Abstract
In many eukaryotes, the introduction of double-stranded RNA (dsRNA) into cells triggers the degradation of mRNAs through a posttranscriptional gene silencing mechanism called RNA interference or RNAi. In this study, we found that endogenous long-dsRNA was substantially more effective at producing interference than endogenous or exogenous short-dsRNA expression in Giardia lamblia. The effects of this interference were not evident in the highly expressed proteins tubulin or the stage-specific cyst wall protein 2. However, long-dsRNA caused potent and specific interference in the medium subunits of adaptins, the RNA-dependent RNA polymerase, and the exogenous GFP. Our results suggest that the ability of dsRNA antisense to inhibit the expression of these specific types of proteins is indicative of a gene-specific mechanism.
The first evidence of RNA interference or an RNAi mechanism in protozoan parasites was obtained from T. brucei (Ngo et al., 1998). Further, it was shown that RNAi was functional in other protozoan trypanosomatids as well as in apicomplexans Plasmodium falciparum and Toxoplasma gondii (see (Militello et al., 2008). Recently, the silencing of parasite cognate genes by the RNAi mechanism was demonstrated in Entamoeba histolytica (Kaur & Lohia, 2004; Solis & Guillen, 2008) and proposed in Giardia lamblia (Prucca et al., 2008; Saraiya & Wang, 2008), thus suggesting that an RNAi pathway exists in these parasites. When exogenous small RNAs (exosRNA) were introduced in mammalian and plant cells, they caused the transient inhibition of gene expression (Hannon, 2002). A similar effect was observed when both endogenous small RNA (endsRNA) and long double stranded RNA (enddsRNA) were stablely produced by a DNA-based vector system in diverse cells (Brummelkamp et al., 2002; Fire et al., 1998; LaCount et al., 2000; Yokota et al., 2003). In this work, we generated exosRNA, endsRNA, and enddsRNA in Giardia lamblia trophozoites, and examined if a differential effect on the mRNA was produced and whether protein expression could also be correlated with a special phenotype.
Cyst wall protein 2 (CWP2), immunoglobulin heavy chain binding protein (BiP), housekeeping alpha tubulin (Tub), the medium subunits of adaptin complex 1 and 2 (μ1 and μ2), the RNA-dependent RNA polymerase (RdRP), and the exogenous enhanced green fluorescence protein (eGFP) were used to test the suppression of targeted mRNA. In vitro synthesis of 21-25nt-small interference exosRNA was carried out by using the HiScribe RNAi Transcription Kit (New England, BioLabs Inc., Ipswich, Massachusetts) following the suggested protocol (Fig. S1). To test the effects of the exogenous small double-stranded RNAs, we introduced 50 μg of exosRNAs per 108 WB1267 G. lamblia trophozoites by electroporation, as previously described for T. brucei (Ngo et al., 1998), and the mRNA levels were assessed after 48 hr. by semiquantitative RT-PCR (sqRT-PCR). For these assays, no significant effects in any of the mRNA tested were found (Table I). For endogenous synthesis of both 25nt-small endsRNA and 600-1000nt-long enddRNA (Fig. S1), a vector in which dsRNA was synthesized from opposing ran promoters under tetracycline (Tet) regulation was used (GenBank accession number GU395185) (Fig. S2) (Touz et al., 2004). Ten micrograms of the different plasmids obtained were transfected into the WB1267 G. lamblia clone by electroporation following the protocol of Yee and Nash (1995). The day after transfection, the puromycin- or neomycin-selecting drugs were added and the cultures cloned by limiting dilution. To test the effects of endsRNA or enddsRNA, 10 μg/ml of tetracycline (final concentration) were added to the medium lacking the selective drug and the parasites were cultivated for 48 hr. As in the case of exosRNAs, no variations in the mRNA levels were observed when endsRNAs were produced (Table I). Due to the fact that sqRT-PCR demonstrated that neither exosRNA nor endsRNA caused a decrease in the amount of the target mRNA, these specific clones were not investigated further.
Table I. Effect of siRNA and dsRNA on targeted gene expression.
| Gene Target | dsRNA | mRNA (% of reduction) | Immunoblotting (% protein reduction) | IFA (% protein reduction) | % reduction on cell growth | % reduction cyst | Effect over Related Proteins |
|---|---|---|---|---|---|---|---|
| BiP | exosRNA | 0.11 | |||||
| endsRNA | 0.21 | ||||||
| enddsRNA | 0.81,2 | - | NE | ND | ND | ND | |
| Alpha tubulin | exosRNA | 0.11 | |||||
| endsRNA | 0.11 | ||||||
| enddsRNA | 0.41,2 | 15 | NE | NE | ND | ND | |
| μ1 | exosRNA | 0.11 | |||||
| endsRNA | 0.51 | ||||||
| enddsRNA | 66.51,2 | 70 | 80 | NE | 83 (Touz et al., 2004) | ESCP, AcPh, CLHa | |
| μ2 | exosRNA | 0.11 | |||||
| endsRNA | 0.51 | ||||||
| enddsRNA | 99.21,2 | 99 | 90 | 68 (Rivero et al., unpublished) | 98 (Rivero et al., unpublished) | LDLb | |
| RdRP | exosRNA | 0.11 | |||||
| endsRNA | 0.11 | ||||||
| enddsRNA | 37.21,2 | ND | 60 | NE | ND | NEc | |
| eGFP | exosRNA | 0.11 | |||||
| endsRNA | 0.21 | ||||||
| enddsRNA | 47.21 | 90 | 80 | NE | ND | NEd | |
| CWP2 | exosRNA | 0.11 | |||||
| endsRNA | 0.31 | ||||||
| enddsRNA | 22.81,2 | 40 | NE | NE | NE | NEe |
Determined by semiquantitative RT-PCR.
Determined by Slot-blot and densitometry analysis using Gel-Pro Analyzer version 4.5.
NE: no effect detected.
ND: not determined.
The encystation-specific cysteine protease ESCP, the acid phosphatase AcPh, and the clathrin heavy chain CLH were analyzed by IFA in μ1-depleted trophozoites, and showed alterations in their subcellular localizations.
Bodipy-LDL was not internalized in μ2-depleted trophozoites (Rivero et al, submitted).
No effect on VSP expression was found. No other proteins were analyzed.
No effect was found on ESCP, BiP, μ2, CWP1, or CWP2 expression or localization.
No effect was found on CWP2, CWP1, or BiP expression or localization.
When enddsRNAs were produced by using the DNA-based vector principle, mild effects on BiP and Tub were seen (Fig. 1A; Table I). However, a reduction in mRNA of 22 to 99 percent was observed for μ1, μ2, RdRP, eGFP, and CWP2 (Figs. 1A; Table I). dsRNA was assessed by sqRT-PCR (Fig. 1B). Similar qualitative results were obtained by Slot-blot and densitometric analysis (Fig. 2A). Twenty-one enddsRNA clones (3 for each protein tested) were analyzed for protein expression by immunoblotting and IFA. In the example shown for immunobloting and IFA, there was an insignificant effect on the BiP, Tub, and CWP2 protein expression. Conversely, a significant suppression on RdRP, μ1, μ2, and eGFP expression was observed (Figs. 2B, 3). We also found that inducible production of gene-specific long dsRNAs in G. lamblia trophozoites resulted in selective degradation of the corresponding mRNAs, thus suggesting that this phenomenon could have been related to genetic interference by dsRNA or to RNA-mediated RNA degradation, as occurred in other cells. To test this last hypothesis, detection of gene silencing-related small RNA species was performed by hybridization accordingly to the protocol reported by Hutvagner et al. (2000) for small RNA (Hutvagner et al., 2000). The results indicated that double-stranded RNA produced by enddsRNA was not efficiently degraded into siRNA (Fig. 4). The proper purification and detection of small RNA was determined by the presence of sRNA from μ1 (control). The cytoplamic endoribonuclease III Dicer is able to produce mature Micro RNAs (miRNAs), a major class of small RNAs that are involved in gene regulation via a translational repression mechanism. In the same way, the G. lamblia Dicer homologous was previously shown to cleave double-stranded RNA (dsRNA) in vitro (Macrae et al., 2006; Prucca et al., 2008). Conversely, our in vivo assays were unable to detect miRNAs after long-dsRNA production thus questioning the ability of G. lamblia Dicer to digest relatively large dsRNA. This finding is consistent with results reported by Saraiya and Wang (2008), suggesting that G. lamblia Dicer is able to digest snoRNAs of lengths ranging from 60 to 160 nts but not longer ones. Further experiments need to be carried out to elucidate the significance of this finding.
FIGURE 1.
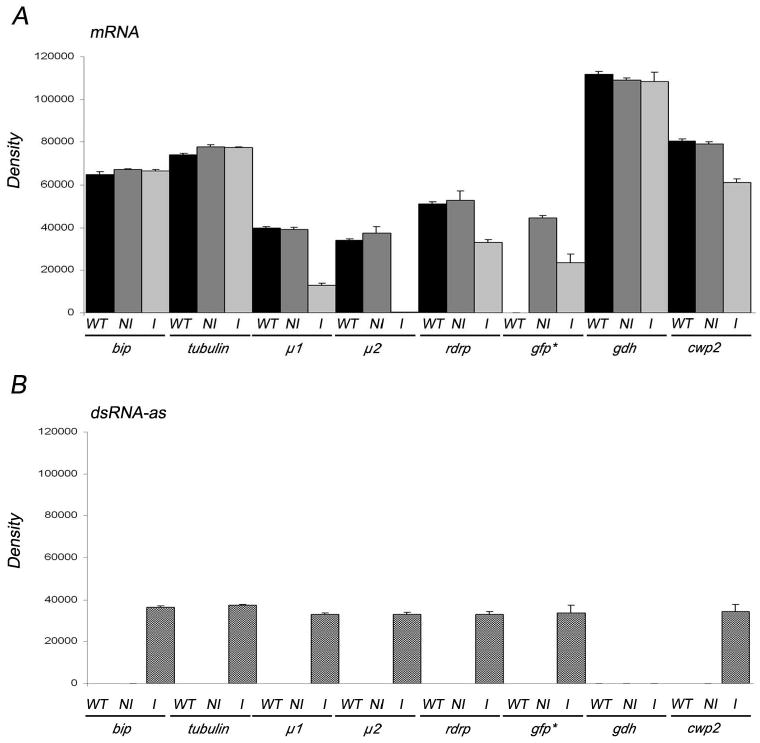
(A) Analysis of gene downregulation by semiquantitative RT-PCR. Bars indicate densitometric assessment of 1 experiment utilized as an example, using 10 ng of total RNA. Total RNA was extracted from wild-type (WT), dsRNA transfected non-induced (NI), or enddsRNA transfected induced (I) growing trophozoites and the One-step RT-PCR kit (Qiagen, Valencia, California) employed for reverse transcription and PCR amplification. Forward and reverse oligonucleotides complementary to the 3′ of each gene tested were used to amplify the endogenous sense mRNA. Only mRNA from μ1, μ2, rdrp, and egfp are significantly reduced after tetracycline induction (I). *mRNA obtained after cotransfection of eGFP and gfp-enddsRNA. gdh mRNAs are shown as control for specificity. Semiquantitative RT-PCR assay using encysting trophozoites reveal that cwp2 mRNAs is slightly reduced comparing induced cells with non-induced or wild-type trophozoites. (B) Bars indicate densitometric assessment of one representative sqRT-PCR experiment. To detect the antisense mRNA from WT, NI, and I cells, a reverse primer was added in the reverse transcription step, followed by the addition of the forward primer for PCR amplification. The antisense RNA from the dsRNA (dsRNAas) vector is only observed in induced trophozoites. No expression of gdh dsRNAas is observed in wild-type or trasfected cells (transgenic μ1dsRNA is shown as example).
FIGURE 2.
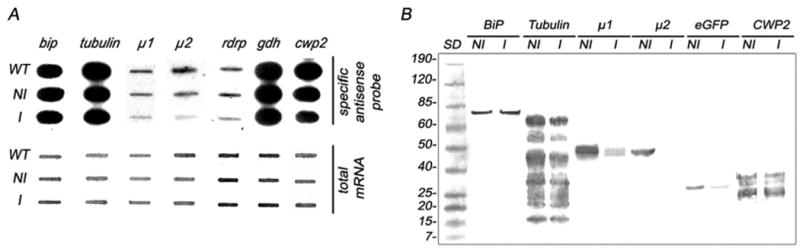
(A) Slot-blot assay using 2 μg of total RNA (top panel). mRNA depletion is observed in μ1, μ2, and rdrp induced trophozoites (I) by testing the 3′ endogenous sequences. gdh antisense probes are shown as controls. cwp2 mRNA is slightly reduced in induced encysting cells. The amount of total RNA for each sample is depicted by staining with methylene blue (bottom panel). (B) Immunoblot assays using anti-HA mAb (SIGMA), 2F5 mAb (Rivero et al., unpublished), and anti-GFP mAb (Santa Cruz Biotechnology, Santa Cruz, CA) to detect μ1-HA, μ2, and eGFP, respectively; show a substantial lower expression of these proteins in induced trophozoites compared with non-induced cells. Uniform BiP, tubulin, and CWP2 protein expression are shown using 9C9, anti-αTubulin (SIGMA), and 7D2 mAbs, respectively.
FIGURE 3.
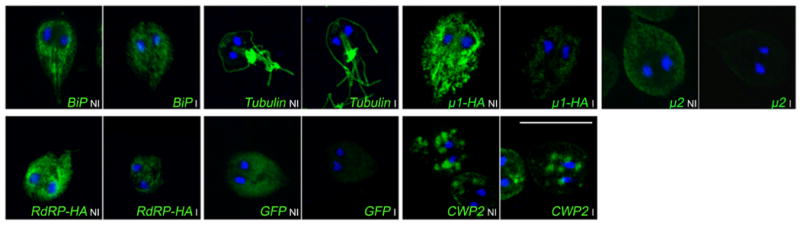
Indirect immunofluorescence assays and confocal microscopy confirms μ1, μ2, RdRP, and eGFP depletion in induced trophozoites. BiP, tubulin, and CWP2 staining in non-induced and induced cells is identical. For μ1, RdRP, and eGFP, double transfections using the expression vector pTubApaHApac and the inducible dsRNAneo were performed. Images were processed in the same way. Bars = 10 μm.
FIGURE 4.
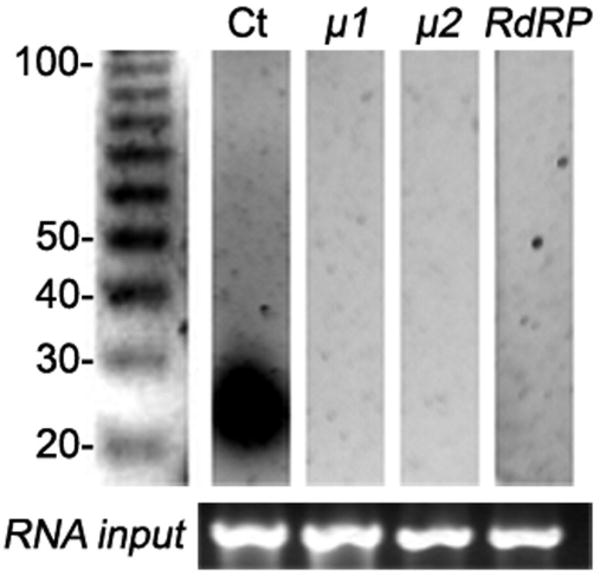
Long foreign dsRNA of μ1, μ2, and RdRP do not produce siRNA. Low-molecular-weight RNAs from the induced trophozoites were electrophoresed in SDS-Urea PAGE, blotted and probed using partially digested in vitro transcribed RNA (Hutvagner et al., 2000). In vitro transcribed 25nt-exoRNA for μ1 was mixed with trophozoite homogenate followed by low-molecular-weight RNA extraction and used as control (Ct). The polyacrylamide gel shows the quality and quantity of RNA (rRNA) by ethidium bromide staining (lower panel). RNA size markers in nucleotides are on the left.
The influence of enddsRNA on G. lamblia growth and differentiation into the cyst form was then examined and it was observed that only the depletion of μ2 caused a significant reduction in cell growth, with manifestations of starvation as occurring after long-term growth in vitro. Moreover, when trophozoites expressing enddsRNA were encysted, we found that both μ1- and μ2-depleted cells showed encystation inhibition. These effects were previously associated with the role of both adaptin subunits in protein trafficking during differentiation (Touz et al., 2004; Rivero et al., 2010). However, because enddsRNA expression did not have a significant effect on BiP, Tubulin, or CWP2 protein expression, in the present study, the role of these proteins during growth and encystation could not be disclosed (see below). This may have been due to the amount of enddsRNA produced being insufficient to inhibit the expression of these particular proteins.
Due to the fact that we were able to produce the downregulation of μ1, μ2, and RdRP, which are involved in identified specific mechanisms, we decided to investigate any possible effects on their related proteins. For μ1, we used specific Ab or the HA-tagged proteins (Fig. S2). The depletion of μ1 produced an arrest of the lysosomal-membrane protein ESCP at the site of sorting and the exclusion of clathrin at this point (Fig. 5) (Touz et al., 2004). However, no effect on the localization of μ2 or on the variant-specific surface protein VSP9B10 was observed compared to control cells (Fig. 5). Another experiment, this time utilizing μ2-depleted cells, showed that this protein was involved in the receptor-mediated endocytosis, thus inhibiting the uptake of the Low-density lipoprotein LDL (Rivero et al., 2010).
FIGURE 5.
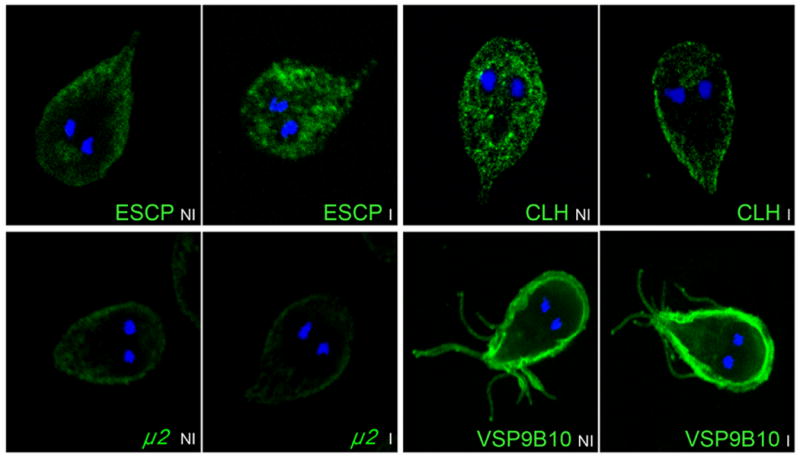
Indirect immunofluorescence assays and confocal microscopy show that ESCP-HA and CLH are retained or excluded from the place of sorting, respectively, when μ1 expression is reduced (ESCPI and CLHI). No alteration in the subcellular localization of μ2 or VSP9B10 is observed in induced cells (μ2I and VSP9B10I). The mAbs anti-HA, 2F5 (Rivero et al., 2010), and anti-9B10 (Nash et al., 2001), were employed as primary Ab to detect ESCP, μ2, and VSP9B10, respectively. GiCLH polyclonal antibody was used for detection of clathrin (Marti et al., 2003).
In wild-type trophozoites, 1 VSP is expressed at any given point in time, except when the trophozoites are going through a switching process. In this regard, it was previously shown that a radical knockdown of RdRP caused by constitutive production of RdRP-specific antisense, resulted in the expression of several vsps transcripts with more than 1 VSP expressed at the same time on the surface of the trophozoite (Prucca et al., 2008). Our results on RdRP downregulation, however, did not show any alteration in the expression pattern compared with controls, when mAbs against 2 different VSPs were used in the WB9B10 clone (Fig. 6). Moreover, RdRP depletion did not produce a significant change in the expression of the mRNA of VSPs, when measured by a consensus probe against the conserved C-terminus of these surface proteins in slot-blotting (Fig. 7A). The fact that we did not observe the changes in VSP behavior as was shown by Prucca et al. (2008), may perhaps be due to the difference in RdRP down regulation using a constitutively expressed antisense (Prucca et al., 2008) vs. Tet-inducible RdRP-enddsRNA (this work). These results evidenced that the success of the protein expression inhibition might include a deep analysis of the target mRNA and the strategy to be used (see below).
FIGURE 6.
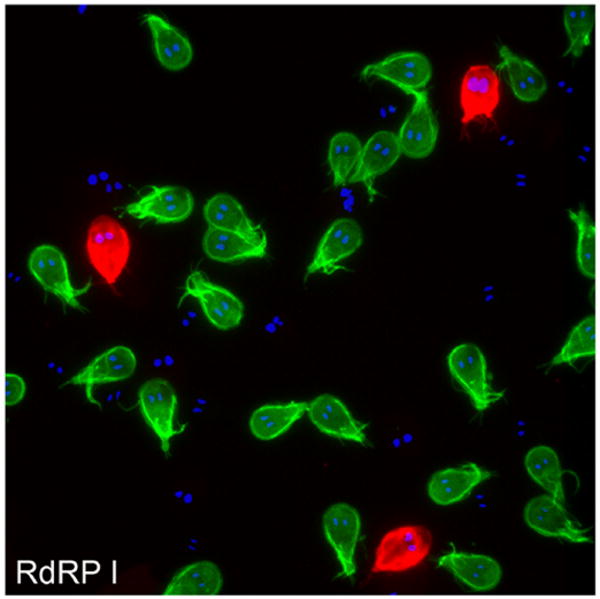
Direct immunofluorescence assays show that after RdRP depletion, VSP9B10 and VSP1267 were detected in different trophozoites using specific 9B10 and 5C1 mAbs, respectively. Bars = 10 μm.
FIGURE 7.
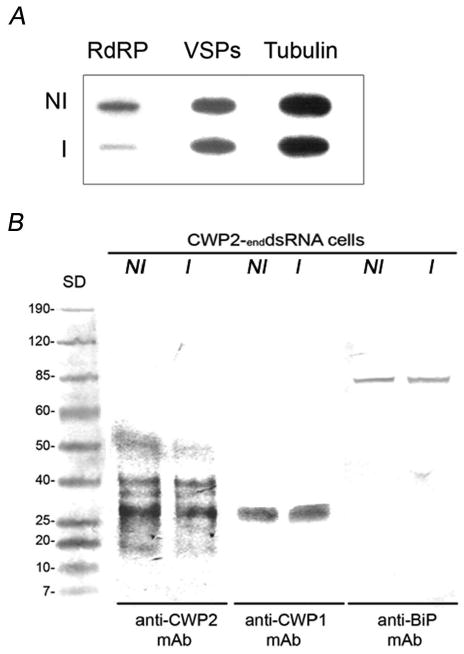
(A) Two μg of total RNA were used for slot-blot assay by using 3′rdrp-, 3′vsp-(CRGDGGBGCCATCGCGGGGATCTC/CGCCTTBCCHCKRCAKATGAACCACCA), or tubulin-antisense probes. NI: total RNA of non-induced transfected trophozoites. I: total RNA of induced transfected trophozoites. Partial reduction on mRNA of RdRP does not have any effect on the amount of vsps mRNA expressed compared with tubulin as control. (B) Immunoblot assays using anti-CWP2 7D2 mAb, anti-CWP1 mAb (Waterborne, Inc. New Orleans, Louisiana), and anti-BiP 9C9 mAb, do not detect substantial changes on the expression of CWP2, CWP1, or BiP (control) in encysting induced trophozoites. NI: non-induced cells. I: induced cells.
Although the effect of CWP2-enddsRNA caused a 22% reduction on its mRNA, no significant effect on CWP2 protein expression could be observed. In addition to the difference in sensitivity between the techniques utilized, we estimate that the reduction in mRNA expression should exceed this 22% in order to observe a substantial change in associated proteins or in the phenotype. This would explain why the CWP2-depleted trophozoites did neither showed significant changes in CWP1 expression (which shares some homology with the mRNA of CWP2) nor any alteration in the rate of cyst production (Fig. 7B; Table I). All results are summarized in Table I.
The present study demonstrates that long-dsRNA, but not small-dsRNA, can be used as a G. lamblia tool to downregulate gene expression. It is possible that, unlike fungi, plants and worms, G. lamblia could not replicate sRNAs and therefore artificial small RNA-directed silencing by transfection does not lead to a long-term effect. To overcome the limited effect of exosRNA in G. lamblia, we developed a DNA-vector-mediated mechanism to express substrates that can be converted into dsRNA in vivo. First, we generated sRNA in situ by transfecting a plasmid encoding two short RNA sequences, which express sense and antisense strands from separate, ran promoters. Our results indicated that endsRNAs did not efficiently inhibit protein expression. These results might be explained by the fact that the endsRNAs expressed failed to recognize the mRNA target since RNAi depends on the sequence that contribute to the unique recognition and precise processing. Subsequently, we utilized a vector system, which combine the dsRNA expression cassette under tetracycline induction and the drug resistance gene on the same plasmid structure. In some cases, we found that the gene transcription regulation was effective but differed from 1 mRNA to another, being less useful for the downregulation of highly expressed proteins such as tubulin, BiP, or CWP2. In addition, the long-dsRNAs induced endogenously by tetracycline were unable to generate siRNA, thus suggesting that G. lamblia did not digest relatively long-dsRNA. Because antisense strategy was shown to be variable depending on the vector used and the target protein (Dan et al., 2000; Prucca et al., 2008; Stefanic et al., 2009; Touz et al., 2005; Touz et al., 2002; Touz et al., 2004), it will be important to determine the conditions that are most appropriate for each special application. Indeed, non-controlled expression of antisense does not generate viable cells when essential proteins are targeted or might even produce controversial results when the target gene product is involved in critical pathways. To overcome these limitations, we designed a vector based on a Tet inducible system that can be used to conditionally regulate the expression of dsRNAs mediating protein knockdown.
Supplementary Material
Acknowledgments
The project was supported by Grant Number R01TW00724 from the Fogarty International Center. The content is solely the responsibility of the authors and does not necessary represent the official views of the Fogarty International Center or the National Institutes of Health. This research was also supported in part by the Agencia Nacional para la Promoción de la Ciencia y Tecnología (FONCyT), the Academy of Science for the Developing World (TWAS), and the National Council for Sciences and Technology (CONICET).
Contributor Information
Maria R. Rivero, Instituto de Investigación Médica Mercedes y Martín Ferreyra. INIMEC – CONICET. Friuli 2434. Córdoba. Argentina
Liudmila Kulakova, W.M. Keck Laboratory for Structural Biology, University of Maryland Biotechnology Institute, Maryland, USA.
Maria C. Touz, Touz. Instituto de Investigación Médica Mercedes y Martín Ferreyra. INIMEC – CONICET. Friuli 2434. 5000. Córdoba. Argentina
Literature Cited
- Ngo H, Tschudi C, Gull K, Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Militello KT, Refour P, Comeaux CA, Duraisingh MT. Antisense RNA and RNAi in protozoan parasites: working hard or hardly working? Molecular and Biochemical Parasitology. 2008;157:117–126. doi: 10.1016/j.molbiopara.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Kaur G, Lohia A. Inhibition of gene expression with double strand RNA interference in Entamoeba histolytica. Biochemical and Biophysical Research Communications. 2004;320:1118–1122. doi: 10.1016/j.bbrc.2004.06.064. [DOI] [PubMed] [Google Scholar]
- Solis CF, Guillen N. Silencing genes by RNA interference in the protozoan parasite Entamoeba histolytica. Methods in Molecular Biology. 2008;442:113–128. doi: 10.1007/978-1-59745-191-8_9. [DOI] [PubMed] [Google Scholar]
- Prucca CG, Slavin I, Quiroga R, Elias EV, Rivero FD, Saura A, Carranza PG, Lujan HD. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature. 2008;456:750–754. doi: 10.1038/nature07585. [DOI] [PubMed] [Google Scholar]
- Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathogens. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Lacount DJ, Bruse S, Hill KL, Donelson JE. Double-stranded RNA interference in Trypanosoma brucei using head-to-head promoters. Molecular and Biochemical Parasitology. 2000;111:67–76. doi: 10.1016/s0166-6851(00)00300-5. [DOI] [PubMed] [Google Scholar]
- Yokota T, Sakamoto N, Enomoto N, Tanabe Y, Miyagishi M, Maekawa S, Yi L, Kurosaki M, Taira K, Watanabe M, Mizusawa H. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. The European Molecular Biology Organization Reports. 2003;4:602–608. doi: 10.1038/sj.embor.embor840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touz MC, Kulakova L, Nash TE. Adaptor protein complex 1 mediates the transport of lysosomal proteins from a Golgi-like organelle to peripheral vacuoles in the primitive eukaryote Giardia lamblia. Molecular Biology of the Cell. 2004;15:3053–3060. doi: 10.1091/mbc.E03-10-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Mlynarova L, Nap JP. Detailed characterization of the posttranscriptional gene-silencing-related small RNA in a GUS gene-silenced tobacco. RNA. 2000;6:1445–1454. doi: 10.1017/s1355838200001096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Rivero MR, Vranych CV, Bisbal M, Maletto BA, Ropolo AS, Touz MC. Biochemical Journal. Immediate Publication; 2010. Adaptor protein 2 regulates receptor-mediated endocytosis and cyst formation in Giardia lamblia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan M, Wang AL, Wang CC. Inhibition of pyruvate-ferredoxin oxidoreductase gene expression in Giardia lamblia by a virus-mediated hammerhead ribozyme. Molecular Microbiology. 2000;36:447–456. doi: 10.1046/j.1365-2958.2000.01863.x. [DOI] [PubMed] [Google Scholar]
- Stefanic S, Morf L, Kulangara C, Regos A, Sonda S, Schraner E, Spycher C, Wild P, Hehl AB. Neogenesis and maturation of transient Golgi-like cisternae in a simple eukaryote. Journal of Cell Science. 2009;122:2846–2856. doi: 10.1242/jcs.049411. [DOI] [PubMed] [Google Scholar]
- Touz MC, Conrad JT, Nash TE. A novel palmitoyl acyl transferase controls surface protein palmitoylation and cytotoxicity in Giardia lamblia. Molecular Microbiology. 2005;58:999–1011. doi: 10.1111/j.1365-2958.2005.04891.x. [DOI] [PubMed] [Google Scholar]
- Touz MC, Gottig N, Nash TE, Lujan HD. Identification and characterization of a novel secretory granule calcium-binding protein from the early branching eukaryote Giardia lamblia. Journal of Biological Chemistry. 2002;277:50557–50563. doi: 10.1074/jbc.M202558200. [DOI] [PubMed] [Google Scholar]
- Nash TE, Luján HT, Mowatt MR, Conrad JT. Variant-Specific Surface Protein Switching in Giardia lamblia. Infection Immunity. 2001;69(3):1922–1923. doi: 10.1128/IAI.69.3.1922-1923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti M, Regos A, Li Y, Schraner EM, Wild P, Muller N, Knopf LG, Hehl AB. An ancestral secretory apparatus in the protozoan parasite Giardia intestinalis. Journal of Biological Chemistry. 2003;278:24837–24848. doi: 10.1074/jbc.M302082200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


