Abstract
Human tissues are an important source of biological material for the discovery of novel biomarkers. Fresh-frozen tissue could represent an ideal supply of archival material for molecular investigations. However, immediate flash freezing is usually not possible, especially for rare or valuable tissue samples such as biopsies. Here, we investigated the compatibility of RCL2/CS100, a non-crosslinking, non-toxic, and non-volatile organic fixative, with shotgun proteomic analyses. Several protein extraction protocols compatible with mass spectrometry were investigated from RCL2/CS100-fixed and fresh-frozen colonic mucosa, breast, and prostate tissues. The peptides and proteins identified from RCL2/CS100 tissue were then comprehensively compared with those identified from matched fresh-frozen tissues using a bottom-up strategy based on nano-reversed phase liquid chromatography coupled with tandem mass spectrometry (nanoRPLC-MS/MS). Results showed that similar peptides could be identified in both archival conditions and the proteome coverage was not obviously compromised by the RCL2/CS100 fixation process. NanoRPLC-MS/MS of laser capture microdissected RCL2/CS100-fixed tissues gave the same amount of biological information as that recovered from whole RCL2/CS100-fixed or frozen tissues. We next performed MALDI tissue profiling and imaging mass spectrometry and observed a high level of agreement in protein expression as well as excellent agreement between the images obtained from RCL2/CS100-fixed and fresh-frozen tissue samples. These results suggest that RCL2/CS100-fixed tissues are suitable for shotgun proteomic analyses and tissue imaging. More importantly, this alternate fixative opens the door to the analysis of small, valuable, and rare target lesions that are usually inaccessible to complementary biomarker-driven genomic and proteomic research.
Introduction
Tissue analyses including histomorphological and immunohistochemical approaches are traditionally used to perform cancer diagnosis or to predict individual cancer patient outcomes. Formalin fixation and paraffin embedding (FFPE) of tissues preserves the cellular, architectural, and morphological details observed by microscopic evaluation of thin tissue sections. This process also stabilizes tissues, allowing for easy storage at room temperature for extensive periods of time. For these reasons, tissue archiving has been performed by FFPE for several decades and is still today the standard processing methodology practiced in histopathology laboratories worldwide. FFPE preserved tissues are therefore the most abundant supply of archival material. High-throughput technologies, such as gene- or protein-tissue arrays, can rapidly screen and identify new biomarkers, which could greatly improve cancer patient management. However, significant obstacles remain in applying these technologies to such clinical samples. Indeed, drawbacks exist concerning FFPE tissue storage and fixation, since protein and nucleic acid integrity are affected by formaldehyde-induced crosslinking 1-4. FFPE alters and fragments nucleic acids, impairing the extraction efficacy and quality of DNA, and more strikingly RNA, thus preventing high quality molecular analyses 5. In addition, as a highly reactive dipolar compound, formalin facilitates the formation of protein-protein crosslinks in vitro and therefore renders FFPE tissues refractory to conventional procedures of protein extraction and subsequent analyses 2, 3. Nevertheless, several procedures to overcome some of these drawbacks have been reported. Thus, some protocols have been shown to generate amplicons of up to 70 base pairs for analysis by real-time PCR 6-8. In addition, using bottom-up proteomic strategies based on nano-reversed phase liquid chromatography coupled with tandem mass spectrometry (nanoRPLC-MS/MS) 9-16 and MALDI-based imaging mass spectrometry 17-19, some proteomic investigations from FFPE archived tissues have been successfully realized. More recently, Nirmalan et al. described a novel protein extraction methodology for western blotting analysis 20. An additional significant drawback is that formalin is an extremely toxic fixative. Repeated exposure via nasal, oral, or dermal routes is a human health risk with outcomes ranging from minor irritation of the eyes, nose, and throat to more severe complications, including dysphagia, bronchitis, pneumonia, and squamous cell carcinoma of the nose and pharynx 21, 22. Consequently, several worldwide regulatory agencies and professional societies have recommended new restrictive occupational exposure limits for formaldehyde.
Unfixed fresh- or snap-frozen tissues could represent a robust alternative for complete molecular analyses, including genomic, transcriptomic, and proteomic analyses, but these samples do not provide sufficient accurate morphological details and may impair histological diagnosis. Further, especially for clinical biopsies, availability may be limited and transport and long-term storage complicated. It is therefore necessary to modernize pathology and propose new alternatives for tissue sample archives. Alternative alcohol-based and formalin-free non-crosslinking tissue fixation procedures, such as Methacarn, UMFIX, or FineFIX, have been shown to be ideal for preserving the morphological details and for DNA and RNA extraction, but the related procedures have been poorly investigated in the proteomic fields 23-28. Ethanol-fixed tissues have also been investigated with relevant results for proteomic investigations 3, 29, 30. In our laboratory, we have previously tested tissue preservation with RCL2/CS100, a new non-crosslinking, non-toxic, and non-volatile fixative, for its effects on morphology as well as DNA and RNA integrity. Interestingly, the histomorphology of RCL2/CS100-fixed paraffin-embedded tissues was found to be similar to that observed from matched formalin-fixed tissues. RCL2/CS100 fixation preserved DNA integrity as demonstrated by successful amplification and sequencing of large DNA amplicons. Similarly, high-quality RNA could be extracted from microdissected breast tumor cells from RCL2/CS100-fixed paraffin-embedded biopsies as assessed by electropherogram profiles and real-time reverse transcription-polymerase chain reaction quantification of various genes 31. More recently, we successfully assessed the compatibility of RCL2/CS100-fixed tissues with several proteomic approaches, including 1- and 2-D gel electrophoresis, immunohistochemistry, western blot, and SELDI MS analyses 1. In the present study, we expanded our investigations and demonstrated that RCL2/CS100-fixed tissues are also suitable for bottom-up proteomics using nanoRPLC-MS/MS and for MALDI-based tissue profiling and imaging mass spectrometry. Since both technologies are very powerful for performing proteomic biomarker discovery investigations, we believe that RCL2/CS100-fixed tissue samples represent an easy-to-use alternative to fresh-frozen tissue, especially for rare and valuable tissue samples.
Experimental sections
Tissue samples and processing
Normal colonic mucosa (n=3), prostate (n=3), and primary breast carcinoma (n=3) tissue samples were acquired by the Department of Pathology (Montpellier). Informed consent was obtained from all patients. To assess the feasibility of peptide identifications from RCL2/CS100-fixed tissues, tissue blocks (1cm3) were cut into two equal parts. From each block, one half was immediately snap-frozen in liquid nitrogen and stored at -80°C. The other was fixed overnight (12h) at 4°C in RCL2/CS100 (volume tumor × 10-20) as recommended by manufacturer (Alphelys, Plaisir, France, www.alphelys.com), dehydrated in eight baths of graded ethanol (30 min each) and two baths of xylene (1 hour each), and paraffin-embedded using a TissueTek VIP automated processor (Bayer HealthCare Diagnosis Division). RCL2/CS100 paraffin-embedded blocks were stored at -20°C.
For direct protein extraction, five 7-μm-thick sections from frozen or RCL2/CS100-fixed tissues from each patient were cut from each block and pooled into a 1.5-ml tube. For the RCL2/CS100-fixed tissues, the sections were de-waxed for 1 hour at 65°C followed by three 5-min extractions in 100% xylene at 65°C prior to washing with 100% ethanol. Tissue samples were air-dried, and the proteins were extracted and analyzed by nanoRPLC-MS/MS as described below.
For tissue microdissection, 7-μm-thick paraffin-embedded breast tissue sections were deparaffinized before staining with haematoxylin in 100% ethanol according to the manufacturer's protocol (Acturus Engineering) and stored in a desiccated container for at least 15 min before laser capture microdissection (LCM). For each analysis, 5,000 cells were laser-captured from the same tissue using the PixCell II LCM system (Acturus Engineering). Microdissection efficiency was assessed by examining cells present on the LCM cap. Microdissected cells were stored at -80°C prior to analysis.
Protein extraction
In order to test buffer conditions that are directly compatible with sequencing grade modified trypsin digestion and analysis using nanoRPLC-MS/MS, 7-μm-thick tissue sections and microdissected cells were processed using different lysis buffers.
Protein extraction from normal colonic mucosa (n=3), prostate (n=3), and primary breast carcinoma (n=3) tissue samples was performed for each patient. Protein extraction was performed in three independent replicates. For 7-μm thick sections, proteins were extracted for 30 min at 95°C followed by 2 hr at 60°C with 60 μl of buffer A (100 mM triethylammonium bicarbonate, 30% acetonitrile), buffer B (50 mM triethylammonium bicarbonate, 0.1% RapiGest SF; Waters, Milford, USA), or buffer C (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% Triton X-100, 1% SDS, 0.5% deoxycholate). Protein extracts were clarified at 15,000 g for 2 min at 4°C and stored at -20°C until analysis. Protein concentrations were measured using the Micro BCA Protein Assay Kit (Pierce Biotechnology, Rockford, IL) according to the manufacturer's protocol. Protein concentrations were measured using 1 or 2 μl of the protein extracts. Forty micrograms of the buffer B samples were incubated with 2 μg of trypsin Gold, mass spectrometry grade (Promega), at 37°C overnight. The reducing agent DTT was added to a final concentration of 10 mM. The samples were reduced for 30 min at 60°C and then alkylated for 30 min at room temperature with 10 mM iodoacetamide. Samples were stored at -20°C until analysis.
For the microdissected cells, films from the LCM caps were removed and transferred into a tube containing 40 μl of buffer B. The cells were spun at 10,000 g for 1 min, incubated at 95°C for 90 min, then cooled on ice for 3 min. Samples were then incubated with 2 μg of trypsin Gold at 37°C overnight. Protein concentrations were measured from 2 μl of sample. The samples were then incubated for 5 min at 95°C with reducing agent and stored at -20°C until analysis.
Statistical analysis of protein recovery yields
Statistical analyses were done using Student's t test and Mann-Whitney U test for comparison of two groups and using one-way ANOVA with Bonferroni multiple comparison post-test and non-parametric ANOVA with Dunn's multiple comparison post-test for comparison of more than two groups using GraphPad InStat (version 3.06). A probability level of P < 0.05 was chosen for statistical significance.
Mass spectrometry analysis of protein extracts
After protein extraction with buffer B, one digested sample for each tissue was clarified at 5,000 rpm for 5 min, vacuum-dried, re-dissolved in 100 μl of 5% acetonitrile (ACN) and 0.5% trifluoroacetic acid (TFA), and purified by activated PepClean™ C-18 Spin Columns according to the manufacturer's protocol (Pierce Biotechnology, Rockford, IL). Peptides were eluted and dried before being reconstituted for nanoRPLC-MS/MS.
Peptides were separated using an Ultimate 3000 nano LC system (Dionex Corporation) coupled to a Probot™ Microfraction Collector (Dionex Corporation). NanoRPLC separations were performed using an Acclaim PepMap™ (C18, 3 μm, 100 Å) 75 μm/15 cm column (Dionex – LC Packing). The mobile phases used were 2% ACN with 0.05% TFA (A) and 80% ACN with 0.05% TFA (B). The gradient elution steps were 0-30% B in 120 min, 30-60% B in 30 min, 60-100% B in 20 min, and then 100% B for an additional 10 min, at a flow rate of 0.3 μl/min. The fractions were mixed directly with the MALDI matrix solution (2 mg/ml α-cyano-4-hydroxycinnamic acid in 70% ACN with 0.1% TFA) at a flow rate of 1.5 μl/min. Forty femtomole of human [Glu1] Fibrinopeptide B (m/z 1570.57) was spiked into each spot as an internal standard. Spotting onto Opti-TOF™ LC/MALDI insert plates (Applied Biosystems) was performed using the Probot spotting device, during 190 min at a speed of 7 s per well.
Opti-TOF™ LC/MALDI insert plates were analyzed using a MALDI TOF/TOF 4800 Proteomics Analyzer mass spectrometer (Applied Biosystems). MS spectra from m/z 700-4,000 were acquired in positive reflector ion mode using 1,500 laser shots. The 10 most abundant peptide precursor ions with signal-to-noise (S/N) ratios greater or equal to 50 were selected for MS/MS analysis using 3,500 laser shots from m/z 300-1,500. Two independent experiments were performed for each tissue. Protein identification was performed as described below.
Data analysis and interpretation
The combined MS/MS spectra obtained from the two independent experiments were searched against the UniProt human proteomic database (uniprot_sprot300108) from the European Bioinformatics Institute using the ProteinPilot™ Software 2.0 and Paragon method (Applied Biosystems, Software revision 50861). Search parameters such as tryptic cleavage specificity, precursor ion accuracy, and fragment ion mass accuracy are built-in functions of the ProteinPilot Software. The unused protein score calculated by ProteinPilot is a measurement of protein identification confidence that takes into account all peptide evidence for a protein and excludes any evidence that is better explained by a higher ranking protein. Proteins with one high confidence (>95%) unique peptide were considered as positively identified. A decoy database was used to estimate the false discovery rate (FDR). The molecular functions and cellular localizations of the unique proteins identified in this study were categorized using the Protein Family Classification System iProClass (http://pir.georgetown.edu/).
Tissue profiling and imaging by MALDI MS
Fresh-frozen sections from colon, prostate, and breast tissues were cut at -15°C at a thickness of 10 μm in a cryostat (CM 3050 S, Leica Microsystems GmbH, Wetzlar Germany) and thaw-mounted on conductive glass slides. For histological evaluation, serial sections were also cut and subsequently stained by haematoxylin and eosin (H&E) using standard protocols. Sections destined to be analyzed by MALDI MS were then rinsed in successive Petri dishes containing 70% and 95% isopropanol solutions for 30 s each to remove the lipid component and improve signal quality 32. The sections were then allowed to dry in a vacuum desiccator for several minutes prior to matrix deposition.
Sections from the matching RCL2-fixed tissue specimens were cut at a thickness of 5 μm using a microtome (HM325A, Utech Products, Inc., Schenectady, NY) and either mounted on MALDI compatible conductive glass slides for subsequent MS analyses or standard microscope slides for staining and histological evaluation. Paraffin removal was accomplished by washes in xylene (100% twice for 10 s) and graded ethanol concentrations (100% twice for 10 s and 95%, 80%, and 70% for 10 s each). The slides were then allowed to fully dry in an oven for 1 h at 65°C.
For tissue profiling and imaging experiments, 20 mg/ml sinapinic acid prepared in 50% acetonitrile and 0.2% trifluoroacetic acid was used as the MALDI matrix. For the profiling studies, matrix was manually deposited on the sections by overlaying two 300-nl droplets at the desired coordinate guided by the histology of each section. For all imaging studies, matrix was automatically deposited on the sections using an acoustic ejection-based Portrait 630 multispotter (Labcyte Inc. Sunnyvale CA) 33-35. In this case, matrix was printed with a 200 μm center-to-center spacing. Each print pattern was repeated for 30 cycles ejecting 1 drop of matrix per cycle. The volume for each drop was estimated to be ∼170 pl.
All sections were analyzed by MALDI-TOF MS using an Autoflex II mass spectrometer (Bruker Daltonics, Billerica MA) operated in the positive linear mode geometry under delayed extraction condition time focused at ∼m/z 15,000. In this case, the best resolutions (M/ΔM measured at full width at half maximum) close to ∼1,000 were attainable for ions at or around m/z 15,000, whereas acceptable resolutions in the range of 600 were obtainable for m/z values in the range of 5,000 to 25,000. The instrument was mass-calibrated in the m/z range from 5,000 to 25,000 using a mixture of standard proteins. Protein profiling data were manually acquired by averaging signals from 10 series of 100 shots (1,000 total shots per spectrum). Imaging MS data were acquired, baseline corrected, and assembled and visualized using the FlexImaging 2.2 software (Bruker Daltonics, Billerica MA). One mass spectrum was acquired per matrix spot. Each spot was analyzed in the same manner by averaging signals from 250 laser shots acquired in 5 series of 50 shots with each series acquired from a different location in a spiral pattern within the spot.
Results
Extraction of proteins by different protocols
The impact of RCL2/CS100 fixation on the ability to extract and identify peptides and proteins by nanoRPLC-MS/MS was undertaken in comparison to similar measurements performed from corresponding frozen tissue specimens. Several types of tissue samples, colonic mucosa, prostate, and breast tissues, were therefore snap-frozen or fixed in RCL2/CS100 before paraffin embedding. We first tested different conditions to improve protein extraction. Two extraction buffers compatible with trypsin digestion and subsequent nanoRPLC-MS/MS analysis (buffers A and B) were compared to a reference buffer used for radioimmunoprecipitation assays (RIPA, buffer C) (Table 1). Quantitative comparisons of extracted proteins from RLC2/CS100 and frozen tissues revealed that buffer B, which contained an acid-cleavable anionic detergent (RapiGest), gave systematically similar protein yields with respect to the reference buffer C, with 80-120% of the protein extracted in the three different tissue samples. By comparison, buffer A, which contained 30% acetonitrile, showed significant lower amounts of protein extracted than buffer C (p < 0.05). For this reason, buffer B was chosen for the subsequent analyses using nanoRPLC-MS/MS. Overall, protein recovery yields obtained from frozen tissues were not found to be significantly different compared to those obtained from RCL2/CS100-fixed tissues, regardless of the extraction protocol used (p > 0.05).
Table 1.
Protein quantities measured from the RCL2/CS100-fixed and frozen colonic mucosa, prostate, and breast tissues using three protein extraction protocols.
| Buffer | Colonic mucosa | Prostate | Breast | |||
|---|---|---|---|---|---|---|
| RCL2 | Frozen | RCL2 | Frozen | RCL2 | Frozen | |
| A | 1.08 ± 0.11 (43%) |
0.83 ± 0.13 (39%) |
0.80 ± 0.11 (41%) |
1.46 ± 0.02 (83%) |
1.28 ± 0.21 (56%) |
0.89 ± 0.15 (51%) |
| B | 2.36 ± 0.27 (94%) |
2.19 ± 0.38 (103%) |
1.65 ± 0.31 (84%) |
1.87 ± 0.32 (106%) |
1.86 ± 0.15 (82%) |
2.03 ± 0.24 (117%) |
| C | 2.51 ± 0.59 (100%) |
2.13 ± 0.22 (100%) |
1.95 ± 0.15 (100%) |
1.76 ± 0.17 (100%) |
2.28 ± 0.24 (100%) |
1.74 ± 0.30 (100%) |
Protein concentrations are μg/μl ± SD. Values in brackets are the percentage of protein quantity compared to buffer C, which was defined as 100%.
Protein identification from frozen tissue and RCL2/CS100-fixed tissue
In order to evaluate the effects of RCL2/CS100 fixation, the numbers of peptides and proteins extracted from fresh-frozen and RCL2/CS100-fixed tissues were compared by LC-MS/MS. A total of 26,522 MS/MS non-empty spectra were generated by nanoRPLC-MS/MS analysis from the three tissue samples and the different preservation conditions. These were searched against the UniProt human proteomic database using ProteinPilot with stringent criteria and a Protscore > 1.3 (C.I. > 95%) cut-off. The number of distinct peptides and identified proteins in each condition are shown in Table 2. For the prostate, breast, and colon tissues, a total of 172, 226, and 309 identified proteins, respectively, were found to be common in both the RCL2/CS100 and frozen conditions. To evaluate a false discovery rate, the MS/MS spectra were searched against a decoy database with all protein sequences shuffled. No protein could be identified. The complete list of the proteins identified can be found in Supplemental Table S1. A representative set of MS/MS spectra of the same peptides consistently identified across the RCL2/CS100-fixed and frozen colonic mucosa, prostate, and breast tissues is presented in Supplemental Figure S1 and shows that proteins can be conclusively identified from these tissues for both preservation conditions. Additionally, we compared five proteins identified from RCL2/CS100-fixed and frozen colonic mucosa tissues on the basis of sequence coverage and number of unique peptides, and overall an excellent correspondence was observed between both tissue preservation conditions (Table 3).
Table 2.
Comparison of the number of unique peptides and proteins identified from RCL2/CS100 and frozen tissues.
| Colonic mucosa | Prostate | Breast | |||||
|---|---|---|---|---|---|---|---|
| RCL2 | Frozen | RCL2 | Frozen | RCL2 | Frozen | LCM | |
| N° of unique peptides | 1,490 | 1,673 | 1,598 | 1,135 | 1,227 | 1,604 | 1,392 |
| N° of unique proteins | 531 | 509 | 377 | 294 | 355 | 468 | 306 |
Table 3.
Comparison of the number of unique peptides identified for the top five proteins identified within RCL2/CS100 and frozen colonic mucosa tissues.
The molecular functions and cellular localizations of all proteins identified in the colonic mucosa, prostate, and breast tissues were classified using the gene ontology classification system (Figure 1). The proteins were associated with every cellular compartment and represented a broad range of molecular functions, including signal transduction, cell proliferation, and transcription. When we compared the gene ontology classification of the proteins identified from RCL2/CS100-fixed tissues with those from frozen tissues, we did not observe any significant bias between both modes of preservation (Figure 1). Finally, we compared the cellular localization of proteins from the same tissues that were found to be different after fresh-frozen or RCL2/CS100 preservation and found no substantial specific bias potentially introduced by the RCL2/CS100 process on protein extraction (Figure 2).
Figure 1.
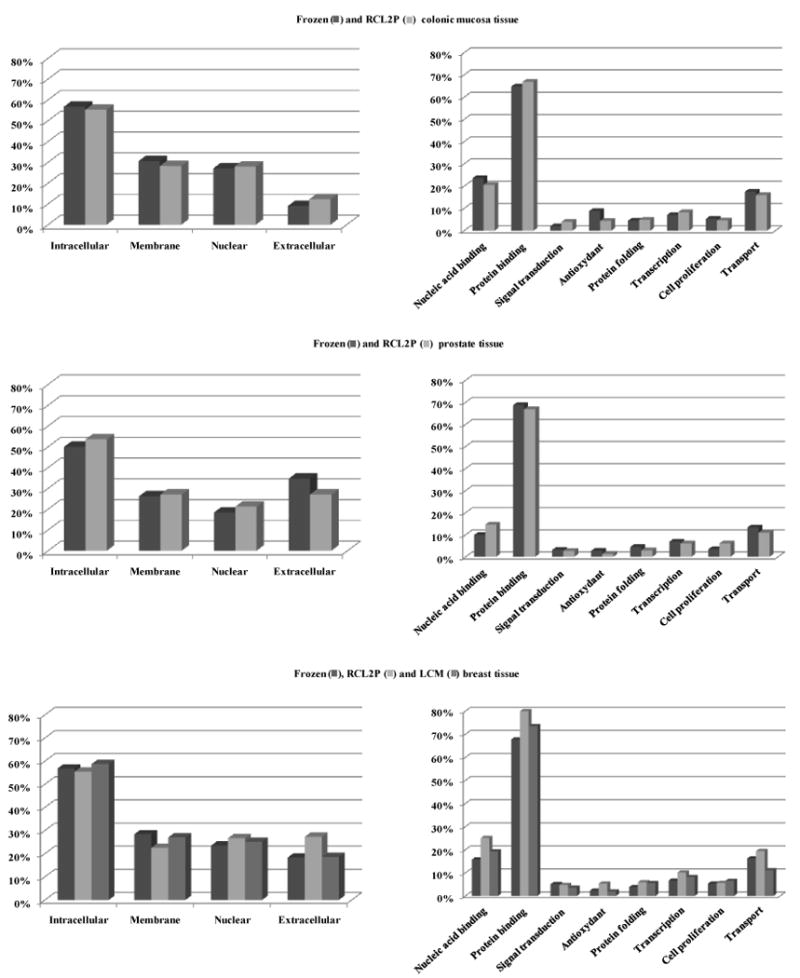
Cellular compartments and molecular functions according to gene ontology classification of the identified proteins from frozen and RCL2/CS100-fixed colonic mucosa, prostate, and breast tissue samples.
Figure 2.
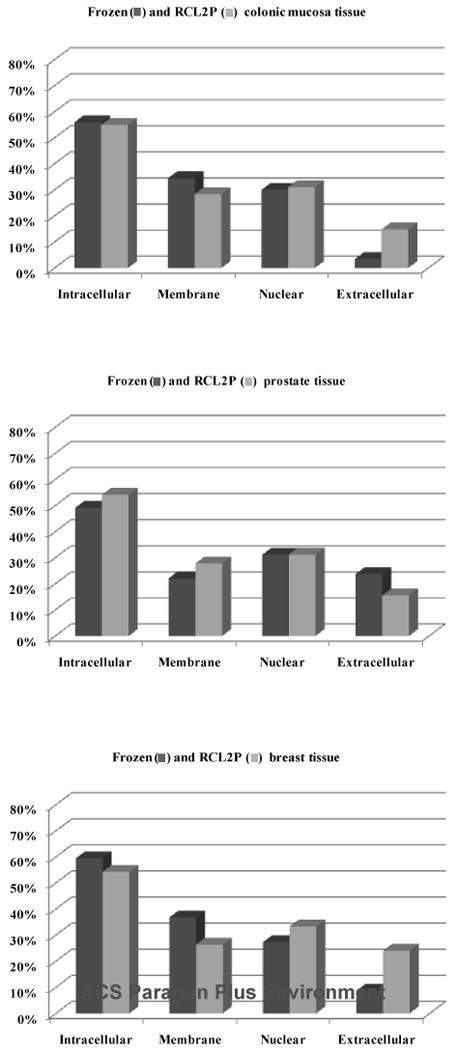
Cellular localizations using gene ontology classification of the identified proteins that were different between the fresh-frozen and RCL2/CS100 preserved samples of the same tissues.
Laser cell microdissection from RCL2/CS100-fixed tissues
One of the technical challenges in the field of cancer is the ability to analyze proteins from relatively complex cell populations. Tumor tissues are highly heterogeneous samples composed of various types of non-tumor cells, such as inflammatory, endothelial, and normal cells. All of these components are homogenized together during protein extraction and can explain the lack of reproducible results due to the variable ratio of cell populations between different samples. Cell enrichment via laser capture microdissection (LCM) solves this problem by allowing cell-specific proteomic analyses. We thus performed LCM to purify or select cancer cells from normal or stromal cells from RCL2/CS100-fixed breast tissues. Areas corresponding to ∼5,000 cells were excised from thin sections and lysed using buffer B. Typically, 14 ± 1.8 μg of proteins were obtained from each of the six LCM analyses. The total number of proteins identified after LCM of cancer cells was similar to the number detected from the global analysis of the breast tumor tissue samples (Table 2); 59% of proteins were identified by two or more unique peptides. Further, using gene ontology classification, we observed that a similar amount of information can be extracted from cells after LCM of RCL2/CS100-fixed breast cancer tissue samples as compared with whole RCL2/CS100-fixed breast tissue samples (Figure 1). Interestingly, among the proteins identified by at least two unique peptides, 12% and 17% for LCM and global RCL2/CS100-fixed tissues, respectively, were found to be unique. Of these, some proteins enriched in the LCM breast samples have been shown to be involved in breast tumorigenesis or be related to regulation of the estrogen receptor (Table 4).
Table 4.
Proteins identified in RCL2/CS100-fixed breast tissue after cell enrichment by LCM that are known to be involved in breast tumorigenesis or related to regulation of the estrogen receptor.
Tissue profiling and imaging by MALDI MS
The proteomic content of thin sections cut from RCL2/CS100-fixed tissue specimens was investigated by MALDI profiling and imaging mass spectrometry. In order to assess the quality of the spectrometric data and to better understand the effect of the preservation procedure, profiles and images obtained from the RCL2/CS100-fixed samples were again compared to those obtained from the fresh-frozen samples. Initial protein profiling measurements were performed on sections from the RCL2/CS100-fixed specimens. After sectioning, paraffin removal, and partial rehydration, MALDI matrix (sinapinic acid) was directly deposited on targeted areas of the tissue sections guided by the histology observed from the serially cut and stained sections. In parallel, sections from the fresh-frozen matched tissues were also cut and spotted under histological guidance with the same matrix solution. After solvent evaporation and matrix crystallization, the samples were analyzed by MALDI-TOF MS using the same instrument settings. MS data was manually acquired in the same manner by averaging signals from 1,000 laser shots from random positions within the matrix spots. Figure 3 presents typical protein profiles obtained from the fresh-frozen and RCL2/CS100-fixed colonic mucosa specimen in the m/z range up to 20,000. Similar results are presented in Supplemental Figure S1, which presents the protein profiles obtained from areas of the cancerous breast and normal prostate fresh-frozen and RCL2/CS100-fixed samples.
Figure 3.
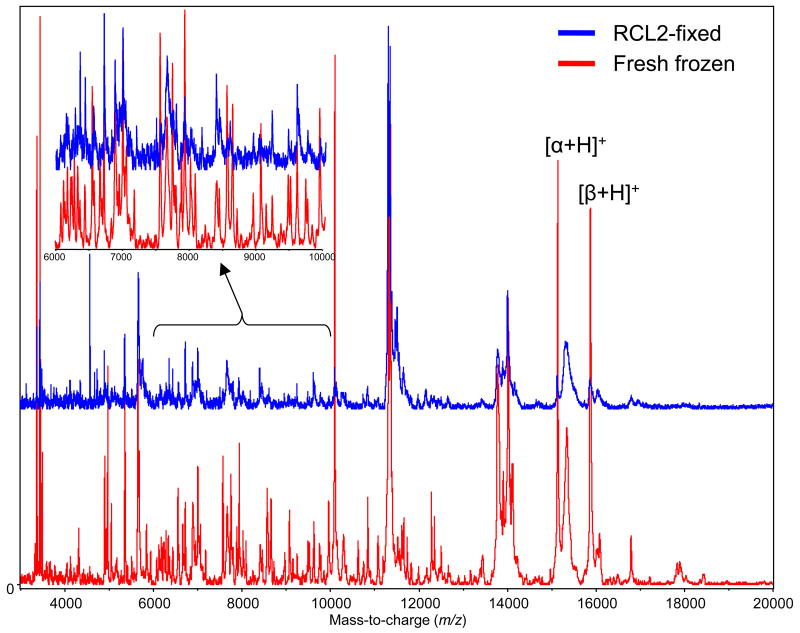
MALDI MS protein profiles acquired from fresh-frozen and RCL2/CS100-fixed colon mucosa sections in the m/z range from 3,000 to 20,000. The inset details signals in the m/z range from 8,000 to 10,000. [α+H]+ and [β+H]+ indicate the hemoglobin alpha and beta chain molecular ions, respectively.
Several general trends were observed. First, the overall quality of the spectra in terms of resolution, signal-to-noise ratio, and overall signal intensity was consistently better for fresh-frozen sections. Therefore, overall number of signals detected from the fresh-frozen sections was increased by roughly a factor of two with respect to those obtained from RCL2/CS100-fixed sections. Second, almost all of the signals observed when analyzing the RCL2/CS100-fixed sections were also observed upon analysis of matched fresh-frozen sections with close to the same relative intensities. A handful of signals, some of which had significant intensities, were however found to be unique to RCL2/CS100-fixation. Another consistent observation from the analysis of RCL2/CS100-fixed tissues sections was a general trend of lower intensity hemoglobin signals (Figures 3 and S1). This can be an added benefit of RCL2/CS100 fixation, because for some highly vascularized tissue types such as lung, overwhelming hemoglobin signals can be observed, leading to evident ion suppression effects.
Imaging MS experiments from fresh-frozen and matched RCL2/CS100-fixed colon sections were also performed (Figure 4). Sections were automatically printed with a 200 μm center-to-center pitch over the areas to be imaged. From the RCL2/CS100-fixed colon section, over 60 meaningful protein images were recovered, whereas from the matched fresh-frozen section, over 200 images were recovered. A large majority of the signals observed in common across both modes of tissue preservation gave very similar molecular images. This is exemplified in Figure 4, which presents imaging MS results for a subset of signals, most of which were common in both sections, along with the corresponding histologies. For all of the proteins presented, excellent agreement was observed between the images obtained from the fresh-frozen and the RCL2/CS100-fixed sections in correlation with the observed histology. The signal at m/z 14210 was found to clearly localize within the colon mucosa, the signal at m/z 4568 was strongly expressed in the sub-mucosa, the signal at m/z 3968 was uniquely expressed in colon circular smooth muscles, and the signal at m/z 3370 correlated well with fat-containing soft tissues. Finally, the signal at m/z 66430, which most probably corresponds to albumin, was abundant in the sub-mucosa from both specimens but was not plotted in the composite density map assembled for the RCL2/CS100-fixed section to avoid congestion.
Figure 4.
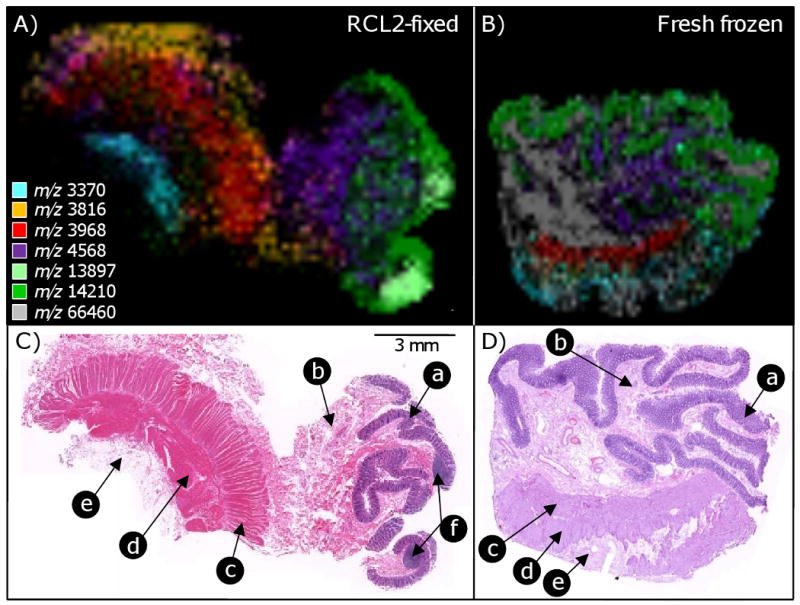
MALDI imaging MS analyses from RCL2/CS100-fixed (A, C) and fresh-frozen (B, D) colon mucosa sections and the corresponding histology. The ion images (A, B) are composite ion density maps obtained from different protein signals each displayed with a different color. These signals (among others) were seen to be significantly enhanced or unique to the different histologies present in the sections (C, D). (a) mucosa, (b) sub-mucosa, (c) circular smooth muscle, (d) longitudinal smooth muscle, and (e) soft tissue (fat).
Discussion
Over the past 20 years, major advances in biotechnology have focused on cataloguing cancer traits at the DNA, RNA, and protein levels. It is now conceivable that large-scale genomic and proteomic investigations will provide novel insights into early detection, diagnosis, prognosis, and monitoring of disease progression. With early diagnosis of cancer, tumor size has become smaller, which limits the availability of biopsy tissue samples for clinical research studies. It is therefore of high importance to find non-toxic tissue fixatives that provide high quality histomorphology and are amenable to genomic and proteomic studies. We have previously demonstrated that RCL2/CS100, a promising new fixative, has great potential for concomitantly performing morphological and molecular analyses on the same tissue sample. In a first study, we showed that RCL2/CS 100 was highly suitable for preservation of tissue morphology, DNA, and most importantly RNA 31. In a second study1, we performed a careful comparison of RCL2/CS100 and formaline fixative using different paraffin-embedded tissue samples and found that no morphological detail and protein degradation were seen by microscopic observation and immunohistochemistry on specimens stored at room temperature for over 8 years supporting its potential use in long-term tissue storage. Our overall goal in this study was to perform a careful evaluation of RCL2/CS100-fixed tissues for proteomic analyses using nanoRPLC-MS/MS and MALDI imaging mass spectrometry to demonstrate that protein analysis from such archival material is feasible and comparable to that recovered from fresh-frozen tissue samples.
The tissue sizes used in this study were approximately 1 cm3. We have notice that tissue thickness upper than 3 cm limits the penetrance of the fixative in the tissue. If the tumor size is upper than 3 cm, the sample should be cut to help fixative penetration in the tissue. For different types of tissues, we first compared the protein recovery yields obtained using three different protein extraction buffers compatible with mass spectrometry analysis. We have shown that buffer B allowed an optimal protein recovery between RCL2/CS100-fixed and frozen tissues. Interestingly, this buffer, containing 0.1% RapiGest, precludes subsequent purification or dilution of the protein extracts prior to trypsin digestion and nanoRPLC-MS/MS analysis, which could compromise the analysis of a small amount of tissue, such as from pinch or core biopsies. Hwang et al. showed a similar result with a RapiGest buffer with an extraction efficiency 77% higher than RIPA buffer, while an acetonitrile buffer extracted 47% of proteins 13. When using buffer B as a common protein extraction procedure, overall protein quantities recovered from fresh-frozen and RCL2/CS100-fixed samples were comparable.
Using a bottom-up proteomic strategy based on nanoRPLC-MS/MS for the same tissue types, 49% to 70% of proteins were found to be common in the RCL2/CS100 and frozen conditions. From both tissue storage conditions, we could identify proteins from several cellular compartments with different cellular functions. Among these, several were found to be tissue-specific, such as mucin-2 and beta catenin for colonic mucosa tissue 36, 37, the prostatic acid phosphatase and clusterin for prostate tissue 38, 39, and periostin and prohibitin 2 for breast tissue 40, 41. Recently, similar analytical approaches have been developed for the proteomic investigation of formalin-fixed paraffin-embedded tissue specimens 9-16. Although these approaches are relevant, one may be concerned by the possibility of introducing biases related to the effects of formalin crosslinking on protein extraction and MS analysis. Specifically, these modifications can be carried by the resulting tryptic peptides and render them unidentifiable using shotgun LC-MS approaches.
We have also demonstrated that for breast cancer cells, the combination of laser cell microdissection with nanoRPLC-MS/MS from RCL2/CS100-fixed tissues produces the same amount of biological information as that recovered from whole RCL2/CS100-fixed tissues. Proteins recovered after LCM have been previously analyzed with different approaches, such as two-dimensional gel electrophoresis 29, 42, surface-enhanced laser desorption/ionization MS 43-45, and LC-MS/MS 9, 46 from fresh-frozen tissues and, in some cases, FFPE tissue samples 9-16. In our protocol, approximately 5,000 tumor cells per RCL2/CS100-fixed tissue sample were sufficient for nanoRPLC-MS/MS analysis. Although it may be possible to analyze a smaller number of cells 45, 47, 48, we have not reduced this number in order to maintain reproducible duplicate or triplicate analyses. In addition, since nanoRPLC-MS/MS analyses of laser captured cells from RCL2/CS100-fixed and frozen tissues gave similar results, ultrasensitive studies and quantitative comparisons of protein expression from selected and targeted tumor cells could be proposed for RCL2/CS100-archived tissues. This approach could be particularly interesting for the identification of potential new tumor markers in certain clinical tissue samples, such as ductal carcinoma in situ of the breast.
We have also demonstrated here the possibility of using RCL2/CS100-fixed tissue specimens for profiling and imaging mass spectrometry analyses. When compared to the results obtained from fresh-frozen tissue sections, the protein profiles recovered were very similar in terms of the protein signals detected and their relative intensities in the spectra. On average, however, signal intensities were lower than those observed from fresh-frozen tissues. The immediate result of this loss of sensitivity is that the overall number of peaks detected from RCL2/CS100-fixed tissue sections is on average a factor of two lower with respect to those observed from fresh-frozen sections. The presence of a reduced signal after RCL2/CS100 fixation, in particular for hemoglobin, can be tentatively explained by a primary infiltration of the fixative within the biopsies via the circulatory system, resulting in its evacuation, or a greater susceptibility of some proteins to migration from the sections during paraffin removal and partial rehydration. From the imaging MS measurements performed from the fresh-frozen and RCL2/CS100-fixed colon tissue samples, a nice agreement of expression was observed for matching histologies. We therefore can conclude that RCL2/CS100-fixed tissue specimens are amenable to profiling and imaging mass spectrometry and generate good quality protein profiles and images. The sum of profiling and imaging MS results from RCL2/CS100-fixed tissues also agrees very well with previous results obtained from ethanol-only preserved tissues 30. Preservation by ethanol dehydration is also a non-crosslinking alcohol-based fixation approach that also offers high quality histology results and from which good quality localized protein information can be obtained by profiling and imaging MS. More generally, the possibility of investigating RCL2/CS100-fixed tissues by profiling and imaging mass spectrometry opens the door to analysis of clinical samples. In effect, diagnostic and prognostic information can be retrieved from protein profiles and images obtained from clinical biopsies as an aid to conventional histopathological evaluations and may help in the choice of a personalized therapeutic regime 32, 49, 50.
Conclusions
We have demonstrated here that tissue samples preserved by RCL2/CS100 fixation are amenable to proteomic studies with “bottom-up” strategies combining intact protein extraction, in-solution trypsin digestion, and subsequent liquid chromatography and mass spectrometry analyses for protein identification and with tissue MALDI imaging mass spectrometry. Consistent with our previous observations, we believe that RCL2/CS100 tissue fixation could thus be proposed as an alternative method for archiving clinical material especially for small, valuable, and rare target lesions that are usually inaccessible for complementary genomic, proteomic, and novel biomarker discovery investigations.
Supplementary Material
Acknowledgments
P.C. and R.M.C. acknowledge funding from the National Cancer Institute (grants # 5R33 CA116123-3, the National Institutes of Health (grant # 5R01 GM058008-10) and the Dept of Defense (grant # W81XWH0510179). J.S. acknowledges funding from the GEFLUC-Montpellier. Laser Capture Microdissection analyses were performed with the LCM platform of the CHU Arnaud de Villeneuve, Dept of Cellular Biology, Montpellier, France. The authors are grateful to Dr X. Rebillard (Clinique Beausoleil, Montpellier, France), and Dr F. Bibeau (CRLC Val d'Aurelle, Dept of Pathology, Montpellier, France), which respectively provided the prostate, and the colon tissue samples used for this study. We are also grateful for F. Boissière and A. Denouel for their technical assistance.
Abbreviations used
- ACN
acetonitrile
- Da
Dalton
- DTT
dithiothreitol
- kDa
kilodalton
- LC
liquid chromatography
- MALDI
matrix-assisted laser desorption/ionization
- MS
mass spectrometry
- m/z
mass-to-charge ratio
- SDS
sodium dodecyl sulfate
- TFA
trifluoroacetic acid
- TOF
time-of-flight
Footnotes
Supporting Information Available: Table listing proteins identified; figure of SDS-PAGE gels of extracted proteins; figure of comparison of selected MS/MS spectra; figure of comparison of the MALDI MS protein profiles. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Bellet V, Boissiere F, Bibeau F, Desmetz C, Berthe M, Rochaix P, Maudelonde T, Mange A, Solassol J. Proteomic analysis of RCL2 paraffin-embedded tissues. J Cell Mol Med. 2008;12(5B):2027–2036. doi: 10.1111/j.1582-4934.2008.00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fetsch PA, Simone NL, Bryant-Greenwood PK, Marincola FM, Filie AC, Petricoin EF, Liotta LA, Abati A. Proteomic evaluation of archival cytologic material using SELDI affinity mass spectrometry: potential for diagnostic applications. Am J Clin Pathol. 2002;118(6):870–6. doi: 10.1309/EJKL-7328-KFPR-56WA. [DOI] [PubMed] [Google Scholar]
- 3.Gillespie JW, Best CJ, Bichsel VE, Cole KA, Greenhut SF, Hewitt SM, Ahram M, Gathright YB, Merino MJ, Strausberg RL, Epstein JI, Hamilton SR, Gannot G, Baibakova GV, Calvert VS, Flaig MJ, Chuaqui RF, Herring JC, Pfeifer J, Petricoin EF, Linehan WM, Duray PH, Bova GS, Emmert-Buck MR. Evaluation of non-formalin tissue fixation for molecular profiling studies. Am J Pathol. 2002;160(2):449–57. doi: 10.1016/S0002-9440(10)64864-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27(22):4436–43. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehmann U, Kreipe H. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods. 2001;25(4):409–18. doi: 10.1006/meth.2001.1263. [DOI] [PubMed] [Google Scholar]
- 6.Cronin M, Pho M, Dutta D, Stephans JC, Shak S, Kiefer MC, Esteban JM, Baker JB. Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol. 2004;164(1):35–42. doi: 10.1016/S0002-9440(10)63093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godfrey TE, Kim SH, Chavira M, Ruff DW, Warren RS, Gray JW, Jensen RH. Quantitative mRNA expression analysis from formalin-fixed, paraffin-embedded tissues using 5′ nuclease quantitative reverse transcription-polymerase chain reaction. J Mol Diagn. 2000;2(2):84–91. doi: 10.1016/S1525-1578(10)60621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Specht K, Richter T, Muller U, Walch A, Werner M, Hofler H. Quantitative gene expression analysis in microdissected archival formalin-fixed and paraffin-embedded tumor tissue. Am J Pathol. 2001;158(2):419–29. doi: 10.1016/S0002-9440(10)63985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bagnato C, Thumar J, Mayya V, Hwang SI, Zebroski H, Claffey KP, Haudenschild C, Eng JK, Lundgren DH, Han DK. Proteomics analysis of human coronary atherosclerotic plaque: a feasibility study of direct tissue proteomics by liquid chromatography and tandem mass spectrometry. Mol Cell Proteomics. 2007;6(6):1088–102. doi: 10.1074/mcp.M600259-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Crockett DK, Lin Z, Vaughn CP, Lim MS, Elenitoba-Johnson KS. Identification of proteins from formalin-fixed paraffin-embedded cells by LC-MS/MS. Lab Invest. 2005;85(11):1405–15. doi: 10.1038/labinvest.3700343. [DOI] [PubMed] [Google Scholar]
- 11.Guo T, Wang W, Rudnick PA, Song T, Li J, Zhuang Z, Weil RJ, DeVoe DL, Lee CS, Balgley BM. Proteome analysis of microdissected formalin-fixed and paraffin-embedded tissue specimens. J Histochem Cytochem. 2007;55(7):763–72. doi: 10.1369/jhc.7A7177.2007. [DOI] [PubMed] [Google Scholar]
- 12.Hood BL, Darfler MM, Guiel TG, Furusato B, Lucas DA, Ringeisen BR, Sesterhenn IA, Conrads TP, Veenstra TD, Krizman DB. Proteomic analysis of formalin-fixed prostate cancer tissue. Mol Cell Proteomics. 2005;4(11):1741–53. doi: 10.1074/mcp.M500102-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Hwang SI, Thumar J, Lundgren DH, Rezaul K, Mayya V, Wu L, Eng J, Wright ME, Han DK. Direct cancer tissue proteomics: a method to identify candidate cancer biomarkers from formalin-fixed paraffin-embedded archival tissues. Oncogene. 2007;26(1):65–76. doi: 10.1038/sj.onc.1209755. [DOI] [PubMed] [Google Scholar]
- 14.Palmer-Toy DE, Krastins B, Sarracino DA, Nadol JB, Jr, Merchant SN. Efficient method for the proteomic analysis of fixed and embedded tissues. J Proteome Res. 2005;4(6):2404–11. doi: 10.1021/pr050208p. [DOI] [PubMed] [Google Scholar]
- 15.Patel V, Hood BL, Molinolo AA, Lee NH, Conrads TP, Braisted JC, Krizman DB, Veenstra TD, Gutkind JS. Proteomic analysis of laser-captured paraffin-embedded tissues: a molecular portrait of head and neck cancer progression. Clin Cancer Res. 2008;14(4):1002–14. doi: 10.1158/1078-0432.CCR-07-1497. [DOI] [PubMed] [Google Scholar]
- 16.Balgley BM, Guo T, Zhao K, Fang X, Tavassoli FA, Lee CS. Evaluation of archival time on shotgun proteomics of formalin-fixed and paraffin-embedded tissues. J Proteome Res. 2009;8(2):917–25. doi: 10.1021/pr800503u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemaire R, Desmons A, Tabet JC, Day R, Salzet M, Fournier I. Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. J Proteome Res. 2007;6(4):1295–305. doi: 10.1021/pr060549i. [DOI] [PubMed] [Google Scholar]
- 18.Ronci M, Bonanno E, Colantoni A, Pieroni L, Di Ilio C, Spagnoli LG, Federici G, Urbani A. Protein unlocking procedures of formalin-fixed paraffin-embedded tissues: application to MALDI-TOF imaging MS investigations. Proteomics. 2008;8(18):3702–14. doi: 10.1002/pmic.200701143. [DOI] [PubMed] [Google Scholar]
- 19.Stauber J, Lemaire R, Franck J, Bonnel D, Croix D, Day R, Wisztorski M, Fournier I, Salzet M. MALDI imaging of formalin-fixed paraffin-embedded tissues: application to model animals of Parkinson disease for biomarker hunting. J Proteome Res. 2008;7(3):969–78. doi: 10.1021/pr070464x. [DOI] [PubMed] [Google Scholar]
- 20.Nirmalan NJ, Harnden P, Selby PJ, Banks RE. Development and validation of a novel protein extraction methodology for quantitation of protein expression in formalin-fixed paraffin-embedded tissues using western blotting. J Pathol. 2009;217(4):497–506. doi: 10.1002/path.2504. [DOI] [PubMed] [Google Scholar]
- 21.Kilburn KH, Warshaw R, Boylen CT, Johnson SJ, Seidman B, Sinclair R, Takaro T., Jr Pulmonary and neurobehavioral effects of formaldehyde exposure. Arch Environ Health. 1985;40(5):254–60. doi: 10.1080/00039896.1985.10545928. [DOI] [PubMed] [Google Scholar]
- 22.Morgan KT. A brief review of formaldehyde carcinogenesis in relation to rat nasal pathology and human health risk assessment. Toxicol Pathol. 1997;25(3):291–307. doi: 10.1177/019262339702500307. [DOI] [PubMed] [Google Scholar]
- 23.Lewis F, Maughan NJ, Smith V, Hillan K, Quirke P. Unlocking the archive--gene expression in paraffin-embedded tissue. J Pathol. 2001;195(1):66–71. doi: 10.1002/1096-9896(200109)195:1<66::AID-PATH921>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 24.Morales AR, Essenfeld H, Essenfeld E, Duboue MC, Vincek V, Nadji M. Continuous-specimen-flow, high-throughput, 1-hour tissue processing. A system for rapid diagnostic tissue preparation. Arch Pathol Lab Med. 2002;126(5):583–90. doi: 10.5858/2002-126-0583-CSFHTH. [DOI] [PubMed] [Google Scholar]
- 25.Shibutani M, Uneyama C, Miyazaki K, Toyoda K, Hirose M. Methacarn fixation: a novel tool for analysis of gene expressions in paraffin-embedded tissue specimens. Lab Invest. 2000;80(2):199–208. doi: 10.1038/labinvest.3780023. [DOI] [PubMed] [Google Scholar]
- 26.Stanta G, Mucelli SP, Petrera F, Bonin S, Bussolati G. A novel fixative improves opportunities of nucleic acids and proteomic analysis in human archive's tissues. Diagn Mol Pathol. 2006;15(2):115–23. doi: 10.1097/00019606-200606000-00009. [DOI] [PubMed] [Google Scholar]
- 27.Takagi H, Shibutani M, Kato N, Fujita H, Lee KY, Takigami S, Mitsumori K, Hirose M. Microdissected region-specific gene expression analysis with methacarn-fixed, paraffin-embedded tissues by real-time RT-PCR. J Histochem Cytochem. 2004;52(7):903–13. doi: 10.1369/jhc.3A6215.2004. [DOI] [PubMed] [Google Scholar]
- 28.Vincek V, Nassiri M, Nadji M, Morales AR. A tissue fixative that protects macromolecules (DNA, RNA, and protein) and histomorphology in clinical samples. Lab Invest. 2003;83(10):1427–35. doi: 10.1097/01.lab.0000090154.55436.d1. [DOI] [PubMed] [Google Scholar]
- 29.Ahram M, Flaig MJ, Gillespie JW, Duray PH, Linehan WM, Ornstein DK, Niu S, Zhao Y, Petricoin EF, 3rd, Emmert-Buck MR. Evaluation of ethanol-fixed, paraffin-embedded tissues for proteomic applications. Proteomics. 2003;3(4):413–21. doi: 10.1002/pmic.200390056. [DOI] [PubMed] [Google Scholar]
- 30.Chaurand P, Latham JC, Lane KB, Mobley JA, Polosukhin VV, Wirth PS, Nanney LB, Caprioli RM. Imaging mass spectrometry of intact proteins from alcohol-preserved tissue specimens: bypassing formalin fixation. J Proteome Res. 2008;7(8):3543–55. doi: 10.1021/pr800286z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delfour C, Roger P, Bret C, Berthe ML, Rochaix P, Kalfa N, Raynaud P, Bibeau F, Maudelonde T, Boulle N. RCL2, a new fixative, preserves morphology and nucleic acid integrity in paraffin-embedded breast carcinoma and microdissected breast tumor cells. J Mol Diagn. 2006;8(2):157–69. doi: 10.2353/jmoldx.2006.050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seeley EH, Oppenheimer SR, Mi D, Chaurand P, Caprioli RM. Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections. J Am Soc Mass Spectrom. 2008;19(8):1069–77. doi: 10.1016/j.jasms.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aerni HR, Cornett DS, Caprioli RM. Automated acoustic matrix deposition for MALDI sample preparation. Anal Chem. 2006;78(3):827–34. doi: 10.1021/ac051534r. [DOI] [PubMed] [Google Scholar]
- 34.Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM. New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. J Proteome Res. 2006;5(11):2889–900. doi: 10.1021/pr060346u. [DOI] [PubMed] [Google Scholar]
- 35.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods. 2007;4(10):828–33. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 36.Gum JR, Jr, Hicks JW, Toribara NW, Siddiki B, Kim YS. Molecular cloning of human intestinal mucin (MUC2) cDNA. Identification of the amino terminus and overall sequence similarity to prepro-von Willebrand factor. J Biol Chem. 1994;269(4):2440–6. [PubMed] [Google Scholar]
- 37.Huiping C, Kristjansdottir S, Jonasson JG, Magnusson J, Egilsson V, Ingvarsson S. Alterations of E-cadherin and beta-catenin in gastric cancer. BMC Cancer. 2001;1:16. doi: 10.1186/1471-2407-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cunha AC, Weigle B, Kiessling A, Bachmann M, Rieber EP. Tissue-specificity of prostate specific antigens: comparative analysis of transcript levels in prostate and non-prostatic tissues. Cancer Lett. 2006;236(2):229–38. doi: 10.1016/j.canlet.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 39.Scaltriti M, Bettuzzi S, Sharrard RM, Caporali A, Caccamo AE, Maitland NJ. Clusterin overexpression in both malignant and nonmalignant prostate epithelial cells induces cell cycle arrest and apoptosis. Br J Cancer. 2004;91(10):1842–50. doi: 10.1038/sj.bjc.6602193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dell'Orco RT, Jupe ER, Manjeshwar S, Liu XT, McClung JK, King R, Hollingsworth A, Lightfoot S, Kern W. Prohibitin: A New Biomarker for Breast Tumors. Breast J. 1997;3(2):85–89. [Google Scholar]
- 41.Shao R, Bao S, Bai X, Blanchette C, Anderson RM, Dang T, Gishizky ML, Marks JR, Wang XF. Acquired expression of periostin by human breast cancers promotes tumor angiogenesis through up-regulation of vascular endothelial growth factor receptor 2 expression. Mol Cell Biol. 2004;24(9):3992–4003. doi: 10.1128/MCB.24.9.3992-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng AL, Huang WG, Chen ZC, Zhang PF, Li MY, Li F, Li JL, Li C, Yi H, Peng F, Duan CJ, Xiao ZQ. Identificating cathepsin D as a biomarker for differentiation and prognosis of nasopharyngeal carcinoma by laser capture microdissection and proteomic analysis. J Proteome Res. 2008;7(6):2415–26. doi: 10.1021/pr7008548. [DOI] [PubMed] [Google Scholar]
- 43.Melle C, Kaufmann R, Hommann M, Bleul A, Driesch D, Ernst G, von Eggeling F. Proteomic profiling in microdissected hepatocellular carcinoma tissue using ProteinChip technology. Int J Oncol. 2004;24(4):885–91. [PubMed] [Google Scholar]
- 44.Nakagawa T, Huang SK, Martinez SR, Tran AN, Elashoff D, Ye X, Turner RR, Giuliano AE, Hoon DS. Proteomic profiling of primary breast cancer predicts axillary lymph node metastasis. Cancer Res. 2006;66(24):11825–30. doi: 10.1158/0008-5472.CAN-06-2337. [DOI] [PubMed] [Google Scholar]
- 45.Wellmann A, Wollscheid V, Lu H, Ma ZL, Albers P, Schutze K, Rohde V, Behrens P, Dreschers S, Ko Y, Wernert N. Analysis of microdissected prostate tissue with ProteinChip arrays--a way to new insights into carcinogenesis and to diagnostic tools. Int J Mol Med. 2002;9(4):341–7. [PubMed] [Google Scholar]
- 46.Guo J, Colgan TJ, DeSouza LV, Rodrigues MJ, Romaschin AD, Siu KW. Direct analysis of laser capture microdissected endometrial carcinoma and epithelium by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom. 2005;19(19):2762–6. doi: 10.1002/rcm.2119. [DOI] [PubMed] [Google Scholar]
- 47.Palmer-Toy DE, Sarracino DA, Sgroi D, LeVangie R, Leopold PE. Direct acquisition of matrix-assisted laser Desorption/Ionization time-of-flight mass spectra from laser capture microdissected tissues. Clin Chem. 2000;46(9):1513–6. [PubMed] [Google Scholar]
- 48.Xu BJ, Caprioli RM, Sanders ME, Jensen RA. Direct analysis of laser capture microdissected cells by MALDI mass spectrometry. J Am Soc Mass Spectrom. 2002;13(11):1292–7. doi: 10.1016/S1044-0305(02)00644-X. [DOI] [PubMed] [Google Scholar]
- 49.Chaurand P, Sanders ME, Jensen RA, Caprioli RM. Proteomics in diagnostic pathology: profiling and imaging proteins directly in tissue sections. Am J Pathol. 2004;165(4):1057–68. doi: 10.1016/S0002-9440(10)63367-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaurand P, Schwartz SA, Reyzer ML, Caprioli RM. Imaging mass spectrometry: principles and potentials. Toxicol Pathol. 2005;33(1):92–101. doi: 10.1080/01926230590881862. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez-Pinilla SM, Sarrio D, Honrado E, Hardisson D, Calero F, Benitez J, Palacios J. Prognostic significance of basal-like phenotype and fascin expression in node-negative invasive breast carcinomas. Clin Cancer Res. 2006;12(5):1533–9. doi: 10.1158/1078-0432.CCR-05-2281. [DOI] [PubMed] [Google Scholar]
- 52.Yoder BJ, Tso E, Skacel M, Pettay J, Tarr S, Budd T, Tubbs RR, Adams JC, Hicks DG. The expression of fascin, an actin-bundling motility protein, correlates with hormone receptor-negative breast cancer and a more aggressive clinical course. Clin Cancer Res. 2005;11(1):186–92. [PubMed] [Google Scholar]
- 53.Jadeski L, Mataraza JM, Jeong HW, Li Z, Sacks DB. IQGAP1 stimulates proliferation and enhances tumorigenesis of human breast epithelial cells. J Biol Chem. 2008;283(2):1008–17. doi: 10.1074/jbc.M708466200. [DOI] [PubMed] [Google Scholar]
- 54.Sakurai-Yageta M, Recchi C, Le Dez G, Sibarita JB, Daviet L, Camonis J, D'Souza-Schorey C, Chavrier P. The interaction of IQGAP1 with the exocyst complex is required for tumor cell invasion downstream of Cdc42 and RhoA. J Cell Biol. 2008;181(6):985–98. doi: 10.1083/jcb.200709076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rayner K, Chen YX, Hibbert B, White D, Miller H, Postel EH, O'Brien ER. Discovery of NM23-H2 as an estrogen receptor beta-associated protein: role in estrogen-induced gene transcription and cell migration. J Steroid Biochem Mol Biol. 2008;108(1-2):72–81. doi: 10.1016/j.jsbmb.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 56.Montagna C, Lyu MS, Hunter K, Lukes L, Lowther W, Reppert T, Hissong B, Weaver Z, Ried T. The Septin 9 (MSF) gene is amplified and overexpressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer Res. 2003;63(9):2179–87. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.