Abstract
The mechanisms by which prions kill neurons and the role of the cellular prion protein in this process are enigmatic. Insight into these questions is provided by the neurodegenerative phenotypes of transgenic mice expressing prion protein (PrP) molecules with deletions of conserved amino acids in the central region. We report here that expression in transfected cells of the most toxic of these PrP deletion mutants (Δ105–125) induces large, spontaneous ionic currents that can be detected by patch-clamping techniques. These currents are produced by relatively non-selective, cation-permeable channels or pores in the cell membrane and can be silenced by overexpression of wild-type PrP, as well as by treatment with a sulfated glycosaminoglycan. Similar currents are induced by PrP molecules carrying several different point mutations in the central region that cause familial prion diseases in humans. The ionic currents described here are distinct from those produced in artificial lipid membranes by synthetic peptides derived from the PrP sequence because they are induced by membrane-anchored forms of PrP that are synthesized by cells and that are found in vivo. Our results indicate that the neurotoxicity of some mutant forms of PrP is attributable to enhanced ion channel activity and that wild-type PrP possesses a channel-silencing activity. Drugs that block PrP-associated channels or pores may therefore represent novel therapeutic agents for treatment of patients with prion diseases.
Keywords: Drug Action, Ion Channels, Mutant, Neurodegeneration, Prions, Patch Clamp
Introduction
Prion diseases are fatal neurodegenerative disorders in which a normal, cell surface glycoprotein called PrPC 2 is converted into PrPSc, a conformationally altered isoform that propagates itself by a molecular templating mechanism (1, 2). Although much is now understood about how infectious prions propagate, the mechanisms by which neurotoxic forms of PrP kill nerve cells remain enigmatic (3). Surprisingly, membrane-bound PrPC appears to play a crucial role in mediating the neurotoxicity of PrPSc. It has been proposed that interactions with PrPSc during the conversion process may alter a normal, physiological activity of PrPC, resulting in the transmission of a neurotoxic signal (4). Thus, identifying the normal function of PrPC is important for understanding the mechanism of prion-induced neurodegeneration. However, the function of PrPC remains unclear because mice lacking this protein display no overt phenotype (5, 6).
Insights into the physiological function of PrPC and how it might be subverted in the disease state have been provided by transgenic mice expressing forms of PrP missing conserved residues in the central region of the protein (7–10). Each of these mice displays a neurodegenerative phenotype that is dose-dependently suppressed by co-expression of wild-type (WT) PrP, suggesting an antagonistic relationship between the normal activity of PrPC and the neurotoxic effects of the deletion mutants. We have found that mice expressing the smallest of the deletions (Δ105–125 or ΔCR for central region) display the most dramatic phenotype, characterized by neonatal lethality with massive degeneration of cerebellar granule neurons and vacuolar degeneration of white matter regions of the brain and spinal cord. ΔCR PrP is neither aggregated nor protease-resistant, and its cellular localization is identical to that of wild-type PrP (10, 11). Thus, the neurotoxicity of ΔCR PrP is likely to be due to an alteration in a physiological activity of PrPC rather than to the formation of PrPSc-like aggregates.
We recently reported the unusual observation that cells expressing ΔCR PrP, as well as other toxic deletion mutants, are hypersensitive to killing by two classes of drugs normally used to select transfected cell lines (12). This effect, which was abolished by membrane depolarization, is likely due to enhanced cellular accumulation of these cationic drugs, possibly down an electrochemical gradient. These results suggested that the mutant PrPs might be altering the activity of membrane channels or transporters responsible for drug uptake.
In the present study, we have employed patch-clamping techniques to test directly for a connection between ΔCR PrP and ion channel activity. We report the surprising observation that PrP molecules carrying deletions or disease-associated point mutations in the central region induce large, spontaneous currents when expressed in a variety of different cell types. We hypothesize that the ion channels or membrane pores underlying these currents play a critical role in the pathological effects of neurotoxic PrP mutants and that they represent a new class of potential drug targets for treatment of prion diseases.
EXPERIMENTAL PROCEDURES
Cells
HEK293 cells (ATCC CRL-1573) were maintained in α-minimum essential medium/Dulbecco's modified Eagle's medium (1:1) supplemented with non-essential amino acids, l-glutamine, 10% fetal bovine serum, penicillin/streptomycin, and 50 μg/ml hygromycin. Stable lines were created by transfecting HEK cells with pcDNA3.1(+)Hygro (Invitrogen) vector alone or with vector containing the cDNA sequence for WT PrP or Δ105–125 (ΔCR) murine PrP (10) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's directions. Clones were selected for 14 days in 200 μg/ml hygromycin. Except where noted (see Fig. 5A), WT PrP contained an epitope tag (L108M/V111M) for the monoclonal antibody 3F4 (13). Point mutations were introduced into WT PrP lacking the 3F4 tag using a QuikChange site-directed mutagenesis kit (Stratagene).
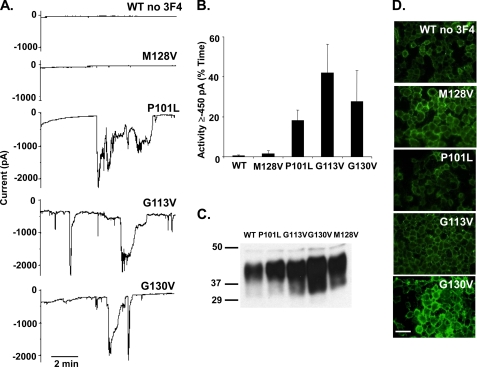
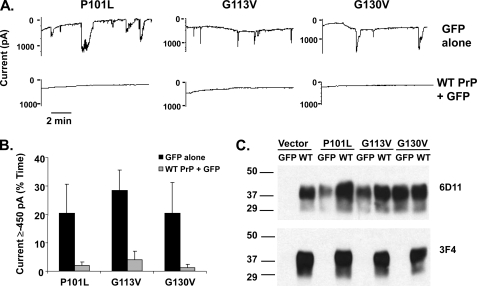
FIGURE 5.
Disease-associated point mutants in the central region of PrP induce spontaneous inward currents. A, whole-cell patch clamp recordings at a holding potential of −80 mV were made from HEK cells expressing WT, M128V, P101L, G113V, or G130V PrP. PrP molecules did not carry a 3F4 tag. B, quantitation of the currents recorded in panel A, plotted as the percentage of total time the cells exhibited inward current ≥450 pA (mean ± S.E., n = 5 cells). C, Western blot showing relative PrP expression levels of each construct. D, surface immunofluorescence staining of PrP on HEK cells expressing the indicated constructs. Scale bar = 50 μm.
N2a cells were maintained in DMEM supplemented with non-essential amino acids, 10% fetal bovine serum, and penicillin/streptomycin. Cells were transiently transfected using Lipofectamine 2000 with pEGFP-N1 (Clontech), along with empty pcDNA3.1(+)Hygro vector or vector encoding WT or ΔCR PrP.
Sf9 cells were maintained in Grace's medium supplemented with lactalbumin hydrolysate, yeastolate, 10% fetal bovine serum, and l-glutamine. Cells were co-transfected using Cellfectin (Invitrogen) with pIB/V5-His (Invitrogen) vector encoding monomeric GFP, along with empty vector or vector encoding WT or ΔCR PrP.
Lentiviruses
Recombinant lentiviruses were constructed according to published procedures (14, 15). cDNAs encoding either enhanced GFP alone or WT PrP followed by an internal ribosomal entry site and enhanced GFP were cloned into the transfer vector pRRLsinCMV. 293T packaging cells were co-transfected with the resulting transfer plasmid, along with the plasmids pMD-Lg, pCMV-G, and RSV-REV. Virus was collected from the medium and concentrated. Viral transductions were performed by incubating cells with purified virus at a multiplicity of infection of 0.5 in medium for 4 h.
Electrophysiology
Whole-cell patch clamp experiments were conducted using standard techniques (16, 17). Pipettes were pulled from borosilicate glass, coated with Sylgard, and polished to an open resistance of 1–10 megaohms. Experiments were conducted at room temperature with the following solutions unless otherwise noted: internal, 140 mm Cs-glucuronate, 5 mm CsCl, 4 mm MgATP, 1 mm Na2GTP, 10 mm EGTA, and 10 mm HEPES (pH 7.4 with CsOH); external, 150 mm NaCl, 4 mm KCl, 2 mm CaCl2, 2 mm MgCl2, 10 mm glucose, and 10 mm HEPES (pH 7.4 with NaOH). Current signals were collected from an Axopatch 200B amplifier and digitized with a Digidata 1330 interface (Axon Instruments) or with an EPC-10 amplifier controlled by PatchMaster acquisition software (HEKA Elektronik) and were saved to disc for analysis with PClamp 9 software.
Western Blotting and Immunofluorescence Staining
Western blots were probed with anti-PrP antibody 6D11 (18) and goat anti-mouse IgG (Pierce) and developed with ECL (GE Healthcare). Samples for each blot were normalized for total protein following measurement with the BCA protein assay (Pierce). Some of the samples in Fig. 3C were deglycosylated by treatment with N-glycosidase F (New England Biolabs, Beverly, MA) according to the manufacturer's directions. Immunofluorescence staining of cell surface proteins was achieved by incubating livings cells on ice with 6D11, fixing in 4% paraformaldehyde in PBS, and labeling with Alexa Fluor 488 goat anti-mouse IgG (Molecular Probes). Cells were viewed with a Nikon TE-2000E inverted fluorescence microscope, and images were captured with MetaMorph software (Molecular Devices, Sunnyvale, CA).
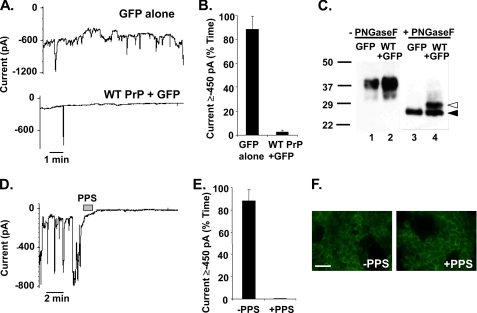
FIGURE 3.
ΔCR PrP-induced current is eliminated by overexpression of WT PrP and by treatment with PPS. A, whole-cell patch clamp recordings were made from HEK cells expressing ΔCR PrP at 48 h after infection with lentivirus encoding GFP alone (upper trace) or WT PrP plus GFP (lower trace). The holding potential was −80 mV. B, quantitation of the currents recorded in panel A, plotted as the percentage of total time the cells exhibited inward current ≥450 pA (mean ± S.E., n = 5 cells). C, cells in panel A were analyzed for PrP by Western blotting. Samples in lanes 3 and 4 were enzymatically deglycosylated with N-glycosidase F (PNGaseF), whereas those in lanes 1 and 2 were not. Deglycosylated WT and ΔCR PrP are identified by white and black arrowheads, respectively. D, whole-cell patch clamp recordings were made from HEK cells expressing ΔCR PrP. During the period indicated by the bar (60 s), the cell was perfused with PPS at 100 μg/ml. E, quantitation of the currents recorded in panel D plotted as the percentage of total time the cells exhibited inward current ≥450 pA (mean ± S.E., n = 5 cells). F, surface immunofluorescence staining of PrP (green) on HEK cells expressing ΔCR PrP, with or without a 2-min treatment with PPS. Scale bar = 50 μm.
RESULTS
Expression of ΔCR PrP Induces Spontaneous Inward Currents
We recorded whole-cell currents from stably transformed clones of HEK cells expressing ΔCR PrP, WT PrP, or empty vector. We observed that cells expressing ΔCR PrP held at −80 mV exhibited inward currents that fluctuated spontaneously over a time period of seconds to minutes, reaching amplitudes of up to several thousand pA (Fig. 1A, upper trace). These currents were absent in cells expressing WT PrP or vector alone (Fig. 1A, two lower traces).
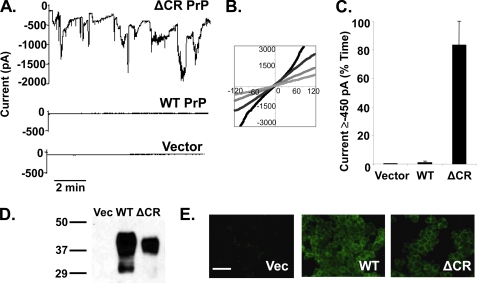
FIGURE 1.
Δ CR PrP induces spontaneous inward currents in HEK cells. A, whole-cell patch clamp recordings were made from HEK cells expressing ΔCR PrP, WT PrP, or vector at a holding potential of −80 mV. B, I-V plots collected at four different times during recording from HEK cells expressing ΔCR PrP. C, current activity recorded from HEK cells expressing vector, WT PrP, or ΔCR PrP was plotted as the percentage of total time the cells exhibited inward current ≥450 pA (mean ± S.E., n = 5 cells) at a holding potential of −80 mV. D, Western blot showing levels of PrP in HEK cells expressing vector (Vec), WT PrP, or ΔCR PrP. Molecular size markers are given in kDa. E, surface immunofluorescence staining of PrP (green) on HEK cells expressing vector, WT PrP, or ΔCR PrP. Scale bar = 50 μm.
We recorded current-voltage (I-V) relationships from ΔCR PrP cells by repeatedly ramping the holding potential from −120 to +120 mV, using an external solution containing NaCl and an internal (patch pipette) solution containing cesium glucuronate. The I-V relations were all linear, although because the current varied over time, the slopes of the curves depended on the magnitude of the current at the time the voltage ramp was applied (Fig. 1B). For each of the I-V curves, however, the reversal potential was near 0 mV (−6 ± 2 mV, n = 4).
To quantify the sporadic current activity, we calculated the percentage of recording time each cell exhibited inward current greater than 450 pA, a cut-off that was selected to discriminate between the ΔCR PrP-induced current and the leakage current present in control cells. Using this criterion, ΔCR PrP cells were active 83 ± 17% (n = 5, mean ± S.E.) of the time when compared with 1 ± 1% (n = 5) and 0 ± 0% (n = 5) for WT PrP and vector controls, respectively (Fig. 1C). The expression level for WT PrP was slightly higher than ΔCR PrP in the HEK cell clones (Fig. 1D), and as expected, both proteins were localized to the cell surface (Fig. 1E).
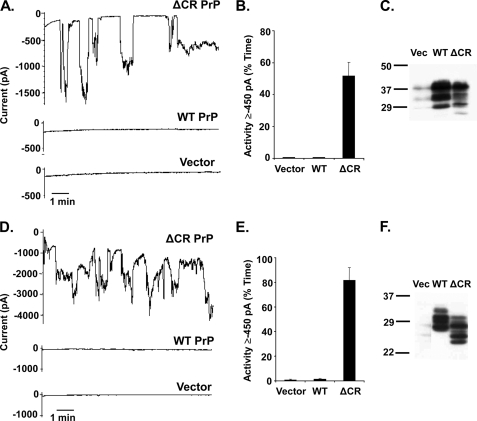
Similar spontaneous currents were observed when ΔCR PrP was expressed in two other cell types, N2a mouse neuroblastoma cells (Fig. 2, A–C) and Sf9 insect cells (Fig. 2, D–F), demonstrating that the channel activity was not restricted to a specific heterologous expression system.
FIGURE 2.
Expression of ΔCR PrP in N2a mouse neuroblastoma cells and Sf9 insect cells produces spontaneous inward currents. A, whole-cell patch clamp recordings at −80 mV of N2a cells 24 h after transfection with plasmids encoding ΔCR PrP, WT PrP, or vector. Transfected cells were recognized by expression of GFP, encoded in a co-transfected plasmid. B, quantitation of the currents shown in panel A, plotted as the percentage of total time the cells exhibited inward current ≥450 pA (mean ± S.E., n = 5 cells). C, Western blot to detect PrP in the N2a cells used in panel A. Vec, vector. D, whole-cell patch clamp recordings at −80 mV of Sf9 cells 36 h after transfection with plasmids encoding ΔCR PrP, WT PrP, or vector. Transfected cells were recognized by expression of GFP, encoded in a co-transfected plasmid. E, quantitation of the currents shown in panel D, plotted as the percentage of total time the cells exhibited inward current ≥450 pA (mean ± S.E., n = 5 cells). F, Western blot to detect PrP in the Sf9 cells used in panel D. PrP expressed in Sf9 cells migrates with a lower Mr than in mammalian cells (HEK or N2a) due to differences between insect and mammalian cells in N-linked glycosylation.
Overexpression of WT PrP or Treatment with a Sulfated Glycosaminoglycan Silences ΔCR PrP-induced Currents
A striking feature of mice expressing ΔCR and other deleted forms of PrP is that the neurodegenerative phenotype can be dose-dependently suppressed by co-expression of WT PrP (8–10). To see whether a similar, antagonistic interaction between WT and ΔCR PrP was apparent in our electrophysiological recordings, we tested whether WT PrP suppressed the spontaneous currents induced by ΔCR PrP. HEK cells stably expressing ΔCR PrP were transduced with lentiviruses encoding both WT PrP and GFP or GFP alone, and patch clamp recordings were made from GFP-positive cells. ΔCR PrP cells receiving the GFP-only virus showed spontaneous inward current 88 ± 11% (n = 5) of the time, similar to untransduced ΔCR PrP cells (Fig. 3, A, upper trace, and B). In contrast, cells receiving the virus encoding WT PrP were active only 3 ± 2% (n = 5) of the time (Fig. 3, A, lower trace, and B). Western blotting confirmed that the latter cells expressed WT PrP in addition to ΔCR PrP and that introduction of the WT protein did not reduce the amount of the mutant protein (Fig. 3C). Thus, WT PrP silences ΔCR PrP-induced currents.
Sulfated glycosaminoglycans have been shown previously to bind PrPC, reduce accumulation of PrPSc, and have therapeutic effect in prion diseases (19–21). We therefore tested the effect of the sulfated glycosaminoglycan pentosan polysulfate (PPS) on ΔCR PrP-induced current in HEK cells. After less than a minute of local perfusion, PPS completely eliminated inward current activity for the remainder of the recording (−PPS 88 ± 11%; +PPS 0 ± 0%, n = 5)(Fig. 3, D and E). PPS had no effect on cells expressing WT PrP or empty vector (data not shown).
PPS has been shown to induce rapid endocytosis of PrPC from the cell surface, an effect that may be connected to its ability to inhibit PrPSc formation (22). To determine whether PPS blocks ΔCR PrP currents by reducing the amount of the mutant protein on the cell surface, cells were treated with PPS for 2 min in recording buffer at room temperature and then surface-stained for ΔCR PrP with anti-PrP antibody (Fig. 3F). Under these conditions, PPS did not cause any apparent change in the amount of ΔCR PrP on the cell surface when compared with untreated cells, suggesting that the inhibitory effect of the glycosaminoglycan does not result from acute changes in localization or trafficking of the mutant protein.
Characterization of ΔCR PrP-induced Currents
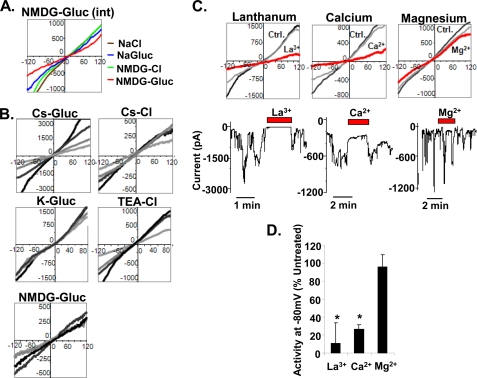
To gain insight into the properties of the ion channel responsible for the ΔCR PrP-induced current, we performed a series of ion substitution experiments to determine which ions can permeate the channel. First, we recorded I-V curves from cells in the presence of several different external solutions, with a constant internal solution containing NMDG and glucuronate (relatively large organic cations and anions, respectively). We observed that in external solutions containing NaCl, sodium glucuronate, NMDG-Cl, or NMDG-glucuronate, the I-V curves were linear and displayed slightly positive reversal potentials (NaCl, +14 ± 1 mV; sodium glucuronate, +15 ± 3 mV; NMDG-Cl, +11 ± 1 mV; NMDG-glucuronate, +9 ± 5 mV, n = 3) (Fig. 4A). Substitution of NMDG for Na+ resulted in a small but statistically significant shift to a less positive reversal potential (p < 0.03, n = 3), whereas there was no difference when glucuronate was substituted for Cl−.
FIGURE 4.
Biophysical properties of ΔCR PrP-induced currents. A, I-V relationships were recorded from HEK cells expressing ΔCR PrP with a constant internal solution containing NMDG-glucuronate (NMDG-Glu (int)) and four different external solutions: NaCl (brown), sodium glucuronate (NaGluc, blue), NMDG-Cl (green), and NMDG-glucuronate (red). The graph shows a set of I-V plots from a representative single cell. B, I-V relationships were recorded from HEK cells expressing ΔCR PrP with a constant external solution containing NaCl and five different internal solutions: cesium glucuronate (Cs-Gluc), potassium glucuronate (K-Gluc), NMDG-glucuronate, Cs-Cl, and TEA-Cl. Each graph shows a representative single cell at four different time points during the recording. C, I-V relationships (upper graphs) were recorded from HEK cells expressing ΔCR PrP under control (Ctrl) solutions (light and dark gray) or in the presence of various multivalent cations (red): La3+ (15 μm), Ca2+ (12 mm), or Mg2+ (10 mm). The lower traces show patch clamp recordings during application of the multivalent cations. D, quantitation of the current activity recorded in the presence of the indicated cations, expressed as the percentage of activity under control conditions (mean ± S.E., n ≥ 3 cells; *, p < 0.05, significantly different from control).
We then recorded from cells with different internal solutions in the presence of a constant external solution containing NaCl. The I-V relationships were linear with internal solutions containing cesium glucuronate, potassium glucuronate, NMDG-glucuronate, Cs-Cl, or TEA-Cl, with the reversal potentials being either slightly negative or slightly positive (cesium glucuronate, −9 ± 2 mV, n = 10; potassium glucuronate, −5 ± 2 mV; NMDG-glucuronate, +11 ± 1 mV; Cs-Cl, −13 ± 2 mV; TEA-Cl, −6 ± 3 mV, n = 5) (Fig. 4B). Substitution of NMDG for Cs+ resulted in a small but statistically significant shift to more positive reversal potentials (p < 0.0001, n = 5), whereas substitution of K+ or TEA for Cs+ or Cl− for glucuronate had no significant effect.
Taken together, these ion substitution data indicate that small, inorganic cations (Na+, K+, Cs+), as well as large, organic cations (NMDG+ and TEA+) are capable of permeating the channel induced by ΔCR PrP, although the permeability to NMDG may be slightly less than that of the other cations. Because the currents recorded in solutions containing either Cl− or glucuronate were similar, the channel is either equally permeant to both anions or impermeant to both.
Next, a set of ions and drugs known to interact with specific classes of channels was added to the external recording solution and tested for their effects on channel activity (Fig. 4, C and D). Raising Ca2+ from 2 to 12 mm reduced activity to 27 ± 5% of control (p < 0.05, n = 5), whereas the addition of 10 mm Mg2+ caused minimal reduction (95 ± 14% of control, p > 0.7, n = 5). These results indicate specific inhibitory effect of Ca2+ that is not shared by all divalent cations. Adding 15 μm lanthanum (La3+), a trivalent cation known to block several kinds of calcium channels (23), reduced activity to 11 ± 23% of control values (p < 0.03, n = 3). In contrast to the effect of PPS, channel blockage by La3+ and Ca2+ was reversible, and spontaneous current activity was restored shortly after local perfusion of the ions was stopped (see current traces in Fig. 4C). TEA, a drug that blocks voltage-gated potassium channels, does not reduce the current and indeed is capable of sustaining the current as well as small, inorganic ions (Fig. 4B). Other drugs tested that did not affect the current include MK-801 (2 μm), which blocks N-methyl-d-aspartic acid (NMDA) receptors, and tetrodotoxin (500 nm), which blocks voltage-gated sodium channels (data not shown).
Disease-associated Point Mutants in PrP Induce Spontaneous Currents
Approximately 15% of the cases of prion disease are caused by dominantly inherited point or insertional mutations in the gene on chromosome 20 that encodes human PrP (24). Because several of the point mutations occur in residues within or near the CR region, we analyzed the mouse homologues of several of them to see whether they induced a channel activity like that associated with the ΔCR deletion. We focused on two mutations flanking the CR region (P101L and G130V) whose human counterparts are associated with Gerstmann-Strässler syndrome and one mutation within the CR region (G113V) whose human equivalent is linked to Creutzfeldt-Jakob disease.
We found that HEK cells stably expressing each of these mutants exhibited spontaneous, inward currents at negative holding potentials (Fig. 5A). Like the currents associated with ΔCR PrP, the currents induced by the point mutants fluctuated randomly, reaching amplitudes of up to several thousand pA. However, the proportion of time that cells displayed spontaneous current activity ≥450 pA was less for the point mutants than for ΔCR: 18 ± 5% for P101L, 42 ± 14% for G113V, and 28 ± 15% for G130V (n = 5) (Fig. 5B) when compared with 83 ± 17% for ΔCR (Fig. 1C). Importantly, M128V, a mutation in mouse PrP that is homologous to a non-pathogenic polymorphism in human PrP, failed to induce any current (2 ± 2%, n = 5), arguing that current activity is specifically associated with disease-causing mutations. Consistent with the conclusion, WT PrP did not induce any current (0 ± 0%, n = 5) regardless of the presence of the 3F4 epitope (L108M/V111M) within the CR region (compare WT PrP in Fig. 5 with WT PrP in Fig. 1).
Western blotting confirmed similar expression levels for WT PrP and the point mutants (Fig. 5C). In addition, all of the mutants could be detected on the cell surface by immunofluorescence staining (Fig. 5D).
Because humans are typically heterozygous for pathogenic PrP mutations, we tested the effect of co-expressing WT PrP with the pathogenic point mutants. Cells stably expressing P101L, G113V, and G130V PrPs were transduced with lentiviruses encoding both WT PrP and GFP or GFP alone, and patch clamp recordings were made from GFP-positive cells (Fig. 6A). Similarly to the ΔCR PrP-induced currents, the currents induced by the point mutants were effectively silenced in cells that co-expressed WT PrP (Fig. 6B). Western blotting with 3F4 antibody confirmed the expression of WT PrP (which carried the 3F4 epitope tag), whereas blotting with 6D11 antibody, which recognizes both WT and mutant PrP, demonstrated that the WT molecule was overexpressed with respect to the mutant molecule (Fig. 6C).
FIGURE 6.
Currents induced by disease-associated point mutants are silenced by overexpression of WT PrP. A, whole-cell patch clamp recordings at a holding potential of −80 mV were made from HEK cells expressing P101L, G113V, or G130V PrP 48 h after infection with lentivirus encoding GFP alone (upper traces) or WT PrP plus GFP (lower traces). B, quantitation of the currents recorded in panel A, plotted as the percentage of total time the cells exhibited inward current ≥450 pA (mean ± S.E., n ≥ 4 cells). Black bars represent GFP alone, and gray bars represent WT PrP plus GFP. C, Western blot showing relative PrP expression levels of each stably transfected cell line after transduction with either WT PrP-encoding or control lentiviruses. Antibody 6D11 recognizes both mutant and WT PrP, whereas antibody 3F4 specifically recognizes WT PrP (which contains the 3F4 epitope).
DISCUSSION
Biophysical Characteristics and Molecular Basis of ΔCR PrP-induced Currents
Expression of ΔCR PrP in several different cell types, including HEK, N2a, and Sf9, results in the induction of spontaneous currents measureable by patch-clamping techniques. These currents, which are inward at negative holding potentials, are highly irregular, fluctuating randomly over a time course of seconds to minutes and reaching amplitudes of several thousand pA. The currents are observed in the presence of small, inorganic cations (i.e. Na+, K+, and Cs+) as well as large, organic cations (i.e. TEA and NMDG) and are inhibited by La3+ and elevated Ca2+. Taken together, these results suggest that the currents associated with ΔCR PrP are produced by relatively non-selective, cation-permeable channels or pores. Consistent with this conclusion, the I-V relationships observed in the presence of all test solutions are linear and have reversal potentials near 0 mV. These cation-permeable channels or pores may be related to the ability of ΔCR PrP to sensitize transfected cells to the killing effects of aminoglycosides and bleomycin-type antineoplastic agents (12), which are known to enter cells via several kinds of ion channels (25–27).
What is the molecular basis of the ΔCR PrP-induced currents? One possibility is that ΔCR PrP enhances the activity of endogenous ion channels already present in the cell types we have analyzed. However, the currents are not characteristic of those associated with known families of voltage-gated sodium, potassium, or calcium channels, based on their biophysical properties, lack of inhibition by tetrodotoxin, persistence in the presence of TEA, and blockage by elevated Ca2+. In addition, HEK cells and Sf9 cells contain few endogenous ion channels (28, 29).
An alternative hypothesis is that ΔCR PrP molecules themselves form cation-permeable channels or pores in the cell membrane. If so, the intermittent nature of the current suggests that these structures assemble randomly over seconds to minutes, possibly reflecting transient associations between ΔCR PrP molecules within the plane of the membrane. The currents are rapidly blocked by PPS, a negatively charged glycosaminoglycan that binds to PrPC and inhibits the formation of PrPSc (19–21), suggesting that PPS binding to ΔCR PrP may interfere with intermolecular associations essential for membrane permeabilization. Other proteins could contribute to the formation of ΔCR PrP-induced channels, but if so, these putative interactors must be ubiquitous given the ability of ΔCR PrP to produce spontaneous currents in both insect and mammalian cells. If ΔCR PrP forms an ion channel, then the structure of the channel must be unusual because this protein lacks a hydrophobic domain sufficiently long to span the lipid bilayer. Although transmembrane forms of PrP have been described (30, 31), the ΔCR deletion (residues 105–125) removes most of the segment (residues 111–135) that serves as the transmembrane anchor in these molecules.
The currents produced by ΔCR PrP may reflect the formation of non-selective membrane pores or localized permeabilization of the lipid bilayer rather than the presence of conventional ion channels. The randomly fluctuating amplitude of the current is reminiscent of the effects produced by certain peptides, including amyloid β and cytolytic toxins, which form pores by transiently integrating into lipid bilayers (32). The peptide human PrP106–126 is toxic to cultured neurons and has been reported to form ion channels in planar lipid bilayers (33–35). However, these channels have different biophysical properties than those associated with ΔCR PrP. These differences, taken together with the fact that residues 106–126 correspond precisely to the region deleted in ΔCR PrP, suggest that the channels or pores induced by the two molecules are distinct. Furthermore, it has recently been reported that PrP106–126 does not interact with artificial membranes under physiological conditions and that association with cellular membranes is likely mediated by binding to PrPC (36, 37). Interestingly, PrP contains an N-terminal positively charged motif (23KKRPK27) shown to function as a protein transduction domain capable of ferrying adjacent polypeptide segments across the lipid bilayer (38). Perhaps the N-terminal region of the glycosylphosphatidylinositol-anchored ΔCR PrP molecule functions as a tethered protein transduction domain, transiently creating pores or channels in the membrane. Regardless of the underlying mechanism, the currents described here represent the first evidence for a channel- or pore-inducing activity associated with a membrane-anchored form of PrP synthesized by cells and found in vivo.
Disease-associated Point Mutants Induce Currents
Familial forms of prion disease are linked to dominantly inherited mutations in the gene encoding PrP, several of which are single amino acid substitutions occurring within or near the CR region (24). Mouse homologues of three of these point mutants (P101L, G113V, and G130V) were found to induce randomly fluctuating currents of several thousand pA, similar to the ΔCR PrP-induced currents. The proportion of time the current is active is lower for the point mutants (20–40% versus ∼80% for ΔCR), suggesting less frequent channel activation or pore formation or lower affinity for an endogenous, membrane-bound interactor.
Our study addresses a long standing conundrum in understanding how PrP mutations cause familial prion diseases. Although some disease-associated mutations destabilize the structure of the PrP molecule, resulting in the formation of potentially neurotoxic, PrPSc-like aggregates, others have little effect on PrP structure and result in a protein that is relatively soluble and protease-sensitive (39–41). Our results suggest that some of these mutations, in particular those within the CR region, induce an ion channel- or pore-inducing activity that contributes to their pathogenicity.
PrP-associated Currents and Neurotoxicity
Based on these results, we hypothesize that the neurotoxicity of PrP molecules carrying certain amino acid deletions and substitutions in the CR region depends on their ability to induce or activate cation-permeable channels or pores in the cell membrane. Supporting this conclusion is our observation that wild-type PrP potently suppresses the currents induced by ΔCR PrP, in parallel with its ability to reverse the neurodegenerative phenotype of Tg(ΔCR) mice (10). The antagonistic effects of WT and ΔCR PrP in both settings argue that the current-inducing activity of ΔCR PrP observed in vitro is mechanistically related to its neurotoxicity in vivo. Whether these effects are mediated by WT PrP itself or by the physiologically generated cleavage fragments such as N1 (42) has yet to be determined. We also find a correlation between current activity and pathogenicity in humans; three disease-associated point mutations in the CR region display current activity, whereas a non-pathogenic polymorphism does not. Interestingly, the currents induced by the point mutations can be silenced completely by overexpression of WT PrP. In a patient who is heterozygous for the mutation, where the WT and mutant PrPs are likely to be expressed in a 1:1 ratio, the amount of WT PrP may be insufficient to completely suppress the currents, resulting in the eventual onset of disease in late adult life.
The currents described here could mediate neuronal death by several possible mechanisms, including excessive influx of Na+ or other cations, depolarization of the membrane potential, or secondary activation of other ion channels. We have found that cerebellar granule cells in Tg(ΔCR) mice die by a novel, non-apoptotic pathway that is reminiscent of certain forms of glutamate-induced cell death (43). This suggests that ΔCR PrP-induced currents may initiate or augment excitotoxic pathways in the cerebellum. There are many precedents for hyperactivity of ion channels playing a role in neurodegeneration (44).
PrP-associated currents may also play a role in the pathogenesis of infectiously acquired prion diseases. The central region of PrP undergoes prominent conformational changes during the transition from PrPC to PrPSc (45), and it is possible that these changes endow the protein with a current-inducing activity similar to that produced by the ΔCR deletion. Such a scenario would be consistent for the essential role of membrane-bound PrPC in transducing the toxic effects of PrPSc (46–48). Oligomers of recombinant PrP have been shown to permeabilize artificial lipid vesicles through interactions involving the central region (49). Interestingly, Laurén et al. (50) recently identified PrPC as a cell surface receptor for amyloid β oligomers, possibly connecting PrP-induced currents and amyloid β synaptotoxicity. This connection, however, remains controversial as another group reported no difference in amyloid β oligomer-mediated impairment of long term memory between wild-type and PrP knock-out mice (51).
There are several previous reports suggesting that wild-type PrPC influences the activity of ion channels. For example, neurons from PrP-null mice have been reported to display various kinds of electrophysiological abnormalities (52–55). It has been suggested that PrPC specifically suppresses the activity of a subclass of NMDA-type glutamate receptors based on the demonstration that PrPC physically interacts with the NR2D subunit of these receptors and the fact that neurons from Prn-p−/− mice showed enhanced NMDA-evoked currents and were more susceptible to glutamate-induced excitotoxic death (56). Taken together with our observation that WT PrP silences the currents induced by ΔCR PrP, the available evidence suggests a possible physiological role for PrPC in regulation of several kinds of ion channels. This activity could explain some of the neuroprotective effects that have been attributed to PrP (reviewed in Ref. 57).
The results presented here suggest a novel therapeutic approach to prion diseases based on inhibiting PrP-related currents. It will be important now to identify compounds that block PrP-induced currents and to test these in animal models of familial and infectious prion diseases. It should be possible to discover such compounds rapidly in high throughput screens using as a cellular assay the ability of ΔCR PrP to sensitize cells to cationic drugs (12).
Acknowledgments
We thank Tim Wilding, Joe Amatrudo, and Jennifer Leubke for advice and technical assistance with patch clamping. We also acknowledge Aimin Li for help with Sf9 cells, Till Voigtlaender and Nada Husic for assistance with lentiviral preparation, and Tania Massignan for generation of several of the HEK cell lines. Lentiviral constructs were created by the Hope Center Viral Vectors Core at Washington University, which is supported by a Neuroscience Blueprint Core grant (Grant P30 NS057105) from the National Institutes of Health.
This work was supported, in whole or in part, by National Institutes of Health Grants NS052526 and NS040975 (to D. A. H.) and NS30888 (to J. E. H.) and by National Institutes of Health Predoctoral Fellowship NS063547 and the Medical Scientist Training Program at Washington University (National Institutes of Health Grant T32GM007200) (to I. H. S.).
- PrP
- prion protein
- PrPC
- cellular isoform of PrP
- PrPSc
- scrapie isoform of PrP
- CR
- central region
- PPS
- pentosan polysulfate
- NMDG
- N-methyl-d-glucamine
- TEA
- tetraethylammonium.
REFERENCES
- 1.Prusiner S. B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prusiner S. B. (ed) (2004) Prion Biology and Diseases, Second Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 3.Chiesa R., Harris D. A. (2001) Neurobiol. Dis. 8, 743–763 [DOI] [PubMed] [Google Scholar]
- 4.Harris D. A., True H. L. (2006) Neuron 50, 353–357 [DOI] [PubMed] [Google Scholar]
- 5.Büeler H., Fischer M., Lang Y., Bluethmann H., Lipp H. P., DeArmond S. J., Prusiner S. B., Aguet M., Weissmann C. (1992) Nature 356, 577–582 [DOI] [PubMed] [Google Scholar]
- 6.Westergard L., Christensen H. M., Harris D. A. (2007) Biochim. Biophys. Acta 1772, 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumann F., Pahnke J., Radovanovic I., Rülicke T., Bremer J., Tolnay M., Aguzzi A. (2009) PLoS One 4, e6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shmerling D., Hegyi I., Fischer M., Blättler T., Brandner S., Götz J., Rülicke T., Flechsig E., Cozzio A., von Mering C., Hangartner C., Aguzzi A., Weissmann C. (1998) Cell 93, 203–214 [DOI] [PubMed] [Google Scholar]
- 9.Baumann F., Tolnay M., Brabeck C., Pahnke J., Kloz U., Niemann H. H., Heikenwalder M., Rülicke T., Bürkle A., Aguzzi A. (2007) EMBO J. 26, 538–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li A., Christensen H. M., Stewart L. R., Roth K. A., Chiesa R., Harris D. A. (2007) EMBO J. 26, 548–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen H. M., Harris D. A. (2009) J. Neurochem. 108, 44–56 [DOI] [PubMed] [Google Scholar]
- 12.Massignan T., Stewart R. S., Biasini E., Solomon I. H., Bonetto V., Chiesa R., Harris D. A. (2010) J. Biol. Chem. 285, 7752–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolton D. C., Seligman S. J., Bablanian G., Windsor D., Scala L. J., Kim K. S., Chen C. M., Kascsak R. J., Bendheim P. E. (1991) J. Virol. 65, 3667–3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolkowicz R., Nolan G. P., Curran M. A. (2004) Methods Mol. Biol. 246, 391–411 [DOI] [PubMed] [Google Scholar]
- 15.Li M., Rossi J. J. (2005) Methods Mol. Biol. 309, 261–272 [DOI] [PubMed] [Google Scholar]
- 16.Huettner J. E., Lu A., Qu Y., Wu Y., Kim M., McDonald J. W. (2006) Stem Cells 24, 1654–1667 [DOI] [PubMed] [Google Scholar]
- 17.Wilding T. J., Zhou Y., Huettner J. E. (2005) J. Neurosci. 25, 9470–9478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pankiewicz J., Prelli F., Sy M. S., Kascsak R. J., Kascsak R. B., Spinner D. S., Carp R. I., Meeker H. C., Sadowski M., Wisniewski T. (2006) Eur. J. Neurosci. 23, 2635–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doh-ura K., Ishikawa K., Murakami-Kubo I., Sasaki K., Mohri S., Race R., Iwaki T. (2004) J. Virol. 78, 4999–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warner R. G., Hundt C., Weiss S., Turnbull J. E. (2002) J. Biol. Chem. 277, 18421–18430 [DOI] [PubMed] [Google Scholar]
- 21.Caughey B., Raymond G. J. (1993) J. Virol. 67, 643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shyng S. L., Lehmann S., Moulder K. L., Harris D. A. (1995) J. Biol. Chem. 270, 30221–30229 [DOI] [PubMed] [Google Scholar]
- 23.Pałasz A., Czekaj P. (2000) Acta. Biochim. Pol. 47, 1107–1114 [PubMed] [Google Scholar]
- 24.Mead S. (2006) Eur. J. Hum. Genet. 14, 273–281 [DOI] [PubMed] [Google Scholar]
- 25.Myrdal S. E., Steyger P. S. (2005) Hear. Res. 204, 170–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcotti W., van Netten S. M., Kros C. J. (2005) J. Physiol. 567, 505–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thornton G., Wilkinson C. R., Toone W. M., Jones N. (2005) Genes Cells 10, 941–951 [DOI] [PubMed] [Google Scholar]
- 28.Thomas P., Smart T. G. (2005) J. Pharmacol. Toxicol. Methods 51, 187–200 [DOI] [PubMed] [Google Scholar]
- 29.Larsen E. H., Gabriei S. E., Stutts M. J., Fullton J., Price E. M., Boucher R. C. (1996) J. Gen. Physiol. 107, 695–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hegde R. S., Mastrianni J. A., Scott M. R., DeFea K. A., Tremblay P., Torchia M., DeArmond S. J., Prusiner S. B., Lingappa V. R. (1998) Science 279, 827–834 [DOI] [PubMed] [Google Scholar]
- 31.Stewart R. S., Drisaldi B., Harris D. A. (2001) Mol. Biol. Cell 12, 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kourie J. I., Shorthouse A. A. (2000) Am. J. Physiol. Cell Physiol. 278, C1063–C1087 [DOI] [PubMed] [Google Scholar]
- 33.Forloni G., Angeretti N., Chiesa R., Monzani E., Salmona M., Bugiani O., Tagliavini F. (1993) Nature 362, 543–546 [DOI] [PubMed] [Google Scholar]
- 34.Lin M. C., Mirzabekov T., Kagan B. L. (1997) J. Biol. Chem. 272, 44–47 [DOI] [PubMed] [Google Scholar]
- 35.Kourie J. I., Culverson A. (2000) J. Neurosci. Res. 62, 120–133 [DOI] [PubMed] [Google Scholar]
- 36.Henriques S. T., Pattenden L. K., Aguilar M. I., Castanho M. A. (2009) Biochemistry 48, 4198–4208 [DOI] [PubMed] [Google Scholar]
- 37.Henriques S. T., Pattenden L. K., Aguilar M. I., Castanho M. A. (2008) Biophys. J. 95, 1877–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wadia J. S., Schaller M., Williamson R. A., Dowdy S. F. (2008) PLoS One 3, e3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riek R., Wider G., Billeter M., Hornemann S., Glockshuber R., Wüthrich K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Apetri A. C., Surewicz K., Surewicz W. K. (2004) J. Biol. Chem. 279, 18008–18014 [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y., Swietnicki W., Zagorski M. G., Surewicz W. K., Sönnichsen F. D. (2000) J. Biol. Chem. 275, 33650–33654 [DOI] [PubMed] [Google Scholar]
- 42.Guillot-Sestier M. V., Sunyach C., Druon C., Scarzello S., Checler F. (2009) J. Biol. Chem. 284, 35973–35986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christensen H. M., Dikranian K., Li A., Baysac K. C., Walls K. C., Olney J. W., Roth K. A., Harris D. A. (2010) Am. J. Pathol. 176, 2695–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lester H. A., Karschin A. (2000) Annu. Rev. Neurosci. 23, 89–125 [DOI] [PubMed] [Google Scholar]
- 45.Peretz D., Williamson R. A., Matsunaga Y., Serban H., Pinilla C., Bastidas R. B., Rozenshteyn R., James T. L., Houghten R. A., Cohen F. E., Prusiner S. B., Burton D. R. (1997) J. Mol. Biol. 273, 614–622 [DOI] [PubMed] [Google Scholar]
- 46.Mallucci G., Dickinson A., Linehan J., Klöhn P. C., Brandner S., Collinge J. (2003) Science 302, 871–874 [DOI] [PubMed] [Google Scholar]
- 47.Brandner S., Isenmann S., Raeber A., Fischer M., Sailer A., Kobayashi Y., Marino S., Weissmann C., Aguzzi A. (1996) Nature 379, 339–343 [DOI] [PubMed] [Google Scholar]
- 48.Chesebro B., Trifilo M., Race R., Meade-White K., Teng C., LaCasse R., Raymond L., Favara C., Baron G., Priola S., Caughey B., Masliah E., Oldstone M. (2005) Science 308, 1435–1439 [DOI] [PubMed] [Google Scholar]
- 49.Chich J. F., Chapuis C., Henry C., Vidic J., Rezaei H., Noinville S. (2010) J. Mol. Biol. 397, 1017–1030 [DOI] [PubMed] [Google Scholar]
- 50.Laurén J., Gimbel D. A., Nygaard H. B., Gilbert J. W., Strittmatter S. M. (2009) Nature 457, 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balducci C., Beeg M., Stravalaci M., Bastone A., Sclip A., Biasini E., Tapella L., Colombo L., Manzoni C., Borsello T., Chiesa R., Gobbi M., Salmona M., Forloni G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collinge J., Whittington M. A., Sidle K. C., Smith C. J., Palmer M. S., Clarke A. R., Jefferys J. G. (1994) Nature 370, 295–297 [DOI] [PubMed] [Google Scholar]
- 53.Maglio L. E., Perez M. F., Martins V. R., Brentani R. R., Ramirez O. A. (2004) Brain Res Mol. Brain Res. 131, 58–64 [DOI] [PubMed] [Google Scholar]
- 54.Herms J. W., Tings T., Dunker S., Kretzschmar H. A. (2001) Neurobiol. Dis. 8, 324–330 [DOI] [PubMed] [Google Scholar]
- 55.Fuhrmann M., Bittner T., Mitteregger G., Haider N., Moosmang S., Kretzschmar H., Herms J. (2006) J. Neurochem. 98, 1876–1885 [DOI] [PubMed] [Google Scholar]
- 56.Khosravani H., Zhang Y., Tsutsui S., Hameed S., Altier C., Hamid J., Chen L., Villemaire M., Ali Z., Jirik F. R., Zamponi G. W. (2008) J. Cell Biol. 181, 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roucou X., Gains M., LeBlanc A. C. (2004) J. Neurosci. Res. 75, 153–161 [DOI] [PubMed] [Google Scholar]