Abstract
The design and synthesis of small molecules that target RNA is immensely important in antibacterial therapy. We had previously reported on the RNA binding of a series of 4,5-disubstituted 2-oxazolidinones that bind to a highly conserved bulge region of bacterial RNA. This biological target T box antitermination system, which is found mainly in Gram-positive bacteria, regulates the expression of several amino acid related genes. In an effort to amplify our library, we have prepared a library of 1,4-disubstituted 1,2,3-triazole analogs that entails an isosteric replacement of the oxazolidinone nucleus. The synthesis of the new analogs was enhanced via copper(I) catalysis of an azide and alkyne cycloaddition reaction. A total of 108 1,4-disubstituted 1,2,3-triazole compounds have been prepared. All compounds were evaluated as RNA binding agents.
Keywords: Bioisosteric replacement, Oxazolidinone antibiotics, T box Antiterminator RNA
The identification of ligands for RNA that modulate transcription has significant potential for the development of new therapeutics. In previous communications, we reported on a novel class of 4,5-disubstituted oxazolidin-2-ones with good binding affinity towards the T box antiterminator RNA system.1 The T box transcription antitermination system regulates many genes in Gram-positive bacteria, including aminoacyl-tRNA synthetase, amino acid biosynthesis and amino acid transport genes. Transcription of these genes requires the interaction of the uncharged cognate tRNA with the 5’ untranslated mRNA leader region, which results in the formation and stabilization of an antiterminator element.2,3 Small ligands that bind to the antiterminator and disrupt the tRNA-antiterminator interaction have the potential to halt the gene expression.
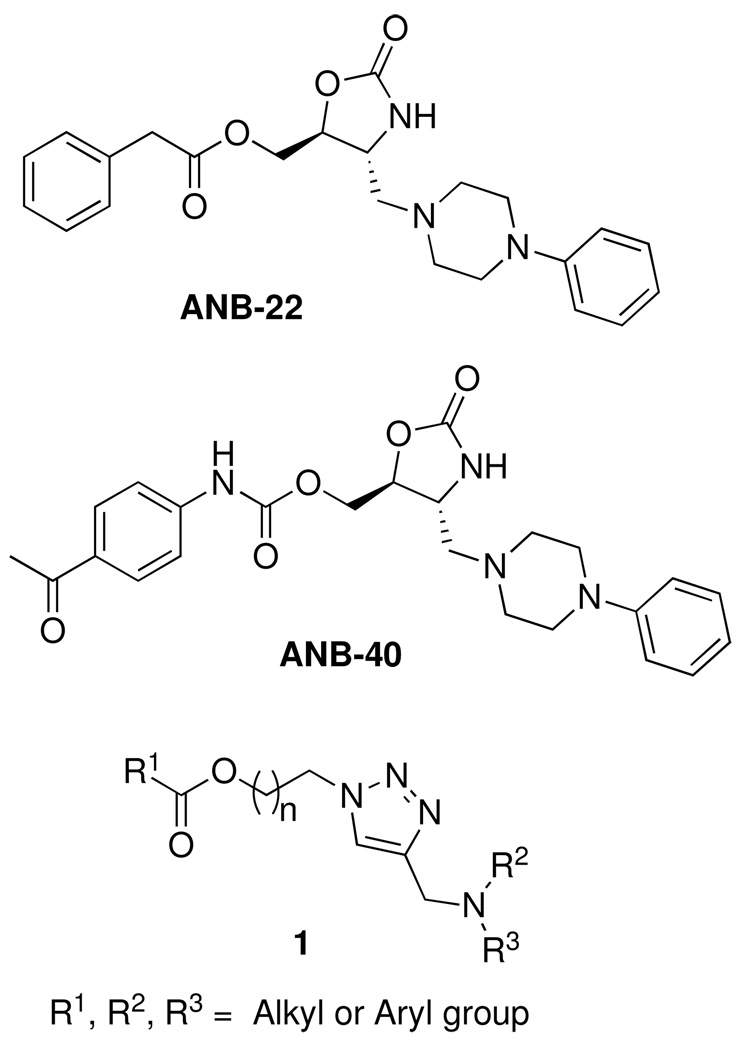
The previously reported oxazolidinones bind the targeted RNA without significant dependence on electrostatic interactions. These compounds also exhibited good binding specificity and affinity for model RNA AM1A relative to C11U.1 More recently, two oxazolidinones, ANB-22 and ANB-40, with improved biological activity have been identified.4 These oxazolidinone leads bind to our model RNA at low micromolar to nanomolar concentrations.4 We were interested in analogs of the oxazolidinone ring that could provide enhanced water solubility and eliminate both stereocenters. Several isosteric analogs to the oxazolidinone ring have been reported with varying degrees of success.5 These include isoxazoline (removes the carbonyl and introduces a C=N double bond),6 benzisoxazolinones (C=C double bond, replacement of sp3 carbon with an N),7 and pyrroles (completely flat aromatic ring system).7 Of these bioisosteric analogs of the oxazolidinone, only the isoxazoline and the benzisoxazolinone had activity similar to the parent compound.
Given the ready availability of 1,4-disubstituted triazoles and the fact that the 1,2,3-triazole ring has been reported to function as an amide bond surrogate,8,9 we wished to examine an appropriately substituted triazole as a potential isosteric replacement for the oxazolidinone ring. Such a bioisosteric replacement would have an advantage in avoiding the stereochemical considerations associated with ANB-22 and ANB-40 as well as provide RNA ligands with potentially improved water solubility. In the context of identifying structurally diverse potential analogs, we desired a library of 1,4-disubstituted 1,2,3-triazole compounds 1 (Figure 1) that incorporated the diversity elements used in the 4,5-disubstituted oxazolidinone compound library. Based on ANB-22 and ANB-40, R1 would be an aliphatic, aryl, or amide group while R3 and R4 would be alkyl groups. This would provide analogs in which key functional groups occupy the same space in both the oxazolidinone and 1,2,3-triazole analogs.
Figure 1.
General structure of the lead oxazolidinone and proposed 1,2,3-triazole compound library
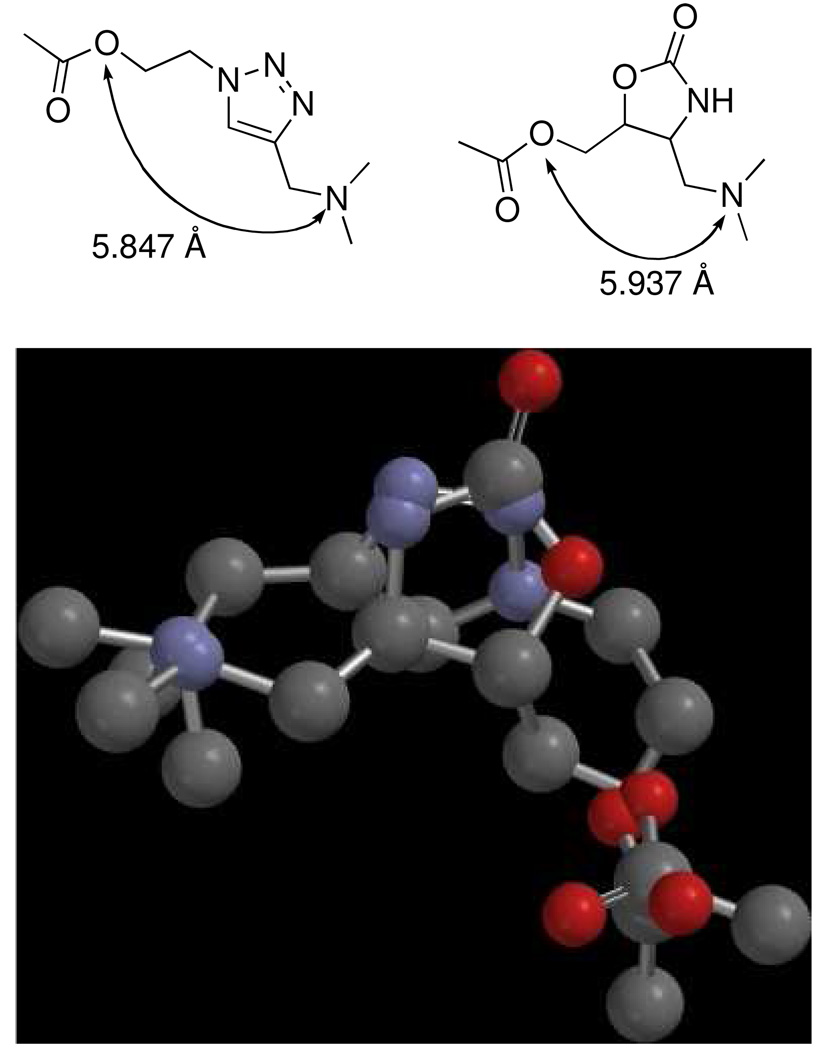
Modeling of a simple analog of triazole 1 (n = 1, R1, R2, R3 = Me) and a similarly substituted oxazolidinone showed good overlap of both the basic amine and the ester.10 The N – acyl oxygen distance in the triazole 1 is 5.847 Å while the N – acyl oxygen distance for the oxazolidinone is 5.937 Å. An overlay of the triazole with the oxazolidinone is shown in Figure 2. One can clearly see the excellent overlap of both basic nitrogens and the ester functionality.
Figure 2.
Overlay of triazole analog and oxazolidinone.
The 1,2,3-triazole scaffolds are generally synthesized via the Huisgen [3+2] cycloaddition reaction of an organic azide and a terminal alkyne.11,12 Though the classic Huisgen 1,3-dipolar cycloaddition reaction provided a regioisomeric mixture of 1,2,3-triazoles, recent advances utilizing Cu(I)-catalysis have added great utility to the [3+2] cycloaddition reaction in the synthesis of more complex, diverse and biologically relevant compounds.13–18 Thus, this Cu(I) variant of the Huisgen 1,3-dipolar cycloaddition reaction enables us to access the desired library of 1,4-disubstituted 1,2,3-triazole compounds in a reliable and efficient manner with only the 1,4-disubstituted regioisomer being produced.
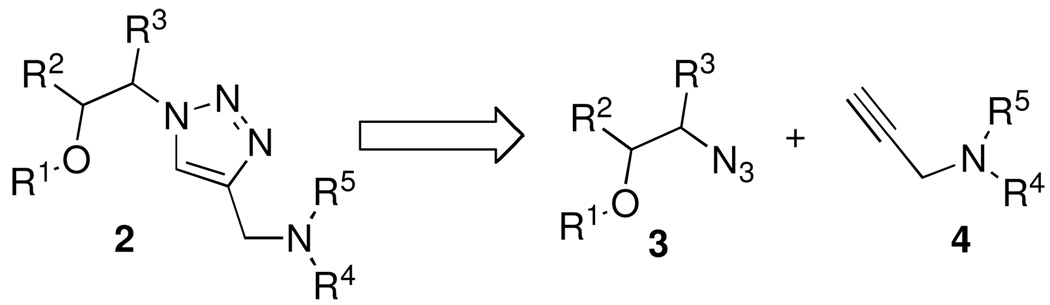
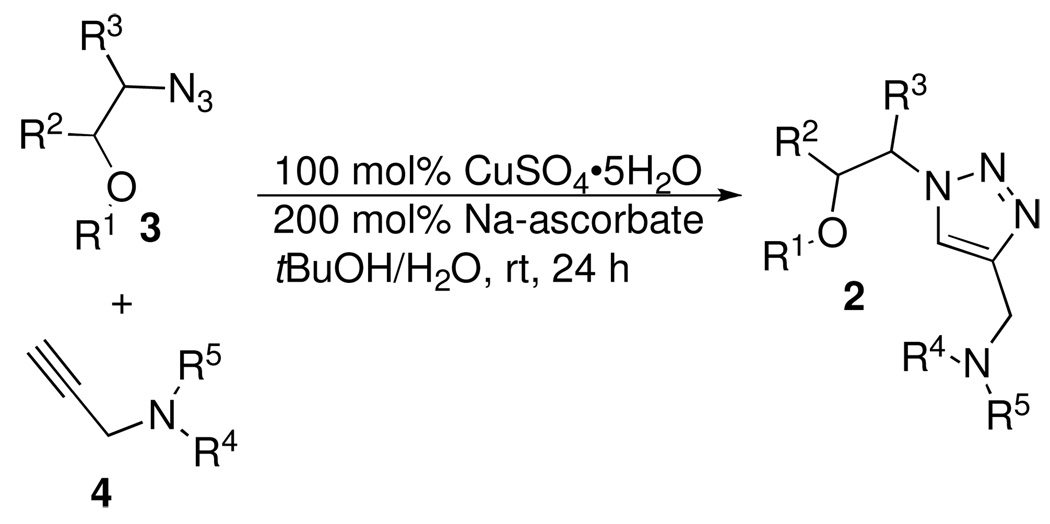
The retrosynthetic analysis and general structure of the 1,4-disubstituted 1,2,3-triazole library (2) and key intermediates are outlined in Figure 3. The triazole 2, will be obtained from an azide (3), alkyne (4) cycloaddition reaction. As we planned to prepare the azide component 3 via ring-opening reactions of an epoxide, we have an opportunity to introduce additional substitutions on the target molecule (e.g. R2 and R3). Such substitution could lead to improved affinity to the RNA target.
Figure 3.
Retrosynthesis and general structure of 1,2,3-triazole library and key intermediates
The azide components were prepared in two or three steps by following three related synthetic routes (Schemes 1, 2 and 3). In general the requisite azide was prepared via the opening of an epoxide with azide. Azidolysis methods have been well documented and azidohydrins in general have been prepared from the ring opening reactions of epoxide with a variety of azide nucleophiles including sodium azide.19
Scheme 1.
Scheme 2.
Scheme 3.
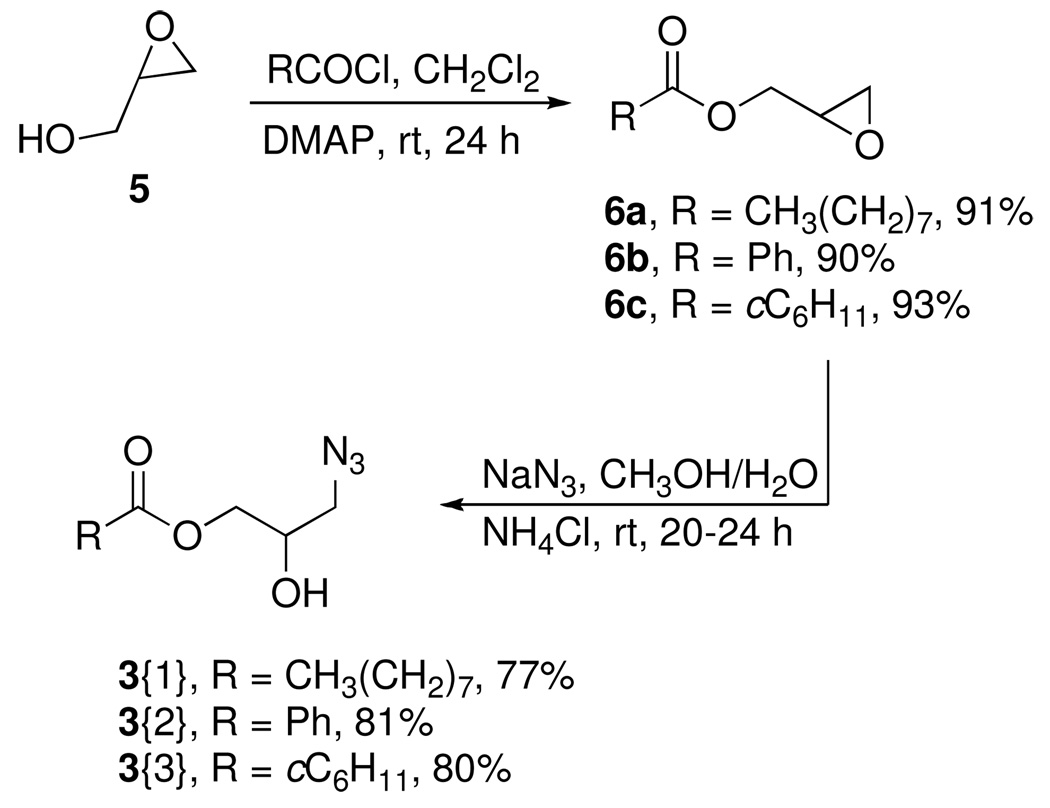
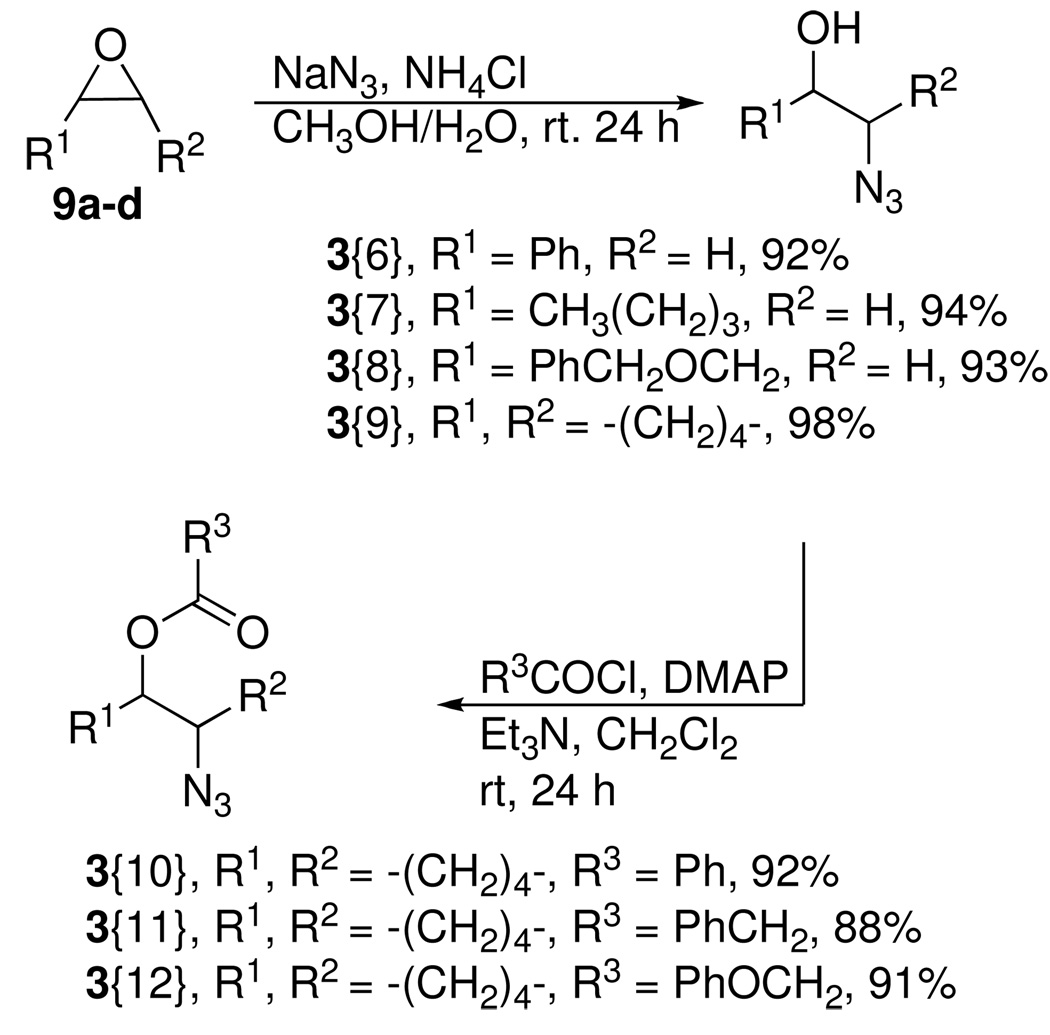
Glycidol 5 was acylated with three acid chlorides to provide the glycidyl esters 6 in good to excellent yields (Scheme 1).20 We were interested in examining methods for epoxide opening by azide that might be amenable for parallel synthesis of the requisite azides. Several epoxide-opening methods including a Ce(III)-mediated epoxide opening using NaN3,21 NaN3 on zeolite,22 NaN3/SiO2 neat,23 NaN3 and 4Å MS were evaluated.24 These conditions gave either complex reaction mixtures or provided very low yields of the expected azidohydrins. Ultimately, the classical protocol, which entailed the use of sodium azide in the presence of NH4Cl was employed to afford the first set of azide diversity elements 3{1–3} in good yield. This provided a series of azides components that were used to generate analogs of the lead ANB-22.
We also wanted to prepare a set of carbamoyl-substituted azides to provide analogs of the lead ANB-40. The synthesis of carbamoyl epoxide 8 from the reaction of glycidol and an isocyanate gave extremely low yields of the expected product. A plausible explanation for the low yield could be attributed to intramolecular ring opening of the epoxide by the neighboring carbamate.25
In view of this, an alternative synthetic route starting from allyl alcohol was developed. Allyl alcohol was treated with benzyl and butyl isocyanate respectively to give the corresponding allylcarbamates (Scheme 2). Epoxidation of the olefin using mCPBA afforded the epoxide 8, in excellent yield. The epoxide ring of 8 was then opened with sodium azide to provide a second set of carbamoyl azide diversity elements 3{4} and 3{5}.
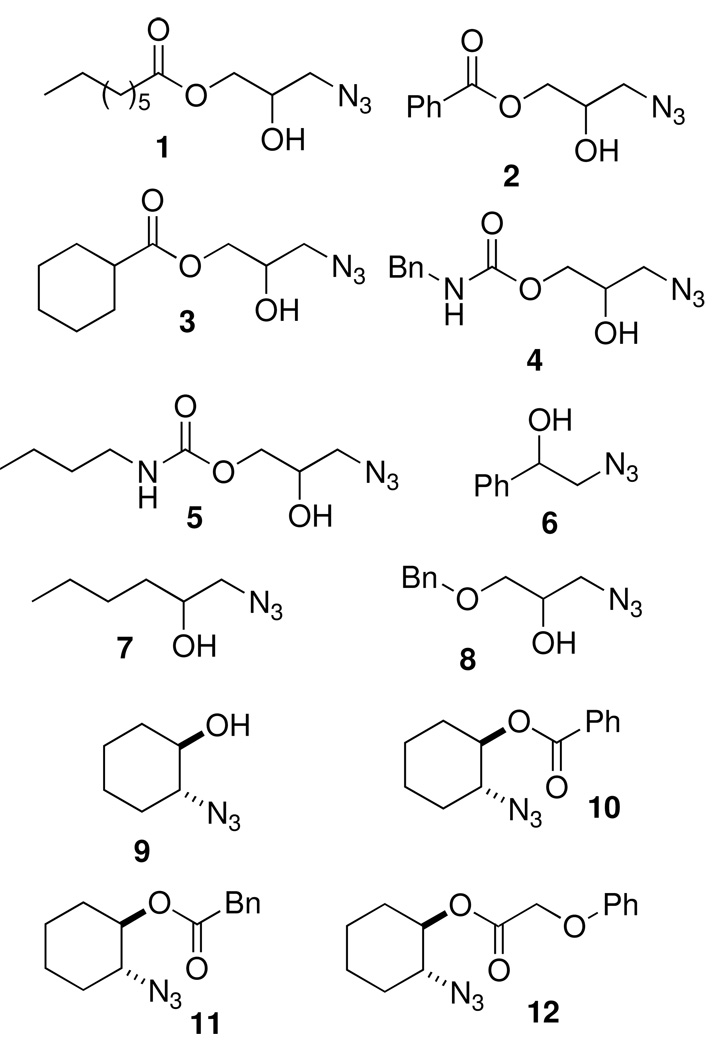
As outlined in Scheme 3, four commercially available epoxides 9a-d were also treated with sodium azide to provide the racemic azidoalcohols 3{6–9} in good to excellent yields. To again provide analogs related to lead ANB-22, one azidoalcohol, racemic trans-2-azidocyclohexanol (3{9}), was acylated with three different acid chlorides to afford the final set of trans-2-azidocyclohexyl ester diversity elements 3{10–12} in good yields. These syntheses provided structural diversification of three different sets of azide components for the 1,4-disubstituted 1,2,3-triazole synthesis (Figure 4).
Figure 4.
Azide diversity elements 3{1–12} employed in the library synthesis
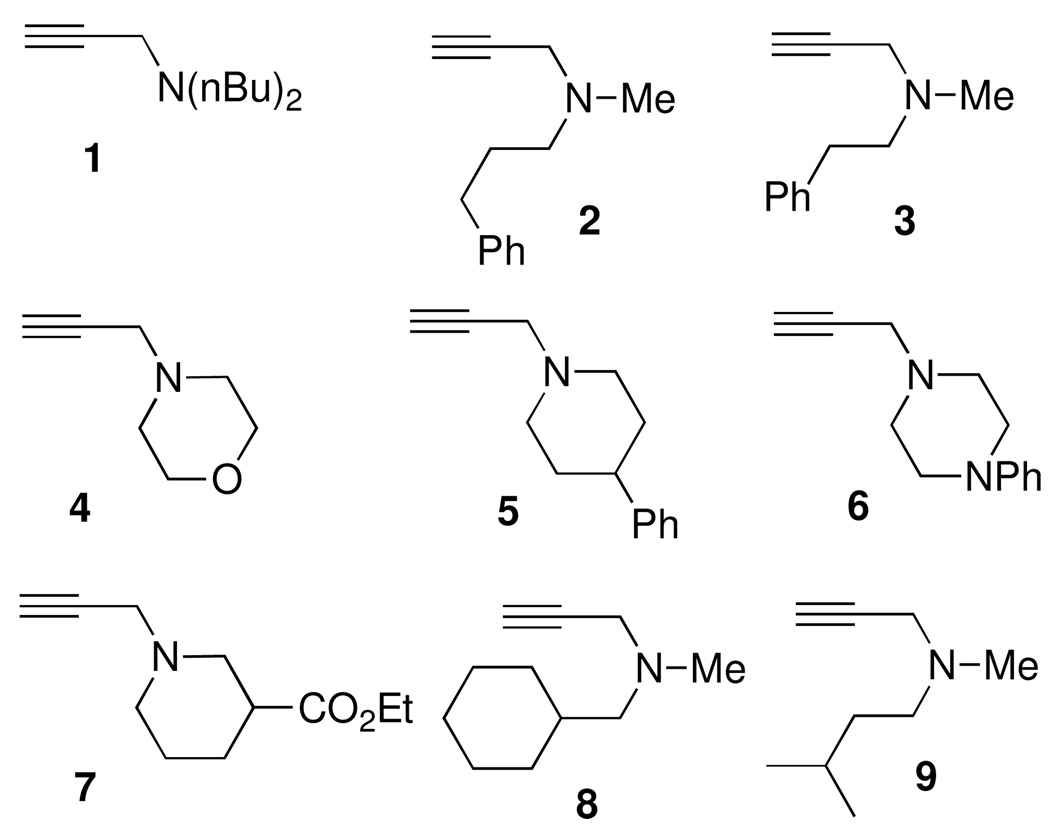
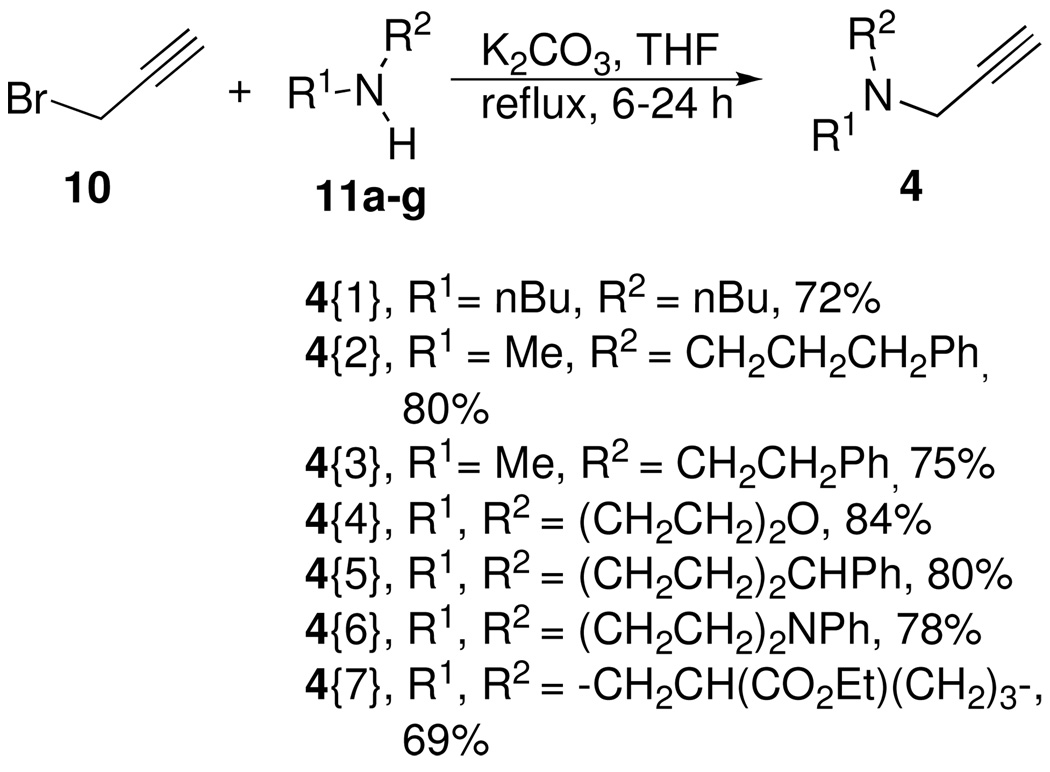
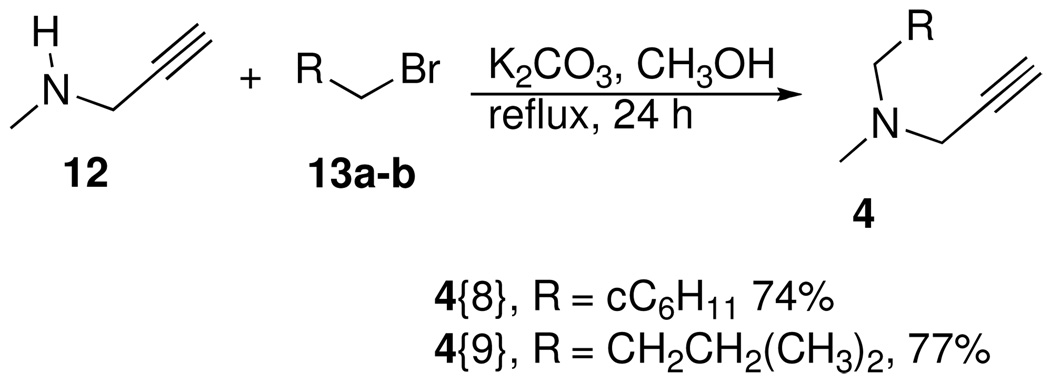
The alkyne diversity elements (Figure 5) were readily accessed in one step via two different N-alkylation reactions in either THF or methanol.26 As outlined in Scheme 4, propargyl bromide was alkylated with a series of secondary amines 11 to provide a set of propargylamine derived alkyne diversity elements, 4{1–7}, in good yields. The modification of Verron et. al. was used for the preparation of 4{4}.27 In a similar manner, N-methyl propargylamine 12 was also alkylated with 2 different alkyl halides, 13a and 13b, to provide the remainder of the alkyne diversity elements 4{8} and 4{9} which were needed for the synthesis of the triazole library (Scheme 5).
Figure 5.
Alkyne diversity elements (4{1–9}) employed in library synthesis
Scheme 4.
Scheme 5.
Having the twelve azide components (3{1–12}, Figure 4) and nine propargylamine derived alkynes (4{1–9}, Figure 5) in hand, the synthesis of the targeted 1,4-disubstituted 1,2,3-triazole analogs of the oxazolidinone RNA binding agents was initiated.
We planned to initially carry out a few cycloaddition reactions to ascertain if the general conditions for the previously reported Cu(I) catalyzed azide/alkyne variant of the Huisgen’s 1,3-dipolar cycloaddition reaction would be a suitable method for preparing our targeted library. A trial cycloaddition reaction was carried out using 3{6} and phenylacetylene following typical Sharpless conditions. These reaction conditions afforded the expected triazole in a yield of 98%. Based on this result a small scouting library of 16 members was prepared using the azides 3{2}, 3{3}, 3{8} and 3{9} and the alkynes 4{1}, 4{2}, 4{5} and 4{6}. In contrast to the trial cycloaddition reaction in which phenylacetylene was used as the alkyne component, these [3+2] cycloaddition reactions using catalytic amounts of CuSO4 afforded very poor yields (~ 20%) of the 1,4-disubstituted 1,2,3-triazole with substantial amount of starting materials. Increasing the temperature and increaing the length of reaction time did not significantly alter the yield. We attributed this decrease in yield to the prescence of a basic amine on the alkyne. The prescence of a basic amine elements in Cu-catalyzed azide-alkyne cycloadditions appears to be quite limited.28–32 While many of these reactions use the catalytic Cu protocols, these methods were unsuccessful in our hands. We had previously observed that 100 mol% of Cu(I) was required for complete consumption of the starting materials in the cycloaddition when basic amine groups were present as part of a library synthesis.33 Further experimental trials using different stoichiometric amounts of CuSO4•5H2O and sodium ascorbate indicated that a full molar equivalent of both CuSO4•5H2O and a 2-fold excess of sodium ascorbate relative to the alkyne to be the optimal loading for the [3+2] cycloaddition reaction. Thus, the library synthesis was carried out as outlined in Scheme 6 using 100 mol% of CuSO4 and 200 mol% of sodium ascorbate.
Scheme 6.
A typical workup procedure for these reactions entailed concentrating the reaction mixture, diluting the residue with CH2Cl2, washing consecutively with a mixture of 1:1 NH4OH/H2O, H2O, brine, drying over MgSO4, filtering and concentrating the filtrate. This process seemed challenging considering the library size. Three different workup procedures were examined in an attempt to discern a relatively simple and quick workup condition for the library preparation.
All three workup conditions required the reaction mixture to be concentrated to at least half its initial volume prior to workup. We first examined a simple filtration of the reaction mixture through poly(vinylpyridine). This method was efficient in cleaning up library members when catalytic amount of Cu(II) salts were used. As the amount of Cu(II) salts was increased to a full equivalent, purification using poly(vinylpyridine) as a copper scavenger became inefficient in removing copper residues from the mixture. This was apparent from the paramagnetic effect of copper in the proton NMR. Consequently, an additional aqueous workup was required to completely remove copper ions. This was time-consuming especially envisaging the size of the library. Adding poly(vinylpyridine) to the reaction mixture and stirring followed by filtration through a phase separator produced similar results as filtration through the poly(vinylpyridine). The final workup procedure involved adding a 1:1 mixture of NH4OH/H2O to the reaction, then adding CH2Cl2 and using a phase separator to cleanly obtain the organic layer. This provided a quick, efficient and simple workup procedure, which afforded the desired product in high yields and excellent purity. As a result, this third workup procedure was adopted for the library.
With the synthetic method and purification scheme optimized all 12 azides (Figure 4) and 9 alkynes (Figure 5) were subjected to the reaction and workup conditions described previously. As depicted in Table 1, yields in the range of 88–96% were obtained for the synthesis of the targeted triazoles. The masses of all 1,4-disubstituted 1,2,3-triazole library members were verified via LC/MS analysis. Purities of 80–96% were obtained for the compound library members based on the integration of the major peak in the HPLC trace.
Table 1.
Yield and purity of the 1,4-disubstituted 1,2,3-triazole library members
| Compd | Yield | Purity | Compd | Yield | Purity | Compd | Yield | Purity | Compd | Yield | Purity |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2{1,1} | 94 | 93 | 2{4,1} | 92 | 100 | 2{7,1} | 91 | 100 | 2{10,1} | 93 | 81 |
| 2{1,2} | 92 | 81 | 2{4,2} | 94 | 98 | 2{7,2} | 91 | 86 | 2{10,2} | 91 | 100 |
| 2{1,3} | 93 | 91 | 2{4,3} | 93 | 95 | 2{7,3} | 94 | 99 | 2{10,3} | 92 | 84 |
| 2{1,4} | 96 | 87 | 2{4,4} | 92 | 84 | 2{7,4} | 95 | 90 | 2{10,4} | 89 | 84 |
| 2{1,5} | 94 | 100 | 2{4,5} | 92 | 100 | 2{7,5} | 93 | 90 | 2{10,5} | 90 | 100 |
| 2{1,6} | 91 | 98 | 2{4,6} | 96 | 80 | 2{7,6} | 96 | 84 | 2{10,6} | 92 | 82 |
| 2{1,7} | 92 | 97 | 2{4,7} | 96 | 97 | 2{7,7} | 91 | 99 | 2{10,7} | 89 | 94 |
| 2{1,8} | 93 | 95 | 2{4,8} | 90 | 84 | 2{7,8} | 93 | 98 | 2{10,8} | 95 | 83 |
| 2{1,9} | 89 | 84 | 2{4,9} | 92 | 81 | 2{7,9} | 89 | 86 | 2{10,9} | 89 | 100 |
| 2{2,1} | 92 | 100 | 2{5,1} | 94 | 96 | 2{8,1} | 89 | 92 | 2{11,1} | 93 | 86 |
| 2{2,2} | 91 | 83 | 2{5,2} | 91 | 99 | 2{8,2} | 95 | 81 | 2{11,2} | 91 | 96 |
| 2{2,3} | 93 | 93 | 2{5,3} | 95 | 94 | 2{8,3} | 91 | 84 | 2{11,3} | 92 | 86 |
| 2{2,4} | 90 | 81 | 2{5,4} | 92 | 100 | 2{8,4} | 92 | 94 | 2{11,4} | 91 | 86 |
| 2{2,5} | 94 | 84 | 2{5,5} | 94 | 100 | 2{8,5} | 95 | 98 | 2{11,5} | 94 | 98 |
| 2{2,6} | 92 | 81 | 2{5,6} | 93 | 80 | 2{8,6} | 96 | 81 | 2{11,6} | 92 | 81 |
| 2{2,7} | 93 | 84 | 2{5,7} | 92 | 92 | 2{8,7} | 91 | 98 | 2{11,7} | 91 | 100 |
| 2{2,8} | 95 | 82 | 2{5,8} | 90 | 85 | 2{8,8} | 93 | 98 | 2{11,8} | 93 | 95 |
| 2{2,9} | 92 | 84 | 2{5,9} | 92 | 84 | 2{8,9} | 93 | 90 | 2{11,9} | 94 | 95 |
| 2{3,1} | 91 | 83 | 2{6,1} | 92 | 94 | 2{9,1} | 92 | 100 | 2{12,1} | 91 | 81 |
| 2{3,2} | 94 | 100 | 2{6,2} | 96 | 84 | 2{9,2} | 92 | 89 | 2{12,2} | 95 | 97 |
| 2{3,3} | 94 | 100 | 2{6,3} | 91 | 84 | 2{9,3} | 92 | 100 | 2{12,3} | 92 | 93 |
| 2{3,4} | 90 | 91 | 2{6,4} | 93 | 100 | 2{9,4} | 92 | 82 | 2{12,4} | 92 | 81 |
| 2{3,5} | 97 | 90 | 2{6,5} | 93 | 89 | 2{9,5} | 94 | 84 | 2{12,5} | 91 | 98 |
| 2{3,6} | 92 | 82 | 2{6,6} | 95 | 80 | 2{9,6} | 95 | 90 | 2{12,6} | 93 | 84 |
| 2{3,7} | 96 | 96 | 2{6,7} | 93 | 98 | 2{9,7} | 92 | 98 | 2{12,7} | 92 | 96 |
| 2{3,8} | 94 | 84 | 2{6,8} | 93 | 94 | 2{9,8} | 92 | 81 | 2{12,8} | 93 | 84 |
| 2{3,9} | 92 | 85 | 2{6,9} | 89 | 95 | 2{9,9} | 91 | 100 | 2{12,9} | 88 | 82 |
Purities > 80% were obtained for all library members based on HPLC analysis.
The 1,4-disubstituted 1,2,3-triazole compounds were tested for their ability to bind to the wildtype antiterminator model RNA AM1A34 and the reduced function variant C11U by using a fluorescence resonance energy transfer (FRET) screening assay.35
The reduced function variant C11U has been used in other ligand-antiterminator RNA binding assays as a specificity control.1,4 The model RNAs were labeled with a fluorescein (donor) on the 3´ end and a rhodamine (acceptor) on U18 in the UUCG loop of AM1A as previously described.1,4 Previous studies correlated changes in FRET with ligand binding affinities.
Prior to examining the library in the FRET-based RNA binding assay, all 108 compounds prepared were examined for both solubility in the assay buffer and interferring fluorescence. This led to the exclusion of 27 compounds from the RNA binding assay.
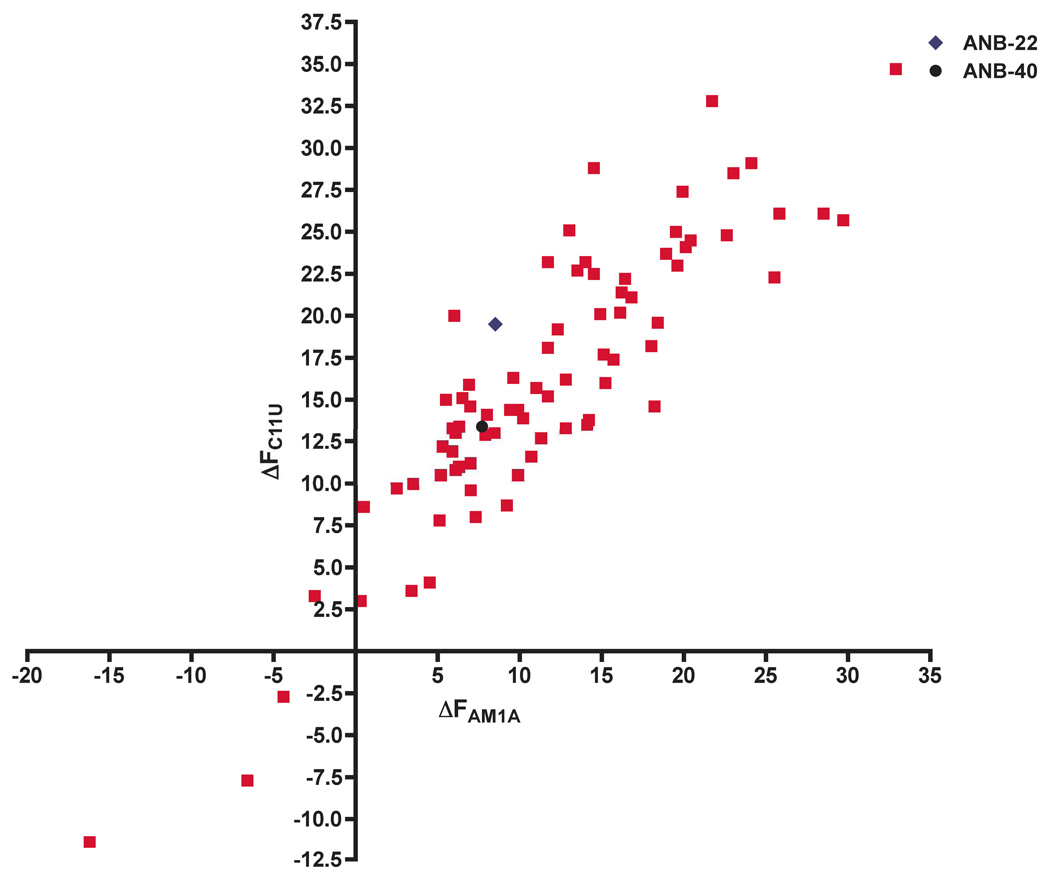
Figure 6 shows all compounds that were evaluated in the FRET assay (including ANB-22 and ANB-40) where the X and Y coordinates represent ΔFAM1A and ΔFC11U respectively.36 The lead compounds ANB-22 and ANB-40 increased the fluorescence intensity of FRET-labeled RNA for both AM1A and C11U significantly. The 11% difference between the signal change of AM1A (8.53%) and C11U (19.53%) nicely correlates with the previous finding that ANB-22 has high binding specificity for these two antiterminator models.4 Gratifyingly a number of compounds are clustered near ANB-22 and ANB-40. Clearly the triazole analogs prepared have retained the RNA affinity of the lead oxazolidinones. As might be expected, compounds with similar side chain substitution as oxazolidinones ANB-22 and ANB-40 (e.g. 2{4,5}, 2{4,6}, 2{12,5}, and 2{12,6}) showed similar fluorescence values as the parent oxazolidinones. A total of forty-nine 1,4-disubstituted 1,2,3-triazole compounds enhanced the AM1A fluorescence intensity more than 8.53%. Furthermore, 2{4,5}, 2{7,4}, 2{8,4} and 2{5,1} showed larger differences of ΔF between AM1A and C11U compared to ANB-22, which indicated higher specificities. Interestingly, 2{10,9} 2{3,8} and 2{1,2} caused a decrease of the fluorescence intensity. One explanation for the decreased fluorescence intensity of 2{10,9}, 2{3,8} and 2{1,2} could be that these compounds bind to the antiterminator in a unique manner that induced an RNA conformational change different from the others.
Figure 6.
FRET screening of 1,4-disubstituted 1,2,3-triazole binding affinity for model RNAs AM1A and C11U. The relative fluorescence intensity change ΔF was calculated by ΔF=[(F−F0)/F0]*100, where F is the fluorescence intensity with ligand and F0 is without ligand at 585 nm upon excitation at 467 nm. All of the compounds were tested at a final concentration of 10 µM.
We have developed useful conditions for the synthesis of amine-substituted 1,2,3-triazoles by using stoichiometric quantities of Cu(I). We have also shown that the 1,2,3-triazole ring can be an effective bioisosteric replacement for an oxazolidinone ring. The 1,4-disubstituted 1,2,3-triazole prepared represents a considerable simplification in both topography and synthesis relative to the oxazolidinone. Further work to determine the structure activity relationships of this class of RNA binding agent and understand the mode of interaction with the RNA target is ongoing and will be reported in due course.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (GM073188) for support of this work. We also thank Ohio University for support of the BioMolecular Innovation and Technology project.
Footnotes
Supporting Information Available
General synthesis of all compounds, HPLC and LCMS data, spectroscopic data and 1H-NMR and 13C NMR characterizations are described. Listing of the fluorescence data for Figure 6. This information is available free via the Internet at http://pubs.acs.org.
References and notes
- 1.Means J, Katz SJ, Anupam R, Nayek A, Hines JV, Bergmeier SC. Bioorg. Med. Chem. Lett. 2006;16:3600–3604. doi: 10.1016/j.bmcl.2006.03.068. [DOI] [PubMed] [Google Scholar]
- 2.Grundy FJ, Rollins SM, Henkin TM. J. Bacteriol. 1994;176:4518–4526. doi: 10.1128/jb.176.15.4518-4526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutierrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. Microbiol. Mol. Biol. Rev. 2009:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anupam R, Nayek A, Green NJ, Grundy FJ, Henkin TM, Means JA, Bergmeier SC, Hines JV. Bioorg. Med. Chem. Lett. 2008;18:3541–3544. doi: 10.1016/j.bmcl.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutchinson DK. Cur. Topics Med. Chem. 2003;3:1021–1042. doi: 10.2174/1568026033452195. [DOI] [PubMed] [Google Scholar]
- 6.Snyder LB, Meng Z, R M, D'Andrea SV, Marinier A, Quesnelle CA, Gill P, DenBleyker KL, Fung-Tomc JC, Frosco M, Martel A, Barrett JF, Bronson J. J. Bioorg. Med. Chem. Lett. 2004;14:4735–4739. doi: 10.1016/j.bmcl.2004.06.076. [DOI] [PubMed] [Google Scholar]
- 7.Barbachyn MR, Cleek GJ, Dolak LA, Garmon SA, Morris J, Seest EP, Thomas RC, Toops DS, Watt W, Wishka DG, Ford CW, Zurenko GE, Hamel JC, Schaadt RD, Stapert D, Yagi BH, Adams WJ, Friis JM, Slatter JG, Sams JP, Oien NL, Zaya MJ, Wienkers LC, Wynalda MA. J. Med. Chem. 2003;46:284–302. doi: 10.1021/jm020248u. [DOI] [PubMed] [Google Scholar]
- 8.Tron GC, Pirali T, Billington RA, Canonico PL, Sorba G, Genazzani AA. Med. Res. Rev. 2008;28:278–308. doi: 10.1002/med.20107. [DOI] [PubMed] [Google Scholar]
- 9.Brik A, Alexandratos J, Lin Y-C, Elder JH, Olson AJ, Wlodawer A, Goodsell DS, Wong C-H. ChemBioChem. 2005;6:1167–1169. doi: 10.1002/cbic.200500101. [DOI] [PubMed] [Google Scholar]
- 10. Molecular modeling was performed with Spartan '08 for Macintosh. Irvine, CA: Wavefunction Inc.; The lowest energy conformer for each compound was obtained using molecular mechanics MMFF base set.
- 11.Huisgen R. In: 1,3-Dipolar Cycloaddition Chemistry. Padwa A, editor. New York: Wiley; 1984. pp. 1–176. [Google Scholar]
- 12.Huisgen R, Knorr R, Moebius L, Szeimies G. Chem. Ber. 1965;98:4014–4021. [Google Scholar]
- 13.Tornøe CW, Christensen C, Meldal M. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 14.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 15.Moses JE, Moorhouse AD. Chem. Soc. Rev. 2007;36:1249–1262. doi: 10.1039/b613014n. [DOI] [PubMed] [Google Scholar]
- 16.Bock VD, Hiemstra H, van Maarseveen JH. Eur. J. Org. Chem. 2006:51–68. [Google Scholar]
- 17.Kolb HC, Sharpless KB. Drug Discovery Today. 2003;8:1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 18.Kolb HC, Finn MG, Sharpless KB. Angew. Chem. Int. Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 19.Scriven E, Turnbull K. Chem. Rev. 1988;88:297–398. [Google Scholar]
- 20.Stamatov SD, Stawinski J. Tetrahedron. 2005;61:3659–3669. [Google Scholar]
- 21.Sabitha G, Babu RS, Rajkumar M, Yadav JS. Org. Lett. 2002;4:343–345. doi: 10.1021/ol016979q. [DOI] [PubMed] [Google Scholar]
- 22.Onaka M, Sugita K, Izumi Y. J. Org. Chem. 1989;54:1116–1123. [Google Scholar]
- 23.Kiasat AR, Kazemi F. Phosphorus, Sulfur and Silicon. 2003;178:2387–2392. [Google Scholar]
- 24.Boruwa J, Borah JC, Kalita B, Barua NC. Tetrahedron Lett. 2004;45:7355–7358. [Google Scholar]
- 25.Langlois N, Moro A. Eur. J. Org. Chem. 1999:3483–3488. [Google Scholar]
- 26.Lambert SJ, Kabalka GW, Knapp FF, Jr, Srivastava PC. J. Org. Chem. 1991;56:3707–3711. [Google Scholar]
- 27.Verron J, Malherbe P, Prinssen E, Thomas AW, Nock N, Masciadri R. Tetrahedron Lett. 2007;48:377–380. [Google Scholar]
- 28.Chittaboina S, Xie F, Wang Q. Tetrahedron Lett. 2005;46:2331–2336. [Google Scholar]
- 29.Yan Z-Y, Zhao Y-B, Fan M-J, Liu W-M, Liang Y-M. Tetrahedron. 2005;61:9331–9337. [Google Scholar]
- 30.Kaval N, Ermolat'ev D, Appukkuttan R, Dehaen W, Kappe CO, Van der Eycken E. J. Comb. Chem. 2005;7:490–502. doi: 10.1021/cc0498377. [DOI] [PubMed] [Google Scholar]
- 31.Collin M-P, Hobbie SN, Bottger EC, Vasella A. Helv. Chim. Acta. 2008;91:1838–1848. [Google Scholar]
- 32.Smith NM, Greaves MJ, Jewell R, Perry MWD, Stocks MJ, Stonehouse JP. Synlett. 2009:1391–1394. [Google Scholar]
- 33.Luesse SB, Wells G, Nayek A, Smith AE, Kusche BR, Bergmeier SC, McMills MC, Priestley ND, Wright DL. Bioorg. Med. Chem. Lett. 2008;18:3946–3949. doi: 10.1016/j.bmcl.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 34.Gerdeman MS, Henkin TM, Hines JV. Nucleic Acids Res. 2002;30:1065–1072. doi: 10.1093/nar/30.4.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Means JA, Hines JV. Bioorg. Med. Chem. Lett. 2005;15:2169–2172. doi: 10.1016/j.bmcl.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 36.A tabular listing of the fluoresence data used to generate Figure 6 can be found in the supplementary information.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.