Abstract
For a long time it has been assumed that the decay of RNA in eukaryotes is mainly carried out by exoribonucleases, which is in contrast to bacteria, where endoribonucleases are well documented to initiate RNA degradation. In recent years, several as yet unknown endonucleases have been described, which has changed our view on eukaryotic RNA metabolism. Most importantly, it was shown that the primary eukaryotic 3′ → 5′ exonuclease, the exosome complex has the ability to endonucleolytically cleave its physiological RNA substrates, and novel endonucleases involved in both nuclear and cytoplasmic RNA surveillance pathways were discovered concurrently. In addition, endoribonucleases responsible for long-known processing steps in the maturation pathways of various RNA classes were recently identified. Moreover, one of the most intensely studied RNA decay pathways—RNAi—is controlled and stimulated by the action of different endonucleases. Furthermore, endoribonucleolytic cleavages executed by various enzymes are also the hallmark of RNA degradation and processing in plant chloroplasts. Finally, multiple context-specific endoribonucleases control qualitative and/or quantitative changes of selected transcripts under particular conditions in different eukaryotic organisms. The aim of this review is to discuss the impact of all of these discoveries on our current understanding of eukaryotic RNA metabolism.
Keywords: endoribonuclease, RNA metabolism, RNA degradation, RNA processing, RNA surveillance, RNA interference
INTRODUCTION
The turnover of RNA is one of the crucial steps in the control of gene expression in archaeal, prokaryotic, and eukaryotic cells (for review, see Kushner 2004; Meyer et al. 2004; Condon 2007; Evguenieva-Hackenberg and Klug 2009). Numerous precisely regulated and complex mechanisms, pathways, and enzymatic activities evolved in the organisms belonging to these three kingdoms of life—which control the levels of particular RNA molecules, ensure their functionality, and carry out their degradation whenever the transcript is incorrect or has simply accomplished its role in the cell. In addition, all primary transcripts are essentially subjected to tightly controlled processing steps to produce mature functional RNA molecules. RNA decay and processing events engage a wide repertoire of proteins, including RNA helicases, poly(A/U) polymerases, and above all RNA-degrading enzymes, namely, exoribonucleases that degrade substrates from either the 3′ or 5′ terminus and endoribonucleases that cleave RNA molecules internally.
A widespread and commonly accepted opinion, which has long persisted among researchers in the RNA field, states that although RNA decay in prokaryotes is initiated by endoribonucleolytic cleavages inside the transcript, eukaryotic RNA turnover relies mostly on the action of exoribonucleases (Kushner 2004; Meyer et al. 2004). The former statement is essentially undeniable since the primary role of endoribonucleases such as RNase E, a major constituent of the Escherichia coli degradosome complex, and its functional equivalents in Gram-positive bacteria, namely, RNase Y and RNases J1/J2, have been substantially proven (Even et al. 2005; Deikus et al. 2008; Carpousis et al. 2009; Shahbabian et al. 2009; Yao et al. 2009). The endoribonucleolytic cuts in bacteria are stimulated by the removal of pyrophosphate from the 5′ end, which in the case of E. coli is carried out by the pyrophosphohydrolase RppH (Celesnik et al. 2007; Deana et al. 2008). The RNA molecules cleaved by endoribonucleases are subsequently degraded by the orchestrated cooperation of exoribonucleolytic activities and accessory enzymes—RNA helicases that unwind secondary structures posing obstacles to the motion of exoribonucleases along the substrate and polyadenylating enzymes such as poly(A) polymerase and PNPase synthesizing poly(A) tails, which act as “landing pads” to facilitate the access of exoribonucleases to the structured substrates (Iost and Dreyfus 2006; Condon 2007; Regnier and Hajnsdorf 2009).
Conversely, RNA decay in eukaryotes has been viewed primarily as an exoribonucleolytic process (Kushner 2004; Meyer et al. 2004). Cytoplasmic mRNA turnover begins with the removal of the poly(A) tail from the 3′ end of the transcript by deadenylases, followed by either the degradation of the substrate in the 3′ → 5′ direction and involving the exosome complex or by decapping and 5′ → 3′ degradation by the Xrn1 exonuclease (Houseley and Tollervey 2009). However, we recently witnessed a wealth of experimental evidence that strongly indicates RNA metabolism processes and protein machineries in prokaryotes and eukaryotes share more common features than previously anticipated. These include, for instance, the discovery of a class of eukaryotic enzymes adding poly(A) tails that, in contrast to those present at the mature 3′ end of mRNAs, exert a destabilizing effect by promoting exoribonucleolytic degradation (Stevenson and Norbury 2006).
Within the past decade, numerous examples have also demonstrated that endoribonucleases are not restricted to bacteria but that they also contribute significantly to RNA turnover in eukaryotes (see Fig. 1). Several endoribonucleases have been found to be implicated in regulating the stability of selected mRNA targets, and their activation is often dependent on specific signals such as stress stimuli. The functions of these enzymes and mechanisms regulating their activities are presented below.
FIGURE 1.
Overview of general RNA metabolism processes in eukaryotic cells involving the action of endoribonucleases and the intracellular localization of the enzymes described in the text. Endonucleolytic cleavages underlie numerous processing events of different RNA classes (rRNA, tRNA, mRNA, snRNA, and snoRNA) occurring in the nuclear compartment, both in the nucleoplasm and the nucleolus. The endoribonucleolytic activity of the exosome Dis3 catalytic subunit participates in the degradation of RNA species such as by-products of rRNA processing and CUTs, while the Swt1 endonuclease is a constituent of RNA surveillance machinery. In the cytoplasm, endonucleases are engaged in RNA quality control pathways dependent on ongoing translation, in the regular turnover of mRNA and in the RNA interference phenomenon. Moreover, Nob1 endonuclease is responsible for the final step of 18S rRNA processing which takes place in the cytoplasm. Finally, in plant chloroplasts, several endoribonucleases are principal enzymes involved in the processing and degradation of organellar transcripts.
Even more importantly, the enzymes displaying endoribonucleolytic activity seem to be involved in virtually all general processes associated with eukaryotic RNA metabolism in various subcellular compartments (see Fig. 1), such as the regular degradation of RNA molecules, RNA surveillance, and RNAi. In addition to the endonucleases that have long been known to be involved in the maturation of tRNA and rRNA (namely, RNases P, Z, and MRP), several new endonucleolytic enzymes responsible for the processing of different RNA classes were recently described (Fig. 1; Table 1).
TABLE 1.
General eukaryotic RNA metabolism factors endowed with endoribonuclease activity belong to several different families and often have a complex domain structure
The eukaryotic endoribonucleases can, in some cases, initiate RNA degradation or act redundantly with exoribonucleases. Alternatively, both classes of nucleases can cooperate with one another in the same pathway. This suggests that RNA helicases, which often play the role of exonuclease cofactors, are in most situations insufficient for enabling effective degradation and that endonucleolytic cleavages fill in the gaps in eukaryotic RNA decay mechanisms. The major aim of this review is to highlight the importance of these recently identified eukaryotic endoribonucleases, present the mechanisms regulating their activity and governing substrate specificity, and discuss their relative contributions to the individual pathways in comparison with exoribonucleases.
ENDONUCLEASES IN NUCLEAR RNA DECAY
Until recently there was no known protein endowed with endonuclease activity involved in nuclear RNA decay, and it was widely assumed that exosome complex exonucleolytic activity supported by RNA helicases and noncanonical poly(A) polymerases was sufficient for progression through RNA secondary structures (Lebreton and Seraphin 2008; Lykke-Andersen et al. 2009). This allows the stripping off of proteins interacting with RNA, which could otherwise stop exonuclease progression. Recently, and essentially at the same time, two different endoribonucleolytic activities responsible for RNA decay in the nucleus were discovered, thereby changing our view on nuclear RNA decay pathways. It was shown that the exosome complex catalytic subunit, Dis3, and a protein described previously as a transcription regulator, Swt1, are active endonucleases (Rother et al. 2006; Lebreton et al. 2008; Schaeffer et al. 2009; Schneider et al. 2009; Skruzny et al. 2009). The endoribonucleolytic activity of both enzymes was ascribed to their domains belonging to the evolutionarily conserved family called PIN (Table 1). PIN domains are ubiquitously present throughout all kingdoms of life (Makarova et al. 1999; Clissold and Ponting 2000), and they were initially identified in a member of the family of proteins participating in the biogenesis of type IV bacterial pili (Wall and Kaiser 1999), and were later suggested as being involved in signaling pathways (Makarova et al. 1999). A subsequent study revealed that PIN domains share some common features with FLAP nucleases, including the T4 bacteriophage RNase H and Fen1 endonucleases (Clissold and Ponting 2000).
The exosome complex was previously thought to be exclusively an exonuclease. The major exoribonucleolytic activity of the nuclear and cytoplasmic exosome forms resides in the RNB domain of the Dis3 subunit (Table 1), which is the processive, magnesium-dependent hydrolase belonging to the RNase II/R enzymatic family (Liu et al. 2006; Dziembowski et al. 2007; Lorentzen et al. 2008). In addition, in the nucleus the exosome is associated with the distributive ribonuclease of the RNase D family, Rrp6 (Allmang et al. 1999; Midtgaard et al. 2006). The additional catalytic role of Dis3 remained undetected until recently, when several groups provided solid evidence to support its novel endoribonucleolytic activity (Lebreton et al. 2008; Schaeffer et al. 2009; Schneider et al. 2009). It is worth emphasizing that the Dis3 PIN domain also has an important structural function since it is indispensable for tethering Dis3 to the ring-shaped catalytically inactive exosome core (Bonneau et al. 2009; Schneider et al. 2009).
The first hint suggesting that Dis3 might have an additional nucleolytic activity was the observation that the exonucleolytic site mutant was still able to degrade synthetic 5′-labeled RNA oligonucleotide substrates in vitro, albeit to a much lesser extent compared with the wild-type protein, and exclusively in the presence of a divalent cation (such as Mn2+, Mg2+, or Zn2+) at a relatively high concentration (1–3 mM) (Lebreton et al. 2008; Schaeffer et al. 2009; Schneider et al. 2009). Manganese was found to be the most efficient cofactor of this activity, corroborating the results obtained for another PIN domain-containing protein, SMG6 (Glavan et al. 2006; see below). However, the optimal metal concentration of 3 mM that stimulated PIN activity in vitro was much higher than the physiological concentration of manganese in the yeast cell (Eide et al. 2005). It thus remains to be explained how the activity of the PIN domain is achieved in vivo, especially when it has to be coordinated with RNB exoribonuclease, which prefers magnesium instead of manganese and at a much lower concentration (in the range of 40–100 μM) (Dziembowski et al. 2007). In contrast, it cannot be excluded that the intracellular environment, by creating suboptimal conditions for PIN activity, provides some means of its regulation, preventing it from triggering the indiscriminate degradation of exosome substrates.
It is worth noting that there are two genes in the human genome that encode proteins with the same domain organization as yeast Dis3: hDIS3 and hDIS3L, and recent studies indicate that, as expected, they both interact and cooperate with the human core exosome (Staals et al. 2010; Tomecki et al. 2010). Interestingly though, they have different intracellular localization patterns: hDIS3 is mainly nuclear while hDIS3L is strictly cytoplasmic, which is reflected by their substrate preferences in vivo (Tomecki et al. 2010). More importantly, biochemical analyses indicate that both proteins are active 3′ → 5′ exonucleases but only hDIS3 displays endonucleolytic activity, which is in agreement with the preservation of the PIN active site in hDIS3 but not in hDIS3L (Staals et al. 2010; Tomecki et al. 2010). These results suggest that in humans the exosome complex endowed with endonucleolytic activity is mainly nuclear.
The functional in vivo analysis of Dis3 PIN catalytic domain mutations was only performed in yeast and revealed that its endonucleolytic activity is involved in the metabolism of typical nuclear exosome substrates (Lebreton et al. 2008; Schaeffer et al. 2009). Furthermore, detailed analysis of the phenotypes of catalytic Dis3 mutations within the PIN and RNB domains at the molecular level provided further evidence that both activities collaborate with one another (for a possible model, see Fig. 2). The accumulation of several physiological exosome targets, namely: (1) a 5′ ETS ribosomal RNA processing by-product, (2) a precursor of 5.8S rRNA, and (3) noncoding NEL025 RNA species, is significantly higher in the double PIN and RNB catalytic domains' mutant than in any of the single mutants. Moreover, at least for the 5′ ETS substrate, these in vivo results were successfully recapitulated in vitro (Lebreton et al. 2008). The combination of two different ribonucleolytic activities within Dis3 might be advantageous for at least two reasons. At the structural level, endonucleolytic cuts introduced by the PIN domain can potentially provide alternative start points for degradation when the exosome gets stuck on secondary structures, and thus enable further delivery of the substrate to the RNB domain active site (Fig. 2A). Second, degradation of a given RNA molecule is probably achieved much more rapidly when the PIN endonuclease and the RNB exonuclease act together. This latter hypothesis is strongly supported by the influence of catalytic mutations in Dis3 PIN and RNB domains on the levels of 5′ ETS and its decay intermediates (Fig. 2B; Lebreton et al. 2008). Furthermore, in addition to the obvious cooperation of the endo- and exoribonuclease activities of Dis3, it seems that they also collaborate with the distributive activity of Rrp6 since some of the molecular phenotypes of catalytic mutations in the Dis3 PIN and RNB domains are further exacerbated in the Δrrp6 background (Lebreton et al. 2008). Therefore, it is clear that the coordinated action of three different catalytic activities of the exosome complex is required for the efficient degradation of its natural substrates.
FIGURE 2.
Possible model of cooperation between exoribonucleolytic and endoribonucleolytic activities of the exosome complex Dis3 catalytic subunit. (A) Structural viewpoint: RNA substrates containing a secondary structure and lacking the 3′ single-stranded extension of sufficient length are unable to reach the Dis3 exoribonuclease active site via the central channel pathway and are partially resistant to exoribonucleolytic degradation. The PIN domain endoribonuclease activity (scissors) may overcome this problem by cleaving within the loop of the hairpin. The resulting upstream cleavage product might be delivered to the RNB catalytic center either through the channel (top), or via an alternative hypothetical path going directly from the PIN domain to the active site of the RNB domain (middle). In parallel, the RNA substrate can be degraded further by the PIN domain endonuclease before accessing RNB exonuclease (bottom). (B) Functional viewpoint: effects of Dis3 RNB exonuclease and PIN endonuclease catalytic mutations. (left) Cooperative action of the two nucleolytic activities in the wild-type leads to the degradation of a given substrate and its decay intermediates arising due to the endonucleolytic cuts. (center) Inactivation of exonuclease activity alone leads to the accumulation of both a full-length substrate and its endo-cleavage intermediates. (right) Inactivation of both exonuclease and endonuclease activities has a synergistic effect on the level of full-length substrates, whereas the cleavage intermediates disappear in the double mutant.
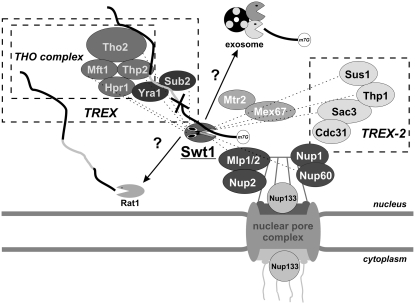
The SWT1 gene was identified because its inactivation causes a synthetically lethal phenotype with the deletion of the gene encoding the Hpr1 protein—a component of the THO/TREX complex, involved in coupling transcription with mRNP assembly and export (Rother et al. 2006; Gonzalez-Aguilera et al. 2008; Skruzny et al. 2009). The SWT1 gene also interacts genetically with genes encoding a functionally related TREX-2 complex, as well as with the MLP1 and NUP60 genes coding for proteins localized in the nuclear pore complex, which participate in perinuclear mRNP surveillance, directly preceding their export to the cytoplasm (Fig. 3; Galy et al. 2004; Vinciguerra et al. 2005; Rougemaille et al. 2008b; Katahira and Yoneda 2009; Skruzny et al. 2009). The inactivation of THO complex subunits results in very interesting molecular phenotypes, demonstrating its important role in mRNA biogenesis and genome stability. A lack of the THO complex, similar to the impairment of the mRNA 3′ end formation, causes mRNA retention near the site of transcription, which can be visualized by in situ hybridization (Rougemaille et al. 2007). Another interesting phenotype caused by THO complex deletion is the formation of large aggregates (called heavy chromatin) composed of transcriptionally active chromatin, nascent transcripts, RNA export machinery, and nuclear pore complexes (Rougemaille et al. 2008a). The Swt1 protein, a novel endonuclease initiating RNA decay in the nucleus, could potentially help to resolve such aggregates. The Swt1 protein was confirmed as displaying manganese cation-dependent endoribonuclease activity, which preferentially cleaves unstructured regions in RNA substrates and does not seem to be strictly sequence specific (Skruzny et al. 2009). Consistent with its predicted role in nuclear RNA quality control, deletion of SWT1 led to the accumulation of polyadenylated RNA in the nucleus and the illegitimate cytoplasmic export of pre-mRNA reporters containing mutations known to impair splicing. Both phenotypes were synergistically enhanced upon the simultaneous deletion of genes coding for the Mlp1 or Nup60 proteins, overseeing the mRNP export process at the nuclear pore. A catalytic mutation in the Swt1 PIN domain caused elevated levels of aberrant pre-mRNAs in the nucleus, suggesting that its endonucleolytic activity is indeed responsible for the removal of such defective molecules, which represent its preferred substrates (Fig. 3). Interestingly, defects in pre-mRNA surveillance were also observed upon the overexpression of functional Swt1 (Skruzny et al. 2009).
FIGURE 3.
The Swt1 endoribonuclease is a novel component of nuclear RNA surveillance machinery. Defective pre-mRNA (for instance, containing the 5′ splice site mutation, represented by the cross) gets trapped in the export-incompetent mRNP with the Tho/TREX complex. Endonucleolytic cleavage of the aberrant transcript by Swt1, concentrated in the proximity of the NPC, might facilitate its subsequent exoribonucleolytic degradation, most probably executed by the exosome and Rat1 5′ → 3′ exoribonuclease. The genetic interactions detected between Swt1 and different components of the Tho/TREX complex, as well as with subunits of the TREX-2 complex, coupling transcription with mRNP assembly and export, and with the NPC-associated proteins involved in the perinuclear RNA surveillance by the retention of unspliced transcripts, are marked with dashed lines.
Neither the catalytic activities nor the regulation of Dis3 or Swt1 have been characterized in detail yet, but we speculate that they must be finely tuned to prevent the destruction of proper transcripts. In the case of the SWT1 gene, expression is maintained at a low level to reduce the chance of interference between the elevated Swt1 PIN endonuclease activity and RNA metabolism events occurring in the nucleus. It is also likely that in some situations Swt1 is transiently recruited and clustered at the nuclear pore complex to perform its catalytic function more efficiently, as demonstrated by localization experiments performed in the nup133Δ background that caused defects in mRNA export (Skruzny et al. 2009).
It seems that Dis3 PIN activity supports the already ongoing degradation by the exosome, whereas Swt1 endonucleolytic cleavage can initiate the destruction process (Fig. 3). It is not yet known what happens to the aberrant transcripts that have been cleaved by Swt1, and whether the exosome, the Rat1 5′ → 3′exonuclease, or another degrading enzyme is responsible for their further digestion also still has to be demonstrated experimentally. It also remains to be investigated whether Swt1 activity could be involved in RNA quality control mechanisms in the nucleus that are different from perinuclear surveillance, and whether it could target substrates other than unspliced pre-mRNA or play any role in the rest of the processes associated with RNA in the nuclear compartment, such as ribosomal RNA processing. Similarly, it has not yet been demonstrated whether the endonucleolytic activity of the exosome complex plays some role in any of the known RNA surveillance pathways operating both in the nuclear and cytoplasmic compartments. Further research will resolve any issues regarding the potentially overlapping functions of both enzymes. In addition, more detailed biochemical experiments should allow greater insight into the mechanisms of their actions and collaborations with other enzymes involved in RNA decay.
THE ROLE OF ENDORIBONUCLEASES IN CYTOPLASMIC RNA SURVEILLANCE MECHANISMS
Endoribonucleolytic activities are not only employed in nuclear RNA quality control but also in mRNA surveillance pathways operating in the cytoplasmic compartment. Mechanisms ensuring the presence of only faultless mRNAs play a fundamental role in the control of eukaryotic gene expression because they guarantee that aberrant transcripts that evaded RNA quality control in the nucleus and were exported to the cytoplasm are degraded before they can produce faulty proteins that could otherwise be harmful to the cell physiology. Three pathways have thus evolved in the cytoplasm through which the cell destroys mRNAs containing specific errors: nonsense-mediated decay (NMD), which eliminates transcripts containing a premature termination codon (PTC); nonstop decay (NSD), which removes the mRNAs lacking a termination codon; and no-go decay (NGD), which is involved in the degradation of mRNA molecules with sequences that induce ribosome stalling during translation (for review, see Doma and Parker 2007; Isken and Maquat 2007; Nicholson et al. 2010).
Nonsense-mediated decay is the best studied process among the cytoplasmic RNA surveillance mechanisms. Transcripts containing a PTC, which could give rise to the C-terminally truncated polypeptides, can be generated in different ways including errors in transcription and pre-mRNA processing, alternative splicing, and mRNA synthesis from the DNA templates containing mutations that lead to the appearance of the nonsense triplet within the open reading frame. In yeast, plants, and metazoa, the major determinant of NMD is the distance between the termination codon and the poly(A) tail. When these two elements are located far from each other the result is the recruitment of principal NMD factors: UPF proteins (Amrani et al. 2006). However, in the case of metazoa the recognition of PTC is more complex and the presence of the exon junction following the termination codon also activates NMD. A specific complex called the exon junction complex (EJC) is deposited 20–24 nucleotides (nt) upstream of the exon–exon junction and interacts with the UPF factors (Le Hir and Séraphin 2008), which allows for PTC sensing during the first round of translation. Moreover, in addition to evolutionarily conserved UPF proteins, SMG factors are also essential for efficient NMD in higher eukaryotes. For both yeast and metazoans, NMD may proceed through the activation of deadenylation-independent decapping and degradation in the 5′ → 3′ direction by the Xrn1 exoribonuclease or through the removal of the poly(A) tail from the 3′ end by deadenylase, followed by mRNA decay in the 3′ → 5′ direction performed by the exosome complex (Isken and Maquat 2007; Nicholson et al. 2010). Additionally, accelerated deadenylation may also activate decapping followed by Xrn1-dependent 5′ → 3′ degradation during NMD in mammalian cells (Fig. 4A; Chen and Shyu 2003; Couttet and Grange 2004).
FIGURE 4.
Endonucleases participate in translation-dependent RNA quality control pathways in the cytoplasm. (A) Nonsense-mediated decay; stalling of the ribosome on a premature termination codon leads to the recruitment of SMG1 and UPF1 NMD factors as a result of signal received from poly(A) binding proteins (PABP) bound to the distant poly(A) tail (pA) by eRF1/eRF3 translation termination factors. PTC recognition in higher eukaryotes is also mediated by the EJC deposited upstream of the exon–exon junction following the stop codon. Phosphorylation of UPF1 by SMG1 with the assistance of UPF2/UPF3 proteins leads to a change in UPF1 conformation and triggers decay of the PTC-containing transcript via the use of SMG5/SMG7 NMD factors through deadenylation and decapping (which might be deadenylation-independent or stimulated by removal of poly[A] tail) followed by the degradation executed by the exosome and Xrn1, respectively. In metazoans, an alternative NMD pathway, dependent on SMG6 endoribonucleolytic cleavage in the proximity of PTC, can be used. Upstream and downstream cleavage products are degraded by the exosome and Xrn1, respectively. (B) No-go decay; ribosome stalling due to the secondary structure in mRNA recruits Dom34/Hbs1 NGD factors homologous with eRF1/eRF3 proteins. This enables ribosome release and leads to the degradation of aberrant mRNA initiated by endoribonucleolytic cleavage in the vicinity of the structure blocking translation. Whether the cut is exerted by Dom34 or occurs through the recruitment of an unknown endonuclease remains to be investigated. Subsequent exoribonucleolytic degradation proceeds in a similar manner to that of the NMD pathway. (C) 18S nonfunctional rRNA decay; a block in translation can also result from the 18S rRNA mutations in the decoding center of the small ribosomal subunit (indicated with the gray cross). Such defective 18S rRNA molecules are subjected to the quality control pathway involving Dom34/Hbs1 factors, most probably in a manner similar to that of NGD, including endonucleolytically mediated initiation.
In recent years, it has been convincingly demonstrated that NMD can be initiated by endoribonucleolytic cleavage in metazoa (but not yeast). This was first shown in Drosophila and then in human cells as an event initiating NMD that takes place in the vicinity of the PTC (Gatfield and Izaurralde 2004; Huntzinger et al. 2008; Eberle et al. 2009). The observed patterns of digestion product accumulation proximal and distal to the cleavage site upon different combinations of RNA interference (RNAi)-mediated depletion of enzymes involved in 5′ → 3′ and 3′ → 5′ degradation, both in Drosophila and in humans, excluded the possibility that exoribonucleases take part in their generation. Instead, it was proposed that the endonuclease activity responsible for the cleavage of PTC-harboring messages resided within the C-terminal PIN domain of the SMG6 protein (see Table 1). Although two different SMG factors, SMG5 and SMG6, possess the PIN domain, only the latter contains three aspartic acid residues that, based on the structural similarity of SMG6 PIN to the FLAP nuclease domain of the T4 bacteriophage RNase H, are predicted to coordinate the divalent cation in the putative catalytic center of the enzyme, which most probably activates the nucleophile attacking the phosphodiester bond (Glavan et al. 2006). Indeed, it was confirmed that human SMG6, but not SMG5, displays manganese-dependent endoribonuclease activity in vitro that is abolished upon the mutation of one of the crucial aspartates (Glavan et al. 2006). Moreover, the SMG6 wild-type protein, but not its mutant form, has the ability to introduce cuts in the circular RNA substrate, verifying that it is a bona fide endoribonuclease (Eberle et al. 2009). In concordance with the results of biochemical assays, it was shown in vivo that the overexpression of a catalytically inactive mutant exerts a dominant-negative effect on NMD and that the exogenously produced mutant SMG6 failed to restore the degradation of mRNAs containing a PTC in the cells depleted of the endogenous enzyme (Glavan et al. 2006; Huntzinger et al. 2008; Eberle et al. 2009). Since these observations were true for both fruit fly and human cells, we assume that the endoribonucleolytic NMD pathway is conserved among metazoans. This mechanism also employs the general mRNA turnover pathways, represented by the exosome and Xrn1 5′ → 3′ exoribonuclease, but instead of degrading the full transcript bearing a PTC, as in the classic model, they promote the decay of digestion products arising because of a prior endoribonucleolytic event, upstream of and downstream from the site of cleavage, respectively (Fig. 4A).
This indisputable existence of two NMD mechanisms raises several important issues. First, from an evolutionary point of view, it is intriguing why metazoans have developed such an alternative NMD pathway cleavage in contrast to yeast, which lacks a SMG6 homolog. Furthermore, it would be interesting to determine whether they represent entirely redundant mechanisms or act in parallel in mammalian cells. It can be hypothesized that one might serve as a backup when the other fails to degrade the PTC-containing transcript. In contrast, if the same substrate can be degraded via either pathway, future research should focus on what factors determine whether the faulty mRNA has to be removed by the classic mechanism, beginning with deadenylation and/or decapping, or the mechanism employing SMG6 endoribonucleolytic activity. One possible explanation could be the mutually exclusive recruitment of the SMG5/7 heterodimer or the SMG6 endonuclease following UPF1 activation. Moreover, although the endonucleolytic pathway seems to be the major one in fruit flies, the same might not necessarily be true in human cells, where the classic pathway seems to predominate (Gatfield and Izaurralde 2004; Huntzinger et al. 2008; Eberle et al. 2009). Therefore, it is vital to understand the real contribution of both mechanisms in different organisms. It is also possible that one of the mechanisms prevails over the other in the particular location of the cytoplasm. In this context, although SMG5/7 proteins were found to accumulate in P-bodies, SMG6 seems to be absent from these cytoplasmic substructures (Unterholzner and Izaurralde 2004). These hypotheses, as well as the biological significance of the reported interaction between human SMG6 and telomerase (Redon et al. 2007), which makes the overall picture of SMG6 functions even more complicated, need to be addressed in future research.
Endoribonucleolytic cleavage initiating the further degradation of substrates by exonucleases is also the hallmark of another translation-dependent cytoplasmic RNA quality control mechanism, namely, NGD. This surveillance pathway targets those mRNAs that possess structures blocking ribosome movement during translation and thereby disabling correct protein synthesis by delaying the elongation step (Fig. 4B; Doma and Parker 2006). The accumulation of degradation intermediates situated 5′ or 3′ with respect to the stem–loop structure inducing ribosome stalling has been observed upon the inactivation of 3′ → 5′ or 5′ → 3′ exoribonucleolytic degradation pathways. The endonucleolytic cleavage sites were found to be located in the proximity of the stem–loop, with some degree of heterogeneity, closely resembling the distribution of SMG6 cleavage sites near the PTC in the endonucleolytically initiated NMD pathway. Interestingly, NGD seems to be initiated by other mRNA sequence and structural elements that impede translation elongation, such as pseudoknots, rare triplets, and the PTC (Doma and Parker 2006), which indicate that in some cases the transcripts containing the PTC might be targeted for degradation via either NMD or NGD. No-go decay has been shown to be strictly dependent on the ongoing translation and the Dom34/Hbs1 dimer of proteins displaying structural similarities to the eRF1/eRF3 translation termination factors (Doma and Parker 2006; Lee et al. 2007; Graille et al. 2008). It was therefore proposed that both of these proteins are recruited to the stalled ribosome and induce the endoribonucleolytic cleavage of defective transcripts (Fig. 4B). The subsequent degradation of two cleavage fragments by exonucleases might be preceded by the release of ribosome, which can occur as in the case of NSD. In this surveillance pathway the Ski7 protein related to eRF3 promotes dissociation of the ribosome stalled at the 3′ terminus of the mRNA that lacks a stop codon (Frischmeyer et al. 2002; van Hoof et al. 2002).
The catalytic activity of the Dom34/Hbs1 dimer is uncertain. It was initially suggested that the Dom34/Hbs1 function might only be required for the stimulation of mRNA cleavage by some endonucleolytic activity residing within the ribosome itself, or for the recruitment of specific endoribonuclease (Doma and Parker 2006; Tollervey 2006). Then, however, it was shown that the archaeal homolog of the yeast Dom34, called Pelota, has an inherent endoribonucleolytic activity (Lee et al. 2007). Detailed analysis of the structures of both Dom34 and Pelota, solved independently using X-ray crystallography, combined with in vitro studies, demonstrated that this activity can be ascribed to the N-terminal domain, which has a three-dimensional architecture resembling that of the Sm and Lsm proteins, including the presence of an RNA-binding fold (Lee et al. 2007; Graille et al. 2008). This might suggest that Dom34-like proteins originated as a result of domain shuffling between eRF1/3-like proteins and the (L)Sm factor, followed by the acquisition of endonuclease activity centered in the domain derived from the latter. The intrinsic endonucleolytic activity of Dom34 against the substrates bearing stable stem–loop structures, which imitate natural NGD targets, supported the idea that it functions as a catalytic component of NGD (Lee et al. 2007). This visionary picture of the involvement of Dom34 in NGD was, however, somewhat complicated by recent findings showing that its in vitro nucleolytic activity, if any, is extremely sensitive to experimental conditions, irrespective of whether Dom34 is tested alone or in a complex with Hbs1 (Passos et al. 2009). The deletion of DOM34 consistently showed no negative effects on endonucleolytic events initiating NGD upon the overexpression of the ribosomal Rps30a protein, which is known to suppress Δdom34 (Davis and Engebrecht 1998; Passos et al. 2009). Moreover, the mutations within the putative Dom34 active site did not abolish the endoribonucleolytic cleavage of the NGD reporter (Passos et al. 2009). In view of these contradictory experimental data, the issue of whether Dom34 is the NGD endonuclease, and perhaps whether some redundant enzyme participates in this RNA quality control mechanism, should be studied more rigorously in the future.
It is noteworthy that besides its still to be unequivocally confirmed enzymatic activity, Dom34 might play another role in NGD, for instance, by increasing the affinity of Hbs1 toward GTP, which most probably promotes ribosome release, similar to the roles of eRF1 and eRF3 (Graille et al. 2008). Therefore, we can speculate that two evolutionarily related complexes evolved in the eukaryotes to determine the fate of mRNA depending on the reason for ribosome pausing. If this is caused by the naturally occurring stop codon, then the mRNA can be either utilized in the subsequent round of translation or subjected to one of the regular mRNA turnover pathways following the translation termination by eRF1/eRF3. Conversely, when the ribosome stalling is because of the secondary-structure blockade, the transcript is rapidly destroyed in NGD by Dom34/Hbs1 and, if Dom34 is inactive, an unknown endonuclease (Fig. 4B). It should be emphasized that NGD is conserved in higher eukaryotes because the Drosophila Dom34 homolog has been implicated in NGD in the insect cells and has the ability to complement Δdom34 in yeast (Passos et al. 2009).
Interestingly, protein-coding transcripts containing barriers that preclude effective translation elongation might not be exclusive substrates for the endoribonucleolytic activity associated with NGD. The deletion of genes encoding either Dom34 or Hbs1 results in the inhibition of another cytoplasmic RNA surveillance pathway, known as 18S nonfunctional rRNA decay (18S NRD), which targets 18S rRNA molecules incorporated in translating ribosomes that have mutations within the decoding center of small ribosomal subunits that impair proper base-pairing between tRNA and mRNA (Fig. 4C; LaRiviere et al. 2006; Cole et al. 2009). Although the NGD endoribonuclease cleavage of 18S NRD substrates has not been demonstrated directly, this would make perfect biological sense since the action of a single endonuclease could then be implicated in the destruction of both mRNAs and rRNAs with defects disabling productive protein synthesis, reducing the need for the evolutionary development of separate degradation pathways. This idea is further supported by a recent finding that NGD induces the degradation of ribosomes stalled because of the presence of the 20S pre-rRNA unprocessed precursor instead of mature 18S rRNA in the small ribosomal subunits. Although such ribosomes are able to complete translation initiation, they cannot progress to the elongation phase (Soudet et al. 2010).
To summarize, both NMD and NGD share at least two common fundamental features. First, they both represent the last line of defense against aberrant transcripts as ultimate quality control mechanisms, acting during the advanced phases of polypeptide chain synthesis. Second, they both employ endoribonucleases to accelerate the decay of their targets by attacking them internally, close to the site of the defect, thereby eliminating the need to remove the protective cap and the poly(A) tail from the termini, which is a prerequisite for exoribonucleolytic degradation. It remains to be investigated whether the other cytoplasmic RNA surveillance mechanisms also utilize the advantages of endonucleolytic cleavage.
ENDORIBONUCLEASES INVOLVED IN RNA PROCESSING
The mRNA turnover and quality control processes are among several processes occurring in eukaryotic cells that utilize endoribonucleolytic activities. Virtually all eukaryotic RNA classes are subject to extensive processing reactions that lead to the generation of functional molecules from their precursors, and many of these processing events rely on endonucleolytic cleavages.
The mRNA 3′ end processing
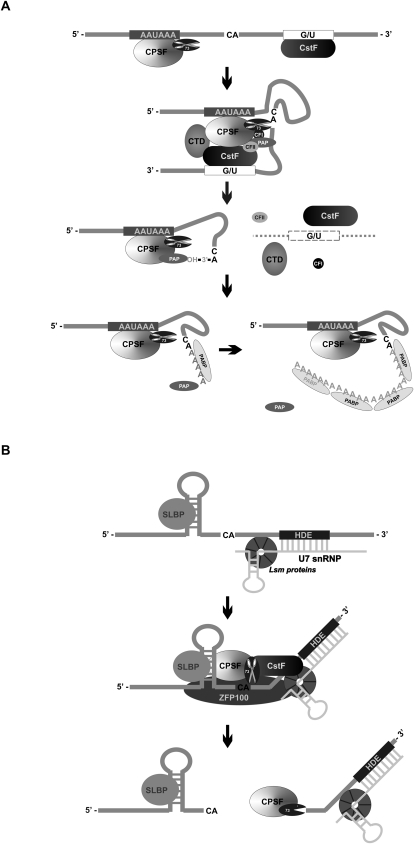
Two of the best-studied phenomena associated with mRNA processing in eukaryotes are the cleavage and polyadenylation reactions that determine the position of the mature 3′ end of the transcript (for reviews, see Zhao et al. 1999; Mandel et al. 2008). The two events are coupled and require the presence of three conserved cis elements in the 3′-UTR and at least six protein factors: CPSF, CstF, CFI, and CFII and poly(A) binding protein (PABP) and poly(A) polymerase (PAP). The reactions are stimulated by RNA polymerase II (RNA pol II), whose C-terminal domain (CTD) couples transcription termination to 3′ end processing and promotes cleavage–polyadenylation complex assembly on pre-mRNA substrates (Fig. 5A; Zhao et al. 1999; Mandel et al. 2008).
FIGURE 5.
CPSF-73: an endoribonuclease critical for the processing of protein-coding transcripts. (A) Poly(A)+ mRNAs. Most protein-coding transcripts undergo coupled cleavage/polyadenylation reactions. The consecutive steps of this process and the roles of individual cis elements in the mRNA sequence and protein factors acting in trans are described in the main text; the 73-kDa CPSF component is an endonuclease belonging to the metallo-β-lactamase family of enzymes, which usually cleave the pre-mRNA after the CA dinucleotide, defining the site of poly(A) tail addition. (B) Metazoan poly(A)- histone mRNAs. These transcripts employ a noncanonical 3′-end processing mechanism dependent on the binding of U7 snRNP, composed of U7 snRNA and the LSm proteins, to the HDE sequence located downstream from the cleavage site. Another element required for histone mRNA processing is the conserved stem-loop upstream of the cleavage site which is bound by the SLBP factor. Cleavage is preceded by an interaction involving SLBP and U7 snRNP, bridged by the zinc-finger protein ZFP100 and utilizing CPSF and possibly also CstF, similarly to the mechanism presented in A. The CPSF-73 subunit is not only responsible for the endonucleolytic cleavage which determines the position of the histone mRNA mature 3′ end, but it might also display 5′ → 3′ exoribonucleolytic activity, allowing for the degradation of the downstream cleavage product.
In humans, the formation of the nuclear polyadenylation complex begins with the attachment of CPSF to the conserved AAUAAA hexanucleotide, which is facilitated by the interaction of CPSF with CstF and binding to the GU-rich region located 10–30 nt downstream from the cleavage site. The CFI and CFII factors, besides participating in the cleavage reaction, allow for the incorporation of PAP into the complex (Fig. 5A). Following cleavage, CstF, CFI, CFII, and RNA pol II CTD dissociate from the complex, and the proximal pre-mRNA fragments remain associated with CPSF and PAP, whereas the distal fragment is rapidly degraded. Despite the presence of CPSF, PAP displays relatively low activity and affinity toward the substrate and initially synthesizes an approximately 10-adenosine-long tail in the distributive manner. The switch to the processive mode of action is dependent on the binding of PABP to this oligoadenylate extension (Fig. 5A). Eventually, 200–300-nt-long poly(A) tails are generated (Zhao et al. 1999; Mandel et al. 2008).
Although this cleavage–polyadenylation mechanism has been well documented, the identity of the endonuclease responsible for the crucial cut in this mRNA maturation pathway remained unknown until recently, when the endoribonucleolytic activity was ascribed to one of the CPSF components, namely, CPSF-73 (Fig. 5A; Table 1; Ryan et al. 2004; Mandel et al. 2006). The CPSF-73 protein is a member of the metallo-β-lactamase family of hydrolytic enzymes, using one or more zinc cations, coordinated by evolutionarily conserved acidic amino acids and histidines, as a cofactor (for review, see Dominski 2007). It was initially demonstrated that the cleavage reaction requires the presence of zinc and that the mutation of residues involved in its binding results in lethality in the case of yeast (Ryan et al. 2004). Later, direct and ultimate proof of the endonucleolytic activity of CPSF-73 was provided by biochemical analyses of recombinant human CPSF-73, showing the cleavage of various pre-mRNAs by a wild-type protein but not by its version with a mutation of one of the amino acids participating in catalysis (Kolev et al. 2008). It can be inferred from the structural data that the two zinc ions coordinated within the catalytic center of the enzyme are bridged by the hydroxide, acting as a nucleophile that attacks the phosphate in the pre-mRNA cleavage site, which would explain their crucial role in this reaction (Mandel et al. 2006).
It is noteworthy that CPSF-73 endonucleolytic activity, possibly along with some other components of the polyadenylation complex, participates in the cleavage of metazoan pre-mRNAs corresponding to histones (Dominski et al. 2005). Histone mRNAs lack poly(A) tails at their 3′ ends since their maturation mechanism is different from the standard cleavage–polyadenylation mechanism and depends on the binding of a U7 snRNP to the sequence located downstream from the cleavage site, called the histone downstream element (HDE) (Fig. 5B; for review, see Dominski and Marzluff 2007). These features suggest that both mechanisms originated from the same ancestor processing pathway that diverged during the course of evolution. Intriguingly, apart from endoribonucleolytic cleavage, which determines the mature 3′ terminus of histone mRNA, CPSF-73 also displays 5′ → 3′ exoribonucleolytic activity that is responsible for the decay of the downstream cleavage product (Fig. 5B; Yang et al. 2009). It has been shown that U7 snRNP binding to HDE is required for the initiation of 5′ → 3′ degradation and that the distance between the 5′ end of the substrate and HDE is a key determinant in whether the enzyme degrades the RNA molecule endo- or exoribonucleolytically (Yang et al. 2009). Therefore, CPSF-73 represents another example of a nuclease that has two distinct enzymatic activities within one polypeptide. However, in contrast to the Dis3 catalytic subunit of the exosome complex, where the two activities are controlled by separate structural domains, for CPSF-73 the exo- and endonucleolytic reactions are catalyzed by the same active center. In this context, it has to be mentioned that other enzymes belonging to the metallo-β-lactamase family, including the eukaryotic DNA nuclease Artemis and RNase J1, which is the major RNA degrading and processing enzymes in Bacillus subtilis, can also function as both endo- and exoribonucleases (Ma et al. 2002; Even et al. 2005; Mathy et al. 2007). In the case of Artemis, its activity was shown to be modified by the absence or presence of a protein kinase interactor (Ma et al. 2002). It was proposed that switching between the two activities might be promoted by the binding of an accessory partner or by covalent modification of the enzyme, which could lead to remodeling of the active site. For RNase J1, it is conceivable that the enzyme detects the presence of the phosphate at the 5′ end of the downstream cleavage fragment following endonucleolysis, and that this “sensing” somehow alternates its activity (Mathy et al. 2007).
The ability of mammalian CPSF-73 to endonucleolytically cleave histone pre-mRNAs is dependent on the formation of a higher-order complex with CPSF-100, which also belongs to the metallo-β-lactamase family, and the mutations in the conserved motifs present in both proteins significantly reduce endoribonucleolytic activity (Kolev et al. 2008). Interestingly, highly homologous proteins, Int11 and Int9, participate in the snRNA 3′ end processing as components of the Integrator complex (Baillat et al. 2005). Although the putative endoribonucleolytic properties of Int11 still await experimental validation, its depletion, as well as the overexpression of the catalytic mutant, causes defects in U1 and U2 snRNA processing. Thus, it can be concluded that different metallo-β-lactamases act as endoribonucleases in the maturation of various classes of RNA (see also Weiner 2005).
The tRNA and snoRNA processing
Endoribonucleases also participate in the processing of eukaryotic small RNA molecules. The tRNAs are processed at their 5′ and 3′ termini by the endonucleolytic removal of extraneous sequences, carried out by the well-characterized site-specific RNase P ribozyme and tRNase Z metallo-β-lactamase (Table 1), respectively (Hartmann et al. 2009). The yeast CPSF-73 homolog, Ysh1, was shown to be involved not only in pre-mRNA processing, but also in the maturation of snoRNA (Garas et al. 2008). The U16 and U86 intron-encoded box C/D snoRNAs in Xenopus laevis, and most probably in other higher eukaryotes, are generated by the XendoU endoribonuclease (Table 1) instead of being synthesized as a consequence of a splicing reaction (Laneve et al. 2003; Gioia et al. 2005). The XendoU endoribonuclease is a manganese-dependent single-strand-specific enzyme showing a preference toward uridines. Intriguingly, it releases products with a 2′–3′ cyclic phosphate and a 5′ hydroxyl, which is very untypical of enzymes using divalent cations as cofactors and rather resembles the properties of secreted enzymes that are not dependent on such cofactors (Laneve et al. 2003; Gioia et al. 2005). Structural analysis indicated that XendoU represents a unique fold with a new type of active site (Renzi et al. 2006). Interestingly, a related enzyme named NendoU is an endoribonuclease involved in the replication cycle of nidoviruses (Ivanov et al. 2004; Posthuma et al. 2006; Ricagno et al. 2006; Nedialkova et al. 2009). The NendoU enzyme is known to participate in nidoviral RNA synthesis; however, the exact substrate for its activity has not yet been identified. Remarkably, the human XendoU homolog PP11 (placental protein 11)—which is specifically expressed in the syncytiotrophoblast and also in several tumors, thereby serving as a useful indicator of carcinogenesis—displays a strikingly similar endoribonucleolytic activity (Laneve et al. 2008). Biochemical experiments performed employing recombinant PP11 revealed that its ability to cleave single-stranded RNA molecules at uridines in a manganese-dependent fashion is strictly dependent on the analogous set of catalytic amino acids (HHK triad), as in the cases of XendoU and NendoU. Despite the extensive in vitro characterization of PP11 endonuclease activity, it has not yet been demonstrated whether it takes part in snoRNA maturation. Therefore, its exact cellular targets and physiological functions in the placenta and the role of its apparently deregulated expression during tumor development still remain to be elucidated.
The processing of rRNA
Endonucleolytic cuts also represent significant steps in rRNA biosynthesis, which accounts for the bulk of transcription and is one of the major and most energy consuming cellular activities in eukaryotes. Around 80 snoRNAs and more than 150 trans-acting proteins, including the RNA helicases that assist in the rearrangement of RNA structures and RNA protein interactions and nucleases—which process longer precursor molecules either endo- or exonucleolytically to release mature rRNAs—participate in ribosome synthesis (Henras et al. 2008). Although most exonucleases involved in pre-rRNA processing have been identified, the identity of endoribonucleases involved in pre-rRNA cleavage events in yeast remains largely unknown, except for the RNase MRP cleaving at site A3 localized in the internal transcribed spacer 1 between 18S rRNA and 5.8S rRNA, and the Rnt1 RNase III-like enzyme (Table 1) that cuts at the B0 site in the hairpin located within the 3′ external transcribed spacer downstream from the 3′ end of mature 25S rRNA (Fig. 6; Lygerou et al. 1994, 1996; Elela et al. 1996; Kufel et al. 1999).
FIGURE 6.
Ribosomal RNA maturation in yeast comprises several processing events controlled by different endoribonucleases. The primary transcript synthesized by the rDNA unit is cleaved by an Rnt1 RNase III–like double-stranded specific endonuclease at the site B0 within the 3′ ETS, followed by cleavage at the site A0 in the 5′ ETS carried out by an as yet unidentified enzyme. The Utp24 endonuclease is most likely responsible for cleavages at sites A1 and A2, removing 5′ ETS and separating further steps of 18S (left) and 5.8S/25S (right) rRNA processing, respectively. The 20S pre-rRNA is converted into the mature 18S rRNA in the cytoplasm due to the action of Nob1 endonuclease at site D, probably in cooperation with the Pfa1/Prp43 and Ltv1 proteins. Processing intermediates encompassing 5.8S and 25S rRNA are trimmed at the 5′ end due to the endoribonucleolytic activity of RNase MRP and Rat1/Xrn1 exoribonucleases. The 5.8S and 25S rRNA maturation pathways are subsequently separated by endonucleolytic cleavage at site C2 and completed by exoribonucleolytic processing.
The synthesis of rRNA mainly occurs in the nucleolus; however, the final steps of ribosome biogenesis take place after the export of immature ribosomal subunits from the nucleus to the cytoplasm (Udem and Warner 1973; Thomson and Tollervey 2010). One of the ultimate events in this complex pathway is the cleavage of the 20S rRNA precursor within pre-40S particles. Interestingly, yet another PIN domain-containing protein, Nob1, which is related to Dis3, Swt1, and SMG6 (Table 1), was recently identified as a long-sought endoribonuclease that cleaves 20S pre-rRNA in the vicinity of its 3′ end, thereby leading to the formation of mature 18S rRNA (Fig. 6; Lamanna and Karbstein 2009; Pertschy et al. 2009).
The Nob1 protein was previously shown to be a component of the pre-40S ribosomal subunit (Fatica et al. 2003). Depletion of Nob1 inhibits cleavage at site D of the 20S 18S rRNA precursor, and this was later largely recapitulated in a strain expressing Nob1 with a mutation of the catalytic aspartate within its PIN domain, strongly indicating that it might be responsible for the conversion of 20S pre-rRNA into 18S rRNA (Fatica et al. 2003, 2004). Direct evidence that Nob1 interacts with the 20S precursor came from a series of in vitro experiments demonstrating the specific binding of recombinant Nob1 to the substrates containing cleavage site D (Lamanna and Karbstein 2009). Importantly, it was also proven that the PIN domain of Nob1 makes contact with the processing site located in the single-stranded region, which is in agreement with the general preference of PIN domain-containing endoribonucleases for unstructured substrates. These observations were largely confirmed by biochemical assays employing Nob1 purified from a native source, showing its ability to cleave the model substrate at the expected site (Pertschy et al. 2009). It is worth emphasizing that, similar to other PIN domains, the endonucleolytic activity was highly dependent on both the presence of manganese and the conserved aspartic acid residue putatively involved in coordination of the divalent cation within the predicted catalytic center.
Unlike Dis3, Smg6, and Swt1 PIN domains, which seem to target a wide palette of substrates, Nob1 is highly specific toward a precisely defined cleavage site. This raises the question of how specificity is achieved. One possibility is because of the interactions between Nob1 and the accessory cofactors. Indeed, it was recently reported that the cleavage of 20S pre-rRNA catalyzed by Nob1 is assisted by the putative complex encompassing the DEAH-box RNA helicase Prp43 and the G-patch protein Pfa1, as well as by Ltv1, a nonessential component of 43S preribosomal particles (Pertschy et al. 2009). Genetic screenings have identified the existence of an interaction network between these four proteins since LTV1 deletion resulted in a synergistic growth phenotype upon additional mutations in PRP43 or PFA1 as an outcome of inhibited 18S rRNA maturation. These effects could be reversed by the overexpression of Nob1 provided that the acidic amino acids crucial for its endoribonucleolytic activity were unchanged. Since the 20S pre-rRNA cleavage under physiological conditions only occurs following the cytoplasmic export of pre-40S ribosomal particles, it is likely that the Prp43-Pfa1 complex acts together with Ltv1 to regulate the activity of Nob1 (Fig. 6). Thus, the role of Nob1 cofactors might be to prevent premature processing at site D and possibly also to restrict its action with respect to other RNA molecules. Taking into account the fact that Prp43 displays RNA helicase activity, it might either displace some factor that blocks the access of Nob1 to its cleavage site or remodel the secondary structure in the substrate after its transport across the nuclear envelope. It can be supposed that, although the region encompassing the Nob1 processing site forms the stable hairpin in the nucleus, it becomes single stranded when the pre-40S ribosomal subunits reach the cytoplasm, which is consistent with the observation made in another report (Lamanna and Karbstein 2009). This enables Nob1 to attack site D, ensuring that the 18S rRNA maturation is properly accomplished. The stage in ribosome assembly when Nob1-mediated processing takes place is not known precisely, but the presence of 20S pre-rRNA in the polysomal fraction of double ltv1 prp43 and ltv1 pfa1 mutants and the ability of Nob1 to interact with polyribosomes suggest that it might even happen as late as after the joining of 60S and 40S subunits or following the initiation of protein synthesis (Pertschy et al. 2009). It was recently shown that the pre-40S ribosomal subunits containing unprocessed 20S pre-rRNA can engage in the initiation of translation (Soudet et al. 2010).
A couple of years ago, another protein containing a PIN domain was implicated in rRNA processing, namely, Utp24 (Bleichert et al. 2006; Table 1). Although no experimental data exist to directly show its nucleolytic activity in vitro, significant in vivo evidence has strongly indicated that it might be one of the still elusive endoribonucleases participating in ribosome biogenesis. The Utp24 protein is enriched in the nucleoli as a component of the SSU processome, involved in A0, A1, and A2 pre-rRNA cleavages, and its depletion leads to a pronounced defect in these initial steps of 40S ribosome subunit maturation, as reflected by the significant impairment of cell growth (Bleichert et al. 2006). The strict requirement for intact acidic residues within the Utp24 PIN domain to enable the rescuing of these phenotypes, in conjunction with the results obtained for all other yeast PIN-domain nucleases in similar experiments, allow us to assume that it is a catalytically active endonuclease responsible for the cleavage of rRNA precursors at sites A1 and/or A2 (Fig. 6).
RNAi: A PROCESS EMPLOYING MULTIPLE ENDORIBONUCLEASES
The RNAi process, a post-transcriptional mechanism silencing gene expression in eukaryotes, largely depends on endoribonucleolytic activities, which are utilized during virtually all phases of the process. Thus, endoribonucleases belonging to different families of enzymes participate in the initiation of RNAi by the generation of siRNA from longer dsRNA molecules. They also promote activation of the RNA-induced silencing complex (RISC) effector complex and are responsible for the siRNA-mediated cleavage of cognate mRNAs in the best studied mode of the RNAi phenomenon.
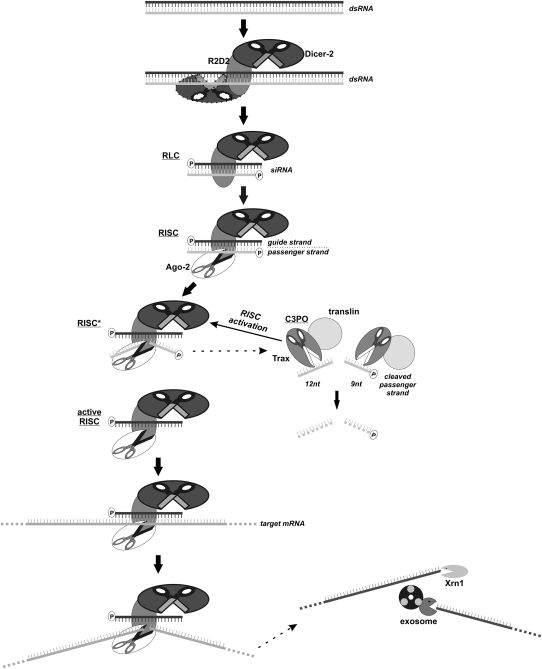
The initiation step is dependent on the activity of Dicer (Table 1), a member of the RNase III family of double-stranded-specific endoribonucleases that cleaves dsRNA substrates to produce 21–25-nt-long siRNA molecules possessing 5′-phosphate and 2-nt 3′ overhangs (Fig. 7). The siRNAs then mediate the sequence-specific silencing of gene expression (Hammond et al. 2000; Zamore et al. 2000; Bernstein et al. 2001). This occurs because of the base pairing of one siRNA strand, the so-called guide strand that is incorporated into the RISC, with complementary mRNA sequences (Martinez et al. 2002). The remaining siRNA strand, known as the passenger strand, is cleaved by the catalytic component of the RISC, the Ago2 protein (Table 1), which “slices” it into 9- and 12-nt fragments (Rand et al. 2005; Leuschner et al. 2006) and then targets the respective mRNA substrate in a similar manner within the site defined by the hybridization of the guide siRNA strand (Fig. 7; Elbashir et al. 2001). The PIWI domain of the Ago2 Slicer responsible for its endoribonucleolytic activity structurally resembles the fold of RNase H, which introduces nicks into the RNA component of RNA/DNA hybrids (Song et al. 2004). The mutations of Ago2 putative catalytic amino acids, located in positions corresponding to the residues involved in two-metal ion catalysis in the case of RNase H, are known to abolish RISC cleavage competence both in vitro and in vivo, which confirms that Ago2 is the source of Slicer endoribonucleolytic activity in the RNAi effector complex (Liu et al. 2004; Meister et al. 2004; Rand et al. 2004). Dicer and Slicer functions and structures have been deeply investigated, and the results of research focused on the role of these enzymes in the initiation and execution of RNAi were described in detail in several recent reviews (Peters and Meister 2007; Tolia and Joshua-Tor 2007; Hutvagner and Simard 2008; Jaskiewicz and Filipowicz 2008; Jinek and Doudna 2009; Nowotny and Yang 2009).
FIGURE 7.
Multiple endoribonucleases participate in different phases of RNA interference in eukaryotic cells. Long dsRNA molecules are processed into 21–25-nt siRNAs by the Dicer RNase III-like enzyme, together with its RNA-binding partner R2D2, leading to the formation of the RISC-loading complex (RLC) which is converted into RISC following the recruitment of Ago-2. The latter enzyme cuts the passenger strand in the RNA duplex, being the first substrate for its endoribonucleolytic activity, into 12- and 9-nt fragments. Cleavage of these RNA species by Trax, a catalytic component of the C3PO endoribonuclease, is a prerequisite for RISC activation, which enables recognition of target mRNA molecules. The targeted transcript is then “sliced” by Ago-2 and finally the RNA fragments located upstream of and downstream from the cleavage site are degraded by the exosome and Xrn1.
Another endoribonuclease, C3PO, composed of translin and Trax (translin-associated factor X) has recently been described as an enzyme that promotes RNAi by removal of the cleavage products of the siRNA passenger strand (Fig. 7; Table 1; Liu et al. 2009). This enzyme was purified from a Drosophila extract in an attempt to identify factors required for assembly of the functional RISC complex. The C3PO greatly enhances RISC-mediated cleavage in vitro. A mutation in the trsn gene, encoding translin, precluded RNAi, suggesting that the in vivo function of C3PO is indispensable for efficient silencing. It has been shown that C3PO enables the transition from the RISC loading complex, containing Dicer-2/R2D2 and siRNA, to the fully active RISC, mainly by facilitating unwinding of the siRNA duplex and the subsequent loading of the guide strand onto Ago2 (Fig. 7). Trax is an active component of the complex and its catalytic activity was found to be magnesium dependent and, as in the case of many other nucleases described herein, required the presence of conserved glutamates and aspartate, which probably coordinate the Mg2+ ion. The in vivo potential of C3PO to stimulate RISC function was shown to rely on intact catalytic residues in Trax. In conclusion, the activity of C3PO can be viewed as a mediator between the generation of duplex siRNA by Dicer and the incorporation of guide siRNA strands into RISC before the mRNA cleavage step executed by Ago2 (Liu et al. 2009).
ENDORIBONUCLEASES IN PLANT CHLOROPLASTS
The metabolism of RNA occurs not only in the nucleus and cytoplasm of eukaryotic cells, but also in the semiautonomous organelles of an endosymbiotic origin, namely, mitochondria and plant chloroplasts (for a recent review, see Schuster and Stern 2009). These compartments contain their own genomes, and the expression of genes in both mitochondria and chloroplasts is regulated to a great extent at the post-transcriptional level. In mitochondria, RNA decay is essentially dependent on polyadenylation and exoribonucleolytic digestion, whereas endoribonucleases such as RNase P and tRNase Z play a role in the processing of polycistronic transcripts and the maturation of different classes of RNA. Mitochondrial endoribonuclease has also been identified, which participates in the RNA editing phenomenon associated with the deletion/insertion of multiple uridines that has so far only been reported in trypanosomatids (Trotter et al. 2005). One of the Arabidopsis mitochondrial PPR proteins has been shown to display endoribonucleolytic activity dependent on the integrity of its C-terminal DYW domain in vitro (Nakamura and Sugita 2008), but the genuine relevance, if any, of this finding to the metabolism of RNA in plant mitochondria remains to be demonstrated in vivo. On the contrary, various steps of chloroplast RNA turnover seem to rely on the action of endoribonucleases.
An ortholog of E. coli RNase E, the principal and best-characterized prokaryotic endonuclease globally regulating gene expression, was recently proposed to initiate the polyadenylation-assisted RNA degradation pathway in Arabidopsis thaliana chloroplasts (Schein et al. 2008). Notably, genes coding for eukaryotic RNase E counterparts are present exclusively in the genomes of algae and higher plants, suggesting that their role is restricted to the regulation of gene expression in chloroplasts. Indeed, Arabidopsis RNase E has only been found in chloroplasts and not in mitochondria, and there is no evidence for the existence of the mitochondrial RNase E-like enzyme (Schein et al. 2008). This indicates that the gene coding for RNase E was incorporated into the plant genome by transferral from a cyanobacterial ancestor of chloroplasts following endosymbiosis. Chloroplast RNase E has a modular organization resembling that of the prokaryotic protein (Table 1) and, based on homology modeling, both enzymes possess structurally similar catalytic sites. Accordingly, A. thaliana RNase E displays nucleolytic activity comparable to the E. coli homolog in that it is a single-stranded-specific endoribonuclease stimulated by the presence of monophosphate at the 5′ end and showing a preference toward AU-rich regions. Both enzymes also have nearly identical substrate specificity (Schein et al. 2008). This functional homology between bacterial and chloroplast RNase E enzymes was reflected in an experiment showing the rescue of the temperature-sensitive mutation of the E. coli rne gene using the expression of plant RNase E (Mudd et al. 2008). Moreover, A. thaliana RNase E is a component of a high molecular weight complex (Schein et al. 2008), but whether it could represent the functional equivalent of the E. coli degradosome complex by incorporating chloroplast PNPase and some RNA helicases remains to be investigated. Nevertheless, this is likely given that the genes encoding PNPase and three other proteins putatively involved in chloroplast RNA metabolism are found to be coexpressed with RNase E, based on the analysis of microarray data sets (Mudd et al. 2008).
It has also been demonstrated that the function of chloroplast RNase E is important for plant physiology since loss-of-function T-DNA insertions resulted in the retardation of chloroplast development and the impairment of autotrophic growth (Mudd et al. 2008). This phenotype is likely to be the consequence of perturbations in organellar RNA metabolism, as exemplified by a severe decrease in the levels of chloroplast ribosomal and messenger RNAs, indicating that RNase E activity might be involved in the degradation and/or processing of these RNA molecules. However, the exact substrates for RNase E endoribonucleolytic activity in plastids have not yet been identified, and it is not known whether this affects chloroplast RNA turnover on a global scale, as in E. coli, or whether it participates in the processing of selected targets within the organelle, which is crucial for the maintenance of its functions.
Plant chloroplasts utilize at least two more endoribonucleases to control their RNA metabolism. Csp41 (Table 1) was identified in spinach as an unspecific enzyme involved in the binding and cleavage of stabilizing stem–loop elements localized within the 3′-UTR of several chloroplast transcripts, and it has been proposed to be a factor controlling mRNA stability in this cellular compartment (Yang et al. 1996; Yang and Stern 1997; Bollenbach and Stern 2003). Concordantly, the turnover of chloroplast mRNA was slowed down in the case of tobacco plants subjected to antisense-mediated csp41 silencing, which pointed toward the conclusion that Csp41 might be responsible for the initiation of regular mRNA degradation by executing a rate-limiting cleavage in the vicinity of the stem–loops and that the secondary structure of the latter might determine how efficient this cleavage would be (Bollenbach et al. 2003). Interestingly, Chlamydomonas reinhardtii Csp41 was identified in a proteomic analysis as a protein associated with plastid ribosomes (Yamaguchi et al. 2003). The functional analysis of its two A. thaliana homologs, Csp41a and Csp41b, revealed that although single T-DNA insertion mutants were indistinguishable from the wild-type plants, the double mutant accumulated increased amounts of 23S rRNA precursors that led to defects in chloroplast polysome assembly (Beligni and Mayfield 2008). This could indicate that Csp41a and Csp41b have partially overlapping functions in the plastid rRNA maturation pathway.
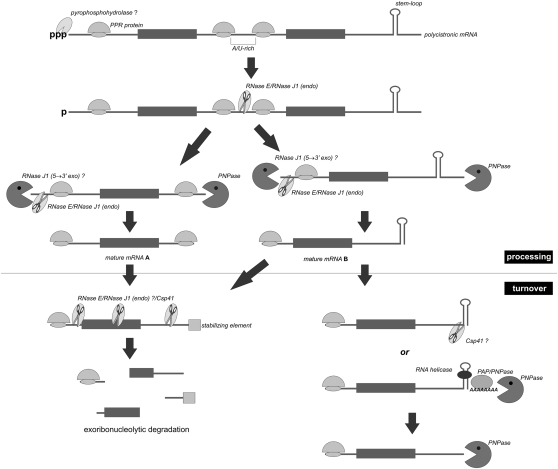
The picture that emerges from the data presented above is that both RNase E and Csp41 are able to target both mRNAs as degradative endoribonucleases (Fig. 8) and rRNAs, playing an important role in their processing. Molecular mechanisms that dictate substrate selection are not yet understood in either case, and it is not known whether accessory factors or, for instance, light activation, might influence the endoribonucleolytic activity and specificity of these enzymes. Moreover, considering that the homolog of B. subtilis RNase J is also likely to be present in the chloroplasts (Bollenbach et al. 2007), it is vital to determine the relative contribution of different endoribonucleases in various aspects of RNA metabolism in the organelle to gain insight into whether they act redundantly on the same substrates or whether their cleavages are entirely independent. It was recently proposed that RNases E and J might be responsible for the initiation of both intercistronic processing and the decay of chloroplast transcripts by cutting in the AU-rich, ribosome-free untranslated regions, which are followed by 3′ → 5′ and 5′ → 3′ exoribonucleolytic trimming until the secondary structure or binding site for a protein—such as one of the PPR factors, abundantly represented in the chloroplasts—is encountered, thereby defining the mature terminus of the RNA (Fig. 8; Pfalz et al. 2009). According to this scenario, the turnover rate of a given transcript would depend on the number of endonucleolytic cleavage sites and their accessibility, as well as on the presence of elements protecting it from the action of exoribonucleases (Fig. 8). This, in turn, would strengthen the general observation that the concerted action of endo- and exoribonucleases is required for efficient RNA degradation in various biological systems.
FIGURE 8.
mRNA processing and turnover in plant chloroplasts are controlled by the concerted action of endo- and exoribonucleases. The event which most likely initiates the processing of polycistronic chloroplast transcripts is the removal of pyrophosphate from the 5′ end, similarly to the case of prokaryotes. This reaction sensitizes the transcript to attack by endoribonucleases such as RNase E and RNase J1, which cleave polycistron at highly accessible single-stranded AU-rich regions. The 5′ and 3′ ends of the pre-mRNAs thus created are subsequently trimmed by endo- and exonucleases until the stable stem-loop structures or sequences bound by PPR motif–containing proteins, defining the positions of mature mRNA termini. The turnover phase of chloroplast transcripts might be initiated by either the combined action of endoribonucleases cleaving within 5′ UTR, coding sequence or 3′ UTR, followed by exoribonucleolytic decay (left branch) and/or through the removal of the 3′ stem-loop which can occur in two ways: 1) endoribonucleolytic cleavage by the Csp41 enzyme, or 2) cooperative destabilization by the RNA helicase supported by repetitive cycles of polyadenylation and exoribonucleolytic degradation (right branch).
CONTEXT-SPECIFIC ENDORIBONUCLEASES
Apart from the general RNA metabolism processes described above, numerous endonucleolytic events play important context-specific roles in the control of RNA degradation in eukaryotes, dictated by the cell type, the particular sequence within the RNA substrate, external signals, or stress conditions, among others, targeting only a subpopulation of the cellular pool of transcripts. Several endoribonucleases involved in these context-specific RNA decay mechanisms have been relatively well characterized. On the other hand, for some eukaryotic proteins displaying endoribonuclease activity, the physiological substrates have yet to be identified. The aim of this section is to briefly present an updated status of the knowledge concerning this diversified group of enzymes with an emphasis on their mode of action, the ways their activities are regulated, and the important roles that they play in controlling different processes in organisms belonging to various eukaryotic taxa.
IRE1: A dual-function enzyme acting at different stages of the unfolded protein response (UPR)
Eukaryotes have developed a number of mechanisms to prevent the accumulation of improperly folded proteins. One of them is the stress-induced unfolded protein response (UPR), in which sensing of excessive amounts of unfolded proteins in the endoplasmic reticulum (ER) leads to the increased production of ER-residing proteins, including chaperones that accelerate correct protein folding and factors facilitating the degradation of misfolded polypeptides (for recent reviews, see Mori 2009; Kohno 2010).
A key enzymatic component of UPR is IRE1, which is definitely the most peculiar context-specific endoribonuclease in eukaryotic cells. The IRE1 enzyme was initially identified as a transmembrane serine/threonine kinase coupling ER sensing to the transcriptional activation of UPR-associated genes in the nucleus following its oligomerization and trans-autophosphorylation (Cox et al. 1993; Mori et al. 1993; Shamu and Walter 1996). The up-regulated expression of genes coding for proteins acting as effectors in UPR is controlled by the Hac1 basic leucine zipper-containing transcription factor that binds to the specific UPRE sequence element present in their promoters (Cox and Walter 1996; Mori et al. 1996; Nikawa et al. 1996). The Hac1 UPR-active form is generated due to an unconventional splicing event involving the excision of an intron located in the vicinity of its mRNA 3′ terminus (Cox and Walter 1996). This process was found to differ from the classical splicing pathway in several aspects, including the presence of noncanonical splice sites flanking the intron, the lack of dependence on the spliceosome function, and the requirement for Rlg1 tRNA ligase (Sidrauski et al. 1996). Moreover, the cleavage takes place on polysome-bound mRNA in the cytosol, unlike classical splicing, which occurs in the nucleus, and intron removal allows for efficient translation of the transcript (Chapman and Walter 1997; Rüegsegger et al. 2001).
Interestingly, in addition to its kinase activity, recombinant IRE1 was shown to be a site-specific endoribonuclease correctly processing Hac1 mRNA at the predicted splice junctions in in vitro assays via its cytoplasmic C-terminal part comprising kinase and kinase extension nuclease (KEN) domains (Mori et al. 1993; Sidrauski and Walter 1997). Moreover, the nuclease activity required adenosine nucleotide as a cofactor and was strictly dependent on the presence of an intact kinase domain, suggesting that cooperation between the two catalytic IRE1 functions is indispensable for proper splicing of the Hac1 transcript (Sidrauski and Walter 1997). More recent studies provided structural explanations for the abovementioned findings. The dimerization of IRE1 N-terminal domains of the two monomers in the ER lumen juxtaposes their cytoplasmic kinase domains in a face-to-face orientation, thereby facilitating trans-autophosphorylation. This in turn facilitates binding of the nucleotide, which stimulates a conformational change reorienting the monomers into a back-to-back configuration in which the active endonucleolytic sites of the KEN domains are exposed (Lee et al. 2008; Ron and Hubbard 2008). Other authors proposed that activation of the IRE1 endonuclease via trans-autophosphorylation may require the assembly of even higher-order oligomers (Korennykh et al. 2009).
The entire splicing reaction was successfully reconstituted in vitro using purified IRE1, tRNA ligase, and a Hac1 pre-mRNA substrate (Sidrauski and Walter 1997). This experimental system allowed detailed mechanistic insights to be gained into the unprecedented reaction, which was reported as largely proceeding in a manner similar to tRNA splicing, despite significant differences between the nucleolytic components of the two pathways. Interestingly, both splice junctions serving as IRE1 cleavage sites adopt a strikingly similar stem–loop conformation, indicating that a secondary structure, rather than a specific primary sequence, might be the major substrate determinant of IRE1-mediated splicing (Gonzalez et al. 1999).
The principal functions of IRE1 in the initiation of UPR, including the absolute requirement for an intact endonuclease activity, seem to be broadly conserved among different eukaryotic taxa, as confirmed by studies in filamentous fungi (Valkonen et al. 2004), fruit flies (Plongthongkum et al. 2007), nematodes (Calfon et al. 2002), plants (Koizumi et al. 2001), and mammals (Tirasophon et al. 1998, 2000; Wang et al. 1998; Yoshida et al. 2001). The substrate for IRE1 in higher eukaryotes is XBP1 mRNA (Yoshida et al. 2001; Calfon et al. 2002; Lee et al. 2002; Plongthongkum et al. 2007). Like in yeast, IRE1-mediated splicing in higher eukaryotes generates an active form of the XBP1 transcriptional factor that contains a basic leucine zipper domain similar to that of Hac1. In both cases this is due to replacement of the C-terminal part of the protein (Sidrauski and Walter 1997; Yoshida et al. 2001; Lee et al. 2002). It must be emphasized though that the effector phase of UPR in mammalian cells is far more complicated than in the case of yeast (for reviews, see Mori 2009; Kohno 2010) and involves other downstream events, such as proteolytic cleavage of the ATF6 transmembrane ER protein, converting it into a potent transcriptional factor that translocates to the nucleus and up-regulates UPR-associated genes through binding to the ERSE promoter sequence (Yoshida et al. 2000) or stimulation of JNK protein kinases (Haze et al. 1999; Urano et al. 2000). The former was demonstrated to intertwine with the generation of XBP1 mRNA by IRE1, in that the up-regulation of the transcript mediated by ATF6 via ERSE precedes its endonucleolytic splicing (Yoshida et al. 2000, 2001; Lee et al. 2002). It has been postulated that by employing various strategies, UPR is executed more efficiently regardless of the amounts of unfolded proteins that have accumulated in the ER, and that the individual contributions of ATF6- and IRE1-mediated pathways may differ depending on the severity of ER stress (Yoshida et al. 2001). Both mechanisms may also act at different stages of UPR, leading to the transcriptional activation of distinct subsets of UPR-associated genes depending on whether refolding and/or degradation of misfolded proteins is required for regaining ER homeostasis (Hampton 2003; Yoshida et al. 2003). This picture is further complicated by the existence of parallel UPR pathways in mammals, involving the PERK/PEK transmembrane kinase in a similar manner to IRE1, but devoid of the nuclease activity that phosphorylates eIF2α and consequently shuts down translation in a global manner upon ER stress (Shi et al. 1998; Harding et al. 2000; Liu et al. 2000).
In addition, while the yeast genome only encodes one IRE1 enzyme, two such proteins seem to exist in higher eukaryotes (Tirasophon et al. 1998; Wang et al. 1998; Koizumi et al. 2001). While one of them, in principle, appears to activate UPR in a similar manner to the yeast Ire1p in human cells (Tirasophon et al. 2000), the other employs a substantially distinct mechanism, characterized by the cleavage of 28S rRNA, leading to the repression of protein synthesis (Iwawaki et al. 2001). A diversification of the functions of the two IRE1 paralogs is also plausible in Arabidopsis, where they display nonoverlapping expression patterns (Koizumi et al. 2001).
Similar to other endoribonucleases described herein, the action of IRE1 is controlled in several different ways. First, dissociation of the ER BiP chaperone from the N-terminal lumenal IRE1 sensing domain is a prerequisite for efficient UPR (Bertolotti et al. 2000; Okamura et al. 2000). Besides this, as mentioned above, IRE1 RNase activity is stimulated upon trans-autophosphorylation and dimerization/oligomerization (Tirasophon et al. 2000; Lee et al. 2008; Ron and Hubbard 2008; Korennykh et al. 2009). On the contrary, IRE1 dephosphorylation mediated by Ptc2 and Dcr2 phosphatases negatively regulates its activity and inhibits UPR (Welihinda et al. 1998; Guo and Polymenis 2006). Interestingly, in mammalian cells trans-autophosphorylation was also proved to be coupled to the self-regulation of mRNA levels by the two IRE1 paralogs (IRE1α and IRE1β) (Tirasophon et al. 2000), although convincing evidence for the direct involvement of IRE1 in cleaving its own messenger was not provided.
The Hac1 mRNA is probably the only target of the unconventional splicing reaction catalyzed by IRE1 in yeast, as revealed by different genome-wide approaches (Niwa et al. 2005). However, as shown in previous studies, IRE1 might play an additional role by influencing the levels of other transcripts, such as its own mRNA and 28S rRNA in higher eukaryotes (Tirasophon et al. 2000; Iwawaki et al. 2001). This indicates that IRE1 could also be a degradative endonuclease. A systematic microarray-based analysis of gene expression profiles in IRE1- and XBP1-depleted Drosophila S2 cells showed that, in addition to the up-regulation of overlapping genes encoding proteins involved in classical UPR, there is a cluster of genes solely regulated by IRE1 via selective repression of ER-targeted mRNAs. Blocking cytoplasmic exonucleolytic degradation pathways allowed for the detection of decay intermediates arising from the endonucleolytic cleavage of the transcripts mediated by IRE1. The degradation was largely dependent on mRNA fragments encoding signal sequences targeting the protein to ER, indicating that it takes place during cotranslational translocation, which may, for instance, bring the target transcript into the vicinity of the IRE1 nuclease domain (Hollien and Weissman 2006). This phenomenon, together with PERK/PEK-induced translational repression, is thought to facilitate the cell's efforts in restoring ER homeostasis by immediately ceasing the production of improperly folded polypeptides. Moreover, it tells us that IRE1 endonucleolytic activity might be important not only at the initial stage of UPR, but also during the early effector phase of the process. It must be also pointed out that the mechanism involving mRNA degradation by IRE1 seems to be conserved in mammals. The very first evidence came from studies demonstrating the attenuated expression of CD59 mRNA due to site-specific cleavage following the overexpression of IRE1, or as a consequence of ER stress (Oikawa et al. 2007). Large-scale follow-up studies revealed that this is also true for a number of mRNAs in mouse fibroblasts (Hollien et al. 2009). Likewise, there are many transcripts in human cells processed by IRE1-mediated cleavage sharing a common consensus site that is conserved in XBP1 mRNA; however, it is not known whether or not they undergo regulated degradation upon ER stress (Oikawa et al. 2010).
In conclusion, two different outcomes of tightly regulated IRE1 endonoribonucleolytic activity, XBP1 mRNA splicing and degradation of transcripts targeted to the ER membrane, are hallmarks of mammalian UPR and represent an unprecedented example of coupling RNA metabolism events to protein quality control in the endoplasmic reticulum. Figure 9 summarizes our current view of the action of IRE1 during UPR.
FIGURE 9.
The IRE1 endoribonuclease is a central component of mammalian UPR pathways. ER stress in mammalian cells results in the accumulation of misfolded proteins which elicits three major responses. The most immediate UPR effector pathway is activation of PERK/PEK which upon dimerization and autophosphorylation phosphorylates eIF2α and leads to translational shutdown. Moreover, the ATF6 transmembrane protein is converted by proteolysis into an active transcription factor which stimulates the expression of genes encoding ER chaperones via binding to ESRE sites in their promoters (top). If these early control mechanisms are sufficient for restoring ER homeostasis, the UPR is completed. Otherwise, ATF6 also activates the transcription of the XBP1 gene, producing an unspliced mRNA. At the same time, BiP dissociates from the lumenal domains (LD) of IRE1 monomers, which is the prerequisite for their dimerization/oligomerization (top). The latter process occurs over a series of events, including trans-autophosphorylation of kinase domains (KD) (which might be counteracted by the action of two phosphatases, namely, Ptc2 and Dcr2), binding of the adenosine nucleotide cofactor and reorientation of oligomer subunits, all of which eventually lead to the activation of IRE1 endoribonucleolytic sites in KEN domains (middle). The active IRE1 nuclease has several functions in the late effector phase of UPR (bottom). Above all, it catalyzes cytoplasmic splicing of XBP1 mRNA together with tRNA ligase, thus enabling its translation into an active transcription factor which up-regulates the expression of genes encoding ER chaperones and enzymes degrading unfolded proteins through their ESRE and UPRE promoter elements. On the other hand, IRE1 is also a degradative endonuclease, trigerring the decay of ER-targeted transcripts. The participation of IRE1 in the degradation of 28S rRNA and the autoregulation of its own mRNA levels is also possible, although it is not known yet whether or not IRE1 cleaves them directly (bottom). All of these RNA metabolism events significantly enhance the abilities of cells to efficiently deal with the presence of improperly folded proteins in the ER.
Endonucleolytic degradation exerted by enzymes having no or little sequence homology with known endoribonucleases
Numerous enzymes catalyzing endoribonucleolytic cleavages in eukaryotic cells share no obvious sequence motifs with the previously discovered endonucleases or even lack the predicted RNA-binding domains and, most importantly, often have other, sometimes strikingly distant biological functions. For instance, aldolases A and C are isoenzymes involved in glycolysis in neurons, which were quite unexpectedly shown to interact specifically with the 68-nt sequence situated at the boundary of the coding sequence and the 3′-UTR of the light neurofilament (NF-L) mRNA, leading to its destabilization (Cañete-Soler et al. 1998a,b, 2005). The modulation of the NF-L mRNA half-life by aldolases appears to involve both displacement of PABP from the poly(A) tail in a competitive manner and an inherent zinc-activated endoribonucleolytic activity of these proteins, stimulated by the presence of a cap at the 5′ end of the transcript (Cañete-Soler et al. 2005; Stefanizzi and Cañete-Soler 2007). Specifically, binding of PABP to the 68-nt instability determinant shields NF-L mRNA from the aldolase endoribonucleolytic activity attack that occurs in the proximity of the 5′ end of the poly(A) tail; interestingly, PABP was found to protect both polyadenylated and deadenylated NF-L transcripts ex vivo, consistent with the idea that it may bind to the body of the transcript rather than to the poly(A) tail (Stefanizzi and Cañete-Soler 2007). It has been speculated that the endonucleolytic degradation of the NF-L transcript by adolases, most probably stimulated by some extracellular signal, might contribute to the proper formation of the neurofilaments that determine the neuronal cell shape (Cañete-Soler et al. 2005). The fundamental issues of whether or not aldolases exploit the same active site for their glycolytic functions and endoribonucleolytic activities, and whether they can target mRNAs other than NF-L in the central nervous system, await further investigation.
Other interesting examples of proteins to which a putative endoribonuclease activity has recently been ascribed are the c-Jun and GCN4 transcription factors (Nikolaev et al. 2010). The first clue suggesting that GCN4 might be able to degrade RNA came with solving the crystal structure of its leucine zipper motif responsible for dimerization in the so-called retro- form, in which the amino acid sequence was inverted with respect to the placement of the N- and C-termini. The structure of this peptide resembled that of the RNase A active site, including a very similar spatial organization of charged residues, critical for RNase A-mediated degradation (Raines 1998; Mittl et al. 2000). More importantly, biochemical assays performed with the use of genuine basic-region leucine zippers of the yeast GCN4 and a human c-Jun proto-oncogene revealed that they are indeed able to cleave RNA substrates. The detected endoribonucleolytic activity was largely sequence unspecific, resistant to the recombinant RNase A inhibitor, and produced fragments that terminated with 2′–3′ cyclic phosphate. Notably, the c-Jun leucine zipper displayed activity irrespective of whether it was tested as a synthetic peptide or in the context of a full-length protein. Mutational analysis performed for the GCN4 leucine zipper disclosed that its catalytic properties are strongly inhibited upon mutations of conserved charged amino acids in the predicted active center (Nikolaev et al. 2010). The intrinsic abilities of GCN4 and c-Jun to degrade RNA in an endonucleolytic manner offer the interesting possibility that transcriptional control executed by these factors is somehow coupled to RNA metabolism. The precise role of these factors in the latter process should be uncovered upon the discovery of their physiological substrates.
It is worth noting that the endoribonucleolytic activity might not be by any means unique to the leucine zipper-containing transcriptional modulators, as was detected more than a decade ago for the peptide corresponding to the C2H2 zinc finger motif of the human male-associated ZFY transcription factor (Lima and Crooke 1999). However, the biological significance of this finding has not yet been explored further.
In addition to aldolases and transcription factors, endoribonuclease activity was also well documented in the case of G3BP being a component of the Ras signal transduction pathway; G3BP is a multifunctional factor that binds to the SH3 domain of RasGAP (GTP-ase activating protein; p120), a negative regulator of Ras (Parker et al. 1996; Gallouzi et al. 1998; for review, see Irvine et al. 2004).
The RasGAP–G3BP interaction occurs at the plasma membrane of proliferating cells, while it does not take place in serum-deprived nondividing cells, most probably due to the hyperphosphorylation of G3BP on serines (Gallouzi et al. 1998). The G3BP protein was found to degrade the RNA fragment derived from the 3′-UTR of c-myc mRNA, but not homoribopolymers, in vitro, indicating that it has a relatively specific endoribonuclease activity. This was shown to be stimulated significantly by serine phosphorylation, and was, in fact, the first endonuclease regulated through this kind of post-translational modification to be identified. In agreement, the RNase activity of G3BP immunoprecipitated from quiescent cells was more pronounced than in the case of its counterpart immunopurified from dividing cells. It was thus postulated that the changes in G3BP phosphorylation are mediated through its interaction with RasGAP (Gallouzi et al. 1998). The tight regulation of ribonucleolytic activity of G3BP being dependent on the proliferative status of the cells suggested that it might be responsible for the fine tuning of levels of transcript(s) encoding factors essential for cell cycle progression, most likely following a response to extracellular stimuli (Gallouzi et al. 1998; Tourrière et al. 2001). Indeed, it was shown that G3BP binds the specific target element within the 3′-UTR of the transcript corresponding to the c-myc proto-oncogene mostly via its C-terminal hnRNP C-type RRM domain and arginine/glycine-rich box (RGG), and that this interaction is a prerequisite for efficient endonucleolytic cleavage of the substrate at the phosphodiester bonds between cytosine and adenosine residues at multiple sites (Tourrière et al. 2001). As G3BP was found to associate with polyadenylated mRNAs (including the c-myc transcript) both in cells expressing and lacking RasGAP, it is likely that its endonuclease activity initiates RNA turnover in a deadenylation-independent manner. Notably, steady-state c-myc mRNA levels in RasGAP-deficient cells exposed to serum were higher during the initial phase of growth factor stimulation than in the case of cells expressing RasGAP, due to the increased half-life. The delay of c-myc mRNA decay in the cells devoid of RasGAP was a consequence of the presence of the G3BP isoform lacking phosphoserine in position 149 (Tourrière et al. 2001). Phosphorylation of this particular serine was shown to be important for the translocation of G3BP from the cytoplasm to the nucleus, leading to the hypothesis that a change of c-myc mRNA stability resulting from the presence of growth factors might partly be an outcome of G3BP relocalization and possible integration into mRNP complexes assembling in the nucleus. The exact role of the nuclear G3BP import upon serine 149 phosphorylation and the mechanism by which RasGAP promotes this post-translational modification definitely deserve further research. Interestingly though, arsenite treatment, which induces specific serine 149 dephosporylation, leads to the accumulation of G3BP in stress granules, providing further evidence that the phosphorylation status of this basic residue is a critical determinant of protein localization within the cell (Tourrière et al. 2003). Finally, it is important to note that although the direct experimental proof showing the involvement of G3BP endoribonucleolytic activity in the control of c-myc transcript stability in vivo has not yet been provided, mostly due to a lack of identifiable catalytic domains that could be subjected to mutational analysis, the enzyme clearly impacts on the turnover of several mRNAs in G3BP knock-out mice. At the organismal level, the function of G3BP is crucial for proper fetal growth and its deficiency leads to neonatal lethality, most probably due to substantial cell death in the central nervous system (Zekri et al. 2005). The detection of changes in the c-myc mRNA turnover rate on G3BP inactivation might prove problematic since there are other endoribonucleases that most likely redundantly participate in the degradation of this particular messenger, including: (1) a polysome-associated endoribonuclease that targets the c-myc open reading frame within the sequence element, referred to as the c-myc coding region stability determinant (CRD), by competing for this site with a CRD-binding protein that exerts a stabilizing effect (Prokipcak et al. 1994; Lee et al. 1998); (2) an APE1 enzyme that also degrades the c-myc transcript by attacking CRD, but that displays distinct biochemical properties (Bergstrom et al. 2006; Barnes et al. 2009; see below); and (3) the PSMA5 subunit of the 26S proteasome (Kulichkova et al. 2010). In the latter case, it is remarkable that the endonucleolytic activity of the proteasomal component seems to be modulated by different external stimuli, similarly to G3BP, and that it is regulated by the phosphorylation of neighboring proteasome subunits, namely, PSMA1 and PSMA3. Both enzymes thus appear to act at the crossroads of signaling pathways and RNA metabolism.
Another endoribonuclease, whose function is still quite enigmatic, is ARD-1 (activator of RNA decay [ARD]). Although ARD-1 is a human protein, it was cloned as a suppressor of the E. coli RNase E mutations (Wang and Cohen 1994). Overexpression of ARD-1 in the mutant E. coli strains was shown to restore site-specific RNase E cleavage in vivo and its sequence inspection revealed the presence of regions similar to the bacterial endonuclease (Wang and Cohen 1994); however, we did not observe any significant similarities between RNase E and ARD-1 in a secondary-structure-based homology search. Interestingly though, biochemical analyses of purified recombinant ARD-1 confirmed that it is able to bind several typical RNase E substrates and cleave them at the same sites within single-stranded AU-rich regions in a magnesium-dependent manner with the generation of 5′-phosphate termini, which is highly reminiscent of RNase E activity (Claverie-Martin et al. 1997). The ARD-1 protein is a translation product of a transcript generated by the alternative splicing of the precursor mRNA that also encodes the NIPP-1 inhibitor of protein phosphate-1 (Chang et al. 1999). It would be of interest to examine whether the alternative splicing of the ARD-1/NIPP-1 messenger might be shifted toward production of the ARD-1 endoribonuclease under specific circumstances or in particular tissue(s). Characterization of ARD-1 substrates is also required to show whether or not it is capable of initiating RNA decay. It is worth mentioning that yet another RNase E-like activity, distinct from ARD-1, was partially purified from human cells that processed E. coli 9S RNA and ompA mRNA at the same sites as the bacterial enzyme and that cross-reacted with antibodies against RNase E (Wennborg et al. 1995). Interestingly, this endoribonuclease was shown to cleave c-myc mRNA at several sites in the 3′-UTR, including two AUUUA pentanucleotides. Whether or not this activity adds to the broad spectrum of enzymes regulating c-myc transcript stability in vivo, as discussed above, remains elusive.
Polysomal ribonuclease 1 (PMR1): An enzyme similar to peroxidases that activates decay of albumin mRNA
Probably the most thoroughly studied endonuclease with no apparent RNase domain is polysomal ribonuclease 1 (PMR1). This enzyme is particularly interesting due to a number of strategies controlling its catalytic function (see below). It was discovered almost two decades ago as a single-stranded-specific, magnesium-independent endonucleolytic activity associated with Xenopus liver polysomes that destabilized albumin mRNA following estrogen induction and regulated the turnover of other mRNAs in hepatocytes in response to hormone stimulation (Pastori et al. 1991b; Pastori and Schoenberg 1993). When purified from its native source to near homogeneity, PMR1 was selective toward albumin transcripts when compared with the other tested substrates and it displayed a clear preference toward the cleavage site encompassing two overlapping APyUGA (where Py stands for either C or U) pentanucleotide motifs (Dompenciel et al. 1995). The PMR1 enzyme has exactly the same sequence specificity in vivo and structural probing revealed that the major, but not exclusive, cleavage site is located in a single-stranded region of a stem–loop structure. Moreover, as PMR1 was found to produce 5′ cleavage fragments with 3′-hydroxyl, it was reasoned that its endonuclease activity might provide entry points for the action of 3′ → 5′ exoribonucleases (Chernokalskaya et al. 1997). Surprisingly, subsequent cloning of the PMR1 cDNA revealed that it belongs to the peroxidase family of proteins, displaying over 50% sequence identity to myeloperoxidase; however, it is devoid of covalently bound heme and N-linked oligosaccharide chains, corroborating its lack of peroxidase activity (Chernokalskaya et al. 1998). Interestingly, PMR1 may also undergo proteolytic processing from a larger precursor protein, leading to the generation of its mature form (Chernokalskaya et al. 1998).
Putative proteolytic processing is just the tip of the iceberg when it comes to the regulatory mechanisms of PMR1 endoribonucleolytic activity. Destabilization of the albumin transcript after estrogen stimulation had been previously found to coincide with the parallel stabilization of vitellogenin mRNA, coding for the egg yolk precursor protein (Brock and Shapiro 1983; Nielsen and Shapiro 1990; Pastori et al. 1991a). As recombinant PMR1 was shown to efficiently trigger vitellogenin mRNA 3′-UTR decay in vitro by attacking it within the APyUGA pentamers (Cunningham et al. 2000), the differential stability of the two transcripts in vivo was initially quite astonishing. This seeming discrepancy was then explained by the selective protection of PMR1 cleavage sites in the 3′-UTR of vitellogenin mRNA, but not of those localized in the coding region of the albumin messenger, by high-affinity vigilin binding (Dodson and Shapiro 1994, 1997; Cunningham et al. 2000).
Another layer of PMR1 activity regulation is linked to its association with polyribosomes. PMR1 was initially shown to be associated with the membrane-bound polysomes containing both albumin and vitellogenin transcripts (Pastori and Schoenberg 1993). The distribution of PMR1 in the polysomal profile changes in response to estrogen stimulation: before hormone treatment it is analogous to the distribution of polysome-bound mRNA, consistent with the finding that PMR1 is a constituent of mRNPs involved in translation, whereas following estrogen induction PMR1 is shifted toward faster sedimenting fractions. The release of PMR1 from polysomes is preceded by a greater than 20-fold increase of its activity toward albumin mRNA (Cunningham et al. 2001). The activation of PMR1 is matched by the presence of albumin transcript decay products arising due to cleavage at consensus sites identified in previous studies (Chernokalskaya et al. 1997, 1998; Hanson and Schoenberg 2001).
The PMR1 enzyme selectively targets its polysome-bound target mRNA through amino acid sequences localized in its N- and C-terminal fragments, which results in the formation of a specific ∼680-kDa complex between PMR1 and its substrate. Both events are a prerequisite for efficient initiation of the endonuclease-mediated decay of the albumin transcript (Yang and Schoenberg 2004). Interestingly, PMR1 was shown to undergo a switch from a latent to an active state owing to the phosphorylation of tyrosine 650 within its C-terminal polysome-targeting domain, which is also crucial for the association of PMR1 with polysomes and formation of the PMR1-albumin mRNA complex (Yang et al. 2004). Post-translational PMR1 modification is mediated by c-Src tyrosine kinase activity in response to the binding of epidermal growth factor (EGF) to its receptor (EGFR) (Peng and Schoenberg 2007).
Additionally, proper PMR1 folding ensuring correctness of the interactions between PMR1 and c-Src is overseen by the Hsp90 chaperone, which transiently protects PMR1 from degradation by the 26S proteasome (Peng et al. 2008). In summary, PMR1 is another endonuclease, in addition to G3BP (see above), whose activity is largely controlled by site-specific phosphorylation and which links mRNA decay to signal transduction pathways. An updated model of the regulation of PMR1 activity referring to most of the molecular events described above is presented in Figure 10.
FIGURE 10.
Model of PMR1-mediated cleavage of albumin mRNA. The PMR1 protein is inherently unstable and unless protected by the Hsp90 chaperone it is subjected to the proteasomal degradation pathway. It is maintained in a latent state until the phosphorylation of tyrosine 650, which can be induced, for instance, following the binding of EGF to its receptor. The activated PMR1 is then recruited to its substrate undergoing translation, most likely with the involvement of some unidentified protein partner (designated here as PMR-BP), thereby leading to formation of the polysome-bound PMR1–albumin mRNA complex. Subsequent cleavage of the albumin transcript is followed by dissociation of PMR1 from polysomes.
A further connection between PMR1 activity and cell signaling was provided by a recent report on the unexpected in vivo interactions of PMR1, irrespective of whether it was complexed with c-Src or associated with a polysome-bound mRNA substrate, with Mena and VASP factors coordinating signal transduction through the actin cytoskeleton (Peng et al. 2009; for reviews on Mena/VASP proteins, see Krause et al. 2003; Kwiatkowski et al. 2003). Most importantly, it was found that the active PMR1, but not its catalytically impaired counterpart, had the ability to stimulate cell motility and colocalized with VASP at the leading edge of the migrating cells, opposite to the site of actin stress fiber accumulation (Peng et al. 2009). Mena/VASP proteins are thus likely to control the intracellular distribution of PMR1 and determine the site of its endonuclease-mediated mRNA decay.
Another theme related to changes in the subcellular localization of PMR1, and also a common feature shared with G3BP, is its accumulation in stress granules following arsenite treatment (Tourrière et al. 2003; Yang et al. 2006). However, while G3BP induces the formation of stress granules following its dephosphorylation and self-aggregation (Tourrière et al. 2003), stress conditions lead to the relocalization of the phospho-PMR1-albumin mRNA 680-kDa complex from polysomes to stress granules in the form of aggregates due to the direct interaction between the N-terminal fragment of endonuclease and the TIA-1 mediators of stress granule assembly (Gilks et al. 2004; Yang et al. 2006). Nevertheless, the issue of whether this stress-induced intracellular PMR1 redistribution is reflected by changes in the albumin transcript half-life remains to be addressed.
Interestingly, the mammalian homolog of PMR1 was implicated in endonucleolytic activity involved in the degradation of normal human β-globin mRNA, and even more significantly in the initiation of the decay of its PTC-containing counterparts during the NMD-like pathway in mouse erythroleukemia (MEL) cells (Bremer et al. 2003). This experimental system allowed for detection of polyadenylated degradation intermediates lacking sequences from the 5′ end of the β-globin transcript. The observed preference for U/G dinucleotides suggested that the enzyme responsible for cleavage might be similar to PMR1. Indeed, both crude and partially purified polysome-containing extracts of MEL cells had an endoribonucleolytic activity displaying the same selectivity for cleavage sites in the in vitro assays employing a synthetic β-globin substrate (Stevens et al. 2002). Furthermore, ectopic expression of catalytically active Xenopus PMR1 in MEL cells destabilized β-globin mRNA and accordingly produced cleavage products similar to those generated by the endogenous enzyme (Bremer et al. 2003). It would be interesting to investigate the relative contribution of endoribonucleolytic NMD initiated by SMG6 (see above) and the pathway involving the PMR1-like enzyme in erythroid cells and other mammalian cell types. The fate of decay products arising due to the activity of the PMR1-like enzyme should also be examined. Difficulties in detection of 5′ cleavage intermediates would suggest that they might be subjected to rapid degradation by the exosome, as in the case of NMD controlled by SMG6. The possible involvement of exosome in the elimination of PMR1 cleavage products is supported by a recent discovery that PMR1, together with the exosome recruited by the AU-rich element-binding protein KSRP, participates in the decay of the parathyroid hormone mRNA in human cells (Nechama et al. 2009). This observation again strengthens the notion that the cooperative action of exo- and endoribonucleolytic activities contributes to RNA metabolic processes in eukaryotic cells.
Zc3h12a: A zinc-finger protein with a nuclease domain displaying distant homology with PIN domains is involved in protection from immune disorders
The Zc3h12a protein, containing the CCCH-type of zinc finger, was recently reported as acting as a regulatory endoribonuclease playing a prominent role in the modulation of the immune response (Matsushita et al. 2009). Mice lacking Zc3h12a grow slower than their wild-type counterparts and die prematurely. This phenotype is further accompanied by acute anemia and the accumulation of plasma cells in lymph nodes and the spleen, as well as an enhanced infiltration of plasma cells in the lung and pancreas. Together with the observed hyperimmunoglobulinemia, these results suggested that Zc3h12a might be involved in protecting mice from immune disorders associated with the increased production of immunoglobulins by plasma cells. A high-throughput investigation of gene expression profiles in wild-type and Zc3h12a-deficient macrophages revealed that a set of genes including those coding for interleukin-6 (IL-6), γ-interferon, and the calcitonin receptor (Calcr) was overexpressed in the latter cells. The Zc3h12a protein was implicated in the post-transcriptional regulation of IL-6 mRNA levels through direct RNA binding and endoribonucleolytic degradation dependent on the conserved N-terminal fragment of the protein, showing some distant sequence similarity to PIN domains (Matsushita et al. 2009). Unlike PIN domain-containing endonucleases, Zc3h12a activity was demonstrated as being most efficient in the presence of Mg2+ rather that Mn2+ ions. On the other hand, similarly to PIN endonucleases, the in vitro catalytic properties and the in vivo function of Zc3h12a strictly rely on the presence of conserved residues located in the negatively charged pocket forming its putative active site, as based on structural modeling. Apart from IL-6 mRNA, Zc3h12a appears to control the levels of IL-12p40 cytokine transcripts and Calcr mRNA (Matsushita et al. 2009). It is likely that the list of mRNAs targeted by the endoribonucleolytic activity of Zc3h12a might soon be expanded, particularly when taking into account the severity of the symptoms in Zc3h12a-deficient mice.
Angiogenin: A member of the RNase A protein family degrading tRNA in the cytosol under stress conditions
Angiogenin is a protein involved in blood vessel growth (for review, see Gao and Xu 2008), for which an endoribonuclease activity was predicted due to its significant homology with the pancreatic ribonuclease A (Fett et al. 1985; Kurachi et al. 1985; Strydom et al. 1985). Similarly to RNase A, angiogenin is a secreted factor but surprisingly it has an additional cytoplasmic function. Angiogenin was shown to act as a weak ribonuclease against rRNA in vitro (Shapiro et al. 1986; Rybak and Vallee 1988) and against tRNA in cell-free lysates following injection into Xenopus oocytes (Saxena et al. 1992). Angiogenin-mediated tRNA degradation has cytotoxic effects due to the shutdown of protein synthesis.
Structural studies in conjunction with mutational analysis and chemical modification approaches provided mechanistic insights into differences between the ribonucleolytic potential of angiogenin and RNase A activities (Shapiro et al. 1987; Acharya et al. 1994; Russo et al. 1994). Although the active site of angiogenin is formed by similar catalytic residues, as in the case of RNase A, it adopts a different spatial arrangement that is primarily caused by the steric hindrance of the pyrimidine-binding pocket resulting from the presence of glutamine in position 117. This may account for the observation that the endoribonucleolytic activity of angiongenin is several orders of magnitude lower when compared with RNase A (Shapiro et al. 1986; Harper and Vallee 1989). Despite the fact that angiogenin is a relatively inefficient endonuclease, its activity seems to be indispensable for effective neovascularization (Shapiro and Vallee 1989, 1992; Shapiro et al. 1989). Notably, the artificial increase of angiogenin catalytic activity by the replacement of its 13 residues with a 15-amino acid-long segment derived from RNase A leads to the loss of its angiogenic properties (Harper and Vallee 1989; Holloway et al. 2002), suggesting that the enzymatic capabilities of angiogenin must be kept low to ensure its proper physiological functions.
More recent research focused on the identification of natural angiogenin substrates. Two groups independently reported on the possible involvement of angiogenin in tRNA cleavage induced by various stress stimuli, such as nutrient deprivation, heat shock, UV irradiation, arsenite treatment, and lowered oxygen concentration (Fu et al. 2009; Yamasaki et al. 2009). In this degradation pathway, angiogenin appears to cleave a subset of mature, but not nascent, tRNAs into 5′ and 3′ halves within the anticodon loop. This event was found to occur in multiple tissues and cell lines (Fu et al. 2009), indicating that the function of angiogenin might not be limited to angiogenesis. Indeed, the biological role of 5′ tRNA-derived small RNAs (referred to as “5′ tiRNAs” by Yamasaki et al. [2009]) generated by angiogenin relies on an eIF2α-phosphorylation-independent global inhibition of protein synthesis in stressed cells that might promote cell survival under unfavorable conditions (Yamasaki et al. 2009). This stress response mechanism is likely to be conserved among eukaryotes, as suggested by the tRNA cleavage observed in yeast and ciliates exposed to different stressors (Lee and Collins 2005; Thompson et al. 2008).
APE1 (apurinic/apyrimidinic endonuclease 1) participates in the decay of c-myc mRNA
As mentioned above, APE1 is another recently identified enzyme that displays endoribonucleolytic activity (Bergstrom et al. 2006; Barnes et al. 2009). It was previously characterized as a multifunctional enzyme and the major component of the DNA base-excision repair pathway (for review, see Tell et al. 2009). Detection of the RNase H activity of APE1 suggested that it might also have broader substrate specificity and cleave RNA targets in addition to DNA molecules (Barzilay et al. 1995b). Indeed, proteomic analyses unequivocally demonstrated that the 35-kDa endonuclease associated with polysomes in the rat liver and responsible for c-myc CRD cleavage is in fact APE1. This activity was abolished following immunodepletion with monoclonal APE1 antibodies. Furthermore, both recombinant APE1 and its counterpart purified from the native source generated similar degradation products from the c-myc CRD substrate due to the cleavages in between the C/A and U/A dinucleotides (Barnes et al. 2009). The endonucleolytic RNA degradation mechanism of APE1 was convincingly verified using several independent techniques (Barnes et al. 2009; Kim et al. 2010a).
The in vivo role of APE1 in regulating c-myc transcript turnover was confirmed by its RNAi-induced depletion, which led to an increase in the c-myc mRNA half-life (Barnes et al. 2009). Remarkably, APE1-directed endoribonuclease degradation appears to be executed by the active site common with its other enzymatic activities (Barzilay et al. 1995a,b; Mol et al. 2000; Chou and Cheng 2003; Barnes et al. 2009). Similarly to the second polysome-associated endonuclease targeting c-myc CRD (Prokipcak et al. 1994; Lee et al. 1998), the catalytic function of APE1 appears to be antagonized by the shielding of its recognition site in c-myc mRNA mediated by CRD-BP (Sparanese and Lee 2007). Biochemical studies performed using recombinant human APE1 support the hypothesis that it may control the stability of other transcripts, including not only mRNAs but also oncogenic miRNAs (Kim et al. 2010b). This idea is further substantiated by the apparent involvement of APE1 in the rRNA quality control process (Vascotto et al. 2009). As with many of the other endoribonucleases described above, a detailed characterization of APE1 RNA substrates is mandatory to fully understand its physiological functions.
Endoribonucleolytic events controlled by undefined enzymatic activities
There are known cases of endoribonucleolytic degradation in eukaryotic cells where the responsible enzyme has not yet been discovered, as exemplified by a recent report on the decay of transcripts containing extinct SIDER-2 retrotransposon elements within their 3′-UTR in Leishmania major (Müller et al. 2010). This intriguing RNA metabolic pathway, so far unique to this unicellular protozoan parasite, is strictly dependent on the site-specific endonucleolytic cut within the so-called signature II motif, which is a highly conserved 79-nt cis-acting sequence of SIDER-2 retrotransposons that has been previously shown to promote the instability of Leishmania mRNA (Bringaud et al. 2007). The endonucleolytically initiated mRNA degradation in SIDER-2-mediated mechanisms was inhibited by both the deletion of signature II motifs and by antisense RNAs hybridizing to the mapped cleavage site (Müller et al. 2010). Remarkably, the rapid decay of short-lived transcripts containing SIDER-2 elements is deadenylation independent and thus significantly distinct from the classic RNA degradation pathway, initiated by the progressive trimming of poly(A) tails.
A somewhat similar mechanism of degradation applies to the α-globin transcript in red blood cells. In this case, a specific sequence within the 3′-UTR is targeted by an unidentified erythroid-enriched endonuclease (ErEN) enzyme (Wang and Kiledjian 2000a). The activity of the ErEN was found to be regulated by the cooperative action of PABP and the mRNA stability α-complex that protects the ErEN cleavage site (Wang et al. 1999; Wang and Kiledjian 2000b). Rather than simply protecting the α-globin transcript from deadenylation, PABP increases the affinity of the α-complex toward a cytosine-rich element (CRE) in the 3′-UTR, which results in the inhibition of ErEN-mediated endoribonucleolytic cutting by competition for the overlapping binding site. In this way, deadenylation leads to disruption of the PABP–α-complex interaction, which in turn exposes the site recognized by ErEN, making it susceptible to cleavage (Wang and Kiledjian 2000a,b). It was proposed that the physiological role of ErEN activity is to ensure the production of equal amounts of α- and β-globins for the assembly of functional hemoglobin tetramers containing two copies of each protein. In addition, ErEN activity could be responsible for the rapid elimination of α-globin transcripts in the final stage of erythrocyte differentiation, irrespective of the presence of poly(A) tails with bound PABPs, due to the naturally diminished levels of α-complex subunits during this process (Liu and Kiledjian 2007).
SUMMARY
This review article focused on the participation of recently identified endoribonucleases in various general and context-specific RNA decay, surveillance, and processing pathways. Further research is needed for many of these novel endonucleases in order to understand their precise roles in the context of whole RNA metabolism—and particularly for nuclear RNA decay—more data are required to understand the interplay between endonucleases and exoribonucleases participating in the same pathways. Moreover, the identification of novel endonuclease families over the last few years strongly suggests that the list is far from being complete and that we can expect new discoveries in the near future.
ACKNOWLEDGMENTS
We are grateful to Joanna Kufel for insightful comments on the manuscript. We also thank Kinga Orlowska for several ideas on the design of the figures. This work was supported by a Polish Ministry of Science and Higher Education grant (N N301 250836) to R.T., a project operated within the Foundation for Polish Science Team Programme co-financed by the EU European Regional Development Fund, Operational Program Innovative Economy 2007-2013, and an EMBO installation grant to A.D.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2237610.
REFERENCES
- Acharya KR, Shapiro R, Allen SC, Riordan JF, Vallee BL 1994. Crystal structure of human angiogenin reveals the structural basis for its functional divergence from ribonuclease. Proc Natl Acad Sci 91: 2915–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P 1999. The yeast exosome and human PM–Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev 13: 2148–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N, Sachs MS, Jacobson A 2006. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol 7: 415–425 [DOI] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123: 265–276 [DOI] [PubMed] [Google Scholar]
- Barnes T, Kim WC, Mantha AK, Kim SE, Izumi T, Mitra S, Lee CH 2009. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res 37: 3946–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilay G, Mol CD, Robson CN, Walker LJ, Cunningham RP, Tainer JA, Hickson ID 1995a. Identification of critical active-site residues in the multifunctional human DNA repair enzyme HAP1. Nat Struct Biol 2: 561–568 [DOI] [PubMed] [Google Scholar]
- Barzilay G, Walker LJ, Robson CN, Hickson ID 1995b. Site-directed mutagenesis of the human DNA repair enzyme HAP1: Identification of residues important for AP endonuclease and RNase H activity. Nucleic Acids Res 23: 1544–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beligni MV, Mayfield SP 2008. Arabidopsis thaliana mutants reveal a role for CSP41a and CSP41b, two ribosome-associated endonucleases, in chloroplast ribosomal RNA metabolism. Plant Mol Biol 67: 389–401 [DOI] [PubMed] [Google Scholar]
- Bergstrom K, Urquhart JC, Tafech A, Doyle E, Lee CH 2006. Purification and characterization of a novel mammalian endoribonuclease. J Cell Biochem 98: 519–537 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2: 326–332 [DOI] [PubMed] [Google Scholar]
- Bleichert F, Granneman S, Osheim YN, Beyer AL, Baserga SJ 2006. The PINc domain protein Utp24, a putative nuclease, is required for the early cleavage steps in 18S rRNA maturation. Proc Natl Acad Sci 103: 9464–9469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach TJ, Stern DB 2003. Secondary structures common to chloroplast mRNA 3′-untranslated regions direct cleavage by CSP41, an endoribonuclease belonging to the short chain dehydrogenase/reductase superfamily. J Biol Chem 278: 25832–25838 [DOI] [PubMed] [Google Scholar]
- Bollenbach TJ, Tatman DA, Stern DB 2003. CSP41a, a multifunctional RNA-binding protein, initiates mRNA turnover in tobacco chloroplasts. Plant J 36: 842–852 [DOI] [PubMed] [Google Scholar]
- Bollenbach T, Schuster G, Portnoy V, Stern D 2007. Polyadenylation, processing and degradation of chloroplast RNA. Top Curr Genet 19: 175–211 [Google Scholar]
- Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E 2009. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell 139: 547–559 [DOI] [PubMed] [Google Scholar]
- Bremer KA, Stevens A, Schoenberg DR 2003. An endonuclease activity similar to Xenopus PMR1 catalyzes the degradation of normal and nonsense-containing human β-globin mRNA in erythroid cells. RNA 9: 1157–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringaud F, Müller M, Cerqueira GC, Smith M, Rochette A, El-Sayed NM, Papadopoulou B, Ghedin E 2007. Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog 3: 1291–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock ML, Shapiro DJ 1983. Estrogen stabilizes vitellogenin mRNA against cytoplasmic degradation. Cell 34: 207–214 [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415: 92–96 [DOI] [PubMed] [Google Scholar]
- Cañete-Soler R, Schwartz ML, Hua Y, Schlaepfer WW 1998a. Stability determinants are localized to the 3′-untranslated region and 3′-coding region of the neurofilament light subunit mRNA using a tetracycline-inducible promoter. J Biol Chem 273: 12650–12654 [DOI] [PubMed] [Google Scholar]
- Cañete-Soler R, Schwartz ML, Hua Y, Schlaepfer WW 1998b. Characterization of ribonucleoprotein complexes and their binding sites on the neurofilament light subunit mRNA. J Biol Chem 273: 12655–12661 [DOI] [PubMed] [Google Scholar]
- Cañete-Soler R, Reddy KS, Tolan DR, Zhai J 2005. Aldolases A and C are ribonucleolytic components of a neuronal complex that regulates the stability of the light-neurofilament mRNA. J Neurosci 25: 4353–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis AJ, Luisi BF, McDowall KJ 2009. Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog Mol Biol Transl Sci 85: 91–135 [DOI] [PubMed] [Google Scholar]
- Celesnik H, Deana A, Belasco JG 2007. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol Cell 27: 79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AC, Sohlberg B, Trinkle-Mulcahy L, Claverie-Martin F, Cohen P, Cohen SN 1999. Alternative splicing regulates the production of ARD-1 endoribonuclease and NIPP-1, an inhibitor of protein phosphatase-1, as isoforms encoded by the same gene. Gene 240: 45–55 [DOI] [PubMed] [Google Scholar]
- Chapman RE, Walter P 1997. Translational attenuation mediated by an mRNA intron. Curr Biol 7: 850–859 [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB 2003. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol 23: 4805–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernokalskaya E, Dompenciel R, Schoenberg DR 1997. Cleavage properties of an estrogen-regulated polysomal ribonuclease involved in the destabilization of albumin mRNA. Nucleic Acids Res 25: 735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernokalskaya E, Dubell AN, Cunningham KS, Hanson MN, Dompenciel RE, Schoenberg DR 1998. A polysomal ribonuclease involved in the destabilization of albumin mRNA is a novel member of the peroxidase gene family. RNA 4: 1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou KM, Cheng YC 2003. The exonuclease activity of human apurinic/apyrimidinic endonuclease (APE1). Biochemical properties and inhibition by the natural dinucleotide Gp4G. J Biol Chem 278: 18289–18296 [DOI] [PubMed] [Google Scholar]
- Claverie-Martin F, Wang M, Cohen SN 1997. ARD-1 cDNA from human cells encodes a site-specific single-strand endoribonuclease that functionally resembles Escherichia coli RNase E. J Biol Chem 272: 13823–13828 [DOI] [PubMed] [Google Scholar]
- Clissold PM, Ponting CP 2000. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr Biol 10: R888–R890 [DOI] [PubMed] [Google Scholar]
- Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ 2009. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell 34: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C 2007. Maturation and degradation of RNA in bacteria. Curr Opin Microbiol 10: 271–278 [DOI] [PubMed] [Google Scholar]
- Couttet P, Grange T 2004. Premature termination codons enhance mRNA decapping in human cells. Nucleic Acids Res 32: 488–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Walter P 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87: 391–404 [DOI] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73: 1197–1206 [DOI] [PubMed] [Google Scholar]
- Cunningham KS, Dodson RE, Nagel MA, Shapiro DJ, Schoenberg DR 2000. Vigilin binding selectively inhibits cleavage of the vitellogenin mRNA 3′-untranslated region by the mRNA endonuclease polysomal ribonuclease 1. Proc Natl Acad Sci 97: 12498–12502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham KS, Hanson MN, Schoenberg DR 2001. Polysomal ribonuclease 1 exists in a latent form on polysomes prior to estrogen activation of mRNA decay. Nucleic Acids Res 29: 1156–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Engebrecht J 1998. Yeast dom34 mutants are defective in multiple developmental pathways and exhibit decreased levels of polyribosomes. Genetics 149: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A, Celesnik H, Belasco JG 2008. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature 451: 355–358 [DOI] [PubMed] [Google Scholar]
- Deikus G, Condon C, Bechhofer DH 2008. Role of Bacillus subtilis RNase J1 endonuclease and 5′-exonuclease activities in trp leader RNA turnover. J Biol Chem 283: 17158–17167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson RE, Shapiro DJ 1994. An estrogen-inducible protein binds specifically to a sequence in the 3′ untranslated region of estrogen-stabilized vitellogenin mRNA. Mol Cell Biol 14: 3130–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson RE, Shapiro DJ 1997. Vigilin, a ubiquitous protein with 14 K homology domains, is the estrogen-inducible vitellogenin mRNA 3′-untranslated region-binding protein. J Biol Chem 272: 12249–12252 [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R 2007. RNA quality control in eukaryotes. Cell 131: 660–668 [DOI] [PubMed] [Google Scholar]
- Dominski Z 2007. Nucleases of the metallo-β-lactamase family and their role in DNA and RNA metabolism. Crit Rev Biochem Mol Biol 42: 67–93 [DOI] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF 2007. Formation of the 3′ end of histone mRNA: Getting closer to the end. Gene 396: 373–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Marzluff WF 2005. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell 123: 37–48 [DOI] [PubMed] [Google Scholar]
- Dompenciel RE, Garnepudi VR, Schoenberg DR 1995. Purification and characterization of an estrogen-regulated Xenopus liver polysomal nuclease involved in the selective destabilization of albumin mRNA. J Biol Chem 270: 6108–6118 [DOI] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B 2007. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol 14: 15–22 [DOI] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55 [DOI] [PubMed] [Google Scholar]
- Eide DJ, Clark S, Nair TM, Gehl M, Gribskov M, Guerinot ML, Harper JF 2005. Characterization of the yeast ionome: A genome-wide analysis of nutrient mineral and trace element homeostasis in Saccharomyces cerevisiae. Genome Biol 6: R77 doi: 10.1186/gb-2005-6-9-r77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T 2001. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J 20: 6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elela SA, Igel H, Ares M Jr 1996. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85: 115–124 [DOI] [PubMed] [Google Scholar]
- Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H 2005. Ribonucleases J1 and J2: Two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res 33: 2141–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evguenieva-Hackenberg E, Klug G 2009. RNA degradation in Archaea and Gram-negative bacteria different from Escherichia coli. Prog Mol Biol Transl Sci 85: 275–317 [DOI] [PubMed] [Google Scholar]
- Fatica A, Oeffinger M, Dlakic M, Tollervey D 2003. Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol Cell Biol 23: 1798–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Tollervey D, Dlakic M 2004. PIN domain of Nob1p is required for D-site cleavage in 20S pre-rRNA. RNA 10: 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fett JW, Strydom DJ, Lobb RR, Alderman EM, Bethune JL, Riordan JF, Vallee BL 1985. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry 24: 5480–5486 [DOI] [PubMed] [Google Scholar]
- Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295: 2258–2261 [DOI] [PubMed] [Google Scholar]
- Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X 2009. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 583: 437–442 [DOI] [PubMed] [Google Scholar]
- Gallouzi IE, Parker F, Chebli K, Maurier F, Labourier E, Barlat I, Capony JP, Tocque B, Tazi J 1998. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: A potential link between signal transduction and RNA stability. Mol Cell Biol 18: 3956–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73 [DOI] [PubMed] [Google Scholar]
- Gao X, Xu Z 2008. Mechanisms of action of angiogenin. Acta Biochim Biophys Sin (Shanghai) 40: 619–624 [DOI] [PubMed] [Google Scholar]
- Garas M, Dichtl B, Keller W 2008. The role of the putative 3′ end processing endonuclease Ysh1p in mRNA and snoRNA synthesis. RNA 14: 2671–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E 2004. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429: 575–578 [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15: 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioia U, Laneve P, Dlakic M, Arceci M, Bozzoni I, Caffarelli E 2005. Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. J Biol Chem 280: 18996–19002 [DOI] [PubMed] [Google Scholar]
- Glavan F, Behm-Ansmant I, Izaurralde E, Conti E 2006. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J 25: 5117–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez TN, Sidrauski C, Dörfler S, Walter P 1999. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J 18: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Aguilera C, Tous C, Gomez-Gonzalez B, Huertas P, Luna R, Aguilera A 2008. The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol Biol Cell 19: 4310–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graille M, Chaillet M, van Tilbeurgh H 2008. Structure of yeast Dom34: A protein related to translation termination factor Erf1 and involved in No-Go decay. J Biol Chem 283: 7145–7154 [DOI] [PubMed] [Google Scholar]
- Guo J, Polymenis M 2006. Dcr2 targets Ire1 and downregulates the unfolded protein response in Saccharomyces cerevisiae. EMBO Rep 7: 1124–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Hampton RY 2003. IRE1: A role in UPREgulation of ER degradation. Dev Cell 4: 144–146 [DOI] [PubMed] [Google Scholar]
- Hanson MN, Schoenberg DR 2001. Identification of in vivo mRNA decay intermediates corresponding to sites of in vitro cleavage by polysomal ribonuclease 1. J Biol Chem 276: 12331–12337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell 5: 897–904 [DOI] [PubMed] [Google Scholar]
- Harper JW, Vallee BL 1989. A covalent angiogenin/ribonuclease hybrid with a fourth disulfide bond generated by regional mutagenesis. Biochemistry 28: 1875–1884 [DOI] [PubMed] [Google Scholar]
- Hartmann RK, Gossringer M, Spath B, Fischer S, Marchfelder A 2009. The making of tRNAs and more—RNase P and tRNase Z. Prog Mol Biol Transl Sci 85: 319–368 [DOI] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell 10: 3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS 2006. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313: 104–107 [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS 2009. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186: 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway DE, Shapiro R, Hares MC, Leonidas DD, Acharya KR 2002. Guest-host crosstalk in an angiogenin-RNase A chimeric protein. Biochemistry 41: 10482–10489 [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2009. The many pathways of RNA degradation. Cell 136: 763–776 [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14: 2609–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ 2008. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol 9: 22–32 [DOI] [PubMed] [Google Scholar]
- Iost I, Dreyfus M 2006. DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res 34: 4189–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine K, Stirling R, Hume D, Kennedy D 2004. Rasputin, more promiscuous than ever: A review of G3BP. Int J Dev Biol 48: 1065–1077 [DOI] [PubMed] [Google Scholar]
- Isken O, Maquat LE 2007. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes Dev 21: 1833–1856 [DOI] [PubMed] [Google Scholar]
- Ivanov KA, Hertzig T, Rozanov M, Bayer S, Thiel V, Gorbalenya AE, Ziebuhr J 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci 101: 12694–12699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T, Hosoda A, Okuda T, Kamigori Y, Nomura-Furuwatari C, Kimata Y, Tsuru A, Kohno K 2001. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat Cell Biol 3: 158–164 [DOI] [PubMed] [Google Scholar]
- Jaskiewicz L, Filipowicz W 2008. Role of Dicer in posttranscriptional RNA silencing. Curr Top Microbiol Immunol 320: 77–97 [DOI] [PubMed] [Google Scholar]
- Jinek M, Doudna JA 2009. A three-dimensional view of the molecular machinery of RNA interference. Nature 457: 405–412 [DOI] [PubMed] [Google Scholar]
- Katahira J, Yoneda Y 2009. Roles of the TREX complex in nuclear export of mRNA. RNA Biol 6: 149–152 [DOI] [PubMed] [Google Scholar]
- Kim SE, Gorrell A, Rader SD, Lee CH 2010a. Endoribonuclease activity of human apurinic/apyrimidinic endonuclease 1 revealed by a real-time fluorometric assay. Anal Biochem 398: 69–75 [DOI] [PubMed] [Google Scholar]
- Kim WC, King D, Lee CH 2010b. RNA-cleaving properties of human apurinic/apyrimidinic endonuclease 1 (APE1). Int J Biochem Mol Biol 1: 12–25 [PMC free article] [PubMed] [Google Scholar]
- Kohno K 2010. Stress-sensing mechanisms in the unfolded protein response: Similarities and differences between yeast and mammals. J Biochem 147: 27–33 [DOI] [PubMed] [Google Scholar]
- Koizumi N, Martinez IM, Kimata Y, Kohno K, Sano H, Chrispeels MJ 2001. Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol 127: 949–962 [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Yario TA, Benson E, Steitz JA 2008. Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation. EMBO Rep 9: 1013–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, Stroud RM, Walter P 2009. The unfolded protein response signals through high-order assembly of Ire1. Nature 457: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB 2003. Ena/VASP proteins: Regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol 19: 541–564 [DOI] [PubMed] [Google Scholar]
- Kufel J, Dichtl B, Tollervey D 1999. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5: 909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulichkova VA, Tsimokha AS, Fedorova OA, Moiseeva TN, Bottril A, Lezina L, Gauze LN, Konstantinova IM, Mittenberg AG, Barlev NA 2010. 26S proteasome exhibits endoribonuclease activity controlled by extra-cellular stimuli. Cell Cycle 9: 840–849 [DOI] [PubMed] [Google Scholar]
- Kurachi K, Davie EW, Strydom DJ, Riordan JF, Vallee BL 1985. Sequence of the cDNA and gene for angiogenin, a human angiogenesis factor. Biochemistry 24: 5494–5499 [DOI] [PubMed] [Google Scholar]
- Kushner SR 2004. mRNA decay in prokaryotes and eukaryotes: Different approaches to a similar problem. IUBMB Life 56: 585–594 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski AV, Gertler FB, Loureiro JJ 2003. Function and regulation of Ena/VASP proteins. Trends Cell Biol 13: 386–392 [DOI] [PubMed] [Google Scholar]
- Lamanna AC, Karbstein K 2009. Nob1 binds the single-stranded cleavage site D at the 3′-end of 18S rRNA with its PIN domain. Proc Natl Acad Sci 106: 14259–14264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laneve P, Altieri F, Fiori ME, Scaloni A, Bozzoni I, Caffarelli E 2003. Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron-encoded small nucleolar RNAs in Xenopus laevis. J Biol Chem 278: 13026–13032 [DOI] [PubMed] [Google Scholar]
- Laneve P, Gioia U, Ragno R, Altieri F, Di Franco C, Santini T, Arceci M, Bozzoni I, Caffarelli E 2008. The tumor marker human placental protein 11 is an endoribonuclease. J Biol Chem 283: 34712–34719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRiviere FJ, Cole SE, Ferullo DJ, Moore MJ 2006. A late-acting quality control process for mature eukaryotic rRNAs. Mol Cell 24: 619–626 [DOI] [PubMed] [Google Scholar]
- Lebreton A, Seraphin B 2008. Exosome-mediated quality control: Substrate recruitment and molecular activity. Biochim Biophys Acta 1779: 558–565 [DOI] [PubMed] [Google Scholar]
- Lebreton A, Tomecki R, Dziembowski A, Seraphin B 2008. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456: 993–996 [DOI] [PubMed] [Google Scholar]
- Lee SR, Collins K 2005. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem 280: 42744–42749 [DOI] [PubMed] [Google Scholar]
- Lee CH, Leeds P, Ross J 1998. Purification and characterization of a polysome-associated endoribonuclease that degrades c-myc mRNA in vitro. J Biol Chem 273: 25261–25271 [DOI] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev 16: 452–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Kim YS, Kim KH, Heo I, Kim SK, Kim O, Kim HK, Yoon JY, Kim HS, Kim DJ, et al. 2007. Structural and functional insights into Dom34, a key component of no-go mRNA decay. Mol Cell 27: 938–950 [DOI] [PubMed] [Google Scholar]
- Lee KP, Dey M, Neculai D, Cao C, Dever TE, Sicheri F 2008. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell 132: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Séraphin B 2008. EJCs at the heart of translational control. Cell 133: 213–216 [DOI] [PubMed] [Google Scholar]
- Leuschner PJ, Ameres SL, Kueng S, Martinez J 2006. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep 7: 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima WF, Crooke ST 1999. Highly efficient endonucleolytic cleavage of RNA by a Cys2His2 zinc-finger peptide. Proc Natl Acad Sci 96: 10010–10015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kiledjian M 2007. An erythroid-enriched endoribonuclease (ErEN) involved in α-globin mRNA turnover. Protein Pept Lett 14: 131–136 [DOI] [PubMed] [Google Scholar]
- Liu CY, Schröder M, Kaufman RJ 2000. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem 275: 24881–24885 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD 2006. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127: 1223–1237 [DOI] [PubMed] [Google Scholar]
- Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, Kinch LN, Grishin NV, Liu Q 2009. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science 325: 750–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E 2008. Structure of the active subunit of the yeast exosome core, Rrp44: Diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell 29: 717–728 [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Mitchell P, Petfalski E, Seraphin B, Tollervey D 1994. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev 8: 1423–1433 [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Allmang C, Tollervey D, Seraphin B 1996. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science 272: 268–270 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S, Brodersen DE, Jensen TH 2009. Origins and activities of the eukaryotic exosome. J Cell Sci 122: 1487–1494 [DOI] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108: 781–794 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI, Koonin EV 1999. Comparative genomics of the Archaea (Euryarchaeota): Evolution of conserved protein families, the stable core, and the variable shell. Genome Res 9: 608–628 [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L 2008. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci 65: 1099–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574 [DOI] [PubMed] [Google Scholar]
- Mathy N, Benard L, Pellegrini O, Daou R, Wen T, Condon C 2007. 5′-to-3′ exoribonuclease activity in bacteria: Role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 129: 681–692 [DOI] [PubMed] [Google Scholar]
- Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, et al. 2009. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Meyer S, Temme C, Wahle E 2004. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit Rev Biochem Mol Biol 39: 197–216 [DOI] [PubMed] [Google Scholar]
- Midtgaard SF, Assenholt J, Jonstrup AT, Van LB, Jensen TH, Brodersen DE 2006. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc Natl Acad Sci 103: 11898–11903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittl PR, Deillon C, Sargent D, Liu N, Klauser S, Thomas RM, Gutte B, Grütter MG 2000. The retro-GCN4 leucine zipper sequence forms a stable three-dimensional structure. Proc Natl Acad Sci 97: 2562–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol CD, Izumi T, Mitra S, Tainer JA 2000. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature 403: 451–456 [DOI] [PubMed] [Google Scholar]
- Mori K 2009. Signaling pathways in the unfolded protein response: Development from yeast to mammals. J Biochem 146: 743–750 [DOI] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, Sambrook J 1993. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74: 743–756 [DOI] [PubMed] [Google Scholar]
- Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T 1996. Signaling from endoplasmic reticulum to nucleus: Transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1: 803–817 [DOI] [PubMed] [Google Scholar]
- Mudd EA, Sullivan S, Gisby MF, Mironov A, Kwon CS, Chung WI, Day A 2008. A 125 kDa RNase E/G-like protein is present in plastids and is essential for chloroplast development and autotrophic growth in Arabidopsis. J Exp Bot 59: 2597–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Padmanabhan PK, Rochette A, Mukherjee D, Smith M, Dumas C, Papadopoulou B 2010. Rapid decay of unstable Leishmania mRNAs bearing a conserved retroposon signature 3′-UTR motif is initiated by a site-specific endonucleolytic cleavage without prior deadenylation. Nucleic Acids Res (in press). doi: 10.1093/nar/gkq349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Sugita M 2008. A conserved DYW domain of the pentatricopeptide repeat protein possesses a novel endoribonuclease activity. FEBS Lett 582: 4163–4168 [DOI] [PubMed] [Google Scholar]
- Nechama M, Peng Y, Bell O, Briata P, Gherzi R, Schoenberg DR, Naveh-Many T 2009. KSRP-PMR1-exosome association determines parathyroid hormone mRNA levels and stability in transfected cells. BMC Cell Biol 10: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova DD, Ulferts R, van den Born E, Lauber C, Gorbalenya AE, Ziebuhr J, Snijder EJ 2009. Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus-wide conservation of a replicative endoribonuclease. J Virol 83: 5671–5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O 2010. Nonsense-mediated mRNA decay in human cells: Mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci 67: 677–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen DA, Shapiro DJ 1990. Insights into hormonal control of messenger RNA stability. Mol Endocrinol 4: 953–957 [DOI] [PubMed] [Google Scholar]
- Nikawa J, Akiyoshi M, Hirata S, Fukuda T 1996. Saccharomyces cerevisiae IRE2/HAC1 is involved in IRE1-mediated KAR2 expression. Nucleic Acids Res 24: 4222–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev Y, Deillon C, Hoffmann SR, Bigler L, Friess S, Zenobi R, Pervushin K, Hunziker P, Gutte B 2010. The leucine zipper domains of the transcription factors GCN4 and c-Jun have ribonuclease activity. PLoS ONE 5: e10765 doi: 10.1371/journal.pone.0010765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Patil CK, DeRisi J, Walter P 2005. Genome-scale approaches for discovering novel nonconventional splicing substrates of the Ire1 nuclease. Genome Biol 6: R3 doi: 10.1186/gb-2004-6-1-r3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M, Yang W 2009. Structural and functional modules in RNA interference. Curr Opin Struct Biol 19: 286–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa D, Tokuda M, Iwawaki T 2007. Site-specific cleavage of CD59 mRNA by endoplasmic reticulum-localized ribonuclease, IRE1. Biochem Biophys Res Commun 360: 122–127 [DOI] [PubMed] [Google Scholar]
- Oikawa D, Tokuda M, Hosoda A, Iwawaki T 2010. Identification of a consensus element recognized and cleaved by IRE1α. Nucleic Acids Res (in press). doi: 10.1093/nar/gkq452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Kimata Y, Higashio H, Tsuru A, Kohno K 2000. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem Biophys Res Commun 279: 445–450 [DOI] [PubMed] [Google Scholar]
- Parker F, Maurier F, Delumeau I, Duchesne M, Faucher D, Debussche L, Dugue A, Schweighoffer F, Tocque B 1996. A Ras-GTPase-activating protein SH3-domain-binding protein. Mol Cell Biol 16: 2561–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos DO, Doma MK, Shoemaker CJ, Muhlrad D, Green R, Weissman J, Hollien J, Parker R 2009. Analysis of Dom34 and its function in no-go decay. Mol Biol Cell 20: 3025–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori RL, Schoenberg DR 1993. The nuclease that selectively degrades albumin mRNA in vitro associates with Xenopus liver polysomes through the 80S ribosome complex. Arch Biochem Biophys 305: 313–319 [DOI] [PubMed] [Google Scholar]
- Pastori RL, Moskaitis JE, Buzek SW, Schoenberg DR 1991a. Coordinate estrogen-regulated instability of serum protein-coding messenger RNAs in Xenopus laevis. Mol Endocrinol 5: 461–468 [DOI] [PubMed] [Google Scholar]
- Pastori RL, Moskaitis JE, Schoenberg DR 1991b. Estrogen-induced ribonuclease activity in Xenopus liver. Biochemistry 30: 10490–10498 [DOI] [PubMed] [Google Scholar]
- Peng Y, Schoenberg DR 2007. c-Src activates endonuclease-mediated mRNA decay. Mol Cell 25: 779–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Liu X, Schoenberg DR 2008. The 90-kDa heat shock protein stabilizes the polysomal ribonuclease 1 mRNA endonuclease to degradation by the 26S proteasome. Mol Biol Cell 19: 546–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Murray EL, Sarkar M, Liu X, Schoenberg DR 2009. The cytoskeleton-associated Ena/VASP proteins are unanticipated partners of the PMR1 mRNA endonuclease. RNA 15: 576–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertschy B, Schneider C, Gnadig M, Schafer T, Tollervey D, Hurt E 2009. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem 284: 35079–35091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters L, Meister G 2007. Argonaute proteins: Mediators of RNA silencing. Mol Cell 26: 611–623 [DOI] [PubMed] [Google Scholar]
- Pfalz J, Bayraktar OA, Prikryl J, Barkan A 2009. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plongthongkum N, Kullawong N, Panyim S, Tirasophon W 2007. Ire1 regulated XBP1 mRNA splicing is essential for the unfolded protein response (UPR) in Drosophila melanogaster. Biochem Biophys Res Commun 354: 789–794 [DOI] [PubMed] [Google Scholar]
- Posthuma CC, Nedialkova DD, Zevenhoven-Dobbe JC, Blokhuis JH, Gorbalenya AE, Snijder EJ 2006. Site-directed mutagenesis of the Nidovirus replicative endoribonuclease NendoU exerts pleiotropic effects on the arterivirus life cycle. J Virol 80: 1653–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokipcak RD, Herrick DJ, Ross J 1994. Purification and properties of a protein that binds to the C-terminal coding region of human c-myc mRNA. J Biol Chem 269: 9261–9269 [PubMed] [Google Scholar]
- Raines RT 1998. Ribonuclease A. Chem Rev 98: 1045–1066 [DOI] [PubMed] [Google Scholar]
- Rand TA, Ginalski K, Grishin NV, Wang X 2004. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc Natl Acad Sci 101: 14385–14389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X 2005. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123: 621–629 [DOI] [PubMed] [Google Scholar]
- Redon S, Reichenbach P, Lingner J 2007. Protein RNA and protein protein interactions mediate association of human EST1A/SMG6 with telomerase. Nucleic Acids Res 35: 7011–7022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier P, Hajnsdorf E 2009. Poly(A)-assisted RNA decay and modulators of RNA stability. Prog Mol Biol Transl Sci 85: 137–185 [DOI] [PubMed] [Google Scholar]
- Renzi F, Caffarelli E, Laneve P, Bozzoni I, Brunori M, Vallone B 2006. The structure of the endoribonuclease XendoU: From small nucleolar RNA processing to severe acute respiratory syndrome coronavirus replication. Proc Natl Acad Sci 103: 12365–12370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S, Egloff MP, Ulferts R, Coutard B, Nurizzo D, Campanacci V, Cambillau C, Ziebuhr J, Canard B 2006. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci 103: 11892–11897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Hubbard SR 2008. How IRE1 reacts to ER stress. Cell 132: 24–26 [DOI] [PubMed] [Google Scholar]
- Rother S, Clausing E, Kieser A, Strasser K 2006. Swt1, a novel yeast protein, functions in transcription. J Biol Chem 281: 36518–36525 [DOI] [PubMed] [Google Scholar]
- Rougemaille M, Gudipati RK, Olesen JR, Thomsen R, Seraphin B, Libri D, Jensen TH 2007. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J 26: 2317–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, Boulay J, Jensen TH, Stutz F, Devaux F, et al. 2008a. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell 135: 308–321 [DOI] [PubMed] [Google Scholar]
- Rougemaille M, Villa T, Gudipati RK, Libri D 2008b. mRNA journey to the cytoplasm: Attire required. Biol Cell 100: 327–342 [DOI] [PubMed] [Google Scholar]
- Rüegsegger U, Leber JH, Walter P 2001. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 107: 103–114 [DOI] [PubMed] [Google Scholar]
- Russo N, Shapiro R, Acharya KR, Riordan JF, Vallee BL 1994. Role of glutamine-117 in the ribonucleolytic activity of human angiogenin. Proc Natl Acad Sci 91: 2920–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Calvo O, Manley JL 2004. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 10: 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak SM, Vallee BL 1988. Base cleavage specificity of angiogenin with Saccharomyces cerevisiae and Escherichia coli 5S RNAs. Biochemistry 27: 2288–2294 [DOI] [PubMed] [Google Scholar]
- Saxena SK, Rybak SM, Davey RT Jr, Youle RJ, Ackerman EJ 1992. Angiogenin is a cytotoxic, tRNA-specific ribonuclease in the RNase A superfamily. J Biol Chem 267: 21982–21986 [PubMed] [Google Scholar]
- Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A 2009. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol 16: 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein A, Sheffy-Levin S, Glaser F, Schuster G 2008. The RNase E/G-type endoribonuclease of higher plants is located in the chloroplast and cleaves RNA similarly to the E. coli enzyme. RNA 14: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Leung E, Brown J, Tollervey D 2009. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res 37: 1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster G, Stern D 2009. RNA polyadenylation and decay in mitochondria and chloroplasts. Prog Mol Biol Transl Sci 85: 393–422 [DOI] [PubMed] [Google Scholar]
- Shahbabian K, Jamalli A, Zig L, Putzer H 2009. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J 28: 3523–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu CE, Walter P 1996. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J 15: 3028–3039 [PMC free article] [PubMed] [Google Scholar]
- Shapiro R, Vallee BL 1989. Site-directed mutagenesis of histidine-13 and histidine-114 of human angiogenin. Alanine derivatives inhibit angiogenin-induced angiogenesis. Biochemistry 28: 7401–7408 [DOI] [PubMed] [Google Scholar]
- Shapiro R, Vallee BL 1992. Identification of functional arginines in human angiogenin by site-directed mutagenesis. Biochemistry 31: 12477–12485 [DOI] [PubMed] [Google Scholar]
- Shapiro R, Riordan JF, Vallee BL 1986. Characteristic ribonucleolytic activity of human angiogenin. Biochemistry 25: 3527–3532 [DOI] [PubMed] [Google Scholar]
- Shapiro R, Weremowicz S, Riordan JF, Vallee BL 1987. Ribonucleolytic activity of angiogenin: Essential histidine, lysine, and arginine residues. Proc Natl Acad Sci 84: 8783–8787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R, Fox EA, Riordan JF 1989. Role of lysines in human angiogenin: Chemical modification and site-directed mutagenesis. Biochemistry 28: 1726–1732 [DOI] [PubMed] [Google Scholar]
- Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC 1998. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translational control. Mol Cell Biol 18: 7499–7509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Walter P 1997. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90: 1031–1039 [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Cox JS, Walter P 1996. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87: 405–413 [DOI] [PubMed] [Google Scholar]
- Skruzny M, Schneider C, Racz A, Weng J, Tollervey D, Hurt E 2009. An endoribonuclease functionally linked to perinuclear mRNP quality control associates with the nuclear pore complexes. PLoS Biol 7: e8 doi: 10.1371/journal.pbio.1000008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Soudet J, Gelugne JP, Belhabich-Baumas K, Caizergues-Ferrer M, Mougin A 2010. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J 29: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparanese D, Lee CH 2007. CRD-BP shields c-myc and MDR-1 RNA from endonucleolytic attack by a mammalian endoribonuclease. Nucleic Acids Res 35: 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Bronkhorst AW, Schilders G, Slomovic S, Schuster G, Heck AJ, Raijmakers R, Pruijn GJ 2010. Dis3-like 1: A novel exoribonuclease associated with the human exosome. EMBO J 29: 2358–2367 doi: 10.1038/emboj.2010.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanizzi I, Cañete-Soler R 2007. Coregulation of light neurofilament mRNA by poly(A)-binding protein and aldolase C: Implications for neurodegeneration. Brain Res 1139: 15–28 [DOI] [PubMed] [Google Scholar]
- Stevens A, Wang Y, Bremer K, Zhang J, Hoepfner R, Antoniou M, Schoenberg DR, Maquat LE 2002. β-Globin mRNA decay in erythroid cells: UG site-preferred endonucleolytic cleavage that is augmented by a premature termination codon. Proc Natl Acad Sci 99: 12741–12746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson AL, Norbury CJ 2006. The Cid1 family of non-canonical poly(A) polymerases. Yeast 23: 991–1000 [DOI] [PubMed] [Google Scholar]
- Strydom DJ, Fett JW, Lobb RR, Alderman EM, Bethune JL, Riordan JF, Vallee BL 1985. Amino acid sequence of human tumor derived angiogenin. Biochemistry 24: 5486–5494 [DOI] [PubMed] [Google Scholar]
- Tell G, Quadrifoglio F, Tiribelli C, Kelley MR 2009. The many functions of APE1/Ref-1: Not only a DNA repair enzyme. Antioxid Redox Signal 11: 601–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E, Tollervey D 2010. The final step in 5.8S rRNA processing is cytoplasmic in Saccharomyces cerevisiae. Mol Cell Biol 30: 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Lu C, Green PJ, Parker R 2008. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14: 2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ 1998. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev 12: 1812–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ 2000. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev 14: 2725–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolia NH, Joshua-Tor L 2007. Slicer and the argonautes. Nat Chem Biol 3: 36–43 [DOI] [PubMed] [Google Scholar]
- Tollervey D 2006. Molecular biology: RNA lost in translation. Nature 440: 425–426 [DOI] [PubMed] [Google Scholar]
- Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. 2010. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J 29: 2342–2357 doi: 10.1038/emboj.2010.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H, Gallouzi IE, Chebli K, Capony JP, Mouaikel J, van der Geer P, Tazi J 2001. RasGAP-associated endoribonuclease G3BP: Selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol 21: 7747–7760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160: 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Trotter JR, Ernst NL, Carnes J, Panicucci B, Stuart K 2005. A deletion site editing endonuclease in Trypanosoma brucei. Mol Cell 20: 403–412 [DOI] [PubMed] [Google Scholar]
- Udem SA, Warner JR 1973. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J Biol Chem 248: 1412–1416 [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16: 587–596 [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D 2000. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287: 664–666 [DOI] [PubMed] [Google Scholar]
- Valkonen M, Penttilä M, Saloheimo M 2004. The ire1 and ptc2 genes involved in the unfolded protein response pathway in the filamentous fungus Trichoderma reesei. Mol Genet Genomics 272: 443–451 [DOI] [PubMed] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC, Parker R 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264 [DOI] [PubMed] [Google Scholar]
- Vascotto C, Fantini D, Romanello M, Cesaratto L, Deganuto M, Leonardi A, Radicella JP, Kelley MR, D'Ambrosio C, Scaloni A, et al. 2009. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol Cell Biol 29: 1834–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra P, Iglesias N, Camblong J, Zenklusen D, Stutz F 2005. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J 24: 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D, Kaiser D 1999. Type IV pili and cell motility. Mol Microbiol 32: 1–10 [DOI] [PubMed] [Google Scholar]
- Wang M, Cohen SN 1994. ard-1: A human gene that reverses the effects of temperature-sensitive and deletion mutations in the Escherichia coli rne gene and encodes an activity producing RNase E-like cleavages. Proc Natl Acad Sci 91: 10591–10595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M 2000a. Identification of an erythroid-enriched endoribonuclease activity involved in specific mRNA cleavage. EMBO J 19: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M 2000b. The poly(A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol Cell Biol 20: 6334–6341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D 1998. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J 17: 5708–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Day N, Trifillis P, Kiledjian M 1999. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol Cell Biol 19: 4552–4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM 2005. E Pluribus Unum: 3′ end formation of polyadenylated mRNAs, histone mRNAs, and U snRNAs. Mol Cell 20: 168–170 [DOI] [PubMed] [Google Scholar]
- Welihinda AA, Tirasophon W, Green SR, Kaufman RJ 1998. Protein serine/threonine phosphatase Ptc2p negatively regulates the unfolded-protein response by dephosphorylating Ire1p kinase. Mol Cell Biol 18: 1967–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennborg A, Sohlberg B, Angerer D, Klein G, von Gabain A 1995. A human RNase E-like activity that cleaves RNA sequences involved in mRNA stability control. Proc Natl Acad Sci 92: 7322–7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Beligni MV, Prieto S, Haynes PA, McDonald WH, Yates JR III, Mayfield SP 2003. Proteomic characterization of the Chlamydomonas reinhardtii chloroplast ribosome. Identification of proteins unique to the 70 S ribosome. J Biol Chem 278: 33774–33785 [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Stern DB 1997. The spinach chloroplast endoribonuclease CSP41 cleaves the 3′-untranslated region of petD mRNA primarily within its terminal stem-loop structure. J Biol Chem 272: 12874–12880 [DOI] [PubMed] [Google Scholar]
- Yang F, Schoenberg DR 2004. Endonuclease-mediated mRNA decay involves the selective targeting of PMR1 to polyribosome-bound substrate mRNA. Mol Cell 14: 435–445 [DOI] [PubMed] [Google Scholar]
- Yang J, Schuster G, Stern DB 1996. CSP41, a sequence-specific chloroplast mRNA binding protein, is an endoribonuclease. Plant Cell 8: 1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Peng Y, Schoenberg DR 2004. Endonuclease-mediated mRNA decay requires tyrosine phosphorylation of polysomal ribonuclease 1 (PMR1) for the targeting and degradation of polyribosome-bound substrate mRNA. J Biol Chem 279: 48993–49002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Peng Y, Murray EL, Otsuka Y, Kedersha N, Schoenberg DR 2006. Polysome-bound endonuclease PMR1 is targeted to stress granules via stress-specific binding to TIA-1. Mol Cell Biol 26: 8803–8813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Sullivan KD, Marzluff WF, Dominski Z 2009. Studies of the 5′ exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol Cell Biol 29: 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Sharp JS, Bechhofer DH 2009. Bacillus subtilis RNase J1 endonuclease and 5′ exonuclease activities in the turnover of ΔermC mRNA. RNA 15: 2331–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K 2000. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol 20: 6755–6767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107: 881–891 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K 2003. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell 4: 265–271 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]
- Zekri L, Chebli K, Tourrière H, Nielsen FC, Hansen TV, Rami A, Tazi J 2005. Control of fetal growth and neonatal survival by the RasGAP-associated endoribonuclease G3BP. Mol Cell Biol 25: 8703–8716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]