguyen YH, Stanbridge EJ. (1997) The QM protein associates with ribosomes in the rough endoplasmic reticulum. Biochemistry 36: 8224–8230 [DOI] [PubMed] [Google Scholar]
- Marty I, Brugidou C, Chartier Y, Meyer Y. (1993) Growth-related gene expression in Nicotiana tabacum mesophyll protoplasts. Plant J 4: 265–278 [DOI] [PubMed] [Google Scholar]
- Mazza CA, Boccalandro HE, Giordano CV, Battista D, Scopel AL, Ballaré CL. (2000) Functional significance and induction by solar radiation of ultraviolet-absorbing sunscreens in field-grown soybean crops. Plant Physiol 122: 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Mills MJ, Gardiner DM, Bryant SV, Stanbridge EJ. (1999) Analysis of the pattern of QM expression during mouse development. Differentation 64: 161–171 [DOI] [PubMed] [Google Scholar]
- Monteclaro FS, Vogt PK. (1993) A Jun-binding protein related to a putative tumor suppressor. Proc Natl Acad Sci USA 90: 6726–6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith TG, Bailey-Serres J. (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen YH, Mills AA, Stanbridge EJ. (1998) Assembly of the QM protein onto the 60S ribosomal subunit occurs in the cytoplasm. J Cell Biochem 68: 281–285 [PubMed] [Google Scholar]
- Nika J, Erickson FL, Hanning EM. (1997) Ribosomal protein L9 is the product of GRC5, a homolog of the putative tumor suppressor Qm in S. cerevisiae. Yeast 13: 1155–1166 [DOI] [PubMed] [Google Scholar]
- Nishimura T, Wada T, Yamamoto KT, Okada K. (2005) The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17: 2940–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh HS, Kwon H, Sun SK, Yang CH. (2002) QM, a putative tumor suppressor, regulates proto-oncogene c-yes. J Biol Chem 277: 36489–36498 [DOI] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, Adám E, Schäfer E, Nagy F, Ulm R. (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul ND, Gwynn-Jones D. (2003) Ecological roles of solar UV radiation: towards an integrated approach. Trends Ecol Evol 18: 48–55 [Google Scholar]
- Penefsky HS. (1977) Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J Biol Chem 252: 2891–2899 [PubMed] [Google Scholar]
- Perkins DN, Pappin DJC, David M, Creasy DM, Cottrell JS. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Piazza P, Jasinski S, Tsiantis M. (2005) Evolution of leaf developmental mechanisms. New Phytol 167: 693–710 [DOI] [PubMed] [Google Scholar]
- Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, Martienssen RA, Byrne ME. (2008) Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 135: 1315–1324 [DOI] [PubMed] [Google Scholar]
- Proud CG. (2007) Signaling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem J 403: 217–234 [DOI] [PubMed] [Google Scholar]
- Revenkova K, Masson J, Koncz C, Afsar K, Jakovleva L, Paszkowski J. (1999) Involvement of Arabidopsis thaliana ribosomal protein S27 in mRNA degradation triggered by genotoxic stress. EMBO J 18: 490–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado A, Sohn EJ, Drakakaki G, Pan S, Swidergal A, Xiong Y, Kang BH, Bressan RA, Raikhela NV. (2010) Auxin-mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis. Plant Physiol 22: 143–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. (2000) Primer3 on the WWW for general users and for biologist programmers. Krawetz SA, Misener S, , Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 20: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Searles PS, Kropp BR, Flint SD, Caldwell MM. (2001) Influence of solar UV-B radiation on peatland microbial communities of southern Argentina. New Phytol 152: 213–221 [Google Scholar]
- Stanbridge E, Farmer A, Mills A, Loftus T, Kongkasuriyachai D, Dowdy S. (1994) Molecular characterization of QM, a novel gene with properties consistent with tumor suppressor function. Cold Spring Harb Symp Quant Biol 59: 573–576 [DOI] [PubMed] [Google Scholar]
- Stapleton AE, Walbot V. (1994) Flavonoids can protect maize DNA from the induction of ultraviolet-radiation damage. Plant Physiol 105: 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szick-Miranda K, Bailey-Serres J. (2001) Regulated heterogeneity in 12-kDa P-protein phosphorylation and composition of ribosomes in maize (Zea mays L.). J Biol Chem 276: 10921–10928 [DOI] [PubMed] [Google Scholar]
- Tron T, Yang M, Dick FA, Schmitt ME, Trumpower BL. (1995) QSR1, an essential yeast gene with a genetic relationship to a subunit of the mitochondrial cytochrome bc1 complex, is homologous to a gene implicated in eucaryotic cell differentiation. J Biol Chem 270: 9961–9970 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2006) Mechanism of leaf-shape determination. Annu Rev Plant Biol 57: 477–496 [DOI] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Mate Z, Adam E, Oakeley EJ, Schafer E, Nagy F. (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Nagy F. (2005) Signalling and gene regulation in response to ultraviolet light. Curr Opin Plant Biol 8: 477–482 [DOI] [PubMed] [Google Scholar]
- Usadel A, Nagel T, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhaeghen R, De Clercq R, Karimi M, Van Montagu M, Hilson P, Van Lijsebettens M. (2006) Leader sequence of a plant ribosomal protein gene with complementarity to the 18S rRNA triggers in vitro cap-independent translation. FEBS Lett 580: 2630–2636 [DOI] [PubMed] [Google Scholar]
- Van Lijsebettens M, Vanderhaeghen R, De Block M, Bauw G, Villarroel R, Van Montagu M. (1994) An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J 13: 3378–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Waterworth WM, Jiang Q, West CE, Nikaido M, Bray CM. (2002) Characterization of Arabidopsis photolyase enzymes and analysis of their role in protection from ultraviolet-B radiation. J Exp Bot 53: 1005–1015 [DOI] [PubMed] [Google Scholar]
- Weijers D, Franke-van Dijk M, Vencken RJ, Quint A, Hooykaas P, Offringa R. (2001) An Arabidopsis Minute-like phenotype caused by a semidominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128: 4289–4299 [DOI] [PubMed] [Google Scholar]
- Wen Y, Shao JZ, Pan XX, Xiang LX. (2005) Molecular cloning, characterization and expression analysis of QM gene from grass carp (Ctenopharyngodon idellus) homologous to Wilms’ tumor suppressor. Comp Biochem Physiol 141: 356–365 [DOI] [PubMed] [Google Scholar]
- West M, Hedges JB, Chen A, Johnson AW. (2005) Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol Cell Biol 25: 3802–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ME, Sussex IM. (1995) Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana. Plant J 8: 65–76 [DOI] [PubMed] [Google Scholar]
- Wintermans JFGH, De Mots A. (1965) Spectrophotometric characteristics of the chlorophylls and their pheophytins in ethanol. Biochim Biophys Acta 109: 448–455 [DOI] [PubMed] [Google Scholar]
- Wool IG. (1996) Extraribosomal functions of ribosomal proteins. Trends Biochem Sci 21: 164–165 [PubMed] [Google Scholar]
- Yao Y, Ling Q, Wang H, Huang H. (2008) Ribosomal proteins promote leaf adaxial identity. Development 135: 1325–1334 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.