Abstract
We previously demonstrated that RhoA-dependent signaling regulates transforming growth factor-β1 (TGF-β1)-induced cytoskeletal reorganization in the human retinal pigment epithelial cell line ARPE-19. Smad pathways have also been shown to mediate TGF-β1 activity. Here, we examined what regulates Rho GTPase activity and tested whether Smad signaling cross-talks with Rho pathways during TGF-β1-induced actin rearrangement. Using small interfering RNAs, we found that NET1, the guanine nucleotide exchange factor of RhoA, is critical for TGF-β1-induced cytoskeletal reorganization, N-cadherin expression, and RhoA activation. In ARPE-19 cells lacking NET1, TGF-β1-induced stress fibers and N-cadherin expression were not observed. Interestingly, in dominant-negative Smad3-expressing or constitutively active Smad7 cells, TGF-β1 failed to induce NET1 mRNA and protein expression. Consistent with these results, both dominant-negative Smad3 and constitutively active Smad7 blocked the cytoplasmic localization of NET1 and inhibited interactions between NET1 and RhoA. Finally, we found that NET1 is a direct gene target of TGF-β1 via Smad3. Taken together, our results demonstrate that Smad3 regulates RhoA activation and cytoskeletal reorganization by controlling NET1 in TGF-β1-induced ARPE-19 cells. These data define a new role for Smad3 as a modulator of RhoA activation in the regulation of TGF-β1-induced epithelial-mesenchymal transitions.
Keywords: Cytokines/TGFβ, Cytoskeleton, Retina, Rho, SMAD Transcription Factor, Rho GEF NET1, Smad3, Actin Rearrangement, Epithelial/Endothelial-Mesenchymal Transitions, Human Retinal Pigment Epithelium
Introduction
Members of the transforming growth factor β (TGF-β)2 superfamily are multifunctional cytokines that regulate cellular processes, including cell-cycle arrest, differentiation, morphogenesis, and apoptosis (1–5). TGF-β promotes extracellular matrix production and suppresses cell proliferation. Morphogenetic responses to TGF-β members include cell migration and epithelial-mesenchymal transitions (EMTs), which are critical during embryogenesis, development of fibrotic diseases, and advanced carcinoma spreading (4–8). The EMT is characterized by disassembly of cell-cell contacts, remodeling of the actin cytoskeleton, and cell-cell separation, resulting in fibroblast-like cells with mesenchymal marker expression and migratory properties (8–11). TGF-β is a major inducer of EMT in development, fibrosis, and carcinogenesis, with different isoforms mediating various effects depending on the specific cellular context (8, 12). Although the β1, β2, and β3 isoforms of TGF-β are present in mammalian tissues and demonstrate similar responses in vitro, their in vivo roles and expression patterns are not uniform. TGF-β1 was first described as an inducer of EMT in normal mammary epithelial cells (13) and has since been shown to mediate EMT in vitro in various epithelial cells, including renal proximal tubular, retinal, lens, and most recently alveolar epithelial cells (14–18).
In EMT-related retinal fibrosis, TGF-β can induce the transformation of retinal pigment epithelial (RPE) cells to myofibroblast-like cells in vitro (16, 19, 20), implicating TGF-β as a key player in the development of proliferative vitreoretinopathy (PVR). Various other growth factors, including platelet-derived growth factor, hepatocyte growth factor, and activin, are also reportedly involved in PVR pathogenesis (21–25). Moreover, TGF-β can induce numerous growth factors, including connective tissue growth factor, platelet-derived growth factor, fibroblast growth factors, vascular endothelial growth factor, and TGF-β1 itself (26, 27). All of these factors play important roles in normal tissue recovery after injury.
Many of the downstream pathways that mediate the effects of TGF-β1 are currently understood. Among the others, a few studies have suggested that the small GTPase Rho and its downstream effector Rho kinase (ROCK) mediate the TGF-β1-induced remodeling of mammary epithelial cell-cell contact (28). This is particularly interesting because Rho is a major cytoskeletal organizer (29–31). Rho regulates actin stress fiber formation by activating ROCK, which phosphorylates LIM kinase, which in turn phosphorylates cofilin. Cofilin binds both actin monomers and polymers and promotes actin filament disassembly; this function is suppressed by cofilin phosphorylation (32). Moreover, Rho can regulate gene expression (33–35). In particular, it is needed for constitutive smooth muscle actin expression in smooth muscle cells (36). We previously showed that TGF-β1 activates RhoA, thereby up-regulating ROCK1, down-regulating cofilin activity, and promoting the actin polymerization that may be responsible for the fibrotic response of RPE cells that can lead to PVR in vivo (15).
Like all GTPases, Rho proteins act as molecular switches by cycling between active (GTP-bound) and inactive (GDP-bound) states. Active GTPases interact with high affinity with one of several downstream effectors to modulate their activity and localization. The activation of Rho GTPases is regulated by specific guanine nucleotide exchange factors (GEFs), which catalyze the exchange of GDP for GTP. More than 60 GEFs for Rho GTPases have been identified in the human genome (37, 38). The NET1 gene, which encodes a specific GEF for Rho, was originally isolated in a tissue culture screen for novel oncogenes using the focus formation assay in NIH 3T3 fibroblasts (39, 40). NET1 encodes a 595-amino acid protein consisting of an N-terminal domain with a series of nuclear localization signals, a DH-PH domain, and a short C-terminal domain carrying a consensus PDZ-binding motif. NET1 contains a nuclear export signal in addition to nuclear import signals, strongly suggesting that it can be stimulated to exit the nucleus and activate cytoplasmic Rho (38).
Critical steps in intracellular TGF-β signaling pathways are mediated by Smad proteins. Briefly, TGF-β initiates its cellular response by binding to its specific receptor, TGF-β receptor II. After ligand binding, TGF-β receptor II activates TGF-β receptor I kinase, which phosphorylates receptor-regulated Smads. These activated receptor-regulated Smads form oligomeric complexes with a common Smad. The oligomeric complexes then translocate into the nucleus, where they regulate target gene transcription either directly by binding to DNA or indirectly by interacting with various cofactors. TGF-β can also stimulate inhibitory Smads, which negatively regulate TGF-β signaling transduction (41, 42). Among mammalian receptor-regulated Smads, Smad2 and Smad3 are specific for TGF-β/activin, whereas Smads 1, 5, and 8 are specific for bone morphogenic protein. Smad4 is the only known common Smad. Smad6 (the preferential inhibitor of bone morphogenic protein signaling) and Smad7 (a potent inhibitor of both TGF-β and bone morphogenic protein signaling) are inhibitory Smads (43–45). Smad-independent signaling transduction pathways are also involved in the biological activities of TGF-β (46, 47).
Because the Smad pathway principally regulates gene expression, it was originally thought that non-Smad effectors signal the rapid or direct effects of TGF-β on the actin cytoskeleton. However, we now understand that Smads are crucial mediators of processes downstream of TGF-β, because they induce dramatic changes in gene expression in epithelial cells (2, 48–50). Furthermore, in a recent microarray screening for microRNAs that are up- or down-regulated by TGF-β in epithelial NMuMG cells, microRNA-155 mediated TGF-β/Smad-induced EMT through the targeting of RhoA (51). These observations led us to hypothesize that the Smad pathway may play an important role in TGF-β1-induced Rho activation by modulating Rho GEFs.
In the present study we focused on the regulatory role of the TGF-β1/Smad pathway in Rho GTPase activation and actin cytoskeletal reorganization. To elucidate the mechanisms by which TGF-β1 regulates cell morphological changes, we used a Smad3 cDNA construct and small interfering RNA (siRNA) to target NET1. We suggest that the Smad signaling pathway plays a critical role in TGF-β1-induced actin rearrangement by regulating the RhoA GTPase. In addition, using an siRNA for NET1, we studied the role of NET1 in the regulation of RhoA and actin rearrangement. Finally, we demonstrated that Smad signaling pathways regulate RhoA during EMTs by modulating NET1 expression and localization. Our results provide evidence for a novel signaling mechanism for TGF-β1-induced cytoskeletal reorganization that is mediated by Smad proteins and Rho GTPases and may regulate EMTs.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Human recombinant TGF-β1 was purchased from R&D Systems (Minneapolis, MN). Specific inhibitors of MEK (PD98059), Akt (Triciribine), and phosphatidylinositol 3-kinase (LY294002) were obtained from Calbiochem. Antibodies used in Western blot analysis and immunocytochemistry include anti-phospho-SMAD2/3 and anti-N-cadherin (Cell Signaling, Beverly, MA), anti-NET1 (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-actin (Sigma). Rhodamine- and Alexa-labeled phalloidin were purchased from Molecular Probes (Eugene, OR). RhoA and Rac1 assay reagents were from Upstate Biotechnology (Temecula, CA).
Cell Culture and Treatments
ARPE-19 cells were obtained from American Type Cell Culture (Rockville, MD) and maintained at 37 °C, 5% CO2 in Dulbecco's modified Eagle's medium/F-12 medium supplemented with 10% fetal bovine serum (Invitrogen) and antibiotics (penicillin-streptomycin solution, Invitrogen). The cells were grown to confluence and deprived of serum for 12 h before use.
The effect of TGF-β1 on cell phenotype was determined by adding recombinant TGF-β1 (10 ng/ml) to growth-arrested cell monolayers at 70% confluence. The cell phenotype was then monitored by phase-contrast microscopy. To test the effect of inhibitors on particular signaling molecules, each inhibitor was added to cells 1 h before TGF-β1 treatment. All experiments were performed under serum-free conditions.
Western Blot Analysis
Cells cultured with or without TGF-β1 on 60-mm dishes were scraped into 300 μl of ice-cold lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 20 μl/ml protease inhibitor mixture; Pharmingen BD Biosciences). Samples were clarified by centrifugation at 13,000 rpm for 5 min at 4 °C and boiled for 5 min with Laemmli sample buffer containing 100 mm NaF. Protein concentrations were determined by the Bradford method (Bio-Rad). Equivalent protein amounts were separated on 10% SDS-polyacrylamide gels and transferred to Immobilon-P polyvinylidene fluoride membranes (Millipore Corp., Bedford, MA). The blots were then hybridized with specific primary antibodies, and antigen-specific signals were detected using horseradish peroxidase-conjugated secondary antibodies and chemiluminescence (Pierce).
Immunoprecipitation
Confluent ARPE-19 cells were lysed in 0.3 ml of radioimmune precipitation assay buffer supplemented with a protease inhibitor mixture (Pharmingen). Cell lysates were incubated with antibodies (2 μg) for 1 h at 4 °C. Protein G-Sepharose (GE Healthcare) was then added, and lysates were incubated for 45 min at 4 °C. Immune complexes were recovered by centrifugation, washed three times with lysis buffer, and boiled in SDS sample buffer. After SDS-PAGE, proteins were transferred to Immobilon-P polyvinylidene fluoride membranes and immunoblotted with the indicated antibodies.
RNA Isolation and Reverse Transcriptase (RT)-PCR of NET1 mRNA
Total mRNA was isolated using Trizol reagent (Invitrogen) per the manufacturer's instructions. mRNA (1 μg) was converted to cDNA using an avian myeloblastosis virus reverse-transcription system (Promega, Madison, WI). A 100-mg aliquot of the resulting cDNA was amplified in a double PCR with 25 ng each of β-actin-specific or NET1-specific primer. PCR products were separated on a 1.2% agarose gel. Primers used were: NET1 forward primer, 5′-GTTCAGCTTCTGGAGGATGC-3′; NET1 reverse primer, 5′-CTTGTGGAACACGTCATTGG-3′; β-actin forward primer, 5′- GACGGGGTCACCCACACTGTGCCCATC TA-3′; β-actin reverse primer, 5′-CTAGAAGCATTTGCGGTGGACGATGGA GG-3′.
Detection of GTP-bound RhoA and Rac1
RhoA activation was assayed as suggested by the manufacturer (Upstate Biotechnology). Briefly, cells were lysed in lysis buffer and incubated with GST-rhotekin-RBD. Pulled-down complexes were washed and subjected to Western blotting with anti-RhoA.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) assays were performed with the EZ-ChIP kit (Upstate Biotechnology, Lake Placid, NY) according to the manufacturer's instructions. Briefly, ARPE-19 cells (1 × 106 cells in 100-mm-diameter plates) were incubated with TGF-β1 or vehicle for 30 min as described above. After cross-linking using 1% formaldehyde at 37 °C for 10 min, ARPE-19 cells were resuspended in SDS lysis buffer (Upstate Biotechnology), and DNA was sheared to small fragments of 200–900 bp by sonication. The supernatant was recovered, diluted, and precleared using protein G-agarose slurry. The recovered supernatant was incubated with either anti-Smad3 monoclonal antibody (ab28379; Abcam, Cambridge, MA) or an isotype control IgG overnight at 4 °C. After 1 h of incubation in the presence of protein G-agarose beads, the immunoprecipitated DNA-protein complexes were washed and eluted from the beads with 1% SDS in 0.1 m NaHCO3. Protein/DNA cross-links were reversed by adding 5 m NaCl at 65 °C for 6 h, and DNA was purified using spin columns. Real-time PCR was carried out on extracted DNA using primers specific for the human NET1 promoter (primers purchased from SABiosciences, Frederick, MD). Primers for human glyceraldehyde-3-phosphate dehydrogenase were purchased from the EZ-ChIP kit (Upstate Biotechnology).
cDNA Constructs and Transient Transfection
An expression vector encoding the dominant-negative form of Smad3 and the empty (control) expression vector were from the UMR cDNA Resource Center. Cells were plated on 60-mm dishes 1 day before transfection. At 30% confluence, the cells were transfected with 2 μg of the appropriate DNA using a 1:1 ratio of DNA/Transfast reagent in serum-free medium followed by 4 h of incubation. Dulbecco's modified Eagle's medium/F-12 medium supplemented with 10% fetal bovine serum was added, and cultures were incubated for an additional 12 h. Transfected cells were then deprived of serum for 3 h before being treated with TGF-β1 (10 ng/ml).
siRNA Gene Silencing
The siRNA sequence used for targeted silencing of NET1 was Silencer pre-designed siRNA (Ambion, Austin, TX). The NET1-targeted oligonucleotide sequences were: 5′-GGAGCCAAGCAAUAAAAGAtt-3′ (sense) and 5′-UCUUUUAUUGCUUGGCUCCtc-3′ (antisense). Silencer negative control #1 siRNA (Ambion) was used as a negative control. The synthetic double-stranded siRNA oligonucleotides were delivered into ARPE-19 cells using different doses of SiPORT reagent (Ambion) according to the manufacturer's recommended protocol. Reduction in NET1 gene expression by NET1 siRNA was measured by RT-PCR 48 h post-transfection.
Phalloidin and Immunofluorescence Staining
Cells were cultured in 4-well multichamber slides (Invitrogen) in serum-free medium for 12 h and then stimulated with recombinant TGF-β1 (10 ng/ml) for up to 2 days. At various time points the cells were rinsed in phosphate-buffered saline for 3 min, fixed in 5% paraformaldehyde for 30 min, and permeabilized with 0.2% Triton X-100 in phosphate-buffered saline for 20 min. For phalloidin staining, cells were incubated for 1 h with rhodamine-labeled phalloidin diluted 1:100. After a 1-h blocking step with 1% BSA/phosphate-buffered saline, the cells were incubated with primary antibodies for 12 h at 4 °C followed by fluorescein isothiocyanate- or Alexa 568-conjugated secondary antibodies (Molecular Probes). Cells were washed with phosphate-buffered saline, mounted with FluorSave reagent (Calbiochem), and analyzed by confocal microscopy (Leica TCS 4D).
RESULTS
TGF-β1-mediated Cell Morphological Changes and RhoA Activation Require Protein Synthesis in ARPE-19 Cells
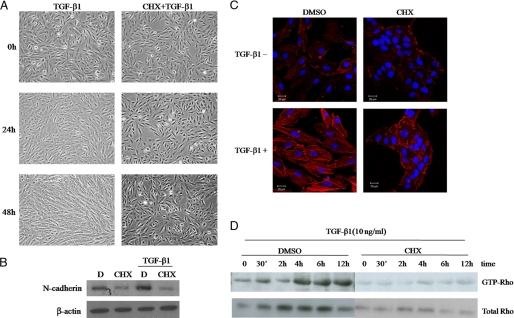
TGF-β1 modulates cell morphological changes in many cell types (52, 53). In the spontaneously immortalized human RPE cell line ARPE-19, TGF-β1 treatment led to dramatic morphological changes after 48 h (Fig. 1A). In contrast with their normal compact appearance, TGF-β1-treated ARPE-19 cells were larger with a less compact cell shape. When ARPE-19 cells were treated with the protein synthesis inhibitor cycloheximide, these dramatic TGF-β1-induced morphological changes were no longer observed (Fig. 1A). Because cycloheximide blocks protein synthesis, this result suggests that TGF-β1 may cause cell morphological changes by inducing the expression of downstream target genes. Furthermore, cycloheximide inhibited TGF-β1-induced N-cadherin expression (Fig. 1B) and stress fiber formation (Fig. 1C).
FIGURE 1.
Protein synthesis is required for TGF-β1-mediated changes in cell morphology and Rho activation. Serum-starved ARPE-19 cells were treated with 10 ng/ml TGF-β1 for 0, 24, or 48 h in the presence or absence of the protein synthesis inhibitor cycloheximide (CHX). D, DMSO. A, the cells were monitored by phase-contrast microscopy at the indicated times. B, shown is a Western blot of N-cadherin under the same treatment conditions as A. C, the actin cytoskeleton was visualized using rhodamine-labeled phalloidin; blue is from Hoechst staining of the nuclei to show all cells. Bar, 20 μm. D, lysed cells were subjected to GST pulldown assays using GST-rhotekin and Western blot analysis using anti-Rho.
Rho GTPases increase stress fiber formation, and we have previously proved that these small GTPases are key mediators of the TGF-β1-induced changes in ARPE-19 cell cytoskeletal organization and F-actin expression (15). To determine whether new protein synthesis is needed to activate RhoA and to elicit cell morphological changes in TGF-β1-induced ARPE-19 cells, we examined the effect of protein synthesis on TGF-β1-induced RhoA activation in ARPE-19 cells using cycloheximide and GST pulldown assays. Both TGF-β1-induced RhoA activation and total RhoA expression were significantly reduced in cycloheximide-treated cells (Fig. 1D), indicating that new protein synthesis is required for TGF-β1-induced cell cytoskeletal reorganization and RhoA activation in ARPE-19 cells.
TGF-β Induces NET1 Expression
Because NET1 acts as a GTPase of Rho and as a specific Rho GEF, we hypothesized that TGF-β1 might up-regulate NET1. To test this, we performed PCR and Western blot analysis on ARPE-19 cells treated with TGF-β1 for different time periods. As shown Fig. 2A, TGF-β1 increased NET1 mRNA expression. The induction started 30 min after TGF-β1 treatment and peaked at 1 h after treatment. NET1 protein levels also increased in a time-dependent manner in TGF-β1-treated cells (Fig. 2B).
FIGURE 2.
TGF-β1 induces NET1 expression in ARPE-19 cells. A, TGF-β1 induces NET1 transcription. Serum-starved ARPE-19 cells were treated with 10 ng/ml TGF-β1 for 0–4 h. After treatment, total RNA was isolated, reverse-transcribed, and amplified by PCR. β-Actin was evaluated as a loading control. B, TGF-β1 induces NET1 protein. Serum-starved ARPE-19 cells were treated with 10 ng/ml TGF-β1 for 0–24 h. Cell lysates were analyzed by Western blotting using a NET1-specific antibody. C, TGF-β1 attenuates cytoplasmic localization of NET1 in ARPE-19 cells. Serum-starved ARPE-19 cells were treated with 10 ng/ml TGF-β1 for 0–6 h. Cells were fixed and stained with anti-NET1 followed by Alexa 488-conjugated rabbit anti-goat and examined by fluorescence microscopy. Bar, 20 μm.
We expected NET1 localization to be important for its role as a Rho GTPase. To test this, we analyzed the intracellular distribution of NET1 in TGF-β1-induced ARPE-19 cells using immunofluorescence. In serum-starved ARPE-19 cells, NET1 protein was not apparent in the cytoplasm (Fig. 2C). However, the cytoplasmic accumulation of NET1 significantly increased within 30 min of TGF-β1 stimulation (Fig. 2C) and had increased exponentially by 6 h (Fig. 2C). These data indicate that NET1 acts directly as a Rho GTPase in the cytoplasm and that TGF-β1 induces NET1 expression and mediates its cytoplasmic localization in ARPE-19 cells.
NET1 Regulates Cell Phenotype and Stress Fiber Formation
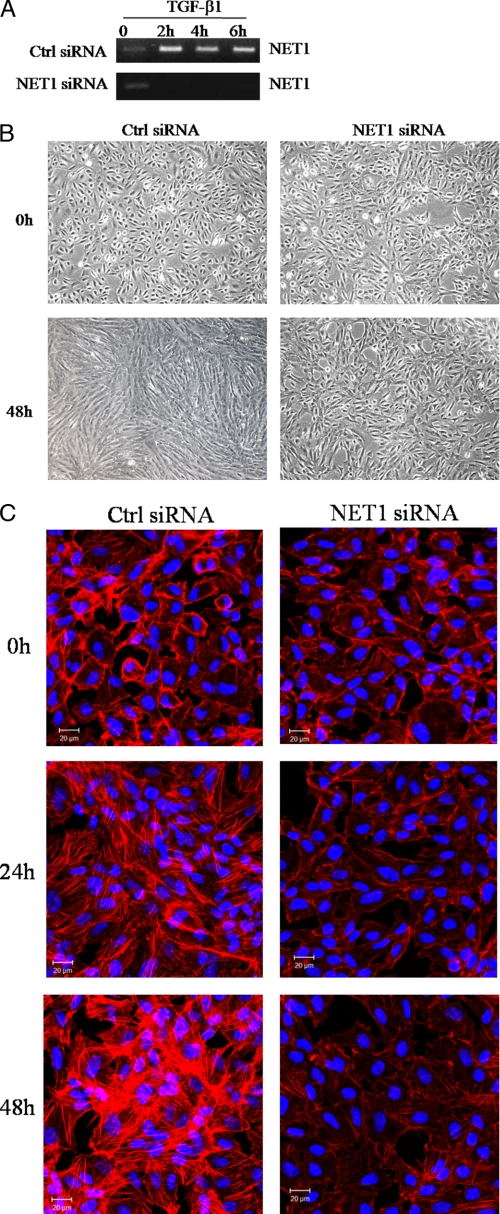
Because NET1 activation was previously shown to be induced by TGF-β1, we hypothesized that NET1 could mediate TGF-β1-induced cell phenotypic changes and stress fiber formation in ARPE-19 cells. To test this hypothesis, the functional effects of NET1 gene knockdown were determined by using siRNA to suppress NET1 gene expression in ARPE-19 cells. Five siRNA duplexes were designed to target each transcript, and gene silencing was confirmed using RT-PCR (data not shown). RT-PCR demonstrated that one siRNA duplex designed to target NET1 strongly reduced NET1 expression (Fig. 3A).
FIGURE 3.
NET1 is critical for TGF-β1-mediated cell morphological changes and stress fiber formation. A, expression of NET1 mRNA was analyzed by RT-PCR at the indicated times. B, NET1 regulates TGF-β1-mediated cell morphological change. ARPE-19 cells were transfected for 12 h with control or NET1-specific siRNA. After transfection, cells were switched to serum-free medium for 3 h and treated with TGF-β1 (10 ng/ml) for 48 h. Cells were monitored by phase-contrast microscopy. C, NET1 knockdown prevents TGF-β1-induced stress fiber formation. ARPE-19 cells were transfected with siRNA and treated with TGF-β1 (10 ng/ml) as described in B. The actin cytoskeleton was visualized by rhodamine-labeled phalloidin, and the blue is from Hoechst staining of the nuclei to show all cells. Bar, 20 μm. Ctrl siRNA, Silencer negative control siRNA.
Cells in which NET1 expression was reduced had significantly fewer TGF-β1-induced cell phenotypic changes compared with cells in which NET1 expression was unperturbed (Fig. 3B). NET1-targeted siRNA duplexes also led to decreases in TGF-β1-induced formation of stress fibers (Fig. 3C). These data demonstrate that the enhanced expression of NET1 is required for TGF-β1-induced cell phenotypic changes and stress fiber formation.
NET1 Induces RhoA Activation
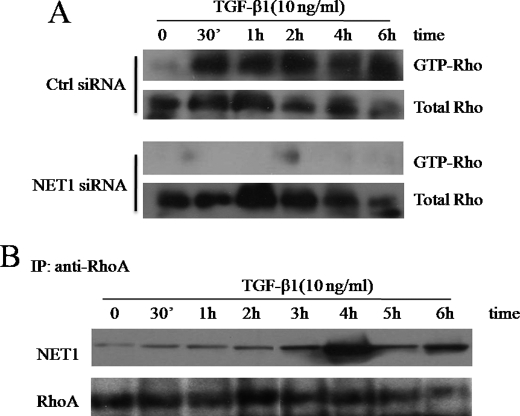
To further explore the putative role of NET1 in TGF-β1-induced ARPE-19 cells, we next studied the effect of RNAi-mediated mRNA down-regulation on RhoA activation. TGF-β1 failed to activate RhoA in NET1-down-regulated ARPE-19 cells (Fig. 4A). The NET1-targeted siRNA duplexes that resulted in 90–100% knockdown in mRNA expression (Fig. 3A) caused a decrease in RhoA activation.
FIGURE 4.
TGF-β1 induces RhoA activation via NET1. A, NET1 induces RhoA activation. ARPE-19 cells were transfected for 12 h with control or NET1-specific siRNA. After transfection, cells were switched to serum-free medium for 3 h and treated with TGF-β1 (10 ng/ml) for 0–6 h. The cells were then lysed, and the levels of active GTP-Rho in TGF-β1-stimulated ARPE-19 cells were determined by GST pulldown assay using GST-rhotekin and Western blot analysis using an anti-Rho antibody. Ctrl, Silencer negative control siRNA. B, TGF-β1 regulates NET1 binding activity to RhoA. ARPE-19 cells were transfected with siRNA and treated with TGF-β1 (10 ng/ml) as described in A and then lysed. Extracts were immunoprecipitated (IP) with anti-human RhoA, and precipitates were subjected to Western blotting using anti-NET1. As a loading control, aliquots of cell extracts were probed with anti-RhoA.
Rho GEFs have an ∼200-amino acid Dbl homology domain that is necessary for binding to the GTPase and stimulating nucleotide exchange activity. To confirm that RhoA is activated by TGF-β1 through this interaction, we analyzed the time course of NET1 binding to RhoA by immunoprecipitation. Interestingly, TGF-β1 increased NET1 binding activity as early as 30 min after treatment, and binding activity reached a maximum at 4 h post-treatment (Fig. 4B). This result indicates that TGF-β1 regulates NET1 binding activity in addition to NET1 production. Taken together, our data indicate that TGF-β1 induces cytoskeletal reorganization and RhoA activation by regulating NET1.
Smad3 Induces Cytoskeletal Reorganization
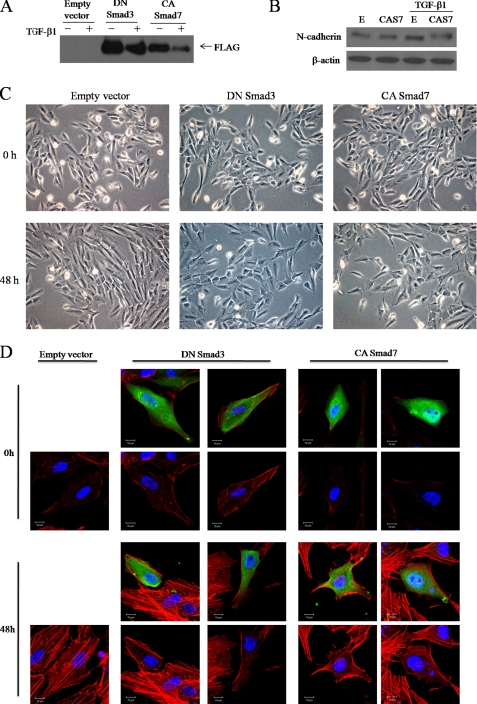
Because our previous study showed that TGF-β1 induces Smad3 phosphorylation (15), we hypothesized that Smad3 influences TGF-β1-induced cytoskeletal rearrangement. To determine whether Smad3 regulates the actin rearrangement caused by TGF- β1 treatment, we transfected FLAG-tagged dominant-negative Smad3 or constitutively active Smad7 into ARPE-19 cells (Fig. 5A). Smad7 is a general antagonist of the TGF-β-induced Smad pathway, so we inhibited the Smad3 pathway by both dominant-negative Smad 3 and overexpression of Smad7. We then treated the cells with TGF-β1 and monitored subsequent morphological changes by phase-contrast microscopy. Dominant-negative Smad3 and constitutively active Smad7 expression substantially suppressed TGF-β1-induced morphological changes in ARPE-19 cells (Fig. 5C). Furthermore, TGF-β1-induced expression of N-cadherin, a mesenchymal marker, was down-regulated in dominant-negative Smad3-expressing cells (Fig. 5B).
FIGURE 5.
Smad3 mediates TGF-β1-induced actin rearrangement. FLAG-tagged dominant-negative (DN) Smad3 and FLAG-tagged constitutively active (CA) Smad7 constructs were transfected into ARPE-19 cells. Empty vector was used as a control. After transfection, cells were incubated for 48 h with medium alone or medium containing 10 ng/ml TGF-β1, examined by phase-contrast microscopy (C), and then lysed and subjected to Western blot analysis using anti-FLAG antibodies (A). B, Western blot of N-cadherin under the same treatment conditions as A is shown. E, empty vector; CAS7, constitutively active Smad7. D, ARPE-19 cells were transfected for 12 h with plasmids expressing DN Smad3 or CA Smad7. After transfection, cells were switched to serum-free medium for 3 h and treated with TGF-β1 (10 ng/ml) for 48 h. Cells were fixed and stained with anti-FLAG followed by Alexa 488-conjugated secondary antibody (green) and stained with rhodamine-labeled phalloidin (red). Blue is Hoechst nuclear staining. Pictures were taken under a Zeiss confocal microscope. Bar, 10 μm.
The above observations led us to hypothesize that the Smad3 pathway may be an important signaling mediator of TGF-β1-induced actin rearrangement. To confirm this, actin cytoskeletal organization was examined by fluorescein-conjugated phalloidin staining of F-actin, and transfected cells were distinguished by FLAG epitope staining. Interestingly, transfection of dominant-negative Smad3 inhibited TGF-β1-induced stress fiber formation (Fig. 5D). Furthermore, inhibition of Smad3 by overexpression of Smad7 dramatically reduced TGF-β1-induced stress fiber formation (Fig. 5D). These observations suggest that the Smad3 pathway plays an important role in the morphological changes and stress fiber formation that are induced in ARPE-19 cells by TGF-β1 treatment.
Smad3 Regulates RhoA
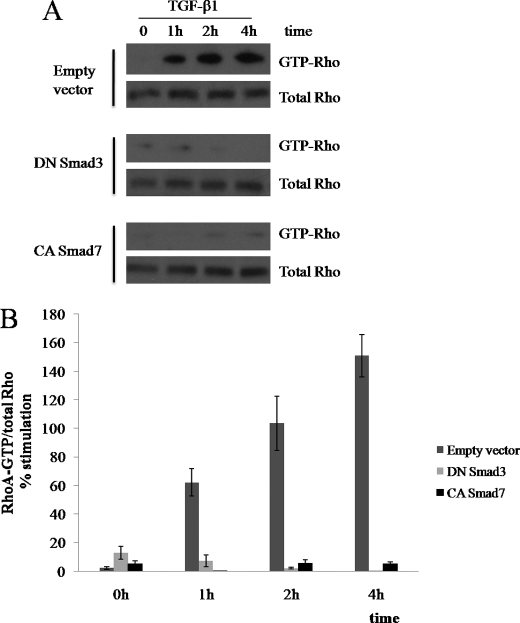
Because Rho GTPases increase stress fiber formation, the results obtained so far suggest that the Smad3 pathway is related to TGF-β1-induced RhoA activation. To test this hypothesis, we transfected dominant-negative Smad3 or constitutively active Smad7 into ARPE-19 cells. Transfected ARPE-19 cells were treated with TGF-β1, and active RhoA levels were examined by GST pulldown assays using GST-rhotekin. TGF-β1 treatment induced a rapid and transient increase in the levels of GTP-bound RhoA in ARPE-19 cells transfected with control vector (Fig. 6A). In contrast, cells transfected with dominant-negative Smad3 or constitutively active Smad7 exhibited substantially suppressed TGF-β1-induced RhoA activation (Fig. 6A). Statistical analysis of individual time points confirmed that RhoA activity significantly decreased dominant-negative Smad3 or constitutively active Smad7-expressing cells although under TGF-β1 treatment (Fig. 6B). These data indicate that Smad3 pathways induce morphological changes and stress fiber formation in ARPE-19 cells by activating RhoA.
FIGURE 6.
Smad3 mediates TGF-β1-induced RhoA activation. ARPE-19 cells were transfected for 12 h with plasmids expressing dominant-negative (DN) Smad3 or constitutively active (CA) Smad7. Empty vector was used as a control. After transfection, cells were switched to serum-free medium for 3 h and treated with TGF-β1 (10 ng/ml) for 0–4 h. A, cells were then lysed, and active RhoA was precipitated with GST-rhotekin. The total levels of RhoA are also shown. B, statistical analysis of TGF-β1-induced RhoA activation. Error bars represent S.D. *, p < 0.01 compared with black bar within same data set as calculated by Student's t test.
Smad3 Regulates RhoA through NET1
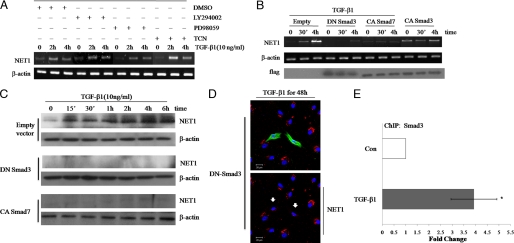
Our finding that NET1 and Smad3 regulate RhoA activity suggests that NET1 acts downstream of the Smad3 pathway in TGF-β1-induced ARPE-19 cells. In addition to Smad, other signaling proteins such as MEK, Akt, and phosphatidylinositol 3-kinase were shown in our previous study to mediate TGF-β1 function in ARPE-19 cells. Thus, we sought to clarify whether these signaling pathways also mediate TGF-β1-induced NET1 expression. To this end, we first used kinase inhibitors to block individual signaling pathways in ARPE-19 cells treated with TGF-β1 and examined NET1 mRNA expression. Inhibitors of MEK, Akt, and phosphatidylinositol 3-kinase did not inhibit the TGF-β1 induction of NET1 (Fig. 7A), suggesting that these pathways are not important for NET1 expression in APRE-19 cells.
FIGURE 7.
Smad3 regulates RhoA activation through NET1. A, MEK, Akt, and phosphatidylinositol 3-kinase signaling do not affect TGF-β1-induced NET1 mRNA expression. Serum-starved ARPE-19 cells were pretreated for 1 h with vehicle (DMSO) or 10 μm MEK, Akt, or phosphatidylinositol 3-kinase inhibitors (PD98059, Triciribine (TCN), or LY294002, respectively) and then treated with 10 ng/ml TGF-β1 for 0, 2, or 4 h. After treatment, total RNA was isolated, reverse-transcribed, and amplified by PCR. β-Actin was evaluated as a loading control. B, Smad3 regulates NET1 expression. ARPE-19 cells were transfected for 12 h with plasmids expressing dominant-negative (DN) Smad3, constitutively active (CA) Smad3, or constitutively active Smad7. Empty vector was used as a control. After transfection, cells were switched to serum-free medium for 3 h and treated with TGF-β1 (10 ng/ml) for 0, 0.5, or 4 h. After treatment, total RNA was isolated, reverse-transcribed, and amplified by PCR and analyzed by Western blot using a FLAG-specific antibody. C, cells were treated as in B, lysed, and analyzed by Western blot using a NET1-specific antibody. D, Smad3 induces cytoplasmic localization of NET1. ARPE-19 cells were transfected with DNA plasmid and treated with TGF-β1 (10 ng/ml) for 48 h as described in B. Cells were fixed and stained with anti-FLAG followed by Alexa 488 (green) and stained with anti-NET1 followed by Alexa 546 (red). Blue is Hoechst nuclear staining. Pictures were taken under a Zeiss confocal microscope. Arrows indicate transfected cells. Bar, 20 μm. E, NET1 is a Smad3 target gene. ChIP analysis was performed using an anti-Smad3 antibody, and quantitative PCR was performed with primers corresponding to the NET1 promoter region. Cells were left untreated or stimulated with TGF-β1 for 30 min. Quantitative ChIP values are expressed as -fold change in site occupancy and represent the average and S.D. from three independent experiments. Con, control.
Next, we sought to determine whether Smad3 positively regulates RhoA to stimulate actin cytoskeletal reorganization or cell transformation through NET1. We used dominant-negative Smad3, constitutively active Smad3, and constitutively active Smad7 vectors with TGF-β1 treatment to determine whether Smad3 induced NET1 in TGF-β1-treated ARPE-19 cells. RT-PCR revealed that NET1 mRNA expression was highly induced by TGF-β1 treatment in ARPE-19 cells transformed with control vector, whereas TGF-β1 failed to induce NET1 mRNA expression in cells expressing either dominant-negative Smad3 or constitutively active Smad7 (Fig. 7B). Cells expressing constitutively active Smad3 showed high levels of NET1 mRNA expression in the absence of TGF-β1 treatment that were similar to those observed in TGF-β1-stimulated cells (Fig. 7B). Similar results were obtained for NET1 protein expression as indicated by Western blotting (Fig. 7C). Finally, we examined the change in NET1 localization after Smad3 inhibition. TGF-β1 induced NET1 production; however, cells with dominant-negative Smad3 had significantly decreased cytoplasmic NET1 accumulation despite TGF-β1 treatment (Fig. 7D). We then addressed whether NET1 is a direct Smad3 target gene. To test this, we used quantitative ChIP analysis to address whether Smads directly target the NET1 promoter. ChIP analysis of ARPE-19 cells revealed an increased interaction of the NET1 promoter with Smad3 after treatment with TGF-β1 (Fig. 7E). This was associated with an increased expression of the gene (Fig. 2A). These results indicate a role for Smad3 in the regulation of NET1 expression in EMT. These findings suggest that Smad3 induces NET1 production. Taken together, our results demonstrate that Smad3 modulates RhoA activation through NET1.
DISCUSSION
As a multifunctional growth factor, TGF-β regulates various biological processes, including cell morphology changes and migration, likely by modulating the expression of downstream target genes. The Smad proteins, which translocate into the nucleus and act as transcription factors (54), are representative factors in the TGF-β-induced signaling pathway. Depending on which Smad protein is activated, the biological effect of TGF-β may vary. The TGF-β/Smad3 pathway is crucial in cutaneous wound repair (55), the epidermal response to ionizing radiation (56), and bleomycin-induced lung fibrosis (57). We previously revealed that TGF-β1 induces human RPE cells to undergo cytoskeletal actin rearrangement via Rho GTPase-dependent pathways that modulate LIM kinase and cofilin activity (15). Here, we confirmed that TGF-β1 strongly induces the Smad3 pathway and that RhoA is not required for TGF-β1-induced Smad3 activation. TGF-β leads to subsequent degradation of RhoA, which causes loss of tight junctions and junction stability in early EMT but also induces activation of RhoA and thereby led to cytoskeletal changes in our study.
Rho family small G proteins control many aspects of cell proliferation, including cell-cycle progression and cytokinesis, by acting as molecular switches, cycling between their active/GTP-bound and inactive/GDP-bound states. Because their capacity to bind GTP in the cell is stimulated by a family of proteins known as Rho GEFs, there has been substantial interest in identifying the regulatory mechanisms that control the activation of individual Rho GEFs. Despite the importance of these factors, the relationship between the Smad and Rho pathways had not previously been elucidated. In the present study we provide evidence supporting an essential role for Smad pathways and the Rho GTPase in TGF-β1-induced cytoskeletal reorganization in ARPE-19 cells. We also show that Smad pathways regulate Rho activation through the Rho GEF NET1.
We explored the signaling pathways that mediate the specific effect of TGF-β1 on changes in stress fiber formation and identified NET1 as a candidate target gene because NET1 is a nuclear Rho GEF that is specific for the RhoA subfamily of small G proteins. NET1 was rapidly induced by TGF-β1 in ARPE-19 cells. TGF-β1 induced stress fiber formation in a RhoA-dependent manner, a process that correlated with the induction of NET1 expression. Furthermore, cycloheximide inhibited not only TGF-β1-induced cytoskeletal reorganization, N-cadherin expression, and stress fiber formation but also RhoA activation. These results provide evidence for the requirement of new protein synthesis in TGF-β1-induced cytoskeletal reorganization and RhoA activation and indicate that this synthesized protein may be NET1. It was originally thought that the ability of NET1 to transform cells was due its active state in the cytosol. Our data also showed that TGF-β1 increased the cytoplasmic localization of NET1. Because we think that NET1 induces cytoplasmic RhoA to stimulate cytoskeletal reorganization, this result is very important for understanding the role of NET1.
Using an RNAi-based approach, NET1 was shown to activate RhoA. NET1 gene knockdown reduced TGF-β1-induced RhoA activation to the level seen in ARPE-19 cells not treated with TGF-β1 (Fig. 4A). Interestingly, NET1 knockdown also decreased the total RhoA expression level, suggesting that in addition to regulating RhoA activation, NET1 may also mediate RhoA transcription. This is the first report that NET1 drives the activation of RhoA in human retinal pigment epithelium. Furthermore, we showed that NET1 acts as an activator through direct binding to RhoA. This finding strengthens the role of RhoA in PVR and elaborates on the biology of NET1, a protein whose role in PVR is not yet fully understood. Further studies are required to define other factors regulating NET1-RhoA complex formation.
Having previously established a role for RhoA in cell morphology and stress fiber formation, we examined the effect of NET1 knockdown on these cellular processes in ARPE-19 cells. As expected, NET1 knockdown significantly reduced TGF-β1-induced cell morphological changes, N-cadherin expression, and stress fiber formation. These data together with previous findings implicating NET1 as an activator of RhoA support a model where TGF-β1-dependent NET1 induction can lead to RhoA activation, which in turn increases stress fiber formation.
Although NET1 is the first gene identified in the TGF-β1 signaling pathway whose function may be directly associated with cytoskeletal reorganization, we suspect that other related genes also mediate TGF-β1-induced cell morphological changes and stress fiber formation. Interestingly, we found that the Smad proteins are involved in TGF-β1-induced actin rearrangement and RhoA regulation. Both dominant-negative Smad3 and constitutively active Smad7 largely blocked the effects of TGF-β1 on RhoA activation, stress fiber formation, N-cadherin expression, and morphological changes. This may reflect a specific requirement for Smad3 in these processes. Furthermore, this result shows the possibility of cross-talk between Rho GTPases and Smad pathways in TGF-β1-induced EMT. Recently, microRNA-155 was shown to mediate TGF-β/Smad pathway-induced EMT through the targeting of RhoA (51). This finding partially supports our hypothesis.
From the rapid time course of NET1 mRNA accumulation (Fig. 2A), we noticed a close correlation between the induction of NET1 expression and the known kinetics of phosphorylation and nuclear accumulation of Smad2 and Smad3 (58, 59). To further explore this possible link, we used dominant-negative Smad3, constitutively active Smad3, and constitutively active Smad7 mutants to determine whether Smad3 overexpression could stimulate endogenous NET1 expression. The result was impressive; whereas Smad3 overexpression largely mimicked the effect of TGF-β1 on NET1 mRNA expression, both dominant-negative Smad3 and constitutively active Smad7 markedly blocked the effect of TGF-β1 on NET1 mRNA and protein expression. In contrast, inhibition of the MEK, Akt, and phosphatidylinositol 3-kinase signaling pathways did not affect NET1 mRNA expression. Using a ChIP approach, we further showed that Smad3 can specifically bind to the NET1 promoter. In agreement with the function of Smad3 proteins in mediating TGF-β1-induced NET1 expression, the ChIP assay revealed that TGF-β1 enhanced the binding of Smad3 to the NET1 promoter. These results indicate that Smad3 is necessary for TGF-β1-induced NET1 expression. Smad3 proteins modulate transcription in collaboration with other co-factors. Currently, it is unclear what other factors work with Smad3 to regulate NET1 expression.
Another interesting finding was that Smad3 regulated distribution of NET1 to the cytoplasm. In cells expressing dominant-negative Smad3, TGF-β1 failed to induce cytoplasmic NET1. This is significant because the ability of NET1 to activate RhoA is thought to be restricted to the cytoplasm. Our results suggest that Smad3 regulates the activity of NET1 by controlling not only mRNA and protein expression but also cytoplasmic localization.
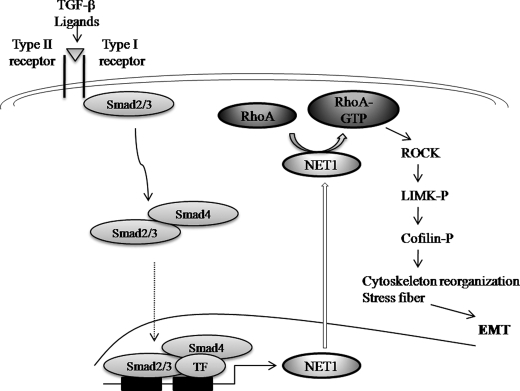
In conclusion, we have demonstrated novel regulatory mechanisms for cytoskeletal reorganization controlled by TGF-β1 and mediated by Smad3. An important finding is the apparent mechanism involving RhoA GTPase activation by the Smad-inducible Rho GEF NET1 in TGF-β1-induced EMT. In this study we have shown that Smad3 regulates RhoA activation through NET1 activity by controlling both mRNA and protein expression as well as cytoplasmic localization of NET1 (Fig. 8). Our results revealed that Smad3 induces both the mRNA and protein expression of NET1 by regulating the NET1 promoter. Further studies are necessary to test whether Smad3 is directly involved in NET1-RhoA complex formation. In ARPE-19 cells, the above mechanisms stimulate actin cytoskeletal reorganization, which is indicative of a transformation to fibrotic cells. The detailed mechanisms underlying Smad-mediated transcriptional regulation of NET1 and Smad3 complex formation with other transcription factors at target genes should next be investigated by cloning and characterization of the NET1 promoter. In addition, further studies will be required to define other factors regulating NET1 binding to RhoA. How TGF-β1 signaling regulates the pluripotency of genes involved in EMT also remains to be elucidated.
FIGURE 8.
Mechanisms of actin cytoskeleton reorganization induced by TGF-β in ARPE-19 cells. Upon stimulation with TGF-β, RhoA can be activated by NET1 through cross-talking with Smad3 to induce actin stress fiber formation during EMT. TF, transcription factor.
Acknowledgment
We are grateful to Dr. J. Oh of the Korea University Graduate School of Medicine (Ansan, Korea) for the constitutively active Smad3 and Smad7 expression vectors.
This work was supported by Center for Biological Modulators of the 21C Frontier R&D Program, Ministry of Education, Science, and Technology Grant CBM34-B3003-01-00-00.
- TGF
- transforming growth factor
- EMT
- epithelial-mesenchymal transition
- RPE
- retinal pigment epithelial
- PVR
- proliferative vitreoretinopathy
- GEF
- guanine nucleotide exchange factor
- siRNA
- small interfering RNA
- RT
- reverse transcriptase
- GST
- glutathione S-transferase
- ChIP
- chromatin immunoprecipitation
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase.
REFERENCES
- 1.Bachman K. E., Park B. H. (2005) Curr. Opin. Oncol. 17, 49–54 [DOI] [PubMed] [Google Scholar]
- 2.Lu J., Wu Y., Sousa N., Almeida O. F. (2005) Development 132, 3231–3242 [DOI] [PubMed] [Google Scholar]
- 3.Massagué J. (1998) Annu. Rev. Biochem. 67, 753–791 [DOI] [PubMed] [Google Scholar]
- 4.Song J. (2007) Cell Res. 17, 289–290 [DOI] [PubMed] [Google Scholar]
- 5.Yang Y., Pan X., Lei W., Wang J., Song J. (2006) Oncogene 25, 7235–7244 [DOI] [PubMed] [Google Scholar]
- 6.Condeelis J., Segall J. E. (2003) Nat. Rev. Cancer 3, 921–930 [DOI] [PubMed] [Google Scholar]
- 7.Gotzmann J., Mikula M., Eger A., Schulte-Hermann R., Foisner R., Beug H., Mikulits W. (2004) Mutat. Res. 566, 9–20 [DOI] [PubMed] [Google Scholar]
- 8.Shook D., Keller R. (2003) Mech. Dev. 120, 1351–1383 [DOI] [PubMed] [Google Scholar]
- 9.Baum B., Settleman J., Quinlan M. P. (2008) Semin. Cell Dev. Biol. 19, 294–308 [DOI] [PubMed] [Google Scholar]
- 10.Guan F., Handa K., Hakomori S. I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7461–7466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hay E. D. (1995) Acta Anat. 154, 8–20 [DOI] [PubMed] [Google Scholar]
- 12.Nawshad A., Lagamba D., Polad A., Hay E. D. (2005) Cells Tissues Organs 179, 11–23 [DOI] [PubMed] [Google Scholar]
- 13.Miettinen P. J., Ebner R., Lopez A. R., Derynck R. (1994) J. Cell Biol. 127, 2021–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasai H., Allen J. T., Mason R. M., Kamimura T., Zhang Z. (2005) Respir. Res. 6, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J., Ko M., Joo C. K. (2008) J. Cell Physiol. 216, 520–526 [DOI] [PubMed] [Google Scholar]
- 16.Saika S., Kono-Saika S., Tanaka T., Yamanaka O., Ohnishi Y., Sato M., Muragaki Y., Ooshima A., Yoo J., Flanders K. C., Roberts A. B. (2004) Lab. Invest. 84, 1245–1258 [DOI] [PubMed] [Google Scholar]
- 17.Saika S., Kono-Saika S., Ohnishi Y., Sato M., Muragaki Y., Ooshima A., Flanders K. C., Yoo J., Anzano M., Liu C. Y., Kao W. W., Roberts A. B. (2004) Am. J. Pathol. 164, 651–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willis B. C., Liebler J. M., Luby-Phelps K., Nicholson A. G., Crandall E. D., du Bois R. M., Borok Z. (2005) Am. J. Pathol. 166, 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casaroli-Marano R. P., Pagan R., Vilaró S. (1999) Invest. Ophthalmol. Vis. Sci. 40, 2062–2072 [PubMed] [Google Scholar]
- 20.Lee S. C., Kwon O. W., Seong G. J., Kim S. H., Ahn J. E., Kay E. D. (2001) Ophthalmic Res. 33, 80–86 [DOI] [PubMed] [Google Scholar]
- 21.Carrington L., McLeod D., Boulton M. (2000) Invest. Ophthalmol. Vis. Sci. 41, 1210–1216 [PubMed] [Google Scholar]
- 22.Cassidy L., Barry P., Shaw C., Duffy J., Kennedy S. (1998) Br. J. Ophthalmol. 82, 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinton D. R., He S., Jin M. L., Barron E., Ryan S. J. (2002) Eye 16, 422–428 [DOI] [PubMed] [Google Scholar]
- 24.Lei H., Hovland P., Velez G., Haran A., Gilbertson D., Hirose T., Kazlauskas A. (2007) Invest. Ophthalmol. Vis. Sci. 48, 2335–2342 [DOI] [PubMed] [Google Scholar]
- 25.Taylor L. M., Khachigian L. M. (2000) J. Biol. Chem. 275, 16709–16716 [DOI] [PubMed] [Google Scholar]
- 26.Holmes A., Abraham D. J., Sa S., Shiwen X., Black C. M., Leask A. (2001) J. Biol. Chem. 276, 10594–10601 [DOI] [PubMed] [Google Scholar]
- 27.Van Obberghen-Schilling E., Roche N. S., Flanders K. C., Sporn M. B., Roberts A. B. (1988) J. Biol. Chem. 263, 7741–7746 [PubMed] [Google Scholar]
- 28.Bhowmick N. A., Ghiassi M., Bakin A., Aakre M., Lundquist C. A., Engel M. E., Arteaga C. L., Moses H. L. (2001) Mol. Biol. Cell 12, 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clements R. T., Minnear F. L., Singer H. A., Keller R. S., Vincent P. A. (2005) Am. J. Physiol. Lung Cell. Mol. Physiol. 288, L294–L306 [DOI] [PubMed] [Google Scholar]
- 30.Hall A. (1998) Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- 31.Morita T., Mayanagi T., Sobue K. (2007) J. Cell Biol. 179, 1027–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vardouli L., Moustakas A., Stournaras C. (2005) J. Biol. Chem. 280, 11448–11457 [DOI] [PubMed] [Google Scholar]
- 33.Broders-Bondon F., Chesneau A., Romero-Oliva F., Mazabraud A., Mayor R., Thiery J. P. (2007) Dev. Dyn. 236, 2555–2566 [DOI] [PubMed] [Google Scholar]
- 34.Mack C. P., Somlyo A. V., Hautmann M., Somlyo A. P., Owens G. K. (2001) J. Biol. Chem. 276, 341–347 [DOI] [PubMed] [Google Scholar]
- 35.Marinissen M. J., Chiariello M., Gutkind J. S. (2001) Genes Dev. 15, 535–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takai Y., Sasaki T., Matozaki T. (2001) Physiol. Rev. 81, 153–208 [DOI] [PubMed] [Google Scholar]
- 37.Schmidt A., Hall A. (2002) Genes Dev. 16, 1587–1609 [DOI] [PubMed] [Google Scholar]
- 38.Schmidt A., Hall A. (2002) J. Biol. Chem. 277, 14581–14588 [DOI] [PubMed] [Google Scholar]
- 39.Chan A. M., Takai S., Yamada K., Miki T. (1996) Oncogene 12, 1259–1266 [PubMed] [Google Scholar]
- 40.Alberts A. S., Treisman R. (1998) EMBO J. 17, 4075–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami G., Watabe T., Takaoka K., Miyazono K., Imamura T. (2003) Mol. Biol. Cell 14, 2809–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakao A., Afrakhte M., Morén A., Nakayama T., Christian J. L., Heuchel R., Itoh S., Kawabata M., Heldin N. E., Heldin C. H., ten Dijke P. (1997) Nature 389, 631–635 [DOI] [PubMed] [Google Scholar]
- 43.Casellas R., Brivanlou A. H. (1998) Dev. Biol. 198, 1–12 [DOI] [PubMed] [Google Scholar]
- 44.Hata A., Lagna G., Massagué J., Hemmati-Brivanlou A. (1998) Genes Dev. 12, 186–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ka S. M., Huang X. R., Lan H. Y., Tsai P. Y., Yang S. M., Shui H. A., Chen A. (2007) J. Am. Soc. Nephrol. 18, 1777–1788 [DOI] [PubMed] [Google Scholar]
- 46.Derynck R., Zhang Y. E. (2003) Nature 425, 577–584 [DOI] [PubMed] [Google Scholar]
- 47.Ng J. (2008) Development 135, 4025–4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kowanetz M., Valcourt U., Bergström R., Heldin C. H., Moustakas A. (2004) Mol. Cell. Biol. 24, 4241–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valcourt U., Kowanetz M., Niimi H., Heldin C. H., Moustakas A. (2005) Mol. Biol. Cell 16, 1987–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zavadil J., Bitzer M., Liang D., Yang Y. C., Massimi A., Kneitz S., Piek E., Bottinger E. P. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6686–6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kong W., Yang H., He L., Zhao J. J., Coppola D., Dalton W. S., Cheng J. Q. (2008) Mol. Cell. Biol. 28, 6773–6784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moustakas A., Stournaras C. (1999) J. Cell Sci. 112, 1169–1179 [DOI] [PubMed] [Google Scholar]
- 53.Lee K. M., Park J., Kim J. H., Yie S. W., Chun G. T., Kim P. H., Choi E. Y. (1999) Cell Biol. Int. 23, 507–517 [DOI] [PubMed] [Google Scholar]
- 54.Massagué J., Wotton D. (2000) EMBO J. 19, 1745–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashcroft G. S., Yang X., Glick A. B., Weinstein M., Letterio J. L., Mizel D. E., Anzano M., Greenwell-Wild T., Wahl S. M., Deng C., Roberts A. B. (1999) Nat. Cell Biol. 1, 260–266 [DOI] [PubMed] [Google Scholar]
- 56.Flanders K. C., Sullivan C. D., Fujii M., Sowers A., Anzano M. A., Arabshahi A., Major C., Deng C., Russo A., Mitchell J. B., Roberts A. B. (2002) Am. J. Pathol. 160, 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao J., Shi W., Wang Y. L., Chen H., Bringas P., Jr., Datto M. B., Frederick J. P., Wang X. F., Warburton D. (2002) Am. J. Physiol. Lung Cell Mol. Physiol. 282, L585–L593 [DOI] [PubMed] [Google Scholar]
- 58.Yingling J. M., Das P., Savage C., Zhang M., Padgett R. W., Wang X. F. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8940–8944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen X., Hu P. P., Liberati N. T., Datto M. B., Frederick J. P., Wang X. F. (1998) Mol. Biol. Cell 9, 3309–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]