Abstract
In winter wheat (Triticum spp.) and barley (Hordeum vulgare) varieties, long exposures to nonfreezing cold temperatures accelerate flowering time (vernalization) and improve freezing tolerance (cold acclimation). However, when plants initiate their reproductive development, freezing tolerance decreases, suggesting a connection between the two processes. To better understand this connection, we used two diploid wheat (Triticum monococcum) mutants, maintained vegetative phase (mvp), that carry deletions encompassing VRN-1, the major vernalization gene in temperate cereals. Homozygous mvp/mvp plants never flower, whereas plants carrying at least one functional VRN-1 copy (Mvp/−) exhibit normal flowering and high transcript levels of VRN-1 under long days. The Mvp/− plants showed reduced freezing tolerance and reduced transcript levels of several cold-induced C-REPEAT BINDING FACTOR transcription factors and COLD REGULATED genes (COR) relative to the mvp/mvp plants. Diploid wheat accessions with mutations in the VRN-1 promoter, resulting in high transcript levels under both long and short days, showed a significant down-regulation of COR14b under long days but not under short days. Taken together, these studies suggest that VRN-1 is required for the initiation of the regulatory cascade that down-regulates the cold acclimation pathway but that additional genes regulated by long days are required for the down-regulation of the COR genes. In addition, our results show that allelic variation in VRN-1 is sufficient to determine differences in freezing tolerance, suggesting that quantitative trait loci for freezing tolerance previously mapped on this chromosome region are likely a pleiotropic effect of VRN-1 rather than the effect of a separate closely linked locus (FROST RESISTANCE-1), as proposed in early freezing tolerance studies.
Exposure to low nonfreezing temperatures, a process known as cold acclimation, increases a plant's freezing tolerance (Thomashow, 1990, 1999). Freezing-tolerant plants that have not been cold acclimated are generally killed at approximately −3°C to −5°C, while cold-acclimated plants can survive much colder freezing temperatures. In addition, increasing the length of the cold acclimation period, up to a point, can also increase freezing tolerance. These two observations suggest that cold acclimation is an active process.
Freezing tolerance is essential for fall-planted temperate cereals (wheat [Triticum spp.], barley [Hordeum vulgare], and rye [Secale cereale]) to survive freezing temperatures during the winter. In contrast, spring-sown genotypes do not require high levels of freezing tolerance, since they are not exposed to the freezing temperatures of winter. One feature that distinguishes winter and spring genotypes is the requirement of the former for a long period (several weeks) at cold temperature to accelerate the transition from the vegetative growth phase to the reproductive growth phase, a process called vernalization. Spring genotypes do not have a vernalization requirement and flower in the absence of the extended low-temperature treatment (for review, see Trevaskis et al., 2007; Distelfeld et al., 2009).
The requirement for exposures to nonfreezing cold temperatures is common to both cold acclimation and vernalization, suggesting a potential connection between these two processes. Winter genotypes maintained under continuous cold, after an initial increase in freezing tolerance, exhibit a progressive decrease in their cold acclimation ability (Fowler et al., 1996a, 1996b; Fowler and Limin, 2004). This progressive decrease inversely parallels the fulfillment of the vernalization requirement. A clear decrease in freezing tolerance occurs after the shoot apical meristem advances to the double ridge stage (Fowler et al., 1996a, 1999; Limin and Fowler, 2006). These studies suggest that a regulatory component of freezing tolerance is linked to a developmental shift between the vegetative and reproductive stages. Limin and Fowler (2006) suggested that the main vernalization gene, VRN-1, which is induced during vernalization, plays an important role in the decrease of the ability to cold acclimate with development.
Early genetic studies also revealed a correlation between growth habit and freezing tolerance; wheat genotypes having a spring growth habit were less freezing tolerant than genotypes having a winter growth habit (Hayes and Aamodt, 1927). Subsequent studies carried out using wheat chromosome substitution lines revealed that homeologous group 5 chromosomes, where VRN-1 is located, have the largest effect (Roberts, 1986). The first major locus affecting freezing tolerance and winter hardiness on homeologous group 5 was designated FROST RESISTANCE-1 (FR-1; Sutka and Snape, 1989). However, since FR-1 cosegregates with VRN-1 in most genetic studies, it is still not clear if FR-1 is an independent gene or just a pleiotropic effect of VRN-1 (Brule-Babel and Fowler, 1988; Sutka and Snape, 1989; Roberts, 1990; Hayes et al., 1993; Francia et al., 2004; Galiba et al., 2009).
More recently, a second locus associated with natural variation in freezing tolerance in wheat and barley was mapped on the long arm of homeologous group 5. This locus, designated FR-2, is approximately 30 centimorgans proximal to VRN-1 and includes a cluster of 11 (or more) C-REPEAT BINDING FACTOR (CBF) genes (Vágújfalvi et al., 2003; Francia et al., 2004, 2007; Miller et al., 2006; Skinner et al., 2006; Knox et al., 2010). The FR-2 CBF gene cluster has surfaced as a major quantitative trait locus (QTL) affecting freezing tolerance in a number of wheat and barley mapping populations (Vágújfalvi et al., 2003; Francia et al., 2004, 2007; Båga et al., 2007).
The role of the CBF genes in freezing tolerance has been studied in detail in Arabidopsis (Arabidopsis thaliana). The CBFs are transcriptional activators that promote the expression of genes whose upstream regulatory sequences harbor the CRT/DRE low-temperature cis-acting DNA regulatory element (Stockinger et al., 1997). Approximately 20% of the Arabidopsis genes whose expression is altered during cold acclimation are directly or indirectly controlled by the CBF transcription factors (Vogel et al., 2005). Direct targets of the CBFs in Arabidopsis include the robustly induced COLD REGULATED (COR) genes (Jaglo-Ottosen et al., 1998). Similar candidate CBF target genes in the cereals, which also harbor CRT/DRE motifs in their upstream regulatory region, include COR14b, DHN5, and DHN8 (Choi et al., 1999; Dal Bosco et al., 2003). Many of these COR genes are induced to higher levels in genotypes exhibiting greater freezing tolerance than in those having lesser freezing tolerance (Houde et al., 1992; Danyluk et al., 1994, 1998; Crosatti et al., 1996; Fowler et al., 1996b; Limin et al., 1997; Grossi et al., 1998; NDong et al., 2002). The use of COR14b as an expression QTL to map loci affecting COR expression levels revealed two major loci, one of which is coincident with VRN-1 and the second one with FR-2 (Vágújfalvi et al., 2000; Francia et al., 2004).
Notably, genotypes carrying the vrn-1 allele for winter growth habit express certain CBF genes at higher levels than genotypes carrying the Vrn-1 allele for spring growth habit (Stockinger et al., 2007). Moreover, once the winter genotypes carrying the vrn-1 allele are vernalized, CBF transcript levels are dampened relative to levels detected in nonvernalized plants (Stockinger et al., 2007). This suggests that VRN-1 somehow acts to repress expression of the CBFs at FR-2 and in turn decrease freezing tolerance.
The molecular isolation of VRN-1 revealed that this gene encodes a MADS box protein similar to the Arabidopsis meristem identity gene APETALA1 (AP1; Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003). The characterization of VRN-1 alleles associated with winter and spring genotypes showed that the primary differences were insertions and deletions in regulatory regions located in the promoter and first intron (Yan et al., 2004; Fu et al., 2005; Pidal et al., 2009). Deletions in the VRN-1 promoter affecting a small region tentatively designated the “VRN box” (Pidal et al., 2009) or large deletions/insertions in the VRN-1 first intron are both associated with spring growth habit (Fu et al., 2005; Pidal et al., 2009). Genotypes with a winter growth habit (vrn-1 allele) show very low VRN-1 transcript levels until plants are vernalized. In contrast, spring genotypes (Vrn-1 allele) constitutively express VRN-1 to high levels. Flowering is initiated once VRN-1 transcripts reach a critical threshold level (Loukoianov et al., 2005).
In addition to vernalization, photoperiod also plays a role in VRN-1 regulation. In photoperiod-sensitive genetic backgrounds, long-day photoperiods enhance VRN-1 transcript accumulation while short-day photoperiods delay transcript accumulation. In both wheat and barley, the delay in the transition to floral initiation in plants grown under short-day photoperiods is associated with increased freezing tolerance (Fowler et al., 2001; Limin and Fowler, 2006). One of the cis-elements responsible for the lack of VRN-1 expression under short days in diploid wheat (Triticum monococcum) is thought to reside within the VRN-1 promoter CArG motif, a binding site for MADS box transcription factors located downstream of the VRN box (Dubcovsky et al., 2006; Pidal et al., 2009). Under short days, diploid wheat (T. monococcum) plants carrying deletions in this CArG motif (Vrn-1f and Vrn-1g alleles) show accumulation of VRN-1 transcripts and a slow transition of the shoot apical meristem to the reproductive stage, whereas plants with intact CArG motifs (e.g. Vrn-1h and vrn-1) show no VRN-1 expression in short days and remain in the vegetative phase (Dubcovsky et al., 2006). Under long days, all accessions with VRN-1 alleles for spring growth habit show accumulation of VRN-1 transcripts and a rapid initiation of the transition to the reproductive stage (Dubcovsky et al., 2006).
T. monococcum mutants with deletions of the VRN-1 gene fail to flower, indicating that this gene is indispensable for the transition to the reproductive phase (Shitsukawa et al., 2007b). Two independently induced nitrogen-ion-beam mutants, designated maintained vegetative phase1 (mvp-1) and mvp-2, were generated in different T. monococcum genetic backgrounds. The deletions in these two mutants encompass the complete VRN-1 gene and several closely linked genes (Distelfeld and Dubcovsky, 2010).
To investigate the role of VRN-1 in freezing tolerance, we made use of the mvp mutants and of natural T. monococcum accessions that differ in their ability to express VRN-1 under short days. We found that freezing tolerance and transcript levels of several CBF and COR genes were higher in the mvp mutants relative to the plants carrying at least one functional VRN-1 copy. However, the expression of VRN-1 under short days was not as effective as under long days to down-regulate COR14b gene transcription. Taken together, these results suggest that VRN-1 transcription is necessary but not sufficient to down-regulate the COR genes.
RESULTS
Effect of the mvp Mutations on Freezing Tolerance
The homozygous mvp-2 mutants (mvp-2/mvp-2) and the plants carrying at least one functional copy of VRN-1 (Mvp-2/−) were identified using a dominant molecular marker for VRN-1 as described in Supplemental Figure S1. Just before the freezing experiments, there were clear differences in apical development between plants from each group. The apices from the mvp-2/mvp-2 mutant were at the vegetative stage, whereas those from the Mvp-2/− plants were already at the double ridge stage (Supplemental Fig. S2).
Significant differences in survival rates were detected between the mvp-2/mvp-2 and Mvp-2/− plants in controlled freezing experiments. Differences between the two genotypic classes were detected at both −9°C and −11°C freezing temperatures (Table I). In the group frozen to −9°C, none of the Mvp-2/− plants survived, whereas 87% of the mvp-2/mvp-2 mutants survived (Table I). In the group frozen to −11°C, none of the Mvp-2/− plants survived, whereas approximately half (46%) of the mvp-2/mvp-2 mutants survived (Table I). In a second freezing experiment performed under slightly different acclimation and freezing conditions (Supplemental Fig. S3), approximately 70% of the mvp-2/mvp-2 mutants survived −12°C freezing temperatures, whereas only 40% of the Mvp-2/− plants survived the same treatment (P = 0.009; Supplemental Fig. S3). In this second experiment, all plants from both genotypic classes were killed at −13°C (Supplemental Fig. S2).
Table I. Survival, relative conductivity, and Fv/Fm in mutant plants (mvp-2/mvp-2= m/m) and plants carrying at least one wild-type VRN-1 allele (Mvp-2/− = M/−).
Sixty-day-old plants were acclimated at 10°C for 19 d, 4°C for 12 d, and –2°C for 12 h before the freezing treatment. P values correspond to factorial ANOVAs including temperature, genotype, and their interactions in the model. Experimental conditions are described in “Materials and Methods.”
| Parameter | Freezing Temperature |
Factorial ANOVA (P between Genotypes) | |||
| −9°C |
−11°C |
||||
| M/− | m/m | M/− | m/m | ||
| Fv/Fm | 0.56 | 0.78 | 0.56 | 0.81 | <0.0001 |
| Relative conductivity (%) | 12.0 | 10.1 | 17.3 | 11.9 | 0.05 |
| No. of plants that regrew per total (%) | 0/50 (0%) | 26/30 (87%) | 0/59 (0%) | 11/24 (46%) | NAa |
All Mvp-2/− plants failed to regrow; therefore, there is no variance within this class to perform an ANOVA. The differences between mvp-2/mvp-2 and Mvp-2/− were obvious.
To evaluate the effect of freezing on the functionality of PSII, the maximum quantum yield of PSII photochemistry was measured by the ratio of variable (Fv) to maximal (Fm) fluorescence in a dark-adapted state, Fv/Fm (Butler and Kitajima, 1975). Fv/Fm ratios taken 2 to 4 h after returning the plants to 20°C paralleled the survival results (high Fv/Fm values indicate low freezing damage). Homozygous mvp-2/mvp-2 mutant plants showed significantly higher Fv/Fm values than the Mvp-2/− plants (Table I; P < 0.0001), in agreement with the greater freezing tolerance of the mvp-2/mvp-2 plants.
Along with the Fv/Fm measurements, samples of the crown and adjoining tissues were collected to measure relative conductivity. This measurement estimates the cellular electrolytes leached from the freeze-damaged tissue as a proportion of the total cellular electrolytes and is based on the principle that the greater the damage to cells from freezing injury, the greater the exosmosis of cellular electrolytes into a water solvent (Dexter, 1956). The Mvp-2/− plants showed higher relative conductivity values than those from the homozygous mvp-2/mvp-2 mutants, both at −9°C (19% increase) and −11°C (45% increase). However, the differences between genotypes were only marginally significant (Table I; P = 0.05), likely due to the limited number of mutant plants sacrificed for relative conductivity measures in this first experiment (three mvp-2/mvp-2 and six Mvp-2/− plants per temperature). A second experiment using leaves from nine plants per genotype-temperature combination confirmed the higher relative conductivity of the Mvp-2/− plants compared with the mvp-2/mvp-2 plants (76% average increase over the three temperatures; P = 0.003; Supplemental Fig. S3).
Taken together, these three sets of data indicate that the presence of the VRN-1 gene in the Mvp-2/− lines is associated with a decrease in freezing tolerance relative to the mvp mutants.
Effect of the mvp Mutations on CBF Transcript Levels
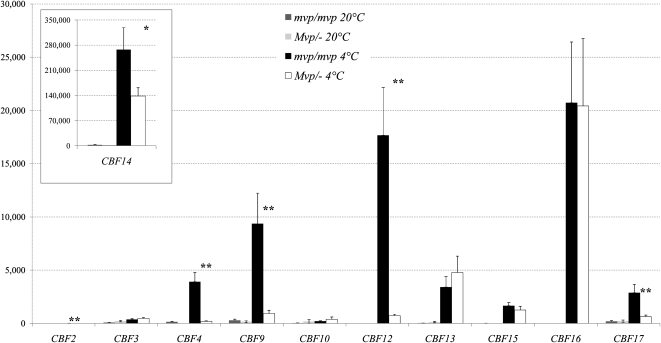
Quantitative reverse transcription (qRT)-PCR was used to compare transcript levels of 11 CBF genes in 4-week-old Mvp-2/− and mvp-2/mvp-2 plants both before (20°C) and after 8 h of cold treatment at 4°C (Fig. 1). With the exception of CBF2, which showed very low levels of expression both at 20°C and 4°C, the other 10 CBF genes showed very low transcript levels at 20°C and were significantly up-regulated after 8 h of cold treatment (P < 0.01). Five CBF genes (CBF2, CBF4, CBF9, CBF12, and CBF17) showed significantly higher expression levels (P < 0.01) in the mvp-2/mvp-2 homozygous mutant plants than in the Mvp-2/− plants (Fig. 1). The same difference was marginally significant for CBF14 (P = 0.03) and not significant for the other CBF genes.
Figure 1.
qRT-PCR analysis of transcript levels of the CBF genes present at the FR-2 locus relative to the ACTIN endogenous control. Samples were collected from leaves of 4-week-old mvp-2/mvp-2 and Mvp-2/− plants (20°C) and again 1 d later at the same time following an 8-h cold treatment at 4°C. Values on the y axis were normalized and calibrated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). The same calibrator was used for all genes, so scales are comparable across genes. Values are averages of eight biological replications ± se. The inset shows CBF14 transcript levels, which were significantly higher than the other genes at this locus. P values for the differences between mvp/mvp and Mvp/− after the cold treatment were calculated using ANOVA and are indicated by asterisks: * P < 0.05, ** P < 0.01.
A second experiment was carried out using 8-week-old Mvp-1/− and mvp-1/mvp-1 plants and, as before, measurements of CBF transcript levels after 8 h at 4°C (Supplemental Fig. S4). However, in this second experiment, no samples were collected at 20°C because of the negligible CBF transcript levels observed in the first experiment at this temperature. In the Mvp-1/− plants, the shoot apical meristems were between the double ridge and terminal spikelet stages, whereas in the mvp-1/ mvp-1 mutants, the shoot apical meristems were in the vegetative stage. As in the 4-week-old Mvp-2 plants used in the previous experiment, the older 8-week-old Mvp-1 plants showed significantly lower transcript levels of CBF2, CBF4, CBF9, CBF12, and CBF17 in the Mvp-1/− plants relative to the homozygous mvp-1/mvp-1 mutants after the cold treatment. Whereas CBF14 transcript level differences between Mvp-2/− and mvp-2/mvp-2 genotypes were marginally significant in the first experiment, in this second experiment the differences in CBF14 transcript levels between Mvp-1/− and mvp-1/mvp-1 were not significant. Curiously, transcript levels of CBF12 and CBF16 were much lower relative to the other genes in the 8-week-old Mvp-1 plants relative to the 4-week-old Mvp-2 plants.
Quantitative PCR measurements of VRN-1 in both experiments revealed high levels of VRN-1 transcripts in the Mvp-1/− and Mvp-2/− plants, which were even higher than those of the highly expressed TRANSLATION ELONGATION FACTOR1 (TEF1) endogenous control gene in both cases. As expected, no VRN-1 transcripts were detected in the homozygous mvp/mvp mutants. Taken together, these results suggest that the presence of VRN-1 (or genes regulated by VRN-1) modulates the response of several CBF genes to cold.
Effect of the mvp Mutations on COR Gene Transcript Levels
Eight hours after transferring 8-week-old plants from 20°C to 4°C, COR14b transcripts were 2-fold higher (P < 0.05) in the homozygous mvp-1/mvp-1 plants than in the Mvp-1/− plants (Supplemental Fig. S4). Two additional experiments, in which plants were exposed to longer periods of cold temperatures, were performed to further characterize the differences in COR14b transcript levels between mutants and nonmutants.
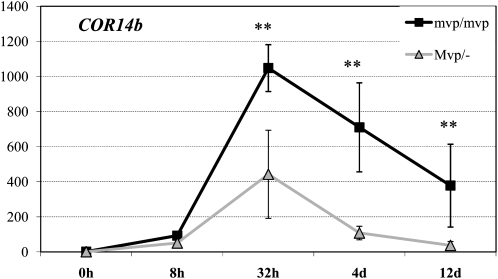
In the first experiment, 4-week-old mvp-2/mvp-2 and Mvp-2/− plants were transferred from room temperature to 4°C and kept at that temperature for 12 d. Leaf samples for RNA analysis were collected on the day prior to the cold treatment and 8 h, 32 h, 4 d, and 12 d after transferring the plants to 4°C. All samples were collected at 2 pm to avoid differences that might be caused by circadian effects. In this qRT-PCR experiment, transcript levels from both genotypes peaked at 32 h and then decayed slowly during the next 11 d. At each of the last three sampling points, the COR14b transcript levels were significantly higher (P < 0.001) in the mvp-2/mvp-2 homozygous plants than in Mvp-2/− plants (Fig. 2).
Figure 2.
qRT-PCR transcript levels of COR14b relative to TEF1 endogenous control. Plants were 4 weeks old at the beginning of the experiment and were exposed to 4°C for 12 d. Values on the y axis were normalized and calibrated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Homozygous mvp-2/mvp-2 plants (null VRN-1) are indicated by black squares and lines, and Mvp-2/− plants (one or two VRN-1 copies) are indicated by gray triangles and lines. Values are averages of eight biological replications in the untransformed scale ± se. P values were calculated using ANOVA of log(n + 1)-transformed values for each time point: ** P < 0.01.
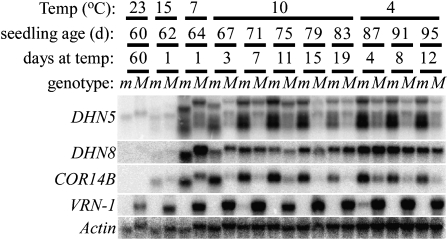
In the second experiment, steady-state transcript levels of COR14b and two additional COR genes (DHN5 and DHN8) were evaluated by RNA-blot analysis using a more gradual decrease in temperatures and longer exposure times to the inductive temperatures (19 d at 10°C followed by 12 d at 4°C, all under long days; Fig. 3). At the beginning of the cold induction, the Mvp-2/− plants were already induced to flower and showed high levels of VRN-1 transcripts, whereas the mvp-2/mvp-2 plants were in the vegetative stage and, as expected from the homozygous deletion, showed no VRN-1 transcripts (Fig. 3).
Figure 3.
Time courses of VRN-1 and COR genes COR14b, DHN5, and DHN8 in mvp-2/mvp-2 plants homozygous for a VRN-1 deletion (m) and Mvp-2/− plants with one or two functional VRN-1 copies (M). The presence of a faint VRN-1 hybridization in the 87-d mutant sample is suspected to be due to cross-contamination.
At all time points after the cold induction, the transcript levels of COR14b and DHN5 were higher in the mvp-2/mvp-2 plants (no VRN-1 transcripts) than in the Mvp-2/− plants (high VRN-1 transcripts). The DHN8 gene did not show this alternate pattern between genotypes (Fig. 3). In mvp-2/mvp-2 homozygous mutants, COR14b and DHN5 levels remained high throughout the sampling time course (Fig. 3).
Taken together, the expression data from these experiments showed that in plants with high VRN-1 transcript levels, several CBF and COR genes are down-regulated. To test if the down-regulation of these genes was a direct effect of the increase in VRN-1 transcript levels or a result of the transition to the reproductive phase, the relationship between VRN-1 and COR14b transcript levels was studied in T. monococcum lines that, under short days, differ in the expression of VRN-1 but show similar delays in the progression to the reproductive phase.
Effect of VRN-1 Transcription on COR14b Transcript Levels under Short Days
T. monococcum lines carrying a “wild-type” vrn-A1 allele and recessive vrn-A2 alleles have a spring growth habit and show no expression of VRN-1 under short days (Dubcovsky et al., 2006). However, T. monococcum lines carrying a 1-bp deletion in the VRN-1 promoter CArG box and an insertion in intron 1 (Vrn-1f allele), or a 34-bp deletion encompassing the complete CArG box (Vrn-1g allele), show high VRN-1 transcript levels under short days (Dubcovsky et al., 2006). These plants show a transition of the shoot apical meristem to the double ridge stage under short days, but further development of the spike is delayed until the plants are transferred to long days (Dubcovsky et al., 2006).
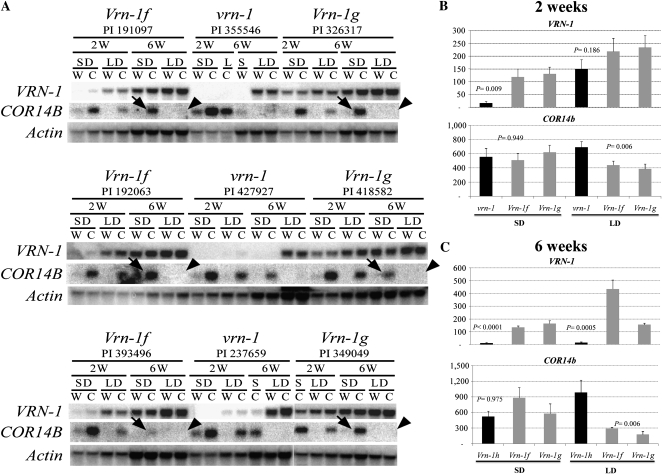
Expression profiling of three independent accessions of T. monococcum lines carrying each of the three genotypes confirmed previously published results (Dubcovsky et al., 2006). Under short days, lines having the Vrn-1f or Vrn-1g allele showed high VRN-1 transcript levels, whereas those having the wild-type vrn-1 allele showed no VRN-1 transcripts (Fig. 4A). As expected, transcript levels of VRN-1 were higher at 6 weeks than at 2 weeks, and COR14b transcripts were high after cold temperatures and nearly absent in plants maintained at warm temperature (Fig. 4A).
Figure 4.
VRN-1 and COR14b transcript levels in a set of T. monococcum lines differing in VRN-1 expression under short days. Abbreviations are as follows: W, warm conditions; C, decrease from 18°C to 6°C (occurring at daybreak); 2W, 2 weeks old; 6W, 6 weeks old; SD, short day; LD, long day; vrn-1, wild type; Vrn-1f, allele with a 1-bp deletion in the CArG box coupled with the VRN-1 intron 1 insertion; Vrn-1g, allele with a 34-bp deletion encompassing the CArG box; Vrn-1h, allele with an insertion in VRN-1 intron 1. The three accessions with the Vrn-1f allele carry the dominant Vrn-2 allele, whereas all the other accessions carry nonfunctional vrn-2 alleles. All the accessions have a spring growth habit. A, mRNA-blot analyses of three genotypes per promoter class (indicated by different PI numbers). Arrows and arrowheads identify the presence and absence, respectively, of COR14b transcripts in 6-week-old plants in the Vrn-1f and Vrn-1g natural mutants under short days and long days. B and C, qRT-PCR validation of VRN-1 and COR14b transcript levels relative to the ACTIN endogenous control at 4°C. Values on the y axes were normalized and calibrated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Lines carrying the wild-type allele (vrn-1) or Vrn-1h (spring, not induced in short days) are indicated by black bars, and lines carrying the Vrn-1f and Vrn-1g alleles (spring, induced in short days) are indicated by gray bars. Values are averages of five biological replications ± se. P values were calculated using contrasts between either vrn-1 or Vrn-1h and the average of the lines carrying the Vrn-1f and Vrn-1g alleles. Samples were collected when the plants were 2 weeks old (B) and 6 weeks old (C).
In the 6-week-old Vrn-1f and Vrn-1g lines grown under short days, COR14b was highly responsive to cold temperatures, despite relatively high VRN-1 transcript levels in these lines (Fig. 4A, arrows). In contrast, when 6-week-old plants of these same genotypes were grown under long-day conditions, the same cold treatment failed to induce COR14b to high levels (Fig. 4A). This result indicates that, under short days, the expression of VRN-1 was not sufficient to down-regulate COR14b.
Analyses of the 2-week-old plants (Fig. 4A) further confirmed the inverse correlation between VRN-1 and COR14b. Under short days, the cold treatments resulted in a strong up-regulation of COR14b (low VRN-1 transcript levels in all genotypes), but under long days, the down-regulation of COR14b was not as strong as in the 6-week-old plants, likely because the VRN-1 transcript levels in 2-week-old plants was not as high as in the 6-week-old plants (Fig. 4A). To confirm these results, this experiment was repeated using qRT-PCR and the same genotypes at the same developmental stage (Fig. 4B). In plants grown under short days and exposed for 32 h to 4°C, COR14b transcript levels showed no significant differences (P = 0.95) between the wild type and Vrn-1 mutant lines (Vrn-1f and Vrn-1g), despite significant differences in VRN-1 transcript levels. In contrast, highly significant differences in COR14b transcript levels (P = 0.006) were detected under long days, in which COR14b transcript levels in the Vrn-1f and Vrn-1g mutant lines were lower than in the wild type (Fig. 4B).
An additional experiment was carried out to compare COR14b expression levels in T. monococcum lines carrying the Vrn-1f and Vrn-1g alleles (early spring growth habit) with those in a T. monococcum accession carrying the Vrn-1h allele (late spring growth habit). The Vrn-1h allele has an almost identical repetitive element insertion in the first intron as Vrn-1f but lacks the CArG box mutation (Dubcovsky et al., 2006). As reported previously, the line with the Vrn-1h allele showed low VRN-1 transcripts under short days and maintained low VRN-1 transcript levels even when plants were grown under long days for 6 weeks. The results presented in Figure 4C confirmed that high VRN-1 transcript levels under short days (Vrn-1f and Vrn-1g) were not sufficient to suppress the induction of COR14b. In contrast, when plants were grown under long days, the Vrn-1h line (low VRN-1 transcript levels) showed a significantly stronger (P = 0.006) induction of COR14b than the Vrn-1f and Vrn-1g mutants (Fig. 4C). As in the previous experiments, both high VRN-1 transcript levels and long days were necessary for the down-regulation of COR14b.
Taken together, the results from this and previous experiments suggest that VRN-1 expression is required to initiate the developmental processes that reduce the ability of COR14b to respond to cold temperatures but that VRN-1 transcription alone is not sufficient to produce this effect.
DISCUSSION
Homozygous mvp/mvp Mutants Show More Freezing Tolerance Than Mvp/− Plants
When the shoot apical meristem of the Mvp/− plants transitions to the reproductive phase, it stops producing new leaves. Therefore, Mvp/− plants are expected to exhibit a reduced ability to generate new leaves after freezing as they transition to the reproductive phase. In contrast, the shoot apical meristem of the mvp/mvp homozygous mutants never transitions to the reproductive phase; thus, their ability to generate new leaves after freezing should not be drastically altered with time.
While changes in the susceptibility of the shoot apical meristem to freezing damage may account for the observed differences in regrowth after freezing, the mvp/mvp homozygous mutants also exhibited greater freezing tolerance in the existing leaves after acclimation than the Mvp/− plants, as suggested by higher Fv/Fm values and lower relative conductivity after the freezing treatment. This suggests that the presence of VRN-1 transcript levels and the concomitant dampening of COR14b induction by cold contributed to a reduction of freezing tolerance in the existing leaves of the Mvp/− plants.
Barley COR14b and the related Arabidopsis COR15 are hydrophilic proteins targeted to the chloroplast stromal compartment (Crosatti et al., 1995). In Arabidopsis, increasing the levels of COR15 results in increased freezing tolerance (Artus et al., 1996). COR15 appears to stabilize membranes from freeze-induced injury, which would account for the reduced electrolyte leakage and higher Fv/Fm values (Artus et al., 1996). In addition, the higher transcript levels of dehydrin genes such as DHN5 (Fig. 3) in the leaves of the mvp/mvp homozygous mutants relative to the Mvp/− plants likely contribute to their improved freezing tolerance. Dehydrins have a highly conserved 15-amino acid segment (the “K segment”) that interacts with acidic phospholipids in lipid vesicles. This interaction results in a conformational change of the protein structure that is hypothesized to stabilize membrane integrity (Koag et al., 2003, 2009).
The Increased Freezing Tolerance of the mvp Mutants Is Likely Caused by the VRN-1 Deletion
The MADS box meristem identity gene VRN-1 plays an essential role in the regulation of the transition between vegetative and reproductive phases, and its deletion results in plants that fail to flower. In Arabidopsis, gene duplications of the VRN-1 homolog that occurred after the monocot-dicot divergence resulted in three paralogous genes, AP1, CAULIFLOWER (CAL), and FRUITFULL (FRU), that have retained partial ability to promote the transition of the vegetative shoot apical meristem to the reproductive phase. Simultaneous deletions of all three genes are required to generate nonflowering Arabidopsis plants (Ferrandiz et al., 2000).
An additional difference between the temperate cereals and Arabidopsis meristem identity genes is their spatial expression profile. VRN-1 transcripts are detected at high levels in the leaves of wheat (Danyluk et al., 2003; Yan et al., 2003; Li and Dubcovsky, 2008), barley (Schmitz et al., 2000; Trevaskis et al., 2003), Lolium (Petersen et al., 2004), and oat (Avena sativa; Preston and Kellogg, 2008), suggesting a similar profile among the temperate cereals. In contrast, the Arabidopsis meristem identity homologs are expressed primarily in the apical meristem and reproductive tissues. AP1 and CAL transcripts are abundant in the induced shoot apical meristem and floral primordia in Arabidopsis but are undetectable or present at much lower levels in some vegetative tissues such as the vascular tissues of cotyledons (Abe et al., 2005). FUL is also expressed primarily in the meristem and floral tissue, but it is also detected in cauline leaves (Teper-Bamnolker and Samach, 2005). In the winter cereals, the expression of VRN-1 in the leaves (and apices) occurs only after vernalization, providing a potentially useful regulatory signal to down-regulate the cold acclimation pathway in this tissue in the spring.
Winter wheat lines exposed to continuous cold temperatures improve their freezing tolerance during the first 3 to 4 weeks of the treatment but then gradually start losing those gains. The inflection point in this freezing tolerance curve coincides with the transition of the shoot apical meristem to the double ridge stage and high VRN-1 transcript levels in the leaves (Danyluk et al., 2003). Limin and Fowler (2006) found that wheat near-isogenic lines for VRN-1 carrying the allele for winter growth habit tolerate freezing temperatures 11°C lower than lines carrying the VRN-1 allele for spring growth habit. The authors also showed that when the same near-isogenic spring lines were grown under short days, which are less promotive of VRN-1 expression than long days, the plants tolerated temperatures 8.5°C colder than the same lines grown under long days (Limin and Fowler, 2006). Based on these results, these authors hypothesized that the expression of VRN-1 might be an important signal to regulate the freezing tolerance pathway.
Previous studies using near-isogenic lines (Limin and Fowler, 2006), and others using QTL mapping for freezing tolerance, all point to the VRN-1 region as important in the regulation of freezing tolerance (Sutka and Snape, 1989; Roberts, 1990; Hayes et al., 1993; Francia et al., 2004; Galiba et al., 2009). However, the recombination points flanking the VRN-1 gene in the lines used in these studies are not known and can encompass large chromosome regions including a large number of genes. The use of deletion mutants in this study provides a more precise delimitation of the chromosome region responsible for the differences in freezing tolerance.
Nonetheless, some caution is still required in the interpretation of the mvp results, since the deletions present in these lines include other genes flanking VRN-1. A screening with probes for multiple genes in the VRN-1 region showed that the mvp deletions include the AGLG1, CYS, and PHYC genes but exclude flanking genes CYB5 and ADA2 (Distelfeld and Dubcovsky, 2010). The exclusion of ADA2 from the deleted regions is relevant because this gene is critical for CBF-mediated transactivation (Stockinger et al., 2001). Based on colinearity with Brachypodium, the mvp deletion is also predicted to include two additional genes, an oligopeptide transporter (Bradi1g08420) and a proteinase inhibitor I9 (Bradi1g08450; Distelfeld and Dubcovsky, 2010). Most of these additional genes are unlikely candidates for the improved freezing tolerance observed in the mvp mutants, with the exception of phytochrome PHYC, since phytochromes B and D have been shown to affect the CBF regulon in Arabidopsis (Franklin and Whitelam, 2007). Thus, we cannot rule out the possibility of the presence of additional genes with an effect on freezing tolerance in the mvp deleted region until it is completely sequenced.
However, the expression studies provided an independent source of evidence pointing to VRN-1 as the best candidate gene for the down-regulation of the cold acclimation response. In all the T. monococcum accessions carrying the VRN-1f or VRN-1g allele, the higher transcript levels of VRN-1 were always associated with a significant down-regulation of COR14b when plants were grown in long-day conditions (Fig. 4, B and C). In addition, larger differences in VRN-1 transcript levels during development were correlated with larger differences in COR14b transcript levels (Fig. 4, B and C). All the expression results presented here support the hypothesis that VRN-1 is the best candidate for the dampening of the cold acclimation response among the genes present within the mvp deletions. We are currently developing TILLING mutants of the VRN-A1 and VRN-B1 genes in tetraploid wheat (Uauy et al., 2009) to provide an independent validation of this hypothesis.
Homozygous mvp Mutants Exhibit Higher Transcript Levels of Several CBF and COR Genes after a Short Cold Treatment
A negative association between VRN-1 and COR genes COR14b and DHN5 (=WCS120) transcript levels has also been reported in previous wheat and barley studies (Vágújfalvi et al., 2000; Danyluk et al., 2003; Knox et al., 2008). In a doubled-haploid barley population segregating for VRN-H1, the lines carrying the recessive vrn-H1 allele showed higher transcript levels of CBF and COR genes than those carrying the dominant Vrn-H1 allele (Stockinger et al., 2007). In addition, lines grown under short days (reduced VRN-H1 levels) showed higher CBF and COR transcript levels than lines grown under long days when plants were transferred to the cold (Stockinger et al., 2007). The reductions in the transcript levels of multiple CBF genes and their downstream COR gene targets in plants with high VRN-1 transcript levels provide a simple explanation for the gradual decrease in freezing tolerance observed after the initiation of the reproductive phase.
The mvp mutants characterized in this study exhibited a similar negative association between VRN-1 and both CBF and COR transcription profiles. Five of the 11 CBF genes tested by qRT-PCR and the COR14b gene showed significantly higher transcript levels 8 h after moving the plants to 4°C in the mvp deletion homozygotes than in those carrying at least one functional copy of VRN-1 (Fig. 1; Supplemental Fig. S4). In plants maintained at 4°C for 12 d, the COR14b transcript levels were still 10-fold higher in the plants homozygous for the mvp deletion (Fig. 2). However, analyses of two additional COR genes, DHN5 and DHN8, showed that not all COR genes respond in the same way. Whereas DHN5 showed the same negative correlation with VRN-1 as COR14b, the DHN8 gene was not significantly affected by the change in the level of VRN-1 transcripts, which indicates that not all COR genes are down-regulated by VRN-1. Similarly, for half of the 11 CBF genes present in the FR-2 cluster, no significant differences in transcript levels were detected between Mvp/− and mvp/mvp lines (Fig. 1; Supplemental Fig. S4), suggesting that cold activation of these genes is not regulated by VRN-1. Thus, it appears that this VRN-1-mediated mechanism may play a role in the regulation of a specific subset of cold-responsive genes.
Allelic Differences in VRN-1 Are Likely Sufficient to Explain Differences in Freezing Tolerances Previously Assigned to a Separate FR-1 Locus
In earlier studies, differences in freezing tolerance mapped to the VRN-1 region in wheat were considered to be the result of a closely linked gene designated FR-1. However, only two studies have reported recombination between FR-1 and VRN-1, and they differ in the relative positions of these two genes, with FR-1 distal to VRN-1 in the initial mapping studies (Galiba et al., 1995, 1997) and proximal to VRN-1 in a later mapping study using deletion lines (Sutka et al., 1999). Although the differences in freezing tolerance across the deletion lines used in the latter study were clear, it is still possible that the reduced freezing tolerance observed in the larger deletion used to map FR-1 to a proximal deletion bin than VRN-1 was the result of the loss of a larger number of genes and an overall reduction in plant vigor rather than the effect of a single FR-1 gene. It is also possible that simultaneous segregation at the linked FR-2 locus, which was not known at the time of these two studies, affected the mapping results.
The improved freezing tolerance and higher transcript levels of CBF and COR genes in the mvp mutants suggest that VRN-1 allelic differences are likely sufficient to explain differences in freezing tolerance previously considered to be the result of a separate FR-1 gene. Therefore, our results support the hypothesis that FR-1 is a pleiotropic effect of VRN-1 rather than a separate gene. This hypothesis is also supported by experiments showing that the repression of VRN-1 by short days in spring wheat genotypes is associated with increased freezing tolerance (Limin and Fowler, 2006) and that VRN-1 transcript levels in the different Triple Dirk near-isogenic lines are inversely correlated with freezing tolerance (Koemel et al., 2004).
VRN-1 Transcription Is Not Sufficient to Promote the Down-Regulation of COR Genes
The experiment using the T. monococcum lines carrying the VRN-1f and VRN-1g alleles (Fig. 4) shows that up-regulation of VRN-1 transcript levels under short days is insufficient to produce a significant down-regulation of COR14b as that observed under long days. Under short days, 6-week-old plants carrying these alleles show high transcript levels of VRN-1 and a transition of the vegetative apex to the double ridge stage. However, under continuous short days, spike development proceeds slowly and stems fail to elongate. Once plants are transferred to long days, genotypes having the VRN-1f and VRN-1g alleles complete their spike development faster and head earlier than genotypes with the wild-type VRN-1 allele due to their more advanced developmental state (Dubcovsky et al., 2006).
When grown under long days, plants carrying the VRN-1f and VRN-1g alleles showed significantly lower levels of COR14b than lines carrying the vrn-1 or Vrn-1h allele. These results were consistent across three independent accessions for each of the VRN-1 alleles, supporting the hypothesis that the differences in COR14b were associated with the differences in the VRN-1 alleles. However, no differences in COR14b were observed among the same genotypes under short days, despite large differences in VRN-1 transcript levels. These results suggest that the down-regulation of the COR14b requires the presence of additional factors that are activated under long days and that require the expression of VRN-1. Taken together, the mvp mutant and VRN-1f and VRN-1g experiments suggest that VRN-1 expression is necessary but not sufficient to down-regulate several COR genes and reduce freezing tolerance in the leaves of wheat plants.
A similar phenomenon has recently been described in Arabidopsis, where the floral activator MADS box gene SOC1 functions as a negative regulator of the cold response pathway through the direct repression of the CBF genes (Seo et al., 2009). In the Columbia wild type, SOC1 was expressed most strongly in leaves but was also detected in vegetative apices, inflorescences, stems of flowering plants, and roots (Lee et al., 2000). A microarray experiment comparing 7-d-old seedlings of a soc1 knockout mutant and a SOC1-overexpressing line with wild-type plants revealed that six COR genes were among the 20 genes most negatively regulated in the SOC1-overexpressing line. In addition, the expression level of the three Arabidopsis CBF genes increased in the soc1 mutants and decreased in SOC1-overexpressing lines, without affecting the transcript levels of the CBF regulatory genes ICE1, HOS1, or ZAT12. A chromatin immunoprecipitation experiment using a SOC1 antibody revealed that the CArG box regions in the CBF promoters were enriched in the SOC1-overexpressing line relative to the soc1 knockout, which suggests that SOC1 negatively regulates cold response through direct repression of the transcription of the CBF genes (Seo et al., 2009). It is interesting that the expression of the wheat homolog of Arabidopsis SOC1, WSOC1, is not affected by vernalization or photoperiod, suggesting different functions in these two species (Shitsukawa et al., 2007a).
Although both SOC1 in Arabidopsis and VRN-1 in the temperate cereals seem to be associated with the down-regulation of the CBF and COR genes in the leaves, the effect of VRN-1 on the CBF genes does not seem to be as direct as the effect of SOC1 in Arabidopsis. The results from the experiments using T. monococcum accessions with differential expression of VRN-1 under short days suggest that additional genes operating downstream of VRN-1 and that are regulated by long days are required to mediate the negative effect of VRN-1 on freezing tolerance. The identification of these downstream genes and the understanding of their regulatory mechanisms could potentially lead to novel strategies to prevent the premature dampening of the cold acclimation pathway in environments where the premature activation of VRN-1 may increase the risk of freezing damage.
Do Temperate Cereals Respond Differently to the Same Cool Temperatures in the Fall and the Spring?
The system described above provides the temperate cereals with the ability to differentiate the same cool temperature in the fall and the spring. A cool temperature in the fall, when plants have low VRN-1 transcript levels, results in the induction of the CBF and the downstream COR genes, initiating the acclimation of the plants to cold temperatures. This is essential in the fall, when cool temperatures are an indication of the approaching freezing temperatures of the winter. The same cool temperature in the spring, when VRN-1 transcript levels in the leaves increase significantly in response to lengthening photoperiod, would result in a significantly lower up-regulation of several CBF and COR genes. Since cool temperatures in spring are generally not a prelude of coming freezing temperatures, a robust up-regulation of the CBF pathway response in the spring would likely not be advantageous for plant survival.
A similar system seems to be operating in Arabidopsis. Arabidopsis soc1 null mutants show increased responsiveness of the CBF genes to cold and improved freeing tolerance, suggesting that low levels of SOC1 transcripts during the fall may favor plant acclimation to cold temperatures (Seo et al., 2009). SOC1 transcript levels increase significantly by the time of the initiation of Arabidopsis flowering (Lee et al., 2000), indicating that high SOC1 transcript levels will be present in the leaves in the spring, down-regulating the CBF genes and their downstream COR targets.
The activation of the CBF regulon has a potentially high energetic cost to plants, since numerous COR genes are up-regulated in the leaves by these transcription factors (Fowler and Thomashow, 2002). In addition, CBF genes have also been shown to repress plant growth (Achard et al., 2008). Therefore, the down-regulation of the CBF genes during the spring has a potential adaptive value, ensuring the plant's rapid development under optimal conditions, and may explain the presence of related systems in Arabidopsis and the temperate cereals.
MATERIALS AND METHODS
Plant Materials
mvp Mutants
Two independent Triticum monococcum mutants (mvp-1 and mvp-2) that remain indefinitely in the vegetative state (Shitsukawa et al., 2007b) were used in this study. Since the two mutants carry similar deletions (Distelfeld and Dubcovsky, 2010), they were alternated among experiments depending on seed supply. Seeds from these mutants were kindly provided by K. Murai. These mutants were generated by ion-beam radiation, and both have large deletions that include VRN-1 (Shitsukawa et al., 2007b) and several flanking genes (Distelfeld and Dubcovsky, 2010). The mvp-1 mutation was generated in the KU104-2 background and the mvp-2 mutation in the KU104-1 background (Shitsukawa et al., 2007b). When grown in the greenhouse under long-day conditions (16-h photoperiods), KU104-2 flowered 10 weeks after planting while KU104-1 flowered 3 weeks later.
Homozygous mvp individuals do not flower and therefore must be maintained in a heterozygous state. Genotyping was carried out using a dominant VRN-1 molecular marker based on a set of three primers that are described in Supplemental Figure S1. Using this assay, the lines carrying one or two functional VRN-1 copies are detected as a single genotypic class, referred throughout the text as Mvp/−.
VRN-1f, VRN-1g, and VRN-1h Alleles
T. monococcum lines with four different VRN-1 alleles were used to test the effect of their differential regulation under short days on COR14b transcript levels. T. monococcum lines PI355546, PI427927, and PI237659 carry the “wild-type” vrn-1 allele and a recessive vrn-2 allele that confers spring growth habit. Lines with the vrn-1 allele showed no expression under short days in previous studies (Dubcovsky et al., 2006). T. monococcum lines PI191097, PI192063, and PI393496 carry the Vrn-1f allele, which has a 1-bp deletion in a CArG box located in the promoter plus an insertion of a repetitive element in the first intron. T. monococcum lines PI326317, PI418582, and PI349049 carry the Vrn-1g allele, which has a 34-bp deletion including the promoter CArG box. Both the Vrn-1f and Vrn-1g alleles confer spring growth habit and show high levels of VRN-1 transcripts under short days (Dubcovsky et al., 2006). T. monococcum accession PI306540 has the VRN-1h allele, which has the same intron 1 insertion as Vrn-1f but lacks the CArG box mutation. This allele is not expressed under short days and confers a spring phenotype but with later flowering than the Vrn-1f and Vrn-1g alleles (Dubcovsky et al., 2006). Although the shoot apical meristem transitions to the double ridge stage, spike development progresses slowly and spikes fail to elongate if these plants are left under short days (Dubcovsky et al., 2006).
Growth, Cold Acclimation, and Freezing Assays
Freezing experiments with the mvp mutants were all carried out using long-day conditions (16 h of light/8 h of dark). Experiments with the T. monococcum lines with different VRN-1 alleles were carried out using both long-day and short-day (8 h of light/16 h of dark) photoperiod cycles.
The RNA-blot analysis and the freezing experiment of the mvp-2 mutants were done in parallel. Seeds collected from Mvp-2/− heterozygotes were grown under cool-white fluorescent lamps in the laboratory at room temperature for 13 d using a light intensity of 50 μmol m−2 s−1. Genotyped seedlings were transplanted to wooden boxes having internal dimensions measuring 42 cm (length) × 22 cm (width) × 14 cm (height) and having 9.5-cm soil depth. The boxes were placed into a Conviron growth chamber (model PGW36; Controlled Environments) for an additional 47 d under cool-white fluorescent and incandescent bulbs using a light intensity of 130 μmol m−2 s−1 and a constant temperature of 23°C. The growth chamber temperature was decreased to 10°C and held at this temperature for 19 d.
Wooden boxes were then transferred to Percival growth chambers (model CU-36L2X; Geneva Scientific) where the seedlings were held at 4°C for 12 d (the PGW36 growth chamber can only cool to +10°C). After cold acclimation, the CU-36L2X growth chamber temperatures were decreased to −2°C and held for 12 h. Ice nucleation was induced by spraying the leaves with ice water. Following 12 h at −2°C, the temperature was then decreased at 1°C h−1 to the target temperatures of −9°C and −11°C and held at these temperatures for 24 h. Afterward, the temperature was returned to +2°C for 12 h. During this time, the plants were kept in the dark. The chamber temperature was then raised to 20°C and returned to a 16-h photoperiod.
Chlorophyll Fluorescence (Fv/Fm)
Chlorophyll fluorescence measurements were made using a hand-held portable pulse amplitude-modulated fluorometer (model OS-30p; Opti Sciences). Fv/Fm measurements were taken 2 to 4 h after the plants were returned to normal growth conditions. Leaves were dark adapted for approximately 10 min prior to taking the measurements.
Relative Conductivity
Conductivity measurements were taken on crown tissue consisting of a 1- to 1.5-cm segment of the white, nonphotosynthetic tissue between the upper photosynthetic green shoot and the primary root. Because the use of the crowns required the destruction of the plant, the number of replications in the first experiment was limited to three mvp/mvp homozygotes and six Mvp/− plants. The second experiment used leaves instead of crown regions and nine plants from the temperature/genotype combination. The electrolyte leakage assay methods are described in detail in the methods used for Supplemental Figure S3. Tubes were shaken for 1 h at 300 rpm before reading the conductivity with an Accumet Basic AB30 electrical conductivity meter (Fisher Scientific). Tubes were then autoclaved for 20 min, cooled to room temperature, and shaken for 1 h at 300 rpm before measuring the total potential conductivity. Values were adjusted by subtracting the conductivity of the deionized water. Relative conductivity represents the adjusted mean ion leakage as a percentage of the total adjusted leakage from frozen-killed samples (for formula, see methods used for Supplemental Fig. S3).
CBF qRT-PCR Experiments
Eight mvp-2/mvp-2 and Mvp-2/− plants and 10 mvp-1/ mvp-1 and Mvp-1/− plants were selected using the VRN-1 molecular marker and were grown in the greenhouse for 4 weeks and 8 weeks, respectively (20°C–25°C, long days). Plants were then transferred to a growth chamber at 4°C for 8 h. RNA samples were collected from leaves from eight mvp-2 and eight Mvp-2/− plants before (20°C) and after (4°C) the cold treatment in the first experiment and from 10 mvp-1 and 10 Mvp-1/− plants only after the cold treatment in the second experiment.
COR14b qRT-PCR Time-Course Experiment
Eight mvp-2 and eight Mvp-2/− plants were selected using the VRN-1 molecular markers and grown in the greenhouse under the same conditions described above. After 4 weeks, when the Mvp-2/− plants were still at the vegetative stage, plants were transferred to 4°C and were kept at this temperature for 12 d at the same light intensity indicated above (long days). Leaf samples for RNA analysis were collected 1 d before the cold treatment and after 8 h, 32 h, 4 d, and 12 d at 4°C. Samples were always collected at 2:00 pm (8 h after the subjective daybreak) to avoid potential differences at different times of the day.
VRN-1/COR14b qRT-PCR Experiment
Five plants with each VRN-1 allele were grown for either 2 or 6 weeks under short- or long-day conditions before transferring to 4°C. RNA samples were collected from leaves after 32 h of cold treatment.
For all qRT-PCR experiments, RNA was extracted using the Spectrum Plant Total RNA Kit (Sigma-Aldrich). First-strand cDNA was synthesized from 1 μg of total RNA using the QuantiTect Reverse Transcription Kit (Qiagen). Primers for qRT-PCR expression analyses are presented in Supplemental Table S1. The TEF1 and ACTIN genes were used as endogenous controls (Distelfeld and Dubcovsky, 2010).
RNA-Blot Analyses
Tissue samples were collected 13 to 14 h after the subjective daybreak from both cold-acclimating plants and nonacclimated plants. Total RNA was isolated using RNeasy Plant Mini kits (Qiagen). Seven micrograms of total RNA was loaded per lane. RNA samples consisted of RNAs pooled from the crown tissue of 10 plants (Mvp-2/− experiment) or five plants (mvp-1 and VRN-1 promoter deletion mutant experiments).
Fragments used as probes were generated by PCR amplification of cloned cDNA inserts. Radiolabeled probes were generated by random priming (Feinberg and Vogelstein, 1983). Overnight hybridizations were at 42°C in 50% formamide, 5× SSC, and 20 mm Na-phosphate buffer, pH 6.8, containing 100 μg mL−1 herring sperm DNA, 1× Denhardt's solution, 0.1% SDS, and 10% dextran sulfate. Three to four 1-h moderate-stringency washes were performed at 62°C to 65°C in 0.2× SSC, 0.05% SDS, and 0.01% Na-pyrophosphate. Images were generated using the Molecular Dynamics Storm840 PhosphorImager (GE Healthcare) and phosphor autoradiography.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Dominant VRN-1 molecular marker.
Supplemental Figure S2. Apices of Mvp-2/− and mvp-2/mvp-2 plants.
Supplemental Figure S3. Comparison of freezing tolerance and relative conductivity between mvp-1/mvp-1 and Mvp-1/− plants.
Supplemental Figure S4. qRT-PCR analysis of transcript levels of the CBF genes.
Supplemental Table S1. Primers used for qRT-PCR.
Supplementary Material
Acknowledgments
We thank Dr. Koji Murai for providing the seeds for the mvp-1 and mvp-2 mutants.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P. (2008) The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin C, Thomashow MF. (1996) Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci USA 93: 13404–13409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Båga M, Chodaparambil SV, Limin AE, Pecar M, Fowler DB, Chibbar RN. (2007) Identification of quantitative trait loci and associated candidate genes for low-temperature tolerance in cold-hardy winter wheat. Funct Integr Genomics 7: 53–68 [DOI] [PubMed] [Google Scholar]
- Brule-Babel AL, Fowler DB. (1988) Genetic control of cold hardiness and vernalization requirement in winter wheat. Crop Sci 28: 879–884 [Google Scholar]
- Butler WL, Kitajima M. (1975) Fluorescence quenching in photosystem II of chloroplasts. Biochim Biophys Acta 376: 116–125 [DOI] [PubMed] [Google Scholar]
- Choi DW, Zhu B, Close TJ. (1999) The barley (Hordeum vulgare L.) dehydrin multigene family: sequences, allele types, chromosome assignments, and expression characteristics of 11 Dhn genes of cv Dicktoo. Theor Appl Genet 98: 1234–1247 [Google Scholar]
- Crosatti C, Nevo E, Stanca AM, Cattivelli L. (1996) Genetic analysis of the accumulation of COR14 proteins in wild (Hordeum spontaneum) and cultivated (Hordeum vulgare) barley. Theor Appl Genet 93: 975–981 [DOI] [PubMed] [Google Scholar]
- Crosatti C, Soncini C, Stanca AM, Cattivelli L. (1995) The accumulation of a cold-regulated chloroplastic protein is light-dependent. Planta 196: 458–463 [DOI] [PubMed] [Google Scholar]
- Dal Bosco C, Busconi M, Govoni C, Baldi P, Stanca AM, Crosatti C, Bassi R, Cattivelli L. (2003) Cor gene expression in barley mutants affected in chloroplast development and photosynthetic electron transport. Plant Physiol 131: 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danyluk J, Houde M, Rassart E, Sarhan F. (1994) Differential expression of a gene encoding an acidic dehydrin in chilling sensitive and freezing tolerant Gramineae species. FEBS Lett 344: 20–24 [DOI] [PubMed] [Google Scholar]
- Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F. (2003) TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol 132: 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danyluk J, Perron A, Houde M, Limin A, Fowler B, Benhamou N, Sarhan F. (1998) Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell 10: 623–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter ST. (1956) The evaluation of crop plants for winter hardiness. Adv Agron 8: 203–239 [Google Scholar]
- Distelfeld A, Dubcovsky J. (2010) Characterization of the maintained vegetative phase (mvp) deletions from einkorn wheat and their effect on VRN2 and FT transcript levels. Mol Genet Genomics 283: 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A, Li C, Dubcovsky J. (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12: 178–184 [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, Yan L. (2006) Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol Biol 60: 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein P. (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 [DOI] [PubMed] [Google Scholar]
- Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF. (2000) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734 [DOI] [PubMed] [Google Scholar]
- Fowler DB, Breton G, Limin AE, Mahfoozi S, Sarhan F. (2001) Photoperiod and temperature interactions regulate low-temperature-induced gene expression in barley. Plant Physiol 127: 1676–1681 [PMC free article] [PubMed] [Google Scholar]
- Fowler DB, Chauvin LP, Limin AE, Sarhan F. (1996a) The regulatory role of vernalization in the expression of low-temperature-induced genes in wheat and rye. Theor Appl Genet 93: 554–559 [DOI] [PubMed] [Google Scholar]
- Fowler DB, Limin AE. (2004) Interactions among factors regulating phenological development and acclimation rate determine low-temperature tolerance in wheat. Ann Bot (Lond) 94: 717–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DB, Limin AE, Ritchie JT. (1999) Low-temperature tolerance in cereals: model and genetic interpretation. Crop Sci 39: 626–633 [Google Scholar]
- Fowler DB, Limin AE, Wang SY, Ward RW. (1996b) Relationship between low-temperature tolerance and vernalization response in wheat and rye. Can J Plant Sci 76: 37–42 [Google Scholar]
- Fowler S, Thomashow MF. (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia E, Barabaschi D, Tondelli A, Laidò G, Rizza F, Stanca AM, Busconi M, Fogher C, Stockinger EJ, Pecchioni N. (2007) Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor Appl Genet 115: 1083–1091 [DOI] [PubMed] [Google Scholar]
- Francia E, Rizza F, Cattivelli L, Stanca AM, Galiba G, Tóth B, Hayes PM, Skinner JS, Pecchioni N. (2004) Two loci on chromosome 5H determine low-temperature tolerance in a Nure (winter) × Tremois (spring) barley map. Theor Appl Genet 108: 670–680 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC. (2007) Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat Genet 39: 1410–1413 [DOI] [PubMed] [Google Scholar]
- Fu D, Szűcs P, Yan L, Helguera M, Skinner J, Hayes P, Dubcovsky J. (2005) Large deletions in the first intron of the VRN-1 vernalization gene are associated with spring growth habit in barley and polyploid wheat. Mol Genet Genomics 273: 54–65 [DOI] [PubMed] [Google Scholar]
- Galiba G, Kerepsi I, Snape JW, Sutka J. (1997) Location of a gene regulating cold-induced carbohydrate production on chromosome 5A of wheat. Theor Appl Genet 95: 265–270 [Google Scholar]
- Galiba G, Quarrie SA, Sutka J, Morgounov A. (1995) RFLP mapping of the vernalization (Vrn1) and frost resistance (Fr1) genes on chromosome 5A of wheat. Theor Appl Genet 90: 1174–1179 [DOI] [PubMed] [Google Scholar]
- Galiba G, Vágújfalvi A, Li C, Soltész A, Dubcovsky J. (2009) Regulatory genes involved in the determination of frost tolerance in temperate cereals. Plant Sci 176: 12–19 [Google Scholar]
- Grossi M, Giorni E, Rizza F, Stanca AM, Cattivelli L. (1998) Wild and cultivated barleys show differences in the expression pattern of a cold-regulated gene family under different light and temperature conditions. Plant Mol Biol 38: 1061–1069 [DOI] [PubMed] [Google Scholar]
- Hayes HK, Aamodt OS. (1927) Inheritance of winter hardiness and growth habit in crosses of Marquis with Minhardi and Minturki wheats. J Agric Res 35: 223–236 [Google Scholar]
- Hayes PM, Blake T, Chen THH, Tragoonrung S, Chen F, Pan A, Liu B. (1993) Quantitative trait loci on barley (Hordeum vulgare L.) chromosome 7 associated with components of winterhardiness. Genome 36: 66–71 [DOI] [PubMed] [Google Scholar]
- Houde M, Dhindsa RS, Sarhan F. (1992) A molecular marker to select for freezing tolerance in Gramineae. Mol Gen Genet 234: 43–48 [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- Koag MC, Fenton RD, Wilkens S, Close TJ. (2003) The binding of maize DHN1 to lipid vesicles: gain of structure and lipid specificity. Plant Physiol 131: 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koag MC, Wilkens S, Fenton RD, Resnik J, Vo E, Close TJ. (2009) The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol 150: 1503–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koemel JE, Jr, Guenzi AC, Anderson JA, Smith EL. (2004) Cold hardiness of wheat near-isogenic lines differing in vernalization alleles. Theor Appl Genet 109: 839–846 [DOI] [PubMed] [Google Scholar]
- Knox AK, Dhillon T, Cheng H, Tondelli A, Pecchioni N, Stockinger EJ. (2010) CBF gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor Appl Genet 121: 21–35 [DOI] [PubMed] [Google Scholar]
- Knox AK, Li C, Vágújfalvi A, Galiba G, Stockinger EJ, Dubcovsky J. (2008) Identification of candidate CBF genes for the frost tolerance locus Fr-Am2 in Triticum monococcum. Plant Mol Biol 67: 257–270 [DOI] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Dubcovsky J. (2008) Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J 55: 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limin AE, Danyluk J, Chauvin LP, Fowler DB, Sarhan F. (1997) Chromosome mapping of low-temperature induced Wcs120 family genes and regulation of cold-tolerance expression in wheat. Mol Gen Genet 253: 720–727 [DOI] [PubMed] [Google Scholar]
- Limin AE, Fowler DB. (2006) Low-temperature tolerance and genetic potential in wheat (Triticum aestivum L.): response to photoperiod, vernalization, and plant development. Planta 224: 360–366 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J. (2005) Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol 138: 2364–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AK, Galiba G, Dubcovsky J. (2006) A cluster of 11 CBF transcription factors is located at the frost tolerance locus Fr-Am2 in Triticum monococcum. Mol Genet Genomics 275: 193–203 [DOI] [PubMed] [Google Scholar]
- NDong C, Danyluk J, Wilson KE, Pocock T, Huner NP, Sarhan F. (2002) Cold-regulated cereal chloroplast late embryogenesis abundant-like proteins: molecular characterization and functional analyses. Plant Physiol 129: 1368–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen K, Didion T, Andersen CH, Nielsen KK. (2004) MADS-box genes from perennial ryegrass differentially expressed during transition from vegetative to reproductive growth. J Plant Physiol 161: 439–447 [DOI] [PubMed] [Google Scholar]
- Pidal B, Yan L, Fu D, Zhang F, Tranquilli G, Dubcovsky J. (2009) The CArG-box in the promoter region of wheat vernalization gene VRN1 is not necessary to mediate the vernalization response. J Hered 100: 355–364 [DOI] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA. (2008) Discrete developmental roles for temperate cereal grass VRN1/FUL-like genes in flowering competency and the transition to flowering. Plant Physiol 146: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DWA. (1986) Chromosomes in Cadet and Rescue wheats carrying loci for cold hardiness and vernalization response. Can J Genet Cytol 28: 991–997 [Google Scholar]
- Roberts DWA. (1990) Identification of loci on chromosome-5A of wheat involved in control of cold hardiness, vernalization, leaf length, rosette growth habit, and height of hardened plants. Genome 33: 247–259 [Google Scholar]
- Schmitz J, Franzen R, Ngyuen TH, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W. (2000) Cloning, mapping and expression analysis of barley MADS-box genes. Plant Mol Biol 42: 899–913 [DOI] [PubMed] [Google Scholar]
- Seo E, Lee H, Jeon J, Park H, Kim J, Noh YS, Lee I. (2009) Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21: 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitsukawa N, Ikari C, Mitsuya T, Sakiyama T, Ishikawa A, Takumi S, Murai K. (2007a) Wheat SOC1 functions independently of WAP1/VRN1, an integrator of vernalization and photoperiod flowering promotion pathways. Physiol Plant 130: 627–636 [Google Scholar]
- Shitsukawa N, Ikari C, Shimada S, Kitagawa S, Sakamoto K, Saito H, Ryuto H, Fukunishi N, Abe T, Takumi S, et al. (2007b) The einkorn wheat (Triticum monococcum) mutant, maintained vegetative phase, is caused by a deletion in the VRN1 gene. Genes Genet Syst 82: 167–170 [DOI] [PubMed] [Google Scholar]
- Skinner J, Szűcs P, von Zitzewitz J, Marquez-Cedillo L, Filichkin T, Stockinger EJ, Thomashow MF, Chen THH, Hayes PM. (2006) Mapping of barley homologs to genes that regulate low temperature tolerance in Arabidopsis. Theor Appl Genet 112: 832–842 [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Mao YP, Regier MK, Triezenberg SJ, Thomashow MF. (2001) Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res 29: 1524–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Skinner JS, Gardner KG, Francia E, Pecchioni N. (2007) Expression levels of barley Cbf genes at the Frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2. Plant J 51: 308–321 [DOI] [PubMed] [Google Scholar]
- Sutka J, Galiba G, Vagujfalvi A, Gill BS, Snape JW. (1999) Physical mapping of the Vrn-A1 and Fr1 genes on chromosome 5A of wheat using deletion lines. Theor Appl Genet 99: 199–202 [Google Scholar]
- Sutka J, Snape JW. (1989) Location of a gene for frost resistance on chromosome 5A of wheat. Euphytica 42: 41–44 [Google Scholar]
- Teper-Bamnolker P, Samach A. (2005) The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17: 2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. (1990) Molecular genetics of cold acclimation in higher plants. Adv Genet 28: 99–131 [Google Scholar]
- Thomashow MF. (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES. (2003) MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA 100: 13099–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemmin MN, Dennis ES, Peacock WJ. (2007) The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci 12: 352–357 [DOI] [PubMed] [Google Scholar]
- Uauy C, Paraiso F, Colasuonno P, Tran RK, Tsai H, Berardi S, Comai L, Dubcovsky J. (2009) A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol 9: 115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vágújfalvi A, Crosatti C, Galiba G, Dubcovsky J, Cattivelli L. (2000) Two loci on wheat chromosome 5A regulate the differential cold-dependent expression of the cor14b gene in frost-tolerant and frost-sensitive genotypes. Mol Gen Genet 263: 194–200 [DOI] [PubMed] [Google Scholar]
- Vágújfalvi A, Galiba G, Cattivelli L, Dubcovsky J. (2003) The cold regulated transcriptional activator Cbf3 is linked to the frost-tolerance gene Fr-A2 on wheat chromosome 5A. Mol Genet Genomics 269: 60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J. (2004) Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Genet 109: 1677–1686 [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. (2003) Positional cloning of wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100: 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.