DBP factors (DNA-binding protein phosphatases) are unique to plants but widely distributed in the plant kingdom. The dual structure of DBP factors suggests that, in addition to directly participating in transcriptional regulation of specific genes by virtue of its DNA-binding capacity, they may also be involved in the regulation of other processes not directly related to gene transcription, particularly in signal transduction pathways. In support of this hypothesis a shuttling mechanism from the nucleus to the cytosol has recently been demonstrated for the dynamic localization of the tobacco (Nicotiana tabacum) NtDBP1 protein (Carrasco et al., 2006). In the model species Arabidopsis (Arabidopsis thaliana) four DBP factors have been identified, with AtDBP1 being the closest structural relative to tobacco NtDBP1, the first member of the family to be isolated (Carrasco et al., 2005). AtDBP1 was found to bind DNA with similar specificity to NtDBP1 and exhibit in vitro Mg2+-dependent protein phosphatase activity as well.
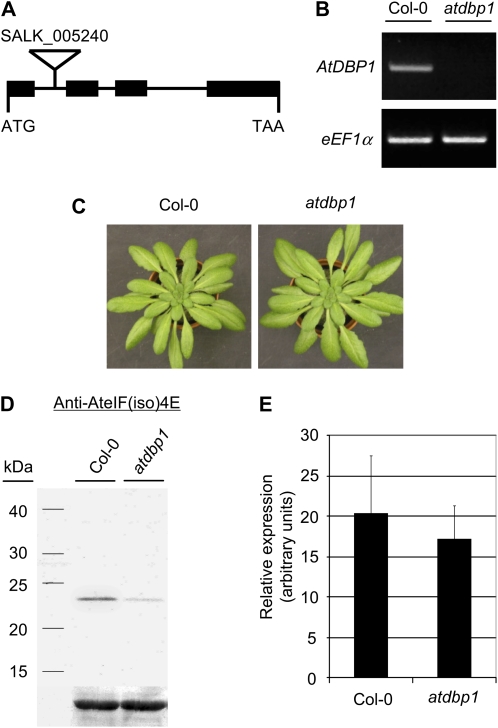
Changes in plant physiology and metabolism that occur as part of developmental programs or in response to environmental stimuli result from specifically modulating gene function. Identifying the factors responsible for this regulation and their mode of action provides a key insight into any biological process under study and is necessary to implement biotechnological strategies directed to the improvement of crop quality and performance. With the aim of investigating the function of DBP factors, a reverse-genetic approach was undertaken using Arabidopsis plants homozygous for a T-DNA insertion in the first intron of the AtDBP1 gene (SALK_005240; Alonso et al., 2003; Fig. 1A). When gene expression was analyzed in the mutant line by reverse transcription (RT)-PCR, AtDBP1 mRNA was found to accumulate at negligible levels as compared to Columbia-0 (Col-0) wild-type plants (Fig. 1B). This remarkable reduction in gene expression does not lead to any major observable effect in plant architecture or growth habit in the mutant plant (Fig. 1C). Loss of AtDBP1 function would be expected to alter gene expression and, eventually, protein accumulation of its targets. Therefore, we analyzed the proteome of the mutant in comparison to that of Col-0 plants in search of proteins showing differential accumulation due to the absence of AtDBP1. Looking at the protein level should enable us to identify both transcriptional and posttranscriptional candidate targets of AtDBP1 function. Among the differential spots detected by two-dimensional gel electrophoresis showing a significant variation (at least 3-fold) between the two genotypes, the translation initiation factor eIF(iso)4E was identified by mass spectrometry and found to accumulate at a lower level in the atdbp1 mutant when compared to Col-0 plants. This observation was verified by western blot using a specific polyclonal antiserum raised against Arabidopsis eIF(iso)4E (Fig. 1D). This antiserum was previously shown to specifically recognize eIF(iso)4E (Duprat et al., 2002). The reduction in eIF(iso)4E abundance caused by loss of AtDBP1 function seems to obey to a posttranscriptional regulatory mechanism, since no significant difference in eIF(iso)4E gene expression between Col-0 and atdbp1 plants was observed at the mRNA level when analyzed by quantitative RT-PCR (Fig. 1E).
Figure 1.
Loss of AtDBP1 function does not compromise plant growth and architecture. A, AtDBP1 gene structure showing localization of the T-DNA insertion in the SALK_005240 (atdbp1) mutant line. B, RT-PCR analysis of AtDBP1 expression in Col-0 and homozygous atdbp1 plants. Amplification of the housekeeping gene eEF1a is shown below as control of RNA loading. C, Comparison of plant morphology and architecture between Col-0 and atdbp1 mutant plants. D, Western blot of leaf extracts using a polyclonal antiserum raised against eIF(iso)4E. Size of molecular markers is indicated on the left. E, RT-PCR analysis of eIF(iso)4E and eEF1a expression in Col-0 and atdbp1 plants. Numbers indicate relative signal intensity of the amplified products referred to Col-0. [See online article for color version of this figure.]
eIF(iso)4E is a plant-specific isoform of eIF4E, and is encoded in Arabidopsis by a single-copy gene. eIF4E isoforms bind the cap structure present at the 5′ end of eukaryotic mRNAs and promote recruitment of additional factors and mRNA circularization, thereby enabling initiation of translation (Browning, 2004). Interestingly, eIF4E and eIF(iso)4E have been selectively implicated as key factors in recessive resistance against potyviruses in many plant species (Robaglia and Caranta, 2006). Mutations that abolish expression of eIF(iso)4E have been reported to confer resistance to Plum pox virus (PPV), Turnip mosaic virus (TuMV), and Lettuce mosaic virus, whereas eIF4E disruption leads to resistance to Clover yellow vein virus (Duprat et al., 2002; Lellis et al., 2002; Sato et al., 2005; Decroocq et al., 2006). The infections caused by potyviruses are responsible for numerous plant diseases that cause important economic losses in production and quality of vegetable and ornamental crops worldwide. The potyviral genome consists of a single-stranded polyA-tailed positive RNA that at the 5′ end lacks a cap structure and instead is covalently bound to a virus-encoded protein termed VPg. Both eIF4E and eIF(iso)4E have been shown to physically interact with the viral protein VPg. The ability of these proteins to interact correlates with virus infectivity (Léonard et al., 2000; Kang et al., 2005), and there is evidence for coevolution between eIF4E and potyviral VPg (Charron et al., 2008).
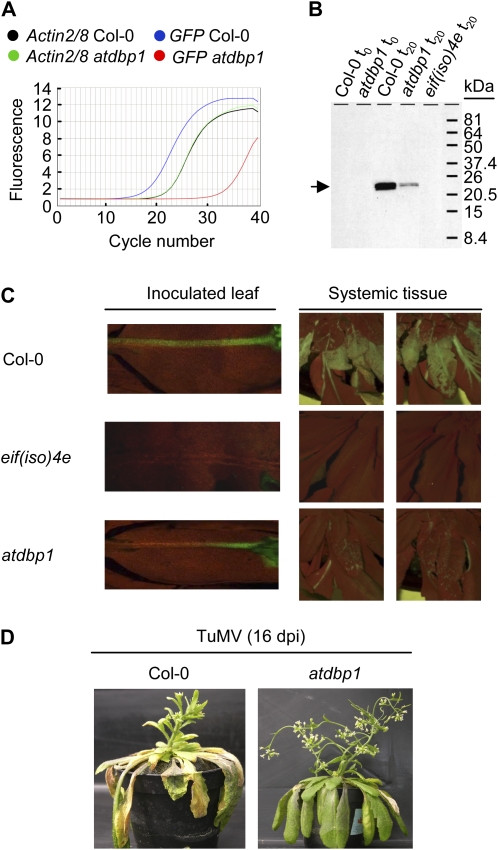
The lower accumulation of eIF(iso)4E in atdbp1 plants prompted us to analyze the response of this mutant to infection by potyviruses. For that, Col-0 and atdbp1 plants were inoculated with a GFP-tagged version of PPV, and the progress of the infection was monitored at different time points both in the inoculated and in noninoculated systemic leaves. Viral accumulation was first analyzed at the RNA level using quantitative RT-PCR (Fig. 2A). Amplification of a GFP-derived product from total RNA isolated from systemic noninoculated leaves 20 d postinoculation (dpi) showed a remarkable and consistent delay in atdbp1 as compared to Col-0 plants, reflecting a roughly 40-fold lower accumulation of viral RNA in the mutant. This observation was further confirmed by western blot by analyzing GFP protein accumulation in systemic leaves of both genotypes. As shown in Figure 2B, accumulation of GFP protein was lower in systemic leaves of the atdbp1 mutant 20 dpi. Moreover, production of GFP during the viral cycle enabled us to visually monitor the infection in situ by fluorescence microscopy. Inspection of GFP distribution at different time points after inoculation revealed that viral spread was delayed in the atdbp1 mutant (Fig. 2C). In Col-0 plants, the virus accumulated in the vascular tissue of the inoculated leaf and moved through the petiole faster than in atdbp1 plants. Furthermore, noninoculated leaves that were already severely infected in Col-0 plants showed only incipient spread of the virus in atdbp1 plants at every time point analyzed after inoculation. As expected, resistance was even more evident in an eif(iso)4e knockout mutant with no detectable accumulation of the virus either in inoculated or in systemic tissue as previously described (Duprat et al., 2002; Lellis et al., 2002). These results suggest that AtDBP1 function is required for successful progression of PPV infection, since although the virus is able to replicate and move, thereby completing its infective cycle, this appears to be impaired. Therefore, we have identified AtDBP1 as a novel host factor contributing to susceptibility to the potyvirus PPV in Arabidopsis. It would be very interesting to find out whether impairment of infection is only due to the reduced level of eIF(iso)4E or whether there is an additional implication of AtDBP1 during infection. Work to answer this question is in progress.
Figure 2.
atdbp1 mutant shows enhanced resistance to infection by two potyviruses. A, Analysis of viral RNA accumulation in Col-0 and atdbp1 plants after inoculation with GFP-tagged PPV. Total RNA from infected noninoculated leaves was analyzed 20 dpi by quantitative RT-PCR using primers specific for the GFP gene. ACTIN2/8 expression was used as a control. Three independent experiments were performed with similar results. B, Western blot of leaf protein extracts before inoculation (t0) and from noninoculated leaves 20 dpi (t20), immunodecorated with a polyclonal antiserum against GFP. Migration of molecular mass markers is indicated on the right. C, Analysis of viral spread in inoculated (12 dpi, left sections) and systemic leaves (20 dpi, right sections) of Col-0 (top) and eif(iso)4e (middle) and atdbp1 (bottom) mutant plants by fluorescence microscopy for GFP detection. D, atdbp1 mutant shows enhanced resistance to TuMV. Symptom development 16 d after TuMV infection in Col-0 versus atdbp1 plants. [See online article for color version of this figure.]
To further characterize the enhanced resistance displayed by the atdbp1 mutant we also analyzed the performance of atdbp1 plants against TuMV infection, which causes more severe symptoms in Col-0 than PPV. Although we still found some variability in symptom development among different plants within the same genotype, both vegetative and inflorescence tissues in atdbp1 plants were again less affected by TuMV infection than in Col-0 (Fig. 2D). These results reinforce the biological relevance of the role of AtDBP1 during infection by potyviruses. Then the question arises as to whether this resistant phenotype was specific to potyviruses. For that reason, we challenged atdbp1 mutant plants with Cucumber mosaic virus (CMV). CMV belongs to a different viral family, Bromoviridae, genus Cucumovirus, and has a segmented genome consisting of three single-stranded positive RNAs, which bear a cap structure at the 5′ end but lack a 3′ poly(A) tail. As shown in Supplemental Figure S1, atdbp1 and Col-0 plants became similarly affected after CMV inoculation. Infection was further analyzed at the molecular level by quantitative RT-PCR and western blot. Both CMV RNAs 2 and 3 and viral protein accumulated at comparable levels in Col-0 and atdbp1 plants, indicating that CMV infection was not hindered by AtDBP1 loss of function. CMV was previously shown to infect eif(iso)4e plants as efficiently as Col-0 (Duprat et al., 2002). Thus, a virus that does not require eIF(iso)4E for infection, also eludes the resistance mechanism exhibited by the atdbp1 mutant. These results further suggest that indeed the reduced accumulation of eIF(iso)4E underlies the enhanced resistance rendered by loss of AtDBP1 function.
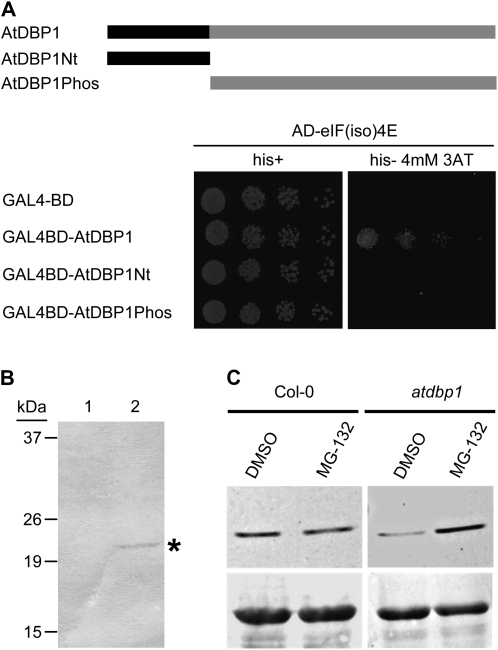
Since compromising AtDBP1 function does not seem to have any significant effect on eIF(iso)4E transcript synthesis and/or stability (Fig. 1E), the reduction in eIF(iso)4E protein accumulation suggests a posttranslational mechanism that might probably involve a direct interaction between AtDBP1 and eIF(iso)4E. Using the yeast (Saccharomyces cerevisiae) two-hybrid system, we demonstrated a specific physical interaction between AtDBP1 and eIF(iso)4E (Fig. 3A) that was confirmed in planta by coimmunoprecipitation (Fig. 3B). Protein extracts derived from Col-0 plants and T1 individuals expressing a translational fusion of AtDBP1 to the hemaglutinin epitope (HA) were subjected to immunoprecipitation with anti-HA antibodies, and the presence of eIF(iso)4E was analyzed in the immunoprecipitate by western blot. As shown in Figure 3A, the interaction required structural integrity of AtDBP1, since truncated forms encompassing the two major domains of this protein failed to interact with eIF(iso)4E. Thus, neither the N-terminal domain of AtDBP1 (AtDBP1Nt), which characteristically supports DNA binding of DBP factors, nor the C-terminal protein phosphatase domain (AtDBP1Phos), was sufficient to mediate interaction with eIF(iso)4E.
Figure 3.
AtDBP1 interacts with and stabilizes eIF(iso)4E. A, Two-hybrid assay. Yeast growth in medium containing (left) and lacking (right) His (his). On top, a scheme of the structure of AtDBP1 and the different modifications of the protein used in the assay is shown. Black and gray boxes represent the N-terminal domain, containing the DNA-binding motif, and the C-terminal protein phosphatase domain, respectively. BD, GAL4-binding domain; AD, GAL4-activation domain; AtDBP1Nt, AtDBP1 N-terminal domain; AtDBP1Phos, AtDBP1 protein phosphatase domain; 3AT, 3-aminotriazol. B, Coimmunoprecipitation of AtDBP1 and eIF(iso)4E. Protein extracts from Col-0 plants (1) and plants expressing AtDBP1 fused to the HA (2) were immunoprecipitated with a polyclonal antiserum against HA and the immunoprecipitated fractions were analyzed by western blot with an anti-eIF(iso)4E antiserum. Asterisk denotes position of the eIF(iso)4E protein. C, AtDBP1 reduces proteasome-mediated degradation of eIF(iso)4E. Western blot using a polyclonal antiserum against Arabidopsis eIF(iso)4E of leaf protein extracts after treatment with 100 μm MG-132. As a control the same amount of the MG-132 solvent (dimethyl sulfoxide) was added. Ponceau-S staining is shown below as loading control.
Since diminished AtDBP1 function resulted in reduced eIF(iso)4E protein accumulation, the interaction with AtDBP1 could stabilize eIF(iso)4E and prevent its degradation. To test this, we analyzed the effect of the proteasome inhibitor MG-132 on eIF(iso)4E protein abundance in Col-0 and in the atdbp1 mutant. Leaves were cut from plants of both genotypes and incubated in the presence of MG-132 as described in Supplemental Materials and Methods S1. Western-blot analysis of protein crude extracts confirmed that proteasome inhibition led to a significant increase in the amount of eIF(iso)4E protein in the atdbp1 mutant background (Fig. 3C). Therefore, when AtDBP1 function is lacking, eIF(iso)4E is more actively degraded via proteasome, suggesting a stabilizing role for the interaction with AtDBP1.
In this work we demonstrate that AtDBP1, a protein-phosphatase 2C of the recently described DNA-binding DBP family, directly interacts with eIF(iso)4E, and that loss of AtDBP1 function results in an increased rate of proteasome-mediated degradation and hence reduced accumulation of eIF(iso)4E. This is a very interesting and suggestive finding, since we have proved a direct relationship between eIF(iso)4E, which plays a determinant role during infection by potyviruses, and AtDBP1. Since viruses strictly depend on the biochemical machinery of the host to accomplish successful infection, absence or misfunction of key host factors may prevent the virus from multiplying and/or systemically moving inside the host (Díaz-Pendón et al., 2004), leading to recessive resistance. Although many host factors must be required by the virus during infection, the survey of both natural and induced resistance alleles in different plant species has repeatedly led to the identification of translation initiation factor eIF4E isoforms as major genetic determinants of resistance to potyviruses (Duprat et al., 2002; Lellis et al., 2002; Nicaise et al., 2003; Gao et al., 2004; Yoshii et al., 2004; Ruffel et al., 2005; Robaglia and Caranta, 2006; Maule et al., 2007). In addition, resistance alleles obtained so far by mutagenesis in the model species Arabidopsis turned out to encode eIF4E or its plant-specific isoform eIF(iso)4E (Duprat et al., 2002; Lellis et al., 2002; Yoshii et al., 2004). Only recently, additional plant factors are being discovered that support infection by potyviruses. Dunoyer et al. (2004) reported on the identification of a Cys-rich protein of unknown function that interacts with VPg and acts as a host ancillary factor in movement of potyviruses. Similarly, a DEAD-box RNA helicase has been found to also interact with potyviral VPg to play a critical albeit uncertain role during infection (Huang et al., 2010). Our results have important implications in terms of eIF(iso)4E function and potyviral infection since we have identified a new host factor playing a role during infection by two potyviruses, namely PPV and TuMV, and, to our knowledge, provide the first evidence for a proteasome-mediated regulation of eIF(iso)4E. Thus, through the unveiling of a biological context for AtDBP1 function, our work sheds light on the molecular interplay and regulation that underlies the plant-potyvirus interaction by involving DBP factors in this complex scenario.
How the observed AtDBP1-mediated eIF(iso)4E regulation is accomplished, and what other molecular components and signals participate in this complex interaction remains to be elucidated. Moreover, down-regulating DBP1 homologs from major crop species potentially represents a novel and secure strategy for engineering plants with a durable resistance to potyviruses. Research on all these aspects becomes our priority for the future.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Loss of AtDBP1 function does not impair CMV infection.
Supplemental Materials and Methods S1. Description of materials and procedures used in this work.
Supplementary Material
Acknowledgments
We thank Sami Irar and Montserrat Pagès (Instituto de Biología Molecular de Barcelona) for providing the resources and the expertise for the proteome analysis, Karen Browning (University of Texas, Austin, TX) for antisera, Christophe Robaglia (Université de la Méditerranée, Marseille, France) and Karen Browning for the eif(iso)4e mutant, Juan Antonio García (Centro Nacional de Biotecnología, Madrid) for PPV strains, Fernando Ponz (Instituto Nacional de Investigación y Tecnología Agraria, Madrid) and Andrew Maule (John Innes Center, Norwich, UK) for TuMV infectious clones, Miguel A. Aranda (Centro de Edafología y Biología Aplicada del Segura, Murcia, Spain) and Fernando Martínez and José Antonio Darós (Instituto de Biología Molecular y Celular de Plantas, Valencia, Spain) for CMV strain and assistance with CMV inoculation, Wei Shen and Linda Hanley-Bowdoin (North Carolina State University, Raleigh, NC) for their help in the proteasome inhibition assays, the Salk Institute Genomic Analysis Laboratory and the European Arabidopsis Stock Center (University of Nottingham, UK) for providing the sequence-indexed Arabidopsis T-DNA insertion mutants, and Ana López (Instituto de Biología Molecular y Celular de Plantas, Valencia, Spain) for critical reading of the manuscript.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Browning KS. (2004) Plant translation initiation factors: it is not easy to be green. Biochem Soc Trans 32: 589–591 [DOI] [PubMed] [Google Scholar]
- Carrasco JL, Ancillo G, Castelló MJ, Vera P. (2005) A novel DNA-binding motif, hallmark of a new family of plant transcription factors. Plant Physiol 137: 602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco JL, Ancillo G, Mayda E, Vera P. (2003) A novel transcription factor involved in plant defense endowed with protein phosphatase activity. EMBO J 22: 3376–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco JL, Castelló MJ, Vera P. (2006) 14-3-3 mediates transcriptional regulation by modulating nucleocytoplasmic shuttling of tobacco DNA-binding protein phosphatase-1. J Biol Chem 281: 22875–22881 [DOI] [PubMed] [Google Scholar]
- Charron C, Nicolai M, Gallois JL, Robaglia C, Moury B, Palloix A, Caranta C. (2008) Natural variation and functional analyses provide evidence for coevolution between plant eIF4E and potyviral VPg. Plant J 54: 56–68 [DOI] [PubMed] [Google Scholar]
- Decroocq V, Sicard O, Alamillo JM, Lansac M, Eyquard JP, García JA, Candresse T, Le Gall O, Revers F. (2006) Multiple resistance traits control Plum pox virus infection in Arabidopsis thaliana. Mol Plant Microbe Interact 19: 541–549 [DOI] [PubMed] [Google Scholar]
- Díaz-Pendón JA, Truniger V, Nieto C, García-Mas J, Bendahmane A, Aranda MA. (2004) Advances in understanding recessive resistance to plant viruses. Mol Plant Pathol 5: 223–233 [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Thomas C, Harrison S, Revers F, Maule A. (2004) A cysteine-rich plant protein potentiates potyvirus movement through an interaction with the virus genome-linked protein VPg. J Virol 78: 2301–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprat A, Caranta C, Revers F, Menand B, Browning KS, Robaglia C. (2002) The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J 32: 927–934 [DOI] [PubMed] [Google Scholar]
- Gao Z, Johansen E, Eyers S, Thomas CL, Ellis THN, Maule AJ. (2004) The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J 40: 376–385 [DOI] [PubMed] [Google Scholar]
- Huang TS, Wei T, Laliberté JF, Wang A. (2010) A host RNA helicase-like protein, AtRH8, interacts with the potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol 152: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BC, Yeam I, Franz JD, Murphy JF, Jahn MM. (2005) The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J 42: 392–405 [DOI] [PubMed] [Google Scholar]
- Lellis AD, Kasschau KD, Whitham SA, Carrington JC. (2002) Loss-of susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr Biol 12: 1046–1051 [DOI] [PubMed] [Google Scholar]
- Léonard S, Plante D, Wittmann S, Daigneault N, Fortin MG, Laliberte JF. (2000) Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J Virol 74: 7730–7737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule AJ, Caranta C, Boulton MI. (2007) Sources of natural resistance to plant viruses: status and prospects. Mol Plant Pathol 8: 223–231 [DOI] [PubMed] [Google Scholar]
- Nicaise V, German-Retana S, Sanjuan R, Dubrana MP, Mazier M, Maisonneuve B, Candresse T, Caranta C, LeGall O. (2003) The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus. Plant Physiol 132: 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia C, Caranta C. (2006) Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci 11: 40–45 [DOI] [PubMed] [Google Scholar]
- Ruffel S, Gallois JL, Lesage ML, Caranta C. (2005) The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol Genet Genomics 274: 346–353 [DOI] [PubMed] [Google Scholar]
- Sato M, Nakahara K, Yoshii M, Ishikawa M, Uyeda I. (2005) Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. FEBS Lett 579: 1167–1171 [DOI] [PubMed] [Google Scholar]
- Shen W, Hanley-Bowdoin L. (2006) Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases. Plant Physiol 142: 1642–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii M, Nishikiori M, Tomita K, Yoshioka N, Kozuka R, Naito S, Ishikawa M. (2004) The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J Virol 78: 6102–6111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.