Abstract
N-terminal truncation of the Escherichia coli ethanolamine ammonia-lyase β-subunit does not affect the catalytic properties of the enzyme (Akita, K., Hieda, N., Baba, N., Kawaguchi, S., Sakamoto, H., Nakanishi, Y., Yamanishi, M., Mori, K., and Toraya, T. (2010) J. Biochem. 147, 83–93). The binary complex of the truncated enzyme with cyanocobalamin and the ternary complex with cyanocobalamin or adeninylpentylcobalamin and substrates were crystallized, and their x-ray structures were analyzed. The enzyme exists as a trimer of the (αβ)2 dimer. The active site is in the (β/α)8 barrel of the α-subunit; the β-subunit covers the lower part of the cobalamin that is bound in the interface of the α- and β-subunits. The structure complexed with adeninylpentylcobalamin revealed the presence of an adenine ring-binding pocket in the enzyme that accommodates the adenine moiety through a hydrogen bond network. The substrate is bound by six hydrogen bonds with active-site residues. Argα160 contributes to substrate binding most likely by hydrogen bonding with the O1 atom. The modeling study implies that marked angular strains and tensile forces induced by tight enzyme-coenzyme interactions are responsible for breaking the coenzyme Co–C bond. The coenzyme adenosyl radical in the productive conformation was modeled by superimposing its adenine ring on the adenine ring-binding site followed by ribosyl rotation around the N-glycosidic bond. A major structural change upon substrate binding was not observed with this particular enzyme. Gluα287, one of the substrate-binding residues, has a direct contact with the ribose group of the modeled adenosylcobalamin, which may contribute to the substrate-induced additional labilization of the Co–C bond.
Keywords: Adenosylcobalamin, Enzyme Catalysis, Enzyme Structure, Vitamin B, X-ray Crystallography, Ethanolamine Ammonia-lyase
Introduction
Adenosylcobalamin (AdoCbl)3 or coenzyme B12 is a naturally occurring organometallic compound that contains a unique Co–C σ bond. It serves as a cofactor for enzymatic radical reactions including carbon skeleton rearrangements, heteroatom eliminations, and intramolecular amino group migrations (1, 2). All of these reactions are initiated by homolysis of the Co–C bond of the enzyme-bound coenzyme, forming an adenosyl radical, and are catalyzed by radical mechanisms. Ethanolamine ammonia-lyase (EAL) (EC 4.3.1.7) or ethanolamine deaminase catalyzes the AdoCbl-dependent conversion of ethanolamine (EA) to acetaldehyde and ammonia (3) (Equation 1).
EAL was first discovered by Bradbeer (4) in choline-fermenting Clostridium sp. This enzyme is essential for the growth of many bacteria on ethanolamine in the presence of exogenous vitamin B12 (5). It is the first enzyme in the ethanolamine-degradative pathway of the bacteria carrying the ethanolamine utilization (eut) operon (6). The eut operon encodes the components of the carboxysome-like ethanolamine utilization microcompartment, a proteinaceous organelle containing the proteins required for ethanolamine degradation (7, 8). Inhibitors of the pathway are of interest because considerable numbers of pathogens that carry the ethanolamine utilization genes are causative agents of food poisoning (9).
Clostridial EAL has been investigated extensively (10), together with diol dehydratase (11), to establish the minimal mechanism of action of AdoCbl. Recombinant enzyme has been used primarily after the cloning of the Salmonella genes encoding EAL (12). Large (α) and small (β) subunits of EAL are encoded by the eutB and eutC genes, respectively, in the eut operon. Overexpression and purification of EAL from Salmonella typhimurium (13) and E. coli (14) have been achieved. The subunit structure was established as α6β6 (13–15). EPR studies have demonstrated that the enzyme binds cobalamin in the base-on mode, that is, with 5,6-dimethylbenzimidazole coordinating to the cobalt atom (14, 16, 17), as suggested from the fact that it does not contain a DXHXXG motif (12). Recent EPR, ENDOR, and electron spin echo envelope modulation (ESEEM) spectroscopy studies have revealed not only the nature of the radical species but also the geometry of reactant centers of this enzyme (18–21). The three-dimensional structure of this enzyme has been constructed from the x-ray structure of diol dehydratase by modeling using spectroscopic results and sequence comparison (22). Despite these intensive biochemical and spectroscopic studies, the detailed reaction mechanism of this enzyme is still unclear because of the lack of complete structural information on the active site. Therefore, the crystal structure of the enzyme has been long awaited to elucidate the radical reaction mechanism. Thus far, x-ray structures of EAL have not yet been available, although the structure of the EutB heterohexamer (α6) has been analyzed recently (Protein Data Bank ID, 2QEZ). To obtain crystals for x-ray analysis, we recently established methods for high level protein expression and simple purification of E. coli wild-type EAL (14). However, purified EAL protein precipitates at high concentrations with a concomitant loss of enzyme activity. Recently, we determined that N-terminal truncations of the β-subunit with the C34S mutation significantly improves EAL stability (14). These data indicate that a short N-terminal sequence is sufficient to change the solubility and stability of the enzyme.
In the present article, we report the crystallization and first x-ray structures of truncated EAL complexed with coenzyme B12 analogs and substrates. Mechanistically important insights obtained from these structures, which may give a hint to design inhibitors for the ethanolamine-degradative pathway of the food poisoning bacteria, are also described here.
EXPERIMENTAL PROCEDURES
Construction of Expression Plasmids
Plasmid pUSI2ENd(EAL), an expression plasmid for wild-type EAL of E. coli, was constructed as described previously (14). For expression of the truncated enzymes EAL(βΔ4–30) and EAL(βΔ4–43), which consist of the α-subunit and the N-terminal His6-tagged β-subunit lacking residues Lysβ4–Alaβ30 and Lysβ4–Cysβ43, respectively, pUSI2ENd(EAL(Δβ4–30) and pUSI2ENd(EAL(Δβ4–43) were constructed from pUSI2ENd(EAL) as follows. The entire region of pUSI2ENd(EAL) excluding Lysβ4–Alaβ30 was amplified by PCR using PfuTurbo DNA polymerase (Agilent Technologies) and 5′-phosphorylated primers b-4-30d6H_f (5′-CATCATCATCATCATCACACCACCAACTGTGCGGCACCGGTGACC-3′) and b-4-30d_r (5′-GCTGCTTTGATCCATGATATGTTATCTCCGCGTCATCAGAAGAAC-3′) (where underlined letters indicate the inserted His6 tag and linker sequence). The resulting blunt-end PCR product was self-ligated with Ligation High premixed T4 DNA ligase reagent (Toyobo) to generate a circular plasmid, pUSI2ENd(EAL(Δβ4–30). pUSI2ENd(EAL(Δβ4–43), which generates the Lysβ4–Alaβ43 deletion mutant, was obtained from the pUSI2ENd(EAL(Δβ4–30) construct using the QuikChange multi-site-directed mutagenesis kit (Agilent Technologies) with the 5′-phosphorylated primer b-43d (5′-CAAAGCAGCCATCATCATCATCATCACGCGCTGGATTTAGGTTCCGCTGAAGCA-3′). Each plasmid was transformed into E. coli JM109 cells.
Expression and Purification
Cultures of E. coli JM109 harboring the expression plasmid pUSI2ENd(EAL(Δβ4–43)) were inoculated to an A600 nm of 0.6. After a 2–3 h incubation at 30 °C, expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 0.5 mm, and growth was continued for 5 h at 30 °C. Cells harvested from 5-liter cultures were suspended in 50 mm potassium phosphate buffer (pH 8.0) containing 20 mm imidazole, 10 mm EA, 5 mm 2-mercaptoethanol, and 1 mm phenylmethanesulfonyl fluoride and disrupted by sonication. Cell lysates were centrifuged at 27,000 × g and applied to a nickel-nitrilotriacetic acid-agarose (Qiagen) column pre-equilibrated with the same buffer. The column was washed with the same buffer containing 40 mm imidazole, and EAL(Δβ4–43) was eluted with the buffer by increasing the imidazole concentration to 250 mm. The eluted protein was concentrated to ∼3 ml with an Amicon stirred pressure cell (Millipore) with a disc membrane (10-kDa cutoff). For the CN-Cbl·EA complex, CN-Cbl powder (Sigma) was added to the protein solution to a final concentration of 2 mm followed by incubation at 30 °C for 30 min. The CN-Cbl-bound enzyme was further purified and buffer-exchanged by size-exclusion chromatography on a Sephacryl SS-500 column (60 × 2.6 cm) pre-equilibrated with buffer (10 mm Tris-HCl buffer (pH 8.0) containing 200 mm KCl, 10 mm EA, 1 mm dithiothreitol, and 20 μm CN-Cbl). Fractions containing the enzyme were pooled and concentrated to 20 mg ml−1 for crystallization. The protein concentration was quantified using the Bio-Rad protein assay kit according to the manufacturer's protocol. For the substrate-free form of EAL, EA was removed prior to the addition of CN-Cbl by the following procedure. Concentrated nickel-nitrilotriacetic acid-purified enzyme was dialyzed against 20 mm potassium phosphate buffer (pH 8.0) containing 0.2 m KCl, 1 mm dithiothreitol, and 200 μm EA to reduce the concentration of substrate. To remove the remaining substrate, AdoCbl was added to the apoenzyme solution to 1% saturation, and the mixture was then incubated at 30 °C for 30 min. For the AdePeCbl·EA complex, size-exclusion chromatography was carried out in the absence of cobalamin. The AdePeCbl-bound enzyme was then prepared in a manner similar to the CN-Cbl complex followed by dialysis against cobalamin-free buffer to remove excess AdePeCbl.
Crystallization and Data Collection
Crystals of EAL(Δβ4–43) were obtained with the sitting drop vapor diffusion method in which 2 μl of protein solution (∼20 mg/ml) was mixed with an equal volume of well solution on a Greiner 96-well CrystalQuick sitting drop plate (Greiner Bio-One, Germany) and equilibrated against 100 μl of the well solution. The final optimized well solution for the CN-Cbl·EA complex crystals was 6.0–7.0% (w/v) polyethylene glycol 6000, 24–26% (v/v) glycerol, 5.0% (v/v) 2-propanol, and 0.1 m HEPES-NaOH (pH 7.0). For the substrate-free form and AdePeCbl·EA complex crystals, the optimized well solution was 6.0–7.0% (w/v) polyethylene glycol 4000, 24–26% (v/v) glycerol, 1.0% (v/v) 2-methyl-2,4-pentanediol, and 0.1 m imidazole-HCl (pH 6.3). To obtain the CN-Cbl·2-amino-1-propanol (PA) complex crystals, substrate-free crystals were harvested and soaked for 30 min in the mother liquor supplemented with 10 mm racemic PA (pH 8.0) as well as each component of the substrate-free final buffer. X-ray diffraction data collections were performed at the SPring-8 (Hyogo, Japan) beamlines BL38B1 for the CN-Cbl·EA complex and BL41XU for the substrate-free form and at the Photon Factory BL-17A beamline (Tsukuba, Japan) for the CN-Cbl·PA and AdePeCbl·EA complexes. Prior to diffraction experiments, the crystals were flash-cooled with a nitrogen gas stream at 100 K. Diffraction data sets were indexed, integrated, and scaled with the HKL2000 program (23). The details of the diffraction experiments are summarized in Table 1. Diffraction data from the CN-Cbl·EA complex crystals were collected up to 2.10 Å for native and 2.80 Å for the single wavelength anomalous dispersion (SAD) data at the peak wavelength of cobalt. The space group was P63, and the unit cell parameters of the native form were a = b = 242.76 Å and c = 76.46 Å. Assuming two (αβ) units (Mr = 79,500 including CN-Cbl) in the asymmetric unit, the Matthews coefficient was estimated to be 4.09 Å3Da−1, corresponding to a solvent content of 70%. The other forms, i.e. the CN-Cbl·PA complex, the AdePeCbl·EA complex, and the CN-Cbl substrate-free form, also had the same space group, similar cell constants, and were diffracted to 2.05–2.25 Å resolution. Details for the data statistics of the x-ray crystallography experiments are shown in supplemental Table S1.
Structure Determination and Refinement
The molecular replacement trial for the CN-Cbl·EA complex using the α6 complex of Listeria monocytogenes EAL (Protein Data Bank ID, 2QEZ) provided one clear solution and interpretable electron densities in the α-subunit and cobalamin regions, but we could not built a model for the β-subunit. An anomalous difference Fourier map of the cobalt-SAD data with the molecular replacement model phases displayed a clear peak at each of the three cobalamin sites in the asymmetric unit. SAD phasing was performed with SHARP (24) using the three cobalt sites followed by density improvement with SOLOMON and DM from the CCP4 suite (25). The density-modified map obtained from the SAD method was not interpretable for the entire molecule. However, combining the molecular replacement and SAD phases dramatically improved the quality of the electron density map (supplemental Fig. S1). The initial model, including both α- and β-subunits and cobalamin, was built with COOT. The model was improved by iterative rounds of refinement using REFMAC5 (26) and model rebuilding using COOT (27) until the Rfree value decreased to less than 30%. At this stage, TLS refinement using REFMAC5 was applied to the model, and several further refinement cycles yielded the final model. For the other complexes, the model of CN-Cbl·EA complex was used as the initial model, and a similar procedure was applied for refinement of the model. Refinement statistics are listed in supplemental Table S1. The model figures were generated with MolScript (28)/Raster3D (29) for Fig. 1B and supplemental Fig. S2C and with CHIMERA (30) for all color other figures.
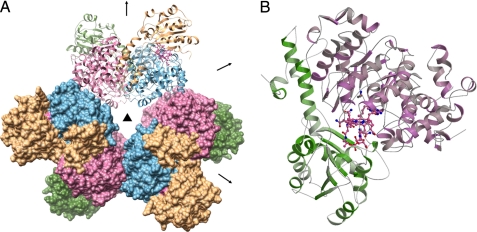
FIGURE 1.
Overall architecture of EAL. A, overall (αβ)6 structure of EAL. One (αβ)2 dimer and the other two dimers are shown as ribbon and surface models, respectively. The α- and β-subunits are shown in pink/sky blue and dark green/khaki, respectively. The crystallographic 3-fold axis and non-crystallographic 2-fold axis are shown by a triangle and arrows, respectively. B, overall fold of the αβ unit of the EAL·AdePeCbl·EA complex. AdePeCbl and EA are shown as ball-and-stick models.
RESULTS AND DISCUSSION
Overall Architecture
E. coli EAL(Δβ4–43) was crystallized in the P63 space group with two α (A and C chains) and two β (B and D chains) subunits in the asymmetric unit. The structures of EAL complexed with CN-Cbl·EA, AdePeCbl·EA, CN-Cbl·PA, and CN-Cbl alone (substrate-free) were refined to crystallographic Rwork/Rfree factors of 0.240/0.266 at 2.10 Å resolution, 0.214/0.244 at 2.25 Å resolution, 0.230/0.269 at 2.05 Å resolution, and 0.248/0.285 at 2.05 Å resolution, respectively (supplemental Table S1). The minimal unit of functional EAL is composed of the α- and β-subunits and a cobalamin cofactor. This composition, the αβ unit, is conserved in most of the other base-on AdoCbl-dependent enzymes except class II ribonucleotide reductase (31). The most striking feature of the overall structure of EAL is that this enzyme is a hexamer of the αβ unit (32–36). In contrast, the other known AdoCbl-dependent enzymes are monomeric or dimeric with respect to the αβ units. EAL has a propeller-like shape when viewed along the crystallographic 3-fold axis (Fig. 1A). Six α-subunits form a ring core, and each β-subunit projects out from its adjacent α-subunit. The adjacent two αβ units are related by a non-crystallographic 2-fold axis perpendicular to the crystallographic 3-fold axis. Each αβ unit has two contact areas with the adjacent αβ units. One is a face-to-face contact and the other is an edge-to-edge contact. The buried surface area of the face-to-face interface (2981 Å2) is considerably larger than that of the edge-to-edge interface (803 Å2). The large difference between the buried surface areas suggests that EAL can be considered a trimer of the (αβ)2 dimer arrangement in which each αβ unit is associated through face-to-face contacts. The (αβ)2 dimer is reminiscent of B12-dependent diol and glycerol dehydratases, although their overall arrangements are different (supplemental Fig. S2A). Interestingly, the edge-to-edge interface corresponds to the interface between the α- and γ-subunits of diol and glycerol dehydratases (supplemental Fig. S2B).
Structure of the αβ Unit
The α-subunit is folded into a (β/α)8 or triose-phosphate isomerase (TIM) barrel-based structure (Fig. 1B), which is similar to other AdoCbl-dependent enzymes (32–36). The C-terminal side of the barrel forms a hollow that accommodates a substrate molecule. The upper side of the corrin ring of a cobalamin molecule faces and closes the hollow. The β-subunit covers the lower part of the cobalamin molecule. The overall fold of the β-subunit is similar to the corresponding subunits or domains of other base-on and base-off types of cobalamin enzymes. Compared with the β-subunit of diol dehydratase, the EAL β-subunit has extra helices in the N-terminal region (residues β44–107) (Fig. 1B and supplemental Fig. S2C), which fill the groove between two adjacent α-subunits within the (αβ)2 dimer and contribute to the face-to-face contact.
Binding of Cobalamin
The cobalamin molecule is bound in the interface of the α- and β-subunits (Fig. 1B), as found in diol and glycerol dehydratases (33, 35), glutamate mutase (34), and lysine 5,6-aminomutase (36). Each side-chain amide group has at least one hydrogen bonding partner. The phosphate group of the nucleotide loop forms a salt bridge with Argβ206. A similar salt bridge between the nucleotide phosphate group and the protein has been found in other base-on type enzymes such as diol dehydratase (Lysβ135) and glycerol dehydratase (Lysβ102). Drennan and co-workers (31) have reported the crystal structures of another base-on type enzyme, class II ribonucleotide reductase, with and without cobalamin. Although only the structure of the cobalamin-free form is available in the Protein Data Bank, they mentioned in their article (31) that Arg33 has contacts with cobalamin. Upon comparison of the figures in that article with the structure of the cobalamin-free form, Arg33 seems to be adjacent to the phosphate group of the nucleotide loop. In contrast, a similar salt bridge with the nucleotide moiety is not present in the known structures of base-off enzymes such as methionine synthase (37), methylmalonyl-CoA mutase (32), glutamate mutase (34), and lysine 5,6-aminomutase (36).
The peak of electron density at the CN− position was not high enough to build the CN group into the final model, which is likely because of the high energy dose of x-ray radiation, as observed previously in diol dehydratase and other cobalamin-dependent enzymes. The bond distances between Co and N3 of 5,6-dimethylbenzimidazole moiety are 2.39 Å (EA-bound form) and 2.31 Å (PA-bound form). These distances are within the range of the bond lengths (2.18–2.50 Å) observed in other CN-Cbl-bound enzymes. They are significantly longer than those in free CN-Cbl and closer to those in cob(II)alamin. It is therefore likely that the Co–CN bond undergoes reductive cleavage during x-ray irradiation. Pyrrole rings A and B, including the b-propionamide side chain and the methyl group on C6 of 5,6-dimethylbenzimidazole, are exposed to solvent (supplemental Fig. S3). The cavity is ∼5 Å in height and ∼15 Å in width. Diol dehydratase also has a similar cavity at the corresponding site. Substrates may enter the active site through this cavity. In addition, the existence of such a cavity might be essential for the release of damaged cofactor, which is mediated by their respective molecular chaperone-like reactivating factors (diol dehydratase-reactivating factor (DDR) for diol dehydratase and EAL-reactivating factor for EAL). The structures of the ADP-bound and nucleotide-free forms of DDR indicate that nucleotide binding drives significant conformational changes in DDR (38), which would promote subunit swapping and lead to destabilization of the intersubunit interactions of diol dehydratase. Movement of the α-subunit of diol dehydratase with respect to the β-subunit upon subunit swapping is estimated to be ∼6 Å based on the DDR·diol dehydratase complex model, which is less than the size of cobalamin (∼10 Å) and therefore not large enough for cobalamin to leave the binding site. However, considering the height of the pre-existing cavity (∼5 Å), the ∼6 Å movement of the α-subunit would form a cavity 11 Å in height, which is comparable with or even larger than the size of cobalamin, thereby allowing the damaged cobalamin cofactor to pass through it.
Adenine Ring-binding Site and Its Vicinity
When the x-ray structure of the EAL·AdePeCbl complex was analyzed, the difference electron density for the adenine ring was clear enough to build its model (supplemental Fig. S4), although the pentamethylene group, particularly at the Co-binding end, was not clear. This might be because the amber light used for crystal centering before x-ray data collection was too bright to keep the Co–C bond intact. Another possibility is that x-ray irradiation broke the bond; the total x-ray dose to the EAL crystal was much higher than that given to the diol dehydratase crystal. The Co–C bond distance obtained was 2.53 Å, which was 0.31 Å longer than that of the diol dehydratase-bound AdePeCbl (39). The flexibility of the pentamethylene group of AdePeCbl enables it to keep its Co–C bond intact even in the substrate-bound enzyme, and therefore the AdePeCbl-bound form should mimic the post-homolysis state. Indeed, diol dehydratase complexed with AdePeCbl and substrate did not change the position and orientation of the adenine ring upon Co–C cleavage by photo-irradiation (39, 40). Therefore, we expected that the adenine ring in the present structure would keep the same position and orientation as that in the intact AdePeCbl-bound enzyme.
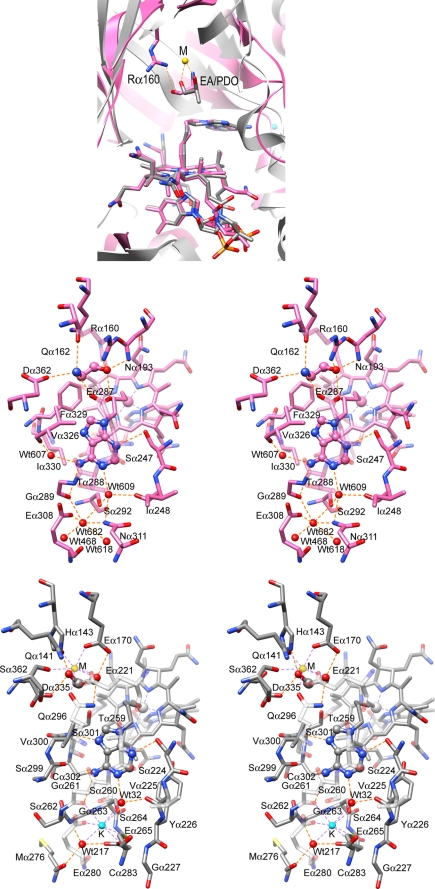
The adenine ring of AdePeCbl shares the same site in complexes with both EAL and diol dehydratase, which sits on the methyl group on C12 of the pyrrole ring C (Fig. 2A). This methyl group is thought to be important for Co–C bond cleavage by propping up the adenine ring (39, 41). As in free AdoCbl, the adenine ring of AdePeCbl is bound to EAL almost parallel to the corrin ring but with the other side facing the pyrrole ring C. This mode of adenine binding is similar to that observed in diol dehydratase. EAL forms fewer hydrogen bonds with the adenine ring than with diol dehydratase. The N1, N3, and 6-NH2 nitrogen atoms of the adenine ring of AdePeCbl have at least one hydrogen bond with EAL, namely N1-Wat609, N3-Serα247, 6-NH2-Glyα289, and 6-NH2-Wat607, whereas N7 and N9 atoms are not involved in hydrogen bonds (Fig. 2B). In diol dehydratase, each nitrogen atom of the adenine-ring except N9 has at least one partner for hydrogen bonding (Fig. 2C), whereas in EAL, a hydrogen bonding partner is also missing for N7 and the 6-NH2-Gα289 hydrogen bond is considerably longer (3.38 Å) than the corresponding 6-NH2-Gα261 hydrogen bond in diol dehydratase (3.10 Å). Moreover, EAL has less van der Waals contacts with the adenine ring than diol dehydratase (10 contacts with Serα247, Gluα287, Thrα288, Glyα289, Valα326, and Pheα329 versus 12 with Serα224, Thrα259, Glyα261, Valα300, and Serα301). These data suggest that EAL interacts with the adenine ring of AdoCbl less strongly than diol dehydratase.
FIGURE 2.
Schematic drawings showing the cobalamin-binding and substrate-binding sites of EAL. A, overall view of the cobalamin-binding and substrate-binding sites of EAL and diol dehydratase superimposed on their corrin rings of AdePeCbl. Detailed structures of the adenine-binding site of the AdePeCbl·EA complex of EAL (B) and diol dehydratase (C) are shown. M, substrate-coordinated metal ion, was recently identified as calcium (59).
Diol dehydratase has a monovalent cation-binding site near the adenine ring-binding site (39). Interestingly, the corresponding site of EAL shares a similar structure with that of diol dehydratase. Indeed, there is an electron density peak at the position corresponding to the monovalent cation site of diol dehydratase. The level of the electron density is the same as that of the oxygen atoms of the adjacent water molecules, suggesting that the peak is likely derived from a water molecule or an ammonium, sodium, or magnesium ion. A potassium ion did not bind to this site, despite the fact that it was present at a significantly high concentration (50–200 mm) throughout the purification, crystallization, and x-ray data collection. The surrounding six atoms are within the typical hydrogen bonding distance range, and the distances are considerably longer than typical coordination distances for Na–O (42) and Mg–O bonds (43): Glyα289 O (2.80 Å), Serα292 OG (3.00 Å), Gluα308 OE1 (2.86 Å), Asnα311 ND2 (3.19 Å), Wat468 (2.65 Å), and Wat618 (2.84 Å), which correspond to Glyα261 O (2.84 Å), Serα264 OG (2.98 Å), Gluα280 OE1 (2.85 Å), Cys283 SG (3.42 Å), Wat217 (3.00 Å), and Glu265 OE1 (2.65 Å) in diol dehydratase, respectively. These coordination distances exclude the possibility that the peak is due to a sodium or magnesium ion. We assigned a water molecule, not an ammonium ion, to the peak because E. coli EAL does not require monovalent cations for activity and ammonium ions were not added throughout the purification and crystallization procedures. Considering the differences in charge of the surrounding amino acid residues between these enzymes, it is possible that the role of the potassium ion in diol dehydratase is substituted by a water molecule in EAL.
Substrate-binding Site
The electron density for the EA molecule looks symmetrical because nitrogen and oxygen atoms are indistinguishable and hydrogen atoms are undetectable by the x-ray diffraction method at the present resolution (2.1–2.3 Å). Addressing this problem, racemic PA, an asymmetric substrate, was also used for crystal structure analysis to unambiguously determine the orientation of substrate. As compared with EA, PA has an extra electron density branched from the C2 atom (supplemental Fig. S5). The best fit model for PA displays a C1-downward orientation with respect to cobalamin. Biochemical experiments by Babior and co-workers (58) showed that both enantiomers of PA bind to the enzyme with different kinetic parameters, a faster kcat and lower Km for the (S)-enantiomer and slower kcat and higher Km for the (R)-enantiomer. Therefore, we expected that the (S)-enantiomer would be predominantly bound to the active site even if the PA-bound crystal was obtained in the presence of racemic compound. However, the electron density map did not clearly indicate which enantiomer was predominantly bound. The present model for the CN-Cbl/PA-bound form was determined as the (S)-enantiomer-bound form, but the crystal may also contain the (R)-enantiomer-bound form.
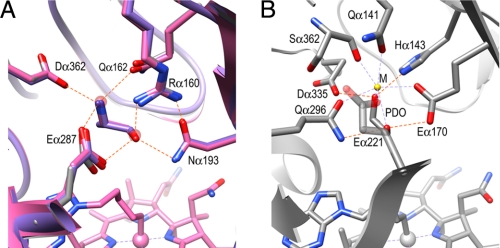
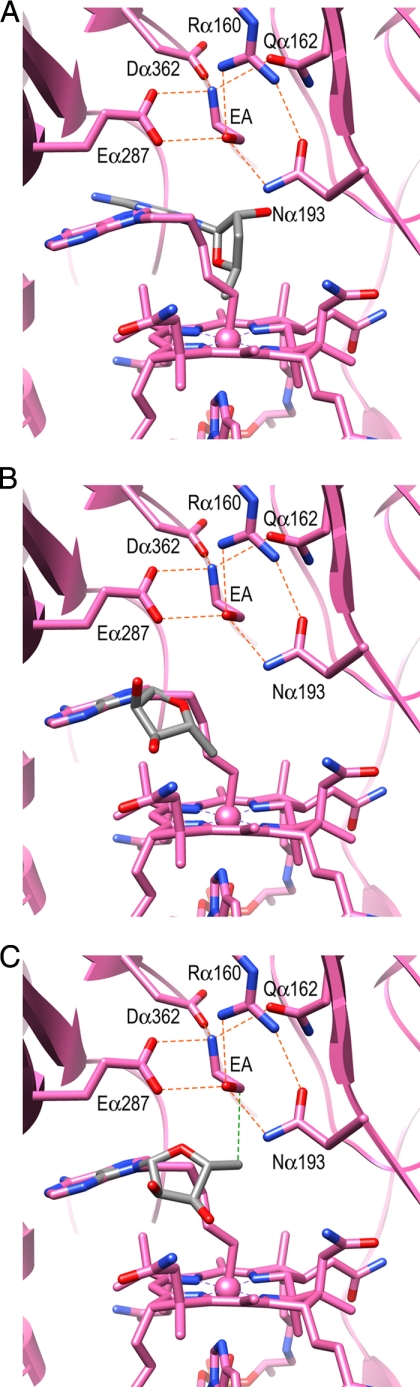
The substrate is bound by six hydrogen bonds with Argα160, Glnα162, Asnα193, Gluα287, and Aspα362. The Gluα287 carboxylate is a bidentate ligand with which two hydrogen bonds with the O1 and N2 atoms of substrate are formed (Fig. 3A). The O1 atom forms three hydrogen bonds with Argα160, Asnα193, and Gluα287, and the N2 atom forms three hydrogen bonds with Glnα162, Gluα287, and Aspα362. These hydrogen bonds are conserved in diol dehydratase, although their chemical characteristics and spatial arrangements are somewhat different. Asnα193 and Gluα287 correspond chemically to Glnα296 and Gluα170 in diol dehydratase, but their positions are reversed (Fig. 3B). Aspα362 corresponds to Aspα335, but Glnα162 is replaced with Hisα143 in diol dehydratase. It should be noted that Argα160 corresponds to the metal ion in diol and glycerol dehydratases, as predicted previously by Sun et al. (22, 44). Argα160 provides a positive charge under neutral and weakly alkaline conditions. Argα160 is located at a typical hydrogen bond distance from the O1 atom of the substrate but at the upper limit for a hydrogen bond with the N2 atom: O1/N2, 2.84/3.24 Å (CN-Cbl·EA); 2.71/3.45 Å (CN-Cbl·PA); 2.98/3.24 Å (AdePeCbl·EA). The direction of N2 with respect to the guanidinium group of Argα160 is also not suitable for hydrogen bonding: N-H–N2 bond angle, 110°; N-H–O1 bond angle, 168°. Thus, Argα160 seems to contribute to substrate binding by hydrogen bonding with the O1 atom without a significant contribution from the Argα160-N2 interaction, which is consistent with the fact that the three available hydrogen bonding positions on the N2 atom are already occupied with the three residues, Glnα162, Gluα287, and Aspα362, mentioned above. In the case of diol dehydratase, both hydroxyl groups of the substrate are coordinated to the metal ion (Figs. 2C and 3B).
FIGURE 3.
Substrate-binding site. A, superimposed models of the EA-bound (pink), (S)-PA-bound (violet), and substrate-free (gray) forms of EAL. Two water molecules bound at the substrate-binding site are shown with a transparent red sphere. B, diol dehydratase. M, substrate-coordinated metal ion, was recently identified as calcium (59).
The guanidinium group of Argα160 is strongly held by hydrophilic interactions with four residues, Glyα191, Asnα193, Tyrα285, and Gluα287. This supports the recent report by Sun et al. (44) that the alanine point mutation of Argα160 completely diminishes the activity of S. typhimurium EAL, and that addition of extrinsic guanidinium significantly restores the activity. The cavity generated by the point mutation could bind a guanidinium ion through which the catalytically active orientation of the bound substrate might be maintained. It was also reported that the Rα160K mutant showed significant activity. A modeling study based on the present structure suggests that the Lys160 substrate distance is only ∼3.5 Å, which is comparable with the wild-type enzyme and shorter than the value estimated of Sun et al. (44).
Protonated State of Substrates
One of the intriguing questions is whether the amino group of the substrate is positively charged (substituted ammonium) or neutral. If protonated, the amino group of the substrate serves as a proton donor for three hydrogen bonds, but if its neutral, then it should be a proton donor for two hydrogen bonds and a proton acceptor for one. Protonation of either Gluα287 or Aspα362 is unlikely because they are in a relatively hydrophilic environment. If the side-chain NH2 group of Glnα162 is directed toward the amino group, it becomes a proton donor, which would lead to the loss of its hydrogen bonds with the SD atom of Metα394, the carboxylate of Aspα399, and the OH atom of Tyr404 (supplemental Fig. S6). These data suggest that Glnα162 is a proton acceptor for the hydrogen bond. In contrast, the proton donor for the corresponding hydrogen bond in diol dehydratase seems to be Hisα143, which has been suggested to assist in the migration and elimination of the O2 hydroxyl group by hydrogen bond donation (45).
Consistency with EPR Studies
Superimposing the structures of EAL and diol dehydratase on their cobalamins indicates that the C1 atom of EA is shifted ∼0.6 Å along the C1–C2 bond of propanediol and that the C2 atom is lifted up toward the carboxylate of Gluα287 and twisted clockwise, as viewed from the cobalamin molecule (Fig. 2A). The Co–C1 and Co–C2 distances of EAL are as follows: CN-Cbl·EA, 8.59 and 9.28 Å; AdePeCbl·EA, 8.53 and 9.19 Å; CN-Cbl·PA, 8.74 and 9.18 Å, respectively, which are comparable with those of diol dehydratase (8.64 and 9.19 Å). The distances between Co(II) and the organic radical from EA and PA were estimated to be 9–10 Å (46, 47) and 10–12 Å (20, 21, 44), respectively, by EPR simulations with S. typhimurium EAL. The organic radicals formed from EA and PA were identified as 2-amino-1-ethanol-l-yl (48) and 2-amino-1-propanol-1-yl radicals, respectively, both of which are C1-centered substrate radicals. Therefore, the Co–C1 distance obtained with E. coli EAL in the present study is in reasonable agreement with the EPR simulations with the S. typhimurium enzyme. A slight discrepancy in their Co–C1 distances by 1–3 Å might be because (i) reacting EAL may undergo a conformational change, (ii) E. coli and S. typhimurium EALs may have minor differences in the position and orientation of enzyme-bound substrate, or (iii) there may be difference in techniques.
Cleavage of the Coenzyme Co–C Bond
One of the central questions to be answered is how the Co–C bond of AdoCbl is cleaved upon its binding to EAL apoenzyme. When the cobalamin moiety of AdoCbl is superimposed on that of the EAL·AdePeCbl complex, the adenine ring of the coenzyme is positioned in a different direction from the adenine ring-binding site and cannot be superimposed on the site without cleaving the Co–C bond (Fig. 4, A and B) as in the case of diol dehydratase (39). The coenzyme adenine ring would be accommodated in the site, because substantial binding energy is obtained from four hydrogen bonds and van der Waals contacts (Fig. 2B). If both the cobalamin moiety and the adenine ring were superimposed on those of the enzyme-bound AdePeCbl with the Co–C bond cleaved and the Co–C distance were kept at a minimum, then the Co–C distance would have to be elongated to 3.5 Å and the Co–C bond would have to lean toward the nitrogen atom of pyrrole ring B to the C5′–Co-N (ring B) bond angle of 61° (84° for free AdoCbl) and the Co–C5′-C4′ bond angle of 150° (124° for free AdoCbl) (Fig. 4B). Therefore, it is evident that marked angular strains and tensile forces induced by tight interactions of the coenzyme with the enzyme cobalamin-binding site and the adenine ring-binding site inevitably break the Co–C bond. Thus, the steric strain model of Co–C bond cleavage for diol dehydratase seems to be applicable to EAL as well. In addition, the AdoCbl-bound model of EAL suggests that Gluα287 has a direct contact with the 2′-hydroxyl group of the ribose moiety (Fig. 4B), which may also contribute to Co–C bond cleavage by stabilizing the post-homolysis state.
FIGURE 4.
AdoCbl-bound model of EAL based on the AdePeCbl-complexed structure. A, without cleavage of the Co–C bond. B, superimposed on the adenine rings of AdePeCbl and AdoCbl so that the Co–C distance is kept at a minimum. C, a model for the hydrogen abstraction step.
Modeling of the Adenosyl Radical in the Hydrogen-abstracting Conformation
On the basis of the AdePeCbl-bound structure, the adenosyl radical in the productive (distal) conformation can be modeled by superimposing its adenine ring on the adenine ring-binding site and by rotating the ribosyl group around the N-glycosidic bond. Warncke and Utada (19) reported the distance between C5′ of 5′-deoxyadenosine and C1 of substrate radical to be 3.2 Å, which was estimated from the substrate radical-5′ methyl group model by electron spin echo envelop modulation (ESEEM) simulation. If the C5′-C1 distance of the model is set at this distance, the conformation obtained seems to be quite reasonable and suitable for hydrogen abstraction from substrates. The pro-S hydrogen atom of the substrate lies almost on the C5′-C1 line (Fig. 4C), which is consistent with the stereochemical study by Diziol et al. (49) using labeled PA. In this conformation, the ribose ether oxygen atom of 5′-deoxyadenosine has direct contacts to the carboxylate of Gluα287 and the hydroxyl group of the substrate with the distances of 2.9 and 3.3 Å, respectively, which are within the O–H-O hydrogen bond distance range. The former contact would generate O–O repulsion because protonation of neither ether oxygen nor carboxylate is likely, and the latter one would contribute to ribosyl rotation by attracting the ribose group through an O–H-O hydrogen bond.
Substrate Switch
The structures of two AdoCbl-dependent enzymes, methylmalonyl-CoA mutase and diol dehydratase, reveal that substrate binding causes significant conformational changes at the overall fold level (50, 51). The bound substrate pushes Glnα336 and Pheα374 toward pyrrole rings A and D of the corrin ring in diol dehydratase. As a result, this enzyme displays movement of cobalamin and the β-subunit with respect to the α-subunit upon substrate binding, which causes the tilting of cobalamin. This is considered responsible for the substrate switch or substrate-induced additional labilization of the Co–C bond (51). To test this possibility, the structure of the substrate-free form of EAL was also analyzed. However, EAL did not show a significant structural difference between the substrate-bound and substrate-free forms at the overall fold level (Fig. 5A). In the active site of the substrate-free form, two water molecules are bound at the positions corresponding to the O1 and N2 atoms of substrate (Fig. 5B). Another major structural change is that the side chain of Gluα287 was almost invisible in the electron density map, which implies that this residue is not fixed at the substrate-binding site. This might be because the bound water molecule has only two hydrogen atoms available for hydrogen bond donation as compared with the N2 of the substrate. Although the Co–C bond of the substrate-free holoenzyme is also distorted and already activated, its dissociation is unfavorable because the energy of adenosyl radical is very high. The fixation of Gluα287 in the presence of substrate would labilize the Co–C bond to some extent by stabilizing the post-homolysis state through direct contact with the ribose 2′-hydroxyl group. This could be the reason that EAL does not need a major structural change for additional labilization of the Co–C bond. The interaction between substrate and the ribose ether oxygen would assist in the access of the adenosyl radical to substrates (Fig. 4C), which could help the hydrogen abstraction from substrate. In the presence of substrate, the equilibrium is shifted in favor of Co–C bond dissociation, because the energy of the substrate radical is much lower than that of the adenosyl radical. The Co–C bond in the substrate-free form of diol dehydratase is also distorted and already activated. Substrate binding increases its steric distortion, which is an entity of the substrate-induced additional labilization of the Co–C bond (51). Apparent substrate triggering of the Co–C bond cleavage would be due to the shift of equilibrium toward the formation of much more stable substrate radical. The Co–C bond cleavage of AdoCbl and 3′,4′-anhydro-AdoCbl by this enzyme in the absence of substrate, but in the presence of potassium ion, has been reported (52–54), although the cleavage rates are much slower than in the presence of substrate.
FIGURE 5.
Structure of the substrate-free form. A, Cα trace models of substrate-free (colored) form and AdePeCbl·EA complex (gray) superimposed on their Cα atoms. B, σA-weighted Fo − Fc omit map at the active site of the substrate-free form.
Proposed Mechanism of Action of EAL
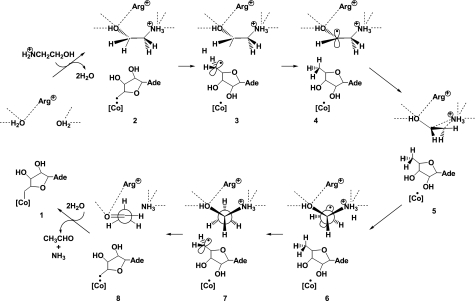
On the basis of the x-ray structures, we have proposed the refined overall mechanism for EAL (Fig. 6). This is consistent with the earlier proposed minimal mechanism (55) but not with the mechanism involving a second intermediate hydrogen carrier (56). When substrate is added, its hydroxyl and amino groups displace the two water molecules in the active site. The Co–C bond of the coenzyme is already activated but not yet cleaved significantly in the resting holoenzyme. The addition of substrate shifts the equilibrium toward the formation of an adenosyl radical and cob(II)alamin (57). The C5′ radical center is far away from the substrate but comes close to the substrate by rotation of the ribosyl moiety around the glycosidic linkage. Then, the radical abstracts the nearest hydrogen atom from substrate C1, forming a substrate-derived radical and 5′-deoxyadenosine. Although there is no direct evidence, the substrate radical could undergo amino group migration through a cyclic transition state analogous with the diol dehydratase reaction. During this step, the positions of the hydroxyl and amino groups would be maintained by interactions with active-site residues. As a result, the C1–C2 bond would turn around, and the new C2 radical center of the product radical would come close to the C5′ methyl group of 5′-deoxyadenosine, resulting in hydrogen back-abstraction. Then, the rearranged product 1-amino-1-ethanol would undergo elimination of ammonia to form the final product acetaldehyde. The products would leave the active site through the displacement by water molecules, which would shift the equilibrium in favor of recombination of the adenosyl radical and cob(II)alamin to regenerate the coenzyme. Fig. 6 was drawn on the basis of the assumption that the amino group of the substrate is protonated. However, this does not rule out the possibility that the protonation state of the substrate might change during reaction.
FIGURE 6.
Proposed overall mechanism of the EAL reaction. [Co], cobalamin; Ade, 9-adeninyl group; Arg+, protonated Argα160.
Supplementary Material
This work was supported by grants from the Global Centers of Excellence (COE) Program (to N. S. and Y. H.) and Grants-in-aid for Scientific Research 18GS0207 and (B)22370061 (to Y. H.) and (B)13480195, 17370038, and Priority Areas 513 (to T. T.) from the Japan Society for the Promotion of Science and the Ministry of Education, Culture, Sports, Science and Technology, Japan. This work was also supported by a grant from the Core Research for Evolutional Science and Technology (CREST) Program of the Japan Science and Technology Agency (to Y. H.) and a grant for natural sciences research assistance from the Asahi Glass Foundation, Tokyo, Japan (to T. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Table S1.
The atomic coordinates and structure factors (codes 3ABO, 3ABQ, 3ABS, and 3ABR) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- AdoCbl
- adenosylcobalamin (coenzyme B12)
- AdePeCbl
- adeninylpentylcobalamin
- CN-Cbl
- cyanocobalamin (vitamin B12)
- DDR
- diol dehydratase-reactivating factor
- EA
- ethanolamine
- EAL
- ethanolamine ammonia-lyase
- PA
- 2-amino-1-propanol
- SAD
- single-wavelength anomalous dispersion.
REFERENCES
- 1.Banerjee R. (1999) Chemistry and Biochemistry of B12, John Wiley & Sons, New York [Google Scholar]
- 2.Dolphin D. (1982) B12, John Wiley & Sons, New York [Google Scholar]
- 3.Bradbeer C. (1965) J. Biol. Chem. 240, 4675–4681 [PubMed] [Google Scholar]
- 4.Bradbeer C. (1965) J. Biol. Chem. 240, 4669–4674 [PubMed] [Google Scholar]
- 5.Chang G. W., Chang J. T. (1975) Nature 254, 150–151 [DOI] [PubMed] [Google Scholar]
- 6.Kofoid E., Rappleye C., Stojiljkovic I., Roth J. (1999) J. Bacteriol. 181, 5317–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sagermann M., Ohtaki A., Nikolakakis K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8883–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penrod J. T., Roth J. R. (2006) J. Bacteriol. 188, 2865–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H., Kristensen D. M., Coleman M. K., Mushegian A. (2009) PLoS One 4, e5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babior B. M. (1975) Acc. Chem. Res. 8, 376–384 [Google Scholar]
- 11.Abeles R. H., Dolphin D. (1976) Acc. Chem. Res. 9, 114–120 [Google Scholar]
- 12.Faust L. R., Connor J. A., Roof D. M., Hoch J. A., Babior B. M. (1990) J. Biol. Chem. 265, 12462–12466 [PubMed] [Google Scholar]
- 13.Faust L. P., Babior B. M. (1992) Arch. Biochem. Biophys. 294, 50–54 [DOI] [PubMed] [Google Scholar]
- 14.Akita K., Hieda N., Baba N., Kawaguchi S., Sakamoto H., Nakanishi Y., Yamanishi M., Mori K., Toraya T. (2010) J. Biochem. 147, 83–93 [DOI] [PubMed] [Google Scholar]
- 15.Wallis O. C., Johnson A. W., Lappert M. F. (1979) FEBS Lett. 97, 196–199 [DOI] [PubMed] [Google Scholar]
- 16.Abend A., Bandarian V., Nitsche R., Stupperich E., Rétey J., Reed G. H. (1999) Arch. Biochem. Biophys. 370, 138–141 [DOI] [PubMed] [Google Scholar]
- 17.Ke S. C., Torrent M., Museav D. G., Morokuma K., Warncke K. (1999) Biochemistry 38, 12681–12689 [DOI] [PubMed] [Google Scholar]
- 18.LoBrutto R., Bandarian V., Magnusson O. T., Chen X., Schramm V. L., Reed G. H. (2001) Biochemistry 40, 9–14 [DOI] [PubMed] [Google Scholar]
- 19.Warncke K., Utada A. S. (2001) J. Am. Chem. Soc. 123, 8564–8572 [DOI] [PubMed] [Google Scholar]
- 20.Canfield J. M., Warncke K. (2002) J. Phys. Chem. B 106, 8831–8841 [Google Scholar]
- 21.Bandarian V., Reed G. H. (2002) Biochemistry 41, 8580–8588 [DOI] [PubMed] [Google Scholar]
- 22.Sun L., Warncke K. (2006) Proteins 64, 308–319 [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24.de La Fortelle E., G., B. (1997) Methods Enzymol. 276, 472–494 [DOI] [PubMed] [Google Scholar]
- 25.Collaborative Computational Project (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 26.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 27.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28.Kraulis J. (1991) J. Appl. Crystallogr. 24, 946–950 [Google Scholar]
- 29.Merritt E. A., Bacon D. J. (1997) Methods Enzymol. 277, 505–524 [DOI] [PubMed] [Google Scholar]
- 30.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 31.Sintchak M. D., Arjara G., Kellogg B. A., Stubbe J., Drennan C. L. (2002) Nat. Struct. Biol. 9, 293–300 [DOI] [PubMed] [Google Scholar]
- 32.Mancia F., Keep N. H., Nakagawa A., Leadlay P. F., McSweeney S., Rasmussen B., Bösecke P., Diat O., Evans P. R. (1996) Structure 4, 339–350 [DOI] [PubMed] [Google Scholar]
- 33.Shibata N., Masuda J., Tobimatsu T., Toraya T., Suto K., Morimoto Y., Yasuoka N. (1999) Structure 7, 997–1008 [DOI] [PubMed] [Google Scholar]
- 34.Reitzer R., Gruber K., Jogl G., Wagner U. G., Bothe H., Buckel W., Kratky C. (1999) Structure 7, 891–902 [DOI] [PubMed] [Google Scholar]
- 35.Yamanishi M., Yunoki M., Tobimatsu T., Sato H., Matsui J., Dokiya A., Iuchi Y., Oe K., Suto K., Shibata N., Morimoto Y., Yasuoka N., Toraya T. (2002) Eur. J. Biochem. 269, 4484–4494 [DOI] [PubMed] [Google Scholar]
- 36.Berkovitch F., Behshad E., Tang K. H., Enns E. A., Frey P. A., Drennan C. L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15870–15875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drennan C. L., Huang S., Drummond J. T., Matthews R. G., Lidwig M. L. (1994) Science 266, 1669–1674 [DOI] [PubMed] [Google Scholar]
- 38.Shibata N., Mori K., Hieda N., Higuchi Y., Yamanishi M., Toraya T. (2005) Structure 13, 1745–1754 [DOI] [PubMed] [Google Scholar]
- 39.Masuda J., Shibata N., Morimoto Y., Toraya T., Yasuoka N. (2000) Structure 8, 775–788 [DOI] [PubMed] [Google Scholar]
- 40.Masuda J., Shibata N., Morimoto Y., Toraya T., Yasuoka N. (2001) J. Synchrotron Radiat. 8, 1182–1185 [DOI] [PubMed] [Google Scholar]
- 41.Toraya T. (2003) Chem. Rev. 103, 2095–2127 [DOI] [PubMed] [Google Scholar]
- 42.Harding M. M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 872–874 [DOI] [PubMed] [Google Scholar]
- 43.Harding M. M. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 401–411 [DOI] [PubMed] [Google Scholar]
- 44.Sun L., Groover O. A., Canfield J. M., Warncke K. (2008) Biochemistry 47, 5523–5535 [DOI] [PubMed] [Google Scholar]
- 45.Kamachi T., Toraya T., Yoshizawa K. (2004) J. Am. Chem. Soc. 126, 16207–16216 [DOI] [PubMed] [Google Scholar]
- 46.Canfield J. M., Warncke K. (2005) J. Phys. Chem. B 109, 3053–3064 [DOI] [PubMed] [Google Scholar]
- 47.Ke S. C. (2003) Biochim. Biophys. Acta 1620, 267–272 [DOI] [PubMed] [Google Scholar]
- 48.Bender G., Poyner R. R., Reed G. H. (2008) Biochemistry 47, 11360–11366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diziol P., Haas H., Rétey J., Graves S. W., Babior B. M. (1980) Eur. J. Biochem. 106, 211–224 [DOI] [PubMed] [Google Scholar]
- 50.Mancia F., Evans P. R. (1998) Structure 6, 711–720 [DOI] [PubMed] [Google Scholar]
- 51.Shibata N., Masuda J., Morimoto Y., Yasuoka N., Toraya T. (2002) Biochemistry 41, 12607–12617 [DOI] [PubMed] [Google Scholar]
- 52.Wagner O. W., Lee H. A., Jr., Frey P. A., Abeles R. H. (1966) J. Biol. Chem. 241, 1751–1762 [PubMed] [Google Scholar]
- 53.Schwartz P. A., Frey P. A. (2007) Biochemistry 46, 7293–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Magnusson O. T., Frey P. A. (2002) Biochemistry 41, 1695–1702 [DOI] [PubMed] [Google Scholar]
- 55.Babior B. M. (1988) Biofactors 1, 21–26 [PubMed] [Google Scholar]
- 56.O'Brien R. J., Fox J. A., Kopczynski M. G., Babior B. M. (1985) J. Biol. Chem. 260, 16131–16136 [PubMed] [Google Scholar]
- 57.Bandarian V., Reed G. H. (2000) Biochemistry 39, 12069–12075 [DOI] [PubMed] [Google Scholar]
- 58.Graves S. W., Fox J. A., Babior B. M. (1980) Biochemistry 19, 3630–3633 [DOI] [PubMed] [Google Scholar]
- 59.Toraya T., Honda S., Mori K. (2010) Biochemistry, in press [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.