Abstract
The majority of mammalian pre-mRNAs contains multiple introns that are excised prior to export and translation. After intron excision, ligated exon intermediates participate in subsequent intron excisions. However, exon ligation generates an exon of increased size, a feature of pre-mRNA splicing that can interfere with downstream splicing events. These considerations raise the question of whether unique mechanisms exist that permit efficient removal of introns neighboring ligated exons. Kinetic analyses of multiple intron-containing pre-mRNAs revealed that splicing is more efficient following an initial intron removal event, suggesting that either the recruitment of the exon junction complex (EJC) to ligated exons increases the efficiency of multiple intron excisions or that the initial definition of splice sites is sufficient to permit efficient splicing of introns neighboring ligated exons. Knockdown experiments show that the deposition of the EJC does not affect subsequent splicing kinetics. Instead, spliceosomal components that are not involved in the initial splicing event remain associated with the pre-mRNA to ensure efficient removal of neighboring introns. Thus, ligated exons do not require redefinition, providing an additional kinetic advantage for exon defined splice sites.
Keywords: alternative splicing, exon definition, U1 snRNP, kinetics, exon junction complex
INTRODUCTION
Pre-mRNA splicing is an essential process in eukaryotic gene expression (Burge et al. 1999). Intronic sequences must be excised and coding exonic sequences are joined in the production of mRNA (Wahl et al. 2009). Inaccuracies in this process have the potential to cause disruptions in cellular functions. However, variation in the process is necessary for increasing proteomic diversity (Maniatis and Tasic 2002). Regulation of pre-mRNA splicing can occur at numerous steps and relies on the coordination of multiple factors. Introns are removed through a series of interactions coordinated by the spliceosome, a large ribonucleoprotein complex. Several protein and RNA particles (small nuclear ribonucleoproteins [snRNPs]), recognize and bind to the 5′ and 3′ splice sites of the intron that is to be removed. Stable interactions of individual snRNPs with the pre-mRNA occur in a stepwise manner and result in conformational changes of the spliceosome, ultimately leading to the excision of the intron and the ligation of exons (Wahl et al. 2009).
Throughout the course of splicing, various proteins associate with the pre-mRNA, changing the RNP composition as the spliceosome transitions from early splice site recognition to intron removal. The complex of proteins formed as a result of intron removal is referred to as the exon junction complex (EJC), a multiprotein complex deposited 20–24 nucleotides (nt) upstream of the exon junction of mRNA (Le Hir et al. 2000). The EJC is noted for its role in directing nonsense-mediated decay (NMD), export, and localization of mRNAs (Le Hir et al. 2001; Tange et al. 2005; Chang et al. 2007), and it contains a stable core (MLN51, Y14, Magoh, and eIF4AIII), as well as several transiently interacting factors (Shibuya et al. 2004; Ballut et al. 2005; Tange et al. 2005). Acting as an anchoring protein, eIF4AIII is at the center of the core and appears to be the only protein contacting the mRNA (Shibuya et al. 2004; Le Hir and Andersen 2008). Direct interactions with MLN51 occur to stabilize mRNA binding, while Y14 and Magoh binding ensures that the core remains locked onto the mRNA (Ballut et al. 2005). The numerous other proteins that interact with the core affect the path of the mRNA through their interactions with various cellular machineries (Shibuya et al. 2004). Interestingly, three of these EJC associated proteins (Pinin, RNPS1, and SRm160) are better known for their involvement in pre-mRNA splicing (Blencowe et al. 1998; Li et al. 2003; Sakashita et al. 2004), but their functions as components of the EJC are not well understood.
The combination of exon and intron sizes of a gene determines how splice sites are recognized (Hertel 2008). Exon definition, which is referred to as splice site recognition across an exon, occurs when exons are small (Berget 1995). Splice sites flanking small introns are recognized through intron definition, which occurs across the intron. The average vertebrate exon size is 137 nt and is separated by long introns (Stamm et al. 2006), which favors recognition through exon definition. Recognition of splice sites is favored when the distance between splice sites is short. As this distance increases, the spliceosome becomes less efficient at recognizing splice sites (Fox-Walsh et al. 2005). For example, exons larger than 300 nt can induce cryptic splicing or exon skipping (Sterner et al. 1996). Constitutively spliced internal exons have an optimal size range of 60–200 nt; if the size falls outside of this range an increase in alternative splicing is induced (P Shepard and KJ Hertel, unpubl.). Recognition of splice sites across the intron also has an optimal range and becomes less efficient when intron length is greater than ∼250 nt (Fox-Walsh et al. 2005). Thus, the exon/intron architecture significantly influences the efficiency of splicing and splice site selection.
Typical vertebrate genes are composed of several small exons separated by large introns, indicating that exon definition is the most common form of splice-site recognition (Stamm et al. 2006). As splicing progresses and multiple introns are removed, ligated exons become increasingly larger. As a consequence, splicing intermediates formed during the processing of the pre-mRNA contain large ligated exons in addition to large introns, creating scenarios where splice sites are separated by large distances regardless of the mode of recognition. However, the neighboring introns of these splicing intermediates must still be excised to complete production of the mRNA. It was previously shown that large exons separated by large introns are inefficiently recognized by the spliceosome (Sterner et al. 1996). Given these observations, it is unclear how the splicing machinery maintains efficiency during multiple intron removal events.
Because multiple intron-containing genes are common in vertebrates, mechanisms must be in place to assist the spliceosome in recognizing and assembling on splice sites of splicing intermediates that are separated by large distances. Here, we evaluated potential mechanisms that promote efficient splicing of multiple intron-containing pre-mRNAs. Our results do not support the hypothesis that EJC-associated splicing factors assist spliceosomal recognition of ligated exons. However, our results show that spliceosomal components not involved in an initial splicing event remain associated with the pre-mRNA, significantly contributing to efficient neighboring intron removal. Thus, ligated exons do not require redefinition, providing an additional kinetic advantage for exon defined splice sites.
RESULTS
Efficient upstream intron removal accelerates subsequent intron removal events
Pre-mRNAs containing multiple exons undergo sequential intron removal events during the course of splicing. Larger exons are formed as intervening introns are removed and remaining exons are ligated. Formation of increasingly larger exons could pose a problem for the spliceosome, which efficiently recognizes exons under 250 nt in length. During the process of exon ligation, new exons are continually formed that exceed this length limitation. Therefore, we anticipate that mechanisms exist to assist the spliceosome in the removal of introns flanking large ligated exons.
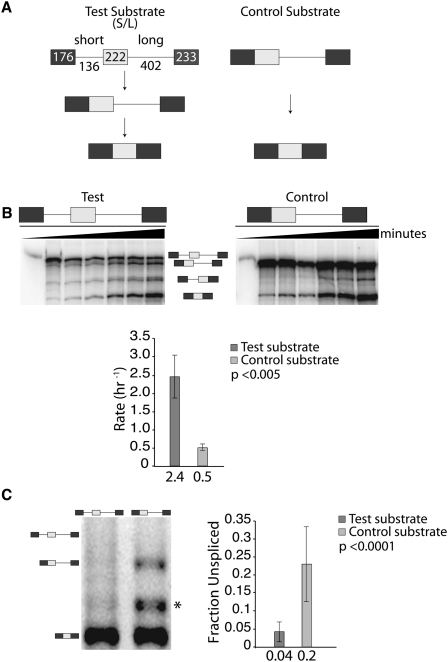
A pair of splicing substrates was created to test intron removal efficiency (Fig. 1A). The test substrate contains an upstream intron that is removed quickly and a downstream intron that is removed slowly. These design features promote the production of a splicing intermediate that contains a long ligated exon. The control substrate contains only the downstream intron and is identical in sequence to the test splicing intermediate. The efficiency of long intron removal can then be compared between substrates—the test having undergone an initial intron removal and the control, which has not—to evaluate the influence a neighboring splicing event has on downstream intron removal. In vitro splicing assays were performed over a 3-h time course to determine the rate of intron removal for the test and control substrates (Fig. 1B). Although the test substrate was designed to undergo an initial short intron removal event, the intermediate for the alternative splicing pathway, initial removal of the long intron, was also observed, albeit at lower frequency. Through monitoring the levels of splicing intermediates it was possible to specifically determine the rate of long intron removal for pre-mRNAs having undergone the short intron removal event first. Analysis of the splicing reactions revealed a fivefold increase in the rate of intron removal in test substrates that underwent a previous intron removal, compared with control substrates (Fig. 1B). Although alternative splicing pathways were observed, such as exon skipping, they were minor and did not significantly affect the calculated rate.
FIGURE 1.
An initial upstream intron removal event increases the efficiency of a downstream intron removal event. (A) Substrates used for analyzing splicing efficiencies. The test substrate (S/L) contains an optimized short upstream intron that is removed quickly and a second, larger intron with delayed removal kinetics. This creates a splicing intermediate containing a long upstream exon and a long intron that is identical in sequence to the control substrate. (B) In vitro analysis showing a 3-h splicing time course. As input control, ∼5% of the starting reaction was loaded on the left lane of each panel. The graph displays the results as the average rate of intron removal based on three biological replicates. The identity of bands detected is indicated between the representative gels. (C) Splicing efficiencies in cell transfection experiments. Representative gel analysis of RT-PCR reactions (left) and combined quantification (right) are shown. The identity of bands detected is indicated on the left side of the representative gel. An asterisk (*) denotes a nonspecific RT-PCR product. The graph represents the average fraction unspliced based on 21 biological replicates. The statistical significance of the differences observed in B and C is defined by the P-value next to each graph.
Splicing efficiency was also analyzed in cell transfection experiments by determining the steady-state levels of spliced products. HeLa cells were transiently transfected with plasmids expressing either the test or control substrates and RNA was isolated from cells 24 h later. Steady-state analysis of mRNA by semiquantitative reverse-transcription polymerase chain reaction (RT-PCR) revealed that test substrates that underwent an initial intron removal were able to undergo removal of subsequent introns five times more efficiently than control substrates that never underwent a prior intron removal (Fig. 1C), consistent with our in vitro analysis. One limitation of steady-state mRNA analyses is the fact that the levels of spliced and unspliced RNA can be influenced by differential stabilities. However, it is also appreciated that pre-mRNAs associated with spliceosomal components are more stable than unassociated splicing substrates (Hicks et al. 2005, 2006), arguing that the test substrate intermediate is more stable than its control counterpart. These considerations suggest that our measurement is a conservative estimate of the steady-state differences. We conclude that pre-mRNAs that underwent a prior upstream intron removal are spliced more efficiently than substrates that did not.
A neighboring downstream intron removal event accelerates upstream intron removal
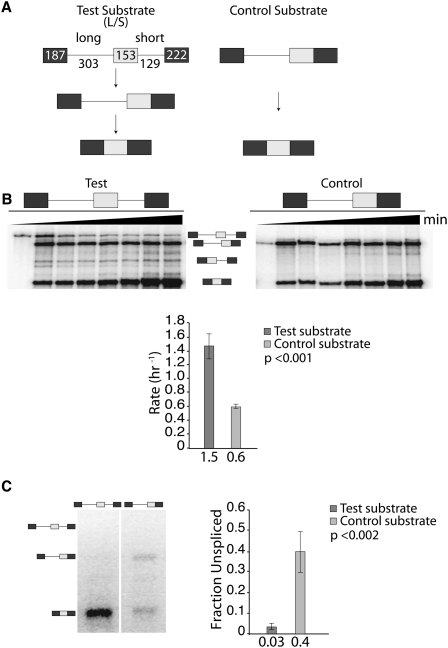
To test the efficiency of an initial downstream intron removal, another pair of splicing substrates was designed (Fig. 2A), similar to that described in Figure 1A, except that a short downstream intron is removed quickly from the test substrate. In vitro splicing reactions using these substrates show that short intron removal is the main splicing pathway for the L/S test substrate, with 74% of the pre-mRNA entering this pathway (Fig. 2B). Kinetic analyses measuring intermediate levels throughout the time course revealed an ∼2.5-fold rate increase in spliced product formation for the test substrate compared with the control (Fig. 2B). For the complementing cell transfection experiments a 13-fold increase in splicing efficiency was observed when the pre-mRNA underwent an initial downstream intron removal event (Fig. 2C). These results demonstrate that the initial removal of an intron increases the efficiency of neighboring intron removal events, regardless of its location within the pre-mRNA. We conclude that mechanisms exist to maintain efficient processing of splicing intermediates.
FIGURE 2.
An initial downstream intron removal increases the efficiency of an upstream intron removal. (A) Substrates used for analyzing splicing efficiencies. The test substrate (L/S) contains an optimized short downstream intron that is removed quickly and a second, larger intron. This creates a splicing intermediate containing a long downstream exon and a long intron that is identical in sequence to the control substrate. (B) In vitro analysis showing a 3-h splicing time course. Approximately 5% of the starting reaction was loaded on the left lane of each panel. The graph displays the results as the average rate of intron removal based on three biological replicates. The identity of bands detected is indicated between the representative gels. (C) Splicing efficiencies in cell transfection experiments. Representative gel analysis of RT-PCR reactions (left) and combined quantification (right) based on three biological replicates are shown. The identity of bands detected is indicated on the left side of the representative gel. The statistical significance of the differences observed in B and C is defined by the P-value next to each graph.
The Influence of the EJC on intron removal
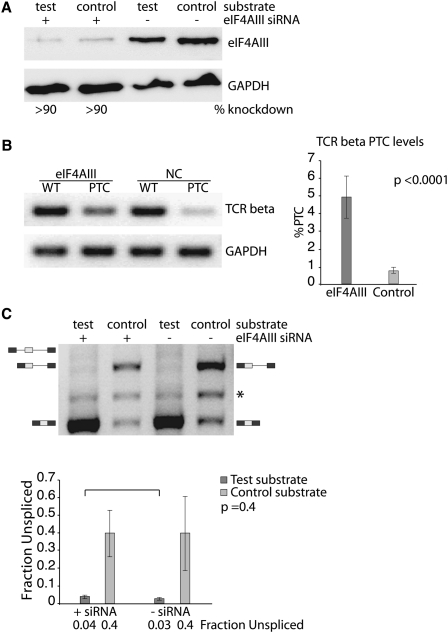
During the course of splicing, the EJC is deposited upstream of exon/exon junctions (Le Hir et al. 2001). It is possible that components of the EJC assist in keeping splicing efficiency high after exon ligation. To determine if our initial observations of multiple intron splicing result from EJC deposition after the upstream intron is removed, we disrupted EJC formation. This was accomplished using siRNA targeting the EJC core component eIF4AIII (Ferraiuolo et al. 2004). This protein is thought to serve as an anchor between the EJC and the RNA. Thus, knockdown of this protein is expected to prevent the remaining EJC components from assembling into a functional complex on the RNA. HeLa cells were transfected with siRNA targeting eIF4AIII, resulting in a reduction greater than 90% in eIF4AIII protein levels (Fig. 3A).
FIGURE 3.
The EJC does not contribute to increased splicing efficiency. (A) Western blot demonstrating eIF4AIII knockdown in HeLa cells. (B) RT-PCR analysis of TCR beta mRNA expression measuring NMD activity. TCR beta mRNA products are compared with GAPDH levels for each sample. The graph on the right shows the quantification of eight independent real-time PCR experiments. The statistical significance of the differences observed is defined by the P-value next to the graph. (C) Splicing efficiencies of S/L test substrates in cell transfection experiments with eIF4AIII protein knockdown. A representative gel shows the results of RT-PCR amplified spliced products (top). The identity of bands detected is indicated on the sides of the representative gel. The asterisk (*) denotes a nonspecific PCR amplification product. The results from three independent experiments are displayed in the graph below as the average fraction unspliced. The statistical significance of the differences observed between siRNA treated and control treated experiments is defined by the P-value next to the graph.
A functional PCR assay using the T-cell receptor beta (TCR beta) minigene (Mühlemann et al. 2001) was used to assess the activity of EJC-sensitive NMD upon eIF4AIII knockdown. The TCR beta reporter gene contains a premature termination codon (PTC) upstream of its last exon/exon junction, which leads to degradation of its mRNA when EJC-enhanced NMD is active. However, when EJC formation or NMD is inhibited, PTC-containing TCR beta mRNAs escape NMD degradation and remain stable. Expression of TCR beta mRNA was used to evaluate the functional effectiveness of eIF4AIII knockdown on EJC assembly based on the expectation that PTC-containing transcripts will be stabilized upon EJC disruption through the inhibition of NMD enhancement. Upon eIF4AIII knockdown we observed a sevenfold increase in the expression of PTC-containing TCR beta transcripts, demonstrating a substantial defect in NMD (Fig. 3B). These results support the expectation that eIF4AIII knockdown is preventing proper EJC assembly on spliced mRNAs.
We next examined the effect of improper EJC assembly on splicing efficiency. The test and control substrates that undergo an initial upstream intron removal event (Fig. 1A) were cotransfected with eIF4AIII siRNA. Importantly, knockdown of eIF4AIII did not result in a significant shift in the steady-state levels of unspliced pre-mRNA of the test and the control substrates (Fig. 3C). We conclude that eIF4AIII knockdown does not affect splicing efficiencies of multiple intron-containing genes, thus, strongly arguing against the notion that EJC components positively influence intron removal.
Spliceosomal components remain associated with splicing intermediates following intron removal
An alternative explanation for the observation that a previous splicing event influences a subsequent splicing event is that spliceosomal components associated with the pre-mRNA during intron removal remain associated with the pre-mRNA. Following intron removal, it is unclear what happens to exon definition components that were associated with the pre-mRNA, but that were not directly involved in the removal of an intron. One possibility is that while an intron is removed, the components initially associated with the pre-mRNA dissociate, leaving the newly ligated exon without any spliceosomal components associated. In such a scenario, a ligated exon would require redefinition to remove neighboring introns. Alternatively, exon definition components remain associated with the pre-mRNA following initial intron removal and are able to participate in subsequent splicing events.
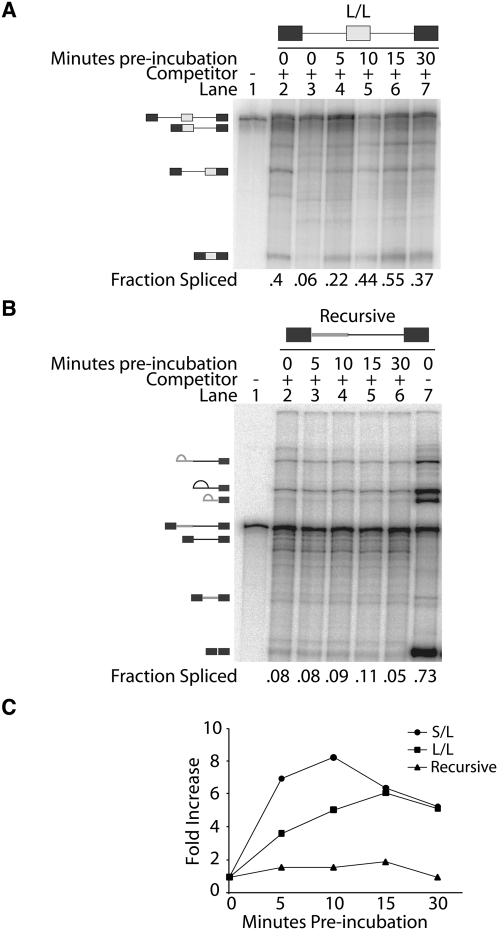
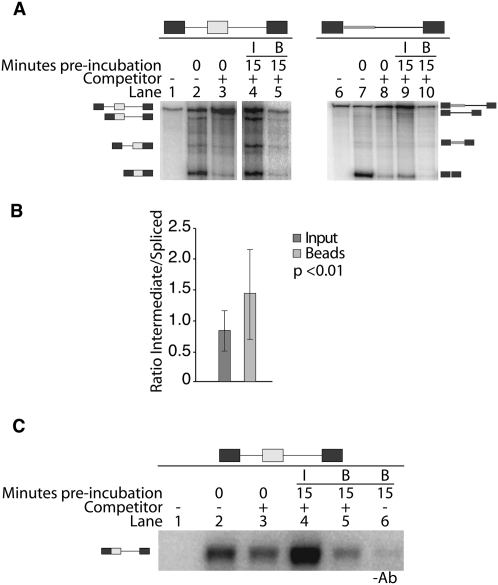
To determine if exon definition components remain associated with the RNA following an initial splicing event, we performed pulse–chase experiments using three-exon splicing substrates (Supplemental Fig. 1). These experiments employed an initial omission of ATP to stall the pre-mRNA in an early exon defined spliceosomal complex (E complex) and to allow the association of U1 and U2 snRNPs with the pre-mRNA. Higher complexes are unable to form in the absence of ATP and therefore splicing does not progress past the E complex (Das et al. 2000; Kotlajich et al. 2009). Chasing these reactions with ATP and unlabeled competitor RNA permits splicing only on splice sites that have already been defined. New spliceosomes are unable to form on the labeled pre-mRNA, instead forming on the excess unlabeled RNA (Lim and Hertel 2004). Thus, a spliced product results only from components already assembled on the pre-mRNA at the time the chase was initiated. This experimental design tests if spliceosomal components remain associated with the RNA following a prior intron removal. An accumulation of splicing intermediates with the first intron removed would suggest that exon definition complexes dissociate from the pre-mRNA while or after the first intron has been removed. The formation of fully spliced products would indicate that exon definition components that are not directly involved in the initial intron removal event remain associated with the pre-mRNA intermediate and are able to participate in subsequent splicing. The results of the pulse–chase experiments clearly show that a fully spliced product is abundantly generated and that its amount increases when the preincubation period is increased (Fig. 4A). Identical outcomes were obtained when analyzing similar pre-mRNA substrates that differ in the length of the introns flanking the internal exon (Fig. 4C; Supplemental Fig. 2). These observations support the model that spliceosomal components initially recruited to the internal exon remain associated with the RNA during and following an initial intron removal.
FIGURE 4.
Splice sites do not require redefinition following an upstream intron removal event. (A) In vitro pulse–chase assay for a substrate that contains two long introns (L/L). The identity of bands detected is indicated on the left side of the representative gel. Approximately 5% of the starting reaction was loaded on the left lane. The reactions were preincubated for the times shown at the top of each lane, followed by a 1.5-h chase incubation. The fraction spliced for each lane is indicated at the bottom of the gel. (B) In vitro pulse–chase assay for the recursive substrate. The identity of bands detected is indicated on the left side of the representative gel. Approximately 5% of the starting reaction was loaded on the left lane. The reactions were preincubated for the times shown at the top of each lane, followed by a 1.5-h chase incubation. The fraction spliced is indicated at the bottom of the gel. (C) Quantification of pulse–chase experiments. The results obtained from at least three independent experiments using two different test substrates (L/L and S/L) and the recursive substrates are shown as fold increase over background levels of spliced product.
To validate these results we created a recursive splicing construct to demonstrate that splice sites that require redefinition are not competent splicing substrates in the presence of competitor RNA. At normal conditions the recursive substrate undergoes two sequential intron removal events, generating an intermediate with a newly formed 5′ splice site after the first intron has been removed (Supplemental Fig. 3). The use of such recursive splicing substrates tests whether splice sites can be redefined in our pulse–chase experiment, with the expectation that the recursive substrate is unable to complete splicing in this assay. As anticipated from the design of the pulse–chase approach, the recursive splicing substrate showed no increase in spliced product formation in the presence of competitor RNA (Fig. 4B,C). These results confirm that the pulse–chase assay is sufficient in preventing the redefinition of splice sites. We conclude that ligated exons do not require redefinition after an initial splicing event. Rather, components of the internal exon definition complex that did not directly participate in initial splicing remain associated with splicing intermediates.
To confirm that spliceosomal components are retained during and following an intron removal event, we performed a U1 70K immunoprecipitation on pulse–chase splicing reactions to pull down RNA species bound to U1 snRNP. As expected, we detect both the pre-mRNA and the splicing intermediate RNA in the immunoprecipitation (Fig. 5A). The presence of the spliced product is a result of a cryptic U1 binding site located in the 3′ terminal exon of the substrate. The presence of the spliced product served as an internal control and was used to determine if the splicing intermediate was enriched in this pulse–chase immunoprecipitation assay. We observed a statistically significant 1.7-fold enrichment (P < 0.01) of the intermediate associated with immunoprecipitated beads (Fig. 5B). U1 70K immunoprecipitation in the presence of the recursive substrate served as a negative control and, as predicted, did not result in measurable pull-down of either the intermediate or the fully spliced product (Fig. 5A).
FIGURE 5.
U1 snRNP is retained on the RNA following an intron removal. (A) Immunoprecipitation of S/L (left) and recursive splicing (right) RNAs during the pulse–chase assay. Splicing reactions were preincubated for 15 min, followed by a 1.5-h chase incubation and subsequent U1 70K immunoprecipitation. Approximately 5% of the starting reaction was loaded as size control (lanes 1,6), and the outcome of splicing efficiencies in the absence (lanes 2,7) or presence (lanes 3,8) of competitor RNA is shown. Input (I) contains 20% of a splicing reaction (lanes 4,9), whereas bead-associated RNAs (B) are derived from a full splicing reaction (lanes 5,10). All lanes shown are from the same gel. (B) Quantification of S/L pulse–chase immunoprecipitation experiments. Results are displayed as the ratio of splicing intermediate to spliced product and were calculated based on eight independent experiments. (C) RT-PCR amplification of immunoprecipitation RNA products. The immunoprecipitated intermediate product was amplified using an exon junction primer paired with an intron primer.
Semiquantitative RT-PCR using an exon junction primer paired with an intron primer specifically amplified the intermediate following immunoprecipitation (Fig. 5C, lane 5). A control immunoprecipitation reaction without a U1 70K antibody resulted in barely detectable levels of intermediate (Fig. 5C, lane 6). Using this assay, we observed a statistically significant threefold increase (P < 0.001) in the signal intensities between the immunoprecipitated intermediate (Fig. 5C, lane 5) and the control immunoprecipitation (Fig. 5C, lane 6), demonstrating low signal contribution from nonspecific binding. The reproducible detection and enrichment of the splicing intermediate in these pulse–chase immunoprecipitation experiments show that U1 snRNP remains associated to the splicing intermediate following the initial intron removal event. In combination with our kinetic analysis, the physical enrichment of U1 snRNP demonstrates that spliceosomal components not engaged in cross-intron pairing remain associated with the pre-mRNA following an initial intron removal event and are competent to participate in subsequent splicing events.
DISCUSSION
The exon/intron architecture of mammalian genes suggests that splice site recognition of multi-intron-containing genes occurs predominantly through exon definition complexes. Due to different kinetic constraints, the complete processing of such genes is expected to proceed through various splicing intermediates containing ligated exons. Given previously documented exon size limitations (Sterner et al. 1996), further processing of splicing intermediates may be restricted by the new dimensions of ligated exons. The results presented here show that the splicing machinery has evolved mechanisms to maintain efficient processing of all introns within a pre-mRNA. Our experiments demonstrated that a neighboring intron removal event increases the efficiency of subsequent splicing. These observations suggest that the order of intron removal may have profound effects on alternative splicing outcomes. Indeed, intron excision is a controlled process following a preferred order that is required for accurate splicing (Lang and Spritz 1987; Gudas et al. 1990; Nasim et al. 1990; Kessler et al. 1993). In the case of human β-globin, rat a-lactalbumin, human preprotachykinin, and murine interleukin-3, highly favored splicing pathways exist that do not follow a strict 5′-to-3′ progression. For example, studies on the processing of human preprotachykinin pre-mRNA showed that a newly formed exon–exon structure is responsible for activating the splicing of the upstream intron, because this prior splicing event repositions and brings into proximity a strong 5′ splice site (Nasim et al. 1990). Importantly, mutations have been shown to disrupt the usual order of intron removal, triggering alternative splicing patterns (Takahara et al. 2002). Thus, changes in the sequence of intron removal affect the outcome of the mRNAs produced, highlighting the importance of maintaining the proper order of intron removal.
Components of the exon definition complex are retained on splicing intermediates
We considered two models to explain our observation that an initial intron removal event increased the efficiency of subsequent intron excisions. EJC formation could cause the rate enhancement, because it is deposited during the splicing reaction onto every ligated exon junction and because it associates with several splicing related factors. The EJC could promote splicing efficiency by retaining splicing factors at sites where they are needed for further intron removal events, thus acting as a splicing enhancer to increase the rate of splicing. However, our knockdown experiments show that the EJC does not play a significant role in maintaining efficient removal of multiple introns. When the master assembler of the EJC, eIF4AIII, was reduced, splicing efficiency was unaltered, even though eIF4AIII reduction resulted in a loss of EJC deposition, as monitored through NMD reporter assays.
An alternative explanation we considered to explain our observations was that spliceosomal components initially recruited to the splice sites of internal exons are retained during intron removal. Retention of previously deposited spliceosomal components would relieve the requirement of newly ligated exons to go through a time consuming exon redefinition step, therefore keeping pre-mRNA intermediates committed to the splicing pathway. Exon definition, the common mode of splice site recognition in humans, occurs when exons are short and flanking introns are long. During exon definition, splice sites are recognized across the exon through the binding of U1 snRNP to the 5′ splice site and U2AF and U2 snRNP binding to the upstream 3′ splice site (Hoffman and Grabowski 1992; Chiara and Reed 1995; Boukis et al. 2004; Sharma et al. 2008). Splicing enhancers and silencers modulate the stability of these complexes, presumably by promoting or interfering with RS domain contacts (Graveley et al. 2001; Boukis et al. 2004). Exon defined splice sites must then transition to pair across the intron to mediate the assembly of the catalytic spliceosome. It is not known how spliceosomal components engaged in exon definition transition to build cross-intron catalytic spliceosomes. During the process of removing an intron, two exon definition complexes come together across the intron to pair the appropriate 5′ and 3′ splice sites. What happens to those factors that are not immediately involved in direct cross-intron interactions remains unclear. For example, the U1 snRNP associated with the 5′ splice site of a downstream exon is essential in establishing the initial exon definition complex. However, its fate during the assembly of the upstream catalytic spliceosome is not known. It could either be used during intron removal, dissociate due to conformational changes during intron removal, or be retained at the 5′ splice site.
Using pulse–chase assays and immunoprecipitation experiments we showed that exon definition components not expected to be involved in cross-intron interactions are retained on the pre-mRNA during the first intron removal event. In the presence of excess competitor RNA, efficient second intron excision was observed, demonstrating that splice sites of exon definition complexes remain associated with functional splicing factors. Two important insights can be derived from these results. First, they show that splicing intermediates do not require redefinition for subsequent intron excisions (Fig. 6). Initial splice site recognition is maintained throughout the process of removing multiple introns. The advantage the splicing machinery gains by holding onto defined splice sites is at least twofold; promoting efficient splicing kinetics, as shown here, and preventing premature degradation of the pre-mRNA transcript. In agreement with this interpretation is the observation that exons flanked by an inefficiently spliced AT–AC intron and an efficiently spliced GT–AG intron are processed faithfully (Wu and Krainer 1996).
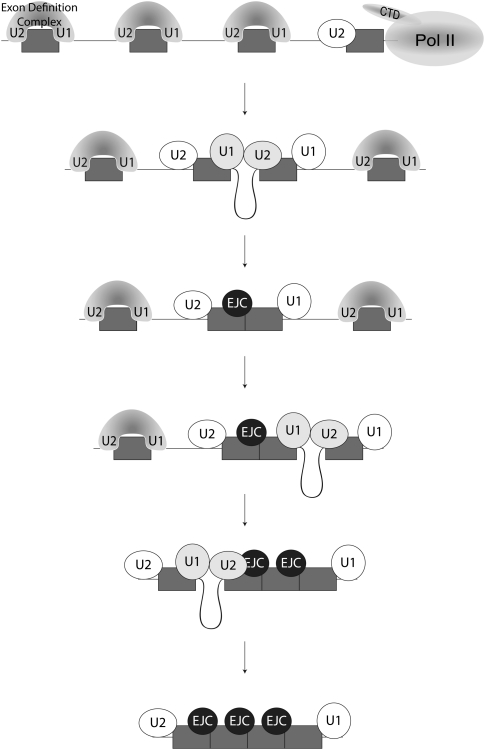
FIGURE 6.
Model for efficient removal of multiple introns. Splice sites are recognized during transcription and spliceosomal components initially associate with the pre-mRNA in exon definition complexes (stretched oval over each exon). A transition to cross-intron interactions occurs during splice site pairing (gray circles) and components previously involved in exon definition interactions remain associated with the pre-mRNA following intron removal (white circles). An EJC (black circle) is deposited during intron removal, but has no measurable effect on the splicing of neighboring introns. However, snRNP retention at the splice sites promotes efficient neighboring intron removal, because ligated exons do not require de novo splice site recognition. The cotranscriptional nature of initial splice site recognition is indicated by the presence of Pol II at the 3′ end of the pre-mRNA.
The demonstration that exon defined splice sites not used during initial splice site pairing remain associated with spliceosomal components also provides clues into the fate of exon definition components during the assembly of cross-intron catalytic spliceosomes. Our results are consistent with the proposal that components of stable exon definition complexes are split to engage in new contacts across the intron. However, this split does not come at the cost of losing splice site definition at the other end of the exon. Rather, spliceosomal recognition of the thus far unpaired splice site is maintained (Fig. 6). How can such rearrangements be envisioned? Once one of the exon defined splice sites commits to splice site pairing, it is likely that the integrity of the cross-exon interaction network is disturbed to allow full engagement across the intron. However, the dissociation rate of snRNPs associated with the other splice site must still be slow enough to maintain an interaction with the splice site, even in the absence of cross-exon networks. It is anticipated that the retention of spliceosomal components occurs following the breaking of cross-exon interactions and before their transition to cross-intron interactions. As such, the primary function of the exon definition complex might only be to ensure the efficient recognition of internal exons. Consistent with this interpretation is the fact that commitment assays that rely on stringent binding competition demonstrate relatively long half-lives of the spliceosomal components that interact with both splice sites (Legrain et al. 1988; Jamison et al. 1992; Kotlajich et al. 2009).
Insights into the composition of exon definition complexes have been provided recently. A proteomic analysis demonstrated that U1 snRNP, U2 snRNP, both subunits of U2AF, SR proteins, and hnRNPs are detected in isolated exon definition complexes (Sharma et al. 2008). These results are consistent with the view that a network of protein interactions exists spanning the exon that is anchored by the stable association of U1 snRNP with the 5′ splice site (Reed 2000). What is less clear is the composition of the exon definition complex at the 3′ splice site. Both U2AF and U2 snRNP are detected, potentially suggesting an intermediate state of stable U2 snRNP association with the branchpoint sequence. Based on these proteomic analyses, it is reasonable to suggest that the factors retained on the pre-mRNA after exon definition complexes transition to splice site pairing complexes are U1 snRNP at the 5′ splice site, verified in this work (Fig. 5), and U2AF or even U2 snRNP at the 3′ splice site.
It has been established that spliceosomal components are deposited onto nascent pre-mRNAs cotranscriptionally by RNA polymerase II (Pol II) (Beyer and Osheim 1988; Lacadie and Rosbash 2005; Listerman et al. 2006; Pandit et al. 2008). The characteristic chromatin immunoprecipitation profiles of analyzed genes suggest that the stable association of snRNPs with the nascent pre-mRNA occurs sequentially, largely following the order at which splice sites were synthesized (Gornemann et al. 2005; Listerman et al. 2006). Depending on the gene, certain introns may also be removed cotranscriptionally, a process highly dependent on the length of the pre-mRNA transcript and the kinetics of spliceosomal assembly, rearrangement, and catalysis (Baurén and Wieslander 1994; Tardiff et al. 2006). Independent of the cotranscriptional nature of splicing catalysis, in vitro analyses demonstrated that the cotranscriptional assembly of splicing factors significantly improved the efficiency of this processing reaction (Das et al. 2006; Hicks et al. 2006), in part by ensuring the expeditious exchange of the nascent pre-mRNA between the transcription and splicing machineries (Hicks et al. 2006). Our observation that splicing factors remain associated with splicing intermediates adds an additional layer of design to ensure efficient processing of multiple-intron-containing genes. The integration of RNA transcription and processing machineries permits fast splice site recognition and assembly of splicing components. Our results show that the process of splice site recognition will only need to be carried out once, because spliceosomal components initially recruited are retained on the pre-mRNA throughout the splicing process. The anticipated net gain of these streamlining mechanisms is to acquire the necessary speeds to prevent nuclear buildup of partially processed pre-mRNAs.
MATERIALS AND METHODS
Splicing constructs
The construction of the test substrate S/L was previously described as (βGskip) (see Supplemental Material in Kotlajich et al. 2009). Briefly, S/L is based on β-globin. β-globin exon 1 (176 nt), intron 1 (136 nt), and exon 2 (222 nt) were fused to a fragment containing β-globin intron 1 and exon 2. The second intron was extended using Drosophila fruitless intronic sequences to a final length of 402 nt. L/S is also based on β-globin and was constructed in a similar manner. A β-globin fragment containing exon 1 (187 nt) and intron 1 was cloned upstream of β-globin exon 1 (153 nt), intron 1 (129 nt), and exon 2 (222 nt). The first intron was extended using Drosophila fruitless intronic sequences resulting in an intron length of 303 nt. The generation of the L/L and S/S substrates was described previously (Fox-Walsh et al. 2005). The matching control substrates were made by amplifying splicing intermediates and subsequent cloning into SP73. The recursive splicing substrate was created by removing the internal exon from the S/L plasmid. Two PCR fragments were generated from this plasmid using primer sets forward 5′-TTTGCTCACATGTTCTTTCCTGCG-3′ with reverse 5′-GTGGCTTCTAGAACCTTGATACCAACCTAAGGGTGGGAAAATAGACCAATAGGC-3′ primers and forward 5′-GCCTATTGGTCTATTTTCCCACCCTTAGGTTGGTATCAAGGTTCTAGAAGCCAC-3′ with reverse 5′-GAGTGCACCATATGGACATATTGTCG-3′ primers. PCR fragments were combined, amplified, and inserted using ExoIII cloning (Tseng 1999) into an SP73 vector digested with AflIII and NdeI.
RNA and splicing reactions
S/L and recursive plasmids were digested with ClaI, the L/S plasmid was digested with BamHI, and L/L and S/S were digested with XhoI. Linearized DNA was in vitro transcribed with T7 RNA polymerase, except for L/L and S/S, which were transcribed with SP6, uniformly labeled with 32P, and gel purified from 7M urea polyacrylamide gels. In vitro splicing reactions were performed in 30% HeLa nuclear extract and carried out as described previously (Lam and Hertel 2002), except using 0.2 U RNasin, 50 mM KCl, and 12 mM Hepes. Splicing time courses were performed up to 3 h to determine the rate of intron removal. The rates for first and second intron removal were determined from the analysis of partially spliced intermediate product formation. A rate equation describing two consecutive splicing steps was used to determine the formation (first intron removal event, k1) and the depletion (second intron removal event, k2) of the splicing intermediate (partally spliced pre-mRNA) according to [intermediate]=[k1/(k2 – k1)] × [A]0 × (e-k1t – ek2t). The fraction of intermediate was calculated for S/L and L/S as (short intron removed RNA)/(unspliced RNA + short intron removed RNA + long intron removed RNA + spliced product) after normalizing to the unspliced RNA size. The fraction spliced for control substrates was calculated as (spliced product)/(unspliced RNA + spliced product). The observed rate of second intron removal (k2) was then compared with the observed rate of product formation from the control series.
Pulse–chase splicing reactions were preincubated in nuclear extract depleted of ATP for 0, 5, 10, 15, or 30 min at 30°C. Following the preincubation, reactions were supplemented with excess cold competitor RNA, ATP, and creatine phosphate, and incubated at 30°C for 90 min.
Cell transfections
HeLa cells were plated to 1.5 × 105 cells/well in six-well plates the day prior to transfection and grown in MEM (Cellgro) supplemented with 10% FBS (v/v), 2 mM glutamine, and 10 mM sodium pyruvate. Lipofectamine 2000 (Invitrogen) was used to transfect 1 μg plasmid following the manufacturer's protocol and cells were harvested 24 h later. Total RNA was extracted using TRIzol (Invitrogen), followed by phenol chloroform extraction and isopropyl alcohol precipitation. DNA contamination was removed using DNase I (Invitrogen). RNA was reverse transcribed using iScript (Bio-Rad). Reactions to detect S/L spliced products used primer sets containing an exon1–exon2 junction forward primer (5′-TAGTAAAGGACAAAGGACAAAAG-3′) combined with a plasmid backbone reverse primer (5′-GTATCTTATCATGTCTGCTCG-3′), or plasmid forward (5′-GCTAACGCAGTCAGTGCTTC-3′) and plasmid reverse primers (5′-GTATCTTATCATGTCTGCTCG-3′). L/S spliced products were detected using a forward primer against exon 1 (5′-ACCGCCCTCGAGCAGCTG-3′) and a reverse primer against an exon 2–exon 3 junction (5′-CTAGCTTTTGTCCTTTGTC-3′). PCR reactions consisted of 10X buffer B without MgCl2 (500 mM KCl and 100 mM HCl), 0.2 mM dNTPs, 15 mM MgCl2, 1 μM each primer, and 0.375 U Taq polymerase. PCR cycles for S/L were: denature 95°C for 3 min, 30 cycles of 95°C for 30 sec, 59°C for 30 sec, and 72°C for 1 min 20 sec, followed by a 5-min extension. L/S PCR conditions were the same except used a 45-sec extension step. The fraction unspliced was calculated as 1 − [spliced/(spliced + unspliced)].
siRNA transfections
HeLa cells were transfected with 200 pmol eIF4AIII siRNA (5′-AGUGGAAUUCGAGACCAGCdTdT-3′) (Ferraiuolo et al. 2004) using lipofectamine 2000. Following a 72-h incubation, cells were cotransfected with another 200 pmol eIF4AIII siRNA with 1 μg plasmid DNA and incubated for 24 h. Cells were harvested, RNA was isolated using TRIzol, and proteins were isolated using RIPA buffer. Protein knockdown was confirmed by Western blot using anti-eIF4AIII (a gift from Dr. Jens Lykke-Andersen, University of California, San Diego) and anti-GAPDH (Santa Cruz Biotechnology) antibodies.
TCR beta transfections
Transfections followed published protocols (Buhler et al. 2006). HeLa cells were plated to 2 × 106 cells in a 10-cm dish. The following day, cells were cotransfected with 5 μg pSUPuro-eIF4AIII plasmid or pSuperior.retro.puro-scrambled plasmid and either pB433 (wt) or pB434 (ter68) plasmids. Following 24-h incubation, media were replaced with media containing puromycin (3μg/mL) and incubated for 30 h. Media were then removed and cells were incubated in the presence of puromycin-free media for 18 h. Cells were harvested and RNA isolated using TRIzol and proteins were isolated using RIPA buffer. TCR beta mRNA was detected using forward primer 5′-GAGAACAGTCAGTCTGGTTC-3′ and reverse primer 5′-CTGACAGCACGGAGAAAG-3′ on 0.1 μg cDNA. PCR reactions consisted of 10X buffer B without MgCl2 (Fisher), 0.2 mM dNTPs, 15 mM MgCl2, 1 μM each primer, and 0.375 U Taq polymerase. PCR cycles were: denature 95°C for 3 min, 25 cycles of 95°C for 30 sec, 54°C for 30 sec, and 72°C for 30 sec, followed by a 5-min extension.
Real-time PCR
Real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad) and 200 nM of each primer: forward 5′-GCGGTGCAGAAACGCTGTA-3′ and reverse 5′-TGGCTCAAACAAGGAGACCTT-3′. Three cDNA dilutions were analyzed in duplicate for each sample during each round of PCR. PCR conditions were 95°C for 3 min, followed by 40 cycles of 95°C for 30 sec and 53°C for 30 sec. Following PCR, a melting curve was performed starting at 55°C, and increasing 0.5°C every 10 sec for 80 cycles.
Immunoprecipitation
Rec-protein G sepharose beads (Invitrogen) were washed with IP150 buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 0.05% Igepal) and rotated at 4°C for 2 h with 4 μg goat U1 snRNP 70K antibody (Santa Cruz Biotechnology). Pulse–chase splicing reactions (the same as described earlier except using 40% nuclear extract) were diluted 1:5 with IP150 buffer. IP150 buffer was added to the bead–antibody mixture to a volume equal to the splicing reaction. Splicing reactions were combined with the bead–antibody mixture and rotated at 4°C for 1 h. Beads were collected by centrifuging at 10,000 rpm for 2 min at 4°C. Beads were washed four times with IP150 buffer and resuspended in 25 μL IP150 buffer. Beads were then treated to proteinase K digest, phenol:chloroform extraction, and ethanol precipitation. Precipitated RNA was divided and either ran on 6% and 8% 7M urea polyacrylamide gel or resuspended in 5 μL of nuclease free water for RT-PCR analysis.
The bands visualized in the polyacrylamide gel were quantified, normalized for size, and used to calculate the ratio of splicing intermediate to spliced product. The reverse transcription reaction consisted of 2.5 μM reverse primer specific for the third exon (5′-CGCGAATTCACGTGC-3′), 1 μL of RNA in a total volume of 5.4 μL. Samples were incubated at 70°C for 5 min and then cooled on ice. An M-MLV mix (5X buffer, 10 mM dNTP, 4U RNasin [Fisher], and 8U M-MLV reverse transcriptase [Promega]) was added to the primer/RNA mix to a total volume of 10 μL and incubated at 37°C for 2 h. PCR to detect the intermediate product was performed using a forward exon–exon junction primer (5′-TAGTAAAGGACAAAGGACAAAAG-3′) with a second intron reverse primer (5′-CTTGTATCTAGATACCATTGGAATTTACGGTT-3′). PCR conditions were 95°C for 3 min, followed by 26 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 1 min, and a final extension of 72°C for 5 min. PCR products were run on a 1.8% agarose gel.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Dr. Jens Lykke-Andersen for providing the eIF4AIII antibody, Dr. Oliver Mühlemann for providing the TCR-beta constructs and eIF4AIII shRNA plasmids, and Dr. Bert Semler for providing the scrambled shRNA plasmid. We are grateful to the members of the Hertel laboratory for helpful comments on the manuscript. HeLa cells were obtained from the National Cell Culture Center. This work was supported by NIH grant GM 62287 (to K.J.H.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2186510.
REFERENCES
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H 2005. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol 12: 861–869 [DOI] [PubMed] [Google Scholar]
- Baurén G, Wieslander L 1994. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell 76: 183–192 [DOI] [PubMed] [Google Scholar]
- Berget SM 1995. Exon recognition in vertebrate splicing. J Biol Chem 270: 2411–2414 [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev 2: 754–765 [DOI] [PubMed] [Google Scholar]
- Blencowe BJ, Issner R, Nickerson JA, Sharp PA 1998. A coactivator of pre-mRNA splicing. Genes Dev 12: 996–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukis LA, Liu N, Furuyama S, Bruzik JP 2004. Ser/Arg-rich protein-mediated communication between U1 and U2 small nuclear ribonucleoprotein particles. J Biol Chem 279: 29647–29653 [DOI] [PubMed] [Google Scholar]
- Buhler M, Steiner S, Mohn F, Paillusson A, Mühlemann O 2006. EJC-independent degradation of nonsense immunoglobulin-μ mRNA depends on 3′ UTR length. Nat Struct Mol Biol 13: 462–464 [DOI] [PubMed] [Google Scholar]
- Burge CB, Tuschl T, Sharp PA 1999. Splicing of precursors to mRNAs by the spliceosome. In The RNA world, 2nd ed (ed. Gesteland RF et al. ), pp. 525–560 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF 2007. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76: 51–74 [DOI] [PubMed] [Google Scholar]
- Chiara MD, Reed R 1995. A two-step mechanism for 5′ and 3′ splice-site pairing. Nature 375: 510–513 [DOI] [PubMed] [Google Scholar]
- Das R, Zhou Z, Reed R 2000. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol Cell 5: 779–787 [DOI] [PubMed] [Google Scholar]
- Das R, Dufu K, Romney B, Feldt M, Elenko M, Reed R 2006. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev 20: 1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo MA, Lee CS, Ler LW, Hsu JL, Costa-Mattioli M, Luo MJ, Reed R, Sonenberg N 2004. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc Natl Acad Sci 101: 4118–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox-Walsh KL, Dou Y, Lam BJ, Hung SP, Baldi PF, Hertel KJ 2005. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc Natl Acad Sci 102: 16176–16181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornemann J, Kotovic KM, Hujer K, Neugebauer KM 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell 19: 53–63 [DOI] [PubMed] [Google Scholar]
- Graveley BR, Hertel KJ, Maniatis T 2001. The role of U2AF35 and U2AF65 in enhancer-dependent splicing. RNA 7: 806–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudas JM, Knight GB, Pardee AB 1990. Ordered splicing of thymidine kinase pre-mRNA during the S phase of the cell cycle. Mol Cell Biol 10: 5591–5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel KJ 2008. Combinatorial control of exon recognition. J Biol Chem 283: 1211–1215 [DOI] [PubMed] [Google Scholar]
- Hicks MJ, Lam BJ, Hertel KJ 2005. Analyzing mechanisms of alternative pre-mRNA splicing using in vitro splicing assays. Methods 37: 306–313 [DOI] [PubMed] [Google Scholar]
- Hicks MJ, Yang CR, Kotlajich MV, Hertel KJ 2006. Linking splicing to Pol II transcription stabilizes pre-mRNAs and influences splicing patterns. PLoS Biol 4: e147 doi: 10.1371/journal.pbio.0040147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BE, Grabowski PJ 1992. U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev 6: 2554–2568 [DOI] [PubMed] [Google Scholar]
- Jamison SF, Crow A, Garcia-Blanco MA 1992. The spliceosome assembly pathway in mammalian extracts. Mol Cell Biol 12: 4279–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler O, Jiang Y, Chasin LA 1993. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol Cell Biol 13: 6211–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlajich MV, Crabb TL, Hertel KJ 2009. Spliceosome assembly pathways for different types of alternative splicing converge during commitment to splice site pairing in the A complex. Mol Cell Biol 29: 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadie SA, Rosbash M 2005. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol Cell 19: 65–75 [DOI] [PubMed] [Google Scholar]
- Lam BJ, Hertel KJ 2002. A general role for splicing enhancers in exon definition. RNA 8: 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang KM, Spritz RA 1987. In vitro splicing pathways of pre-mRNAs containing multiple intervening sequences? Mol Cell Biol 7: 3428–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain P, Seraphin B, Rosbash M 1988. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol Cell Biol 8: 3755–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Andersen GR 2008. Structural insights into the exon junction complex. Curr Opin Struct Biol 18: 112–119 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Moore MJ, Maquat LE 2000. Pre-mRNA splicing alters mRNP composition: Evidence for stable association of proteins at exon-exon junctions. Genes Dev 14: 1098–1108 [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ 2001. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20: 4987–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Lin RI, Lai MC, Ouyang P, Tarn WY 2003. Nuclear Pnn/DRS protein binds to spliced mRNPs and participates in mRNA processing and export via interaction with RNPS1. Mol Cell Biol 23: 7363–7376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SR, Hertel KJ 2004. Commitment to splice site pairing coincides with A complex formation. Mol Cell 15: 477–483 [DOI] [PubMed] [Google Scholar]
- Listerman I, Sapra AK, Neugebauer KM 2006. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol 13: 815–822 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Tasic B 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418: 236–243 [DOI] [PubMed] [Google Scholar]
- Mühlemann O, Mock-Casagrande CS, Wang J, Li S, Custodio N, Carmo-Fonseca M, Wilkinson MF, Moore MJ 2001. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol Cell 8: 33–43 [DOI] [PubMed] [Google Scholar]
- Nasim FH, Spears PA, Hoffmann HM, Kuo HC, Grabowski PJ 1990. A sequential splicing mechanism promotes selection of an optimal exon by repositioning a downstream 5′ splice site in preprotachykinin pre-mRNA. Genes Dev 4: 1172–1184 [DOI] [PubMed] [Google Scholar]
- Pandit S, Wang D, Fu XD 2008. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol 20: 260–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr Opin Cell Biol 12: 340–345 [DOI] [PubMed] [Google Scholar]
- Sakashita E, Tatsumi S, Werner D, Endo H, Mayeda A 2004. Human RNPS1 and its associated factors: A versatile alternative pre-mRNA splicing regulator in vivo. Mol Cell Biol 24: 1174–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL 2008. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat Struct Mol Biol 15: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T, Tange TØ, Sonenberg N, Moore MJ 2004. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol 11: 346–351 [DOI] [PubMed] [Google Scholar]
- Stamm S, Riethoven JJ, Le Texier V, Gopalakrishnan C, Kumanduri V, Tang Y, Barbosa-Morais NL, Thanaraj TA 2006. ASD: A bioinformatics resource on alternative splicing. Nucleic Acids Res 34: D46–D55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DA, Carlo T, Berget SM 1996. Architectural limits on split genes. Proc Natl Acad Sci 93: 15081–15085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara K, Schwarze U, Imamura Y, Hoffman GG, Toriello H, Smith LT, Byers PH, Greenspan DS 2002. Order of intron removal influences multiple splice outcomes, including a two-exon skip, in a COL5A1 acceptor-site mutation that results in abnormal pro-α1(V) N-propeptides and Ehlers-Danlos syndrome type I. Am J Hum Genet 71: 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange TØ, Shibuya T, Jurica MS, Moore MJ 2005. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA 11: 1869–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Lacadie SA, Rosbash M 2006. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly post-transcriptional. Mol Cell 24: 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H 1999. DNA cloning without restriction enzyme and ligase. Biotechniques 27: 1240–1244 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wu Q, Krainer AR 1996. U1-mediated exon definition interactions between AT-AC and GT-AG introns. Science 274: 1005–1008 [DOI] [PubMed] [Google Scholar]