Abstract
Background and Aims
To assess the number and phylogenetic distribution of large-scale genome duplications in the ancestry of Actinidia, publicly available expressed sequenced tags (ESTs) for members of the Actinidiaceae and related Ericales, including tea (Camellia sinensis), were analysed.
Methods
Synonymous divergences (Ks) were calculated for all duplications within gene families and examined for evidence of large-scale duplication events. Phylogenetic comparisons for a selection of orthologues among several related species in Ericales and two outgroups permitted placement of duplication events in relation to lineage divergences. Gene ontology (GO) categories were analysed for each whole-genome duplication (WGD) and the whole transcriptome.
Key Results
Evidence for three ancient WGDs in Actinidia was found. Analyses of paleologue GO categories indicated a different pattern of retained genes for each genome duplication, but a pattern consistent with the dosage-balance hypothesis among all retained paleologues.
Conclusions
This study provides evidence for one independent WGD in the ancestry of Actinidia (Ad-α), a WGD shared by Actinidia and Camellia (Ad-β), and the well-established At-γ WGD that occurred prior to the divergence of all taxa examined. More ESTs in other taxa are needed to elucidate which groups in Ericales share the Ad-β or Ad-α duplications and their impact on diversification.
Keywords: Paleopolyploidy, Actinidiaceae, Ericales, Actinidia, Camellia, kiwi, genome duplication, dosage balance
INTRODUCTION
The importance of polyploidy, or whole genome duplication, in plant evolution has been long recognized by researchers. Nearly 35 % of flowering plants are of recent polyploid provenance and at least 15 % of angiosperm speciation events are caused by whole genome duplication (Wood et al., 2009). Using recently developed genomic approaches, several ancient genome duplication events have also been inferred during the evolution of flowering plants (Blanc and Wolfe, 2004; Cui et al., 2006; Barker et al., 2008; Soltis et al., 2009). Recent polyploids are easy to detect by changes in chromosome numbers, genome size and gene copy number compared with progenitors, but ancient polyploids, or paleopolyploids, are much harder to identify because diploidization, gene loss and chromosomal rearrangements erode the signal. Despite the obfuscating action of these forces, paleopolyploidy may still be inferred by recognition of homoeologous chromosomes or large bursts of gene duplication (Barker and Wolf, 2010).
Although completely sequenced and assembled nuclear genomes provide the ultimate resource for inferring paleopolyploidy, the identification of peaks of gene duplications in expressed sequenced tags (ESTs) provides an economical method to survey ancient polyploidy. The thousands of ESTs available for many plants provide a useful ‘snapshot’ of each genome. Large-scale duplication events lead to a punctuated, dramatic increase in the number of duplicated genes (Lynch and Connery, 2000; Blanc and Wolfe, 2004). The resulting excess of paralogues of a particular age produces a peak in the age distribution of duplications across gene families within a genome. This approach has been successfully employed in a variety of plants including wheat (Blanc and Wolfe, 2004), maize (Blanc and Wolfe, 2004; Schlueter et al., 2004), Solanum (Schlueter et al., 2004; Blanc and Wolfe, 2004; Cui et al., 2006), Populus (Sterck et al., 2005), Arabidopsis (Blanc and Wolfe, 2004; Maere et al., 2005; Barker et al., 2009), lettuce (Barker et al., 2008) and sunflower (Barker et al., 2008). Researchers have begun to elucidate the phylogenetic position of paleopolyploidizations in relation to lineage divergence by combining genomic and phylogenetic methods in the Fabaceae (Pfeil et al., 2005), Compositae (Barker et al., 2008) and Brassicales (Bowers et al., 2003; Schranz and Mitchell-Olds, 2006; Tang et al., 2008; Barker et al., 2009).
The genus Actinidia, well known as kiwifruit, contains 76 species of climbing plants originating mainly in China (Huang and Ferguson, 2007). Over the past three decades, kiwifruit has developed into an important horticultural cash crop and a fruit industry worldwide (Huang and Ferguson, 2007). Recently, a collection of 132 577 ESTs in Actinidia were sequenced and publicly released (Crowhurst et al., 2008). Comparative cytological studies among the three extant genera of the Actinidiaceae – Saurauia (x = 13), Clematoclethra (x = 12) and Actinidia (x = 29) – suggest that Actinidia is a paleotetraploid derived from an ancestor with x = 14 (He et al., 2005). However, no other study has confirmed this hypothesis. Here, publicly available sources of ESTs are used for Actinidia, several related genera, such as Camellia and Diospyros in the Ericales and Populus and Vitis as outgroups to (a) identify ancient genome duplications in Actinidia and other Ericales, (b) place genome duplications onto the current phylogeny, (c) analyse the gene ontology (GO) patterns of retained duplicates from putative whole-genome duplications (WGDs).
MATERIALS AND METHODS
Unigene assembly
EST collections of three Actinidia species (A. chinensis, 47 380 ESTs; A. deliciosa, 57 752 ESTs; A. eriantha, 12 648 ESTs), Camellia sinensis (10 431 ESTs) and Diospyros kaki (9475 ESTs) were downloaded from GenBank. Simulations (Cui et al., 2006) have indicated that species with 10 000 or more ESTs are sufficient for inferring ancient polyploidy, and the analysed Actinidia and Camellia data are beyond this threshold. Annotated coding sequences (cds) from the whole genome sequences of Populus trichocarpa (v1·1, http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html) (Tuskan et al., 2006) and Vitis vinifera (v1, http://www.genoscope.cns.fr/spip/Vitis-vinifera-whole-genome.html) (Jaillon et al., 2007) were downloaded from their project sites. For the EST reads, vector and low quality sequences were removed using Seqclean with the UniVec contaminant database (http://www.ncbi.nlm.nih.gov/VecScreen/UniVec.html). Contigs were assembled for each EST collection by TGICL with default settings (Quackenbush et al., 2000), and a unigene file containing assembled contigs and singletons was created. Raw reads and assembled unigenes are available at http://msbarker.com and http://biotorrents.net.
Gene family construction and Ks calculation
For each annotated cds or assembled unigene collection, gene families were identified and their duplications, in terms of substitutions per synonymous site (Ks), was calculated. Duplicate pairs were identified as sequences that demonstrated 40 % sequence similarity over at least 300 bp from a discontinguous all-against-all MegaBLAST (Zhang et al., 2000; Ma et al., 2002). Pairs of genes containing identifiable transposable elements were removed from the analysis because duplication resulting from transposition may obscure a signal from paleopolyploidy. To reduce the possibility that identical genes are represented in the data set, but missed by the TGICL clustering because of alternative splicing, all Ks values from one member of a duplicate pair with Ks = 0 were removed. Further, to reduce the multiplicative effects of multicopy gene families on Ks values, phylogenies for each gene family were constructed by single linkage clustering (Blanc and Wolfe, 2004), and node Ks values calculated. Node Ks values <2 were used in subsequent analyses.
Calculations of the synonymous divergence among gene copies were based upon protein-guided DNA alignments of gene families. Each duplicated gene was searched against all plant proteins available on GenBank (Wheeler et al., 2007) using BLASTX (Altschul et al. 1997). Best-hit proteins were paired with each gene at a minimum cutoff of 30 % sequence similarity over at least 150 sites. Genes that did not have a best-hit protein at this level were removed before further analyses. To determine reading frame and generate estimated amino acid sequences, each gene was aligned against its best-hit protein by Genewise 2.2.2 (Birney et al., 1996). Using the highest scoring Genewise DNA–protein alignments, custom Perl scripts were used to remove stop and ‘N’-containing codons and produce estimated amino acid sequences for each gene. Amino acid sequences for each duplicate pair were then aligned using MUSCLE 3·6 (Edgar, 2004). The aligned amino acids were subsequently used to align their corresponding DNA sequences using RevTrans 1·4 (Wernersson and Pedersen, 2003). Ks values for each duplicate pair were calculated using the maximum likelihood method implemented in codeml of the PAML package (Yang 1997) under the F3x4 model (Goldman and Yang, 1994).
Identification of paleopolyploidy
Two statistical tests were employed to identify significant features in the age distribution. A bootstrapped K-S goodness-of-fit test was used (Cui et al., 2006) to assess if the overall age distributions deviated from a simulated null. Taxa that significantly deviated from the null were then analysed using a mixture model. A mixture model of normal distributions was fit to the age distribution data by maximum likelihood using the EMMIX package (McLachlan et al., 1999). Peaks produced by paleopolyploidy are expected to be approximately Gaussian (Schlueter et al., 2004; Blanc and Wolfe, 2004), and the mixture model identifies the number of normal distributions and their position(s) that best explain the observed age distributions. For the mixture model analyses, one to ten normal distributions were fitted to the data with 1000 random starts and 100 k-mean starts. The Bayesian Information Criterion (BIC) was used to select the best model because it more strongly penalizes increasing the number of model parameters than the Akaike Information Criterion and should be more robust against fitting insignificant distributions.
Phylogenetic placement of ancient duplications and age estimation
To place duplications in relative phylogenetic context and account for substitution rate heterogeneity among members of the Ericales, Ks values for each lineage were corrected using relative rate corrections based on Ks branch length ratios. A representative of each Ericales lineage with genomic data was included along with two outgroups (Populus and Vitis) to calculate Ks branch lengths of orthologues across a constrained topology in PAML. Putative orthologues were identified as reciprocal best blast hits among these taxa with at least 300 bp alignment overlap (Table S2 in Supplementary data, available online). Using these orthologues, Ks branch lengths were calculated for each gene in the Ericales ingroup across a constrained topology (Anderberg et al., 2002). For each orthologue phylogeny, the ratios of branch lengths for Actinidia deliciosa and Camellia sinensis versus Diospyros kaki were calculated. The mean ratio over all orthologues for each lineage was applied as a relative rate correction to the Ks values for their respective taxa. To assess if duplications occurred after lineage divergence, one-sided Wilcoxon Rank Sum tests were used to compare the distribution of rate-corrected duplications versus the distribution of rate-corrected lineage divergences from each orthologue phylogeny.
Ages for ancient genome duplications were estimated from the mean synonymous divergence of Camellia and Actinidia. Using the maximum likelihood age estimate for the divergence time of Camellia and Actinidia from Wikström et al. (2001) – 71 MYA – and the mean number of synonymous substitutions among the nuclear orthologues found above, the synonymous substitution rate per million years was calculated as in Gaut and Doebley (1997). The age of each Ericales ancient genome duplication was then estimated from the data of Actinidia deliciosa, the species with the most complete data set, using this calibrated substitution rate based on the median peak Ks from the mixture model analyses.
Gene retention pattern analyses
GO annotations of the Actinidia (Actinidia chinensis, A. deliciosa, A. eriantha, A. arguta, A. hemsleyana, A. polygama and A. setosa) transcriptome from bud, fruit, leaf, petal, root and stem were obtained through discontiguous MegaBlast searches against Arabidopsis thaliana cds from TAIR (Initiative TAG, 2000) for the best hit with at least 100 bp aligned and an e value of 1e – 10. To ensure comprehensive coverage of the Actinidia transcriptome, all EST reads for each Actinidia species were pooled into a single assembly with TGICL, analysed with the above duplication pipeline, and annotated with the TAIR cds. To identify significant differences among GO annotations, chi-square tests with P values computed from 100 000 Monte Carlo simulations were conducted in R (R Development Core Team, 2005). When chi-square tests were significant (P < 0·05), GO categories with residuals >|2| were implicated as major contributors to the significant chi-square statistic. This statistical framework was used to evaluate differences in GO category representation patterns between paleologues (paralogues derived from paleopolyploidy) and non-paleologues (paralogues not derived from paleopolyploidy) in Actinidia. Expander (Shamir et al., 2005) was employed to cluster different GO category patterns of normalized number of paleologues and non-paleologues using the Complete Linkage Clustering (default options). Boundaries for each whole genome duplication were defined by the mixture model results. Duplications in the region of overlap between two distributions were assigned to a particular whole genome duplication based on their probability assignment from the mixture model analysis.
RESULTS
Age distributions of gene duplications
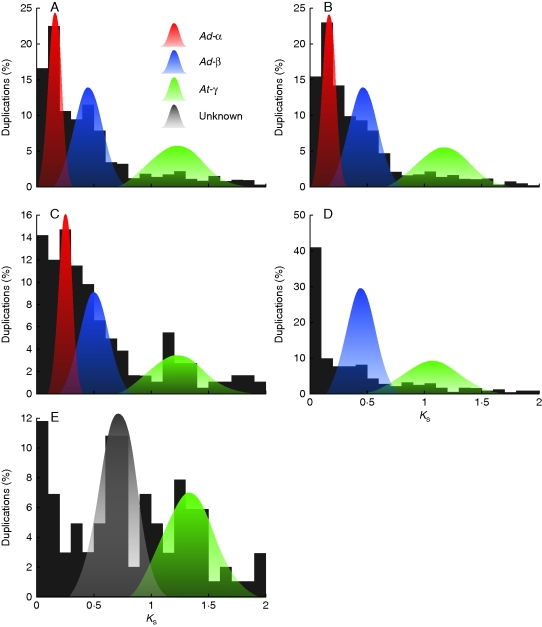
A total of 2916 gene duplications younger than Ks = 2 were inferred across the total data set of 49 715 assembled unigenes (Table S1 in Supplementary data, available online). The histograms of duplication ages for each Ericales species analysed demonstrated evidence of at least one large-scale duplication (Fig. 1). Consistent with this observation, the K-S goodness-of-fit test rejected the null model of no large duplications for each taxon examined (P = 0). Subsequent mixture model analyses identified multiple peaks in the analysed Ericales taxa (Fig. 1, Table 1 and Table S1). In the genus Actinidia, the duplication distributions of A. chinensis, A. deliciosa and A. eriantha each contained evidence of three peaks of similar synonymous divergences. For example, in A. chinensis these peaks are located at median Ks of 0·13777, 0·4221 and 1·1923 (Fig. 1 and Table S1). The ΔBIC values indicate that models including these peaks describe the age distributions significantly better than models that lack these duplications (Tables 2 and S1). Similarly, evidence was found of two peaks in Camellia with Ks medians at 0·379355 and 1·038 (Tables 1 and S1). Although the mixture model for Diospyros does not identify significant peaks – probably because there are relatively few data – two peaks are apparent in the histogram near Ks = 0·7 and Ks = 1·3 (Fig. 1). The mixture model also identified one or two components in the duplication-rich initial peaks (<0·1) of all taxa that are likely to reflect variation in birth and death rates of tandem, small-scale, segmental duplications, or alleles.
Fig. 1.
Histograms of the age distribution of gene duplications from (A) Actinidia chinensis, (B) Actinidia deliciosa, (C) Actinidia eriantha, (D) Camellia sinensis and (E) Diospyros kaki. Shaded distributions represent mixture model fits of inferred whole genome duplications: red = Ad-α, blue = Ad-β, green = putative At-γ, and grey = possible duplication.
Table 1.
Rate-corrected mixture model medians of Actinidia and Camellia paleopolyploidizations
| Rate-corrected paleopolyploidization |
|||||
|---|---|---|---|---|---|
| Species | Relative rate (% Ks) | Ad-α | Ad-β | At-γ | BIC |
| Actinidia chinensis | 102 | 0·135069 | 0·413824 | 1·168922 | 350·9 |
| Actinidia deliciosa | 102 | 0·155794 | 0·418020 | 1·099020 | 229·9 |
| Actinidia eriantha | 102 | 0·184069 | 0·443118 | 1·205196 | 169·5 |
| Camellia sinensis | 89 | 0·426242 | 1·166292 | −17·8 | |
Table 2.
Mixture model ΔBIC values for Actinidia and Camellia age distributions without (w/o) the inferred paleopolyploidizations
| w/o (Ad-α) | w/o (Ad-β) | w/o (At-γ) | |
|---|---|---|---|
| Actinidia chinensis | 1183·54 | Fit by all models | 13·87 |
| Actinidia deliciosa | 745·40 | Fit by all models | 47·98 |
| Actinidia eriantha | 97·54 | Fit by all models | 14·41 |
| Camellia sinensis | NA | 380·68 | 8·58 |
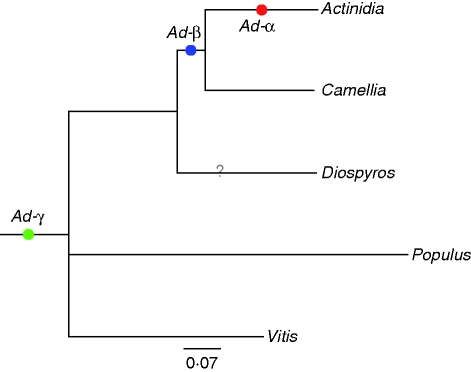
To resolve the number and phylogenetic placement of large-scale genome duplications, rate-corrected duplications were placed on the mean phylogeny of orthologues in Ericales (Fig. 2 and Tables 2 and S2). A total of 37 nuclear orthologues were identified among the analysed taxa. Mean ratios of Ks branch lengths for Actinidia and Camellia versus the Diospyros are 1·02 and 0·89, respectively (Table 2). Taking into account this rate heterogeneity, the mean rate-corrected divergence between Actinidia and Camellia was calculated as Ks = 0·4172, whereas Diospyros and these two genera diverged at Ks = 0·5247. After correcting the duplication peak medians with the appropriate ratio, the peaks in Actinida, named Ad-α and Ad-β, are centred at Ks = 0·16 and Ks = 0·42, respectively. Similarly, Camellia contains a peak centred at Ks = 0·42 after rate correction. Considering the placement of these duplications on the Ericales phylogeny, the youngest peak, Ad-α, occurred after the divergence of Actinidia and Camellia (U-test P < 1e – 5), but Ad-β most likely occurred prior to the divergence of these two lineages (U-test P = 1; Tables 1 and S2 and Fig. 2). The current estimates also place Ad-β after the divergence of Actinidia and Camellia from the clade containing Diospyros (U-test P <1e – 5). However, Diospyros may have a duplication near Ks = 0·75 that could be Ad-β, and additional data are needed to resolve better the incidence and position of duplications in this part of the phylogeny. Further, both Actinidia and Camellia have an older peak centred at Ks = 1·13 and Ks = 1·17, respectively, well before divergence of the Ericales and Populus or Vitis. This peak is also apparent in the histogram of Diospyros at Ks = 1·3. Taking into account the antiquity of this age range, this peak is likely to correspond to At-γ, an ancient polyploidy shared by most eudicots (Vision et al., 2000; Bowers et al., 2003; De Bodt et al., 2005; Cui et al., 2006; Jaillon et al., 2007; Barker et al., 2008, 2009; Lyons et al., 2008).
Fig. 2.
Phylogeny of Ericales taxa and related Rosid outgroups displaying inferred paleopolyploidizations. Branch lengths are mean Ks values from 37 nuclear orthologues (see Table S2, available online). Coloured dots indicate inferred paleopolyploidizations placed in relation to lineage divergence base on the rate corrections. The ‘?’ represents an ambiguous paleopolyploidization inferred from Diospyros ESTs.
Age estimates of the ancient genome duplications are consistent with the phylogenetic placements. Based on the mean synonymous divergence across the 37 nuclear orthologue phylogenies for Actinidia and Camellia (Table S2) a synonymous substitution rate of 2·81 × 10−9 is estimated. Based on the peak medians for A. deliciosa, the species whose orthologues were used to calculate the substitution rate, the ages of the Ericales genome duplications were calculated. Ad-α is estimated to have occurred approx. 28·3 MYA, whereas Ad-β is estimated to have occurred nearly 75·9 MYA.
Comparison of gene retention and loss patterns
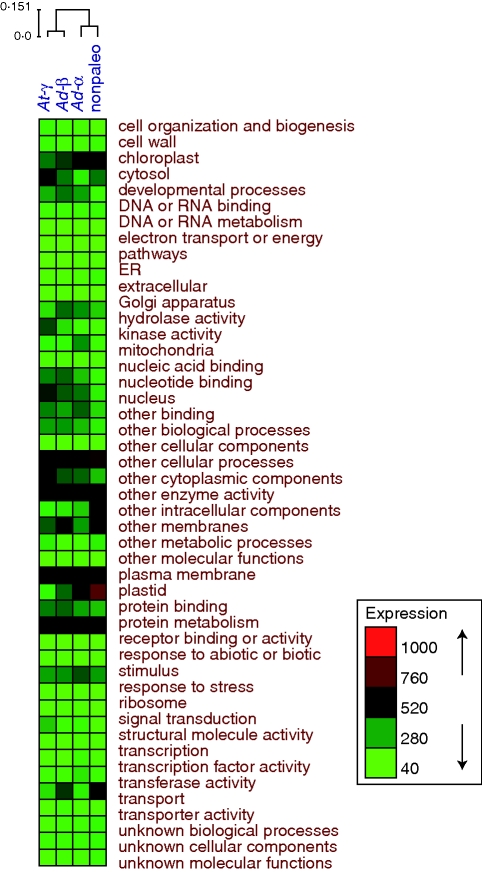
The GO patterns of genes retained in duplicate varied among the different large-scale duplication events. For the pooled Actinidia EST assembly, the GO patterns of genes retained in duplicate from the Ad-α, Ad-β and At-γ duplications were significantly different from each other (χ2 = 497·2, P = 1e – 5; Table S3). Hierarchical clustering of normalized GO slim data patterns showed that non-paleopolyploid duplications and Ad-α duplicates were grouped together while duplicates from Ad-β and At-γ were grouped together although they were all significantly different from each other (Fig. 3). The most consistent pattern observed was enrichment of ‘plastid’ and ‘chloroplast’ GO slim categories in the non-paleopolyploid and Ad-α duplicates with significant under-representation of these categories in duplicates from Ad-β, and At-γ. However, compared with the non-paleologues, analyses found that the pooled paleologues from all three duplications were enriched for the ‘developmental processes’, ‘hydrolase activity’ and ‘kinase activity’ GO categories with ‘electron transport’ and ‘structural molecular activity’ under-represented (Fig. 3 and Table S3).
Fig. 3.
GO annotations of Actinidia non-paleologues and paleologues. The colour enrichment represents the normalized unigene number of each GO slim category in the pooled Actinidia non-paleologues and paleologues. The GO slim category patterns among non-paleologues and paleologues were revealed by complete linkage hierarchical clustering. Boxes reflect relative level of GO category representation from low (green) to high (red).
DISCUSSION
Based on cytological analyses, botanists have long suggested that many plants have experienced ancient genome duplications and genomic data are now able to test these hypotheses. In angiosperms, a long-standing estimate for the original base chromosome number (x = 7) is much smaller than the accepted base numbers of many genera (Pires and Hertweck, 2008). One plausible explanation for this observation is past polyploidy. A classic example of this situation occurs in the Actinidiaceae where the genus Actinidia has long been suspected to have a polyploid ancestry (Huang and Ferguson, 2007). Previous cytological analyses of the base numbers for the three extant genera of the Actinidiaceae – Saurauia (x = 13), Clematoclethra (x = 12) and Actinidia (x = 29) – indicated that Actinidia was likely to be a paleotetraploid (He et al., 2005). The observation of peaks in the history of gene duplications in species of Actinidia is consistent with this hypothesis and provides the first genomic evidence of ancient polyploidy in the genus. The age estimate for Ad-α, 28·3 MYA, is consistent with this duplication being restricted to the Actinidiaceae but suggests it might be older than the genus. Additional data from the other two genera of the Actinidiaceae are needed to test further if the Ad-α duplication is restricted to the ancestry of Actinidia as expected from chromosomal analyses or is shared by more members of the family.
Recent cytological research on Actinidia and its relatives (reviewed in Huang and Ferguson, 2007) provides an excellent opportunity to evaluate genomic approaches for detecting ancient genome duplications. Examples such as Actinidia are critical because chromosomal diploidization has obscured evidence of past polyploidy in many groups. As in the Heliantheae (Barker et al., 2008), the same bioinformatic tools combined with modest amounts of transcriptome data sufficiently recovered evidence of paleopolyploidy consistent with previous research. Future phylogenomic analyses of the Actinidiaceae to place Ad-α on the phylogeny more precisely will provide a further test of this approach. Considering the decreasing cost of transcriptome sequencing and the coming explosion of genomic data, these natural examples are critical for selecting the best combination of data and methods for inferring ancient polyploidy across the eukaryote phylogeny.
Combined with ESTs of other genera from the Ericales available on GenBank evidence was found that Actinidia and Camellia have an older shared duplication, Ad-β. The rate-corrected phylogeny suggests that Ad-β occurred after the divergence of Diospyros from the clade containing Actinidia and Camellia. However, a peak near this position is apparent in the duplication distribution of Diospyros, but it is not clear if this is a whole genome duplication or Ad-β because there are relatively few data points and the signal is noisy. The estimated age of Ad-β, 75·9 MYA, is also inconclusive because Diospyros diverged from the clade containing Actinidia and Camellia at nearly the same time (Wikström et al., 2001). Additional transcriptome data from Diospyros and other Ericales are needed to confirm the location of Ad-β. It is worth noting that the currently proposed position of Ad-β is within the ancestry of a distinct clade of families that includes the Theaceae, Symplocaceae, Styracaceae, Diapensiaceae, Ericaceae and Actinidiaceae (Geuten et al., 2004). A recent study of the floral regulatory genes APETALA3 (AP3) and PISTILLATA (PI) genes found that the PI gene duplicated before the divergence of basal asteroid Ericales families but this duplication had not been shown in AP3 lineage (Viaene et al., 2009). It is possible this PI gene duplication is the result of Ad-β, but it cannot be ruled out that it was produced by random small-scale or segmental duplications. More genomic data from this clade are needed to examine which other families share the Ad-β duplication. These data will provide a valuable resource to test if this and possibly other paleopolyploidizations are correlated with the diversification of the Ericales as well as the association of duplications with shifts in floral regulatory gene counts and morphology.
In angiosperms, previous analyses have found the biased retention of some functional gene classes after large-scale duplication events. In particular, dosage-sensitive gene categories, such as those involved in signal transduction and transcriptional regulation, were preferentially retained after the three whole genome duplication events within the ancestor of Arabidopsis thaliana, whereas there was biased loss of these genes after small-scale duplication events (Blanc and Wolfe, 2004; Maere et al., 2005). These patterns of duplicate gene retention and loss have been described as support for the dosage-balance hypothesis (Birchler et al., 2007; Edger and Pires, 2009). According to this hypothesis, the stoichiometry of dosage-sensitive gene products must be maintained for the proper functioning of signalling networks or macromolecular complexes, especially those associated with regulatory processes (Edger and Pires, 2009). Although analyses of Arabidopsis paleologue support this hypothesis, results from other plant lineages have been less consistent with it. For example, the paleologues of the Compositae were over enriched for the GO categories ‘structural components’ and ‘cellular organization’ (Barker et al., 2008). In the moss Physcomitrella patens, GO and pathway analyses of the duplicated genes reveal different biases of gene retention compared with seed plants, and enriched GO categories all belong to the KEGG ontology (KO) class ‘metabolism’ (Rensing et al., 2007).
In the current study of Actinidia, no broad pattern of paleologue retention emerges from analyses of each duplication. Only non-paleologues and Ad-α shared some similarity of GO slim pattern in which the ‘chloroplast’ and ‘plastid’ categories are enriched whereas in Ad-β and At-γ these were reduced. However, the GO categories of non-paleologues and Ad-α may be confounded by the difficulty of separating genes from these two distributions because of the young age of Ad-α. A broad pattern of duplicate retention may also not emerge if the Actinidia transcriptome was not sequenced to sufficient depth to reveal a pattern. However, similar numbers of pooled transcriptome reads did reveal a consistent pattern among the Compositae (Barker et al., 2008). An alternative explanation is that independent loss of paralogues derived from each duplication event in Actinidia obscures an overall pattern and it may be more appropriate to consider all paleologues. In this case, when compared with non-paleologues, all the paleologues (pooled from the three WGDs) have GO categories such as ‘developmental processes’, ‘hydrolase activity’, ‘kinase activity’ over-represented. This result is much more consistent with previous analyses from Arabidopsis (Blanc and Wolfe, 2004; Maere et al., 2005) and the predictions of the dosage-balance hypothesis (Birchler et al., 2007; Edger and Pires, 2009). This result also suggests that previous analyses which did not support the dosage-balance hypothesis for paleologue retention, particularly Barker et al. (2008), should be re-evaluated in this manner. However, the significant GO category consistency of paleologues retained across the multiple duplications in the Compositae suggests that the result would not be drastically different (Barker et al., 2008). Regardless, future analyses of paleologue retention should examine the overall paleologues as well as the genes retained from individual duplications to provide a more complete picture of duplicate retention biases.
Future research on the Ericales, and Actinidia in particular, provides an outstanding opportunity to understand better the consequences of ancient genome duplication. Additional genomic from other Ericales will permit more precise placement of the ancient duplication events and a critical evaluation of whether Ad-β is associated with the K-T boundary as suggested for other duplications by Fawcett et al. (2009). Deeper transcriptome sequencing is needed in Actinidia to make phylogenetic and functional genomic comparisons of each gene family in the enriched GO categories among species of Actinidia and to reveal the fates of those duplicated genes in groups of Ericales which share the ancient duplications. Considering that ancient genome duplications account for approx. 75 % of duplicate genes in Actinidia and there is variation in ploidy among species and cultivars, the genus provides a unique opportunity to understand how paleologues may uniquely contribute to domestication, the evolution of morphological complexity, and the diversification of this unique group of Ericales.
SUPPLEMENTARY DATA
Supplementary Material
ACKNOWLEDGEMENTS
We thank The Horticultural and Food Research Institute of New Zealand and Max-Planck-Institute of Molecular Plant Physiology of Germany for providing the ESTs to the public. We also thank Professor Ying Wang from Wuhan Botanical Garden of CAS for productive discussions. M.S.B. is supported by the Natural Sciences and Engineering Research Council of Canada CREATE Training Program in Biodiversity Research and a Young International Scientist Fellowship from the Chinese Academy of Science (2009Y2BS3). T.S. and H.W.H. are supported by a key initiative grant of the Chinese Academy of Sciences (KSCX2-YW-N-061) and National Science Foundation of China grant (30771479) and the International Partnership Program for Creative Research Teams jointly funded by Chinese Academy of Sciences and State Administration of Foreign Experts Affairs. We also acknowledge Key Laboratory of Plant Resource Conservation and Sustainable Utilization, Chinese Academy of Sciences.
LITERATURE CITED
- Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderberg AA, Rydin C, Källersjö M. Phylogenetic relationships in the order Ericales s.l.: analyses of molecular data from five genes from the plastid and mitochondrial genomes. American Journal of Botany. 2002;89:677–687. doi: 10.3732/ajb.89.4.677. [DOI] [PubMed] [Google Scholar]
- Barker MS, Wolf PG. Unfurling fern biology in the genomics age. BioScience. 2010;60:177–185. [Google Scholar]
- Barker MS, Vogel H, Schranz ME. Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biology and Evolution. 2009;1:391–399. doi: 10.1093/gbe/evp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, Kane NC, Matvienko M, et al. Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Molecular Biology and Evolution. 2008;25:2445–2455. doi: 10.1093/molbev/msn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Thompson J, Gibson T. PairWise and SearchWise: finding the optimal alignment in a simultaneous comparison of a protein profile against all DNA translation frames. Nucleic Acids Research. 1996;24:2730–2739. doi: 10.1093/nar/24.14.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. The Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Yao H, Chudalayandi S. Biological consequences of dosage dependent gene regulatory systems. Biochemica et Biophysica Acta. 2007;1769:422–428. doi: 10.1016/j.bbaexp.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. Unraveling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Crowhurst RN, Gleave AP, Macrae EA, et al. Analysis of expressed sequence tags from Actinidia: applications of a cross species EST database for gene discovery in the areas of flavor, health, color and ripening. BMC Genomics. 2008;9:351+. doi: 10.1186/1471-2164-9-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Wall KP, Leebens-Mack JH, et al. Widespread genome duplications throughout the history of flowering plants. Genome Research. 2006;16:738–749. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Maere S, Van de Peer Y. Genome duplication and the origin of angiosperms. Trends in Ecology and Evolution. 2005;20:591–597. doi: 10.1016/j.tree.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acid Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, Pires JC. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Research. 2009;17:699–717. doi: 10.1007/s10577-009-9055-9. [DOI] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proceedings of the National Academy of Sciences of the USA. 2009;106:5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proceedings of the National Academy of Sciences of the USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Molecular Biology and Evolution. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Geuten K, Smets E, Schols P, et al. Conflicting phylogenies of balsaminoid families and the polytomy in Ericales: combining data in a Bayesian framework. Molecular Phylogenetics and Evolution. 2004;31:711–729. doi: 10.1016/j.ympev.2003.09.014. [DOI] [PubMed] [Google Scholar]
- He ZC, Li JQ, Cai Q, Wang Q. The cytology of Actinidia, Saurauia and Clematoclethra (Actinidiaceae) Botanical Journal of the Linnean Society. 2005;147:369–374. [Google Scholar]
- Huang HW, Ferguson AR. Genetic resources of kiwifruit: domestication and breeding. Horticultural Reviews. 2007;33:1–121. [Google Scholar]
- Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Lynch M, Connery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Lyons E, Pedersen B, Kane J, et al. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with Rosids. Plant Physiology. 2008;148:1772–1781. doi: 10.1104/pp.108.124867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan G, Peel D, Basford K, Adams P. The EMMIX software for the fitting of mixtures of normal and t-components. Journal of Statistical Software. 1999;4:2. [Google Scholar]
- Ma B, Tromp J, Li M. PatternHunter: faster and more sensitive homology search Bioinformatics. 2002;18:440–445. doi: 10.1093/bioinformatics/18.3.440. [DOI] [PubMed] [Google Scholar]
- Maere S, Bodt SD, Raes J, et al. Modeling gene and genome duplications in eukaryotes. Proceedings of the National Academy of Sciences of the USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeil B, Schlueter J, Shoemaker R, Doyle J. Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Systematic Biology. 2005;54:441–454. doi: 10.1080/10635150590945359. [DOI] [PubMed] [Google Scholar]
- Pires JC, Hertweck KL. A renaissance of cytogenetics: studies in polyploidy and chromosomal evolution. Annals of the Missouri Botanical Garden. 2008;95:275–281. [Google Scholar]
- Quackenbush J, Liang F, Holt I, Pertea G, Upton J. The TIGR gene indices: reconstruction and representation of expressed gene sequences. Nucleic Acid Research. 2000;28:141–145. doi: 10.1093/nar/28.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team R. A language and environment for statistical computing, reference index version 2xx (2005) Vienna: R Foundation for Statistical Computing; 2005. ISBN 3-900051-07-0. Available from: http: //wwwR-projectorg . [Google Scholar]
- Rensing SA, Ick J, Fawcett JA, et al. An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens. BMC Evolutionary Biology. 2007;7:130. doi: 10.1186/1471-2148-7-130. doi:10.1186/1471-2148-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter JA, Dixon P, Granger C, et al. Shoemaker RC mining EST databases to resolve evolutionary events in major crop species. Genome. 2004;47:868–876. doi: 10.1139/g04-047. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Mitchell-Olds T. Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. The Plant Cell. 2006;18:1152–1165. doi: 10.1105/tpc.106.041111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir R, Maron-Katz A, Tanay A, et al. EXPANDER: an integrative program suite for microarray data analysis. BMC Bioinformatics. 2005;6:232. doi: 10.1186/1471-2105-6-232. doi:10.1186/1471-2105-6-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, et al. Polyploidy and angiosperm diversification. American Journal of Botany. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Sterck L, Rombauts S, Jansson S, Sterky F, Rouze P, Peer YVD. EST data suggest that poplar is an ancient polyploid. New Phytologist. 2005;167:165–170. doi: 10.1111/j.1469-8137.2005.01378.x. [DOI] [PubMed] [Google Scholar]
- Tang H, Wang X, Bowers JE, Ming R, Alam M, Paterson AH. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Research. 2008;18:1944–1954. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, et al. The genome of black cottonwood Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Viaene T, Vekemans D, Irish VF, et al. Pistillata: duplications as a mode for floral diversification in (basal) asterids. Molecular Biology and Evolution. 2009;26:2627–2645. doi: 10.1093/molbev/msp181. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Wernersson R, Pedersen AG. RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acid Research. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acid Research. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikström N, Savolainen V, Chase MW. Evolution of the angiosperms: calibrating the family tree. Proceedings of the Royal Society. B: Biological Sciences. 2001;268:2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences of the USA. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Computer Applications in the Biosciences: CABIOS. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. Journal of Computational Biology. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.