Abstract
Thermococcales (phylum Euryarchaeota) are model organisms for physiological and molecular studies of hyperthermophiles. Here we describe three new plasmids from Thermococcales that could provide new tools and model systems for genetic and molecular studies in Archaea. The plasmids pTN2 from Thermococcus nautilus sp. 30-1 and pP12-1 from Pyrococcus sp. 12-1 belong to the same family. They have similar size (∼12 kb) and share six genes, including homologues of genes encoded by the virus PAV1 from Pyrococcus abyssi. The plasmid pT26-2 from Thermococcus sp. 26-2 (21.5 kb), that corresponds to another plasmid family, encodes many proteins having homologues in virus-like elements integrated in several genomes of Thermococcales and Methanococcales. Our analyses confirm that viruses and plasmids are evolutionary related and co-evolve with their hosts. Whereas all plasmids previously isolated from Thermococcales replicate by the rolling circle mechanism, the three plasmids described here probably replicate by the theta mechanism. The plasmids pTN2 and pP12-1 encode a putative helicase of the SFI superfamily and a new family of DNA polymerase, whose activity was demonstrated in vitro, whereas pT26-2 encodes a putative new type of helicase. This strengthens the idea that plasmids and viruses are a reservoir of novel protein families involved in DNA replication.
INTRODUCTION
Plasmids are especially abundant in the domains Archaea and Bacteria [for a recent monography, see ref. (1)]. Their size ranges from a very small plasmid with a single gene, such as the plasmid pRQ7 from the hyperthermophilic bacterium Thermotoga (2), to large megaplasmids whose size rival those of bacterial or archaeal chromosomes (3,4). Plasmids can use different replication strategies (mostly rolling-circle mode for the smallest ones and theta mode for the others) and a variety of replication origins (5). In Archaea, plasmids from Sulfolobales (thermoacidophilic members of the phylum Crenarchaea) have been especially well characterized (6,7). Two plasmid families have been identified. The first one is the family of pRN-type plasmids (with sizes ranging mainly from 5 up to 14 kb) (6,8,9). The second one corresponds to rather large conjugative plasmids of 24 to 36 kb, pNOB8 and its relatives (10,11). All pRN-type plasmids encode two DNA binding proteins (CopG and PlrA) that could be involved in the regulation of copy number and gene expression (12,13) and a large protein, dubbed RepA, involved in plasmid replication. The RepA protein of pRN1 is a multifunctional enzyme whose N-terminal domain harbours DNA primase/polymerase activities and the C-terminal domain corresponds to a DNA helicase of the SFIII superfamily (14,15). The primase/polymerase of pRN1 is the prototype of a new DNA polymerase family whose members are encoded by various archaeal and bacterial plasmids and some bacterial viruses (15). The structural characterization of the N-terminal domain of the pRN1 DNA polymerase revealed that its catalytic domain is evolutionary related to those of archaeal and eukaryotic DNA primases (16). Two plasmids of the pRN family lack homologue of pRN1 RepA: one of them, pXZ1, encodes instead a large protein of similar size but without sequence similarity to pRN1 RepA (17), whereas another one, pTAU4, encodes a MCM helicase instead of a Rep protein (8). An interesting member of the pRN family is pSSVx, a virus–plasmid hybrid that coexists intracellularly with the fusellovirus SSV1 and can be packaged into viral particles (18,19). Besides typical pRN proteins, pSSVx encodes two SSV1 proteins probably involved in plasmid packaging. Several pRN-type plasmids have been found integrated into tRNA genes in the chromosomes of different Sulfolobales (18). The site-specific integrase that promotes their insertion is homologous to the integrases of the SSV1 and other fuselloviruses which can also integrate in the chromosomes of Sulfolobus species (20). The insertion site recognized by SSV-type integrases is located in the coding part of their own gene and the integration event provokes its disruption. The plasmid pXZ1 also encodes an SSV-type integrase and coexists in Sulfolobus cell with a fusellovirus (SSV4) but, unlike pSSVx, pXZ1 cannot be packaged into viral particle (17). The SSV1 virus, as well as pSSVx and pRN1 has been used to develop genetic tools for Sulfolobus species, including shuttle vectors to express foreign proteins in Sulfolobales (21,22).
The Sulfolobus conjugative plasmids of the pNOB8-type encodes ∼40–50 proteins (10,11). Ten proteins are conserved in all these plasmids and 80% of the remaining proteins are common to at least two plasmids (11). Most of these proteins are of unknown function. Their conserved genes are grouped in two regions: one of them includes two genes encoding proteins with low similarity to TraG and TraE proteins involved in bacterial plasmids conjugation, as well as several proteins with membrane domains that are supposed to be involved in DNA transfer. The second region contains the genes encoding homologues of the pRN-type plasmids, CopG and PrlA proteins, and an integrase gene. This integrase represents a novel family of integrases, the pNOB8-type, that are widely distributed in archaeal genomes (10). Unlike the SSV-type, pNOB8-type integrases never bear their insertion site inside the sequence of their genes and consequently these genes are never disrupted after integration. Several pNOB8-type plasmids encode transposases, and one of them (pKEF9) also encodes a pRN-like RepA protein. A new member of the pNOB8 conjugative plasmid family, pAH1, has been recently discovered in an Acidianus species that belongs to the order Sulfolobales (20). Interestingly, the replication of pAH1 is inhibited by infection by the lipothrixvirus AFV1, revealing a new type of plasmid/virus functional interaction (20).
All plasmids presently isolated from Sulfolobales are larger than 5 kb. Although their mode of replication has not yet been determined experimentally, they probably use the theta replication mode, since none of them encode the Rep protein typical for rolling-circle replication. In contrast, all known plasmids from Thermococcales (hyperthermophilic and anaerobic members of the phylum Euryarhaea) are small (<4 kb) and replicate via the rolling circle (RC) mode. This has been experimentally demonstrated for the plasmid pGT5 from Pyrococcus abyssi (23–25) and it is most likely the replication mode for the related plasmid pTN1, isolated from Thermococcus nautilus (26). Both plasmids encode large homologous proteins related to Rep proteins that initiate RC replication. These two RC initiator proteins, Rep75 and Rep74, respectively, form a new family of RC Rep proteins and are distantly related to transposases encoded by bacterial IS (IS91, IS1294 and IS801 families) that replicate themselves via the RC mode (26). A third small plasmid, called pRT1, from Pyrococcus sp. JT1, has been sequenced and tentatively described as a RC plasmid (27). However, its putative Rep protein shows no clear sequence similarity with known RC Rep proteins (26). The pGT5 and pTN1 plasmids have been used to construct shuttle vectors for P. abyssi and T. kodakaraensis, respectively (28,29). The pTN1-based shuttle vector for T. kodakaraensis has been engineered into an expression vector for tagged proteins that can be used to isolate protein complexes formed in vivo (28). The recent development of efficient transformation methods and gene knock-out strategies for T. kodakaraensis (28,30) clearly emphasize how useful the vectors based on Thermococcus plasmids could be.
During our characterization of the plasmid pTN1, we noticed that the strain T. nautilus contains additionally a larger plasmid of ∼13 kb, that we have called pTN2. Screening for further plasmids in our collection of Thermococcales strains (31), we then isolated a plasmid of similar size, called pP12-1, from a Pyrococcus strain (sp. 12-1) and a larger plasmid (∼20 kb) in a Thermococcus isolate (sp. 26-2) that we called pT26-2. We present here the analysis of the sequences of these three plasmids, focusing on the annotation of their proteins and on their evolutionary relationships with archaeal viruses and cellular hosts. Most of the predicted proteins of these three new plasmids are either ORFans or have only closely related homologues in viruses or integrated virus-like elements from Thermococcales and/or Methanococcales. They do not encode typical Rep proteins for RC replication, but new types of DNA replication proteins with distant homologues encoded by Sulfolobus pRN-type plasmids, indicating that they probably replicate by the theta mode. In particular, the Rep protein of pTN2 and pP12-1 appears to be related to the Rep protein of the Sulfolobus plasmids pXZ1 and pTIK4. Interestingly, the N-terminal domain of these Rep proteins corresponds to a new family of DNA polymerase (demonstrated here experimentally for the pTN2 Rep protein), whereas the pT26-2 plasmid encodes probably a new type of helicases. These discoveries are in good agreement with the idea that plasmids and viruses are a reservoir of novel protein families involved in DNA replication.
MATERIALS AND METHODS
Strains cultivation
Thermococcus nautilus sp. 30-1, Pyrococcus sp. 12-1 and Thermococcus sp. 26-2, were isolated from single colonies by plating on Gelrite enrichment cultures obtained from fragments of chimneys collected at deep sea hydrothermal vents (−2630 m) located in the East Pacific ocean (31). Cells were grown in Zillig’s broth (ZB) made anaerobic by addition of Na2S (0.1 mg/l), with sulphur flowers S° (Fisher Scientific) as previously described (31). The cultures were incubated in Penny’s flasks at 85°C for Thermococcus sp. 26-2 and T. nautilus, or 95°C for Pyrococcus sp. 12-1.
Plasmid isolation
Plasmids were obtained by alkaline extraction as described previously (26) from 50 ml culture of T. nautilus 30-1, Pyrococcus 12-1 or Thermococcus 26-2 cells that had been grown until stationary phase. Plasmids were sequenced by Fidelity Systems, USA. The complete sequences of pTN2, pP12-1 and pT26-2 have, respectively, been submitted to the GenBank database under accession numbers GU056177, GU056178 and GU056179.
Sequence analysis
ORFs were identified with a minimum of 50 amino acids length and with one of the initiation codons (ATG, GTG, CTG, TTG). The exact position of each initation codon was then checked individually (and manually modified if required) depending on the position of putative Shine–Dalgarno sequence located upstream (Tables 1–3). BLAST and PSI-BLAST searches were performed in the NCBI non-redundant (nr) databank using the following web sites: http://www.ncbi.nlm.nih.gov/BLAST/ and http://www-archbac.u-psud.fr/genomics/GenomicsToolbox.html/. Hydrophobic regions were detected using TMpred and TMHMM prediction programs available at the web site http://www.expasy.org/tools/. Searches for specific putative domains or sites were performed using Interproscan located at the website http://www.ebi.ac.uk/Tools/InterProScan/.
Table 1.
Annotation of pTN2 plasmid from the strain T. nautilus 30/1
| ORF | Putative protein | Size (kDa) | ORF position (start–stop) | RBS and start codon | Homologue in pP12-1 plasmid | Percentage identity, length of BLAST alignment (aa) | Putative motives and function |
|---|---|---|---|---|---|---|---|
| 1 | tn2-1p | 66.1 | 1–1710 | aggagggggtggtcggGTG | p12-1p | 69%, 569 | UvrD/Rep/PcrAsuperfamily I type helicase |
| 2 | tn2-2p | 104.9 | 1785–4622 | ggggggatttgtATG | – | – | Putative coiled-coil domains |
| 3 | tn2-3p | 20.3 | 4841–5365 | tgggggatgacgATG | p12-14p | 46%, 173 | |
| 4 | tn2-4p | 15.2 | 5590–5979 | gggtggtgtcgtaattATG | – | – | Putative helix–turn–helix domain |
| 5 | tn2-5p | 11.2 | 5983–6279 | aggggaggtgtaaccgATG | – | – | |
| 6 | tn2-6p | 26.4 | 6285–6959 | agaggtgtaaaacaaATG | – | – | |
| 7 | tn2-7p | 19.5 | 6959–7477 | aggaggggttatgATG | p12-9p | 41%, 41 | |
| 8 | tn2-8p | 43 | 7542–8648 | tggaggtgccgagcGTG | p12-10p | 46%, 367 | ATP binding site, that is related to ABC transporters |
| 9 | tn2-9p | 16 | 9184–9597 | agggggtactcataATG | – | – | |
| 10 | tn2-10p | 6.7 | 9551–9730 | cggaggaggataATG | – | – | |
| 11 | tn2-11p | 18.4 | 9763–10 251 | tggaggtgatgtaaATG | – | – | |
| 12 | tn2-12p | 106.8 | 10 248–4 | agggggtgaaagtATG | p121-17p | 55%, 922 | DNA polymerase/primase activities |
Table 2.
Annotation of pP12-1 plasmid from the strain Pyrococcus sp. 12/1
| ORF | Putative protein | Size (kDa) | ORF position (start–stop) | RBS and start codon | Homologue in pTN2 plasmid | Percentage identity, length of BLAST alignment (aa) | Putative motives and function |
|---|---|---|---|---|---|---|---|
| 1 | p12-1p | 66.7 | 1–1710 | aggagggggaggcctgATG | tn2-1p | 69%, 569 | UvrD/Rep/PcrA superfamily I type helicase |
| 2 | p12-2p | 20.3 | 1721–2257 | gaggtgggagaaATG | – | – | Zinc finger motif |
| 3 | p12-3p | 6.9 | 2260–2439 | gaggagggttgattATG | – | – | |
| 4 | p12-4p | 17.1 | 2423–2782 | gggaggggatatggGTG | – | – | |
| 5 | p12-5p | 22.9 | 2736–3305 | tggtggttatgaaggATG | – | – | |
| 6 | p12-6p | 10.0 | 3302–3565 | – | – | – | Putative transmembrane domains |
| 7 | p12-7p | 19.9 | 3685–4194 | agagggcaaagttagagtgagATG | – | – | |
| 8 | p12-8p | 13.0 | 4221–4553 | aagaggtgagaaggaTTG | – | – | |
| 9 | p12-9p | 18.8 | 4531–5019 | ggaggggaggagtATG | tn2-7p | 41%, 41 | |
| 10 | p12-10p | 43.0 | 5211–6323 | gaggggtgtagaATG | tn2-8p | 46%, 367 | ATP binding site, that is related to ABC transporters |
| 11 | p12-11p | 23.4 | 6350–6937 | aggaggcaataaaATG | – | – | Putative motives of 3′–5′ exonucleases |
| 12 | p12-12p | 7.1 | 6981–7175 (c) | aggaggtggtgcaaATG | – | – | Putative transmembrane domains |
| 13 | p12-13p | 7.1 | 7227–7400 | gaggtgccgccgtATG | – | – | |
| 14 | p12-14p | 19.9 | 7479–7988 (c) | gtgtggtggtgatATG | tn2-3p | 46%, 173 | |
| 15 | p12-15p | 20.0 | 8564–9106 | gtggtgcaccccATG | – | – | Putative transcription regulator related to the CopG family |
| 16 | p12-16p | 15.6 | 9103–9513 | cgggggtgtccccATG | – | – | |
| 17 | p12-17p | 103.1 | 9510–1 | agggggtgagagcATG | tn2-12p | 55%, 922 | DNA polymerase/primase activities |
Table 3.
Annotation of pT26-2 plasmid from the strain Thermococcus sp. 26-2
| ORF | Putative protein | Size (kDa) | ORF position (start–stop) | RBS and start codon | Closest homologue in integrated elements | Putative motives and function |
|---|---|---|---|---|---|---|
| 1 | t26-1p | 42.8 | 1–1059 | tggtgggccaaaGTG | TKV2 (68%, 282) | Integrase related to SSV1 integrase family |
| 2 | t26-2p | 6.8 | 1078–1257 | aggaggtgagcggaggATG | TKV2 et 3 (74%, 58) | |
| 3 | t26-3p | 6.7 | 1460–1636 | agggcgggcacgcccgcggcgATG | TGV1 (46%, 63) | |
| 4 | t26-4p | 12.6 | 1633–1992 | aggtggtggcagtATG | TKV2 (63%, 119) | Putative transmembrane domain |
| 5 | t26-5p | 68.8 | 2098–3984 | aggaggtggtctaaGTG | TKV2 et 3 (79%, 406) | Putative hydrophobic domains at extremities – Hit with carboxypeptidase regulatory domain in Interproscan |
| 6 | t26-6p | 81.5 | 3989–6208 | aggaggtgataacATG | TKV2 et 3 (41%, 622) | Putative transmembrane domains |
| 7 | t26-7p | 18.0 | 6241–6738 | agggggtgatgaattGTG | TKV2 et 3 (43%, 172) | Putative transmembrane domains |
| 8 | t26-8p | 12.1 | 6750–7094 | ctgaggtgaaaaacaATG | PHV1 (75%, 114) | Putative membrane protein |
| 9 | t26-9p | 8.7 | 7084–7335 | aggagggagggactATG | PHV1 (65%, 78) | Putative transmembrane domains |
| 10 | t26-10p | 29.2 | 7339–8145 | agggggtgtttgaagaATG | TGV1 (75%, 264) | Putative transmembrane domain |
| 11 | t26-11p | 18.5 | 8160–8642 | gaggtggactgagATG | TGV1 (76%, 160) | Putative leucine zipper motif–Putative transmembrane domain |
| 12 | t26-12p | 8.1 | 8811–9020 (c) | agggaggtgtactcttgATG | – | |
| 13 | t26-13p | 27.9 | 9114–9878 | aggggtgaggagaATG | TKV2 (44%, 264) | Putative coiled-coil domain–Putative transmembrane domains |
| 14 | t26-14p | 50.2 | 10 037–11 338 | cggaggtgcgggttagATG | TGV1 (95%, 433) | Putative ATPase, AAA superfamily (weak similarities with RuvB helicase) |
| 15 | t26-15p | 27.0 | 11 349–12 044 | gaggtggtgaagATG | TKV3 (92%, 231) | |
| 16 | t26-16p | 11.8 | 12 046–12 369 | tggagggtgtgatATG | TGV1 (81%, 104) | |
| 17 | t26-17p | 8.0 | 12 480–12 680 | cggtggggctgATG | TKV3 (89%, 66) | |
| 18 | t26-18p | 21.5 | 12 677–13 228 | tggtggtggaATG | TKV3 (88%, 183) | Weak similarities with bacterial plasmid transfer factor TraG |
| 19 | t26-19p | 31.7 | 14 271–13 438 (c) | cggaggagggcccgagggATG | TKV3 (62%, 280) | Putative transmembrane domains |
| 20 | t26-20p | 15.8 | 14 426–14 842 | AccacggtgagagctcaATG | TKV3 (65%, 138) | Putative transcription regulator, family SpoVT/AbrB |
| 21 | t26-21p | 6.4 | 15 343–15 504 | aggtggttccaATG | TKV1 (34%, 52) | |
| 22 | t26-22p | 81.2 | 15 519–17 624 | gaggcgagctctATG | TKV3 (49%, 707) | Homologous to RepA proteins of plasmids pTIK4 and pORA1 from Sulfolobus neozealandicus— Putative ATPase |
| 23 | t26-23p | 9.1 | 17 814–18 062 | cggaggagggtgaaagcATG | TKV3 (82%, 80) | |
| 24 | t26-24p | 13.0 | 18 059–18 406 | aggagggaggaggaATG | – | |
| 25 | t26-25p | 7.5 | 18 420–18 614 | cgggggtgagggcATG | TKV1 (66%, 63) | |
| 26 | t26-26p | 14.1 | 18 611–18 976 | tggggggtcgctcATG | TKV1 (63%, 121) | |
| 27 | t26-27p | 6.6 | 18 973–19 143 | tggaggtggtttcATG | TKV1 (75%, 54) | |
| 28 | t26-28p | 21.7 | 19 143–19 685 | cgggggtgggagctgATG | TKV1 (42%, 182) | |
| 29 | t26-29p | 9.5 | 19 692–19 931 | gaggggaggtgacggggcGTG | – | |
| 30 | t26-30p | 13.1 | 19 931–20 275 | aggagcgtgATG | TKV2 (51%, 80) | |
| 31 | t26-31p | 24.7 | 20 363–20 998 | gggtggttcggaaATG | PHV1 (45%, 155) | Putative resolvase, related to family pfam00239 and to PinR (COG1961) |
| 32 | t26-32p | 19.8 | 20 995–21 495 | aggaggaaggtgtATG | PHV1 (39%, 166) |
Phylogenetic analyses
Homologous sequences were recovered by BLAST searches, and multiple alignment were performed with the selected sequences using MUSCLE program (32). Only homologous positions were used to build unrooted maximum likelihood trees using PHYML (33). The JTT model of amino acid substitution was choosen, and a gamma correction with four discrete classes of sites was used. The alpha parameter and the proportion of invariable sites were estimated. The robustness of trees was tested by non-parametric bootstrap analysis using PHYML.
In silico identification of integrated mobile element
Mobile elements integrated in cellular genome were identified as Cluster of Atypical Genes (CAGs) according to Cortez et al. (41). Briefly, archaeal genomes were analysed with a species-specific Markov model in order to obtain the list of atypical genes. We then searched for atypical genes that cluster together, since these may be recently integrated foreign elements. We used a sliding window of 10 ORFs that moved along the genome sequence. A CAG was defined when seven or more ORFs in that window showed an atypical composition. To define CAGs families, Blast searches were performed with all the ORFs contained in our CAGs with an e-value of 10e−20. We then generated several topological networks of CAGs by drawing a line between pairs of CAGs that share a defined number of ORFs (from two up to six). The graphical representation was obtained with the Cytoscape program (http://www.cytoscape.org).
Cloning, expression and purification of tn2-12p
The coding sequence of tn2-12p was amplified by PCR from plasmidic DNA isolated from the strain T. nautilus 30/1, and the amplified fragment was cloned in a pET21 (Novagen) plasmid allowing fusion of a 6His tag at 3′-end. Expression was done at 37°C using the Escherichia coli Rosetta (DE3) pLysS strain (Novagen) and the 2xYT medium (BIO 101 Inc.). When the cell culture reached an OD600 nm of 0.8, induction at 15°C was performed overnight with 0.5 mM IPTG (Sigma). Cells were harvested by centrifugation and resuspended in buffer A (20 mM Tris–HCl pH 7.5, 200 mM NaCl, 5 mM β-mercaptoethanol). Cell lysis was completed by sonication and the lysate was heated for 20 min at 70°C before centrifugation at 20 000 rpm for 20 min. The soluble fraction was loaded on a Ni–NTA column (Qiagen Inc.) equilibrated with buffer A. The protein was eluted with imidazole and subsequently loaded on a heparin column (GE Healthcare) equilibrated against buffer A′ (20 mM Tris Tris–HCl pH 7.5, 50 mM NaCl, 5 mM β-mercaptoethanol). Elution was performed using a gradient between buffer A′ and B (20 mM Tris Tris–HCl pH 7.5, 1 M NaCl, 5 mM β-mercaptoethanol). The tn2-12p protein was eluted at ∼0.9 M NaCl. Eluted fractions were pooled and loaded on a Superdex200 column (Amersham Pharmacia Biotech) equilibrated against buffer A supplemented with 10 mM β-mercaptoethanol. The homogeneity of the protein sample was checked by SDS–PAGE.
DNA polymerase assay
Two different 5′-labelled primer-template systems were used (see legend of Figure 2). The standard polymerase assay contained 10 nM primer-template substrate, 2.5–15 µM of tn2-12p (see legend of Figure 2) in 10 mM Tris–HCl pH 9, 1 mM DTT, 50 mM KCl, 0.1% Triton X-100, 50 mM MgCl2 and 0.2 mM dNTPs. The protein was diluted in 50 mM Tris–HCl pH 8, 1 mM DTT, 100 mM NaCl, 0.1 mM EDTA. The significant amount of protein required for the optimal polymerization reaction could be explained by a relatively low stability of the isolated protein. The reactions were allowed to proceed for 20 min at 70 or 80°C and were analysed by denaturating PAGE.
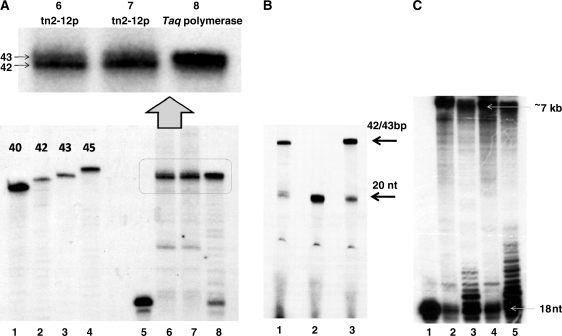
Figure 2.
tn2-12p is a DNA polymerase. (A) Lanes 1–4: 5′-32P oligonucleotides of 40, 42, 43 and 45 mers. Lane 5: 5′-32P labelled 20 nt oligonucleotide CGAACCCGTTCTCGGAGCAC, no protein added. The recombinant protein tn2-12p (2.5 and 5 μM, lanes 6 and 7, respectively) or Taq polymerase (Promega, 0.04 U/μl, lane 8) were incubated with this short primer-template substrate hybridized to 42-nt template TTCTGCACAAAGCGGTTCTGCAGTGCTCCGAGAACGGGTTCG. Primers are extended in the presence of 0.2 mM dNTPs during 20 min at 70°C. The buffer used for the polymerization reaction is described in ‘Materials and Methods’ section. Extension products were analysed on the 16% denaturating polyacrylamide gel. Magnification: two very close bands of 42 and 43 bp are clearly visible. (B) The recombinant protein tn2-12p (15 μM, lanes 1 and 2) or Taq polymerase (Promega, 0.02 U/μl, lane 3) were incubated with a short primer-template substrate (5′-32P labelled 20 nt oligonucleotide CGAACCCGTTCTCGGAGCAC hybridized to 42 nt template TTCTGCACAAAGCGGTTCTGCAGTGCTCCGAGAACGGGTTCG). Primers are extended in the presence of 0.2 mM dNTPs (lanes 1 and 3) or 0.2 mM NTPs (lane 2) during 20 min at 70°C. In the control reaction with NTPs (lane 2) no primer elongation was observed. The buffer used for the polymerization reaction is described in ‘Materials and Methods’ section. Extension products were analysed on the 16% denaturating polyacrylamide gel. (C) The primer extension activity of tn2-12p was assayed at 70°C (lanes 1–3) and 80°C (lanes 4 and 5). A 5′-32P-labelled 18-nt primer (GTAAAACGACGGCCAGTG) was hybridized with ssDNA of M13. This primer-template substrate (10 nM) were incubated for 20 min either without any proteins (lane 1), either with 10 μM of tn2-12p (lanes 2 and 4) or with 0.05 U/μl of Taq polymerase (lanes 3 and 5) in the presence of 0.2 mM dNTPs. The buffer used for the polymerization reaction is described in ‘Materials and Methods’ section. Extension products were analysed on the 16% denaturating polyacrylamide gel.
RESULTS
Isolation of three new plasmids from Thermococcales
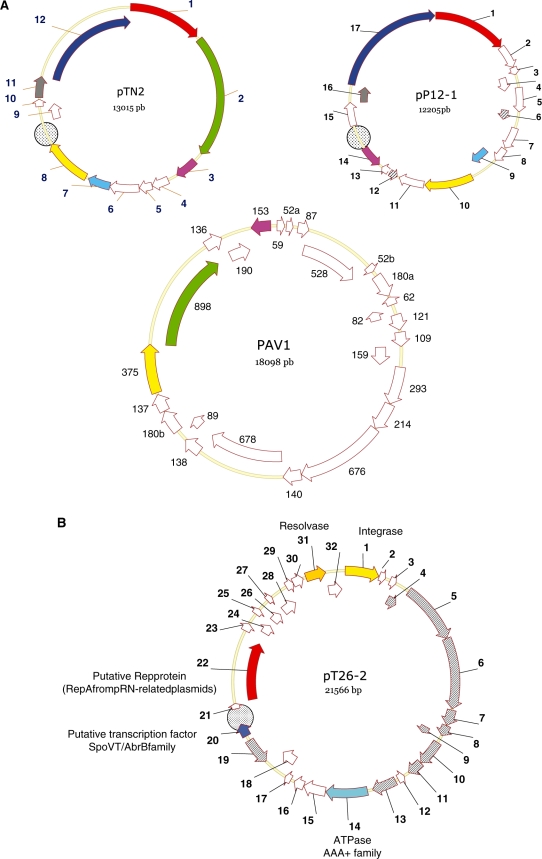
Plasmids pTN2, pP12-1 and pT26-2 were isolated from three strains of Thermococcales that were purified from samples collected at three different deep-sea hot vents located in the East-Pacific ridge during the AMISTAD cruise (31). The strain 30-1, which harbours the plasmid pTN2, has been tentatively described as the type strain of a new species, T. nautilus (26). The strain 26-2, that harbours the plasmid pT26-2 is closely related to T. nautilus sp. 30-1 by 16S rRNA analysis and could belong to the same species, although it exhibits a very different RAPD profile (31). Finally, the strain 12-1, which harbours the plasmid pP-12-1 belongs to a RAPD group that includes a Pyrococcus species (isolate 32-4) and will be thereafter called Pyrococcus sp. 12-1. The three plasmids are circular and their sizes are 13 015, 12 205 and 21 566 bp, respectively. The plasmids sequences have a GC content of ∼47.5, 44.6 and 49.6% for pTN2, pP12-1 and pT26-2, respectively, which are close to GC% of Thermococcales chromosomal DNA (40–50%). By in silico analysis, we identified 12, 17 and 32 putative ORFs in pTN2, pP12-1 and p26-2, respectively (see ‘Materials and Methods’ section). Although pTN2 was isolated from a Thermococcus species and pP12-1 from a Pyrococcus species, they turned out to be evolutionary related, since they share six homologous genes. In contrast, although plasmids pTN2 and pT26-2 are present in two closely related strains (30-1 and 26-2), they turned out to be completely unrelated to each other.
All ORFs of pTN2 and pP12-1 are located on the same DNA strand with only two exceptions detected in pP12-1. Similarly, 30 of the 32 pT26-2 ORFs are transcribed in the same direction (Figure 1). We predicted the position of the replication origin of these three plasmids by performing cumulative GC skew analysis (34). This method is based on the general observation that GC content usually differ between the leading and lagging strands of replication forks (35). The cumulative GC skews graphics for the three plasmids show GC frequency inversion producing V-like curves. Strikingly, the minima (blue arrows, Supplementary Figure S1) are located in the larger intergenic regions for each of the three plasmids (circles, Figure 1). These intergenic regions are among the most AT-rich of the three plasmids and contain many direct and inverted repeats; features that are general characteristics of plasmid replication origins (Supplementary Figure S2). Interestingly, the predicted origin regions of pP12-1 and pT26-2 are located close to ORFs (p12-14p, p12-12p and t26-19p) that are transcribed in the direction opposite to most other ORFs encoded by these plasmids (Figure 1).
Figure 1.
Schematic representation of the three new plasmids. (A) pTN2 and pP12-1 plasmid maps were drawn at the same scale together with PAV1 genome. ORFs are numbered and represented as arrows. ORFs encoding homologous proteins have the same colour. White ORFs do not have detectable homologues among these three genomes. (B) pT26-2 plasmid map with ORFs numbered and represented as arrows. Coloured ORFs encode proteins with expected activity or function. (A and B) Hachured ORFs harbour putative hydrophobic segments. Circles indicate large intergenic regions including putative replication origins.
The pTN2 plasmid family
The two plasmids pTN2 and pP12-1 share six homologous proteins (Figure 1A). They can thus be considered as members of a new plasmid family (thereafter dubbed the pTN2 family) that is presently only found in Thermococcales. The plasmids pTN2 and pP12-1 share in particular two large proteins whose genes are contiguous and located downstream of the putative plasmid replication origins. The first ones (tn2-1p in pTN2 and p12-1p in P12-1, ∼66 kDa) are homologous to helicases of the superfamily I (SFI). Indeed, conserved motives of this family (36) are also found in the sequences from these two plasmids (alignment in Supplementary Figure S3). This family includes the well characterized bacterial helicases UvrD, PcrA and Rep (37). They are widespread in the three domains of life, with many representatives in most bacterial and several euryarchaeal phyla, as well as in a few eukaryotic ones. We have built a phylogenetic tree of this helicase family with a selection of sequences from the three domains (at least one sequence per represented phylum, Supplementary Figure S4). In this tree, the closest relatives of the pTN2 and pP12-1 helicases are encoded in the genomes of T. gammatolerans and T. onnurineus. The four helicases from Thermococcales form a monophyletic group with seven helicases from Haloarchaea. Overall, the archaeal SFI helicases tree is not congruent with the archaeal phylogeny based on ribosomal proteins or RNA polymerase subunits (38,39). The archaeal sequences form five monophyletic groups and Archaea of the same orders are often present in different groups (Supplementary Figure S4). The SFI helicase of the thaumarchaeon Cenarchaeum symbiosum is grouped with some Methanococcales (phylum Euryarchaea). Furthermore, several groups of eukaryotic helicases (from plants and protists) are interspersed between archaeal groups, whereas helicases from fungi form a monophyletic group with sequences from Methanomicrobiales and Thermoplasmatales. This phylogeny is difficult to interpret because lengths are highly variable, probably inducing phylogenetic artefacts. Interestingly, the ‘cellular’ homologue of pTN2-type helicases in T. gammatolerans is located in an integrated virus-like element, TGV2 (40). In Methanococcus maripaludis strains C6 (MMC6V1) and C7 (MMC7V2), the genes encoding pTN2-type helicases are located in predicted integrated elements of plasmid/virus origin that have been identified in silico, based on their dinucleotide sequence composition [see ‘Materials and Methods’ section and ref. (41)]. These observations suggest that some archaeal, and possibly eukaryotic SFI helicases, originated from viruses and plasmids and were transferred independently into various lineages, explaining the incongruence between their phylogeny, and the classical archaeal and eukaryotic phylogenies. The frequent grouping of Archaea from different phyla in the same clade furthermore suggests that horizontal gene transfers of genes encoding these helicases also sometimes occurred in Archaea. The bacterial helicases of the SFI helicase family are quite well separated from archaeal and eukaryotic ones, with the exception of the helicase from the bacterium Aquifex aeolicus that branches with the group mixing some helicases from Archaea and Fungi. Bacterial UvrD-like helicases form a monophyletic group (with two representatives in the eukaryotes Ostreococcus, a probable case of gene transfer from a mitochondrial or plastid genome). However, as in the case of Archaea, Bacteria from the same phylum are often dispersed in different parts of the tree, suggesting that the genes encoding these helicases have been also sometimes transferred between bacterial phyla.
The second large ORFs conserved between pTN2 and pP12-1 (tn2-12p and p12-17p, respectively) encodes proteins of 107 and 103 kDa, respectively. Psi-BLAST searches using tn2-12p as query gave no significant result, whereas similar searches using p12-17p gave significant matches with two very similar proteins from M. voltae A3 (MvolDRAFT_1375 and MvolDRAFT_1398). Interestingly, we noticed among hits of the first PSI-BLAST iterations the putative Rep protein of the recently described plasmid pXZ1 from Sulfolobus islandicus (17). BLAST search with the sequence of the pXZ1 Rep protein then retrieved the Rep protein of the plasmid pTIK4 from S. neozelandicus (8). We have been able to align manually the N-terminal regions of tn2-12p and p12-17p with those of the two proteins of M. voltae and the Rep proteins of pXZ1 and pTIK4 (Supplementary Figure S5). We then noticed that these proteins exhibit a conserved DhD motif, known to be present in the active sites of many DNA polymerases, primases and/or nucleotidyl transferases (42). Although this similarity was minimal, we decided to express the tn2-12p protein in E. coli to test if this protein exhibits a DNA polymerase activity in vitro. The purified recombinant tn2-12p protein was first incubated with dNTPs and 5′-labelled 20 nt DNA primer hybridized to a complementary 42 nt DNA template. As expected if our prediction was correct, the primer was efficiently extended up to the full-length of the template (Figure 2A and B). The major fraction of the polymerization products was, as expected, represented by a 42-bp-long fragment but we also observed a minor DNA fragment of 43 bp. In the same conditions, the Thermus aquaticus (Taq) DNA polymerase synthesized also two subfamilies of DNA fragments: a major one composed of 43 bp and a second one (minor) having only 42 bp. It is well known that Taq DNA polymerase has a non-template-dependent terminal transferase activity and is able to add an additional non-template-directed nucleotide to the 3′-ends of a blunt-ended DNA fragment via this terminal transferase-like activity (43,44). This result indicates that the tn2-12p protein has also a nucleotidyl transferase activity which will be described in details elsewhere (Desnoues et al., manuscript in preparation). The DNA polymerizing activity of tn2-12p requires the presence of dNTPs and a DNA template (data not shown) and its activity is not stimulated in the presence of ATP (as was observed for another archaeal polymerase coded by the plasmid pRN1, 14). No extension was observed when the dNTPs were replaced by NTPs. The protein tn2-12p is able to extend the primer up to several kilobases at 20 min of the reaction times and the obtained DNA shows a pattern close to that observed in the similar experiment with the Taq polymerase. This result indicates that tn2-12p can produce long extension products without the help of additional proteins.
The DNA polymerase activity associated to tn2-12p is consistent with the idea that tn2-12p and the homologous protein p12-17p correspond to the Rep proteins of the plasmids pTN2 and pP12-1, respectively. In the case of pRN1, the DNA polymerase domain of RepA is located in the N-terminal part of the protein and is fused to a helicase domain (SFIII) in C-terminal. In the case of the pTN2 and pP12-1 Rep proteins, Psi-BLAST analysis suggests that the DNA polymerase domain is also located in their N-terminal part (which contains the two conserved aspartate residues probably present in the active site). They also indicate that, as in the case of pRN1-type RepA, these polymerase domains are fused to large C-terminal domains. However, Psi-BLAST analysis using these domains as queries retrieved no significant hit in databases.
In both pTN2 and pP12-1, the rep genes form a replication cassette with the contiguous helicase gene. Interestingly, these replication cassettes are located upstream the large intergenic regions predicted to be the replication origin of these plasmids by GC skew analysis (Figure 1A and Supplementary Figure S1). Several small putative ORFs are located between the origin regions of pTN2 and pP12-1 and the beginning of the rep genes. One of them (tn2-11p and p12-16p) is conserved in both plasmids, indicating that they should encode bona fide proteins involved in the regulation of plasmid replication. In pP12-1, the protein p12-15p, which is located close to the putative replication origin, belongs to the CopG family of transcriptional regulators. This protein could play a role in the regulation of the Rep protein expression, similarly to the CopG-like proteins whose genes are located upstream of the rep gene in Sulfolobus pRN-type plasmids (45).
Three other proteins shared by pTN2 and pP12-1 (tn12-3p/p12-14p, tn2-7/pP12-9p and tn2-8p/pP12-10p) are encoded by genes whose order is not conserved between the two plasmids (Figure 1A). Interestingly, two of them are homologous to proteins encoded by the virus PAV1 from P. abyssi (Figure 1A). This lemon-shaped virus, with a double-stranded DNA genome of ∼16 kb, is presently the only known virus that infects an hyperthermophilic euryarchaeon (46). The tn2-8p and p12-10p proteins (∼42 kDa) are homologous to the ORF375 protein of PAV1, whereas the tn2-3p and p12-14p proteins (∼20 kDa) are homologous to the ORF153 protein of PAV1.
BLAST searches with proteins tn2-8p, p12-10p or ORF375 as queries retrieved significant sequence similarities with proteins encoded in the genomes of M. voltae A3 and Methanosarcina barkeri, in a DNA fragment from an environmental sample enriched for virus sequences (47) and in several plasmids from haloarchaea (pNG4027, pNG1017, pNG2044 and pNG3054). See Supplementary Figure S6 for an alignment of these homologous proteins. In M. voltae, the homologue of tn2-8p/p12-10p, MvolDRAFT_1394, is located in a large region of ∼60 genes located between a tRNA and a cluster of CRISPR associated Cas genes. All these genes encode uncharacterized proteins, except for three integrase genes, two putative transcriptional regulators, a resolvase gene and, strikingly, two genes encoding homologues of the pTN2 and pP12-1 Rep-DNA polymerases (MvolDRAFT_1375 and 1398). This region thus most likely corresponds to integrated virus and/or plasmids. In M. barkeri, the homologue of tn2-8p/p12-10p is located close to a gene encoding a putative integrase (Mbar_A2247) next to a tRNA gene, indicating that this protein is also encoded by an integrated mobile element. This suggests that tn2-8p/p12-10p are the prototype of a new protein family specific for viral/plasmid. Visual inspection of alignment of these proteins revealed a typical Walker type motif in their N-terminal extremity (Supplementary Figure S6). Indeed, Psi-BLAST iterations retrieved weak hits with many proteins annotated as putative ATPases (including ABC transporters).
The proteins tn2-3p, p12-14p and PAV1 ORF153 (alignment in Supplementary Figure S7) have only one detectable homologue in databases, a protein of unknown function (previously ORFans) present in the genome of T. barophilus TERMP_2062. However, weak hits were obtained with various proteins from the three domains of life, including a protein encoded by the plasmid pURB500 from M. maripaludis C5.
Interestingly, the plasmid pTN2 (but not pP12-1) encodes a large protein (105 kDa), with a predicted coiled-coil domain, tn2-2p, that has also a homologous encoded by the virus PAV1, ORF898 (Figure 1A). The gene encoding this protein is located downstream the one encoding the SFI helicase, suggesting that it could be involved in DNA replication. Unfortunately, this protein has no detectable homologue in database and BLAST searches retrieved no significant hits that could have suggested putative function.
The plasmids pTN2 and pP12-1 encode several small putative proteins that are specific for each plasmid (5 and 11, respectively). Only four of them (including the CopG-like protein previously mentioned, pP12-15p) have detectable homologue and/or motives in databases. The protein tn2-4p exhibits weak similarities with helix–turn–helix proteins annotated as putative transcriptional regulator. The protein p12-11p has a distant homologue in the genome of a Methanococcales, Methanocaldococcus fervens AG86 (Mefer_1580). These two proteins harbour weak similarities in N-terminal with the epsilon subunits of bacterial DNA polymerase III (DnaQ) (data not shown). Interestingly, we could indeed detect three putative exonuclease motives (ExoI, ExoII, ExoIII; 48) in the sequence of p12-11p (alignment in Supplementary Figure S8) suggesting that this protein could harbour a 3′–5′ exonuclease activity. It will be interesting to determine if this protein is used for proof-reading by the DNA polymerase activity associated to the Rep protein of the pP12-1 plasmid. Finally, the protein pP12-2p harbours a C2H2-type zinc-finger motif and is homologous to a hypothetical protein (MJECL27) encoded by an extra-chromosomal element present in Methanocaldococcus jannaschii.
The plasmid pT26-2
The plasmid pT26-2 is unrelated to pTN2 and pP12-1 and can be considered as the prototype of a new family of archaeal plasmids (thereafter dubbed the pT26-2 family). Only four of the 32 predicted proteins of pT26-2 have biological function and/or biochemical activity that can be predicted (Figure 1B): an integrase of the SSV type, a serine recombinase and two proteins with Walker type motives (i.e. putative ATPases).
The closest homologues of the pT26-2 integrase (t26-1p) are encoded in the genomes of T. gammatolerans (TGAM_0651) and of T. kodakaraensis (TK0381) (71 and 68% identity, respectively). Three other homologues are present in T. kodakaraensis (TK0073, TK0614, TK1342), two in P. horikoshii, (PH1200 PH1863) and one in Aeropyrum pernix (APE_0716.1). More distantly related integrases are encoded by SSV viruses or by mobile elements integrated in the genome of several Sulfolobales. The homologues of the pT26-2 integrase in the genomes of Thermococcales are located at the border of the virus like integrated elements TKV2, TKV3 (49) and TGV1 (40). These elements contain clusters of genes encoding proteins homologous to other pT26-2 proteins, delineating a family of integrated elements related to this plasmid (see next section for their detailed analysis).
The plasmid pT26-2 encodes a putative nucleotide hydrolase (t26-14p) with Walker type motives in N-terminal that has no close homologues, except in virus-like elements present in Thermococcales and Methanococcales (see below) but exhibit weak similarities with proteins whose known biological function can be relevant for plasmid/virus physiology. Hence, this protein exhibits weak similarities to the RuvB protein of Dyctioglomus thermophilus. RuvB is a helicase involved in branch migration during resolution of Holliday-junction by the RuvABC complex, suggesting that t26-14p could be involved in DNA pumping, i.e. in DNA transfer during putative conjugation.
The protein t26-22p turned out to be especially interesting. The closest homologue of this protein is located in the integrated element TKV3 of T. kodakaraensis. Remarkably, this protein has been replaced by the replicative helicase MCM in the integrated element TKV1, in an otherwise conserved cluster of six genes conserved between pT26-2 and TKV1 (Figure 3). This suggests that t26-22p could be a helicase. This protein is also homologous and closely related to the C-terminal domains of two RepA proteins encoded by two pRN-type plasmids from S. neozealandicus pORA1 and pIT3 (8) and to two proteins from S. islandicus (alignment in Supplementary Figure S10). These domains are fused in N-terminal to a DNA polymerase/primase of the pRN1 type. Strikingly, in that arrangement, the homologues of t26-22p replace the SFIII helicase of the pRN1 RepA protein (50), again suggesting a helicase function for t26-22p. Psi-BLAST searches with the protein t26-22p as query did not retrieved known helicases. However, after aligning t26-22p and homologous proteins of unknown functions present in databases, we noticed the presence of a Walker type motif located in the central region of these proteins. Furthermore, after third iterations, we retrieve many AAA+ ATPases present in the three domains of life. All these data strongly suggest that t26-22p is the replicative helicase of pT26-2 and can be considered as the prototype of a new family of replicative DNA helicases distantly related to the superfamily of AAA+ ATPases. Unfortunately, we did not succeed purifying this protein to test its putative helicase activity. Interestingly, Psi-BLAST using t26-22p sequence as query retrieved in the fourth iteration the C-terminal domain of the hypothetical protein MvolDRAFT_1375 from M. voltae A3. The N-terminal domain of this protein is precisely homologous to the primase/polymerase domain of RepA proteins encoded by pTN2, pP12-1, pXZ1 and pTIK4 previously described. This fusion between a pTN2 type polymerase and a pT26-2 type helicase observed in a plasmid/virus integrated in the genome of a methanogen illustrates the domain swapping that has occurred between polymerase and helicase domains in the course of plasmid evolution.
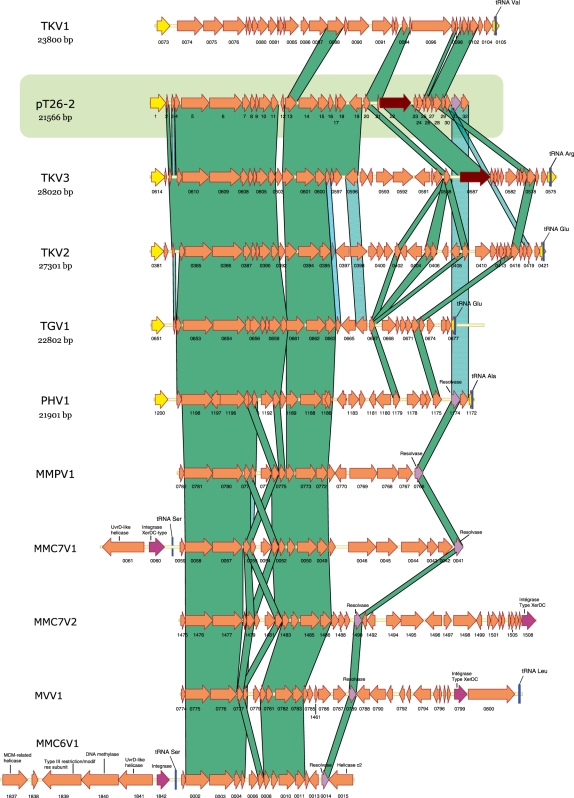
Figure 3.
Schematic alignment of pT26-2 genome with integrated elements in Thermococcales and Methanococcales genomes. Thermococcus kodakaraensis virus-related regions (TKV1, TKV2 and TKV3), P. horikoschii (PHV1), T. gammatolerans (TGV1), M. maripaludis strain S2 (MMPV1), M. maripaludis strain C7 (MMC7V1 and MMC7V2) and strain C6 (MMC6V1), and M. voltae (MVV1). ORFs are indicated as arrows. Yellow ORFs represent genes of fragments of genes encoding an integrase related to the SSV1 integrase, and blue bars indicate tRNA genes. Brown ORFs share sequence similarities with RepA proteins encoded by the Sulfolobus plasmids pTIK4 et pORA1 (t262-22 and TK0587). Pink ORFs are homologous with genes encoding integrases of XerCD family, whereas mauve ORFs are homologous to genes encoding resolvases. Among the integrated elements represented here, ORFs encoding homologous proteins with pT26-2 are linked together with green bands.
As in the case of the pTN2 and pP12-1 Rep proteins, the gene encoding the t26-22p putative helicase of pT26-2 is located just upstream of the large intergenic region identified as the putative replication origin of this plasmid by GC skew analysis (Figure 1B and Supplementary Figure S1). We have expressed in E. coli the protein t26-22p but the protein turned out to be insoluble, preventing us to test its activity.
Finally, the fourth pT26-2 protein with predictable biochemical function, t26-31p, belongs to a huge family of serine recombinases widespread in Archaea and Bacteria but missing in Eukarya (51). The most closely related homologues of t26-31p are found in archaeal genomes, both in Euryarchaeota and in Crenarchaeota. Homologues in Thermococcales and Methanococcales are present within the integrated elements (virus-like) related to pT26-2 (see below).
Several putative pT26-2 proteins have no clear-cut homologue with known function in databases but gave interesting hits in BLAST analyses. The protein t26-18p exhibits some weak similarities with bacterial proteins annotated as TraG (plasmid transfer factors) suggesting a role in DNA transfer (Supplementary Figure S9). The protein t26-20p, located immediately adjacent to the putative replication origin, exhibits similarities with transcription factors of the SpoVT/AbrB family (52), suggesting that it could play a role in plasmid copy number regulation. Several proteins of pT26-2, whose genes are clustered in one half of the plasmid, contain hydrophobic domains (Figure 1B), suggesting that they could be membrane bound proteins or part of complex architectural elements. These include two large proteins (t26-5p and t26-6p, 68 and 81 kDa, respectively) shared by pT26-2 and all related elements integrated in the genomes of Thermococcales and Methanococcales (see below). Analysis of t26-5p using Interproscan detected a central domain annotated as carboxypeptidase regulatory domain and two short putative hydrophobic domains at both extremities. This corresponds to the description of metalloproteases of the SSF49464 family that exhibit a central carboxypeptidase regulatory domain surrounded by two transmembrane domains. All these observations suggest that t26-5p could be a membrane protease that can hydrolyse glycoproteins. The neighbouring protein t26-6p contains five short hydrophobic domains (three in the N-terminal part and two in the C-terminal part). We have recently solved the structure of a recombinant t26-6p protein lacking these hydrophobic domains (53). The protein has an elongated shape and is composed of three domains which all correspond to novel folds. Interestingly, homologues of the putative membrane protein t26-13p are located immediately upstream those of the putative ATPase t26-14p (sharing weak similarities with RuvB helicase) in all integrated elements related to pT26-2. This suggests that t26-14p and t26-13p could work together to pump DNA through the membrane, and raises the possibility that pT26-2 is either a conjugative plasmid or encodes remnants of a viral apparatus for DNA injection and/or packaging. These proteins could also work together with the putative ATPase t26-18p which, as was previously mentioned, exhibits weak similarities with plasmid transfer factors (Supplementary Figure S9).
A family of pT26-2 related elements in genomes of Thermococcales and Methanococcales
Clusters of genes encoding homologues of several pT26-2 proteins are present in the genomes of several Thermococcales and Methanococcales species (Figure 3). Two of these clusters are located in the previously described TKV2 and TKV3 virus-like elements from T. kodakaraensis (49) and TGV1 from T. gammatolerans (40). We identified a new closely related integrated virus-like element (IE) in the genome of P. horikoshii (thereafter named PHV1). The plasmid pT26-2 shares 20, 19, 17 and 17 homologous genes with TKV3, TKV2, TGV1 and PHV1, respectively (Figure 3). In all cases, the 3′-end of elements are located adjacent to a tRNA gene and the two extremities of the elements are bordered by the two parts of a splitted gene encoding the SSV-type integrase homologous to the pT26-2 integrase. The N-terminal moieties of these genes are located in 3′ of the elements (adjacent to the tRNA gene) whereas their C-terminal moieties are located in 5′, as expected for integration mediated by SSV1-type integrases. We also identified clusters of genes homologous to pT26-2 in the genomes of several mesophilic Methanococcales (M. maripaludis strains S2, C6 and C7, M. voltae, Figure 3). Importantly, the syntheny of genes homologous between pT26-2 and integrated elements in Thermococcales is maintained in clusters present in Methanococcales. As illustrated in Supplementary Figure S11A, both in Thermococcales and Methanococcales, these clusters (horizontal green bars) are located in larger regions (containing between ∼40 and 100 ORFs) that contain many atypical genes (Supplementary Figure S11A, vertical blue bars), i.e. genes whose sequence composition differ from those of genes conserved in the genome of all Methanococcales (invisible white bars in the figure) [see ref. (41) for the definition of clusters of atypical genes (CAGs)]. These regions include integrase genes (black arrows), tRNA genes (vertical pink bars) and serine recombinases (resolvases) related to those of pT26-2. All these observations indicate that the clusters of pT26-2 homologous genes in the genomes of Thermococcales and Methanococcales are located within bona fide integrated ‘virus-like elements’. These integrated elements will be called thereafter MMPV1, MMC6V1, MMC7V1, MMC7V2 and MMV1 to follow the nomenclature adopted for IE from Thermococcales. As previously mentioned, MMC7V1 and MMC6V1 IE include a gene encoding a pTN2-type helicase. In addition, MMC6V1 also include a homologue of the cellular replicative helicase MCM.
The plasmid pT26-2 also harbours seven genes that have homologues in TKV1, a third integrated element of T. kodakaraensis. Six of them are located in the region located just upstream the C-terminal integrase moiety of pT26-2 and have homologues in the corresponding region of pT26-2 related elements present in Thermococcales (but not in Methanococcales). The genome of pT26-2 and related integrated elements can thus be divided in two parts, a highly conserved 5′ region which includes genes that are present in all integrated elements of the pT26-2 family present in Thermococcales, including seven genes also present in pT26-2 related integrated elements present in Methanococcales (dubbed core genes thereafter, i.e. homologous of t26-5p-6p-7p-11p-13p-14p and -15p) and a variable 3′ region that includes both ORFans and genes of mixed origins (TKV1 and/or TKV2/TKV3). This suggests that a relatively ancient recombination event occurred between a TKV1-like element and the ancestor of the pT26-2 and TKV2/V3 related elements.
The variable region of pT26-2 includes the genes for the putative helicase t26-22p, the putative transfer proteins t26-18p and t26-20p, and the serine recombinase t26-31p, whereas the core genes include the large proteins t26-5p and t26-6p. Interestingly, further homologues of these last two proteins are encoded in the genomes of several species of M. maripaludis that lack the other core genes of the pT26-2 family. In order to get insight into the evolution of the pT26-2 family, we have performed a phylogenetic analysis of a concatenation of six core proteins (the homologues of the core gene t26-7p was removed from this phylogeny because they include several paralogues). The topology of the resulting tree (Supplementary Figure S12) was similar to those of the tree that we previously constructed using only the two core proteins t26-5p and t26-6p (53). As shown in Supplementary Figure S12, family members from Thermococcales and Methanococcales are clearly separated. The phylogeny obtained is roughly congruent with the archaeal species phylogeny if the root is located between these two orders, as Thermococales and Methanococcales each form monophyletic groups. Among Thermococcales, pT26-2 and integrated elements from Thermococcus species form a monophyletic group, separated from PHV1 of P. horikoshii. These data suggest that pT26-2 and related integrated elements have co-evolved with their hosts and diverged from an ancestor that already infected Archaea before the divergence between Methanococcale and Thermococcales.
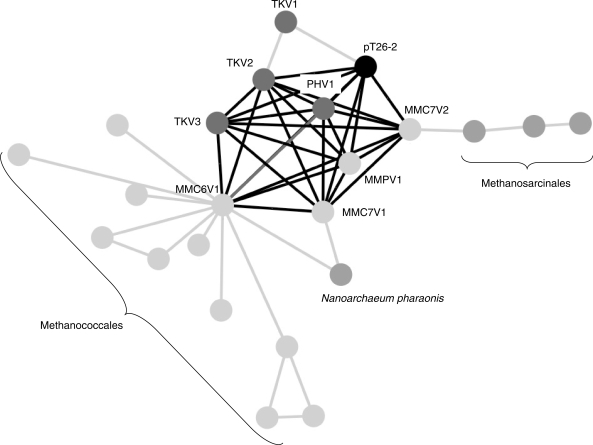
To get clue about possible more distant evolutionary relationships between members of the pT26-2 family and other plasmids and virus-like elements integrated in archaeal genomes (previously identified as CAGs, 41), we have draw different networks in which two elements are linked to each other by a line if they share either three, four or five genes (threshold) and a bold line if they share the seven core genes of the pT26-2 family. Interestingly, members of the pT26-2 family become linked to more and more CAGs or to free plasmids when the threshold was reduced from five to two (Figure 4 and S11B-E). Whereas pT26-2 was only linked to CAGs or plasmids from Thermococcales and Methanococcales with a threshold of five, it became also linked to Methanosarcinales with a threshold of four. Finally, when the threshold was relaxed to three, CAGs from Thermoplasmatales and Halobacteriales were recruited into the network of plasmid/CAG interactions and several large families are clearly delineated (plasmids from halobacterial were removed from this analysis because their high number introduced a bias). This analysis thus defines a huge superfamily of mobile elements loosely related to pT26-2 that encompass the whole phylum of Euryarchaea. Sulfolobales (phylum Crenarchaea) only entered into the network when the threshold was reduced to two.
Figure 4.
CAGs families. In this figure, each circle correspond to either a plasmid (pT26-2) or to an integrated element identified as a cluster of atypical genes (41). Each link between two elements means that they share at least five homologous genes (see ‘Materials and Methods’ section for further details). The plasmid pT26-2 is in black, and the other Thermococcales integrated elements are in dark grey (TKV1, TKV2, TKV3, PHV1). The integrated elements sharing core genes with pT26-2 (described in the text), are linked by bold lines. Integrated elements from Methanococcales are represented in light grey whereas integrated elements from other phyla (Methanosarcinales and Nanoarchaea) are represented in grey.
DISCUSSION
All plasmids previously isolated from Thermococales were small RC plasmids (23–25,54). Here we report the isolation, sequencing and characterization of three new plasmids from Thermococcales that are larger and probably replicate via the theta mode. The plasmids pTN2 and pP12-1 encode a DNA polymerase fused to a domain of unknown function. These proteins are homologous to the RepA protein of the plasmids pTIK4 from S. neozelandicus and pXZ1 from S. solfataricus (17) and to proteins encoded by integrated elements present in genomes of Methanococcales. They form therefore a new family of archaeal RepA proteins common to mobile elements present in both Euryarchaea and Crenarchaea. The DNA polymerase domain of these RepA proteins have no detectable sequence similarities with those of previously known DNA polymerases, including the DNA polymerase discovered by Georg Lipps in pRN1 (14), indicating that these proteins could be the prototypes of a new DNA polymerase family. As in the case of the pRN1 polymerase, the DNA polymerase of pTN2 exhibits a primase activity (not shown, manuscript in preparation) and could be used for the initiation as well as for the elongation step of plasmid DNA replication. The DNA polymerase domain of the Rep proteins of pTN2 and pP12-1 are linked in C-terminal to a large domain of unknown function. In pRN-type plasmids, the DNA polymerase domain of the Rep proteins is fused in C-terminal to a helicase domain. This is probably not the case for pTN2 and pP12-1 since their C-terminal domain do not contain detectable ATP binding site. Indeed, the genes encoding the Rep-DNA polymerase in pTN2 and pP12-1 are contiguous to the genes encoding a putative SF1 helicase that is probably involved, together with the Rep proteins, in plasmid replication. It is tempting to speculate that the C-terminal domains of the pTN2 and pP12-1 Rep proteins bear an origin binding activity since the combination of such activity with their DNA polymerase/primase activities and the unwinding activity of the associated helicase SFI would reconstitute a complete DNA replication system for the plasmids.
The plasmid pT26-2 does not seem to encode a DNA polymerase and probably recruits a cellular DNA polymerase. Interestingly, this plasmid encodes a putative replicative helicase that would also correspond to a new family of replication protein. It is striking that the polymerase/helicase cassette of pTN2/pP12-1 and the putative replicative helicase of pT26-2 are both located downstream of the largest intergenic region present on each of the three plasmids. These regions are predicted to be replication origins by cumulative GC skew analysis and include many direct and inverted repeats, reminiscent of the ‘iterons’ typical of bacterial plasmid replication origins (55).
The three new plasmids of Thermococcales described here could become interesting models to study plasmid replication both in vitro and in vivo in hyperthermophilic archaea. Furthermore, the identification of their replication cassettes will help to use these plasmids in future vector construction for T. kodakaraensis, which is becoming the model species for genetic and molecular studies in hyperthermophilic archaea (28,30,56). It should be interesting for instance to express different genes from different compatible plasmids in the same strain for complementation studies. We already know that pTN1 and pTN2 are compatible, since they both are present in the same strain of T. nautilus (26). It should be also possible to use the SSV-type integrase of pT26-2 to insert groups of foreign genes in Thermococcales. Conversely, the genetic tools already available for T. kodakaraensis should help to analyse the biological functions of the proteins encoded by the three plasmids described here if, as in the case of pTN1, they can be propagated in T. kodakaraensis.
The plasmid pT26-2 encodes many homologues of proteins encoded by ‘virus-like elements’ present in Thermococcales and Methanococcales, including a SSV-type integrase, thus defining a large family of plasmid and integrated elements that probably predated the separation of these two archaeal orders. The pT26-2 family is characterized by the presence of seven ‘core genes’ present in all members of the family. Whereas pTN2 and pP12-1 can be considered as cryptic plasmids, the evolutionary relationships between pT26-2 and virus-like elements present in Thermococcales (TKV2/3, TGV1, PHV1) and Methanococcales (MMPV1, MMC7V1/2, MVV1, MMC6V1) suggests that pT26-2 plasmid could be a conjugative plasmid or a viral genome (or derived from such elements). Indeed, pT26-2 encode hydrophobic proteins that are clustered in one half of the plasmid, suggesting that they could be involved in the formation of protein complexes involved in DNA transfer, either for conjugation or viral infection. The putative glycoprotease activity of the large and conserved membrane protein t26-5p also suggests that pT26-2 and related elements indeed have the ability to destroy and/or modify the cell envelope to allow DNA transfer either during conjugation or viral infection. Krupovic and Bamford (57) have shown that one of the genes present in TKV4 encodes a capsid protein of the PRD1-adenovirus type (with a double-jelly roll fold), suggesting that TKV4 is a bona fide virus. In contrast, we could not detect gene encoding putative capsid proteins in pT26-2 or related elements. We also did not detect virus particles in cultures of T. nautilus, or in cultures of the Thermococcales containing related elements (54). These strains produce virus-like vesicles and some intracellular DNA is strongly associated to vesicles of T. gammatolerans (54) but we failed to specifically detect DNA from pT26-2 or related elements in these vesicles, nor proteins encoded by these elements (data not shown).
The origin and mode of evolution of plasmids and viruses, as well as their position in the tree of life, remain controversial topics (58–63). It is sometimes assumed that these biological entities essentially evolve by recruiting cellular genes (62). However, in contradiction with this view, homologues of cellular genes are usually rare in viral genomes and plasmids, with the exception of cellular genes that are present in integrated viruses and/or plasmids (58–60). Indeed, none of the proteins encoded by the three plasmids studied here has cellular homologues, except for genes mostly present within plasmid and/or viruses integrated in cellular genomes. The putative SFI helicases of pTN2 and pP12-1 are homologous to bacterial UvrD-type helicases, however, they are more closely related to putative helicases present in archaeal genomes which are most likely of viral origin (Supplementary Figure S4). This indicates that none of the proteins encoded by the three plasmids studied here correspond to a bona fide cellular gene that has been recently captured from a host cell.
The presence of a high proportion of ORFans among plasmid and viral genes is another specific feature that cannot be explained easily in the traditional view of virus/plasmid evolution. The question of the origin and the nature of cellular and viral ORFans has been raised repeatedly (64,65). Most of them encode short proteins, and it has been argued sometimes that these ORFs do not encode bona fide proteins. The three plasmids studied here also encode a relatively large proportion of small ORFs. We can assume that most of them are real genes because they are conserved either between pTN2 and pP12-1, or between pT26-2 and the virus-like elements present in the genomes of Thermococcales and Methanococcales. The characterization of a large family of pT26-2 related elements present in the genomes of Thermococcales and Methanococcales illustrates our recent observation, made at larger scale, that viral/plasmid genes represent a large reservoir of genes that constantly invade bacterial and archaeal genomes (41).
Plasmids and viruses often encode large proteins that are involved in genome replication or in the formation of structural apparatus, such as virions or DNA transferring machinery. These genes, that have either no cellular homologues or only distantly related ones can be considered as viral specific proteins (59), or virus hallmark protein (60). The three plasmids studied here encode several examples of such proteins. This is the case for instance of the two large proteins conserved in all members of the pT26-2 family (t26-5p and t26-6p). Structural analysis indeed confirmed the uniqueness of one of them, the protein t26-6p, which contains three novel folds (53). Many of the large proteins encoded by plasmids and DNA viruses are involved in DNA replication, repair recombination and/or integration, and this is indeed the case in the present study. These proteins are usually conserved in all members of the same plasmid/viral family (as the integrases of the pT26-2 family or the Rep-DNA polymerases and the putative SFI helicases of the pTN2 family) but they can be also shared by different plasmid families (as in the case of the pTN2-type helicase SFI, the DNA polymerase/primase or else the SSV1-like integrases).
To explain the existence of many proteins involved in DNA replication, repair or recombination among viral specific proteins, it has been suggested that DNA and DNA-manipulating enzymes originated first in an ancient virosphere, and that only a subset of these proteins were later on transferred into modern cellular lineages (61,66,67). This hypothesis predicts that many new types and families of viral-specific DNA-manipulating enzymes remain to be discovered in the plasmid/viral world. The present work fulfils this prediction since, by studying only three plasmids, we discovered a new family of DNA polymerase and probably a new family of helicases. It gives us the exiting expectation that more new families of enzymes involved in DNA metabolism await to be discovered in systematic analyses of yet uncharacterized proteins encoded by plasmids and viruses.
Interestingly, plasmids and related viruses of Thermococcales studied here have no close homologues encoded by bacterial or eukaryotic viruses/plasmids but only in archaeal plasmids and viruses, thus confirming that the three domains have largely independent viral reservoirs (68). The pTN2 and pP12-1 plasmids share three genes with the virus PAV1 from P. abyssi, which reminds the presence of SSV1-like genes in the genome of the Sulfolobus pRN-type plasmids pSSVx (18). A homologue of a PAV1 gene has also been detected recently in TKV4 from T. kodakaraensis (57), showing that gene exchange between PAV1 and plasmids has not been limited to the pTN2 family. Finally, the pTN2 and pP12-1 putative SFI helicase is also present in the virus-like element TGV2 from T. gammatolerans. All these observations suggest that viruses and plasmids infecting and/or present in Thermococcales share a common pool of genes that can be shifted from one genome to the other by recombination. The same phenomenon has been observed in Bacteria. For instance, Krupovic and Bamford (69) have shown that some viruses of the Corticoviridae family, whose prototype is the bacteriovirus PM2, use a replication machinery of plasmid origin instead of the viral DNA replication machinery of PM2.
It is often assumed that the evolution of plasmid and viruses cannot be reconstructed because they are constantly involved in horizontal gene transfer between various cellular lineages (62). However, a recent phylogenomic analysis of 16 genomes of T4-related viruses identified a conserved core of 24 genes that all (but one) exhibit a similar phylogenetic pattern, indicating that these viruses mainly evolved vertically (70). Similarly, phylogenetic analysis of the core proteins of the pT26-2 family is congruent with those of Thermococcales (using Methanococcales as outgroup, see Supplementary Figure S12). It has been recently observed that viruses and their hosts exhibit similar biogeographic patterns in genomic studies focusing on the archaeal species S. islandicus (71). This shows that cells and their associated viruses and plasmids co-evolved at the level of species. The analysis of the proteins encoded by the three plasmids from Thermococcales studied here suggests that co-evolution occurred at the order level as well. Indeed, the most closely related homologues of the pTN2, pP12-1 and pT26-2 proteins are mostly found in viruses, plasmids or integrated elements of Thermococcales. If Thermococcales proteins are removed from the analysis, the most closely related homologues are frequently found in Methanococcales, which are closely related to Thermococcales in phylogenetic trees of Archaea based on ribosomal proteins or RNA polymerase subunits (38,39). Our large-scale analysis of the relationship between the pT26-2 family and other plasmids or integrated elements (CAGs) present in Archaea confirms this view, since the network of evolutionary interactions between these elements overlaps with the phylogenetic pattern of the archaeal domain (Figure 4 and Supplementary Figure S11B). These results show that gene transfers between viral, plasmids and cellular lineages have not obliterated the phylogenetic signal that testifies for their co-evolution with their hosts. This has important implications for the current debate about the inclusion of viruses in the tree of life. Indeed, although viruses and plasmids cannot be placed in a tree based on universally conserved proteins, they can be probably included as companions in a universal tree of life based on the evolution of cellular species. We think that further systematic analyses of free or integrated plasmids and viruses in all archaeal, bacterial and eukaryotic groups are now mandatory in order to draw a comprehensive tree of life unifying all types of living entities present in the biosphere.
ACCESSION NUMBERS
pTN2, pP12-1 and pT26-2 have respectively been submitted to the GenBank database under accession numbers GU056177, GU056178 and GU056179.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence Nationale de la Recherche with grant GIS Génomique Marine (ANR-05-BLAN-0408-01); PhD grant from the Ministère de l’Enseignement supérieur et de la Recherche to N.S. ARC (Association pour la Recherche contre le Cancer). Funding for open access charge: Institut Pasteur.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Lipps G, editor. Plasmids Current Research and Future Trends. Norfolk, UK: Caaister Academic Press; 2008. p. 259. [Google Scholar]
- 2.Harriott OT, Huber R, Stetter KO, Betts PW, Noll KM. A cryptic miniplasmid from the hyperthermophilic bacterium Thermotoga sp. strain RQ7. J. Bacteriol. 1994;176:2759–2762. doi: 10.1128/jb.176.9.2759-2762.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soppa J. From genomes to function: haloarchaea as model organisms. Microbiology. 2006;152:585–590. doi: 10.1099/mic.0.28504-0. [DOI] [PubMed] [Google Scholar]
- 4.Baliga NS, Bonneau R, Facciotti MT, Pan M, Glusman G, Deutsch EW, Shannon P, Chiu Y, Weng RS, Gan RR, et al. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 2004;14:2221–2234. doi: 10.1101/gr.2700304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipps G. Archaeal plasmids. In: Lipps G, editor. Plasmids Current Research and Future Trends. Norfolk, UK: Caaister Academic Press; 2008. pp. 25–50. [Google Scholar]
- 7.Lipps G. Plasmids and viruses of the thermoacidophilic crenarchaeote Sulfolobus. Extremophiles. 2006;10:17–28. doi: 10.1007/s00792-005-0492-x. [DOI] [PubMed] [Google Scholar]
- 8.Greve B, Jensen S, Phan H, Brugger K, Zillig W, She Q, Garrett RA. Novel RepA-MCM proteins encoded in plasmids pTAU4, pORA1 and pTIK4 from Sulfolobus neozealandicus. Archaea. 2005;1:319–325. doi: 10.1155/2005/159218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipps G. Molecular biology of the pRN1 plasmid from Sulfolobus islandicus. Biochem. Soc. Trans. 2009;37:42–45. doi: 10.1042/BST0370042. [DOI] [PubMed] [Google Scholar]
- 10.Greve B, Jensen S, Brugger K, Zillig W, Garrett RA. Genomic comparison of archaeal conjugative plasmids from Sulfolobus. Archaea. 2004;1:231–239. doi: 10.1155/2004/151926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erauso G, Stedman KM, van de Werken HJ, Zillig W, van der Oost J. Two novel conjugative plasmids from a single strain of Sulfolobus. Microbiology. 2006;152:1951–1968. doi: 10.1099/mic.0.28861-0. [DOI] [PubMed] [Google Scholar]
- 12.Lipps G, Ibanez P, Stroessenreuther T, Hekimian K, Krauss G. The protein ORF80 from the acidophilic and thermophilic archaeon Sulfolobus islandicus binds highly site-specifically to double-stranded DNA and represents a novel type of basic leucine zipper protein. Nucleic Acids Res. 2001;29:4973–4982. doi: 10.1093/nar/29.24.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipps G, Stegert M, Krauss G. Thermostable and site-specific DNA binding of the gene product ORF56 from the Sulfolobus islandicus plasmid pRN1, a putative archael plasmid copy control protein. Nucleic Acids Res. 2001;29:904–913. doi: 10.1093/nar/29.4.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipps G, Rother S, Hart C, Krauss G. A novel type of replicative enzyme harbouring ATPase, primase and DNA polymerase activity. EMBO J. 2003;22:2516–2525. doi: 10.1093/emboj/cdg246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipps G. The replication protein of the Sulfolobus islandicus plasmid pRN1. Biochem. Soc. Trans. 2004;32:240–244. doi: 10.1042/bst0320240. [DOI] [PubMed] [Google Scholar]
- 16.Lipps G, Weinzierl AO, von Scheven G, Buchen C, Cramer P. Structure of a bifunctional DNA primase-polymerase. Nat. Struct. Mol. Biol. 2004;11:157–162. doi: 10.1038/nsmb723. [DOI] [PubMed] [Google Scholar]
- 17.Peng X. Evidence for the horizontal transfer of an integrase gene from a fusellovirus to a pRN-like plasmid within a single strain of Sulfolobus and the implications for plasmid survival. Microbiology. 2008;154:383–391. doi: 10.1099/mic.0.2007/012963-0. [DOI] [PubMed] [Google Scholar]
- 18.Arnold HP, She Q, Phan H, Stedman K, Prangishvili D, Holz I, Kristjansson JK, Garrett R, Zillig W. The genetic element pSSVx of the extremely thermophilic crenarchaeon Sulfolobus is a hybrid between a plasmid and a virus. Mol. Microbiol. 1999;34:217–226. doi: 10.1046/j.1365-2958.1999.01573.x. [DOI] [PubMed] [Google Scholar]
- 19.Stedman KM, She Q, Phan H, Arnold HP, Holz I, Garrett RA, Zillig W. Relationships between fuselloviruses infecting the extremely thermophilic archaeon Sulfolobus: SSV1 and SSV2. Res. Microbiol. 2003;154:295–302. doi: 10.1016/S0923-2508(03)00074-3. [DOI] [PubMed] [Google Scholar]
- 20.Basta T, Smyth J, Forterre P, Prangishvili D, Peng X. Novel archaeal plasmid pAH1 and its interactions with the lipothrixvirus AFV1. Mol. Microbiol. 2009;71:23–34. doi: 10.1111/j.1365-2958.2008.06488.x. [DOI] [PubMed] [Google Scholar]
- 21.Albers SV, Jonuscheit M, Dinkelaker S, Urich T, Kletzin A, Tampe R, Driessen AJ, Schleper C. Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl. Environ. Microbiol. 2006;72:102–111. doi: 10.1128/AEM.72.1.102-111.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berkner S, Grogan D, Albers SV, Lipps G. Small multicopy, non-integrative shuttle vectors based on the plasmid pRN1 for Sulfolobus acidocaldarius and Sulfolobus solfataricus, model organisms of the (cren-)archaea. Nucleic Acids Res. 2007;35:e88. doi: 10.1093/nar/gkm449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erauso G, Marsin S, Benbouzid-Rollet N, Baucher MF, Barbeyron T, Zivanovic Y, Prieur D, Forterre P. Sequence of plasmid pGT5 from the archaeon Pyrococcus abyssi: evidence for rolling-circle replication in a hyperthermophile. J. Bacteriol. 1996;178:3232–3237. doi: 10.1128/jb.178.11.3232-3237.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsin S, Forterre P. A rolling circle replication initiator protein with a nucleotidyl-transferase activity encoded by the plasmid pGT5 from the hyperthermophilic archaeon Pyrococcus abyssi. Mol. Microbiol. 1998;27:1183–1192. doi: 10.1046/j.1365-2958.1998.00759.x. [DOI] [PubMed] [Google Scholar]
- 25.Marsin S, Forterre P. The active site of the rolling circle replication protein Rep75 is involved in site-specific nuclease, ligase and nucleotidyl transferase activities. Mol. Microbiol. 1999;33:537–545. doi: 10.1046/j.1365-2958.1999.01498.x. [DOI] [PubMed] [Google Scholar]
- 26.Soler N, Justome A, Quevillon-Cheruel S, Lorieux F, Le Cam E, Marguet E, Forterre P. The rolling-circle plasmid pTN1 from the hyperthermophilic archaeon Thermococcus nautilus. Mol. Microbiol. 2007;66:357–370. doi: 10.1111/j.1365-2958.2007.05912.x. [DOI] [PubMed] [Google Scholar]
- 27.Ward DE, Revet IM, Nandakumar R, Tuttle JH, de Vos WM, van der Oost J, DiRuggiero J. Characterization of plasmid pRT1 from Pyrococcus sp. strain JT1. J. Bacteriol. 2002;184:2561–2566. doi: 10.1128/JB.184.9.2561-2566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santangelo TJ, Cubonova L, Reeve JN. Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 2008;74:3099–3104. doi: 10.1128/AEM.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucas S, Toffin L, Zivanovic Y, Charlier D, Moussard H, Forterre P, Prieur D, Erauso G. Construction of a shuttle vector for, and spheroplast transformation of, the hyperthermophilic archaeon Pyrococcus abyssi. Appl. Environ. Microbiol. 2002;68:5528–5536. doi: 10.1128/AEM.68.11.5528-5536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato T, Fukui T, Atomi H, Imanaka T. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 2005;71:3889–3899. doi: 10.1128/AEM.71.7.3889-3899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lepage E, Marguet E, Geslin C, Matte-Tailliez O, Zillig W, Forterre P, Tailliez P. Molecular diversity of new Thermococcales isolates from a single area of hydrothermal deep-sea vents as revealed by randomly amplified polymorphic DNA fingerprinting and 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 2004;70:1277–1286. doi: 10.1128/AEM.70.3.1277-1286.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 34.Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 36.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-funciton relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 37.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 38.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 39.Brochier C, Forterre P, Gribaldo S. An emerging phylogenetic core of Archaea: phylogenies of transcription and translation machineries converge following addition of new genome sequences. BMC Evol. Biol. 2005;5:36. doi: 10.1186/1471-2148-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zivanovic Y, Armengaud J, Lagorce A, Leplat C, Guerin P, Dutertre M, Anthouard V, Forterre P, Wincker P, Confalonieri F. Genome analysis and genome-wide proteomics of Thermococcus gammatolerans, the most radioresistant organism known amongst the Archaea. Genome Biol. 2009;10:R70. doi: 10.1186/gb-2009-10-6-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cortez D, Forterre P, Gribaldo S. A hidden reservoir of integrative elements is the major source of recently acquired foreign genes and ORFans in archaeal and bacterial genomes. Genome Biol. 2009;10:R65. doi: 10.1186/gb-2009-10-6-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iyer LM, Koonin EV, Leipe DD, Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005;33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark JM. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 1988;16:9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mole SE, Iggo RD, Lane DP. Using the polymerase chain reaction to modify expression plasmids for epitope mapping. Nucleic Acids Res. 1989;17:3319. doi: 10.1093/nar/17.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berkner S, Lipps G. Characterization of the transcriptional activity of the cryptic plasmid pRN1 from Sulfolobus islandicus REN1H1 and regulation of its replication operon. J. Bacteriol. 2007;189:1711–1721. doi: 10.1128/JB.01586-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geslin C, Gaillard M, Flament D, Rouault K, Le Romancer M, Prieur D, Erauso G. Analysis of the first genome of a hyperthermophilic marine virus-like particle, PAV1, isolated from Pyrococcus abyssi. J. Bacteriol. 2007;189:4510–4519. doi: 10.1128/JB.01896-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 48.Bernad A, Blanco L, Lazaro JM, Martin G, Salas M. A conserved 3′-5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 49.Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 2005;15:352–363. doi: 10.1101/gr.3003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanchez M, Drechsler M, Stark H, Lipps G. DNA translocation activity of the multifunctional replication protein ORF904 from the archaeal plasmid pRN1. Nucleic Acids Res. 2009;37:6831–6848. doi: 10.1093/nar/gkp742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 52.Asen I, Djuranovic S, Lupas AN, Zeth K. Crystal structure of SpoVT, the final modulator of gene expression during spore development in Bacillus subtilis. J. Mol. Biol. 2009;386:962–975. doi: 10.1016/j.jmb.2008.10.061. [DOI] [PubMed] [Google Scholar]
- 53.Keller J, Leulliot N, Soler N, Collinet B, Vincentelli R, Forterre P, van Tilbeurgh H. A protein encoded by a new family of mobile elements from Euryarchaea exhibits three domains with novel folds. Protein Sci. 2009;18:850–855. doi: 10.1002/pro.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soler N, Marguet E, Verbavatz JM, Forterre P. Virus-like vesicles and extracellular DNA produced by hyperthermophilic archaea of the order Thermococcales. Res. Microbiol. 2008;159:390–399. doi: 10.1016/j.resmic.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 55.Chattoraj DK. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol. 2000;37:467–476. doi: 10.1046/j.1365-2958.2000.01986.x. [DOI] [PubMed] [Google Scholar]
- 56.Santangelo TJ, Cubonova L, Matsumi R, Atomi H, Imanaka T, Reeve JN. Polarity in archaeal operon transcription in Thermococcus kodakaraensis. J. Bacteriol. 2008;190:2244–2248. doi: 10.1128/JB.01811-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krupovic M, Bamford DH. Archaeal proviruses TKV4 and MVV extend the PRD1-adenovirus lineage to the phylum Euryarchaeota. Virology. 2008;375:292–300. doi: 10.1016/j.virol.2008.01.043. [DOI] [PubMed] [Google Scholar]
- 58.Forterre P. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 2006;117:5–16. doi: 10.1016/j.virusres.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 59.Forterre P. The two ages of the RNA world, and the transition to the DNA world: a story of viruses and cells. Biochimie. 2005;87:793–803. doi: 10.1016/j.biochi.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 60.Koonin EV, Senkevich TG, Dolja VV. The ancient Virus World and evolution of cells. Biol. Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forterre P, Prangishvili D. The origin of viruses. Res. Microbiol. 2009;160:466–472. doi: 10.1016/j.resmic.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 62.Moreira D, Lopez-Garcia P. Ten reasons to exclude viruses from the tree of life. Nat. Rev. Microbiol. 2009;7:306–311. doi: 10.1038/nrmicro2108. [DOI] [PubMed] [Google Scholar]
- 63.Raoult D, Forterre P. Redefining viruses: lessons from Mimivirus. Nat. Rev. Microbiol. 2008;6:315–319. doi: 10.1038/nrmicro1858. [DOI] [PubMed] [Google Scholar]
- 64.Yin Y, Fischer D. On the origin of microbial ORFans: quantifying the strength of the evidence for viral lateral transfer. BMC Evol. Biol. 2006;6:63. doi: 10.1186/1471-2148-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yin Y, Fischer D. Identification and investigation of ORFans in the viral world. BMC Genomics. 2008;9:24. doi: 10.1186/1471-2164-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Forterre P. The origin of DNA genomes and DNA replication proteins. Curr. Opin. Microbiol. 2002;5:525–532. doi: 10.1016/s1369-5274(02)00360-0. [DOI] [PubMed] [Google Scholar]
- 67.Forterre P. Displacement of cellular proteins by functional analogues from plasmids or viruses could explain puzzling phylogenies of many DNA informational proteins. Mol. Microbiol. 1999;33:457–465. doi: 10.1046/j.1365-2958.1999.01497.x. [DOI] [PubMed] [Google Scholar]
- 68.Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: a unifying view. Nat. Rev. Microbiol. 2006;4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- 69.Krupovic M, Bamford DH. Putative prophages related to lytic tailless marine dsDNA phage PM2 are widespread in the genomes of aquatic bacteria. BMC Genomics. 2007;8:236. doi: 10.1186/1471-2164-8-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Filee J, Tetart F, Suttle CA, Krisch HM. Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proc. Natl Acad. Sci. USA. 2005;102:12471–12476. doi: 10.1073/pnas.0503404102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Held NL, Whitaker RJ. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ. Microbiol. 2009;11:457–466. doi: 10.1111/j.1462-2920.2008.01784.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.