Abstract
Pancreatic cancer is the fourth leading cancer causing deaths in the USA, with more than 30,000 deaths per year. The overall median survival for all pancreatic cancer is 6 months and the 5-year survival rate is less than 10%. This dismal outcome reflects the inefficacy of the chemotherapeutic agents, as well as the lack of an early diagnostic marker. A protein known as prion (PrP) is expressed in human pancreatic cancer cell lines. However, in these cell lines, the PrP is incompletely processed and exists as pro-PrP. The pro-PrP binds to a molecule inside the cell, filamin A (FLNa), which is an integrator of cell signaling and mechanics. The binding of pro-PrP to FLNa disrupts the normal functions of FLNa, altering the cell’s cytoskeleton and signal transduction machineries. As a result, the tumor cells grow more aggressively. Approximately 40% of patients with pancreatic cancer express PrP in their cancer. These patients have significantly shorter survival compared with patients whose pancreatic cancers lack PrP. Therefore, expression of pro-PrP and its binding to FLNa provide a growth advantage to pancreatic cancers. In this article, we discuss the following points: the biology of PrP, the consequences of binding of pro-PrP to FLNa in pancreatic cancer, the detection of pro-PrP in other cancers, the potential of using pro-PrP as a diagnostic marker, and prevention of the binding between pro-PrP and FLNa as a target for therapeutic intervention in cancers.
Keywords: Creutzfeldt-Jakob disease, cytoskeleton, filamin A, glycosylphosphatidylinositol anchor, pancreatic ductal cell adenocarcinoma, peptide signal sequence, pre-pro-prion, prion, pro-prion, signal transduction, transmissible spongiform encephalopathy
From scrapie to prion
Transmissible spongiform encephalopathy (TSE) is a group of subacute, fatal and infectious neurodegenerative diseases in animals and humans [1]. Scrapie, a form of TSE in sheep and goats, has been an endemic in the UK since the 1750s, and has been known to be transmissible since the early 1900s [2]. For decades, owing to its atypical features, the etiologic agent of TSE remained baffling. In 1967, Griffith proposed that the TSE agent might be a self-replicating protein [3]. In 1982, Prusiner and his associates isolated and characterized the pathogen, which they named proteinaceous infectious particle or scrapie prion (PrPSc) (Table 1) [4–6]. Subsequently, it was found that PrPSc is an aberrant, rogue isoform of a normal cellular prion protein (PrP) [7]. Prusiner proposed that all prion diseases share a common pathogenic mechanism, which is based on the conformational conversion of the normal PrP into the infectious and pathogenic, PrPSc [8]. It is thought that the accumulation of PrPSc in the brain then causes neuropathology. Since then, prion diseases have been used synonymously with TSE. Prion disease, such as Creutzfeldt–Jakob disease (CJD) in humans is rare, with an incidence of approximately one case/million people/year [1]. Owing to its rarity, over the years, human prion disease has not received much attention. However, in 1996, a new human prion disease known as variant CJD (v-CJD) gained widespread attention and caused grave public health concerns. It was thought that v-CJD was caused by the transmission of bovine spongiform encephalopathy (mad cow disease) from cattle to humans, owing to the consumption of meats from affected cattle [9]. Since peaking in 2000, the cases of v-CJD have dropped significantly over the years. Therefore, it appears that the original hysteria of a v-CJD epidemic has not materialized.
Table 1.
Key terms in prion biology.
| Term | Definition |
|---|---|
| GPI anchor | A GPI anchor that some proteins use to attach to the outer membrane leaflet of a cell |
| Pre-pro-PrP | A precursor of the mature prion protein that contains the N-terminal peptide signal sequence as well as the C-terminal GPI anchor peptide signal sequence |
| Pro-PrP | A precursor of the mature prion protein, in which the N-terminal peptide sequence has been removed but that retains the C-terminal GPI anchor peptide sequence. This sequence is removed prior to the addition of the GPI anchor |
| PrP | Mature prion protein, which is a highly conserved, widely expressed glycoprotein that is tethered to the outer cell surface by a GPI anchor |
| PrPSc | Scrapie prion, an abnormal, rogue isoform of the normal PrP that is pathogenic and infectious |
| Filamin A | A cytolinker, which links cell surface receptors to actin filaments. Filamin A is an integrator of the cell signaling and cell mechanical force |
GPI: Glycosylphosphatidylinositol; PrP: Prion protein; PrPSc: Scrapie prion protein.
Normal PrP
The human prion gene, PRNP, is located on chromosome 20, at 20p13, with a three-exon structure. The third exon contains the entire open reading frame of the protein. At a protein level, the PrP is a glycosylphosphatidylinositol (GPI)-anchored, highly conserved and ubiquitously expressed glycoprotein [10,11].
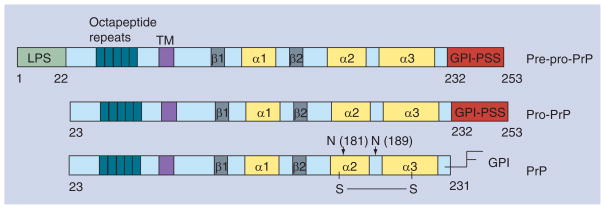
Human PrP is first synthesized as a pre-pro-PrP polypeptide of 253 amino acids (Figure 1). The first 22 amino acids at the N-terminus contain the leader peptide sequence. The last 22 amino acids at the C-terminus contain the GPI anchor peptide signal sequence (GPI-PSS). Both of these sequences are removed in the endoplasmic reticulum and, thus, are absent from the mature PrP.
Figure 1. The synthesis and processing of PrP from pre-pro-PrP to pro-PrP to mature, glycosylated, GPI-anchored PrP.
GPI: Glycosylphosphatidylinositol; LPS: Leader peptide signal sequence; N: N-linked glycans; PrP: Prion protein; PSS: Peptide signal sequence; S–S: Disulfide bridge; TM: Putative transmembrane domain.
Mature, human PrP has 209 amino acids from residue 23–231, and can arbitrarily be divided into three major domains. The N-terminal domain encompasses the first 90 amino acids, and has a conserved motif of five repeating octapeptides. NMR studies have revealed that the N-terminal domain of PrP is flexible, and lacks noticeable secondary structure. The central domain is located between amino acid 110 and 130. On the other hand, the C-terminal region contains a well-defined, globular domain that has two potential N-linked glycosylation sites and a disulfide bridge [12,13]. The protein backbone of a mature PrP has a molecular mass of approximately 23 kDa. The addition of two N-linked glycans, and a GPI anchor completes the maturation of GPI-anchored PrP, which has a molecular mass of approximately 34–39 kDa. Despite the fact that PrP is a relatively small protein, the synthesis, processing and transit of PrP are complex, cell-context dependent and not completely understood [14].
Since the cloning of PRNP, more than 30 PrP-binding partners have been identified. However, the normal physiologic function of PrP remains elusive [15–17]. Cell-surface PrP has been reported to function as an adhesion molecule. This hypothesis is supported by findings that demonstrate that cell surface PrP binds glycosaminoglycan [18,19], laminin receptor [20], selectin [21] and neural cell-adhesion molecule (NCAM) [22]. These molecules are important in cell–cell as well as cell–matrix interactions. This interpretation is further strengthened by a recent finding demonstrating that PrP is involved in the proliferation as well as the distribution of desmosomal-associated proteins, such as desmoglein in human and mouse enterocytes [23].
The octapeptide-repeat region of PrP binds divalent cations, such as Cu2+ and Zn2+ [24,25]. Binding of Cu2+ enables PrP to function as an antioxidant in a cell context-dependent manner [24]. Thus, PrP functions as a metal transporter and plays a role in regulating oxidative stress [24,25]. In addition, PrP has also been reported to interact with stress-inducible protein [26], plasminogen [27], heat shock proteins [28], dystroglycan [29], glypican 1 [30], Grb2 [31] and tubulin [32], as well as lipids [33] and nucleic acids [34]. However, the biological significance of these interactions is not clear.
Accumulated evidence suggests that cell surface PrP also participates in signal transduction [35]. PrP is present on the cell surface in lipid rafts or caveolae, which are microdomains on the cell membrane that are important in organizing signal-transducing platforms [36]. Supporting this interpretation is evidence demonstrating that PrP colocalizes and interacts with caveolin 1 – a major component of the caveolae [37–39]. Caveolin 1 can positively or negatively impact multiple signal-transducing pathways, such as G protein-coupled receptors, MAPK/ERK, PKC, as well as Src family kinases, such as Fyn and Src [37,40]. It appears that PrP can participate in numerous signal-transducing pathways either by binding directly to the signaling molecule or by interacting with caveolin 1.
Prion protein plays a role in apoptosis in a cell context-dependent manner. PrP regulates the activity of p53 and sensitizes neurons to apoptotic stimuli [41]. On the other hand, PrP has also been reported to have antiapoptotic function. For example, neuronal cell lines from Prnp−/− mice are more susceptible to apoptosis than similar cell lines from wild type, Prnp+/+ mice [42]. In a cAMP/PKA-dependent manner, PrP provides protective signals to neurons [43]. PrP inhibits the functions of Bax and, thus, protects cells against Bax-mediated apoptosis [44]. PrP also protects neurons against metal-induced toxicity (e.g., manganese) and cell death by interfering with its transport [45]. In a human breast cancer cell lines, PrP protects the cells from TNF-induced cell death [46]. To unravel these seemingly contradictory findings, it has been recently suggested that PrP functionality is pathway-dependent; PrP is proapoptotic during endoplasmic reticulum stress and antiapoptotic during oxidative stress-induced cell death [47]. PrP also binds amyloid-β oligomers, which then cause synaptic dysfunction and contributes to neurodegeneration in Alzheimer’s disease [48]. Another study suggests that whether PrP is neuroprotective also depends on the underlying pathogenic mechanism [49].
While most of the PrP-interacting partners bind to the N-terminus or the C-terminus globular domain of PrP, the central region of PrP, from residues 113–128, also contain a stretch of highly conserved, small hydrophobic amino acids. This region can function as a transmembrane domain in cell lines [50]. Furthermore, expression and accumulation of this aberrant PrP isoform in transgenic mice causes neurodegeneration [51]. However, whether this region is important in the normal biology of PrP is not clear.
Adding to the complexity is the recent realization that cell-surface PrP is proteolytically cleaved by multiple proteases, such as ADAM10 and TACE [52]. Some of the truncated PrP species are present on the cell surface, and may be important in prion functionality as well as the pathogenesis of prion disease. In addition to the cell surface, a GPI-anchored PrP is detected in the nucleus of proliferation cells [23]. However, the function of the GPI-anchored PrP in the nucleus is not clear. PrP is also detected in body fluids, such as blood [53], milk [54] and urine [55]. The PrP species detected in blood and urine have been reported to be infectious, indicating the presence of PrPSc [53,55]. Most of the normal PrP species detected in the circulation are truncated PrP species lacking a portion of the C-terminal globular domain.
At present, it is difficult to conceive how a relatively small protein can interact with so many partners that are present in different cellular compartments. It should be noted that despite all the important functions that have been attributed to PrP, a Prnp−/− mouse is apparently normal without any overt aberrant phenotype [56]. Therefore, the normal physiologic function of PrP remains an enigma.
The GPI anchor
Prion protein is a GPI-anchored protein [8]. It is not clear why some proteins are GPI anchored while others are not. It has been estimated that approximately 0.5–1% of the mammalian genome encodes for GPI-anchored proteins [57–59]. The biosynthesis of the GPI anchors and their attachment to proteins are complex, and protein- and cell-context dependent [57–59]. At least 24 genes have been identified to be important in this process. The common core structure of the GPI anchor is synthesized in the endoplasmic reticulum in a stepwise mechanism. The first step is the transfer of N-acetyl-glucosamine (GlcNAc) from UDP-GlcNAc to phosphatidylinositol (PI) to yield GlcNAc-PI. This reaction is catalyzed by a α1–6 GlcNAc transferase complex, which is composed of seven gene products: PIG-A, PIG-C, PIG-H, GPI-1, PIG-Y, PIG-P and DPM2. The second step is the de-N-acetylation by PIG-L to generate GlcN-PI. Subsequently, three mannose residues are sequentially added. The last step in GPI anchor processing is attachment of the already assembled GPI structure, en bloc, to the newly synthesized pro-protein in a transamidase reaction. This attachment occurs concurrently with the cleavage of the C-terminal GPI-PSS, at a site known as the ω site [57–59]. The ω residue for mammalian proteins is confined to amino acids glycine, serine, cysteine, alanine, aspartic acid and asparagine. There is no other obvious motif in the GPI-PSS that signals the transamidase reaction.
In general, the GPI-PSS contains between 15 and 25, small hydrophobic amino acids, similar to a typical transmembrane domain. Substitution of a single amino acid at the ω site of Qa2, a normally GPI-anchored protein, prevents its GPI anchor modification [60]. Nonetheless, Qa2 is still present on the cell surface, as an integral membrane protein; in this case, the Qa2 uses the GPI-PSS as a surrogate transmembrane domain. Using an in vitro GPI anchor assembly assay, Chen et al. reported that the GPI-PSS of PrP is intrinsically inefficient (14%) compared with nine other GPI-PSS (ranging from 24 to 60%) in accepting the GPI anchor [61]. However, the significance of this finding is not known.
Interestingly, it has been reported that exchanging the GPI-PSS of NCAM for the GPI-PSS of carcinoembryonic antigen generates a mature protein that has a NCAM external domain, but carcinoembryonic antigen-like biological properties [62–64]. Based on these findings, it is postulated that the GPI-PSS posses cryptic biological information that specifies the addition of a particular functional GPI anchor that, ultimately, determines the function of the mature protein. It is interesting to note that while the coding region of the human PRNP and mouse Prnp is approximately 85% conserved, their GPI-PSSs are identical. On the other hand, their N-terminal peptide sequences, which are also deleted prior to maturation, share only 60% identity [65]. The significance of this conservation is not known.
Accumulated evidence suggests that aberrations in the GPI-anchor assembly pathway may contribute to oncogenesis. mRNAs for some genes in the GPI-anchor pathway, such as PIG-T, PIG-U and GPAA1, are upregulated in bladder, breast, and head and neck cancers [66,67]. Overexpression of PIG-T and GPAA1 in 3T3 cells enhances their invasiveness in vitro and growth in vivo. Conversely, downregulation of PIG-T and GPAA1 in breast cancer cell lines inhibits their growth [68]. Increased GPAA1 expression is associated with hepatocarcinoma’s poor cellular differentiation and poor prognosis [69]. The underlying mechanisms causing these effects are not known. The oncogene, KRAS, is frequently mutated in human cancers [70,71]. In yeast, Ras regulates the function of a complex, GPI–N-acetylglucosaminyltransferase, which catalyzes the first step in GPI anchor biosynthesis, transferring a GLcNAC from UDP-GlcNAc to an acceptor PI in the endoplasmic reticulum. Furthermore, the GTP-bound Ras is associated with the GPI–N-acetylglucosaminyltransferase complex and inhibited its activity in vivo [72]. It is worth noting that some of the genes essential for GPI anchor modification are located in chromosomal regions, which are amplified, deleted or mutated in human cancers. For example, in human pancreatic cancer 8q24.3 (GPAA-1), 9p13.3 (PIG-O) and 14q11–24 (PIG-H) were amplified, while 16p13.3 (PIG-Q), 17p13.1–12 (MPDU-1), 1p36 (PIG-V) and 21q22.2 (PIG-P) were deleted [73–75]. Some of these chromosomal abnormalities may have impacted the integrity of the GPI anchor pathway genes.
Since PrP has been associated with cellular survival, proliferation and differentiation, aberrant PrP function may contribute to tumorigenesis. It is reported that PrP is overexpressed in human gastric cancers and contributes to their resistance to chemotherapeutic agents [76]. An expression microarray study found that PRNP is also overexpressed in human colorectal cancers [77]. PRNP is one of the 25 genes that are overexpressed in pancreatic cancer cell lines [78]. However, the mechanism by which overexpression of PrP promotes tumorigenesis is not clear. In 2007, we initiated a study to screen for PrP expression in a panel of human tumor cell lines, starting with pancreatic cancers.
PrP in pancreatic ductal cell adenocarcinoma
The majority of the human pancreatic cancers are ductal cell adenocarcinoma (PDAC), which is the fourth leading cause of cancer deaths in the USA [79,80]. Patients with PDAC have dismal prognosis as the overall median survival is 6 months and the 5-year survival rate is less than 10%. Progression of human PDAC correlates with a series of histological changes from a flat, columnar epithelium to a papillary, mucinous epithelium, with increasing loss of cellular architecture and with nuclear atypia [81–83]. These lesions are referred to as pancreatic intraepithelial neoplasia (PanIN). PanIN is divided into three stages: PanIN-1, -2 and -3. These morphological changes are accompanied by increasing numbers of genetic alterations.
Genome-wide aberrations have impeded pinpointing the culprit genes in the etiology of human PDAC. Nevertheless, there is significant progress in identifying the molecular mechanisms underlying PDAC carcinogenesis. The most common genetic lesions found in human PDAC are mutations in KRAS, TP53, DPC4 and CDNK2A [84–86]. It is now generally accepted that the KRAS mutation is one of the earliest, and most important genetic lesion in the development of PDAC; the majority of PDAC cases have a mutation in codon 12 of KRAS, substituting a glycine with aspartate, valine or arginine.
Our research has demonstrated that PrP is expressed in a panel of human PDAC cell lines (n = 7) [87]. However, in contrast to normal PrP, which is glycosylated and GPI anchored, in the PDAC cell lines, PrP is neither glycosylated nor GPI anchored; it exists as pro-PrP retaining its GPI-PSS (Figure 1). This deficiency is not caused by a global failing of the GPI anchor machinery in the PDAC cell lines; two normally GPI-anchored proteins, CD55 and flotillin 1 remain GPI anchored in the PDAC cell lines. Despite lacking a GPI anchor, the pro-PrP is present on the PDAC cell surface, using the GPI-PSS as a transmembrane domain. Our immunoblotting results with multiple anti-PrP monoclonal antibodies (mAbs) suggest that pro-PrP is the only PrP species detected in the PDAC cell lines. However, some other PrP species, such as glycosylated or GPI-anchored PrP, may be present in these cells but at levels that are under the detection limit of immunoblots.
To further study the biological functions of pro-PrP, we attempted to identify proteins in the PDAC cell lines that physically interact with pro-PrP and, thus, could be copurified with PrP. While many proteins did copurify with anti-PrP mAbs as well as control mAbs, one protein, with a relative molecular mass of 280 kDa, consistently copurified only with anti-PrP mAbs but not with control mAbs. Mass spectrometry analysis revealed that this protein is filamin A (FLNa). FLNa is an actin-binding cytolinker, which connects cell surface molecules to the actin filaments of the cytoskeleton and, thus, integrates signaling events with cellular mechanical forces [88–90]. Upon binding F-actin filaments, FLNa promotes high-angle branching of actin filaments to maintain a 3D orthogonal network, which is important in cell-shape maintenance as well as cell migration. FLNA is mapped to chromosome Xq28 [89,90]. Two transcript variants, encoding different isoforms, have been found for FLNA. In males, FLNa deficiency caused by a null mutation is embryonic lethal. In females, depending on the nature and local of the mutation, it causes several developmental syndromes involving neuronal, skeletal and connective tissues [89,90]. The Flna−/− mouse is nonviable owing to widespread developmental defects in the heart and vascular structures [91,92]. Whether abnormality in FLNA contributes to tumorigenesis is not known; patients with FLNA mutations are not known to have a higher incident of cancers.
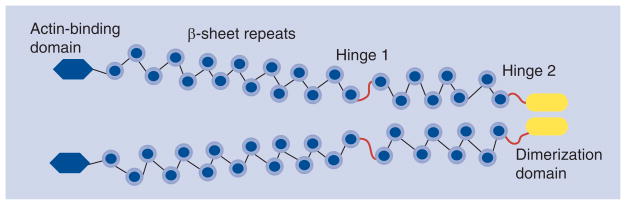
Native FLNa is a homodimer; each subunit contains a F-actin-binding domain at the N-terminus followed by 24 β-sheet Ig-like domains at the C-terminus [89–91] (Figure 2). More than 40 proteins have been reported to interact with FLNa. These molecules include signal-transducing molecules, other scaffolding proteins and adhesion molecules, as well as growth factor receptors [89–91]. All FLNa-interacting partners bind to the C-terminal Ig-like domains from domain 10 to domain 24 of FLNa. Domain 24 is the dimerization domain and is essential for optimal FLNa function [88–90].
Figure 2. Dimeric filamin A.
At the N-terminus of each monomeric unit, there is an actin-binding domain, followed by 24 β-sheet Ig-like domain. Domain 24 is the self-association domain. The two hinge regions provide flexibility to permit high-angle binding of the F-actin filament.
The atomic structure of the interface between the FLNa Ig-like domain and some of the common FLNa binding partners has been resolved [93–95]. These FLNa-binding partners share a conserved hydrophobic amino-acid motif. Unexpectedly, ClustalW alignment suggests that the GPI-PSS of pro-PrP contains a FLNa-binding motif. Binding of FLNa to pro-PrP was first demonstrated by in vitro pull-down assays using recombinant FLNa and recombinant pro-PrP. FLNa only binds pro-PrP but not mature PrP, which lacks the GPI-PSS. Second, in the PDAC cell lines, pro-PrP and FLNa are also copurified by immunoprecipitation in both directions. Third, PrP and FLNa colocalize in the PDAC cell lines, as revealed by immunofluorescent staining and observed in a confocal microscope [87].
The presence of an FLNa-binding motif in the GPI-PSS appears to be specific for PrP. We examined 14 GPI-PSS from other normally GPI-anchored proteins and we found that only the GPI-PSS of PrP has the FLNa-binding motif. Therefore, even if some other normally GPI-anchored proteins also exist as proproteins, retaining their GPI-PSS, in the PDAC cell lines, they are not expected to bind FLNa. The reason that a FLNa-binding motif is present in a normally discarded region of PrP is not known.
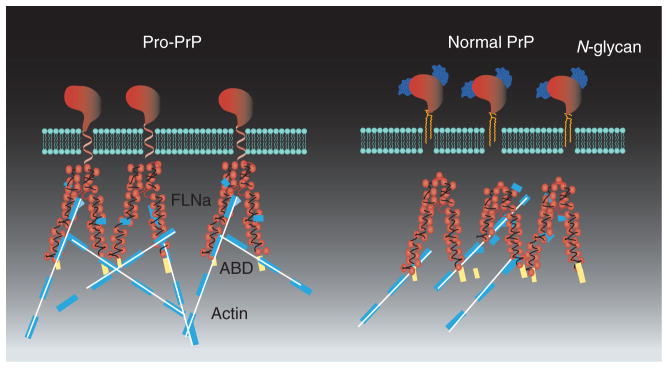
More recent studies reveal that pro-PrP has multiple binding sites at the C-terminal Ig-like domains of FLNa, including domains 10, 16, 17, 18, 20, 21 and 23 [Li CY, Yu S, Nakamura F et al., Pro-prion promotes melanoma cells spreading and migration by binding to FLNa and modulates its interaction with integrin β1. Submitted]. This finding is not unexpected because the Ig-like domains are highly conserved. However, whether all these binding sites are available for pro-PrP binding in native, dimeric, FLNa is not known. On the other hand, we found that the last five amino acids at the C-terminal end of the PrP GPI-PSS are critical for FLNa binding. Removal of these five amino acids completely eliminates its FLNa-binding capacity. These results suggest that the GPI-PSS of PrP is able to transverse the membrane bilayer, and some of the amino acids are exposed in the cytosolic side of the membrane, enabling it to bind FLNa (Figure 3).
Figure 3. Cell surface pro-PrP and normal, glycosyated, GPI-anchored PrP.
Cell surface pro-PrP is using the GPI-PSS as a transmembrane domain. The GPI-PSS transverses the membrane and interacts with FLNa. The ABD of FLNa binds actin filaments. The normal PrP is GPI-anchored and glycosylated.
ABD: Actin-binding domain; GPI: Glycosylphosphatidylinositol; PrP: Prion protein; PSS: Peptide signal sequence.
Consequences of binding between pro-PrP & FLNa
To study the contribution of pro-PrP to PDAC biology, we downregulated PrP expression in these cell lines using PrP-specific shRNA. Reducing PrP expression in the PDAC cell lines did not alter the expression level of FLNa; however, it did drastically change the spatial distribution of FLNa [87]. In control cells, FLNa is mainly present close to the inner-membrane leaflet in the leading edges. In PrP downregulated cells, FLNa is rare in the leading edges, and more concentrated in the cytosol. It appears that without PrP, FLNa is unable to reach the inner membrane leaflet.
As expected, downregulation of PrP also alters the organization of the actin filaments. In vitro, the PrP downregulated cell lines proliferate more slowly and are less invasive than control cells. In vivo, their growth in nude mice is inhibited. Thus, binding of pro-PrP to FLNa enables the PDAC cell lines to proliferate faster, to be more invasive in vitro and to have a more rapid growth rate in vivo. The precise mechanism by which binding of pro-PrP alters the function of FLNa is not clear. Binding of pro-PrP may physically remove FLNa from its normal environment and, thus, render it unable to interact with its normal partners. Alternatively, binding of pro-PrP may compete for binding sites on FLNA that are normally occupied by its interaction partners. Furthermore, expression of pro-PrP may also disrupt the normal function of GPI-anchored PrP, which then contributes to the biology of the PDAC cell lines.
At the molecular level, PrP downregulated cell lines have reduced levels of p-cofilin-1, a critical regulator of the actin filament polymerization. On the other hand, the levels of p-Rac1, a Rho-GTPase; p-ERK-1/2 and p-MEK-1, two serine/threonine kinases in the MAPK pathway; and p-Fyn, a Src family tyrosine kinase, are markedly increased in the PrP downregulated cells. Therefore, reducing the expression of PrP in the PDAC cell lines appears to have effects on multiple signal transduction pathways. More than 40 proteins have been reported to bind FLNa [88–90]. It is almost certain that binding of pro-PrP to FLNa will have rippling effects on the binding of FLNa to some of its binding partners, such as integrins, which are known to play critical roles in cellular adhesion, invasion and migration.
The reason why the GPI-PSS of PrP is not removed in the PDAC cell lines is not known. The GPI anchor-modification process is complex, and protein- and cell-type dependent [57–59]. A mutation in the PRNP gene is not the cause; sequencing of PRNP in six PDAC cell lines revealed no mutation. It should be noted that the GPI-PSS of PrP is intrinsically inefficient compared with other GPI-anchored proteins [61]. Thus, a slight defect in the GPI anchor assembly machinery in PDAC may have a more dramatic effect on PrP than other GPI-anchored proteins with a more efficient GPI-PSS, such as CD55, which is GPI-anchored in the PDAC cell lines. Pro-PrP is a precursor of mature PrP. In cell lines that express a GPI-anchored PrP, pro-PrP is undetectable. Therefore, in these cell lines, either the processing of pro-PrP or the removal of unprocessed pro-PrP is very efficient. On the other hand, a defect in the quality-control system in the endoplasmic reticulum or in the removal of the unprocessed pro-PrP, presumably by the proteasomal degradation machinery, may also contribute to the accumulation of pro-PrP.
In the PDAC cell lines, PrP is also not glycosylated. It is known that the presence of the N-linked glycans on PrP is not required for GPI anchor modification [96]. On the other hand, the presence of a GPI anchor has been reported to influence the glycosylation of Thy-1, a GPI-anchored protein [97]. Thus, failure to remove the GPI-PSS may cause PrP to not be glycosylated. The lack of N-linked glycans may then alter the metabolism or transit of pro-PrP, contributing to its accumulation in the PDAC cell lines.
PrP is a poorer prognosis marker in human PDAC
We also determined whether our findings in PDAC cell lines have clinical relevance. For this purpose, we investigated the expression of PrP in normal human pancreas, in pancreas with pancreatitis, in pancreas with preneoplastic lesions, such as PanIN-1, -2 and -3, and in pancreas with PDAC by immunohistochemcial staining. In normal human pancreas, only islet cells demonstrated PrP immunoreactivity; neither acinar nor ductal epithelial cells stained for PrP. PrP was also undetectable in the duct cells in chronic pancreatitis, and PanIN-1 and -2. Approximately 13% PanIN-3 specimens showed weak PrP staining. Among the 83 PDAC cases examined, 34 (41%) showed strong PrP staining. All PDAC tumor cells reacted strongly with the anti-GPI-PSS antiserum. Thus, as in the PDAC cell lines, PrP exists as pro-PrP in human PDAC lesions [88].
Most interestingly, we found that the expression of pro-PrP is associated with poorer clinical outcome. Patients with intratumoral pro-PrP had a median survival time of 360 days, while patients without PrP expression in their tumors had a mean survival time of over 1000 days. Furthermore, this association is independent of other factors, such as age and gender of the patient, as well as the size or differentiation stage of the tumor. It is clear that PrP expression is not essential for PDAC initiation because only 41% of the PDAC cases have detectable PrP. However, the presence of PrP is associated with poorer clinical outcome, suggesting that, similar to PDAC cell lines, PDAC tumors with PrP have a growth advantage and, thus, are more aggressive.
Little is known about the regulation of PrP expression, either at the gene or protein level in the pancreas. PRNP has been reported to be upregulated in BxPC 3, Capan 1 and five other PDAC cell lines [78]. However, other gene profiling studies have not identified PRNP as a contributing factor in human PDAC [73–75,98]. The reasons for these discrepancies are not known. More than 90% of PDAC cases have a KRAS mutation and inactivation of CDKN2A, but, only 41% of the PDAC cases have detectable PrP. Therefore, it is unlikely that a mutation in KRAS or inactivation of CDNK2A by itself is responsible for the activation of PrP expression in PDAC. On the other hand, gene profiling is much more sensitive than immunohistochemcial staining; it is possible that immunohistochemical staining may not be able to detect PDAC cases that express low levels of PrP. Alternatively, other genetic anomalies, such as mutation in TP53 and/or DPC-4, which are found in approximately 50% of PDAC cases, must have occurred to facilitate the expression of PrP in PDAC. It is interesting to note that in the promoter region of PRNP, there is a potential p53 binding site [99], and PrP regulates p53 function [33].
So far, only resectable PDAC cases have been studied. Nonresectable PDAC is rarely available. If our interpretation that expression of PrP provides PDAC cells with growth advantages is correct and PrP-positive PDAC is more aggressive, then we will expect a larger proportion of the nonresectable PDAC to express PrP.
PrP & FLNa in human melanomas & other tumors
Filamin A is an integrator of cell signaling and mechanics and, thus, participates in many biological responses [88–90]. However, despite its importance in cellular biology, FLNa is dispensable for cell-autonomous survival. Some human melanoma cell lines, such as M2 and -3 do not express FLNa [100]. These cells lack actin fiber bundles and, thus, are impaired in their cellular migration in vitro. This phenotype is revised in A7 cells, which are derived from M2 cells after the transfection of human FLNa [101]. This pair of isogenic cell lines has been used extensively to study the functions of FLNa [102–105]. Biological responses, such as signal transduction and cellular migration, observed in A7 cells but not in M2 cells, have been attributed to the functions of FLNa [101–105].
More recently, we found that both M2 and A7 cells express pro-PrP. Similar to the PDAC cell lines, in A7 cells, pro-PrP binds FLNa; binding of pro-PrP to FLNa modulates cellular behaviors, such as cell spreading and migration [Li CY, Yu S, Nakamura F et al., Pro-prion promotes melanoma cells spreading and migration by binding to FLNa and modulates its interaction with integrin β1. Submitted]. Reduction of pro-PrP expression in A7 cells inhibits A7 cell spreading and migration. Therefore, in A7 cells, FLNa does not act alone. It is the binding between pro-PrP and FLNa that influences the function of FLNa in A7 cells.
Whether PrP is a marker in human melanoma tumorigenesis has never been studied. In normal human skin, only epithelial cells and sporadic mononuclear cells within the dermis demonstrated weak PrP immunoreactivity [106]. However, it is known that not all melanoma cell lines express FLNa [100,101]. Interestingly, the patterns of FLNa expression in melanocytes in nevi and in melanomas vary, depending on the location of the lesion – whether it is in the epidermis or dermis. It was postulated that FLNa might be important in cell–cell adhesion, and in tumor cell motility during tissue invasion from the epidermis to the dermis [107]. Therefore, immunostaining for pro-PrP and FLNa in melanoma biopsies may provide new insights into the role these molecules play in human melanoma tumorigenesis.
Hepatocarcinoma (HCC) is rare in the USA, but is one of the most common tumors worldwide. The majority of HCC cases are secondary to either a viral infection (hepatitis B or C viruses), or cirrhosis caused by toxic agents or alcohol consumption [108]. A PrP species with molecular mass similar to pro-PrP is detected in five human HCC cell lines. All five HCC cell lines also express FLNa. Small-cell lung carcinoma is generally thought to be a neuroendocrine tumor originating from neuroendocrine cells in bronchi [109]. In three human small-cell carcinoma cell lines, FLNa, but not PrP, was undetected. This observation is consistent with our finding in patient biopsies; neuroendocrine tumors originating from the lung did not express PrP (not shown). On the other hand, neither PrP nor FLNa was detected in two human neuroblastoma cell lines. The numbers of cell lines we have studied were rather small, nevertheless, these findings suggest that the contribution of PrP and FLNa to tumor biology does not apply to all tumor types.
Binding of pro-PrP to FLNa as a potential target for therapeutic intervention in human cancers
The PrP binds nucleic acid [38]. When a recombinant PrP (rPrP) is mixed with an expression plasmid in vitro, the rPrP/DNA complex is taken up by cells – leading to gene expression – in a Ca2+-dependent manner [110]. Interestingly, rPrP, by itself, is also taken up by cells. However, cells do not take up rPrPΔKKRPK, which lacks the KKRPK motif. When mixed with DNA, a pentapeptide KKRPK, but not KKKKK, is sufficient for DNA internalization and expression. Recently, we found that if the cells were incubated with the peptide for a longer time (>1 h), the peptide can enter the cells without Ca2+ [Li CY, Yu S, Nakamura F et al., Pro-prion promotes melanoma cells spreading and migration by binding to FLNa and modulates its interaction with integrin β1. Submitted].
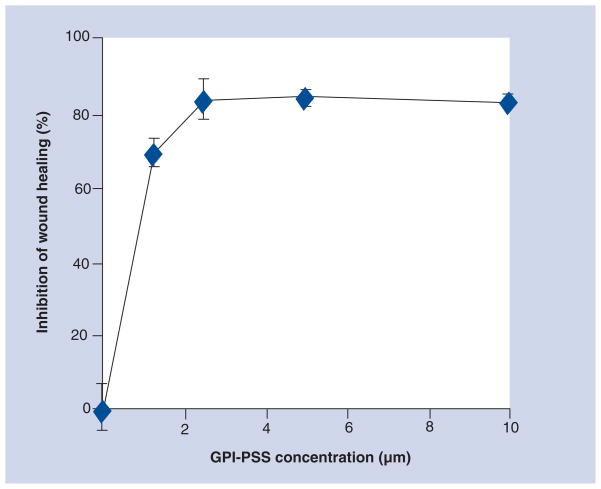
We have previously reported that the in vitro copurification of pro-PrP and FLNa was inhibited in the presence of a synthetic peptide corresponding to the GPI-PSS of PrP [87]. Based on the aforementioned findings, we hypothesized that if we link a KKRPK motif to the N-terminus of the PrP GPI-PSS, the peptide, KKRPK-PrP-GPI-PSS, might be able to enter cells, and compete for the binding of pro-PrP to FLNa. Indeed, the synthetic peptide is able to block the migrations of melanoma cell line A7 [Li CY, Yu S, Nakamura F et al., Pro-prion promotes melanoma cells spreading and migration by binding to FLNa and modulates its interaction with integrin β1. Submitted], as well as a hepatocarcinoma cell line (Figure 4) in a wound-healing assay. The inhibition is peptide-concentration dependent. These results provide the ‘proof of priciple’ that the binding of FLNa to pro-PrP could be a target of therapeutic intervention in human cancers that have both pro-PrP and FLNa.
Figure 4. Inhibition of hepatocarcinoma cell migration with the KKRPK-PrP-GPI-PSS synthetic peptide.
Various concentrations of the synthetic peptide were added at the beginning of the wound-healing assay. At 24 h after the assay, the areas of the wound were imaged on a microscope equipped with a digital camera system, and quantified. Inhibition of cell migration was determined by comparing the healed area of nontreated cells with the healed area of cells treated either with the KKRPK-PrP-GPI-PSS peptide or the KKRPK-control peptide.
GPI: Glycosylphosphatidylinositol; PrP: Prion protein; PSS: Peptide signal sequence.
Conclusion & future perspective
Our finding that a protein, which was infamous owing to the role it plays in mad cow disease, is involved in human pancreatic cancer is totally unexpected. We hypothesize that the fatal attraction between pro-PrP to FLNa is the underlying mechanism by which PrP-positive PDAC cells have a growth advantage. As an integrator of cell signaling and mechanics, many proteins have been reported to bind FLNa. It is almost certain that binding of pro-PrP would have disrupted the interaction between FLNa and some of these proteins. Identifying these proteins will provide insights into the mechanisms by which expression of pro-PrP modulates tumor cell biology. In addition, identifying the underlying mechanisms that cause the retention of the GPI-PSS on PrP in cancer cell lines will help us understand the cell biology of the GPI-anchor modification pathway and the roles it plays in tumor biology. Furthermore, since high levels of soluble PrP are detected in the culture supernatants of the PDAC cell lines [88], therefore, soluble pro-PrP may be present in the circulation or body fluid of patients with PDAC. Detection of pro-PrP may provide an early and noninvasive method for detecting PDAC. In addition, prevention of the interaction between pro-PrP and FLNa could provide a novel target for therapeutic intervention in PDAC. Finally, our finding that pro-PrP is also detected in some other tumors, such as melanoma and hepatocarcinoma, suggests that the aberrant processing of prion is not only limited to pancreatic cancer. On the other hand, the absence of either PrP or FLNa in other tumors, such as small-cell lung carcinomas and neuroblastomas, suggests that an interaction between pro-PrP and FLNa do not occur in all tumor types.
Executive summary.
Prion protein exists as the prion pro-protein in pancreatic cancer cell lines
Instead of being a glycosylated and glycosylphosphatidylinositol (GPI)-anchored protein, the prion protein (PrP) in pancreatic ductal cell adenocarcinoma (PDAC) cell lines are neither glycosylated nor GPI-anchored.
The GPI peptide signal sequence of PrP contains a filamin A (FLNa)-binding motif and, thus, binds FLNa.
The fatal attraction between pro-PrP and FLNa disrupts the normal functions of FLNa and, thus, provides the PDAC cells with a growth advantage.
Expression of PrP is a poorer prognosis marker in human pancreatic cancer
In normal human pancreas, only islet cells have detectable PrP. PrP is not detectable in precancerous lesions such as PIN-I and -2.
Approximately 13% of PIN-3 cases have detectable PrP. The frequency is increased to approximately 40% in PDAC. As in the PDAC cell lines, the PrP in the tumor is pro-PrP.
PDAC patients, whose PDAC tumors had PrP, had a much shorter survival.
Detection of pro-PrP may provide an early noninvasive diagnostic marker for PDAC.
Prp & FLNa in human melanomas & other tumors
Pro-PrP is detected in some but not all human tumor cell lines: melanoma (n = 3), hepatocarcinoma (n = 5).
Small-cell lung carcinomas and neuroblastomas do not express PrP.
Some melanomas do not express FLNa.
Binding of pro-PrP to FLNa as a potential target for therapeutic intervention
A PrP-GPI peptide signal sequence synthetic peptide with cell-penetrating capacity blocks the interaction between PrP and FLNa and, thus, inhibits the migration of tumor cells in a wound-healing assay.
Footnotes
Financial & competing interests disclosure
This work was partially supported by a grant from NIH. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
For reprint orders, please contact: reprints@futuremedicine.com
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Asher DM, Gibbs CJ, Jr, Gajdusek DC, et al. Pathogenesis of subacute spongiform encephalopathies. Ann Clin Lab Sci. 1976;6(1):84–103. [PubMed] [Google Scholar]
- 2.Greig JR. Scrapie in Sheep. J Comp Pathol. 1950;60(4):263–266. doi: 10.1016/s0368-1742(50)80024-3. [DOI] [PubMed] [Google Scholar]
- 3▪.Griffith JS. Self-replication and scrapie. Nature. 1967;215:1043–1044. doi: 10.1038/2151043a0. JS Griffith, a mathematician who had never worked on scrapie, suggested three mechanisms for the replication of the scrapie agent; one of the mechanisms was that the agent was a protein and could self-replicate. [DOI] [PubMed] [Google Scholar]
- 4▪▪.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. The naming of the scrapie agent as scrapie prion. [DOI] [PubMed] [Google Scholar]
- 5▪▪.Bolton DC, McKinley MP, Prusiner SB. Identification of a protein that purifies with the scrapie prion. Science. 1982;218:1309–1311. doi: 10.1126/science.6815801. Identification of the infectious agent as a protein. [DOI] [PubMed] [Google Scholar]
- 6.Prusiner SB, Groth DF, Bolton DC, Kent SB, Hood LE. Purification and structural studies of a major scrapie prion protein. Cell. 1984;38:127–134. doi: 10.1016/0092-8674(84)90533-6. [DOI] [PubMed] [Google Scholar]
- 7▪.Oesch B, Westaway D, Wälchli M, et al. A cellular gene encodes scrapie PrP 27–30 protein. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. Demonstrates that the scrapie agent is encoded by a host gene. [DOI] [PubMed] [Google Scholar]
- 8▪▪.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;9:13363–13383. doi: 10.1073/pnas.95.23.13363. Short article on prions and transgenic mouse models of prions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9▪.Smith PG, Cousens SN, d’Huillard Aignaux JN, Ward HJ, Will RG. The epidemiology of variant Creutzfeldt–Jakob disease. Curr Top Microbiol Immunol. 2004;284:161–191. doi: 10.1007/978-3-662-08441-0_7. Describes the rise and fall of variant Creutzfeldt–Jakob disease, its link to bovine spongiform encephalopathy and the potential public health concern of variant Creutzfeldt–Jakob disease. [DOI] [PubMed] [Google Scholar]
- 10.Brockes JP. Topics in prion cell biology. Curr Opin Neurobiol. 1999;9:571–577. doi: 10.1016/S0959-4388(99)00016-1. [DOI] [PubMed] [Google Scholar]
- 11.Harris DA. Cellular biology of prion diseases. Clin Microbiol Rev. 1999;12:429–444. doi: 10.1128/cmr.12.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donne DG, Viles JH, Groth D, et al. Structure of the recombinant full-length hamster prion protein PrP(29–231): the N-terminus is highly flexible. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zahn R, Liu A, Lührs T, et al. NMR solution structure of the human prion protein. Proc Natl Acad Sci USA. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegde RS, Rane NS. Prion protein trafficking and the development of neurodegeneration. Trends Neurosci. 2003;26:337–339. doi: 10.1016/S0166-2236(03)00143-7. [DOI] [PubMed] [Google Scholar]
- 15.Behrens A, Aguzzi A. Small is not beautiful: anatogonizing functions for the prion protein PrPc and its homologue Dpl. Trends Neurosci. 2002;25(3):150–154. doi: 10.1016/s0166-2236(00)02089-0. [DOI] [PubMed] [Google Scholar]
- 16.Caughey B, Braon GS. Prions and their partners in crime. Nature. 2006;443:803–810. doi: 10.1038/nature05294. [DOI] [PubMed] [Google Scholar]
- 17.Chiesa R, Harris DA. Fishing for prion protein function. PLoS Biol. 2009;7(3):e75. doi: 10.1371/journal.pbio.1000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caughey B, Brown K, Raymond GJ, Katzenstein GE, Thresher W. Binding of the protease sensitive form of PrP to sulfated glycosaminoglycans and Congo red. J Virol. 1994;68:4107–4111. doi: 10.1128/jvi.68.4.2135-2141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan T, Wong BS, Liu T, Li R, Petersen RB, Sy MS. Cell-surface PrP interacts with glycosaminoglycans. Biochem J. 2002;368:81–90. doi: 10.1042/BJ20020773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rieger R, Edenhofer F, Lasmézas CI, Weiss S. The human 37-kDa laminin receptor precursor interacts with the prion protein in eukaryotic cells. Nat Med. 1997;3(12):1383–1388. doi: 10.1038/nm1297-1383. [DOI] [PubMed] [Google Scholar]
- 21.Li C, Wong P, Pan T, et al. Normal cellular prion protein is a ligand of selectins: binding requires LeX but is inhibited by sLeX. Biochem J. 2007;406:333–341. doi: 10.1042/BJ20061857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitt-Ulms G, Legname G, Baldwin MA, et al. Binding of neural cell adhesion molecules (N-CAMs) to the cellular prion protein. J Mol Biol. 2001;314(5):1209–1225. doi: 10.1006/jmbi.2000.5183. [DOI] [PubMed] [Google Scholar]
- 23.Morel E, Fouquet S, Strup-Perrot C, et al. The cellular prion protein PrP is involved in the proliferation of epithelial cells and in the distribution of junction-associated proteins. PLoS ONE. 2008;3:e3000. doi: 10.1371/journal.pone.0003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown DR, Qin K, Herms JW, et al. The cellular prion protein binds copper in vivo. Nature. 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 25.Millhauser GL. Copper and the prion protein: methods, structure, function, and disease. Annu Rev Phys Chem. 2007;58:299–320. doi: 10.1146/annurev.physchem.58.032806.104657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zanata SM, Lopes MH, Mercadante AF, et al. Stress-inducible protein 1 is a cell surface ligand for cellular prion that triggers neruoprotection. EMBO J. 2002;21:3307–3316. doi: 10.1093/emboj/cdf325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellis V, Daniels M, Misra R, Brown DR. Plasminogen activation is stimulated by prion protein and regulated in a copper dependent manner. Biochem. 2002;41:6891–6896. doi: 10.1021/bi025676g. [DOI] [PubMed] [Google Scholar]
- 28.Edenhofer F, Rieger R, Famulok M, Wendler W, Weiss S, Winnacker EL. Prion protein PrPc interacts with chaperones of the Hsp60 family. J Virol. 1996;70:4724–4728. doi: 10.1128/jvi.70.7.4724-4728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keshet GI, Bar-Peled O, Yaffe D, Nudel U, Gabizon R. The cellular prion protein colocalizes with the dystroglycan complexes in the brain. J Neurochem. 2000;75:1889–1897. doi: 10.1046/j.1471-4159.2000.0751889.x. [DOI] [PubMed] [Google Scholar]
- 30.Mani K, Cheng F, Havsmark B, Jönsson M, Belting M, Fransson LA. Prion, amyloid-β-derived Cu(II) ions, or free Zn(II) ions support S-nitroso-dependent autocleavage of glypican-1 heparan sulfate. J Biol Chem. 2003;278(40):38956–38965. doi: 10.1074/jbc.M300394200. [DOI] [PubMed] [Google Scholar]
- 31.Lysek DA, Wuthrich K. Prion protein interaction with the C-terminal SH3 domain of Grb2 studied using NMR and optical spectroscopy. Biochem. 2004;43:10393–10399. doi: 10.1021/bi0494828. [DOI] [PubMed] [Google Scholar]
- 32.Nieznanski K, Nieznanska H, Skowronek KJ, Osiecka KM, Stepkowski D. Direct interaction between prion protein and tubulin. Biochem Biphys Res Commun. 2005;334:403–411. doi: 10.1016/j.bbrc.2005.06.092. [DOI] [PubMed] [Google Scholar]
- 33.Klein TR, Kirsch D, Kaufmann R, Riesner D. Prion rods contain small amounts of two host sphingolipids as revealed by thin-layer chromatography and mass spectrometry. Biol Chem. 1998;379(6):655–666. doi: 10.1515/bchm.1998.379.6.655. [DOI] [PubMed] [Google Scholar]
- 34.Nandi PK. Interaction of HuPrP106–126 with nucleic acid. Arch Virol. 1997;142:2537–2545. doi: 10.1007/s007050050261. [DOI] [PubMed] [Google Scholar]
- 35.Mouillet-Richard S, Ermonval M, Chebassier C, et al. Signal transduction through prion. Science. 2000;289:1925–1928. doi: 10.1126/science.289.5486.1925. [DOI] [PubMed] [Google Scholar]
- 36.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 37.Parat MO. The biology of caveolae: achievements and perspectives. Int Rev Cell Mol Biol. 2009;273:117–162. doi: 10.1016/S1937-6448(08)01804-2. [DOI] [PubMed] [Google Scholar]
- 38.Harmey JH, Doyle D, Brown V, Rogers MS. The cellular isoform of the prion protein, PrPC is associated with caveolae in mouse neuroblastoma (N2a) cells. Biochem Biophy Res Comm. 1995;25:753–759. doi: 10.1006/bbrc.1995.1723. [DOI] [PubMed] [Google Scholar]
- 39.Toni M, Spisni E, Griffoni C, et al. Cellular prion protein and caveolin-1 interaction in a neuronal cell line precedes fyn/erk1/2 signal transduction. J Biomed Biotechnol. 2006;5:69469. doi: 10.1155/JBB/2006/69469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okamoto T, Schlegel A, Scherer PE, Lisanti MP. Caveolins, a family of scaffolding proteins for organizing “preassembled signaling complexes” at the plasma membrane. J Biol Chem. 1998;273:5419–5422. doi: 10.1074/jbc.273.10.5419. [DOI] [PubMed] [Google Scholar]
- 41.Paitel E, Fahraeus R, Checler F. Cellular prion protein sensitizes neurons to apoptotic stimuli through Mdm2-regulated and p53-dependent caspase 3-like activation. J Biol Chem. 2003;278(12):10061–10066. doi: 10.1074/jbc.M211580200. [DOI] [PubMed] [Google Scholar]
- 42.Kuwahara C, Takeuchi AM, Nishimura T, et al. Prions prevent neuronal cell line death. Nature. 1999;400:225–226. doi: 10.1038/22241. [DOI] [PubMed] [Google Scholar]
- 43.Chiarini LB, Freitas AR, Zanata SM, Brentani RR, Martins VR, Linden R. PrPC transduces neuroprotective signals. EMBO J. 2002;21:317–326. doi: 10.1093/emboj/cdf324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bounhar Y, Zhang Y, Goodyer CG, LeBlanc A. Prion protein protects human neurons against Bax-mediated apoptosis. J Biol Chem. 2001;276:39145–39149. doi: 10.1074/jbc.C100443200. [DOI] [PubMed] [Google Scholar]
- 45.Choi CJ, Anantharam V, Saetveit NJ, Houk RS, Kanthasamy A, Kanthasamy AG. Normal cellular prion protein protects against manganese-induced oxidative stress and apoptotic cell dead. Toxicol Sci. 2007;98:495–509. doi: 10.1093/toxsci/kfm099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diarra-Mehrpour M, Arrabal S, Jalil A, et al. Prion protein prevents human breast carcinoma cell line from tumor necrosis factor-induced cell death. Cancer Res. 2004;64:719–727. doi: 10.1158/0008-5472.can-03-1735. [DOI] [PubMed] [Google Scholar]
- 47.Anantharam V, Kanthasamy A, Choi CJ, et al. Opposing role of prion protein in oxidative stress- and ER stress-induced apoptotic signaling. Free Radic Biol Med. 2008;45(11):1530–1541. doi: 10.1016/j.freeradbiomed.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laurén J, Gimbel DA, Nygaard HB, et al. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β oligomers. Nature. 2009;457:1128–1132. doi: 10.1038/nature07761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steele AD, Zhou Z, Jackson WS, et al. Context dependent neuroprotective properties of prion protein (PrP) Prion. 2009;3:240–249. doi: 10.4161/pri.3.4.10135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hay B, Barry RA, Lieberburg I, Prusiner SB, Lingappa VR. Biogenesis and transmembrane orientation of the cellular isoform of the scrapie prion protein. Mol Cell Biol. 1987;7:914–920. doi: 10.1128/mcb.7.2.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hegde RS, Mastrianni JA, Scott MR, et al. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 52.Vincent B, Paitel E, Saftig P, et al. The disintegrins ADAM10 and TACE contribute to the constitutive and phorbol ester-regulated normal cleavage of the normal cellular prion protein. J Bio Chem. 2001;276:37743–37746. doi: 10.1074/jbc.M105677200. [DOI] [PubMed] [Google Scholar]
- 53.Brown P. Creutzfledt–Jakob disease: reflections on the risk from blood product therapy. Haemophilia. 2007;13:33–40. doi: 10.1111/j.1365-2516.2007.01572.x. [DOI] [PubMed] [Google Scholar]
- 54.Franscini N, El Gedaily A, Matthey U, et al. Prion protein in milk. PLoS ONE. 2006;1:e71. doi: 10.1371/journal.pone.0000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaked GM, Shaked Y, Kariv-Inbal Z, Halimi M, Avraham I, Gabizon R. A protease-resistant prion protein isoform is present in urine of animals and human affected with prion diseases. J Biol Chem. 2001;276(34):31479–31482. doi: 10.1074/jbc.C100278200. [DOI] [PubMed] [Google Scholar]
- 56.Büeler H, Fischer M, Lang Y, et al. Normal development and behavior of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 57.Maeda Y, Ashida H, Kinoshita T. CHO glycosylation mutants: GPI anchor. Methods Enzymol. 2006;416:182–205. doi: 10.1016/S0076-6879(06)16012-7. [DOI] [PubMed] [Google Scholar]
- 58.Ikezawa H. GPI-anchored proteins. Biol Pharm Bull. 2002;25:409–417. doi: 10.1248/bpb.25.409. [DOI] [PubMed] [Google Scholar]
- 59.Orlean P, Menon AK. Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J Lipid Res. 2007;48:993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]
- 60▪.Waneck GL, Stein ME, Flavell RA, et al. Conversion of a GPI-anchored protein to an integral membrane protein by a single amino acid mutation. Science. 1998;241:697–699. doi: 10.1126/science.3399901. Demonstrating that the glycosylphosphatidylinositol (GPI) peptide signal sequence (PSS) can function as a transmembrane domain. [DOI] [PubMed] [Google Scholar]
- 61▪▪.Chen R, Knez JJ, Merrick WC, Medof ME. Comparative efficiencies of C-terminal signals of native GP-anchored proproteins in conferring GPI-anchoring. J Cell Biochem. 2001;84:68–83. doi: 10.1002/jcb.1267. Demonstrates that in comparison to other GPI-PSS, the GPI-PSS of the prion protein is intrinsically inefficient in accepting the GPI anchor. [DOI] [PubMed] [Google Scholar]
- 62▪▪.Screaton RA, DeMarte L, Dráber P, Stanners CP. The specificity for the differentiation blocking activity of carcinoembryonic antigen resides in its GPI anchor. J Cell Biol. 2000;150:613–626. doi: 10.1083/jcb.150.3.613. Demonstrates that the GPI-PSS of a GPI-anchored protein has cryptic information and, ultimately, determines the functionality of the protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nicholson TB, Stanners CP. Identification of a novel functional specificity signal within the GPI anchor signal sequence of carcinoembryonic antigen. J Cell Biol. 2007;177:211–218. doi: 10.1083/jcb.200701158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naghibalhossaini F, Yoder AD, Tobi M, Stanners CP. Evolution of a tumorigenic property conferred by glycophosphatidyl-inositol membrane anchors of carcinoembryonic antigen gene family members during the primate radiation. Mol Biol Cell. 2007;18:1366–1374. doi: 10.1091/mbc.E06-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wopfner F, Weidenhöfer G, Schneider R, et al. Analysis of 27 mammalian and 9 avaian PrPs reveal high conservation of flexible regions of the prion protein. J Mol Biol. 1999;289:1163–1178. doi: 10.1006/jmbi.1999.2831. [DOI] [PubMed] [Google Scholar]
- 66▪▪.Guo Z, Linn JF, Wu G, et al. CDC91L1(PIG-U) is a newly discovered oncogene in human bladder cancer. Nat Med. 2004;10:374–381. doi: 10.1038/nm1010. First demonstration that a gene involved in the GPI-anchor pathway is an oncogene. [DOI] [PubMed] [Google Scholar]
- 67.Jiang WW, Zahurak M, Zhou ZT, et al. Alteration of GPI transamidase subunits in head and neck squamous carcinoma. Mol Cancer. 2008;6:74–80. doi: 10.1186/1476-4598-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu G, Guo Z, Chatterjee A, et al. Over expression of GPI transamidase subunits GPI glycan class T and/or GPI anchor attachment 1 induces tumorigenesis and contributes to invasion in human breast cancer. Cancer Res. 2006;66:9829–9836. doi: 10.1158/0008-5472.CAN-06-0506. [DOI] [PubMed] [Google Scholar]
- 69.Ho JC, Cheung ST, Patil M, Chen X, Fan ST. Increased expression of GPI-anchor attachment protein 1 (GPAA1) is associated with gene amplification in hepatocellular carcinoma. Int J Cancer. 2006;119:1330–1337. doi: 10.1002/ijc.22005. [DOI] [PubMed] [Google Scholar]
- 70.Malumbres M, Barbacid M. Ras oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–460. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 71.Wennerberg K, Rossman KL, Der CJ. The Ras super-family at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 72▪.Sobering AK, Watanabe R, Romeo MJ, et al. Yeast Ras regulates the complex that catalyzes the first step in GPI-anchor biosynthesis at the ER. Cell. 2004;117:637–648. doi: 10.1016/j.cell.2004.05.003. First demonstration that Ras regulates GPI anchor synthesis. [DOI] [PubMed] [Google Scholar]
- 73.Holzmann K, Kohlhammer H, Schwaenen C, et al. Genomic DNA-chip hybridization reveals a higher incidence of genomic amplification in pancreatic cancer than conventional comparative genomic hybridization leads to identification of novel candidate genes. Cancer Res. 2004;64:4428–4433. doi: 10.1158/0008-5472.CAN-04-0431. [DOI] [PubMed] [Google Scholar]
- 74.Nowak NJ, Gaile D, Conroy JM, et al. Genome-wide aberrations in pancreatic adenocarcinoma. Cancer Genet Cytogenet. 2005;161:36–50. doi: 10.1016/j.cancergencyto.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 75.Aguirre AJ, Brennan C, Bailey G, et al. High-resolution characterization of the pancreatic adenocarcinoma genome. Proc Nat Acad Sci USA. 2004;101:9067–9072. doi: 10.1073/pnas.0402932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liang J, Pan YL, Ning XX, et al. Overexpression of PrPC and its antiapoptosis function in gastric cancer. Tumor Biol. 2006;27:84–91. doi: 10.1159/000092488. [DOI] [PubMed] [Google Scholar]
- 77.Antonacopoulou AG, Grivas PD, Skarlas L, et al. POLR2F, ATP6V0A1 and PRNP expression in colorectal cancer: new molecules with prognostic significance? Anticancer Res. 2008;28:1221–1227. [PubMed] [Google Scholar]
- 78▪▪.Han H, Bearss DJ, Browne LW, Calaluce R, Nagle RB, Von Hoff DD. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 2002;62:2890–2896. First report demonstrating that PRNP is over expressed in a panel of human pancreatic cancer cell lines. [PubMed] [Google Scholar]
- 79.Maitra A, Hruban RH. Pancreatic cancer. Ann Rev Pathol. 2008;3:157–188. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jemal A, Murray T, Ward E, et al. Cancer Statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 81.Warshaw AL, Fernándezdel Castillo C. Pancreatic carcinoma. N Engl J Med. 1992;326:455–465. doi: 10.1056/NEJM199202133260706. [DOI] [PubMed] [Google Scholar]
- 82.Hruban RH, Adsay NV, Albores-Saavedra J, et al. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol. 2001;25:579–586. doi: 10.1097/00000478-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 83.Hruban RH, Wilentz RE, Maitra A. Identification and analysis of precursors to invasive pancreatic cancer. Methods Mol Med. 2005;103:1–13. doi: 10.1385/1-59259-780-7:001. [DOI] [PubMed] [Google Scholar]
- 84.Yamano M, Fujii H, Takagaki T, Kadowaki N, Watanabe H, Shirai T. Genetic progression and divergence in pancreatic carcinoma. Am J Pathol. 2000;156:2123–2133. doi: 10.1016/S0002-9440(10)65083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85▪▪.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. Review on the genetic and molecular biology of the development of the pancreas, the underlying molecular defects in pancreatic cancers and transgenic mouse models of pancreatic cancers. [DOI] [PubMed] [Google Scholar]
- 86.Rustgi AK. The molecular pathogenesis of pancreatic cancer: clarifying a complex circuitry. Genes Dev. 2006;20:3049–3053. doi: 10.1101/gad.1501106. [DOI] [PubMed] [Google Scholar]
- 87▪▪.Li C, Yu S, Nakamura F, et al. Binding of pro-prion to filamin A disrupts cytoskeleton and correlates with poor prognosis in pancreatic cancer. J Clin Invest. 2009;119(9):2725–2736. doi: 10.1172/JCI39542. First to demonstrate that the prion pro-protein (pro-PrP) is expressed in human pancreatic cancer, pro-PrP binds filamin A and expression of PrP is a poorer prognosis marker in a subset of patients with pancreatic cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stossel TP, Condeelis J, Cooley L, et al. Filamins as integrators of cell mechanics and signaling. Nat Rev Mol Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 89.Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signaling. Nat Cell Biol. 2004;6:1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- 90.Robertson SP. Filamin A: phenotypic diversity. Curr Opin Genet Dev. 2005;15:301–307. doi: 10.1016/j.gde.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 91.Hart AW, Morgan JE, Schneider J, et al. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum Mol Genet. 2006;15:2457–2467. doi: 10.1093/hmg/ddl168. [DOI] [PubMed] [Google Scholar]
- 92.Feng Y, Chen MH, Moskowitz IP, et al. Filamin A is required for cell–cell contact in vascular development and cardiac morphogenesis. Proc Nat Acad Sci USA. 2006;103:19836–19841. doi: 10.1073/pnas.0609628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kiema T, Lad Y, Jiang P, et al. The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006;21:337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 94.Nakamura F, Pudas R, Heikkinen O, et al. The structure of the GPIb-filamin A complex. Blood. 2006;107:1925–1932. doi: 10.1182/blood-2005-10-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J Cell Biol. 2007;179:1011–1025. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cancellotti E, Wiseman F, Tuzi NL, et al. Altered glycosylated PrP proteins can have different neuronal trafficking in brain but do not acquire scrapie-like properties. J Biol Chem. 2005;280:42909–42918. doi: 10.1074/jbc.M509557200. [DOI] [PubMed] [Google Scholar]
- 97.Devasahayam M, Catalino PD, Rudd PM, Dwek RA, Barclay AN. The glycan processing and site occupancy of recombinant Thy-1 is markedly affected by the presence of a GPI-anchor. Glycobiology. 1999;9:1381–1387. doi: 10.1093/glycob/9.12.1381. [DOI] [PubMed] [Google Scholar]
- 98.Bashyam MD, Bair R, Kim YH, et al. Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer. Neoplasia. 2005;7:556–562. doi: 10.1593/neo.04586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guillot-Sestier MV, Sunyach C, Druon C, Scarzello S, Checler F. The α-secretase-derived N-terminal product of cellular prion, N1 displays neuroprotective function, in vitro and in vivo. J Biol Chem. 2009;284(51):35973–35986. doi: 10.1074/jbc.M109.051086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Byers HR, Etoh T, Doherty JR, Sober AJ, Mihm MC., Jr Cell migration and actin organization in cultured human primary, recurrent, cutaneous and metastatic melanoma. Am J Pathol. 1991;139:423–435. [PMC free article] [PubMed] [Google Scholar]
- 101.Cunningham CC, Gorlin JB, Kwiatkowski DJ, et al. Actin-binding protein requirement for cortical stability and efficient locomotion. Science. 1992;255:325–327. doi: 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- 102.Glogauer M, Arora P, Chou D, Janmey PA, Downey GP, McCulloch CAG. The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J Biol Chem. 1998;273:1689–1698. doi: 10.1074/jbc.273.3.1689. [DOI] [PubMed] [Google Scholar]
- 103.He H-J, Kole S, Kwon Y-K, Crow MT, Bernier M. Interaction of filamin A with the insulin receptor alters insulin-dependent activation of the mitogen-activated protein kinase pathway. J Biol Chem. 2003;278:27096–27104. doi: 10.1074/jbc.M301003200. [DOI] [PubMed] [Google Scholar]
- 104.Sasaki A, Masuda Y, Ohta Y, Ikeda K, Watanabe K. Filamin associates with Smads and regulates transforming growth factor beta signaling. J Biol Chem. 2001;276:17871–17877. doi: 10.1074/jbc.M008422200. [DOI] [PubMed] [Google Scholar]
- 105.Zhang M, Breitwieser GE. High affinity interaction with filamin A protects against calcium-sensing receptor degradation. J Biol Chem. 2005;280:11140–11146. doi: 10.1074/jbc.M412242200. [DOI] [PubMed] [Google Scholar]
- 106.Pammer J, Weninger W, Tschachler E. Human keratinocytes express cellular prion-related protein in vitro and during inflammatory skin diseases. Am J Pathol. 1998;5:1353–1358. doi: 10.1016/S0002-9440(10)65720-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bouffard D, Duncan LM, Howard CA, Mihm MC, Jr, Byers HR. Actin binding protein expression in benign and malignant melanocytic proliferations. Hum Pathol. 1994;25:709–714. doi: 10.1016/0046-8177(94)90305-0. [DOI] [PubMed] [Google Scholar]
- 108.Mínguez B, Tovar V, Chiang D, Villanueva A, Llovet JM. Pathogenesis of hepatocellular carcinoma and molecular therapies. Curr Opin Gastroenterol. 2009;25:186–194. doi: 10.1097/MOG.0b013e32832962a1. [DOI] [PubMed] [Google Scholar]
- 109.Jackman DM, Johnson BE. Small-cell lung cancer. Lancet. 2005;366:1385–1396. doi: 10.1016/S0140-6736(05)67569-1. [DOI] [PubMed] [Google Scholar]
- 110.Yin S, Fan X, Yu S, Li C, Sy MS. Binding of recombinant but not endogenous prion protein to DNA causes DNA internalization and expression mammalian cells. J Biol Chem. 2008;283:25446–25454. doi: 10.1074/jbc.M800814200. [DOI] [PMC free article] [PubMed] [Google Scholar]