Abstract
Background
Comparative phylogeography recently performed on the mitochondrial cytochrome oxidase I (mtCOI) gene from seven deep-sea vent species suggested that the East Pacific Rise fauna has undergone a vicariant event with the emergence of a north/south physical barrier at the Equator 1-2 Mya. Within this specialised fauna, the tube-dwelling polychaete Alvinella pompejana showed reciprocal monophyly at mtCOI on each side of the Equator (9°50'N/7°25'S), suggesting potential, ongoing allopatric speciation. However, the development of a barrier to gene flow is a long and complex process. Secondary contact between previously isolated populations can occur when physical isolation has not persisted long enough to result in reproductive isolation between genetically divergent lineages, potentially leading to hybridisation and subsequent allelic introgression. The present study evaluates the strength of the equatorial barrier to gene flow and tests for potential secondary contact zones between A. pompejana populations by comparing the mtCOI gene with nuclear genes.
Results
Allozyme frequencies and the analysis of nucleotide polymorphisms at three nuclear loci confirmed the north/south genetic differentiation of Alvinella pompejana populations along the East Pacific Rise. Migration was oriented north-to-south with a moderate allelic introgression between the two geographic groups over a narrow geographic range just south of the barrier. Multilocus analysis also indicated that southern populations have undergone demographic expansion as previously suggested by a multispecies approach. A strong shift in allozyme frequencies together with a high level of divergence between alleles and a low number of 'hybrid' individuals were observed between the northern and southern groups using the phosphoglucomutase gene. In contrast, the S-adenosylhomocysteine hydrolase gene exhibited reduced diversity and a lack of population differentiation possibly due to a selective sweep or hitch-hiking.
Conclusions
The equatorial barrier leading to the separation of East Pacific Rise vent fauna into two distinct geographic groups is still permeable to migration, with a probable north-to-south migration route for A. pompejana. This separation also coincides with demographic expansion in the southern East Pacific Rise. Our results suggest that allopatry resulting from ridge offsetting is a common mechanism of speciation for deep-sea hydrothermal vent organisms.
Background
Many population genetic models have been proposed to describe how individuals disperse between geographically separated localities including n-island models [1,2], stepping-stone [3] or isolation-by-distance [4,5] models, and have subsequently been adapted to a metapopulation context to integrate population dynamics through time (i.e. local extinction and recolonisation rates; reviewed in [6,7]). Nevertheless, a physical barrier to dispersal can disrupt the relationship between genetic differentiation and migration rate: all models show that the establishment of a long-term physical barrier to dispersal abruptly reduces or stops gene flow between populations and leads to a rapid change in allele frequencies on either side of the barrier that may be dampened or accelerated depending on the type of selection at specific loci. This often leads to the fixation of mutations (divergence) and reciprocal monophyly for many loci, provided that no lineage extinction or population admixture occurs [8]. Secondary contact events can produce genetic incompatibilities and clinal allelic distributions whose shapes depend primarily on the strength of selection against hybrids [9].
Deep-sea hydrothermal vents represent a patchily distributed habitat (sometimes separated by thousands of kilometres) suggestive of a one-dimensional stepping-stone model over 60 000 kilometres of globe-encircling ridge crests [10]. Because of their dependence on 'hot' sulphidic fluids from vent chimneys, deep-sea hydrothermal vent species display an island-like distribution along ridges. This habitat distribution raises questions about vent species' dispersal among sites and the putative occurrence of physical barriers to dispersal [11,12]. It has been hypothesised that hydrothermal vent species disperse primarily in their local neighbourhood [13], as in the Kimura and Weiss [3] one-dimensional stepping-stone model, and thus follow an isolation-by-distance model [11]. However, in most cases, allozyme studies have failed to detect isolation-by-distance among populations of hydrothermal vent species (e.g. [14,15]), suggesting that an n-island model [1] is a better description of gene flow in vent systems.
Amongst vent species, the deep-sea hydrothermal vent polychaete Alvinella pompejana, which only lives at the top of hydrothermal-vent chimneys of the East Pacific Rise (EPR) between 38°S and 27°N (Guaymas Basin), has been described as a pioneer species because it is the first species to colonise new hot-sulphide edifices [16]. The larval development of this tube-dwelling worm is still largely unknown, but large yolk-rich eggs (150-200 μm), suggest that larval development is lecithotrophic and could be delayed in cold (1.5-2°C) water away from vent fields [12]. This worm species reproduces via internal fertilization and has nearly continuous gametogenesis, maximising its reproductive effort [17]. During mating, spermatozoids are transferred to the female in a spermatecae, and multiple paternity is possible [18]. This kind of reproductive mode is known to strongly favour larval retention in the immediate surrounding of the source population, which may be advantageous in unstable and patchy environments [19]. Given this set of reproductive characteristics and the recurrent extinction and recolonisation of vent edifices, Jollivet et al. [20,21] have proposed that this species follows a propagule-flux model of dispersal (i.e. (re)colonisation of a previously extinct or new vent site from the closest populations, [22]). A hydrodynamic model predicts that effective migration rates - accounting for A. pompejana's larval biology - cannot be maintained across vent fields if larvae travel more than 15-30 days in the water column [12]. Nevertheless, using allozymes this species lacks any clear genetic structure from latitudes 9°50'N to 21°N despite a microgeographical population differentiation at the scale of vent fields (i.e. tens of kilometres) [23]. These results suggest that the potential for larvae to disperse may be higher than previously thought provided that no physical barrier to gene flow exists along the ridge system [24]. Accordingly, Pradillon et al. [25] reported an experiment where A. pompejana embryos can arrest their development in cold water, potentially delaying their metamorphosis during dispersal from vents. To better explain the absence of isolation-by-distance in the northern EPR, three hypotheses have been proposed: (1) allozymes lack the power to detect genetic differentiation between populations; (2) balancing selection - due to the homogeneity of vent conditions along the whole ridge system - maintains homogeneous allozyme frequencies; or (3) displacement of habitat patches over geological times promote metapopulation dynamics, thereby maintaining gene flow [24]. The first hypothesis was tested by Audzijonyte and Vrijenhoek [26] who suggested that undersampling biased some of the previous analyses. The second hypothesis was supported by Piccino et al. [27] who observed balancing selection on the phosphoglucomutase (PGM) locus with one particularly thermostable allele being more frequent in young populations. Interestingly, this study also showed allele frequencies were homogenous at the metapopulation scale, proving support for the third hypothesis. The third hypothesis was tested by a model which showed that bursts of colonisation events following the emergence of new vent sites along with population reconnections were sufficient to prevent genetic differentiation between distant sites given limited dispersal of A. pompejana larvae [21]. The model predictions were however, based on the possibility that new vent sites could occur across physical barriers such as transform faults.
Recently, genetic divergence of nearly 1% was observed at the mitochondrial cytochrome c oxidase subunit I (mtCOI) gene between the northern and southern EPR populations of A. pompejana [28,29], suggesting the occurrence of an impermeable equatorial barrier to gene flow. Furthermore, Plouviez et al. [29] demonstrated that this barrier affected most EPR vent fauna 1 to 2 Mya, giving credence to the role of transform faults in provoking vicariant events between vent communities and allopatric speciation. In the present study, an extended analysis of allozyme and mtCOI haplotype distributions over the geographic range of the species, including the southern EPR, was performed to better address the ability of allozymes to detect genetic differentiation and to discriminate between the three hypotheses described above. Three additional nuclear genes, including the PGM locus, were used to evaluate the strength of this equatorial barrier to gene flow and to test for potential secondary contact. We also attempted to quantify migration and/or introgression rates between the previously isolated northern and southern EPR populations to disentangle the effects of selective and demographic processes contributing to the genetic break.
Methods
Collection
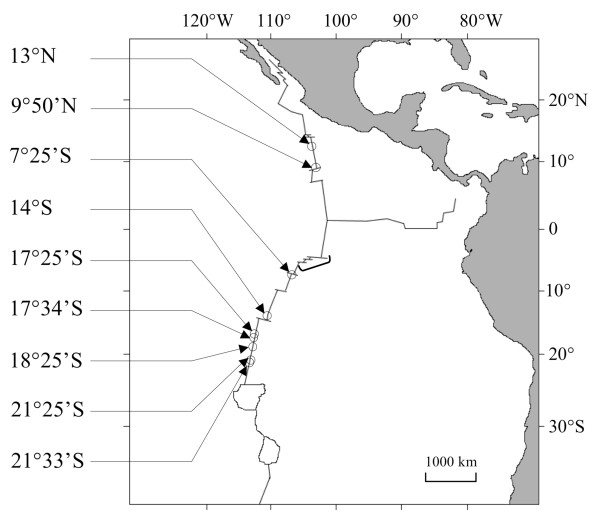
Alvinella pompejana polychaetes were collected with the manned submersible Nautile during two oceanographic cruises in 1999 (HOPE) and 2002 (PHARE) at northern EPR sites (Figure 1) and one cruise in 2004 (BIOSPEEDO) at southern EPR sites from latitudes 7°25'S to 21°33'S. Onboard (R/V L'Atalante), samples were measured at the fourth setigerous segment, sexed, and cut in two: the cephalic portion was frozen immediately at - 80°C for allozyme analyses and the body was preserved in 95% ethanol for DNA analyses.
Figure 1.
Map of the south-east Pacific and Alvinella pompejana sampling locations along the East Pacific Rise. Dots represent sampled hydrothermal vent fields. The bracket between 9°50'N and 7°25'S indicates the Quebrada, Discovery and Gofar transform faults from north to south, respectively.
Allozyme genotyping
Allozymes, encoded by nine enzyme systems, were examined from seven hydrothermal vent field samples (Table 1), following the protocols of Pasteur et al. [30]. Proteins from each frozen gill tissue were extracted according to the method in Piccino et al. [27]. Allozyme electrophoreses were conducted on 12% starch gels using three different buffer systems: (1) Tris-citrate pH 8.0 (TC 8.0) for phosphoglucomutase (PGM, E.C. 5.4.2.2), mannose phosphate isomerase (MPI, E.C. 5.3.1.8), 6-phosphogluconate dehydrogenase (6PGD, E.C. 1.1.1.44) and glucose phosphate isomerase (GPI, E.C. 5.3.1.9); (2) Tris-HCl pH 8.5/8.2 (THCl 8.5) for triose phosphate isomerase (TPI, E.C. 5.3.1.1), leucine amino peptidase (LAP, 3.4.11.1) and hexokinase-1 (HK, 2.7.1.1); and (3) Tris-citrate pH 6.7/6.3 (TC 6.7) for acid phosphatase (ACP, E.C. 3.1.3.2) and isocitrate dehydrogenase (IDH, E.C. 1.1.1.42).
Table 1.
Location of individuals and sample size for allozymes (nallo), mtCOI (nmtCOI), and nuclear genes.
| Vent field | Geographic position | Depth (m) | nallo | nmtCOI | nGlobX | nSAHH | nPGM | nRFLP |
|---|---|---|---|---|---|---|---|---|
| 13°N | 12°43-50'N | 2560-2700 | 40 | 67 | 19 | 17 | 20 | 74 |
| 103°53-57'W | 10 | 11 | 12 | |||||
| 9°50'N | 9°31-51'N | 2500-2585 | 0 | 10 | 7 | 20 | 8 | 0 |
| 104°15-18'W | 4 | 15 | 6 | |||||
| 7°25'S | 7°25'S | 2735-2752 | 17 | 14 | 14 | 13 | 15 | 12 |
| 107°47-49W | 9 | 8 | 11 | |||||
| 14°S | 13°59'S | 2623-2632 | 15 | 20 | 16 | 16 | 15 | 16 |
| 112°29'W | 9 | 14 | 11 | |||||
| 17°25'S | 17°25'S | 2578-2590 | 34 | 30 | 15 | 16 | 10 | 15 |
| 113°12'W | 10 | 9 | 8 | |||||
| 17°34'S | 17°35-36'S | 2591-2597 | 19 | 36 | 18 | 15 | 12 | 17 |
| 113°15'W | 12 | 10 | 10 | |||||
| 18°25'S | 18°26-37'S | 2636-2680 | 52 | 29 | 17 | 17 | 14 | 14 |
| 113°23-24'W | 11 | 14 | 10 | |||||
| 21°25'S | 21°25'S | 2804-2831 | 12 | 30 | 16 | 16 | 13 | 22 |
| 114°16-17'W | 12 | 11 | 10 | |||||
| 21°33'S | 21°33'S | 2771-2846 | 23 | 25 | 14 | 20 | 17 | 11 |
| 114°16-18'W | 10 | 14 | 13 |
nallo: number of individuals genotyped for allozymes, nmtCOI: number of individuals sequenced for the mtCOI gene, nGlobX, nSAHH, nPGM: number of recaptured sequences for each nuclear gene using a maximum of two consensus sequences (alleles) per individual with the number of recaptured individuals given in bold, nRFLP: number of genotyped individuals for PGM RFLPs.
DNA sequencing
Genomic DNA was extracted using a CTAB extraction procedure following the protocol described in Plouviez et al. [29]. A. pompejana mtCOI sequences from this previous study and the sequences of the outgroup A. caudata were obtained with the 'universal' LCOI and HCOI primers [31] using the previously described sequencing protocol. Specific primers (additional file 1) were designed to amplify partial exon-intron sequences of three nuclear genes, either identified during transcriptome sequencing (the S-adenosylhomocysteine hydrolase (SAHH) and globin X (GlobX) genes [32]), or from the complete phosphoglucomutase gene sequence in A. pompejana previously obtained using degenerate primers (D.J., unpublished data). A non-synonymous change was discovered in exon 4 of PGM and specifically targeted in subsequent analyses. Sizes of the DNA fragments and their exon-intron structures (boundaries) are given in additional file 2.
Polymerase chain reactions (PCR) and cloning were performed using the mark-recapture (MR) method developed by Bierne et al. [33] with 4-nucleotide tails added to the 5' end of each primer. Each individual was amplified by PCR separately with a different combination of 5'-tailed primers. PCR were conducted in a 25 μl volume that included 1X buffer (supplied by manufacturer), 2 mM MgCl2, 0.05 mM (0.25 mM for PGM) of each dNTP, 0.48 μM (0.4 μM for PGM) of each primer (additional file 1), 0.5 U of Taq polymerase (Thermoprime plus), 5 μl of template DNA and sterile H2O. Thermal cycling parameters used an initial denaturation step at 94°C for 3 min, followed by 40 cycles at 94°C for 30 s, Ta for 20 s and 72°C for 2 min 30 s, before a final 10 min extension at 72°C. As per the MR protocol, eight PCR-products were pooled together per cloning experiment with two cloning experiments per vent locality (16 individuals per sampled site). PCR products were isostochiometrically mixed, purified using QIAQuick™ columns, and ligated into a pGEM-T vector (pGEM-T cloning kit, Promega, Madison, WI, USA). For each cloning experiment, 32 positive clones were sequenced on both strands at Genoscope (Evry) with universal plasmid primers (SP6 and T7), leading to a total number of approximately 3500 nuclear sequences. Sequences from all genes were proofread using CodonCode Aligner 2.0.6 http://www.codoncode.com/aligner/. Sequence alignments were initially performed with ClustalW [34] in BioEdit version 6.0.6 [35] and adjusted manually. For nuclear genes, in vitro recombinants between individuals from the same cloning set were detected by their abnormal combination of 5'-tails and removed from the dataset. Multiple recaptures also allowed us to discard intra-individual in vitro recombinants (PCR recombination of heterozygous alleles) and artefactual/somatic mutations.
Detecting a putative barrier to gene flow
Linkage disequilibrium between allozyme loci was examined for each locality and for the complete dataset using Genetix version 4.05.2 [36] to detect redundant information. For each locality, allele frequencies were then calculated for each locus and departures from Hardy-Weinberg equilibrium were estimated using the Weir and Cockerham [37]'s estimator f and tested with 1000 permutations. To detect putative barriers to gene flow, genetic differentiation was estimated along the EPR from a multilocus Fst as calculated using Weir and Cockerham's θ [37], adding populations one at a time from the most southern locality (21°33'S) to 13°N. At each step, the F index was tested with 1000 permutations of the dataset. Exact G test was performed using Genepop version 4.0.10 [38] for all populations.
Isolation-by-distance was tested along the whole EPR using a Mantel Spearman test [39] with 5000 permutations using Genepop version 4.0.10 [38]. Because previous studies [28,29] detected a barrier to gene flow across the Equator for the mtCOI marker, this test was also performed without the 13°N population. To better understand population structure, the number of genetically-differentiated groups (K) was evaluated using individual assignments to these a priori groups based on their multilocus allozyme genotypes from an admixture model with a 150 000 burn-in length period followed by 50 000 MCMC repetitions. The most appropriate number was obtained by testing K values from 1 to 15 using the Structure program (version 2.2) [40]. Convergence of the Markov chain simulations was checked for stationarity by monitoring ten iterations.
Average recapture rates were 0.60, 0.74 and 0.63 for GlobX, SAHH and PGM genes, respectively. For all statistical analyses based on recaptured sequences, one consensus allele was randomly selected per individual to avoid any putative bias due to the cloning method. Allele diversity Hd, nucleotide diversity πN, and Watterson's theta θW were estimated for northern and southern EPR sites separately with DNAsp 4.10.3 [41]. For each gene, allele networks were constructed using Network 4.5.1.0 [42] to search for divergence across the Equator (all recaptured alleles were used). Genetic differentiation indexes (øst) were computed using Arlequin 3.1 [43] from nucleotide sites by grouping populations from either side of the Equator and departures from zero (no differentiation) were tested using 1000 permutations.
Because most coalescent-based approaches impose strong assumptions on the absence of recombination, recombinants were detected by the Hudson and Kaplan [44]'s four-gamete test, using DNAsp 4.10.3, and discarded from the dataset for the IMa and Sweep_bott analyses.
Detection of gene flow between the two geographic groups of A. pompejana using an isolation-with-migration model
Population sizes, migration rates, and the time of population splitting were estimated using the isolation-with-migration model (IMa software, [45]) by grouping populations from each part of the geographical barrier (i.e. 9°50'N and 13°N on one side and the remaining southern populations on the other side). This model allowed us to calculate posterior density probabilities of these three parameters assuming that migration could still occur after the splitting event of an ancestral population into two descendant populations. To check convergence of the Markov chain Monte Carlo (MCMC) with the true stationary distribution, multiple runs were performed using different starting points and autocorrelation between parameter values was assessed over the course of the runs. Upper bounds of the uniform prior distributions used for the final run were: q = 30 (prior population size parameters), m = 1 (migration rates) and t = 2 (divergence time). A swapping procedure was also used to enhance the mixing of chains using a geometric heating scheme with 15 parallel chains. Inheritance scalars were assigned to 1 for nuclear genes (autosomal) and 0.25 for mtCOI to adjust for their expected population sizes. The Hasegawa-Kishino-Yano model [46] was chosen to allow for multiple substitutions at the same site.
To better estimate the date since the splitting event, IMa parameters of divergence between populations (tIMa) were converted into years. Because mutation rates have not yet been estimated for alvinellid polychaetes, with the exception of the mtCOI gene [47] which is known to have a higher mutation rate compared to nuclear genes, a geometric mean was derived from mtCOI, as well as PGM and SAHH for which a mutation rate has been obtained from the hydrothermal vent gastropod Lepetodrilus fucensis across the Blanco transform fault [48] and the hydrothermal vent mussels Bathymodiolus thermophilus across the Easter microplate [49], respectively. The GlobX gene was not used because there is no adequate calibration for its mutation rate. Before calculating the geometric mean, an average mutation rate was assessed per locus per generation accounting for different lengths of the three genes (mtCOI 507 bp, PGM 940 bp, SAHH 531 bp), assuming one generation per year. The geometric mean U = 0.64 × 10-6, close to the value previously found using eight genes (mtCOI and 7 nuclear genes, U = 0.63 × 10-6) on B. thermophilus [49], was then used to recalibrate the divergence time from IMa to time in years (T = tIMa /U).
Detecting putative 'hybrids' between the north and south EPR using the PGM gene
Because substantial divergence has been observed between the northern and southern EPR clades at the nuclear PGM gene, fixed mutations were used to detect and locate 'hybrids' between these two geographic regions (heterozygous individuals with one allele from the northern clade and one allele from the southern clade or individuals sampled in a geographic region with alleles typical of the clade of the other region). A restriction fragment length polymorphism (RFLP) analysis combined with genotyping insertion-deletions (indels) was performed to create a composite PGM genotype for each individual. Among the four clades observed, clade 3 displayed two restriction sites for the RsaI enzyme whereas the three remaining clades only had one. Clade 4 displayed two restrictions sites for SspI whereas the three remaining clades have three sites. Clades 1 and 2 differed by several fixed substitutions for which no restriction site was observed and the presence of a 9 bp indel. RsaI and SspI digestions were performed on PGM PCR products following manufacturer's protocols and the presence/absence of the indel was genotyped using new nested primers defined from each side of the indel. Individuals were amplified in 15 μl solutions that included 1X buffer (supplied by manufacturer), 1.25 mM MgCl2, 0.2 mM each dNTP, 0.1 μM of fluorescent-labelled (IRD700™) forward primer, 0.27 μM of unlabelled forward primer, 0.33 μM of unlabelled reverse primer (additional file 1), 0.5 U of Taq polymerase (Thermoprime plus), 4 μl of template DNA and sterile H2O. Thermal cycling parameters used an initial denaturation at 94°C for 2 min, followed by 30 cycles at 94°C for 30 s, annealing temperature (Ta, see additional file 1) for 30 s and 72°C for 25 s, before a final 10 min extension at 72°C. PCR products were then electrophoresed on a denaturing acrylamide 41 cm gel in a Li-Cor NEN Global IR2 DNA analyzer.
Detecting demographic and/or selective events from coalescence trees
Because gene genealogies contain information on demographic and/or selective events, neighbour-joining trees (distances obtained by the maximum composite likelihood) were performed using Mega 4.0.2 [50], separately on northern and southern EPR populations and rooted with an A. caudata sequence for non-ambiguous portions of each gene. Departures from neutral expectations were assessed by estimating Tajima (D, [51]), Fu and Li (F*, [52]) and Fay and Wu (H, [53]) indexes in each group of populations to test for putative demographic and/or selective events. A multi-locus HKA test [54] was computed for each group with one sequence of A. caudata used as an outgroup using the HKA software (J. Hey's web page: http://lifesci.rutgers.edu/~heylab/HeylabSoftware.htm#HKA). This test compares divergence and polymorphism at several loci to determine if any loci display a departure from neutral evolution. McDonald Kreitman tests (MK, [55]) were also performed to test for selection. We tested for the fit of either a bottleneck or a selective sweep model for all four loci (including mtCOI) in northern and southern EPR populations (excluding introgressed alleles) using Sweep_bott [56] with 100 000 first-step iterations, 1 000 000 second-step iterations and 20 optimisation processes with a theta range from 1 to 30. This test compares three distinct coalescent models (including a null constant-size model: M1) using likelihood ratio tests (LRTs) and estimates whether the strength and the time since bottleneck are sufficiently similar between loci to suggest a historical bottleneck (M2 model). If there is a departure from the M1 model but no detected bottleneck, then the selective sweep model is considered as more probable. A strong selective sweep at a single locus can significantly reduce the fit to a bottleneck model resulting in false rejection of a bottleneck hypothesis. To better discriminate the bottleneck and the sweep models, departure from the M1 model was tested individually for each locus (called M3 model). If all loci showed a departure from M1 model, then a bottleneck would occur even if the sweep model was found more probable using the multilocus approach. If only a few loci gave a departure from this model, then the bottleneck was ruled out and the selective sweep model selected.
Results
Equatorial barrier to gene flow
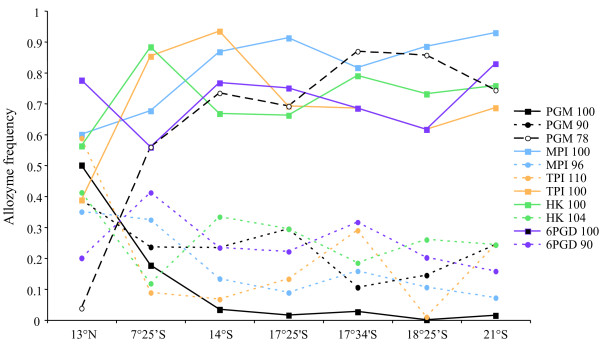
Linkage disequilibria were not significant for any pair of loci in any population (p > 0.05 using the Garnier-Géré and Dillman [57] test), indicating that each locus behaves independently. The number of alleles ranged from 3 (GPI, IDH, ACP, LAP, HK) to 5 (TPI, see: additional file 3), however most loci displayed only one to 3 alleles at moderate frequency (> 5% in the total population) and were shared among all populations. In contrast, rare alleles were often private (additional file 3). Common alleles reversed in frequency at the TPI locus across the Equator, with the 13°N site showing opposite trends in allele frequencies compared to the 7°25'S site (Figure 2). A similar result was observed at the PGM locus but with a nearly complete replacement of allele 100 by allele 78 and the frequency of allele 90 slightly decreasing from north to south. Significant heterozygote deficiencies (Fis > 0, see additional file 3) were not linked to any particular locus and were not systematically observed at any location. However, a more detailed examination of Fis values at loci with two or three common alleles indicated that the highest values were found at 14°S and 17°25'S.
Figure 2.
Allozyme frequency clines along the East Pacific Rise. Frequencies of the most frequent alleles found in A. pompejana populations for PGM, MPI, TPI, HK and 6PGD allozyme loci are plotted according to latitude. Each colour represents a distinct allozyme locus. Individuals from 9°50'N were not used in the analysis simply because they have been only preserved in alcohol.
The multilocus estimate of Weir and Cockerham's θ [37] over all populations was not significantly different from zero (θ = 0.097, p > 0.05, 1000 permutations) but Fst significantly differed from zero with the Robertson and Hill's correction ([58], RH = 0.060, p < 0.05, 1000 permutations). The exact G test for genic differentiation was significant for the overall populations (p < 0.05). Tests for isolation-by-distance showed a significant correlation between the genetic distance of allozymes θ/(1-θ) and geographic distance across the entire EPR region (Mantel Spearman test: p = 0.014) but was not significant when the 13°N site was removed from the dataset (p = 0.115). Partitioning genetic structure using the Bayesian clustering method of Pritchard et al. [40] identified three genetically distinct groups present in different proportions within each vent field. The first group was primarily made up of individuals from 13°N, whereas the second group clustered individuals mostly from 7°25'S-14°S, and the third group clustered 17°25'S to 21°33'S individuals.
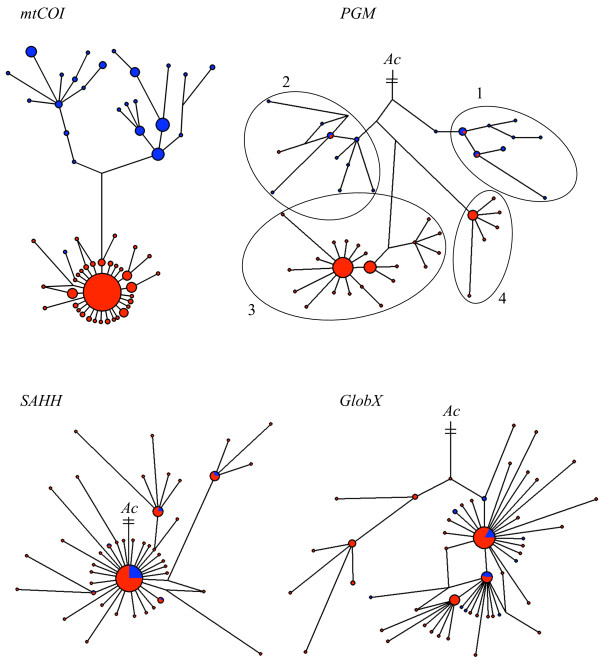
DNA sequence variation among the three nuclear genes (SAHH, GlobX and PGM) and mtCOI were analysed in the northern and southern populations of A. pompejana across the Equator (Table 2, Genbank accession numbers: HM183084-HM183493). As shown by Plouviez et al. [29], the network of mtCOI haplotypes displayed two reciprocally monophyletic clades (~1% divergence) in the northern and southern EPR, respectively (Figure 3). Similarly, the PGM locus possessed two sets of two divergent clades that were geographically structured (average northern/southern EPR divergence = 0.76%): clades 1 and 2 were mainly found in the northern EPR, whereas clades 3 and 4 were distributed in the southern EPR (Figure 3). Populations from the northern and southern EPR were significantly different from each other at this locus (øst = 0.18, p < 0.01). Although populations appeared to be genetically differentiated across the Equator at the GlobX locus (øst = 0.04, p < 0.05), these populations were genetically homogeneous across regions at the SAHH locus (øst value = 0.00, p > 0.05).
Table 2.
Nucleotide sequence variation in Alvinella pompejana.
| Gene | EPR locality | n | Hd (SD) | πN (SD) | θW (SD) | D | F* |
|---|---|---|---|---|---|---|---|
| mtCOI | |||||||
| northern | 108 | 0.911 (0.014) | 0.009 (0.000) | 0.014 (0.004) | -0.983NS | -2.164 NS | |
| southern | 210 | 0.618 (0.040) | 0.002 (0.000) | 0.015 (0.004) | -2.564*** | -4.330** | |
| GlobX | |||||||
| northern | 14 | 0.890 (0.060) | 0.003 (0.000) | 0.0034 (0.0017) | -0.872NS | -1.504NS | |
| southern | 73 | 0.860 (0.031) | 0.0054 (0.001) | 0.012 (0.004) | -1.721+ | -4.107** | |
| SAHH | |||||||
| northern | 26 | 0.471 (0.119) | 0.002 (0.001) | 0.004 (0.002) | -1.989* | -2.7696* | |
| southern | 80 | 0.604 (0.064) | 0.0031 (0.001) | 0.011 (0.003) | -2.202** | -4.484** | |
| PGM | |||||||
| northern | 18 | 0.921 (0.030) | 0.0056 (0.001) | 0.007 (0.003) | -0.693NS | -1.437 NS | |
| southern | 74 | 0.725 (0.055) | 0.0042 (0.001) | 0.009 (0.003) | -1.836* | -3.950** |
n, number of sequences; Hd, Haplotype diversity, πn nucleotide diversity; θw, Watterson's theta per site from the number of segregating sites, D, Tajima's D index; F*, the Fu and Li index; SD, standard deviation; NS: P > 0.10, +: P < 0.10, *: P < 0.05, **: P < 0.01, ***: P < 0.001.
Figure 3.
Median-joining networks of the mtCOI and nuclear genes. Blue and red labels represent individuals sampled from the northern (9°50'N and 13°N) and southern (7°25'S-21°33'S) EPR, respectively. Sizes of haplotype circles and lengths of connecting lines are proportional to the number of sequences studied and the number of mutations that separate two linked haplotypes, respectively. Nuclear genes are rooted by a consensus outgroup sequence of A. caudata (connecting line between A. pompejana and the A. caudata outgroup is not representative of sequence differences due to the high number of mutations).
Testing an isolation model with migration between the two geographic groups of A. pompejana
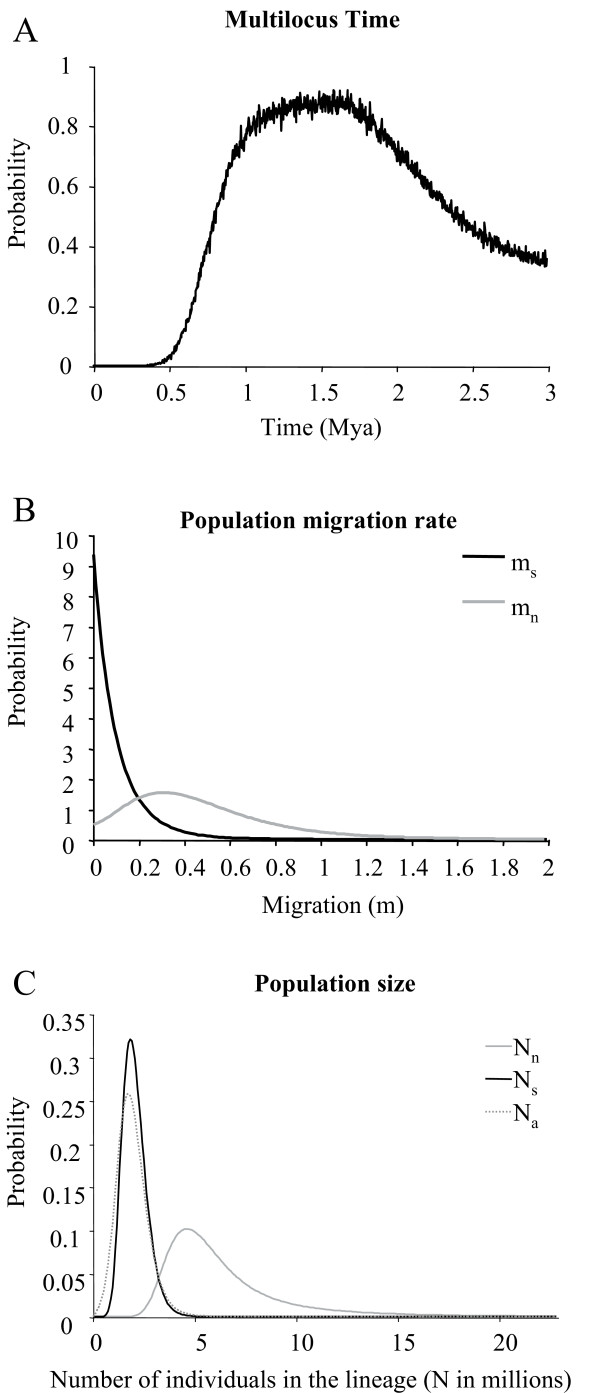
The isolation-with-migration (IM) model of Hey and Nielsen [45] was used to test the occurrence of historical gene flow between northern and southern populations of A. pompejana or recent hybridisation and introgression following secondary contact. The estimated IM parameters are presented in additional file 4. Using all four genes, the split between the two groups of populations showed the highest likelihood values at divergence times of around 1-2 Mya (Figure 4A). IMa calculates migration rate based on coalescent analysis by looking from the present backward in time reversing the direction of migration when looking forward in time. The magnitude of migration from the north to the south (mn) and from the south to the north (ms) shows that gene flow occurred predominantly from the north to the south with no migration being detected in the opposite direction (Figure 4B).
Figure 4.
Marginal posterior probability distribution for parameters estimated by the isolation-with-migration model. Marginal posterior probabilities are estimated for (A) divergence time between the northern and southern EPR, (B) the forward-in-time migration rates between the northern and southern EPR, where ms represents south-to-north migration and mn north-to-south and (C) effective population sizes of the ancestral population (Na) and the two sister populations (i.e. Nn and Ns for the northern EPR population and the southern EPR population, respectively). Values estimated using IMa and 90% highest posterior density intervals of these parameters are presented in additional file 4.
Single-locus estimates of migration also exhibited asymmetry from the north to the south, although the parameters for PGM, SAHH, and GlobX were not significantly different from zero on the basis of their 90% highest posterior density (HPD) interval. This was clearly suggested by the allele networks for PGM and GlobX (Figure 3) for which a few alleles from northern clades were sampled in the southern EPR region. The mean time of migration events from the north to the south ranged from 25 000 to 300 000 years ago (Table 3), indicating that introgression occurred well after the initial isolation (~1.6 Mya) but not recently. The mean time of migration events (mtime) at PGM was, however, ten times lower than the two other loci suggesting more recent introgression (~23 000 years) at this locus.
Table 3.
Number (m#) and mean time (mtime) of migration events, obtained from IM analyses.
| Locus | m#s | m#n | mtimes (My) | mtimen (My) |
|---|---|---|---|---|
| mtCOI | 0 | 0 | - | - |
| SAHH | 0 | 0-7 | - | 0.347 |
| GlobX | 0 | 0-7 | - | 0.388 |
| PGM | 0 | 1 | - | 0.023 |
Forward in time, labels (s) and (n) correspond to the south-to-north and north-to-south orientation of migration, respectively. For SAHH and GlobX genes, m# values correspond to the range of the posterior probability distribution without considering m = 0.
Detection of hybrids at PGM between northern and southern populations
Based on the PGM network (Figure 3), we detected three individuals sampled on the southern EPR with at least one allele from a northern clade (two from 14°S and one from 7°25'S), but with the southern mitochondrial type. Owing to fixed substitutions and indels between the four PGM clades, all individuals collected at these sites were genotyped by combining RFLP and intron length polymorphisms. By comparing RFLP/indel genotypes and mitochondrial haplotypes of each individual, four 'hybrids' (33.3%) between the northern and southern groups were detected at 7°25'S, two (16.7%) at 14°S and only one (1.7%) at 13°N. No 'hybrids' were found at the other latitudes.
Detecting demographic changes and/or selective effects
Tajima (D) and Fu and Li (F*) indexes were significantly negative for all loci in the south, but only SAHH displayed significant values in the north. All genes exhibited departures from neutral equilibrium between drift and mutation in the southern EPR, but not in the northern EPR. Similarly, for each nuclear gene, Fay and Wu tests were not significantly different from the neutral expectation in the northern populations, but were significantly negative in the southern populations.
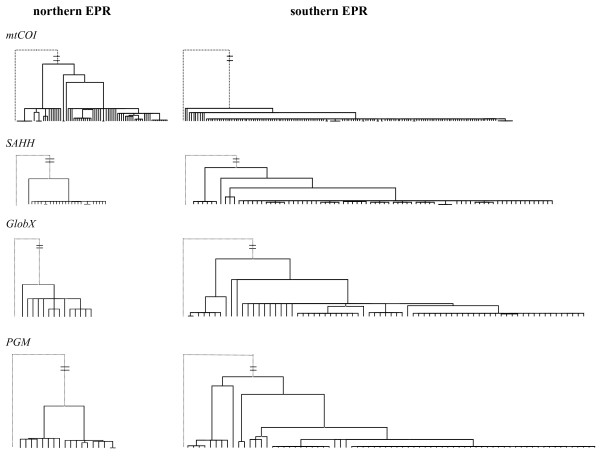
The PGM neighbour-joining tree displayed a topology similar to a gene experiencing balancing selection (i.e., the maintenance of 'old' lineages), whereas trees from the two other nuclear genes (SAHH and GlobX) had more star-like topologies typical of a selective sweep (Figure 5). The HKA multilocus test was significant using all genes (probability from χ2 distribution < 0.00), indicating that at least one gene has experienced positive selection (or gene hitchhiking). After excluding mtCOI, the HKA test became nonsignificant (p χ2 > 0.05), suggesting this gene could be experiencing strong purifying selection or a sweep. However, McDonald-Kreitman tests failed to reject neutral expectations at any locus (p > 0.05) when comparing A. pompejana polymorphism to the divergence of the A. caudata outgroup.
Figure 5.
Ultrametric neighbour-joining topologies of four genes in A. pompejana in the northern and southern EPR. The outgroup A. caudata (dotted lines) was used to root the phylogenetic trees that were constructed from the most well-aligned parts of the four studied genes.
The Sweep_bott analyses performed on northern and southern groups of populations using the four loci are summarised in Table 4. When the analysis was performed over all genes, LRTs indicated that the bottleneck (M2) and selective sweep (M3) models both showed a significantly better fit with the dataset than the constant-size (M1) model. However, it was not possible to discriminate between the M2 and M3 hypotheses. When the analysis was performed on each gene individually, LRTs for M3 were all significantly better when compared with the M1 model for the southern populations, suggesting that a bottleneck might have occurred in this region. When the same analysis was done on the northern populations, LRTs for M3 were only significant for the SAHH gene, indicating that in addition to a bottleneck this gene may have undergone a selective sweep, at least in the northern populations. In effect, the M2 and M3 models are not mutually exclusive if a gene has experienced a moderate sweep in a bottlenecked population.
Table 4.
MLEs and likelihood ratio tests computed using Sweep_bott software for bottleneck and selective models.
| Multilocus | Single-locus | ||||
|---|---|---|---|---|---|
| mtCOI | GlobX | SAHH | PGM | ||
| northern EPR | |||||
| M1 MLE | -99.718 | -37.728 | -11.171 | -14.341 | -36.475 |
| M2 MLE | -92.660 | ||||
| M3 MLE | -90.460 | -35.171 | -9.712 | -11.170 | -34.407 |
| LRT | M2-M1: 14.116 | ||||
| p < 0.005 | |||||
| M3-M1: 18.516 | 5.114 | 2.918 | 6.342 | 4.136 | |
| p < 0.025 | (NS) | (NS) | p < 0.025 | (NS) | |
| M3-M2: 4.400 | |||||
| (NS) | |||||
| southern EPR | |||||
| M1 MLE | -159.367 | -4.149 | -57.112 | -38.064 | -60.042 |
| M2 MLE | -153.211 | ||||
| M3 MLE | -139.745 | -0.290 | -52.196 | -30.557 | -56.704 |
| LRT | M2-M1: 12.312 | ||||
| p < 0.005 | |||||
| M3-M1: 39.243 | 7.718 | 9.830 | 15.014 | 6.677 | |
| p < 0.005 | p < 0.025 | p < 0.01 | p < 0.005 | p < 0.005 | |
| M3-M2: 23.282 | |||||
| p < 0.005 | |||||
M1, neutral model; M2, bottleneck model; M3, selective sweep model; NS, non-significant. Significances of the likelihood ratio tests (LRT) were tested according to a χ2 table with a df equal to the difference of the number of parameters estimated for each model (df = 2, 8 and 6 for M2-M1, M3-M1 and M3-M2 multilocus LRTs, respectively, and df = 1 for the M3-M1 single-locus LRT).
Discussion
Emergence and strength of the equatorial barrier to gene flow
A genetic break at the mtCOI gene between the northern and southern EPR populations has been identified in previous studies by using a large number of hydrothermal vent species [28,29]. For some species, such as the gastropod Lepetodrilus ovalis and the bivalve Bathymodiolus thermophilus, a clinal distribution of the northern and southern mtCOI haplotypes was observed and attributed to the occurrence of a secondary contact zone between the northern and southern EPR [29]. In contrast, A. pompejana exhibited a clear equatorial break at this locus, raising the question of the permeability of this barrier to gene flow. The strength of the break between the northern and southern populations of A. pompejana appears to have significantly affected both allozyme and nuclear loci despite the recent formation of the barrier. Although the SAHH gene exhibited very little differentiation across this barrier, both the GlobX and PGM loci displayed a clear geographic separation of alleles. Divergence between northern and southern clades was estimated at ~1-2 Mya using the IMa multilocus estimation with very low migration rates. This estimate is in agreement with the shared vicariant event estimated at ~1.3 Mya from a series of vent species using approximate Bayesian computation on the mtCOI gene [29]. This isolation may be the consequence of the formation of multiple transform faults ~1-2 Mya in the region (Figure 1, [59-61]) reinforced by a strong transverse deep-sea current at the Equator [62].
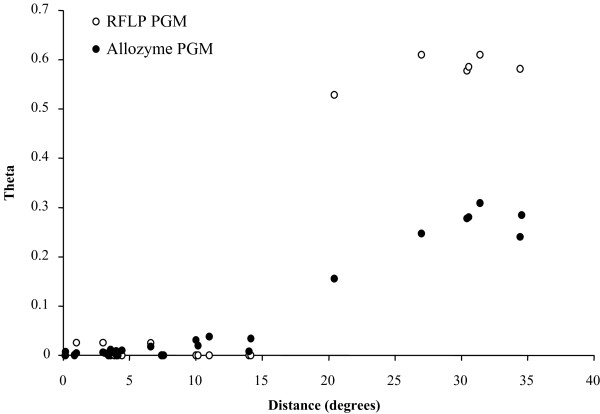
Slight, but significant differentiation at six allozyme loci across the equatorial barrier indicates that 1-2 My may not be sufficient to affect the net charge state of allozymes to create divergence, but likely reduced migration rates enough to modify allozyme frequencies by random genetic drift. This is particularly obvious in the distributions of allozymes and RFLP variants at the PGM locus (see Figure 6). Comparing patterns of genetic differentiation between PGM allozymes and DNA sequences indicated that allozymes are less sensitive at detecting the break. Moreover, the nearly complete replacement of the most frequent allele (100) in the northern populations by allele 78 in the south suggested that genetic drift is the main force acting to separate these populations at this locus. This equatorial break contrasts with the complete lack of genetic differentiation previously observed along thousands kilometres of vent fields on both sides of the EPR ([18], along the southern EPR in the present study) and confirms that isolation-by-distance by itself is not a valid argument to explain the observed decrease of gene flow for A. pompejana. In the absence of physical barriers, the species is able to disperse farther than expected based on moderate inter-field distances and the high bottom-current velocities [12]. This may be explained by either a developmental arrest of embryos in cold abyssal water masses and subsequent delay in metamorphosis [25], or the displacement of vent sites due to the movement of hydrothermal activity along the ridge axis, allowing bursts of colonisation by reconnecting previously isolated populations [21].
Figure 6.
Sensitivity of RFLP and allozyme PGM markers for detecting the equatorial genetic break. The distance θ is plotted against the geographic distance between hydrothermal vent localities. The RFLP dataset shows a clearer departure from the isolation-by-distance model's expectation, indicating that markers to better detect the equatorial genetic break.
Questions addressing potential migration during an ongoing allopatric isolation process may be explored in two ways, depending on the primary cause for separation of populations (transform faults or bottom water circulation): (1) is there a progressive isolation of the north and south populations of A. pompejana by the offsetting of ridge segments at the Equator? or (2) is there emergence of a secondary contact between previously isolated populations due to the relaxation (i.e. attenuation of the transverse circulation) of the barrier?
Permeability of the barrier
Variation in the magnitude of the genetic differentiation among loci has been widely reported during the process of allopatric speciation [63] and is commonly detected across barriers to gene flow [64-66]. This is mainly attributable to: (1) the sensitivity of the molecular method used to detect polymorphism, which in turn depends on the time scale at which the isolation is observed (i.e. the accumulation of non-synonymous mutations that change the net charge of a protein is a long process) [67], (2) the stochasticity of the coalescence process [68], and (3) the formation of a genetic barrier when secondary contacts occur [9,49,69]. Fitting an isolation-with-migration model [45] between the northern and southern EPR populations help to estimate both historical and contemporary migration and better understand the role of the equatorial barrier on the isolation process. Even if the estimated migration rates were not significantly different from zero, the shape of the posterior probability distribution of mn (north to south) was shifted in comparison to ms (see Figure 4B) and suggests that the barrier is porous. While migration was weak or absent at mtCOI, all nuclear genes displayed an asymmetric number of migration events across the Equator from north to south (Table 3), suggesting that allele migration can occur between the northern and southern groups. This was confirmed by detecting a few intermediate individuals at 7°25'S and 14°S between the northern and southern PGM groups.
The fact that gene flow is virtually absent for at least two loci (mtCOI and PGM) raises the question of whether the barrier is becoming increasingly impermeable to dispersal with time, or whether it represents a semi-permeable genetic barrier following recent secondary contact. In the first case, one can expect that most "migrating" alleles (i.e. alleles typifying one lineage found in the other one) would coalesce deeply in the evolutionary tree as they will represent incomplete lineage sorting. In the other case (emergence of a tension zone), all "migrating" alleles should coalesce shortly in the coalescence tree, leading to very low mtimes (times at which gene flow was maximum).
Mean times of migration at the three nuclear genes indicated that gene exchanges are probably not recent and may have been maximal ~20 000 to 300 000 years ago and support the hypothesis of incomplete lineage sorting and ancestral variation [see [49,70]], favouring an increasingly stronger barrier scenario. However, the high level of gene flow heterogeneity across both nuclear and mitochondrial loci favours the notion of a semi-permeable barrier to gene flow as previously defined by Harrison [71]. Natural selection is expected to prevent gene flow in regions of the genome linked to genetic incompatibilities and to generate interloci differences in migration rates [72]. To this extent, the restriction of 'hybrid' individuals as estimated from the PGM RFLP screening to a narrow zone located between 7°25'S-14°S could be indicative of the occurrence of a genetic barrier with differential selection against hybrids [69,73,74]. However, it may also simply reflect the fact that very few individuals are able to cross the geographic barrier by chance maintaining low frequencies according to the migration/drift equilibrium. Currently, the present number of nuclear loci is too small to strongly indicate the presence of a genetic barrier. Given that the vicariant event was quite recent (1-2 Mya), genetic incompatibilities and effective selection against hybrids may not have had time to arise supporting a model of progressive isolation of A. pompejana populations.
Demographic expansion in the southern EPR hypothesis and evidence for a possible selective sweep at the SAHH gene
A comparative phylogeographic approach involving seven vent species using the mtCOI gene (including A. pompejana) revealed the occurrence of a concomitant demographic expansion in the southern EPR within the last 0.5 My [29]. In the present study, southern populations (but not necessarily northern populations) displayed significantly negative Tajima's D and Fu and Li statistics for all genes. Moreover, the bottleneck model (M2) was significantly better than the constant-size model (M1) in describing the genealogies associated with the southern populations. Assuming a similar distribution of nucleotide diversity across the studied genes (as expected for a demographic event such as expansion [54,56]), these results are in agreement with an expansion of southern populations, as hypothesised by Plouviez et al. [29]. Multiple extinction and recolonisation events possibly caused by a higher rate of tectonic rearrangements and eruptive phases in the southern EPR [75,76], would have favoured the recurrence of bottlenecks through time as previously seen from the long-term monitoring of targeted vent fields since 1991 [77,78]. This also fits well with the drastic change of allozyme frequencies at the PGM locus on each side of the barrier as genetic drift may be greatly enhanced by recurrent bottlenecks and/or founder effects [79,80]. However, IMa results provided a complete reverse scenario (Figure 4C) suggesting that only the northern populations increased in size since the population splitting. Although unexpected, this finding may be due to several causes: (1) the very high proportion of divergent allelic forms found in the northern populations, a situation typifying an 'old' diversified lineage, (2) the nearly equal sample sizes used in the analysis by suppressing both obvious recombinant alleles and randomly-selected alleles in the south, and (3) the possible non-neutral behaviour of at least one loci (i.e. the SAHH). The two former explanations seem unlikely. In the first case, the expectation would be an ancestral population size nearly similar to that of the northern one. In the second case, the Sweep_bott analysis (based on the same sequence dataset) would have validated the bottleneck scenario in the northern populations. The detection of a bottleneck in the southern populations does not preclude the possible additional effect of a selective sweep at any specific locus. The SAHH locus was the only locus that conformed to a selective sweep model on both sides of the EPR with virtually no genetic differentiation across the expected barrier, if an initial isolation step in allopatry is assumed. An advantageous allele (or a neutral allele linked to a gene under selection by hitch-hiking) is more likely to cross a genetic barrier and to spread within populations of the recipient lineage if the allele is advantageous for both lineages (e.g. [81,82]). Analyses of nucleotide polymorphism at this locus for northern and southern populations of Alvinella caudata, the most closely-related and syntopic species to A. pompejana, indicated a clear fixed divergence of alleles across the equatorial barrier analogous to divergences observed at both the mtCOI and the PGM loci in A. pompejana (S.P., unpublished data). This suggests that the lack of lineage sorting found in A. pompejana at the SAHH locus is probably not the result of a slow evolutionary rate at this locus but rather a case of high migration rate across the barrier. The spread of an allele across a permeable barrier is expected to be related to the magnitude of the selective advantage [83], suggesting that the sweep may be recent or very strong, even if the Fay and Wu test is only significantly negative in the southern EPR. Moreover, Faure et al. [49] proposed that diversifying selection helped to accelerate the evolution toward reciprocal monophyly at this specific gene in the Atlantic deep-sea vent mussels, suggesting that the SAHH gene may be indeed very sensitive to selection or linked to a potential positively selected gene.
Conclusion
In summary, our multilocus study of A. pompejana populations confirmed the presence of an equatorial barrier, which plays an important role in structuring populations of the species A. pompejana, as proposed by Plouviez et al. [29]. This geographic barrier appears to be still permeable to migration. However, low migration rates that would occur primarily from north to south may also indicate that the barrier is becoming more and more impermeable suggesting that transform faults are a likely mean to promote allopatric speciation along oceanic ridges. In addition, discriminating a bottleneck model from a selective sweep model through the comparison of mtCOI and nuclear genes strongly supports the southern demographic expansion hypothesis previously suggested by a multispecies analysis using mtCOI only. Combining multilocus and multispecies analyses therefore allows us to propose that allopatry is probably one of the major forces promoting speciation and biodiversity within the vent fauna along the one-dimensional framework of oceanic ridges.
Authors' contributions
SP carried out sequence acquisition and RFLP/indel genotyping as well as the molecular genetic analyses and drafted the manuscript. DLG genotyped individuals using allozymes. OL annotated the cDNA library of A. pompejana. FHL organised two of the three oceanographic cruises, helped sample specimens and provided helpful elements of discussion. DJ designed the study, carried out sampling, performed analyses with SP and edited the manuscript. All authors have read and approved the final manuscript.
Authors' information
This study is part of SP's PhD thesis on the comparative phylogeography of deep-sea hydrothermal vent species along the East Pacific Rise (EPR). Her research involves multiple species and markers to examine speciation and demographic processes along the EPR. DLG is lab technician in charge of genotyping and biochemical analyses. OL works on comparative genomics and bioinformatics. FHL has broad knowledge of vent ecology and has spent most of his career working on the physiology of deep-sea hydrothermal vent species. DJ is a population geneticist working on the evolution and dispersal of deep-sea hydrothermal vent species. He has recently developed a genomic approach to better understand the role of gene adaptation in the evolution of vent species.
Supplementary Material
Primers designed for DNA amplification on Alvinella pompejana. Table of sequences for forward and reverse primers used in the study.
Size and exon-intron structure of the four sequenced genes. Schematic drawings of the gene portion used in which exonic regions are represented by boxes.
Allele frequencies, expected and observed heterozygosities and Fis estimated from enzyme loci across vent fields. Table of allozyme frequencies and heterozygosities for each sampled population.
Estimates and 90% Highest Posterior Density (HPD) intervals of demographic parameters from IMa multilocus analysis. Table of 90% HPD intervals for population sizes, migration rates and time since population splitting estimated from the IMa Bayesian computations.
Contributor Information
Sophie Plouviez, Email: sophie.plouviez@duke.edu.
Dominique Le Guen, Email: leguen@sb-roscoff.fr.
Odile Lecompte, Email: lecompte@igbmc.fr.
François H Lallier, Email: lallier@sb-roscoff.fr.
Didier Jollivet, Email: jollivet@sb-roscoff.fr.
Acknowledgements
We thank the chief scientists and 'Nautile' crews for their technical support and effort during our oceanographic expeditions: PHARE2002, HOPE99 and BIOSPEEDO2004. We are very grateful to Stéphane Hourdez and Baptiste Faure for collecting and sorting alvinellid polychaetes and to Grant Pogson, Carolyn Engel-Gauthier, Thomas Schultz and the two anonymous referees for their valuable comments and editorial suggestions on the manuscript. We would like to thank Christophe Caron and Erwan Corre for their help in bioinformatics. We are also truly indebted to the sequencing genomic plateforms GENOMER (Station Biologique de Roscoff, France) and GENOSCOPE (Evry, France) for the sequencing of the mtCOI fragment and our clone collection, respectively. This work was supported by the GDR Ecchis, the ANR-06-BDV-005 (Deep Oases: coord. Daniel Desbruyères) and ANR-05-BLAN-0407 (Alvi_Stress_Adapt: coord. Didier Jollivet). S. Plouviez was supported by a PhD grant from the Université Pierre et Marie Curie.
References
- Wright S. Evolution in mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The distribution of gene frequencies in populations. Proc Nat Acad Sci USA. 1937;23:307–320. doi: 10.1073/pnas.23.6.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Weiss GH. Stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics. 1964;49:561–576. doi: 10.1093/genetics/49.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malécot G. Quelques schémas probabilistes sur la variabilité des populations naturelles. Ann Univ Lyon Sci. 1950;13:37–60. [Google Scholar]
- Harrison S, Hastings A. Genetic and evolutionary consequences of metapopulation structure. Trends Ecol Evol. 1996;11:180–183. doi: 10.1016/0169-5347(96)20008-4. [DOI] [PubMed] [Google Scholar]
- Pannell JR, Charlesworth B. Effects of metapopulation process on measures of genetic diversity. Philos Trans R Soc Lond. 2000;355:1851–1864. doi: 10.1098/rstb.2000.0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CW, Collins TM. In: Molecular Approaches to Ecology and Evolution. DeSalle R, Schierwater B, editor. Birkhäser Verlag Basel; 1998. Beyond area relationships: extinction and recolonization in molecular marine biogeography; pp. 297–321. [Google Scholar]
- Barton NH, Hewitt GM. Analysis of hybrid zones. Annu Rev Ecol Syst. 1985;16:113–148. doi: 10.1146/annurev.es.16.110185.000553. [DOI] [Google Scholar]
- Schlager W. Earth's layers, their cycles and Earth system science. Austrian J Earth Sci. 2008;101:4–16. [Google Scholar]
- Vrijenhoek RC. Gene flow and genetic diversity in naturally fragmented metapopulations of deep-sea hydrothermal vent animals. J Hered. 1997;88:285–293. doi: 10.1093/oxfordjournals.jhered.a023106. [DOI] [PubMed] [Google Scholar]
- Chevaldonné P, Jollivet D, Vangriesheim A, Desbruyères D. Hydrothermal-vent alvinellid polychaete dispersal in the eastern Pacific .1. Influence of vent site distribution, bottom currents, and biological patterns. Limnol Oceanogr. 1997;42:67–80. doi: 10.4319/lo.1997.42.1.0067. [DOI] [Google Scholar]
- Lutz RA. Dispersal of organisms at deep-sea hydrothermal vents: a review. Oceanol Acta. 1988;8(Suppl):23–29. [Google Scholar]
- Black MB, Lutz RA, Vrijenhoek RC. Gene flow among Vestimentiferan tube worm (Riftia pachyptila) populations from hydrothermal vents of the Eastern Pacific. Mar Biol. 1994;120:33–39. [Google Scholar]
- Craddock C, Hoeh WR, Lutz RA, Vrijenhoek RC. Extensive gene flow among Mytilid (Bathymodiolus thermophilus) populations from hydrothermal vents of the Eastern Pacific. Mar Biol. 1995;124:137–146. doi: 10.1007/BF00349155. [DOI] [Google Scholar]
- Jollivet D. PhD thesis. Université de Bretagne occidentale, IFREMER centre de Brest; 1993. Distribution et évolution de la faune associée aux sources hydrothermales profondes à 13°N sur la dorsale du Pacifique oriental: le cas particulier des polychètes Alvinellidae. [Google Scholar]
- Faure B, Chevaldonné P, Pradillon F, Thiébaut E, Jollivet D. Spatial and temporal dynamics of reproduction and settlement in the Pompeii worm Alvinella pompejana (Polychaeta: Alvinellidae) Mar Ecol Prog Ser. 2007;348:197–211. doi: 10.3354/meps07021. [DOI] [Google Scholar]
- Pradillon F, Gaill F. Oogenesis characteristics in the hydrothermal vent polychaete Alvinella pompejana. Inv Reprod Dev. 2003;43:223–235. [Google Scholar]
- Byers JE, Pringle JM. Going against the flow: retention, range limits and invasions in advective environments. Mar Ecol Prog Ser. 2006;313:27–41. doi: 10.3354/meps313027. [DOI] [Google Scholar]
- Jollivet D, Dixon LRJ, Desbruyères D, Dixon DR. Ribosomal (rDNA) variation in a deep sea hydrothermal vent polychaete, Alvinella pompejana, from 13 degrees N on the East Pacific Rise. J Mar Biol Ass UK. 1998;78:113–130. doi: 10.1017/S0025315400039989. [DOI] [Google Scholar]
- Jollivet D, Chevaldonné P, Planque B. Hydrothermal-vent alvinellid polychaete dispersal in the eastern Pacific. 2. A metapopulation model based on habitat shifts. Evolution. 1999;53:1128–1142. doi: 10.2307/2640817. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Gene flow and genetic drift in a species subject to frequent local extinction. Theor Pop Biol. 1977;12:253–262. doi: 10.1016/0040-5809(77)90045-4. [DOI] [PubMed] [Google Scholar]
- Jollivet D, Desbruyères D, Bonhomme F, Moraga D. Genetic differentiation of deep-sea hydrothermal vent Alvinellid populations (Annelida, Polychaeta) along the East Pacific Rise. Heredity. 1995;74:376–391. doi: 10.1038/hdy.1995.56. [DOI] [Google Scholar]
- Jollivet D. Specific and genetic diversity at deep-sea hydrothermal vents: An overview. Biodiv Conserv. 1996;5(12):1619–1653. doi: 10.1007/BF00052119. [DOI] [Google Scholar]
- Pradillon F, Shillito B, Young CM, Gaill F. Deep-sea ecology - Developmental arrest in vent worm embryos. Nature. 2001;413:698–699. doi: 10.1038/35099674. [DOI] [PubMed] [Google Scholar]
- Audzijonyte A, Vrijenhoek RC. When gaps realy are gaps: statistical phylogeography of hydrothermal vent invertebrates. Evolution. in press . [DOI] [PubMed]
- Piccino P, Viard F, Sarradin PM, Le Bris N, Le Guen D, Jollivet D. Thermal selection of PGM allozymes in newly founded populations of the thermotolerant vent polychaete Alvinella pompejana. Proc R Soc Lond. 2004;271:2351–2359. doi: 10.1098/rspb.2004.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado L, Lutz R, Vrijenhoek R. Distinct patterns of genetic differentiation among annelids of eastern Pacific hydrothermal vents. Mol Ecol. 2004;13:2603–2615. doi: 10.1111/j.1365-294X.2004.02287.x. [DOI] [PubMed] [Google Scholar]
- Plouviez S, Shank TM, Faure B, Daguin-Thiébaut C, Viard F, Lallier FH, Jollivet D. Comparative phylogeography among hydrothermal vent species along the East Pacific Rise reveals vicariant processes and population expansion in the south. Mol Ecol. 2009;18:3903–3917. doi: 10.1111/j.1365-294X.2009.04325.x. [DOI] [PubMed] [Google Scholar]
- Pasteur N, Pasteur G, Bonhomme F, Catalan J, Britton-Davidian J. Manuel technique de génétique par électrophorèse de protéines. Paris: Lavoisier; 1987. [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech. 1994;3:294–299. [PubMed] [Google Scholar]
- Gagnière N. PhD thesis. Université de Strasbourg, IGBMC Laboratoire de Bioinformatique et de Génomique Intégratives; 2009. Développement d'une suite logicielle pour l'analyse et l'annotation intégrative automatiques de transcrits et de protéines. Application aux banques d'ADNc de l'annélide polychète Alvinella pompejana. [Google Scholar]
- Bierne N, Tanguy A, Faure M, Faure B, David E, Boutet I, Boon E, Quere N, Plouviez S, Kemppainen P, Jollivet D, Boudry P, David P. Mark-recapture cloning: a straightforward and cost-effective cloning method for population genetics of single copy nuclear DNA sequences in diploids. Mol Ecol Notes. 2007;7:562–566. doi: 10.1111/j.1471-8286.2007.01685.x. [DOI] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal-W - improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Belkir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. Genetix 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, Adaptations, UMR 5000, Université Montpellier 2; 2004. http://www.genetix.univ-montp2.fr/genetix/genetix.htm [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population-structure. Evolution. 1984;38:1358–1370. doi: 10.2307/2408641. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP, version 1.2: population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. http://www.fluxus-engineering.com [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evolutionary Bioinformatics. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA-sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc Nat Acad Sci USA. 2007;104:2785–2790. doi: 10.1073/pnas.0611164104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano TA. Dating of the human ape splitting by a molecular clock of mitochondrial-DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Chevaldonné P, Jollivet D, Desbruyères D, Lutz RA, Vrijenhoek RC. Sister-species of eastern Pacific hydrothermal vent worms (Ampharetidae, Alvinellidae, Vestimentifera) provide new mitochondrial COI clock calibration. Cah Biol Mar. 2002;43:367–370. [Google Scholar]
- Johnson SB, Young CR, Jones WJ, Warèn A, Vrijenhoek RC. Migration, isolation, and speciation of hydrothermal vent limpets (Gastropoda; Lepetodrilidae) across the Blanco transform fault. Biol Bull. 2006;210:140–157. doi: 10.2307/4134603. [DOI] [PubMed] [Google Scholar]
- Faure B, Jollivet D, Tanguy A, Bonhomme F, Bierne N. Speciation in the deep sea: multi-locus analysis of divergence and gene flow between two hybridizing species of hydrothermal vent mussels. PLoS One. 2009;4(8):e6485. doi: 10.1371/journal.pone.0006485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XY, Li WH. Statistical test of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Galtier N, Depaulis F, Barton NH. Detecting bottlenecks and selective sweeps from DNA sequence polymorphism. Genetics. 2000;155:981–987. doi: 10.1093/genetics/155.2.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier-Géré P, Dillmann C. A computer-program for testing pairwise linkage disequilibria in subdivided populations. J Hered. 1992;83:239–239. doi: 10.1093/oxfordjournals.jhered.a111204. [DOI] [PubMed] [Google Scholar]
- Robertson A, Hill WG. Deviations from Hardy-Weinberg proportions: sampling variances and use in estimation of inbreeding coefficients. Genetics. 1984;107:703–718. doi: 10.1093/genetics/107.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kureth CL, Rea DK. Large-scale oblique features in an active transform-fault, the Wilkes fracture-zone near 9-degrees-S on the East Pacific Rise. Mar Geophys Res. 1981;5:119–137. [Google Scholar]
- Naar DF, Hey RN. Speed limit for oceanic transform faults. Geology. 1989;17:420–422. doi: 10.1130/0091-7613(1989)017<0420:SLFOTF>2.3.CO;2. [DOI] [Google Scholar]
- Francheteau J, Armijo R, Cheminee JL, Hekinian R, Lonsdale P, Blum N. 1 Ma East Pacific Rise oceanic-crust and uppermost mantle exposed by rifting in Hess Deep (Equatorial Pacific-Ocean) Earth Planet Sci Lett. 1990;101:281–295. doi: 10.1016/0012-821X(90)90160-Y. [DOI] [Google Scholar]
- Reid JL. On the total geostrophic circulation of the South-Pacific ocean: flow patterns, tracers, and transports. Progr Oceanogr. 1997;39:263–352. doi: 10.1016/S0079-6611(97)00012-8. [DOI] [Google Scholar]
- Lewontin RC, Krakauer J. Distribution of gene frequency as a test of theory of selective neutrality of polymorphisms. Genetics. 1973;74:175–195. doi: 10.1093/genetics/74.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson GH, Mesa KA, Boutilier RG. Genetic population-structure and gene flow in the Atlantic cod Gadus morhua - a comparison of allozyme and nuclear rflp loci. Genetics. 1995;139:375–385. doi: 10.1093/genetics/139.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CT, Tsaur SC, Wu CI. The phylogeny of closely related species as revealed by the genealogy of a speciation gene, Odysseus. Proc Nat Acad Sci USA. 2000;97:5313–5316. doi: 10.1073/pnas.090541597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton RE, Harrison RG. Nuclear gene genealogies reveal historical, demographic and selective factors associated with speciation in field crickets. Genetics. 2003;163:1389–1401. doi: 10.1093/genetics/163.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnucks P. Efficient genetic markers for population biology. Trends Ecol Evol. 2000;15:199–203. doi: 10.1016/S0169-5347(00)01825-5. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Turelli M. Stochasticity overrules the "three-times rule": Genetic drift, genetic draft, and coalescence times for nuclear loci versus mitochondrial DNA. Evolution. 2003;57:182–190. doi: 10.1111/j.0014-3820.2003.tb00229.x. [DOI] [PubMed] [Google Scholar]
- Barton N, Bengtsson BO. The barrier to genetic exchange between hybridizing populations. Heredity. 1986;57:357–376. doi: 10.1038/hdy.1986.135. [DOI] [PubMed] [Google Scholar]
- Nosil P, Harmon LJ, Seehausen O. Ecological explanations for (incomplete) speciation. Trends Ecol Evol. 2009;24:145–156. doi: 10.1016/j.tree.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Harrison RG. Hybrid zones: windows on evolutionary processes. Oxf Surv Evol Biol. 1990;7:69–128. [Google Scholar]
- Wang RL, Wakeley J, Hey J. Gene flow and natural selection in the origin of Drosophila pseudoobscura and close relatives. Genetics. 1997;147:1091–1106. doi: 10.1093/genetics/147.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, Hewitt GM. The genetic-basis of hybrid inviability in the grasshopper Podisma pedestris. Heredity. 1981;47:367–383. doi: 10.1038/hdy.1981.98. [DOI] [Google Scholar]
- Kruuk LEB, Baird SJE, Gale KS, Barton NH. A comparison of multilocus clines maintained by environmental adaptation or by selection against hybrids. Genetics. 1999;153:1959–1971. doi: 10.1093/genetics/153.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier M-H. The ultrafast East Pacific Rise: instability of the plate boundary and implications for accretionary processes. Philos Trans R Soc Lond. 1997;355:341–367. doi: 10.1098/rsta.1997.0012. [DOI] [Google Scholar]
- Fouquet Y, Auzende JM, Ballu V, Batiza R, Bideau D, Cormier MH, Geistdoerfer P, Lagabrielle Y, Sinton J, Spadea P. Variability of the present-day hydrothermal activity along an ultra-fast spreading ridge - East Pacific Rise between 17- degrees-C and 19-degrees-S (Naudur Cruise) C R Acad Sci Ser A. 1994;319:1399–1406. [Google Scholar]
- Haymon RM, Fornari DJ, Edwards MH, Carbotte S, Wright D, Macdonald KC. Hydrothermal vent distribution along the East Pacific Rise crest (9-degrees-09'- 54'N) and its relationship to magmatic and tectonic processes on fast-spreading midocean ridges. Earth Planet Sci Lett. 1991;104:513–534. doi: 10.1016/0012-821X(91)90226-8. [DOI] [Google Scholar]
- Tunnicliffe V, Embley RW, Holden JF, Butterfield DA, Massoth GJ, Juniper SK. Biological colonization of new hydrothermal vents following an eruption on Juan de Fuca ridge. Deep-Sea Res Part I. 1997;44:1627. doi: 10.1016/S0967-0637(97)00041-1. [DOI] [Google Scholar]
- Gilpin M. The genetic effective size of a metapopulation. Biol J Linn Soc. 1991;42:165–175. doi: 10.1111/j.1095-8312.1991.tb00558.x. [DOI] [Google Scholar]
- Leberg PL. Effects of population bottlenecks on genetic diversity as measured by allozyme electrophoresis. Evolution. 1992;46:477–494. doi: 10.2307/2409866. [DOI] [PubMed] [Google Scholar]
- Piálek J, Barton NH. The spread of an advantageous allele across a barrier: The effects of random drift and selection against heterozygotes. Genetics. 1997;145:493–504. doi: 10.1093/genetics/145.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure MF, David P, Bonhomme F, Bierne N. Genetic hitchhiking in a subdivided population of Mytilus edulis. BMC Evol Biol. 2008;8:164–178. doi: 10.1186/1471-2148-8-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morjan CL, Rieseberg LH. How species evolve collectively: implications of gene flow and selection for the spread of advantageous alleles. Mol Ecol. 2004;13:1341–1356. doi: 10.1111/j.1365-294X.2004.02164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers designed for DNA amplification on Alvinella pompejana. Table of sequences for forward and reverse primers used in the study.
Size and exon-intron structure of the four sequenced genes. Schematic drawings of the gene portion used in which exonic regions are represented by boxes.
Allele frequencies, expected and observed heterozygosities and Fis estimated from enzyme loci across vent fields. Table of allozyme frequencies and heterozygosities for each sampled population.
Estimates and 90% Highest Posterior Density (HPD) intervals of demographic parameters from IMa multilocus analysis. Table of 90% HPD intervals for population sizes, migration rates and time since population splitting estimated from the IMa Bayesian computations.