Abstract
Coronary arteries supply the heart with oxygen and nutrients, and coronary artery disease is the leading cause of death. Elucidating the program of coronary artery development could aid understanding of the disease and lead to new treatments, but many aspects of the process, including their developmental origin, remain obscure. Current models posit that coronary arteries form by de novo assembly of endothelial tubes from progenitor cells in the proepicardium, a tissue that spreads over and contributes to the developing heart. Here, we use histological and clonal analysis in mice, and cardiac organ culture, to show that coronary vessels arise instead from angiogenic sprouts of the sinus venosus, the major vein that returns circulating blood to the embryonic heart. Sprouting venous endothelial cells dedifferentiate as they migrate over and invade the myocardium. Invading cells differentiate into arteries and capillaries, whereas cells remaining on the surface redifferentiate into veins. These results show that some differentiated venous cells retain developmental plasticity, and suggest that position-specific cardiac signals trigger their dedifferentiation and conversion into coronary arteries, capillaries, and veins. Understanding this novel developmental reprogramming process and identifying the endogenous signals should suggest more natural ways of engineering coronary bypass grafts and revascularizing the heart.
Introduction
There has been tremendous progress over the past two decades elucidating general principles of blood vessel development and factors that control it1–4, especially during tumor angiogenesis5,6. Comparatively little is known, however, about the cellular origins and developmental programs of many of our most important vessels and vascular beds, such as the coronary arteries that supply the heart muscle (myocardium) and its pacemaker7–9. Seven million people die each year of coronary artery disease and its sequealae, myocardial infarction and cardiac arrest, making it the leading cause of death worldwide10. Tens of millions of others manage the disease medically or undergo major interventions such as angioplasty or coronary bypass surgery10. Elucidating how coronary vessels arise during development and are maintained in adult life, and how they are remodeled under pathological conditions such as arteriosclerosis, should further our understanding of the disease. It could also lead to new treatments that stimulate vessel growth and new ways of engineering bypass grafts with the flow properties and durability of healthy young vessels.
The development of coronary arteries has been studied in a variety of animals for over a century. Early anatomical studies in humans and other mammals suggested that they bud from the aorta11–14. More recent experiments with chick-quail chimeras argued against this and suggested instead that they arise from the proepicardium, a transitory structure in the embryo that contacts and spreads over the developing heart to form its epithelial covering (epicardium) and several internal tissues15–17. The chick experiments and subsequent studies led to the current textbook view that coronary vessels form from proepicardial cells that undergo an epithelial-to-mesenchymal transition, and then differentiate into isolated endothelial progenitors that assemble de novo (“vasculogenesis”) into endothelial tubes7,18. However, not all data from chick is easily reconciled with this model9,19, and recent lineage tracing experiments in mouse show that, although the proepicardium gives rise to myocardial stroma and vascular smooth muscle, it gives rise to few, if any coronary endothelial cells20–23. Thus, the origin of coronary artery endothelial cells, among the most medically important cells in the body, remains an enigma.
Here, we use histological and clonal analysis in mouse, and cardiac organ culture, to investigate the origins and early development of coronary arteries. The results show that coronary arteries do not derive from the proepicardium but from endothelial sprouts of the sinus venosus, the venous inflow tract of the embryonic heart, plus a small contribution from endocardium lining the cardiac chambers. The patterns of migration and marker expression of the venous sprouts suggest a model in which local signals in the developing heart induce angiogenic outgrowth, dedifferentiation, and stepwise conversion into coronary arteries, capillaries, and veins.
Histological analysis of coronary development
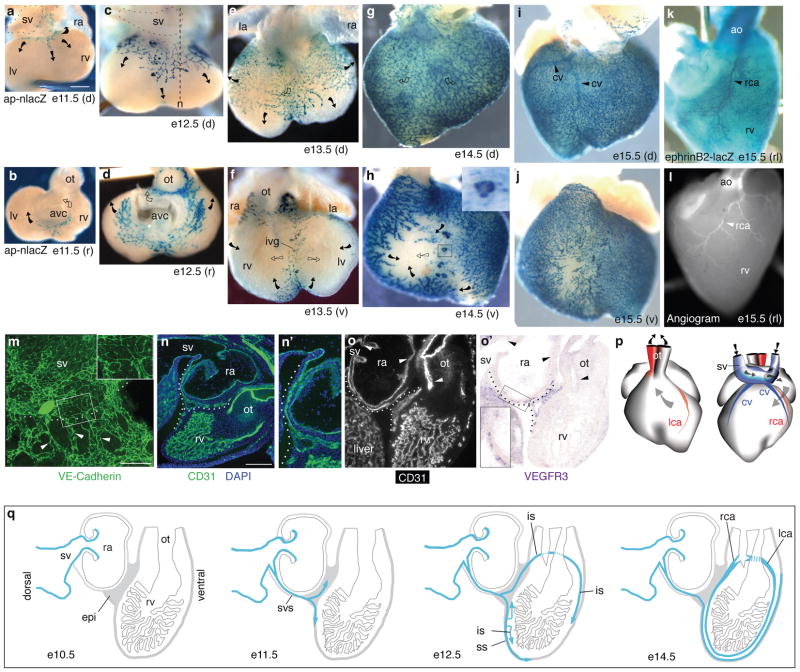
We carried out a thorough anatomical and histological analysis of coronary vessel development during mouse embryogenesis using endothelial markers (Figs. 1 and S1). Because canonical endothelial markers also label endocardial cells, complicating the analysis (Fig. S2), we used an apelin-nlacZ knock-in mouse strain24 that selectively expresses nuclear beta-galactosidase in coronary endothelial cells but not endocardium (Fig. S3). X-gal staining showed an expanding vascular plexus on the heart beginning at embryonic day (e) 11.5 (Fig. 1a–j), indistinguishable from that detected by CD31 (PECAM-1) immunostaining (Fig. S2)25,26. The plexus originated on the dorsal cardiac surface (Fig. 1a), and expanded around the atrioventricular canal (Fig. 1b) to reach the outflow tract (Fig. 1d,f), and invade the ventral interventricular groove (Fig. 1f). It also expanded caudally (Fig. 1c,e), populating the entire dorsal surface by e14.5 (Fig. 1g); during this period (e12.5–14.5), dorsal plexus sprouts invaded the underlying myocardium (Fig. S1). Larger bore coronary arteries appeared within the myocardium at e14.5 and subsequently matured (Fig. 1k,l,p). By e15.5, the entire ventral surface was also covered with plexus (Fig. 1j), and coronary veins began to appear on the dorsal surface (Fig. 1i). There were also isolated clusters of endothelial cells surrounding blood cells including erythrocytes (Ter119+), platelets (CD41+), and leukocytes (CD45+), which appeared transiently at the interventricular groove (Fig. 1h inset and Fig. S2); these were noted previously and called “blood islands”14,27, but their origins and developmental fate are unknown (see below).
Figure 1. Coronary vessels sprout from the sinus venosus.
a–j. X-gal stained apelin-nlacZ hearts from embryonic days indicated, shown in dorsal (d), rostral (r), and ventral (v) views. Coronary vessels (blue nuclei) originate near the sinus venosus (sv, outlined in a,c) and spread dorsally (a, c, e, g) and around the atrioventricular canal (avc) (b, d) and outflow tract (ot) (d) to the ventral interventricular groove (ivg) (f). Arrows indicate direction of new growth on the surface (filled) or in deeper layers (open). Inset in h is close up of boxed blood island-like structure. la, left atrium; ra, right atrium; rv, right ventricle; lv, left ventricle. Scale bar (for a–l), 200 μm. k. X-gal-stained e15.5 ephrinB2-lacZ heart (right lateral (rl) view) showing right coronary artery (rca, arrowhead). ao, aorta. l. Angiogram of e15.5 heart showing right coronary artery (rca, arrowhead). m. Frontal section through e11.5 heart immunostained for VE-cadherin. Coronary sprouts (green, arrowheads) are continuous with the SV (above dashed line). Inset is close up of boxed region showing junction between SV and coronary sprouts. Scale bar, 100 μm. n. Sagittal section (approximate section plane shown in c) through e12.5 heart stained for CD31 (green) and with nuclear dye DAPI (blue). Coronary sprout (dotted line) extends from the sv and courses around right atrium (ra) to reach right ventricle (rv). n’. Close-up of coronary sprout in n. o,o’. Adjacent sections from e11.5 heart immunostained for CD31 (o) and probed for VEGFR3 mRNA (purple) (o’). Coronary vessels (dotted line) express VEGFR3, an angiogenic sprout marker, whereas vessels formed by vasculogenesis (sv, atria, outflow tract) do not (arrowheads). Inset in o’ is close-up of boxed region. Scale bar (for n,o,o’), 200 μm. p. Schematic of right (rca) and left (lca) coronary arteries and coronary veins (cv) at e15.5. Arrows show blood flow direction. q. Schematic sagittal sections showing coronary sprout (blue) progression during development. epi, epicardium; svs, sinus venosus sprouts; ss, superficial sprouts; is, invading sprouts.
Close inspection of the site where the plexus originated (Fig. 1a,m,n,n’) suggested that the source of endothelial cells is the sinus venosus (SV). Vessels extending from the SV expressed apelin and three other angiogenic sprout markers28–31 (VEGFR3, Delta-like 4, and PDGF-B) (Fig. 1o, o’, and data not shown), supporting an angiogenic origin. These sprouts grew through the inflow tract myocardium and onto the cardiac surface, traversing a stereotyped path as they extended to invest the heart (Fig. 1n,q). Aside from these sprouts and adjacent liver plexus, we did not detect any endothelial cells in or associated with the proepicardium (Fig. S4), nor did we detect any proepicardial cells that became SV or other endothelial cells by Tbx-18–Cre lineage tracing (Fig. S5). We also detected only extremely rare VEGFR2+CD31− cells, putative endothelial progenitors (“angioblasts”) that are abundant around vessels forming by vasculogenesis in other body regions32, in the proepicardium or heart wall (Fig. S6). Thus, coronary vessel progenitors appear to arise from SV angiogenic sprouts, not from the proepicardium or other vasculogenic sources.
Analysis of coronary origins in vitro
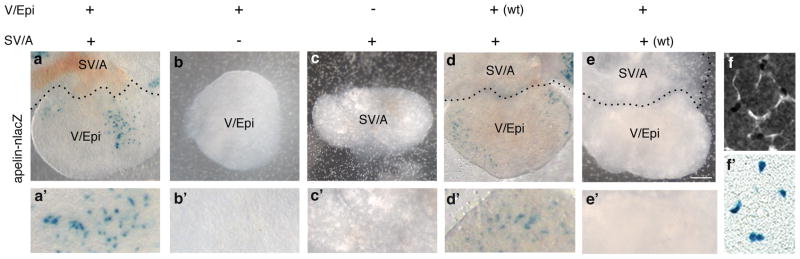
We tested the SV sprouting model in two ways. First, the origins and requirements for coronary sprouting were analyzed in a cardiac organ culture system26 modified to allow tissue recombination experiments (see Methods). Developing hearts were isolated from apelin-nLacZ mice at e10.5, after the proepicardium has spread over the heart surface to form the epicardium but before any coronary sprouts are present. Hearts were either left intact or dissected to separate the ventricles with their epicardial covering (V/Epi) from SV and atria (SV/A), and then cultured for 72 hours and stained for coronary endothelial markers (apelin-nlacZ or CD31) to determine if coronary vessels had formed (Fig. 2). All of the dissected chambers (n=39) appeared healthy and continued to beat throughout the culture period, as did control hearts that were left intact (n=24) (Movies S1-4). However, whereas coronary sprouts formed in 22 of 24 (92%) intact control hearts, none developed when the V/Epi (n=30) or SV/A (n=9) was cultured alone (Fig. 2a–c, f,f’). In tissue recombination experiments in which a dissected SV/A from apelin-nlacZ transgenic mice was recombined with a wild-type (non-transgenic) V/Epi, coronary vessels formed on the V/Epi and expressed the apelin-nlacZ transgene (Fig. 2d; 13 of 20, 65%). By contrast, no coronary vessels expressing the transgene developed when a wild type SV/A was cultured with an apelin-nlacZ V/Epi (Fig. 2e; n=5). Hence, apelin-nlacZ+ coronary vessels arise from the dissected SV/A tissue, not the epicardial cells or other proepicardial derivatives in the dissected V/Epi, but they require signals provided by the V/Epi for sprouting and outgrowth.
Figure 2. Analysis of coronary vessel sprouting in vitro.
a–e. Intact heart (a) or dissected sinus venosus/atria (SV/A) or ventricle/epicardium (V/Epi) as indicated from e10.5 apelin-nlacZ embryos (or wild type (wt) where noted) cultured for 3 days at 37°C and stained with X-gal (blue nuclei) to show coronary development. Coronary sprouts formed only on intact hearts (a) or when dissected SV/A were recombined with V/Epi (d). Vessels arise from SV/A because no X-gal+ sprouts were detected when SV/A was from wild type (non-transgenic) embryo (e). a’–e’ Close up views. f,f’. Close-up of a cultured intact heart as in a stained for CD31 (f) and with X-gal (f’), confirming lacZ-positive (blue) nuclei are in coronary vessels. Scale bar, 200 μm.
Clonal analysis of coronary development
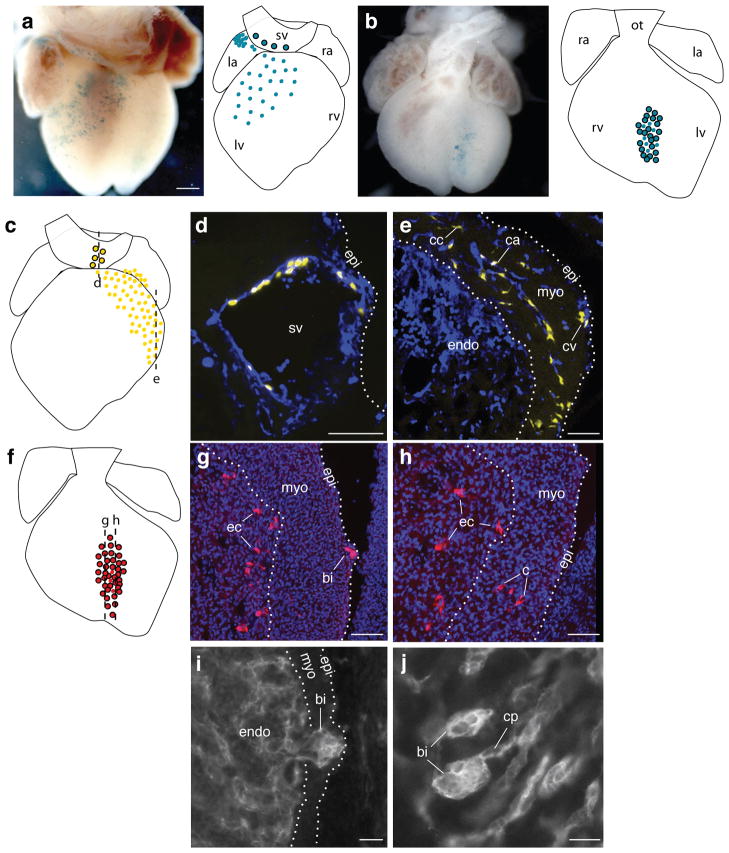
We next tested the SV sprouting model by clonal analysis, which allowed us to map the origin and determine the proliferation rate, outgrowth pathway, and fate of coronary endothelial progenitors in vivo. We used a VE-Cadherin-CreER transgene that expresses a tamoxifen-inducible Cre recombinase33, in combination with Cre-dependent marker genes (see Methods), to permanently label individual endothelial cells and their descendants. Recombination was induced by tamoxifen administration between e7.5 and e9.5 and analyzed 4–7 days later. Limiting doses of tamoxifen were used, and hearts with a single cluster of marked cells (putative clone) or two or three well-isolated clusters were analyzed (Fig. 3a,b). A multi-color Cre-dependent marker similar to Brainbow34 was used to confirm clonality of clusters (Fig. 3c–h). The endothelial cell proliferation rate estimated from the clonal analysis (Table S1) was 11±5 hours.
Figure 3. Clonal analysis of coronary artery development.
a–h. Micrographs and schematics of VE-cadherin-CreER-induced coronary clones at e13.5 (b,f) and e14.5 (a,c) marked with Rosa-lacZ (a,b) or Multi-color (c–h), in which marked cells permanently express one of three different fluorescent reporters. Colored dots in schematics represent 10 cells in clone. Outlined dots are sister cells in SV or endocardium. a. Coronary clone (no. 24 in Table S1) with sister cells in sinus venosus (sv). Scale bar (for a,b), 200 μm. b. Coronary clone (no. 34) with sister cells in ventricular endocardium. c–e. Schematic (c) and two sections (d,e, dashed lines in c) of a coronary clone (no. 25) with sister cells in sinus venosus (d) and coronary artery (ca), coronary capillary (cc), and coronary vein (cv) (e). CD31 immunostaining (blue) shows all endothelial and endocardial cells. f–h. Schematic (f) and sections (g,h) of coronary clone (no. 28) with sister cells in coronary vessels (c), endocardial cells (ec), and adjacent blood island (bi). Nuclei are DAPI-stained (blue). Scale bars (d,e,g,f), 100 μm. i, j. Sections of e11.5 (i) and e12.5 (j) hearts immunostained for CD31 showing blood island-like structures budding from endocardium (endo) into myocardium (myo) (i) and joining the coronary vascular plexus (cp) (j). Scale bars, 25 μm. epi, epicardium; la, left atria; lv, left ventricle; ot, outflow tract; ra, right atria; rv, right ventricle.
The clonal analysis (Table S1, Fig. S7) established two key aspects of the SV sprouting model. First, because coronary endothelial clones were consistently obtained following early (e7.5) induction of VE-Cadherin-CreER, coronary artery progenitors must arise from differentiated (VE-Cadherin+) endothelial cells present at this age. This is the result expected from the SV sprouting model, and it argues against a proepicardial origin because no VE-Cadherin+ cells other than SV sprouts and liver plexus are associated with the proepicardial organ (Fig. S4). Second, most coronary artery cells are clonally related to sinus venosus cells. 25 of 34 coronary endothelial clones, and all 20 that contained more than 85 coronary endothelial cells, had labeled sister cells within the SV (Fig. 3a, c–e; Table S1, Fig. S7). These clones spanned the coronary plexus from the SV at the dorsal side of the heart, where plexus formation begins, to the growing front of the plexus (Fig S7, type I). The longer the period between clone induction and analysis, the farther the clones extended from the SV, consistent with an SV origin. At the latest times examined, clones included endothelial cells of coronary arteries, veins, and capillaries, demonstrating that a single labeled cell can be reprogrammed to form not just coronary artery but all three types of coronary endothelial cells (Fig. 3e; Table S1).
The clonal analysis revealed a minor secondary source of coronary endothelial cells. The nine coronary endothelial clones that did not contain any SV sister cells each contained sister cells in a patch of ventricular (8 of 9) or atrial (1 of 9) endocardium underlying the clone (Fig. 3b, f–h; Table S1). The presence of blood island-like structures in four of the ventricular clones, and their location near the interventricular groove, indicate that they correspond to the endothelial “blood islands” noted above in the histological analysis (Figs. 1h and S2). Thus, this population of endocardium-derived coronary endothelial cells may form by endocardial budding through the myocardium, pinching-off and forming endothelial spheres with entrapped blood cells (“blood islands”) that later join the SV-derived plexus. In support of this, we identified putative intermediates in which endocardial buds were seen penetrating the myocardium (Fig. 3i) and “blood islands” were caught joining the coronary plexus (Fig. 3j).
Marker expression during venous reprogramming
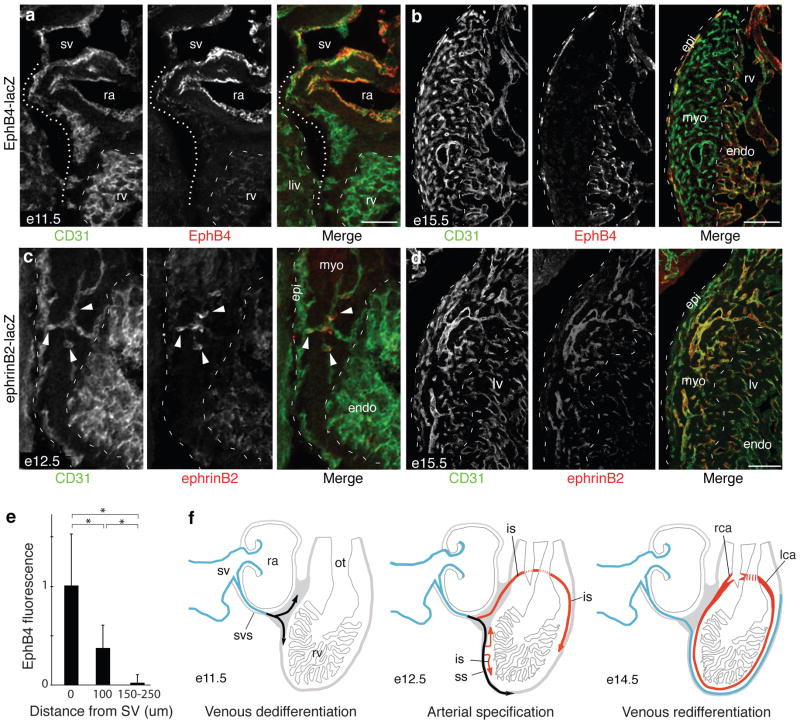
The above experiments establish that coronary artery progenitors arise primarily from VE-Cadherin+ cells that sprout from the SV and migrate over the heart and into the myocardium as they become coronary arteries. Because the path is stereotyped, we could analyze molecular transitions during the process. Marker analysis showed that sprouting SV cells, like other SV cells, are initially differentiated (EphB4+, Coup-TF2+) venous cells. But as they extend out from the SV, expression of the markers rapidly declines and venous identity is lost (Figs. 4a, e, and S8). A day later (e12.5), vessels that have invaded the myocardium begin to express ephrinB2-lacZ (Fig. 4c)35 and other arterial markers (Dll4, Hey1, Notch4, and Depp) (Fig. S9) and subsequently assemble into mature coronary arteries (Figs. 4d, S9). Vessels that remain superficial turn on EphB4-lacZ (Fig. 4b)35 and other venous markers (VEGFR3, Np2, Aplnr, Coup-TF2) as they form mature coronary veins (Fig. S9).
Figure 4. Downregulation of venous markers and induction of arterial markers during coronary artery development.
a. Sagittal section through heart of e11.5 EphB4-lacZ embryo immunostained for CD31 (green) and β-gal (EphB4, red). Coronary sprouts (dotted line) downregulate EphB4 as they migrate away from sinus venosus (sv). ra, right atria; rv, right ventricle; endo, endocardium; myo, myocardium; liv, liver. Scale bar, 100 μm. b. e15.5 EphB4-lacZ heart. Vessels on surface (epicardium, epi) have re-acquired EphB4 expression. Scale bar, 200 μm. c. Sagittal section through e12.5 ephrinB2-lacZ heart. Coronary sprouts that have invaded myocardium (green, arrowheads) upregulate artery/capillary marker ephrinB2 (red), whereas surface vessels (green, dashed line at left) do not. d. e15.5 ephrinB2-lacZ heart. EphrinB2 is expressed by all arteries and capillaries within myocardium but not by surface vessels. Scale bar, 100 μm. e. Quantification of EphB4 expression in coronary sprouts budding from sinus venosus (sv) at e11.5. Values are from double-stained hearts as in (a), normalized to CD31 staining at same position. Error bars, s.d. *, p<0.002 by Students t-test. f. Schematic showing changes in venous (blue) and arterial (red) marker expression during coronary development; black indicates dedifferentiated venous cells. svs, sinus venosus sprouts; ss, superficial sprouts; is, invading sprouts.
Discussion
Our results resolve the century-old enigma of the origin of coronary arteries and demonstrate two surprising sources of progenitors and a novel program of arteriogenesis. The major source is differentiated venous endothelial cells of the SV, which sprout onto the developing heart. There they dedifferentiate and spread to form the coronary plexus, and subsequently redifferentiate and remodel into coronary arteries, capillaries, and veins (Fig. S10). A minor secondary source is the endocardium, from which cells separate to form blood islands and then join the coronary plexus near the interventricular septum. Our data do not exclude rare contributions from other sources, or distinct origins in other species such as chick.
Coronary sprouts follow a specific outgrowth path, and dedifferentiation and specific redifferentiation events occur at stereotyped times and positions. This suggests that local signals along the outgrowth path function not only as sprout inducers and guidance cues, but also serve to sequentially deprogram SV venous endothelial cells and reprogram them to coronary artery, venous, and capillary fates (Fig. S10). Our results demonstrate that differentiated venous cells can give rise to arterial vessels36, and we speculate that other organ-specific vascular beds, such as those of the retina and kidney, form in a similar manner: by developmental reprogramming of differentiated venous cells. Indeed, at least some venous cells in other parts of the body appear to retain plasticity because they can give rise to lymphatic vessels during development4. Furthermore, experimental manipulations such as flow reversal37 and transplantation38,39 have been shown to alter the identity of embryonic venous cells, and adult saphenous veins used in coronary bypass grafts downregulate venous markers40. However, venous grafts do not acquire a full arterial phenotype40, which likely contributes to their lower success rate compared to arterial grafts41,42. Identification of the endogenous signals that control coronary artery development, especially the dedifferentiation and reprogramming factors, would begin to elucidate the molecular basis of this novel developmental process and suggest more natural ways of inducing new coronary vessels and engineering bypass grafts.
Methods summary
Wild type (CD1) and transgenic marker, Cre recombinase, and Cre reporter mouse strains are described in Full Methods. Staged embryos and hearts from timed pregnancies (morning of vaginal plug designated day e0.5) were dissected and fixed in 4% paraformaldehyde and stored in PBS. Fixed tissues were left intact or sectioned, and then processed for whole mount histochemistry (X-gal), immunohistochemical staining, indirect or direct immunofluorescence, and RNA in situ hybridization with digoxigenin-labeled antisense probes as described in Full Methods. Specimens were imaged on a stereomicroscope (whole mount tissue) or fluorescence compound or confocal microscopes (tissue sections); images were digitally captured and processed.
For organ culture, dissected hearts from e10.5 or e11 wild type or apelin-nlacZ embryos were placed on polycarbonate filters at the air-liquid interface of DMEM media with supplements. Some hearts were cultured intact; others were dissected into SV/atria and ventricle and the fragments cultured alone or recombined with an SV/atria contacting a ventricle. Cultures were maintained at 37°C and 5% CO2 for 72 hours, then fixed and subjected to whole mount X-gal staining to detect apelin-nlacZ+ vessels.
For clonal analysis, VE-cadherin-CreER mice were crossed to Rosa-lacZ or Multi-color Cre recombination reporters. At e7.5, 8.5 or 9.5, dams were injected intraperitonealy with low doses of tamoxifen to activate CreER and induce rare recombination events, generating single or well-separated clones of endothelial cells that constitutively express β-gactosidase (Rosa-lacZ) or one of three different fluorescent proteins (Multi-color). At e13.5 or 14.5, embryos were dissected, fixed, and stained to visualize the marked endothelial clones: embryos carrying Rosa-lacZ were stained with X-gal and the endothelial-specific antibody anti-CD31, and Multi-color embryos were sectioned and stained with DAPI or anti-CD31 and analyzed with fluorophore-specific filters.
Methods
Animals
CD1 mice (Charles River Laboratories) were used for wild-type analysis. EphB4-lacZ (Jackson Laboratories strain B6.129S7-Ephb4tm1And/J), ephrinB2-lacZ (B6.129S7-Efnb2tm2And/J), VE-cadherin-Cre (B6. Cg-Tg(Cdh5-cre)7Mlia/J), the Cre reporter strains Rosa-lacZ (B6.129S4-Gt(ROSA)26Sortm1Sor/J) and Rosa-YFP (B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J), apelin-nlacZ24, VE-cadherin-CreERT2 33, and Tbx18-Cre22 mice have been described. The “Multi-color” Cre reporter mice contain a transgene that constitutively expresses GFP, and in the presence of Cre recombinase is randomly recombined once to express instead one of three other fluorescent proteins: mCherry, mOrange, or Cerulean, similar to the Brainbow system34. Details of this reporter will be described elsewhere (H. U. and I. W., in prep).
Immunohistochemistry
Staged embryos and hearts generated from timed pregnancies, where the morning of the vaginal plug was designated day e0.5, were dissected and fixed in 4% paraformaldehyde for 1–2 hours and stored in phosphate-buffered saline (PBS) at 4°C. Whole mount hearts were either incubated with X-gal (Sigma) to visualize β-galactosidase activity or processed for immunohistochemistry as described43. Tissue sections were prepared by cryoprotecting fixed tissue in 20% sucrose at room temperature for 1 hour or overnight at 4°C, snap freezing in optical cutting temperature compound (OCT, Tissue Tek), and sectioning with a Leica CM3050 S cryostat. Sections (20 or 80μm) were washed in PBS and incubated with primary antibodies diluted in blocking solution (5% goat serum, 0.5% TritonX-100 in PBS) at room temperature for 1–3 hours or overnight at 4°C. Sections were then washed in PBS and incubated with fluorescent-conjugated secondary antibodies for one hour at room temperature. Antibodies used were: anti-CD31 (1:100 dilution, BD Pharmingen), anti-VEGFR2 (1:100 dilution, BD Pharmingen), fluorescein isothiocyanate (FITC)-coupled anti-CD34 (1:100 dilution, BD Pharmingen), anti-Wt1 (undiluted, DAKO), anti-beta-galactosidase (1:500, Immunology Consultants Laboratory), anti-α4 integrin (1:100, BD Pharmingen), anti-VE-cadherin (1:100, BD Pharmingen), anti-GFP (1:500, Abcam), and Alexa-555- and -488-conjugated secondary antibodies (1:250, Molecular Probes).
Coronary artery angiogram
E15.5 coronary arteries were perfused by injecting FITC-conjugated tomato lectin (Vector) into the left ventricle of dissected embryos and visualized immediately afterward by fluorescence stereomicroscopy.
In situ hybridization
Tissue sections (20μm) were fixed in 4% paraformaldehyde/PBS for 10 minutes at room temperature, washed three times in PBS, and acetylated in 1.3% triethanolamine (Sigma), 0.25% acetic anhydride (Sigma) for 10 minutes at room temperature. After another three washes in PBS, sections were blocked with hybridization solution (50% formamide [Sigma], 5X saline sodium citrate solution [SSC], 5X Denhardts [Invitrogen], 0.5 mg/mL sperm DNA [Invitrogen], 0.25 mg/mL yeast tRNA [Invitrogen]) for 1 hour at room temperature in a chamber hydrated with 50% formamide, 5X SSC. Digoxigenin (DIG)-labeled (Roche) antisense probes or their sense controls were diluted to 400 ng/mL in hybridization buffer, denatured at 80°C for five minutes, cooled and added to slides, which were incubated overnight at 58°C in a Model 241000 hybridization oven (Boekel Scientific). After hybridization, slides were washed with 2X SSC for 30 minutes at room temperature, 0.2X SSC three times for 40 minutes at 65°C, and tris-buffered saline containing 0.1% Tween-20 (TBST) for 5 minutes at room temperature. Sections were then blocked with 5% sheep serum (Invitrogen)/TBST and incubated overnight with alkaline phosphatase-conjugated anti-DIG antibodies (Roche) diluted in blocking solution. Signal was detected by NBT-BCIP (Roche) histochemistry according to the manufacturer’s instructions. Adjacent sections were stained with anti-CD31 antibodies.
Organ cultures
Heart cultures were carried out as described26 with the following modifications. Embryonic hearts were from either wild type or apelin-nlacZ embryos. The atria and attached sinus venosus were left intact or dissected from the ventricle with fine-point forceps. Intact hearts or separated SV/atria (SV/A) or ventricles were cultured dorsal side up at the air-liquid interface on 8 mm Millicell Cell Culture Insert Filters (Millipore). For tissue recombination experiments, SV/A tissue was placed adjacent to the ventricle at the position where the original SV/A was removed. Cultures were maintained at 37°C and 5% CO2 in DMEM media supplemented with 2 μg/ml heparin, 10% fetal calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM L-glutamine. After 72 hours, explants were fixed with 4% paraformaldehyde and subjected to whole mount X-gal staining. Some stained explants were then sectioned and immunostained with anti-CD31 antibodies to confirm that X-gal+ nuclei were coronary endothelial cells. Control littermates were fixed immediately after dissection but before culturing, and then immunostained to assess coronary vessel (anti-CD31) and epicardial (anti-α4 integrin) coverage at the beginning of the culture period.
Clonal analysis
VE-cadherin-CreER mice were crossed to either the Rosa-lacZ or Multi-color Cre reporters. At e7.5, 8.5 or 9.5, dams were injected intraperitonealy (25G needle) with tamoxifen (Sigma), generally 0.25–0.5 mg, dissolved in 100 μl corn oil. These low doses of tamoxifen were selected to induce rare recombination events and generate single or well-separated clones. Embryos were dissected at e13.5 or 14.5. For lacZ-labeled clones, embryos were fixed and stained with X-gal as above, and those containing sparsely labeled cells were further analyzed. Hearts with just a single cluster or two or three well-isolated clusters of cells were sectioned, stained with an endothelial-specific antibody (anti-CD31) before cells were counted and identities assigned by marker (CD31) expression and their location and morphology. Clones marked with Multi-color reporters were fixed, sectioned, and stained with either DAPI or CD31 as above and analyzed using fluorescent filters specific to each fluorophore. Endothelial cell division rate for each clone was estimated from final cell counts, assuming that recombination occured between 6–48 hours44,45 after tamoxifen injection.
Imaging
Samples were imaged on a Leica MZ16FA stereomicroscope (whole mount tissue) or Zeiss Axiophot upright fluorescence microscope using Axiovision software (version 4.2) or on a Leica Sp2 confocal microscope using LCS software. Adobe Photoshop was used to adjust image levels and process image overlays.
Supplementary Material
Acknowledgments
We thank Drs. Thomas Quertermous, Luisa Iruela-Arispe, and Sylvia M. Evans for mouse strains, Krasnow lab members for input and comments, and C. Breitweiser and M. Petersen for help preparing figures. K.R. was supported by an NIH Developmental Biology and Neonatal Training Grant. M.A.K. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author contributions
K.R. designed and performed all experiments. K.R. and M.A.K. analyzed the experiments and wrote the manuscript. H.U. and I.L.W provided the Multi-color reporter mice and advised on its use.
References
- 1.Folkman J, Haudenschild C. Angiogenesis in vitro. Nature. 1980;288:551–556. doi: 10.1038/288551a0. [DOI] [PubMed] [Google Scholar]
- 2.Leung DW, Cachianes G, Kuang WJ, Goeddel DV, Ferrara N. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science. 1989;246:1306–1309. doi: 10.1126/science.2479986. [DOI] [PubMed] [Google Scholar]
- 3.Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- 4.Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol. 2007;8:464–478. doi: 10.1038/nrm2183. [DOI] [PubMed] [Google Scholar]
- 5.Ferrara N, Kerbel RS. Angiogenesis as a therapeutic target. Nature. 2005;438:967–974. doi: 10.1038/nature04483. [DOI] [PubMed] [Google Scholar]
- 6.Folkman J. Angiogenesis. Annu Rev Med. 2006;57:1–18. doi: 10.1146/annurev.med.57.121304.131306. [DOI] [PubMed] [Google Scholar]
- 7.Majesky MW. Development of coronary vessels. Curr Top Dev Biol. 2004;62:225–259. doi: 10.1016/S0070-2153(04)62008-4. [DOI] [PubMed] [Google Scholar]
- 8.Lavine KJ, Ornitz DM. Shared circuitry: developmental signaling cascades regulate both embryonic andadult coronary vasculature. Circ Res. 2009;104:159–169. doi: 10.1161/CIRCRESAHA.108.191239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smart N, Dube KN, Riley PR. Coronary vessel development and insight towards neovascular therapy. Int J Exp Pathol. 2009;90:262–283. doi: 10.1111/j.1365-2613.2009.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.http://www.who.int/healthinfo/global_burden_disease/2004_report_update/en/index.html
- 11.Lewis FT. The question of sinusoids. Anat Anz. 1904;25:261–279. [Google Scholar]
- 12.Grant RT. Development of the cardiac coronary vessels in the rabbit. Heart. 1923;13:261–271. [Google Scholar]
- 13.Bennett HS. The development of the blood supply to the heart in the embryo pig. Am J Anat. 1936;60:27–54. [Google Scholar]
- 14.Hutchins GM, Kessler-Hanna A, Moore GW. Development of the coronary arteries in the embryonic human heart. Circulation. 1988;77:1250–1257. doi: 10.1161/01.cir.77.6.1250. [DOI] [PubMed] [Google Scholar]
- 15.Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with in growth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- 16.Manner J. Does the subepicardial mesenchyme contribute myocardioblasts to the myocardium of the chick embryo heart? A quail-chick chimera study tracing the fate of the epicardial primordium. Anat Rec. 1999;255:212–226. doi: 10.1002/(sici)1097-0185(19990601)255:2<212::aid-ar11>3.3.co;2-o. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Pomares JM, et al. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. Int J Dev Biol. 2002;46:1005–1013. [PubMed] [Google Scholar]
- 18.Kirby ML. Cardiac Development. Oxford University Press; 2007. [Google Scholar]
- 19.Poelmann RE, Gittenberger-de Groot AC, Mentink MM, Bokenkamp R, Hogers B. Development of the cardiac coronary vascular endothelium, studied with antiendothelial antibodies, in chicken-quail chimeras. Circ Res. 1993;73:559–568. doi: 10.1161/01.res.73.3.559. [DOI] [PubMed] [Google Scholar]
- 20.Merki E, et al. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci U S A. 2005;102:18455–18460. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilm B, Ipenberg A, Hastie ND, Burch JB, Bader DM. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development. 2005;132:5317–5328. doi: 10.1242/dev.02141. [DOI] [PubMed] [Google Scholar]
- 22.Cai CL, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou B, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheikh AY, et al. In vivo genetic profiling and cellular localization of apelin reveals a hypoxia-sensitive, endothelial-centered pathway activated in ischemic heart failure. Am J Physiol Heart Circ Physiol. 2008;294:H88–98. doi: 10.1152/ajpheart.00935.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kattan J, Dettman RW, Bristow J. Formation and remodeling of the coronary vascular bed in the embryonic avian heart. Dev Dyn. 2004;230:34–43. doi: 10.1002/dvdy.20022. [DOI] [PubMed] [Google Scholar]
- 26.Lavine KJ, et al. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 2006;20:1651–1666. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiruma T, Hirakow R. Epicardial formation in embryonic chick heart: computer-aided reconstruction, scanning, and transmission electron microscopic studies. Am J Anat. 1989;184:129–138. doi: 10.1002/aja.1001840204. [DOI] [PubMed] [Google Scholar]
- 28.Hellstrom M, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 29.Kidoya H, et al. Spatial and temporal role of the apelin/APJ system in the caliber size regulation of blood vessels during angiogenesis. EMBO J. 2008;27:522–534. doi: 10.1038/sj.emboj.7601982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tammela T, et al. Blocking VEGFR-3 suppresses angiogenic sprouting and vascular network formation. Nature. 2008;454:656–660. doi: 10.1038/nature07083. [DOI] [PubMed] [Google Scholar]
- 31.Suchting S, et al. The Notch ligand Delta-like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc Natl Acad Sci U S A. 2007;104:3225–3230. doi: 10.1073/pnas.0611177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drake CJ, Fleming PA. Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood. 2000;95:1671–1679. [PubMed] [Google Scholar]
- 33.Monvoisin A, et al. VE-cadherin-CreERT2 transgenic mouse: a model for inducible recombination in the endothelium. Dev Dyn. 2006;235:3413–3422. doi: 10.1002/dvdy.20982. [DOI] [PubMed] [Google Scholar]
- 34.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 35.Lavine KJ, Long F, Choi K, Smith C, Ornitz DM. Hedgehog signaling to distinct cell types differentially regulates coronary artery and vein development. Development. 2008;135:3161–3171. doi: 10.1242/dev.019919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swift MR, Weinstein BM. Arterial-venous specification during development. Circ Res. 2009;104:576–588. doi: 10.1161/CIRCRESAHA.108.188805. [DOI] [PubMed] [Google Scholar]
- 37.le Noble F, et al. Flow regulates arterial-venous differentiation in the chick embryo yolk sac. Development. 2004;131:361–375. doi: 10.1242/dev.00929. [DOI] [PubMed] [Google Scholar]
- 38.Moyon D, Pardanaud L, Yuan L, Breant C, Eichmann A. Plasticity of endothelial cells during arterial-venous differentiation in the avian embryo. Development. 2001;128:3359–3370. doi: 10.1242/dev.128.17.3359. [DOI] [PubMed] [Google Scholar]
- 39.Othman-Hassan K, et al. Arterial identity of endothelial cells is controlled by local cues. Dev Biol. 2001;237:398–409. doi: 10.1006/dbio.2001.0383. [DOI] [PubMed] [Google Scholar]
- 40.Kudo FA, et al. Venous identity is lost but arterial identity is not gained during vein graft adaptation. Arterioscler Thromb Vasc Biol. 2007;27:1562–1571. doi: 10.1161/ATVBAHA.107.143032. [DOI] [PubMed] [Google Scholar]
- 41.Goldman S, et al. Long-term patency of saphenous vein and left internal mammary artery grafts after coronary artery bypass surgery: results from a Department of Veterans Affairs Cooperative Study. J Am Coll Cardiol. 2004;44:2149–2156. doi: 10.1016/j.jacc.2004.08.064. [DOI] [PubMed] [Google Scholar]
- 42.Sabik JF, 3rd, Lytle BW, Blackstone EH, Houghtaling PL, Cosgrove DM. Comparison of saphenous vein and internal thoracic artery graft patency by coronary system. Ann Thorac Surg. 2005;79:544–551. doi: 10.1016/j.athoracsur.2004.07.047. discussion 544–551. [DOI] [PubMed] [Google Scholar]
- 43.Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching programme of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayashi S, McMahon AP. Efficient Recombination in Diverse Tissues by a Tamoxifen-Inducible Form of Cre: A Tool for Temporally Regulated Gene Activation/Inactivation in the Mouse. Developmental Biology. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 45.Zovein AC, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.