Abstract
The BNIP3 subfamily of BH3-only proteins consists of BNIP3 and BNIP3-like (BNIP3L) proteins. These proteins form stable homodimeration complexes that localize to the outer membrane of the mitochondria after cellular stress. This promotes either apoptotic or non-apoptotic cell death such as autophagic cell death. While the mammalian cells contain both members of this subfamily, the genome of C. elegans codes for a single BNIP3 ortholog, ceBNIP3 that shares homology in the transmembrane domain and a conserved region close to the BH3 domain of mammalian BNIP3 protein. The cell death activities of BNIP3 and BNIP3L are determined by either the BH3 domain or the C-terminal transmembrane domain. The transmembrane domain of BNIP3 is unique since it is capable of autonomous stable dimerization and contributes to mitochondrial localization of BNIP3. In knock-out mouse models, BNIP3L was demonstrated to be essential for normal erythrocyte differentiation and hematopoietic homeostasis whereas BNIP3 plays a role in cellular responses to ischemia/reperfusion injury in the heart. Both BNIP3 and BNIP3L play a role in cellular responses to stress. Under hypoxia, both BNIP3 and BNIP3L expression levels are elevated and contribute to hypoxia induced cell death. In addition, these proteins play critical roles in disease states. In heart disease, both BNIP3 and BNIP3L play a critical role in cardiomycyte cell death following ischemic and non-ischemic injuries. In cancer, expression of BNIP3 and BNIP3L is down regulated by promoter hypermethylation or by homozygous deletion of the gene locus in certain cancers whereas their expression was increased in other cancers. In addition, BNIP3 expression has been correlated with poor prognosis in some cancers. The results reviewed here suggest BNIP3 and BNIP3L may be novel therapeutic targets for intervention due to their pathological roles in regulating cell death in disease states.
Keywords: BNIP3, BNIP3L, apoptosis, autophagy, cancer, cardiomyopathies
Introduction
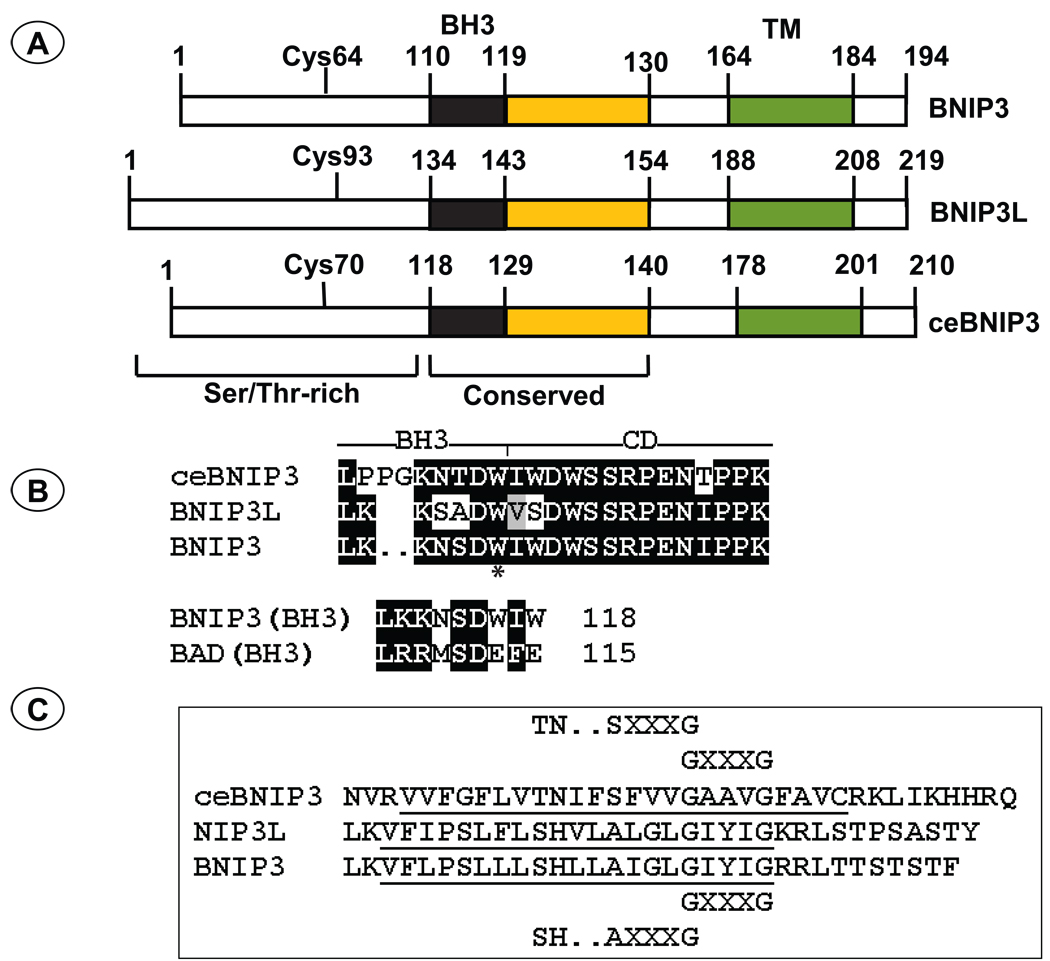
The first member of this subfamily of BH3-only proteins, BNIP3 (Bcl-2/E1B-19K interacting protein 3; initially called Nip3) was isolated by yeast two hybrid screening using E1B-19K as the bait (Boyd et al., 1994). BNIP3 was shown to be a mitochondrial protein that also interacted with other BCL-2 family anti-apoptotic proteins, BCL-2, BCL-xL and EBV-BHRF1 (Boyd et al., 1994; Theodorakis et al., 1996). BNIP3 resembles other BCL-2 family proteins by virtue of a characteristic mitochondrial targeting C-terminal trans-membrane (TM) domain and by the presence of a BH3 domain (Yasuda et al., 1998b). A second member of this family, BNIP3L which shares about 55% sequence similarity to BNIP3 was identified by data bank searches (variously named BNIP3L (Matsushima et al., 1998), B5 (Ohi et al., 1999), BNIP3α (Yasuda et al., 1999), NIX (Chen et al., 1999), BNIP3h (Farooq et al., 2001), hereafter called BNIP3-like). Like BNIP3, BNIP3L (designated B5) was also identified in a two hybrid screen with the E1B-19K bait (Ohi et al., 1999). BNIP3 maps in human chromosome 10q26.3 while BNIP3L maps in 8p21. Homology searches of the C. elegans sequences identified a BNIP3 ortholog with 21% amino acid homology to BNIP3 in the locus of CEC14F5.1 (Cizeau et al., 2000; Yasuda et al., 1998a). The homology between ceBNIP3 and the mammalian members includes a highly conserved 19-amino acid sequence encompassing the BH3 domain and the trans-membrane domain (Fig. 1). However, the BH3 domain of ceBNIP3 is somewhat divergent from those of the mammalian BNIP3 and BNIP3L. These proteins are also characterized by the presence of PEST sequences at the N-terminal region. Data bank analysis has revealed conservation of BNIP3 related sequences in most animal species (Aouacheria et al., 2005). Unlike most other BH3-only proteins, the cell death activities of BNIP3 and BNIP3L are controlled by the BH3 domain as well as the transmembrane domain causing both apoptotic and necrotic cell death (Chen et al., 1997; Kubasiak et al., 2002; Vande Velde et al., 2000; Yasuda et al., 1998b). It appears that BNIP3 and BNIP3L may constitute important mitochondrial sensors for various stress stimuli. The expression of BNIP3 is strongly activated by hypoxia and is the direct transcriptional target for HIF-1α (Bruick, 2000; Kothari et al., 2003). While BNIP3L was shown to be activated by hypoxia (Fei et al., 2004; Sowter et al., 2001), hypoxia-independent activation has also been observed in specific cell types such as cadiomyocytes (Galvez et al., 2006; Yussman et al., 2002). Both BNIP3 and BNIP3L appear to play important roles in cell death induced by ischemic stress of cardiomycytes and neuronal cells. Additionally, they also appear to play a role in tumor progression as well as in tumor suppression. This review summarizes the critical aspects of the biological activities of BNIP3 subfamily proteins.
Fig. 1. Domain structure of BNIP3 subfamily members.
(A). The BH3 domain, the conserved domain (CD), the trans-membrane domain (TM), and the conserved Cys residue implicated in stabilization of the BNIP3 homodimers are indicated. The coordinates of ceBNIP3 are based on the ORF described by Yasuda et al (Yasuda et al., 1998a). The ORF described by Cizeau et al (Cizeau et al., 2000) contains an extra 11-amino acid region at the N-terminus. (B). The amino acid sequence of the BH3 domain and the conserved domain are indicated. A conserved Trp residue within the BH3 domain of all BNIP3 related proteins is indicated by (*). In the bottom panel the BH3 domain of BNIP3 is compared with the BH3 domain of BAD to highlight the Ser residue. (C). The amino acid sequences of the TM domain and the sequence elements that mediate homodimerization are shown.
Domains of BNIP3 family members
BH3 domain
BNIP3 was shown to possess a functional BH3 domain based on mutational analysis and domain substitution studies (Yasuda et al., 1999). First, a deletion within the BH3-like domain of BNIP3 was shown to abolish the interaction of BNIP3 with E1B-19K and BCL-2. Secondly, a 16-amino acid region of BNIP3 corresponding to the 16-amino acid α-helical region of BCL-xL and BAK containing the BH3 domain (Sattler et al., 1997) or a 9-amino acid region of BNIP3 corresponding to the core BH3-domain in BAX was shown to functionally substitute for the BH3 domain of BAX in the transient cell death assays and in interaction with BCL-xL. Similarly, deletion of a 7-amino acid sequence corresponding to the core BH3 domain of BNIP3L, diminished its interaction with BCL-2 and the cell death activity (Imazu et al., 1999). The sequence of ceBNIP3 corresponding to the BH3-domain of BNIP3 and BNIP3L is divergent (Cizeau et al., 2000; Yasuda et al., 1998a). A deletion of the BH3 domain of ceBNIP3 did not significantly alter its cell death activity in mammalian cells (Cizeau et al., 2000). Similarly, deletion of the BH3 domain in BNIP3 failed to alters its cell death activity in epithelial derived cells (Vande Velde et al 2000). In general, the BH3 domains of BNIP3 subfamily members appear to be weak, in comparison with other strongly pro-apoptotic members of the BH3-only proteins, with regard to their affinity for pro-survival proteins and may contribute to the delayed cell death activities of these proteins. The BH3 domains of all BNIP3 family proteins contain a Trp residue instead of D/E residues in the BH3 domain of most other BH3-only members (Aouacheria et al., 2005). The possibility that the Trp residue may regulate selectivity in interaction with pro-survival proteins remains to be investigated. Additionally, it is possible that the activity of the BH3 domain, at least in the case of BNIP3, might be regulated by phosphorylation. It should be noted that the BH3 domain of BNIP3 resembles the BH3 domain of BAD that has been known to be phosphorylated by the survival kinases such as PKA induced by growth factors (Datta et al., 2000; Zhou et al., 2000).
TM domain
The TM domains of BNIP3 family members are unique among the various BCL-2 family members and appear to contribute to their death promoting activity. When BNIP3 was identified, it was shown to be localized in outer membrane of the mitochondria (Boyd et al., 1994). Subsequently, it was shown that the C-terminal transmembrane domain was responsible for targeting BNIP3 to mitochondria, since a BNIP3 mutant lacking the TM domain localized dispersedly throughout the cell and the BNIP3 TM conferred mitochondrial localization to GFP (Yasuda et al., 1998b). The TM domain of BNIP3L is highly homologous to the TM domain of BNIP3 and was shown to be functionally similar (Chen et al., 1999; Imazu et al., 1999; Yasuda et al., 1999). Greenberg’s group first reported that BNIP3, when analyzed by SDS-PAGE migrated predominantly as a 60 kD dimer in addition to the 30 kD monomer (Chen et al., 1997). Deletion of the TM domain abolished dimer formation, suggesting a critical role for the TM domain in detergent-stable dimerization. The TM domain of BNIP3L (Chen et al., 1999; Imazu et al., 1999) and ceBNIP3 (Cizeau et al., 2000) functioned similarly in promoting stable dimerization. The TM domains of BNIP3 and BNIP3L were also shown to mediate heterodimerization among these proteins (Ohi et al., 1999). Due to the high stability of BNIP3 dimers, the BNIP3 TM domain has become a favored model to study biophysical aspects of dimerization of integral membrane proteins by lateral interactions between transmembrane α helices (Metcalf et al., 2007; Sulistijo et al., 2003; Sulistijo and MacKenzie, 2006). Isolated TM domain of BNIP3 was shown to dimerize strongly in membranes and in detergents (Sulistijo et al., 2003).
The BNIP3 TM domain contains a GXXXG motif which is a critical determinant for dimerization of other membrane proteins (Russ and Engelman, 2000). Additionally, BNIP3 also contains a tandem AXXXG motif. These tandem motifs and electrostatic interactions involving an adjoining His (173) and Ser (172) have been shown to be important for stabilization of the BNIP3 dimer (Sulistijo et al., 2003; Sulistijo and MacKenzie, 2006). The side chain of the His has been proposed to hydrogen bond with the side chain of Ser across the dimer interface. These sequence features are conserved in BNIP3L. Interestingly, ceBNIP3 also contains two tandem SXXXG and GXXXG motifs. Since small residues such as Ala and Ser could substitute for the G residue (Sulistijo and MacKenzie, 2006), it appears that the two GXXXG-related motifs present in ceBNIP3 could also contribute to stable dimerization. The conserved feature of highly stable dimerization in BNIP3 family members suggests a functional significance. However, the precise role of dimerization in the pro-death activity of BNIP3 remains unresolved.
Several studies (see below) have shown that the deletion of the entire TM domain (ΔTM) abolished the pro-death activity of all three members of the BNIP3 family. It is possible that this functional defect of the ΔTM mutants might be the result of their inability to localize in the mitochondria. It appears that the unique stable dimerization activity of BNIP3 is not critical for its cell death activity. Substitution of the TM domain of BNIP3 with heterologous TM domain of BCL-2 that targeted BNIP3 to mitochondria as well as other cytosolic membrane locations such as the ER, did not affect the cell death activity and the ability to heterodimerize with BCL-2 and BCL-xL (Ray et al., 2000). Such a chimeric construct was also shown to induce loss of mitochondrial permeability transition and increased reactive oxygen species (Kim et al., 2002). An amino acid substitution mutant in the His residue (His→Ala) of the TM domain implicated in stabilization of BNIP3 dimers did not significantly alter the death promoting activity under hypoxic and acidotic conditions (Frazier et al., 2006). However, the same mutation was shown to abolish the cell death activity of BNIP3L by another group (Kubli et al., 2008). These authors also reported that the homodimer of BNIP3 was stabilized by the disulfide linkage mediated by a conserved N-terminal Cys (Cys64) residue. The above summarized results suggest that the TM domain of BNIP3 is important for its pro-death activity and for specific targeting of BNIP3 to mitochondria. However, the pro-death activity could not be attributed to the unique property of BNIP3 TM domain. This conclusion was supported by the observation that substitution of the BNIP3 TM for the TM domain of pro-survival protein EBV-BHRF1 did not diminish the pro-survival function of BHRF1 (Yasuda et al., 1998b).
N-terminal and conserved domains
The N-terminal regions of the BNIP3 subfamily members are divergent. However, all three members of the subfamily are characterized by the presence of PEST sequences that are associated with proteins that are subjected to high turnover by proteasome mediated degradation (Rogers et al., 1986). Although no other notable sequence motifs in the N-terminal 49-amino acid region of BNIP3 can be identified, it was shown to be important for heterodimerization with BCL-2 (along with the TM domain) while the interaction with BCL-xL was shown to require either the N-terminal or the C-terminal region (Ray et al., 2000). The precise mode of such interactions with BCL-2/BCL-xL remains to be resolved. Although the role of the N-terminal region of BNIP3 in cell death activity has not been examined, deletion of the N-terminal region (39-aminoacids) of BNIP3L (which lacks any homology to BNIP3) was reported to enhance the cell death activity (Ohi et al., 1999).
All three members of the BNIP3 subfamily contain a highly conserved domain immediately adjacent to the BH3 domain. Although the high degree of such homology between vertebrate proteins (BNIP3 and BNIP3L) and invertebrate protein (ceBNIP3) suggests a conserved function, mutational analyses of this domain have not yielded any functional clues. Deletion of this domain along with the BH3 domain did not significantly alter the pro-cell death activity of BNIP3 in certain in vitro cell death assays (Ray et al., 2000).
Regulation of expression of BNIP3 family members
BNIP3 was identified as one of the most prominent hypoxia responsive genes by subtractive hybridization studies (Bruick, 2000). The promoter of BNIP3 was found to contain a hypoxia response element (HRE) that suggested BNIP3 was a direct target for HIF-1. This study also observed a modest (~ 2-fold) activation of BNIP3L under hypoxia. Since no other member of the BCL-2 family was found to be activated by the hypoxic stress, it was suggested that BNIP3 and BNIP3L might be key players in cell death induced under hypoxic injury such as ischemia. Subsequently, overexpression of HIF-1α was shown to induce expression of BNIP3 and cell death (Guo et al., 2001), which strengthened the conclusion that BNIP3 was a target for HIF-1α. Harris and colleagues also reported transcriptional activation of BNIP3 and BNIP3L in several carcinoma cell lines exposed to hypoxia (Sowter et al., 2001). In these studies, the role of HIF-1α was further demonstrated using a cell line that lacked the tumor suppressor pVHL (a negative regulator of HIF-1α) in which BNIP3 was found to be overexpressed and that reintroduction of pVHL reduced BNIP3 expression. Additionally, Sowter et al observed elevated expression of BNIP3 and BNIP3L in the perinecrotic regions (which result from hypoxia) of human tumors. Many subsequent reports have confirmed that BNIP3 was one of the early hypoxia responsive genes and showed that such activation might contribute to several pathological processes such as neuronal and myocardial ischemic cell death (Graham et al., 2004; Kubasiak et al., 2002; Regula et al., 2002). It was also suggested that hypoxia induced tumor necrosis via BNIP3 expression may contribute to the development of certain solid tumors (see below).
In addition to HIF-1α, other transcription factors also significantly contribute to BNIP3 expression under hypoxia. The zinc-finger transcription factor Pleomorphic adenomas gene-like 2 (PLAGL2) (which is expressed in response to hypoxia or iron deficiency) was reported to activate BNIP3 resulting in apoptotic death of cultured fibroblasts and neuroblastoma cells (Mizutani et al., 2002). The effect of PLAGL2 was shown to be independent of HIF-1 since PLAGL2 activated the BNIP3 promoter that lacked a HRE. It is possible that both HIF-1 and PLAGL2 may independently and additively activate BNIP3 during hypoxia. Although the precise site with which PLAGL2 interacts on the BNIP3 promoter has not been determined, it appears that it may interact with one of the several GC-rich boxes of the BNIP3 promoter.
While probing the mechanism of cell death in Rb null fetal liver during mouse embryo development (which experiences ischemia), Macleod and colleagues discovered that loss of pRb resulted in derepression of BNIP3 expression and non-apoptotic cell death (Tracy et al., 2007). These results suggested a survival promoting activity of pRb in fetal liver cells. They further identified that BNIP3 was a direct target for transcriptional repression by pRb/E2F and that pRb reduced the expression of BNIP3 during hypoxia resulting in BNIP3-dependent autophagy and cell survival. They showed that under nonstress conditions the BNIP3 promoter was associated with E2F4 (a repressive member of the E2F family) and under hypoxic stress conditions the transcription factor E2F1 bound to the BNIP3 promoter displacing E2F4. ChIP analysis revealed that recruitment of E2F1 also accompanied recruitment of pRb resulting in reduction of the BNIP3 promoter activity, suggesting a functional cooperation between pRb and HIF-1α in reducing the level of BNIP3 expression. Thus, this study illuminated a novel function for the pRb tumor suppressor in regulating the levels of BNIP3 expression under stress conditions that would be a determining factor in autophagic cell survival vs cell death. Schneider, Kirshenbaum and their colleagues have shown that an activity of adenovirus E1A that liberates E2F transcription factors from pRb family proteins was able to induce proliferation of adult rat cardiac myocytes and also showed that exogenous introduction of E2F1 via a recombinant adenovirus vector induced cell proliferation and apoptosis (Agah et al., 1997; Kirshenbaum et al., 1996). More recently Kirshenbaum and colleagues have shown that apoptosis induced by E2F1 was a consequence of direct transcriptional activation of BNIP3 through E2F1 binding with the BNIP3 promoter (Yurkova et al., 2008). The possibility that E2F1 may function in concert with HIF-1α to activate BNIP3 in ventricular myocytes remains to be investigated.
A study by Sandri and coworkers, who analyzed the target genes for the transcription factor, FoxO3 (which contributes to skeletal muscle atrophy via autophagy) identified BNIP3 as a FoxO3 target gene, along with other autophagy related genes such as LC3 and certain ubiquitin ligases (Mammucari et al., 2007). They reported binding of FoxO3 to the promoters of BNIP3 and BNIP3L in fasting muscles cells by ChIP analysis and increased expression of BNIP3 and BNIP3L. Depletion of FoxO3 in fasting cells reduced the level of BNIP3 expression and transfection of FoxO3 in HEK293 cells activated expression of endogenous BNIP3 supporting the conclusion that FoxO3 activated BNIP3 in these cells. Thus, BNIP3 appears to be the direct target of transcription factors that induce apoptosis or autophagy such as PLAGL2, E2F1 and FoxO3, in addition to HIF-1. Although expression of BNIP3 strikingly regulated by hypoxia in tissues such as heart, appears to be constitutive in certain tissues such as liver and kidney (Galvez et al., 2006). It is possible that transcription factors such as Sp1 that recognize the GC boxes of the BNIP3 and BNIP3L promoters contribute to their constitutive basal expression. In addition to transcriptional repressors such as pRb and E2F4, and hypermethylation of the Bnip3 promoter that have been shown to down regulate Bnip3 expression, an NF-κB-dependent mechanism of transcriptional silencing of Bnip3 has also been identified (Baetz et al., 2005; Shaw et al., 2008; Shaw et al., 2006). In contrast to the well known transcriptional activation function of NF-κB, the studies by Kirshenbaum and colleagues showed that the binding of the p65 subunit of NF-κB to the Bnip3 promoter resulted transcriptional repression in rat ventricular myocytes. The repression activity of p65 was dependent on recruitment of HDAC1. Under non-stress conditions, NF-κB was found to be associated with the Bnip3 promoter, preventing association of E2F-1 to an adjacent E2F-binding site (Shaw et al., 2008). Under hypoxia, when the level of NF-κB was low, expression of Bnip3 was activated as result of E2F-1 binding.
As mentioned above modest activation of BNIP3L was observed when cultured cells were exposed to hypoxia (Bruick, 2000; Sowter et al., 2003), consistent with the presence of potential HREs in the BNIP3L promoter (Aerbajinai et al., 2003; Fei et al., 2004; Galvez et al., 2006). However, expression of BNIP3L appears to be more constitutive than that of BNIP3 in several cellular contexts. BNIP3, along with BAK was identified as one of the pro-apoptotic target genes for the immediate-early growth response gene Egr-2 in the PTEN-induced apoptotic pathway (Unoki and Nakamura, 2003). Exogenous expression of EGR2 was shown to directly activate BNIP3L and BAK in a variety of human cancer cells, including those harboring mutations in p53 and PTEN genes. BNIP3L can be cooperatively activated by HIF-1 and p53 under hypoxia resulting in p53-dependent apoptosis under hypoxia (Fei et al., 2004). In addition to HIF-1, other transcription factors also have been reported to activate BNIP3L. In cardiac myocytes BNIP3L was shown to be activated by factors that activate PKCα via the transcription factor Sp1 (Galvez et al., 2006). Therapeutic blockade of the activities of the transmembrane tyrosine kinase receptors EGFR (ErbB1) and HER-2 (ErbB2) has been reported to activate the expression of BNIP3L via the transcription factor FoxO3 (Real et al., 2005). As pointed out above, BNIP3L was shown to be transcriptionally activated by FoxO3 in fasting muscle cells (Mammucari et al., 2007). Interestingly, ceBnip3 has also been identified as a transcriptional target for daf-16, the worm homolog of FoxO (Murphy et al., 2003)
In addition to transcriptional regulation of Bnip3 and Bnip3L, the levels of transcripts of these genes were also reported to be regulated at post-transcriptional level in some instances. In murine macrophages treated with sublethal doses of anthrax lethal toxin, there was destabilization of Bnip3 and Bnip3L mRNAs (Ha et al., 2007). Although the mechanism of down-regulation Bnip3/Bnip3L mRNAs was not known, it was suggested that phosphorylation of proteins that regulate the mRNA levels via binding to AU-rich elements in the 3’-untranslated regions might play a role.
Cell death and autophagy induced by BNIP3 family members
Although BNIP3 was identified and cloned prior to BIK (the founding member of the BH3-only proteins), its cell death promoting activity remained unresolved since BNIP3 overexpression in common cell lines under normal culture conditions did not result in profound changes in cell viability. Subsequent studies showed that overexpression of BNIP3 caused a delayed cell death activity (Chen et al., 1997; Yasuda et al., 1998b). A similar cell death activity was also observed in cells transfected with BNIP3L (Imazu et al., 1999). The cell death observed in transient transfection based assays was shown to be dependent on the BH3 domain (Imazu et al., 1999; Yasuda et al., 1998b) or the transmembrane domain (Chen et al., 1999; Ray et al., 2000). Although the cell death induced by transient overexpression of BNIP3 was referred to as apoptotic in these publications, those assays were not sensitive enough to discriminate between different modes of cell death.
In subsequent studies Greenberg and coworkers observed that overexpression of BNIP3 caused a form of cell death that resembled necrosis that was not inhibited by the pancaspase inhibitor zVAD-fmk or the caspase agonist baculovirus p35 (Vande Velde et al., 2000). Further, BNIP3-mediated cell death was observed in mouse embryo fibroblasts that individually lacked Apaf-1, caspase-3 and caspase-9. Ultrastructural studies of cells undergoing BNIP3-inducd death revealed extensive cytoplasmic vacuoles that resembled autophagosomes. Subsequent studies by Kondo and colleagues showed that malignant glioma cells treated with the anticancer agents sphingolipid ceramide (Daido et al., 2004) and arsenic trioxide (Kanzawa et al., 2005) underwent cell death that was accompanied by autophagic signatures such as association of the microtubule-associated protein light-chain 3 (LC3) in autophagosomes. Several recent studies have further substantiated that one mode of BNIP3-induced cell death may involve autophagy (Azad et al., 2008; Hamacher-Brady et al., 2007; Mammucari et al., 2008; Tracy et al., 2007; Zhang et al., 2008).
The evidence linking BNIP3 to autophagy and autophagic cell death appears to be strong. Although these activities of BNIP3 appear to be related to strong mitochondrial localization of BNIP3, the precise mechanism by which BNIP3 promotes autophagy and autophagic death remains to be clarified in detail. Like BNIP3, other BH3-only members and BH3 mimetics have also been reported to induce autophagy and autophagic death (Abedin et al., 2007; Maiuri et al., 2007; Rashmi et al., 2008; Yanagisawa et al., 2003; Zhaorigetu et al., 2008). The pharmacological BH3 mimetic ABT737 was shown to competitively inhibit interaction of Beclin-1 with BCL-2/BCL-xL (Maiuri et al., 2007). In cotransfection experiments, BNIP3 was shown to compete with Beclin-1 for interaction with BCL-2 (Zhang et al., 2008). These results suggest that liberating Beclin-1 from BCL-2/BCL-xL may be one of the mechanisms by which BH3-only members, including BNIP3, may promote autophagy. Since loss of mitochondrial permeability transition appears to induce autophagy (Elmore et al., 2001), BNIP3 may also induce autophagy indirectly as a consequence of such mitochondrial injury. It appears that prolonged BNIP3 expression or acute overexpression beyond an autophagic survival threshold may result in autophagic cell death. It was recently reported that prolonged exposure of several apoptosis-competent cancer lines to hypoxia induced autophagy and cell death in a BNIP3-dependent manner (Azad et al., 2008). The onset of autophagy and resulting cell death was dependent on BNIP3 as depletion of BNIP3 reduced autophagy. These results suggested that activation of BNIP3 by hypoxia alone could contribute to autophagy and autophagic death, independent of apoptosis.
In neonatal rat cardiac myocytes exposed to hypoxia and acidotic conditions, BNIP3 was shown to cause a form of cell death that resembled apoptosis since the plasma membrane remained intact without significant loss of ATP accompanied by the opening of the mitochondrial permeability transition pore and extensive DNA-laddering (Kubasiak et al., 2002). Nonetheless, such cell death was shown to be independent of caspase activation. Although the role of the BH3 domain in this unique mode of cell death was not examined, the TM domain was shown to be essential. In a study that involved a cardiac myocyte cell line exposed to ischemia and reperfusion (I/R) injury, an apoptotic cell death was suppressed by overexpression of the dominant negative version of BNIP3 (BNIP3ΔTM), or by shRNA-mediated down regulation of BNIP3 or by zVAD-fmk (Hamacher-Brady et al., 2007). These results suggested that the endogenous BNIP3 induced caspase-dependent apoptotic cell death under I/R stress. Studies employing Bak−/−/Bak−/− double knockout (DKO) mouse embryo fibroblasts (MEF), revealed that BNIP3-mediated cell death was dependent on these pro-apoptotic BCL-2 family members (Kubli et al., 2007). Overexpression of BNIP3 in wt MEFs caused rapid loss of mitochondrial transmembrane potential, increased production of reactive oxygen species and cell death. These activities of BNIP3 were deficient in Bax/Bak DKO MEF and were observed upon reintroduction of Bax and Bak, suggesting a Bax/Bak dependency for BNIP3 cell death activity. Like other prototypic pro-apoptotic stimuli, overexpression of BNIP3 was shown to induce conformational activation of BAX and BAK leading to permeabilization of the mitochondrial outer membrane, release of cytochrome c and mitochondrial fragmentation. Although the overall features of BNIP3-induced cell death in MEFs resembled features common to caspase-dependent and –independent cell death, the specific involvement of caspases were not examined in these studies. Thus, it appears that BNIP3 may induce both apoptotic and non-apoptotic cell death, depending on the context of cell death stimulation or on the cell types studied. However, it also appears that the extent of apoptotic cell death induced by BNIP3 and BNIP3L is generally weaker than BH3-only members such as BIK, and BIM.
BNIP3 and BNIP3L in cancer
A hallmark of solid tumors is that they are poorly oxygenated in the interior of the tumor mass due to insufficient vascularization. The hypoxic regions of the tumors are characterized by excessive apoptotic and necrotic cell death. It is widely believed that the hypoxic stress in the tumor microenvironment contributes to the evolution of malignant tumor cells with increased invasive and angiogenic potentials. Tumor cells under hypoxic conditions experience a high incidence of genome instability and increased mutation rate (Reynolds et al., 1996). Among the tumor cells in the hypoxic area variant tumor cells often arise and are resistant to apoptosis (Graeber et al., 1996). Tumor hypoxia has been reported to be a negative prognostic factor in a number of tumor sites. Both clinical and experimental studies have suggested a positive correlation between tumor hypoxia and increased metastatic efficiency and resistance to radiotherapy and chemotherapy resulting in tumor relapse. Although several cellular factors are up-regulated by hypoxia, BNIP3 may significantly contribute to the oncogenic evolution of cancer cells. In situ hybridization studies have revealed that BNIP3 was highly expressed in perinecrotic regions of human breast tumors compared to normal tissues (Sowter et al., 2001). In an analysis of clinical samples of ductal carcinoma in situ (DCIS) of the breast, expression of Bnip3 was found to be associated with high-grade tumors while expression of Bnip3L did not reveal such a correlation (Sowter et al., 2003). These studies suggested that expression of Bnip3 might be associated with high grade and necrotic phenotype of breast DCIS. An immunohistochemical analysis of more than 100 non-small cell lung cancer clinical samples revealed that expression of BNIP3 was closely linked with HIF-1α and was strongly correlated with poor prognosis (Giatromanolaki et al., 2004).
Expression of BNIP3 was also found to be associated with expression of other hypoxia-induced growth factors such as VEGF, bFGF and PEDGF (Giatromanolaki et al., 2004). Although it is counterintuitive why a gene that promotes cell death is overexpressed in high-grade tumors, it is possible the survival promoting activities of various cytokines induced during hypoxia may suppress the cell death activity of BNIP3 in these specific cancers. In this context, it has been shown that the cell death activity of BNIP3 is inhibited by growth factors such as EGF and IGF in epithelial cells (Kothari et al., 2003). The conflicting signals imposed by the cell death factor (i.e., BNIP3) and the survival factors (such as VEGF, bFGF, etc) may allow selection of tumor cells that survive chronic Bnip3 overexpression. Other regulatory mechanisms in these ‘high-grade tumors’ may also modulate the cell death activity of BNIP3. For example, an analysis of a number of lung cancers with the worst prognosis revealed nuclear localization of BNIP3 (Giatromanolaki et al., 2004). Similarly, in about 80% of the primary glioblastoma multiforme tumors BNIP3 was predominantly localized in the nucleus (Burton et al., 2006). Such nuclear localization of BNIP3 during tumor progression may constitute another mechanism by which tumors under hypoxia may overcome BNIP3-induced cell death. This notion is consistent with the results that elevated levels of BNIP3 expression in invasive carcinoma of the breast was associated with good survival outcome and shorter disease free survival in pre-invasive DISC (Tan et al., 2007).
Inactivation of BNIP3 expression may constitute a critical step in tumor progression in several human cancers such as the cancer of the pancreas. An analysis of pancreatic adenocarcinomas and pancreatic cancer cell lines revealed that the expression of BNIP3 was inactivated by hypermethylation of the BNIP3 promoter (Okami et al., 2004) (Abe et al., 2005). In nine of nine pancreatic carcinomas BNIP3 was down regulated compared to normal pancreatic tissues despite up-regulation of other hypoxia-responsive genes such as glucose transporter-1 and insulin-like growth factor-binding protein 3 (Okami et al., 2004). The BNIP3 promoter was found to be methylated in the CpG island near the transcription initiation site of the BNIP3 promoter. In pancreatic cancer cell lines hypoxia-inducible BNIP3 expression was restored by treatment with the demethylating agent 5-aza-deoxycytidine (5-aza-dC) (Abe et al., 2005; Okami et al., 2004). A microarray analysis revealed that in certain pancreatic ductal adenocarcinomas, down-regulation of BNIP3 was correlated with up-regulation of S100 family proteins such as S100A2 and S100A4 (Mahon et al., 2007). Specific depletion of the metastasis promoting member of the S100 family, S100A4 activated BNIP3 expression resulting in apoptosis of pancreatic cancer cell lines. An apparent need to down regulate BNIP3 during tumor progression was demonstrated in studies that investigated the link between BNIP3 expression and metastasis of mouse breast cancer cell lines (Manka et al., 2005). Depletion of BNIP3 in a non-metastatic cell line (4T07), blocked cell death and increased clonogenic survival under hypoxic culture conditions in vitro, besides increased tumor size enabling metastasis to multiple organs in mice injected with the depleted cells (Manka and Millhorn, 2006). A report by these authors also raised the possibility that in metastatic cells that still expressed residual levels of BNIP3, the activity of BNIP3 might be impaired as a result of post-translational modifications such as addition of O-linked GlcNAc (Manka and Millhorn, 2006). The effect of post-translational modifications including O-linked GlcNAc on BNIP3 function remains to be investigated. The above report highlights the need for a systematic investigation on the role of post-translational modification of BNIP3 on its cell death activity. Thus, it appears that transcriptional silencing of BNIP3 via promoter hypermethylation or through up-regulation of transcriptional regulators such as S100A4 or post-translational protein modifications may play a critical role in pancreatic cancer progression and that BNIP3 may be a tumor suppressor in the pancreas. Analyses of several pancreatic cancer cell lines revealed that their relative chemoresistance to first-llne anti-pancreatic cancer drugs such as gemcitabine and 5-flurouracil was linked to the lack of BNIP3 expression (Akada et al., 2005; Erkan et al., 2005; Ishida et al., 2007) and loss of BNIP3 expression also correlated with worsened prognosis of pancreatic ductal adenocarcinomas (Erkan et al., 2005). These results suggest that anti-cancer drugs that may activate expression of BNIP3 in these cancers might constitute an important treatment modality.
In addition to pancreatic cancers, BNIP3 was also reported to be down regulated by promoter hypermethylation and histone deacetylation in colorectal cancers (Bacon et al., 2007; Murai et al., 2005b), gastric cancers (Murai et al., 2005b) and in hematopoietic cancers (Murai et al., 2005a). Reversal of these epigenetic marks by treatment with 5-aza-dC or trichostatin A resulted in re-expression of BNIP3 and conferred hypoxia-inducible cell death in cancer cell lines. These results reinforce the notion that down-regulation of BNIP3 is an important epigenetic mechanism that contributes to tumor progression in multiple cancers. However, it raises the question why BNIP3 expression was associated with high-grade pre-invasive cancers of the breast. As in the case of pancreatic cancers (Erkan et al., 2005), down-regulation of BNIP3 expression or functional sequestration by nuclear localization may be a late and essential stage in tumor progression that may confer survival advantage to invasive cancers. In contrast, during early pre-invasive stages BNIP3 up-regulation as a result of hypoxic tumor environment and chronic tumor necrosis may drive tumor progression. A detailed discussion on the role of BNIP3 in multiple human cancers can also be found elsewhere (Mellor and Harris, 2007).
Our understanding of the role of BNIP3L in cancer is somewhat limited compared to that of BNIP3. Certain studies with human tumor cell lines and clinical samples suggest that BNIP3L may function as a tumor suppressor. An RT-PCR analysis of several lung cancer cell lines and clinical samples showed down regulation of Bnip3L (Sun et al., 2004). A gene expression profiling study that involved implantation of human pancreatic carcinoma cells in SCID mice indicates that BNIP3L was down regulated in the liver metastases and tumor invasion front of the tumor compared to primary tumors (Niedergethmann et al., 2007). A genome wide analysis of CpG methylation in human hepatocellular carcinomas identified selective inactivation of BNIP3L and BNIP3 was correlated with poor prognosis (Calvisi et al., 2007). A genome wide Gene Chip (GeneChip 500K) analysis of prostate cancer cell lines and clinical samples revealed homozygous deletion of BNIP3L (Liu et al., 2008). Although the above reports suggested that down-regulation of BNIP3L might be important for tumor progression, in addition to down-regulation of other tumor suppressor genes, the specific involvement of BNIP3L was not investigated. A convincing study by El-Deiry and colleagues indicated that Bnip3L was a transcriptional target for the tumor suppressor p53 under hypoxia (Fei et al., 2004). Interestingly, other known pro-apoptotic p53 targets such as Puma, Bax and p21 were not induced under hypoxia. Specific depletion of BNIP3L in a non-tumorigenic human osteosarcoma (U2OS) conferred tumorigenic property in mouse xenografts. These studies also revealed that BNIP3L contributed to the p53-dependent tumor suppression of U2OS cells. More studies would be required to ascertain whether the BNIP3L plays a tumor promoting role in early stages of tumor development, like BNIP3.
BNIP3 and BNIP3L in cell differentiation
Gene expression profiling studies have identified an increase in BNIP3 expression during differentiation of rat oligodentrocytes (Itoh et al., 2003). Interestingly, the increase in BNIP3 occurred in parallel with Bcl-xL, suggesting that BNIP3 and BCL-xL may simultaneously regulate cell survival and cell death during differentiation. Up-regulation of BNIP3 was also reported during mouse chondrocyte differentiation (Chen et al., 2005). In the chondrocyte system, also the increase in BNIP3 expression during late stages of differentiation was accompanied by other genes implicated in cell proliferation. In human umbilical cord and bone marrow mesenchymal stem cells (which exist under hypoxic environment and differentiate into multiple cell lineages) BNIP3 and the cytokine VEGF were prominently expressed in response to hypoxia (Martin-Rendon et al., 2007). Immunohistochemistry analysis of several hypoxia responsive markers during osteoclast differentiation also revealed up-regulation of BNIP3 and VEGF (Knowles and Athanasou, 2008). Although these reports suggest a role for BNIP3 (in concert with certain survival factors) in cell differentiation, the precise role of BNIP3 in this process remains to be investigated.
In contrast, genetic studies using Bnip3L−/− mouse model indicate that BNIP3L plays an important role in erythrocyte differentiation. Although BNIP3L is constitutively expressed in several tissues, a gene profiling study indicated that expression of BNIP3L and BCL-xL was increased during erythropoietin (Epo)-induced differentiation of human erythroblasts (Aerbajinai et al., 2003). Bnip3L null mice exhibited anemia accompanied by striking enlargement of spleen and erythroblast hyperplasia in the spleen and bone marrow (Diwan et al., 2007a). These results suggested that the accumulation of erythroblasts in the absence of BNIP3L might be the consequence of reduced cell death. This conclusion was supported by in vitro studies in which the erythroid cells cultured from the spleen of Bnip3L null mice were shown to be hypersensitive to Epo treatment (which activates BCL-xL-dependent survival signaling) and resistant to multiple cell death stimuli. Thus, these studies revealed that BNIPL-mediated cell death that opposes the Epo-induced erythroblast survival was important for normal erythrocyte production in conjunction with pro-survival proteins such as BCL-xL. The same group of investigators also showed that the erythrocytes from Bnip3L−/− mice were morphologically abnormal, suggesting the requirement of BNIP3L for the maintenance of cell number and for the elimination of abnormal erythrocytes during erythropoiesis (Diwan et al., 2008a). Erythroid cells undergo selective elimination of nuclei and organelles during terminal differentiation (Holm et al., 2002). BNIP3L was shown to be essential for the clearance of mitochondria during the terminal stage of erythrocyte maturation (Schweers et al., 2007). BNIP3L was shown to mediate this process via mitochondrial autophagy (Sandoval et al., 2008; Zhang and Ney, 2008). This activity was linked to the ability of BNIP3L to induce loss of ΔΨm that resulted in mitochondrial sequestration in the autophagasomes. Since defective autophagy is linked to anemia in humans (Kent et al., 1966), extrapolation of the results obtained with Bnip3L−/− mouse model should provide valuable information for intervention with this disorder.
BNIP3 and BNIP3L in cardiomyocyte death
Death of cardiomyocytes following ischemic and non-ischemic injury has been shown to contribute to heart failure (Narula et al., 1996; Olivetti et al., 1997). Both BNIP3 and BNIP3L have been prominently implicated in cardiomycyte elimination as a result of these injuries. Acute ischemia has been shown to upregulate BNIP3 expression and its insertion in the mitochondrial membrane (Graham et al., 2004; Regula et al., 2002). Expression of the dominant negative form of BNIP3 (BNIP3ΔTM) inhibited myocyte death and prevented membrane insertion of endogenous BNIP3 (Regula et al., 2002), suggesting a direct involvement of BNIP3. Further, transduction of a cell permeable version of BNIP3ΔTM (tagged with the HIV-1 Tat peptide) into whole heart conferred protection against I/R injury and improved cardiac function (Hamacher-Brady et al., 2007). In addition to hypoxia-mediated transcriptional activation of BNIP3, the acidotic condition that occurs during ischemia was also reported to contribute to BNIP3-dependent death of myocytes (Graham et al., 2004). A feature of myocardial infarction is the ischemic injury that is followed by sudden reoxygenation (known as ischemia/reperfusion, I/R) that exacerbates the damage to the myocardium due to the oxidation of proteins and generation of reactive oxygen species. A recent report indicated that BNIP3 was ‘activated’ through enhanced homodimerization mediated by the TM domain and a conserved N-terminal Cys residue (Cys64) and membrane insertion, suggesting that BNIP3 functions as a redox sensor during I/R injury (Kubli et al., 2008). Thus, it appears that hypoxia-induced overexpression and subsequent BNIP3 activation by homodimerization and mitochondrial insertion is required for BNIP3-mediated myocyte death. This was highlighted by studies with transgenic mice in which forced cardiac overexpression of BNIP3 increased myocyte cell death (Diwan et al., 2007b). As discussed earlier, BNIP3-induced myocyte death appears to be both autophagic as well as apoptotic. Studies with the Bnip3−/− mouse model indicated that myocyte death following I/R was a delayed response (Diwan et al., 2007b). The reader is directed to the Chapter by Dorn and Kirshenbaum as well as a recent review (Shaw and Kirshenbaum, 2008) for a more detailed discussion on this subject.
Our current understanding of the role of BNIP3L in cardiomyocyte death is primarily based on elegant studies by Dorn and his collaborators who employed transgenic as well as knockout mouse models of BNIP3L (Diwan et al., 2008b; Yussman et al., 2002). A microarray analysis in a genetic experimental cardiac hypertrophy mouse model (cardiac specific overexpression of the heterotrimeric G protein, Gq that recapitulates essential features of cardiac hypertrophy induced by pressure overload) by Dorn and colleagues revealed up-regulation of BNIP3L in the myocardium (Yussman et al., 2002). Transgenic mice that overexpressed Bnip3L in the heart died as neonates (6 to 7 days after birth) and exhibited features of cardiac hypertrophy and apoptosis. Importantly, the hearts of transgenic lines that overexpressed a splice variant of Bnip3L lacking the C-terminal ten amino acids (a dominant negative form of Bnip3L) were normal. Overexpression of the short isoform of Bnip3L in the double transgenic mice delayed early Gq-mediated death and prevented the symptoms of cardiomyopathy. In contrast to the severe neonatal phenotype of the Bnip3L transgenic mice, the phenotype was mild in adults when expressed from an inducible promoter (Syed et al., 2004). However, the adult phenotype was severe when combined with Gq overexpression (which by itself exhibited mild adult phenotype) suggesting a synergy between specific hypertrophic factors that influence post-natal cardiac growth and factors such as BNIP3L that regulate cardiomyocyte cell death in heart failure. Cardiac specific deletion of Bnip3L by Cre-mediated deletion provided a high degree of protection against cardiomyopathies induced by Gq overexpression or by pressure overload (Diwan et al., 2008b). These results suggested that cell death mediated by BNIP3L is a critical determinant in hypertrophic heart failure. Thus, the results from the animal models suggest that BNIP3 and BNIP3L may be important therapeutic targets in myocardial salvage during pathological heart failures. It remains to be determined whether BNIP3L also becomes activated in adult heart during pathological stimuli that lead to heart failure. The animal models generated by the Dorn group would also be valuable to study interesting questions such as the factors that restrain the cell death activity in the adult heart and the mode of BNIP3L activation.
Role of BNIP3 family in other diseases and pathological cell death
Attenuation of BNIP3’s ability to induce cell death has been implicated in inducing synovial hyperplasia in rheumatoid arthritis (RA). BNIP3 is up-regulated in synoviocytes, likely due to hypoxia present in arthritic joints and plays a role in the pathogenesis of arthritis (Kammouni et al., 2007). Under hypoxic conditions, fibroblast-like synoviocytes in culture induced BNIP3 expression, which resulted in BNIP3-induced cell death. However, BNIP3 up-regulation in synoviocytes from RA patients failed to induce cell death. This is likely due to the presence of pro-inflammatory cytokines in RA patients since treatment with TNF-α and IL-1β blocked BNIP3 induced cell death in fibroblast-like synoviocytes (Kammouni et al., 2007).
BNIP3 has been shown to play a role in neuronal cell death after a stroke. BNIP3 is up-regulated in neurons after focal brain ischemia in a rat model of stroke and in cultured primary neurons exposed to hypoxia. After blockage of the middle cerebral arteries for 48 hrs, BNIP3 was expressed in both cytoplasm and nuclei. Furthermore, inhibition of BNIP3 by siRNA protected the neurons from hypoxia-induced cell death. Nuclear localization of BNIP3 has also been found in neurons in response to ischemia. Moreover, neurons that had higher levels of BNIP3 in the nucleus were more resistant to cell death, but the mechanism of this localization is still unknown (Schmidt-Kastner et al., 2004).
BNIP3 has also been implicated to play a role in neuronal cell death in Alzheimer’s disease. Amyloid β-plaques are a hallmark of Alzheimer’s disease that can induce caspase-independent cell death in neurons. Amyloid-β treatment of cultured rat cortical neurons resulted in an increase in BNIP3 expression through HIF-1α stabilization. Knock-down of BNIP3 blocked amyloid-induced cell death (Zhang et al., 2007). This implicates BNIP3 in neuronal cell death in Alzheimer’s. The role of BNIP3L is less well understood in these diseases and should be the focus of future research.
Both BNIP3 and BNIP3L have been shown to play a role in macrophage cell death induced by anthrax lethal toxin (Ha et al., 2007). A genome-wide analysis of transcripts in murine macrophages that were spontaneously resistant to the toxin showed down-regulation of mRNAs for Bnip3 and Bnip3L compared to susceptible cells. Overexpression of BNIP3 and BNIP3L in resistant cells conferred toxin sensitivity. Inhibition of the p38 MAPK pathway by the toxin was linked to mRNA destabilization of these genes. Since pharmacological inhibitors of p38 resulted in resistance to the toxin, the use of such inhibitors was suggested as an adjunctive therapy for anthrax toxemia, in addition to antibiotic regimens against Bacillus anthracis.
Concluding Remarks
BNIP3 is now recognized as one of the most prominent hypoxia activated genes. The role of hypoxia in up-regulation of BNIP3L appears to be context dependent. This mode of regulation has led to a significant body of investigation on the role of these molecules in hypoxia modulated pathologies such as cancer and cardiomyopathies. In cancer, they appear to function as tumor promoters during development of solid tumors that experience hypoxic environment and chronic tumor necrosis. However, they function as tumor suppressors during the tumor progression phase. There is also strong evidence linking them to mitochondrial homeostasis and mitochondrial elimination during erythrocyte differentiation. Both BNIP3 and BNIP3L appear to mediate their function through autophagy and apoptosis-dependent mechanisms. The mechanisms by which they mediate cell death still remain to be clarified in detail. Future studies will be required to elucidate the role of different signaling pathways and post translational modifications of BNIP3 and BNIP3L that modulate their activity. The mouse knockout models have proved valuable in elucidating the roles of BNIP3 and BNIP3L in cardiomyopathies and erythrocyte differentiation. These animal models would be expected to be important for future investigations on the role of these proteins in oncogenesis and in evaluation of potential therapeutic molecules.
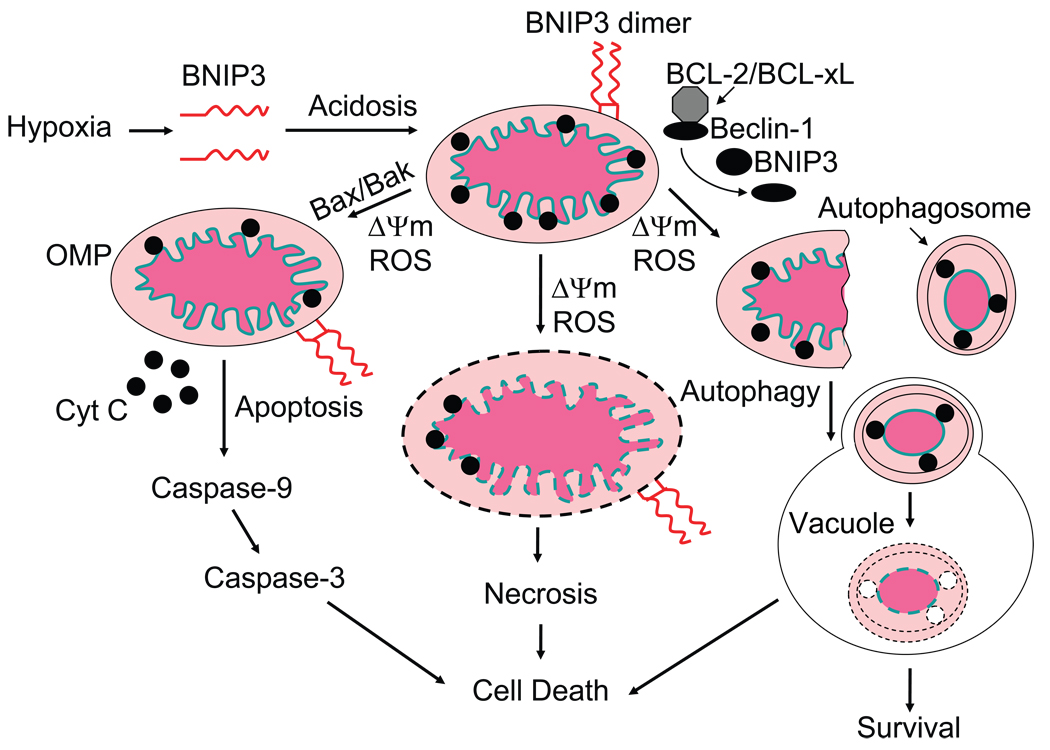
Fig. 2. Model for BNIP3-mediated mitochondrial damage and cell death.
Hypoxia-induced activation of BNIP3 expression and the resulting acidotic condition are shown to mediate stable homodimerization and mitochondrial membrane insertion of BNIP3 resulting in loss of ΔΨm and generation of ROS. As a consequence of the mitochondrial damage, cells are shown to undergo three different fates, BAX/BAK-dependent apoptosis, autophagy (by the release of Beclin-1 from the BCL-2/BCL-xL complex) and possibly necrosis.
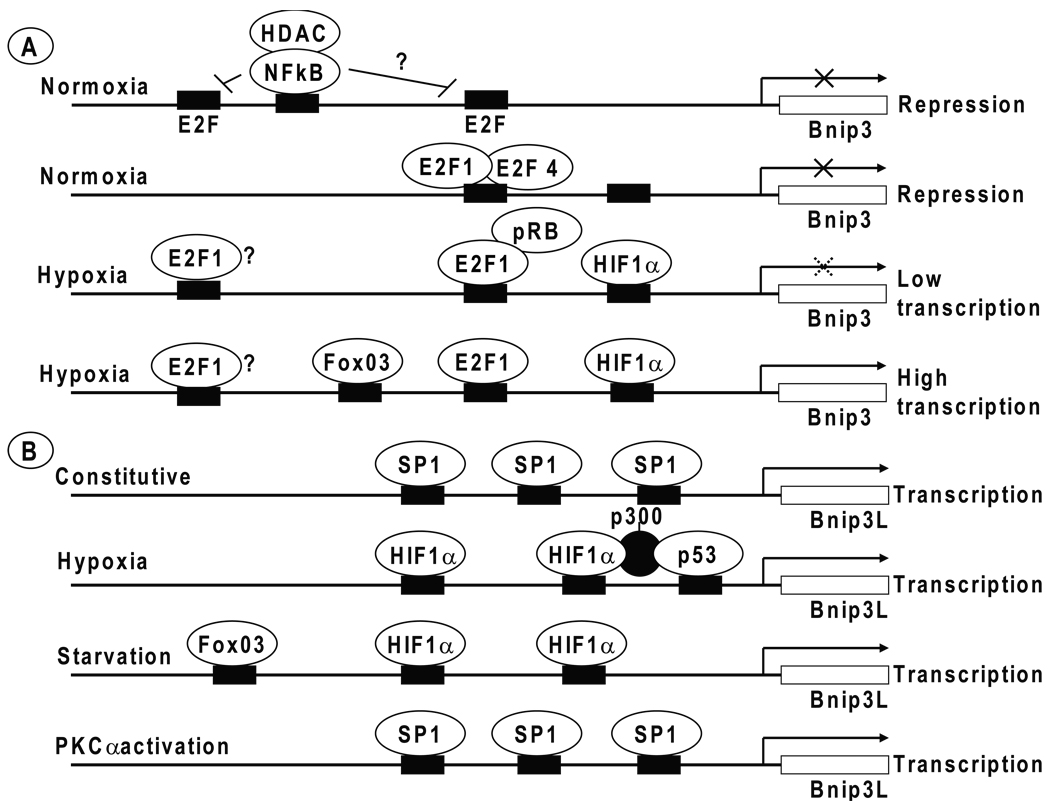
Fig. 3. Transcriptional regulation of Bnip3 and Bnip3L.
(A) The transcription of the Bnip3 gene is depicted to be repressed under normoxic conditions by transcription factors NFκB and E2F4. Under hypoxia, the activity of HIF1α is shown to be moderated by the negative regulatory effects of pRb and NFκB. Hypoxia-induced high level transcription appears to be mediated by the combined action of E2F1 and/or FoxO3 with HIF1α. (B) In contrast to Bnip3, expression of Bnip3L is constitutive in several tissues and is shown to be mediated by transcription factors such as Sp1. Under hypoxic conditions, p53 is shown to cooperatively activate transcription with HIF1α. Similarly, under conditions of starvation, the transcription is shown to be activated by FoxO3, possibly in cooperation with HIF1α. Under specific conditions such as cardiac hypertrophy that results in PKCα activation, the transcription is indicated to be activated by Sp1. The context under which the various transcription factors may regulate transcription of Bnip3 and Bnip3L is discussed in the text. The binding sites for various transcription factors are indicated by solid bars (not all sites are shown).
Acknowledgement
GC received grant support from the National Cancer Institute grants CA-33616, CA-116262 and CA-73803. SBG was supported by a grant from Canadian Institutes for Health Research MOP-64330 and is a Manitoba Research Chair supported by the Manitoba Health Research Council. We thank Lorrie Kirshenbaum for comments on this review.
References
- Abe T, Toyota M, Suzuki H, Murai M, Akino K, Ueno M, et al. Upregulation of BNIP3 by 5-aza-2'-deoxycytidine sensitizes pancreatic cancer cells to hypoxiamediated cell death. J Gastroenterol. 2005;40:504–510. doi: 10.1007/s00535-005-1576-1. [DOI] [PubMed] [Google Scholar]
- Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- Aerbajinai W, Giattina M, Lee YT, Raffeld M, Miller JL. The proapoptotic factor Nix is coexpressed with Bcl-xL during terminal erythroid differentiation. Blood. 2003;102:712–717. doi: 10.1182/blood-2002-11-3324. [DOI] [PubMed] [Google Scholar]
- Agah R, Kirshenbaum LA, Abdellatif M, Truong LD, Chakraborty S, Michael LH, et al. Adenoviral delivery of E2F-1 directs cell cycle reentry and p53-independent apoptosis in postmitotic adult myocardium in vivo. J Clin Invest. 1997;100:2722–2728. doi: 10.1172/JCI119817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akada M, Crnogorac-Jurcevic T, Lattimore S, Mahon P, Lopes R, Sunamura M, et al. Intrinsic chemoresistance to gemcitabine is associated with decreased expression of BNIP3 in pancreatic cancer. Clin Cancer Res. 2005;11:3094–3101. doi: 10.1158/1078-0432.CCR-04-1785. [DOI] [PubMed] [Google Scholar]
- Aouacheria A, Brunet F, Gouy M. Phylogenomics of life-or-death switches in multicellular animals: Bcl-2, BH3-Only, and BNip families of apoptotic regulators. Mol Biol Evol. 2005;22:2395–2416. doi: 10.1093/molbev/msi234. [DOI] [PubMed] [Google Scholar]
- Azad MB, Chen Y, Henson ES, Cizeau J, McMillan-Ward E, Israels SJ, et al. Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving BNIP3. Autophagy. 2008;4:195–204. doi: 10.4161/auto.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon AL, Fox S, Turley H, Harris AL. Selective silencing of the hypoxia-inducible factor 1 target gene BNIP3 by histone deacetylation and methylation in colorectal cancer. Oncogene. 2007;26:132–141. doi: 10.1038/sj.onc.1209761. [DOI] [PubMed] [Google Scholar]
- Baetz D, Regula KM, Ens K, Shaw J, Kothari S, Yurkova N, et al. Nuclear factor-kappaB-mediated cell survival involves transcriptional silencing of the mitochondrial death gene BNIP3 in ventricular myocytes. Circulation. 2005;112:3777–3785. doi: 10.1161/CIRCULATIONAHA.105.573899. [DOI] [PubMed] [Google Scholar]
- Boyd JM, Malstrom S, Subramanian T, Venkatesh LK, Schaeper U, Elangovan B, et al. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 1994;79:341–351. doi: 10.1016/0092-8674(94)90202-x. [DOI] [PubMed] [Google Scholar]
- Bruick RK. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci U S A. 2000;97:9082–9087. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton TR, Henson ES, Baijal P, Eisenstat DD, Gibson SB. The pro-cell death Bcl-2 family member, BNIP3, is localized to the nucleus of human glial cells: Implications for glioblastoma multiforme tumor cell survival under hypoxia. Int J Cancer. 2006;118:1660–1669. doi: 10.1002/ijc.21547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvisi DF, Ladu S, Gorden A, Farina M, Lee JS, Conner EA, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713–2722. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Cizeau J, Vande Velde C, Park JH, Bozek G, Bolton J, et al. Nix and Nip3 form a subfamily of pro-apoptotic mitochondrial proteins. J Biol Chem. 1999;274:7–10. doi: 10.1074/jbc.274.1.7. [DOI] [PubMed] [Google Scholar]
- Chen G, Ray R, Dubik D, Shi L, Cizeau J, Bleackley RC, et al. The E1B 19K/Bcl-2-binding protein Nip3 is a dimeric mitochondrial protein that activates apoptosis. J Exp Med. 1997;186:1975–1983. doi: 10.1084/jem.186.12.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Fink T, Zhang XY, Ebbesen P, Zachar V. Quantitative transcriptional profiling of ATDC5 mouse progenitor cells during chondrogenesis. Differentiation. 2005;73:350–363. doi: 10.1111/j.1432-0436.2005.00038.x. [DOI] [PubMed] [Google Scholar]
- Cizeau J, Ray R, Chen G, Gietz RD, Greenberg AH. The C. elegans orthologue ceBNIP3 interacts with CED-9 and CED-3 but kills through a BH3- and caspase-independent mechanism. Oncogene. 2000;19:5453–5463. doi: 10.1038/sj.onc.1203929. [DOI] [PubMed] [Google Scholar]
- Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004;64:4286–4293. doi: 10.1158/0008-5472.CAN-03-3084. [DOI] [PubMed] [Google Scholar]
- Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB, et al. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- Diwan A, Koesters AG, Capella D, Geiger H, Kalfa TA, Dorn GW., 2nd Targeting erythroblast-specific apoptosis in experimental anemia. Apoptosis. 2008a;13:1022–1030. doi: 10.1007/s10495-008-0236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwan A, Koesters AG, Odley AM, Pushkaran S, Baines CP, Spike BT, et al. Unrestrained erythroblast development in Nix−/− mice reveals a mechanism for apoptotic modulation of erythropoiesis. Proc Natl Acad Sci U S A. 2007a;104:6794–6799. doi: 10.1073/pnas.0610666104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwan A, Krenz M, Syed FM, Wansapura J, Ren X, Koesters AG, et al. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest. 2007b;117:2825–2833. doi: 10.1172/JCI32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwan A, Wansapura J, Syed FM, Matkovich SJ, Lorenz JN, Dorn GW., 2nd Nix-mediated apoptosis links myocardial fibrosis, cardiac remodeling, and hypertrophy decompensation. Circulation. 2008b;117:396–404. doi: 10.1161/CIRCULATIONAHA.107.727073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. Faseb J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- Erkan M, Kleeff J, Esposito I, Giese T, Ketterer K, Buchler MW, et al. Loss of BNIP3 expression is a late event in pancreatic cancer contributing to chemoresistance and worsened prognosis. Oncogene. 2005;24:4421–4432. doi: 10.1038/sj.onc.1208642. [DOI] [PubMed] [Google Scholar]
- Farooq M, Kim Y, Im S, Chung E, Hwang S, Sohn M, et al. Cloning of BNIP3h, a member of proapoptotic BNIP3 family genes. Exp Mol Med. 2001;33:169–173. doi: 10.1038/emm.2001.29. [DOI] [PubMed] [Google Scholar]
- Fei P, Wang W, Kim SH, Wang S, Burns TF, Sax JK, et al. Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell. 2004;6:597–609. doi: 10.1016/j.ccr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Frazier DP, Wilson A, Graham RM, Thompson JW, Bishopric NH, Webster KA. Acidosis regulates the stability, hydrophobicity, and activity of the BH3-only protein Bnip3. Antioxid Redox Signal. 2006;8:1625–1634. doi: 10.1089/ars.2006.8.1625. [DOI] [PubMed] [Google Scholar]
- Galvez AS, Brunskill EW, Marreez Y, Benner BJ, Regula KM, Kirschenbaum LA, et al. Distinct pathways regulate proapoptotic Nix and BNip3 in cardiac stress. J Biol Chem. 2006;281:1442–1448. doi: 10.1074/jbc.M509056200. [DOI] [PubMed] [Google Scholar]
- Giatromanolaki A, Koukourakis MI, Sowter HM, Sivridis E, Gibson S, Gatter KC, et al. BNIP3 expression is linked with hypoxia-regulated protein expression and with poor prognosis in non-small cell lung cancer. Clin Cancer Res. 2004;10:5566–5571. doi: 10.1158/1078-0432.CCR-04-0076. [DOI] [PubMed] [Google Scholar]
- Graeber TG, Osmanian C, Jacks T, Housman DE, Koch CJ, Lowe SW, et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- Graham RM, Frazier DP, Thompson JW, Haliko S, Li H, Wasserlauf BJ, et al. A unique pathway of cardiac myocyte death caused by hypoxia-acidosis. J Exp Biol. 2004;207:3189–3200. doi: 10.1242/jeb.01109. [DOI] [PubMed] [Google Scholar]
- Guo K, Searfoss G, Krolikowski D, Pagnoni M, Franks C, Clark K, et al. Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death Differ. 2001;8:367–376. doi: 10.1038/sj.cdd.4400810. [DOI] [PubMed] [Google Scholar]
- Ha SD, Ng D, Lamothe J, Valvano MA, Han J, Kim SO. Mitochondrial proteins Bnip3 and Bnip3L are involved in anthrax lethal toxin-induced macrophage cell death. J Biol Chem. 2007;282:26275–26283. doi: 10.1074/jbc.M703668200. [DOI] [PubMed] [Google Scholar]
- Hamacher-Brady A, Brady NR, Logue SE, Sayen MR, Jinno M, Kirshenbaum LA, et al. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- Holm TM, Braun A, Trigatti BL, Brugnara C, Sakamoto M, Krieger M, et al. Failure of red blood cell maturation in mice with defects in the high-density lipoprotein receptor SR-BI. Blood. 2002;99:1817–1824. doi: 10.1182/blood.v99.5.1817. [DOI] [PubMed] [Google Scholar]
- Imazu T, Shimizu S, Tagami S, Matsushima M, Nakamura Y, Miki T, et al. Bcl-2/E1B 19 kDa-interacting protein 3-like protein (Bnip3L) interacts with bcl-2/Bcl-xL and induces apoptosis by altering mitochondrial membrane permeability. Oncogene. 1999;18:4523–4529. doi: 10.1038/sj.onc.1202722. [DOI] [PubMed] [Google Scholar]
- Ishida M, Sunamura M, Furukawa T, Akada M, Fujimura H, Shibuya E, et al. Elucidation of the relationship of BNIP3 expression to gemcitabine chemosensitivity and prognosis. World J Gastroenterol. 2007;13:4593–4597. doi: 10.3748/wjg.v13.i34.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Itoh A, Pleasure D. Bcl-2-related protein family gene expression during oligodendroglial differentiation. J Neurochem. 2003;85:1500–1512. doi: 10.1046/j.1471-4159.2003.01795.x. [DOI] [PubMed] [Google Scholar]
- Kammouni W, Wong K, Ma G, Firestein GS, Gibson SB, El-Gabalawy HS. Regulation of apoptosis in fibroblast-like synoviocytes by the hypoxia-induced Bcl-2 family member Bcl-2/adenovirus E1B 19-kd protein-interacting protein 3. Arthritis Rheum. 2007;56:2854–2863. doi: 10.1002/art.22853. [DOI] [PubMed] [Google Scholar]
- Kanzawa T, Zhang L, Xiao L, Germano IM, Kondo Y, Kondo S. Arsenic trioxide induces autophagic cell death in malignant glioma cells by upregulation of mitochondrial cell death protein BNIP3. Oncogene. 2005;24:980–991. doi: 10.1038/sj.onc.1208095. [DOI] [PubMed] [Google Scholar]
- Kent G, Minick OT, Volini FI, Orfei E. Autophagic vacuoles in human red cells. Am J Pathol. 1996;48:831–857. [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Cho JJ, Ha J, Park JH. The carboxy terminal C-tail of BNip3 is crucial in induction of mitochondrial permeability transition in isolated mitochondria. Arch Biochem Biophys. 2002;398:147–152. doi: 10.1006/abbi.2001.2673. [DOI] [PubMed] [Google Scholar]
- Kirshenbaum LA, Abdellatif M, Chakraborty S, Schneider MD. Human E2F-1 reactivates cell cycle progression in ventricular myocytes and represses cardiac gene transcription. Dev Biol. 1996;179:402–411. doi: 10.1006/dbio.1996.0270. [DOI] [PubMed] [Google Scholar]
- Knowles HJ, Athanasou NA. Hypoxia-inducible factor is expressed in giant cell tumour of bone and mediates paracrine effects of hypoxia on monocyte-osteoclast differentiation via induction of VEGF. J Pathol. 2008;215:56–66. doi: 10.1002/path.2319. [DOI] [PubMed] [Google Scholar]
- Kothari S, Cizeau J, McMillan-Ward E, Israels SJ, Bailes M, Ens K, et al. BNIP3 plays a role in hypoxic cell death in human epithelial cells that is inhibited by growth factors EGF and IGF. Oncogene. 2003;22:4734–4744. doi: 10.1038/sj.onc.1206666. [DOI] [PubMed] [Google Scholar]
- Kubasiak LA, Hernandez OM, Bishopric NH, Webster KA. Hypoxia and acidosis activate cardiac myocyte death through the Bcl-2 family protein BNIP3. Proc Natl Acad Sci U S A. 2002;99:12825–12830. doi: 10.1073/pnas.202474099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubli DA, Quinsay MN, Huang C, Lee Y, Gustafsson AB. Bnip3 Functions as a Mitochondrial Sensor of Oxidative Stress during Myocardial Ischemia and Reperfusion. Am J Physiol Heart Circ Physiol. 2008 doi: 10.1152/ajpheart.00552.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubli DA, Ycaza JE, Gustafsson AB. Bnip3 mediates mitochondrial dysfunction and cell death through Bax and Bak. Biochem J. 2007;405:407–415. doi: 10.1042/BJ20070319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xie CC, Zhu Y, Li T, Sun J, Cheng Y, et al. Homozygous deletions and recurrent amplifications implicate new genes involved in prostate cancer. Neoplasia. 2008;10:897–907. doi: 10.1593/neo.08428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon PC, Baril P, Bhakta V, Chelala C, Caulee K, Harada T, et al. S100A4 contributes to the suppression of BNIP3 expression, chemoresistance, and inhibition of apoptosis in pancreatic cancer. Cancer Res. 2007;67:6786–6795. doi: 10.1158/0008-5472.CAN-07-0440. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. Embo J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Schiaffino S, Sandri M. Downstream of Akt: FoxO3 and mTOR in the regulation of autophagy in skeletal muscle. Autophagy. 2008;4:524–526. doi: 10.4161/auto.5905. [DOI] [PubMed] [Google Scholar]
- Manka D, Millhorn DE. A potential molecular link between aerobic glycolysis and cancer. Cell Cycle. 2006;5:343–344. doi: 10.4161/cc.5.4.2474. [DOI] [PubMed] [Google Scholar]
- Manka D, Spicer Z, Millhorn DE. Bcl-2/adenovirus E1B 19 kDa interacting protein-3 knockdown enables growth of breast cancer metastases in the lung, liver, and bone. Cancer Res. 2005;65:11689–11693. doi: 10.1158/0008-5472.CAN-05-3091. [DOI] [PubMed] [Google Scholar]
- Martin-Rendon E, Hale SJ, Ryan D, Baban D, Forde SP, Roubelakis M, et al. Transcriptional profiling of human cord blood CD133+ and cultured bone marrow mesenchymal stem cells in response to hypoxia. Stem Cells. 2007;25:1003–1012. doi: 10.1634/stemcells.2006-0398. [DOI] [PubMed] [Google Scholar]
- Matsushima M, Fujiwara T, Takahashi E, Minaguchi T, Eguchi Y, Tsujimoto Y, et al. Isolation, mapping, and functional analysis of a novel human cDNA (BNIP3L) encoding a protein homologous to human NIP3. Genes Chromosomes Cancer. 1998;21:230–235. [PubMed] [Google Scholar]
- Mellor HR, Harris AL. The role of the hypoxia-inducible BH3-only proteins BNIP3 and BNIP3L in cancer. Cancer Metastasis Rev. 2007;26:553–566. doi: 10.1007/s10555-007-9080-0. [DOI] [PubMed] [Google Scholar]
- Metcal DG, Law PB, DeGrado WF. Mutagenesis data in the automated prediction of transmembrane helix dimers. Proteins. 2007;67:375–384. doi: 10.1002/prot.21265. [DOI] [PubMed] [Google Scholar]
- Mizutani A, Furukawa T, Adachi Y, Ikehara S, Taketani S. A zinc-finger protein, PLAGL2, induces the expression of a proapoptotic protein Nip3, leading to cellular apoptosis. J Biol Chem. 2002;277:15851–15858. doi: 10.1074/jbc.M111431200. [DOI] [PubMed] [Google Scholar]
- Murai M, Toyota M, Satoh A, Suzuki H, Akino K, Mita H, et al. Aberrant DNA methylation associated with silencing BNIP3 gene expression in haematopoietic tumours. Br J Cancer. 2005a;92:1165–1172. doi: 10.1038/sj.bjc.6602422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai M, Toyota M, Suzuki H, Satoh A, Sasaki Y, Akino K, et al. Aberrant methylation and silencing of the BNIP3 gene in colorectal and gastric cancer. Clin Cancer Res. 2005b;11:1021–1027. [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Narula J, Haider N, Virmani R, DiSalvo TG, Kolodgie FD, Hajjar RJ, et al. Apoptosis in myocytes in end-stage heart failure. N Engl J Med. 1996;335:1182–1189. doi: 10.1056/NEJM199610173351603. [DOI] [PubMed] [Google Scholar]
- Niedergethmann M, Alves F, Neff JK, Heidrich B, Aramin N, Li L, et al. Gene expression profiling of liver metastases and tumour invasion in pancreatic cancer using an orthotopic SCID mouse model. Br J Cancer. 2007;97:1432–1440. doi: 10.1038/sj.bjc.6604031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi N, Tokunaga A, Tsunoda H, Nakano K, Haraguchi K, Oda K, et al. A novel adenovirus E1B19K-binding protein B5 inhibits apoptosis induced by Nip3 by forming a heterodimer through the C-terminal hydrophobic region. Cell Death Differ. 1999;6:314–325. doi: 10.1038/sj.cdd.4400493. [DOI] [PubMed] [Google Scholar]
- Okami J, Simeone DM, Logsdon CD. Silencing of the hypoxia-inducible cell death protein BNIP3 in pancreatic cancer. Cancer Res. 2004;64:5338–5346. doi: 10.1158/0008-5472.CAN-04-0089. [DOI] [PubMed] [Google Scholar]
- Olivetti G, Abbi R, Quaini F, Kajstura J, Cheng W, Nitahara JA, et al. Apoptosis in the failing human heart. N Engl J Med. 1997;336:1131–1141. doi: 10.1056/NEJM199704173361603. [DOI] [PubMed] [Google Scholar]
- Rashmi R, Pillai SG, Vijayalingam S, Ryerse J, Chinnadurai G. BH3-only protein BIK induces caspase-independent cell death with autophagic features in Bcl-2 null cells. Oncogene. 2008;27:1366–1375. doi: 10.1038/sj.onc.1210783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray R, Chen G, Vande Velde C, Cizeau J, Park JH, Reed JC, et al. BNIP3 heterodimerizes with Bcl-2/Bcl-X(L) and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J Biol Chem. 2000;275:1439–1448. doi: 10.1074/jbc.275.2.1439. [DOI] [PubMed] [Google Scholar]
- Real PJ, Benito A, Cuevas J, Berciano MT, de Juan A, Coffer P, et al. Blockade of epidermal growth factor receptors chemosensitizes breast cancer cells through up-regulation of Bnip3L. Cancer Res. 2005;65:8151–8157. doi: 10.1158/0008-5472.CAN-05-1134. [DOI] [PubMed] [Google Scholar]
- Regula KM, Ens K, Kirshenbaum LA. Inducible expression of BNIP3 provokes mitochondrial defects and hypoxia-mediated cell death of ventricular myocytes. Circ Res. 2002;91:226–231. doi: 10.1161/01.res.0000029232.42227.16. [DOI] [PubMed] [Google Scholar]
- Reynolds TY, Rockwell S, Glazer PM. Genetic instability induced by the tumor microenvironment. Cancer Res. 1996;56:5754–5757. [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- Schmidt-Kastner R, Aguirre-Chen C, Kietzmann T, Saul I, Busto R, Ginsberg MD. Nuclear localization of the hypoxia-regulated pro-apoptotic protein BNIP3 after global brain ischemia in the rat hippocampus. Brain Res. 2004;1001:133–142. doi: 10.1016/j.brainres.2003.11.065. [DOI] [PubMed] [Google Scholar]
- Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J, Kirshenbaum LA. Molecular regulation of autophagy and apoptosis during ischemic and non-ischemic cardiomyopathy. Autophagy. 2008;4:427–434. doi: 10.4161/auto.5901. [DOI] [PubMed] [Google Scholar]
- Shaw J, Yurkova N, Zhang T, Gang H, Aguilar F, Weidman D, et al. Antagonism of E2F-1 regulated Bnip3 transcription by NF-kappaB is essential for basal cell survival. Proc Natl Acad Sci U S A. 2008;105:20734–20739. doi: 10.1073/pnas.0807735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J, Zhang T, Rzeszutek M, Yurkova N, Baetz D, Davie JR, et al. Transcriptional silencing of the death gene BNIP3 by cooperative action of NF-kappaB and histone deacetylase 1 in ventricular myocytes. Circ Res. 2006;99:1347–1354. doi: 10.1161/01.RES.0000251744.06138.50. [DOI] [PubMed] [Google Scholar]
- Sowter HM, Ferguson M, Pym C, Watson P, Fox SB, Han C, et al. Expression of the cell death genes BNip3 and NIX in ductal carcinoma in situ of the breast; correlation of BNip3 levels with necrosis and grade. J Pathol. 2003;201:573–580. doi: 10.1002/path.1486. [DOI] [PubMed] [Google Scholar]
- Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001;61:6669–6673. [PubMed] [Google Scholar]
- Sulistijo ES, Jaszewski TM, MacKenzie KR. Sequence-specific dimerization of the transmembrane domain of the "BH3-only" protein BNIP3 in membranes and detergent. J Biol Chem. 2003;278:51950–51956. doi: 10.1074/jbc.M308429200. [DOI] [PubMed] [Google Scholar]
- Sulistijo ES, MacKenzie KR. Sequence dependence of BNIP3 transmembrane domain dimerization implicates side-chain hydrogen bonding and a tandem GxxxG motif in specific helix-helix interactions. J Mol Biol. 2006;364:974–990. doi: 10.1016/j.jmb.2006.09.065. [DOI] [PubMed] [Google Scholar]
- Sun JL, He XS, Yu YH, Chen ZC. [Expression and structure of BNIP3L in lung cancer] Ai Zheng. 2004;23:8–14. [PubMed] [Google Scholar]
- Syed F, Odley A, Hahn HS, Brunskill EW, Lynch RA, Marreez Y, et al. Physiological growth synergizes with pathological genes in experimental cardiomyopathy. Circ Res. 2004;95:1200–1206. doi: 10.1161/01.RES.0000150366.08972.7f. [DOI] [PubMed] [Google Scholar]
- Tan EY, Campo L, Han C, Turley H, Pezzella F, Gatter KC, et al. BNIP3 as a progression marker in primary human breast cancer; opposing functions in in situ versus invasive cancer. Clin Cancer Res. 2007;13:467–474. doi: 10.1158/1078-0432.CCR-06-1466. [DOI] [PubMed] [Google Scholar]
- Theodorakis P, D'Sa-Eipper C, Subramanian T, Chinnadurai G. Unmasking of a proliferation-restraining activity of the anti-apoptosis protein EBV BHRF1. Oncogene. 1996;12:1707–1713. [PubMed] [Google Scholar]
- Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27:6229–6242. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoki M, Nakamura Y. EGR2 induces apoptosis in various cancer cell lines by direct transactivation of BNIP3L and BAK. Oncogene. 2003;22:2172–2185. doi: 10.1038/sj.onc.1206222. [DOI] [PubMed] [Google Scholar]
- Vande Velde C, Cizeau J, Dubik D, Alimonti J, Brown T, Israels S, et al. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol Cell Biol. 2000;20:5454–5468. doi: 10.1128/mcb.20.15.5454-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H, Miyashita T, Nakano Y, Yamamoto D. HSpin1, a transmembrane protein interacting with Bcl-2/Bcl-xL, induces a caspase-independent autophagic cell death. Cell Death Differ. 2003;10:798–807. doi: 10.1038/sj.cdd.4401246. [DOI] [PubMed] [Google Scholar]
- Yasuda M, D'Sa-Eipper C, Gong XL, Chinnadurai G. Regulation of apoptosis by a Caenorhabditis elegans BNIP3 homolog. Oncogene. 1998a;17:2525–2530. doi: 10.1038/sj.onc.1202467. [DOI] [PubMed] [Google Scholar]
- Yasuda M, Han JW, Dionne CA, Boyd JM, Chinnadurai G. BNIP3alpha: a human homolog of mitochondrial proapoptotic protein BNIP3. Cancer Res. 1999;59:533–537. [PubMed] [Google Scholar]
- Yasuda M, Theodorakis P, Subramanian T, Chinnadurai G. Adenovirus E1B-19K/BCL-2 interacting protein BNIP3 contains a BH3 domain and a mitochondrial targeting sequence. J Biol Chem. 1998b;273:12415–12421. doi: 10.1074/jbc.273.20.12415. [DOI] [PubMed] [Google Scholar]
- Yurkova N, Shaw J, Blackie K, Weidman D, Jayas R, Flynn B, et al. The cell cycle factor E2F-1 activates Bnip3 and the intrinsic death pathway in ventricular myocytes. Circ Res. 2008;102:472–479. doi: 10.1161/CIRCRESAHA.107.164731. [DOI] [PubMed] [Google Scholar]
- Yussman MG, Toyokawa T, Odley A, Lynch RA, Wu G, Colbert MC, et al. Mitochondrial death protein Nix is induced in cardiac hypertrophy and triggers apoptotic cardiomyopathy. Nat Med. 2002;8:725–730. doi: 10.1038/nm719. [DOI] [PubMed] [Google Scholar]
- Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang J, Ney PA. NIX induces mitochondrial autophagy in reticulocytes. Autophagy. 2008;4:354–356. doi: 10.4161/auto.5552. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yang X, Zhang S, Ma X, Kong J. BNIP3 upregulation and EndoG translocation in delayed neuronal death in stroke and in hypoxia. Stroke. 2007;38:1606–1613. doi: 10.1161/STROKEAHA.106.475129. [DOI] [PubMed] [Google Scholar]
- Zhaorigetu S, Wan G, Kaini R, Jiang Z, Hu CA. ApoL1, a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy. 2008;4 doi: 10.4161/auto.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XM, Liu Y, Payne G, Lutz RJ, Chittenden T. Growth factors inactivate the cell death promoter BAD by phosphorylation of its BH3 domain on Ser155. J Biol Chem. 2000;275:25046–25051. doi: 10.1074/jbc.M002526200. [DOI] [PubMed] [Google Scholar]