Abstract
Polycomb group (PcG) proteins are transcriptional repressors, which regulate proliferation and cell fate decisions during development, and their deregulated expression is a frequent event in human tumours. The Polycomb repressive complex 2 (PRC2) catalyzes trimethylation (me3) of histone H3 lysine 27 (K27), and it is believed that this activity mediates transcriptional repression. Despite the recent progress in understanding PcG function, the molecular mechanisms by which the PcG proteins repress transcription, as well as the mechanisms that lead to the activation of PcG target genes are poorly understood. To gain insight into these mechanisms, we have determined the global changes in histone modifications in embryonic stem (ES) cells lacking the PcG protein Suz12 that is essential for PRC2 activity. We show that loss of PRC2 activity results in a global increase in H3K27 acetylation. The methylation to acetylation switch correlates with the transcriptional activation of PcG target genes, both during ES cell differentiation and in MLL-AF9-transduced hematopoietic stem cells. Moreover, we provide evidence that the acetylation of H3K27 is catalyzed by the acetyltransferases p300 and CBP. Based on these data, we propose that the PcG proteins in part repress transcription by preventing the binding of acetyltransferases to PcG target genes.
INTRODUCTION
Polycomb group (PcG) proteins are transcriptional repressors that play an essential role in cell fate decisions during development (1,2). They exist in two distinct multiprotein Polycomb repressive complexes (PRCs) namely, PRC1 and PRC2 (2). The PRC2 complex contains the three PcG proteins EZH2, EED and SUZ12, and the SET domain of EZH2 catalyzes the di- and trimethylation (me2/me3) of histone H3K27 (3–6). In contrast to PRC2, which is a well-defined complex, the PRC1 complex is in reality not a single complex, but a multitude of complexes containing different PcG proteins. PRC1 catalyzes the ubiquitylation (Ubi) of histone H2A mainly through the ubiquitin E3 ligase activity of RING1B (1,2). The PRC1 and PRC2 complexes share a large number of common target genes and the majority of these genes encode for important developmental regulators (7–9). Consistent with this, the core subunits of PRC2 (Ezh2, Eed and Suz12) and PRC1 (Ring1b) are essential for mouse embryonic development at early postimplantation stages (10–13). PRC1 recruitment to target genes is dependent on the activity of the PRC2 complex, and it has been suggested that this could involve the specific binding of the chromodomain proteins of the PRC1 complex to H3K27me3 (3,5,14,15). Importantly, increased expression of different subunits of PRC2 (EED and EZH2) and PRC1 (BMI1) as well as translocations of the SUZ12 gene locus, are frequent events in human cancers (16–20). Moreover, increased PcG levels can contribute to transformation in vitro (EZH2, BMI1, CBX7 and CBX8) and in vivo (BMI1 and CBX7), supporting the notion that PcG proteins have oncogenic properties (16–18,21,22).
Despite recent results have provided substantial new knowledge regarding the biochemical and biological functions of PRC1 and PRC2, several aspects regarding the mechanisms by which the PcGs control transcription have not been addressed yet. This includes the molecular mechanisms by which H3K27me3 maintains transcriptional repression, as well as the mechanisms that regulate the activation of target genes upon loss of PcG binding. To obtain insights into the functional consequences of H3K27me3 loss, we have performed mass spectrometry on histones stably isotope labeled with amino acids in cell culture (SILAC) purified from both WT and Suz12 KO embryonic stem (ES) cells and quantified the global levels of histone modifications in the presence or absence of H3K27me3. By this approach, we have shown that the loss of H3K27me3 results in increased levels of H3K27Ac. Further experiments demonstrated that these increased levels of H3K27Ac are specifically dependent of the PRC2 activity and that increased H3K27Ac levels are located at the promoters of PcG target genes. Moreover, we show that the increase in H3K27Ac levels correlates with PcG displacement from promoters during both ES cell differentiation and upon MLL-AF9 transduction of hematopoietic stem and progenitor cells (HSPC). Finally, we provide evidence that both histone acetyltransferases (HAT), p300 and Cbp play an important role in histone H3K27Ac. Based on these results, we propose that preventing H3K27 acetylation is an important part of the mechanism by which PRC2 represses transcription.
MATERIALS AND METHODS
Cell culture and cell line generation
All ES cells were cultured on 0.1%/1× PBS gelatinized Tissue Culture (TC) plates (Nunc) in Glasgow media (Sigma) supplemented with 15% FBS (Hyclone), Penicillin/Streptomycin (P/S) (Gibco), Glutamax (Gibco), Non-Essential Amino Acids (Gibco), Sodium-Pyruvate (Gibco), β-mercaptoethanol (Gibco) and leukemia inhibitory factor.
For SILAC labelling: to obtain full incorporation of heavy isotope, the previously described Suz12−/− mouse ES cells (23) were cultured for 6 days in SILAC DMEM (Sigma) containing 15% dialyzed FBS (Gibco), P/S (Gibco), NEAA (Gibco), Pyruvate (Gibco), 50 mM β-mercaptoethanol, d-glucose (3.5 g/l), (105 mg/l; final 0.802 mM), ESGRO LIF (107 U/ml; Chemicon), 0.802 mM l-Leucine (Sigma), 0.398 mM l-Arginine (Sigma) and 0.798 mM l-Lysine (Sigma). Lys8 isotope (Cambridge Isotopes, CNLM-291) was used for the Suz12−/− ES cells.
Ezh2 conditional (loxP/loxP) ES cells were generated by TC expansion of the inner cell mass outgrowths of Ezh2 loxP/loxP E3.5 embryos (blastocysts) (24) as described previously (25). Ezh2−/− ES cells were generated by transient expression of CRE recombinase in Ezh2 loxP/loxP ES cells using Lipofectamine 2000 (Invitrogen) transfection reagent following manufacturer’s instructions. Ezh2−/− clones were identified by PCR genotyping and subsequentely expanded in TC as described above.
Suz12−/− rescued ES cells were generated by inactivation of the gene-trap cassette by transient CRE expression. Suz12−/− rescued ES cell clones were generated following the same procedure described for the Ezh2−/− ES cells. Positive ES cell clones were identified by western blot analysis using a specific Suz12 antibody (Santa Cruz).
Eed−/− cells (26), Gal4-EZH2 293T cells were described previously (27). MLL-AF9 hematopoietic stem cells (HSPCs) were generated as described previously (28). Cell transduction was modified using Retronectin instead of spinoculation as described previously (29). Viruses were produced using a pMSCV MLL-AF9 retroviral expression vectors described elsewhere (30). TC of FDCP-mix cells and differentiation into granulocytes was performed as described previously (31).
siRNA
siRNA oligos were purchased from Sigma. The different target sequences are available in Supplementary Table S1. siRNA oligos were delivered into ES cells using Lipofectamine 2000 (Invitrogen) following manufacturer’s instructions.
Histone purification and mass spectrometry
Histones were purified with the histone purification kit (Active Motif) according to the manufacturer’s instruction. Briefly, cells were lysed in extraction buffer at 4°C overnight on rotating platform. Cleared lysates were neutralized by addition of 5× neutralizing buffer and loaded on pre-equilibrated column packed with purification resin. Columns were extensively washed with histone wash buffer and histones were eluted in 0.5 ml fractions using elution buffer. Purified histones were separated on a SDS-PAGE 15% acrylamide gel and visualized by Coomassie staining. The concentration of purified histones was measured using Q-bit (Invitrogen). Heavy and light amino acid-labeled histones were mixed in a 1 : 1 ratio and a total of 200 μg histone mixture was separated by reverse phase-high performance liquid chromatography (RP-HPLC) using a C18 column (250 × 2 mm, Jupiter, 300 Å; Phenomenex, Torrance, CA, USA) on an Akta-Basic system (GE healthcare). The A buffer consisted of 0.06% trifluoroacetic acid (TFA) in ddH2O. The HPLC gradient, made of B buffer [0.04% TFA + 90% acetonitrile (MeCN, Sigma)], increased from 5–35% in 10 min, 35–60% in 60 min and 60–90% in 2 min. A detailed description of tandem mass spectrometry and data analysis of the purified histones is described elsewhere (32). Briefly, samples were digested with the endoprotease ArgC (Calbiochem) and the peptide mixtures were analyzed by easyLC (Proxeon, Odense, Denmark) interfaced to LTQ-Orbitrap (ThermoFisher Scientific, Bremen, Germany). The raw data from LTQ-Orbitrap was converted to mgf files using Proteome Discoverer 1.0 software (ThermoFisher Scientific). Database searching was performed against a custom-made database containing mouse histones retrieved from Uniprot using Mascot Daemon version 2.1.0 (Matrix Science).
ChIP analysis
ChIP analyses were performed as described (33). Briefly, cells were fixed in 1% formaldehyde/1× PBS for 10 min. Then they were blocked with 0.125 M glycine for 5 min, washed extensively in 1× PBS, collected in SDS buffer, pelleted and re-suspended in IP buffer. Samples were sonicated with the Diagenode Bioruptor in 1.5 ml for 8 min at high power and chromatin sonication controlled on 2% agarose gels. The DNA was sonicated to ∼700–400 bp in all experiments. For each IP, ∼1 mg of chromatin was used. Primary antibodies were incubated overnight at 4°C on a rotating platform. To each sample, 30 µl of 50% slurry of protein A-Sepharose (Amersham) beads were added for 2–3 h. Beads were washed three times in 150 mM wash buffer and one time in 500 mM wash buffer. Beads (and input samples) were resuspended in 120 µl of 0.1% SDS, 0.1 M NaHCO3 buffer and de-cross-linked at 65°C for a minimum of 3 h. DNA was purified using Qiagen PCR purification kit following the manufacturer’s instruction and eluted in 200 µl of H2O. Eluted material of 1–2 µl was used for each real-time quantitative PCR (qPCR) reaction.
Antibodies
For western blot, the following antibodies were used: rabbit-anti H3K27me3 (Cell Signaling Technology, 9733S); rabbit-antiH3K27Ac (Upstate/Millipore, 07-360); rabbit-antiH3K4me3 (Cell Signaling Technology, 9751S); rabbit-anti Histone H3 (Abcam, ab1791); goat-anti Suz12 (Santa Cruz, sc-46264); rabbit-anti β-Tubulin (Santa Cruz, sc-9104); rabbit-anti H3K9Ac (Upstate, 06-942); rabbit-anti H3K9me3 (Upstate, 07-442); rabbit-anti H3K14Ac (Abcam, ab46984); rabbit-anti Gal4 (Santa Cruz, sc-510); rabbit-anti P300 (Santa Cruz, sc-585); rabbit-anti CBP (Santa Cruz, sc-369); mouse-anti Vinculin (Abcam, ab18058); and mouse-anti Ezh2 as described previously (34).
For ChIP assays the following antibodies were used: rabbit-anti H3K27me3 (Cell Signaling Technology, 9733S); rabbit-antiH3K27Ac (Abcam, ab4729); rabbit-anti H3K9Ac (Upstate, 06-942); rabbit-anti acetyl H3 (Upstate, 17-615); rabbit-anti Suz12 (Cell Signaling Technology, 3737); rabbit-anti GAL4 (Santa Cruz, sc-510); and rabbit-anti EZH2 as described previously (12).
RNA extraction and expression analyses
RNA was purified from cells using the RNeasy extraction kit (Qiagen) following the manufacturer’s instructions. cDNA was prepared with TaqMan Reverse Transcriptase kit (Applied Biosystems) following manufacturer’s instructions. Oligo-dT retro-transcribed RNA of ∼5 ng was used in each real-time qPCR reaction.
Real-time qPCR primers
Primers sequences for both expression and ChIP analyses are available in Supplementary Table S2.
Quantification of western blot analyses
Intensities of western blot bands were determined using ImageJ software (rsbweb.nih.gov/ij/). Quantifications for each experiment are calculated as the average of the intensities of the H3K27Ac/H3 ratio calculated on increasing exposures.
RESULTS
With the goal of achieving insights into the mechanisms of PRC2-dependent transcriptional regulation, we decided to investigate if the trimethylation of H3K27 influences other posttranslational modifications of the histones. To do this, we used Suz12 KO mouse ES cells as a model system for global loss of H3K27me3 and quantified by SILAC mass spectrometry, the relative changes of a large number of other histone modifications. We grew Suz12 KO ES cells (12) for 6 days (equal to approximately 15 population doublings) in a media that contained heavy-isotope-labeled lysine (Lys8) and Suz12 WT ES cells in media containing light-isotope-labeled lysine (Lys0) (Figure 1A, left panel). The histones were purified from these cells and analyzed by nanoLC-tandem mass spectrometry to determine the relative abundance of all histone modifications in the two cell lines. The technical aspect and the overall results of this analysis are described in a separate manuscript (32). Interestingly, this analysis showed that the most significant posttranslational change of the histones in the Suz12 KO ES cells, apart from the global loss of H3K27me2 and H3K27me3 [Figure 1A and (32)], was a significant increase of H3K27Ac (Figure 1A, right panel). This observation could suggest the existence of a posttranslational switch between the acetylation and methylation of H3K27 controlled by PRC2.
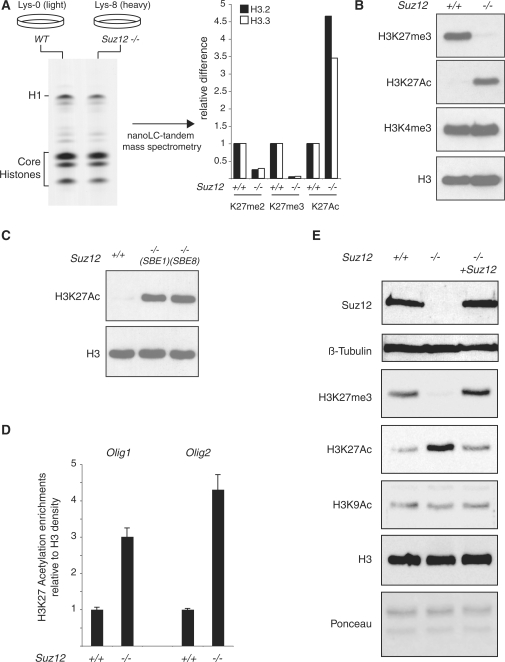
Figure 1.
Loss of Suz12 induces H3K27 hyperacetylation. (A) (left panel) Coomassie-blue staining of SILAC-labeled histones purified from light-isotope-labeled (Lys-0) WT ES cells and heavy-isotope-labeled (Lys-8) Suz12 KO ES cells. Nanolc-tandem mass spectrometry quantification of the K27 methylation and acetylation levels of H3.2 and H3.3 in WT and Suz12 KO ES cells (right panel). (B) Western blot analyses of histones purified from WT and Suz12 KO ES cells using the indicated antibodies. H3 is presented as loading control. (C) Western blot analyses of histones purified from WT and two independent Suz12 KO ES cell lines using the indicated antibodies. H3 is presented as loading control. (D) ChIP analysis of the Olig1 and Olig2 promoter in WT and Suz12 KO ES cells using an H3K27Ac-specific antibody. H3K27Ac signal is normalized to histone density using an H3-specific antibody. (E) Western blot analyses of total protein extracts obtained from WT and Suz12 KO ES cell before and after CRE expression using the indicated antibodies. β-tubulin, H3 and Ponceau staining are presented as loading controls.
To validate the mass spectrometry results, we performed western blot analysis using antibodies specific for H3K27me3 and H3K27Ac on histones purified from WT and Suz12 KO ES cells. In agreement with the mass spectrometry data, loss of Suz12 results in a significant increase of global H3K27Ac and a loss of H3K27me3 (Figure 1B). This result was confirmed in an independently isolated Suz12 KO ES cell line (12) (Figure 1C), strongly suggesting that the increased H3K27Ac levels are a specific consequence of Suz12 loss.
These findings highlight the possibility that the switch between H3K27 methylation and acetylation may play a role in the transcriptional activation that follows displacement of PcG proteins from promoters. Furthermore, they may suggest that preventing H3K27 acetylation could be part of the mechanism by which PRC2 controls transcription. In order to obtain further evidence that this switch occurs specifically at the promoters of PRC2 target genes, we tested the H3K27Ac levels at the Olig1 and Olig2 promoters (8) in WT and Suz12 KO ES cells by ChIP analysis. Consistent with the results presented in Figure 1B and C, loss of Suz12 results in a specific increase of H3K27Ac of the Olig1 and Olig2 promoters (Figure 1D).
The global increase of H3K27Ac in Suz12 KO ES cells suggests that the PcG proteins antagonize an H3K27 acetyltransferase activity. Thus, we tested if re-expression of Suz12 in the KO ES cells could restore wild type levels of H3K27 posttranslational modifications. To do this, we inactivated the gene-trap cassette (12) by CRE-mediated excision of the splice-acceptor site situated upstream of the β-galactosidase-neomycin cassette. Western blot analysis of Suz12 KO ES cells before and after CRE expression demonstrated that the inactivation of the gene-trap cassette restores physiological level of Suz12 expression (Figure 1E). Importantly, western blot analyses on histones purified from the same cells demonstrated that the re-expression of Suz12 restores global H3K27me3 levels and, at the same time, decreases H3K27Ac to levels comparable to those observed in WT ES cells (Figure 1E). Taken together, these results strongly suggest that the PcG proteins prevent H3K27 acetylation of target genes.
Next, we wanted to analyze if loss of other components of the PRC2 complex also leads to increased global levels of H3K27Ac. To do this, we analyzed the H3K27me3 and H3K27Ac levels in ES cell lines lacking different components of PRC2. Western blot analyses of histones purified from either WT or Suz12, Eed and Ezh2 KO ES cells demonstrated that all the PcG subunits of the PRC2 complex are essential for H3K27 trimethylation (Figure 2A). Moreover, loss of H3K27me3 leads to a global increase of H3K27Ac in all the different PRC2 KO ES cell lines (Figure 2A). Finally, ChIP analyses in Suz12 and Eed KO ES cells showed that H3K27Ac is specifically increased at the Olig2 promoter when compared to WT ES cells (Figure 2B), confirming the results presented in Figure 1B and D.
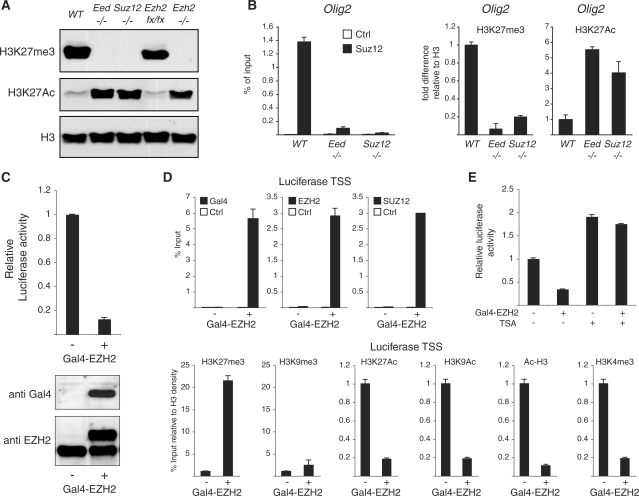
Figure 2.
PRC2 activity regulates H3K27Ac levels. (A) Western blot analyses of histones purified from WT, Eed−/−, Suz12−/−, Ezh2 conditional (Ezh2 loxP/loxP) and Ezh2−/− ES cells using the indicated antibodies. H3 is presented as loading control. (B) ChIP analysis of the Olig2 promoter in WT, Suz12 and Eed KO ES cells using the indicated antibodies. Signals are normalized to histone density using an H3-specific antibody. (C) Western blot analyses using the indicated antibodies and luciferase activity of 293T cells containing a stable integration of a heterologous Gal4/luciferase reporter construct before and after Gal4-EZH2 expression. (D) ChIP analysis of the luciferase TSS in the cells presented in (C) using the indicated antibodies. Gal4, EZH2 and SUZ12 enrichments are presented as percentage of input while the different histone modifications signals are normalized to histone density using an H3-specific antibody.
To directly analyze if PRC2 recruitment to target genes excludes H3K27Ac, we took advantage of a reporter system developed in our laboratory that combines the integration of a heterologous luciferase reporter construct containing five Gal4 DNA binding sites with the stable expression of a Gal4-EZH2 fusion protein (27). As previously reported (27), Gal4-EZH2 expression leads to a strong repression of luciferase activity (Figure 2C). Moreover, ChIP analysis using Gal4-, EZH2- and SUZ12-specific antibodies demonstrated that EZH2 binding to the artificial promoter recruits endogenous components of the PRC2 complex (Figure 2D, upper panel). Importantly, PRC2 recruitment to the luciferase promoter correlates with increased H3K27me3 levels and a significant decrease of H3K27Ac (Figure 2D, lower panel). Moreover, EZH2 recruitment to the luciferase promoter does not lead to an enrichment of H3K9me3, but to a loss of H3K9 acetylation and possibly of other acetylated H3 residues as indicated by the decrease in global H3 acetylation (see Figure 2D; Ac-H3 ChIP measuring K14/K9 acetyl H3). Consistent with previous publications (27,33), EZH2 recruitment also correlates with a strong decrease of H3K4me3 (Figure 2D). In order to understand the contribution of lysine de-acetylation in EZH2-mediated transcriptional repression, we treated the cells presented in Figure 2C with the HDAC inhibitor Trichostatin-A (TSA). As shown in Figure 2E, 6 h treatment with TSA abolished the repressive activity of EZH2. All together, these data show that PRC2-mediated trimethylation of H3K27 is sufficient to displace and/or prevent acetylation of histone H3 at PcG target genes.
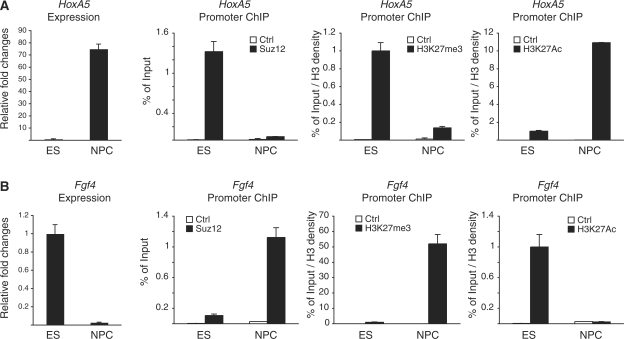
The differentiation of ES cells to neural precursor cells (NPC) leads to the displacement of the PcG proteins from ∼50% of their target genes and to their recruitment to a similar number of other target genes (35). In both situations, binding of PcG proteins correlates with repressed transcription, whereas loss of PcG binding correlates with transcriptional activation (35). To analyze if H3K27 acetylation is involved in the transcriptional activation of PcG target genes, we differentiated ES cells into NPCs, and characterized PcG binding and H3K27 modifications by ChIP analyses. We focused on two genes whose expression changes in opposite direction during differentiation. One gene, Hoxa5, is repressed in ES cells and transcribed in NPC cells (Figure 3A, left panel). The other gene, Fgf4, is expressed at high levels in ES cells and silenced in NPC cells (Figure 3B, left panel). To analyze PRC2 recruitment and the modification status of H3K27 in these two conditions, we performed ChIP analysis using Suz12-, H3K27me3- and H3K27Ac-specific antibodies in ES and NPC cells. Consistent with Hoxa5-specific NPC expression, the Suz12 binding and the H3K27me3 levels at the Hoxa5 promoter are strongly reduced in NPC cells (Figure 3A). Moreover, loss of PRC2 activity in NPC is associated with a strong increase of H3K27Ac levels that correlates with Hoxa5 transcriptional activation (Figure 3A). In contrast, Suz12 and H3K27me3 are not found associated with the Fgf4 promoter in ES cells (Figure 3B). Importantly, lack of PcG activity at Fgf4 promoter correlates with a strong enrichment of H3K27Ac and with high expression of Fgf4 in ES cells (Figure 3B). Differentiation of ES cells to NPC leads to a strong repression of Fgf4 transcription that correlates with the recruitment of Suz12, the loss of H3K27Ac and the enrichment of H3K27me3 (Figure 3B). Together, these data provide strong evidence for a competition between H3K27me3 and H3K27Ac in regulating gene expression during ES cell differentiation.
Figure 3.
Regulation of H3K27me3 and H3K27Ac during ES cell differentiation. (A and B) qPCR expression (left panels) and ChIP analyses (right panels) of the Hoxa5 and Fgf4 promoters in ES and NPC cells using the indicated antibodies. Suz12 enrichments are presented as percentage of input while H3K27Ac and H3K27me3 signals are normalized to histone density using an H3-specific antibody.
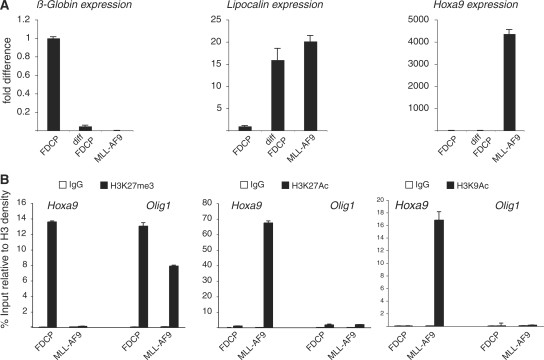
Homeotic genes (HOX) are the best-characterized PcG target genes. HOX genes play an essential role in the regulation of normal development (36). Moreover, deregulation of HOX expression has been linked to the development of different forms of human cancer (37). For example, HOXA9 overexpression in HSPC is important for HSPC immortalization (38), and the specific activation of HOXA9 expression is a feature of several leukemic fusion proteins including MLL-AF9 (39–41). To investigate if the increased expression of Hoxa9 in immortalized HSPC involves transcriptional mechanism similar to the one described above for ES cell differentiation, we compared MLL-AF9 immortalized c-kit+ HSPC with a multipotent hematopoietic progenitor cell line FDCP-mix [the FDCP-mix cells were chosen to allow the expansion in TC of normal hematopoietic progenitors (31)]. As previously reported (28,42), MLL-AF9 immortalized HSPCs are blocked at the progenitor stage of the granulocytic differentiation pathways as confirmed by the expression of the granulocytic marker Lipocalin (Figure 4A). Lipocalin expression is silenced in the FDCP-mix cells and is activated to similar levels as in MLL-AF9 expressing cells when induced to differentiate into granulocytes (Figure 4A). Importantly, Hoxa9 expression was specifically detected in the MLL-AF9 expressing cells, but not in the differentiating FDCP-mix cells demonstrating the direct role of MLL-AF9 in Hoxa9 transcriptional activation (Figure 4A).
Figure 4.
Regulation of H3K27me3 and H3K27Ac target gene binding in MLL-AF9 HSPCs and FDCP-mix cells. (A) β-Globin, Lipocalin and Hoxa9 qPCR expression analyses in FDCP-mix cells before and after granulocytic differentiation and in MLL-AF9-expressing HSPC. (B) ChIP analyses of Hoxa9 and Olig1 promoters in FDCP-mix cells and MLL-AF9-expressing HSPCs using the indicated antibodies. H3K27Ac, H3K9Ac and H3K27me3 signals are normalized to histone density using an H3-specific antibody.
To compare the effect of MLL-AF9 on the posttranslational modification of H3K27 on the Hoxa9 promoter, we performed ChIP analyses using H3K27me3- and H3K27Ac-specific antibodies in undifferentiated FDCP-mix cells and in MLL-AF9 expressing HSPC. In agreement with our previous observations, MLL-AF9 expression correlates with a specific loss of H3K27me3 from the Hoxa9 promoter (Figure 4B). Importantly, this loss correlates with a strong increase of H3K27Ac and, consistent with the data presented in Figure 2D, with an increase in H3K9Ac (Figure 4B). Taken together, these data show that MLL-AF9 can compete for PcG binding to the Hoxa9 promoter and suggests that the molecular switch between H3K27me3 and H3K27Ac might play a role in Hoxa9 expression.
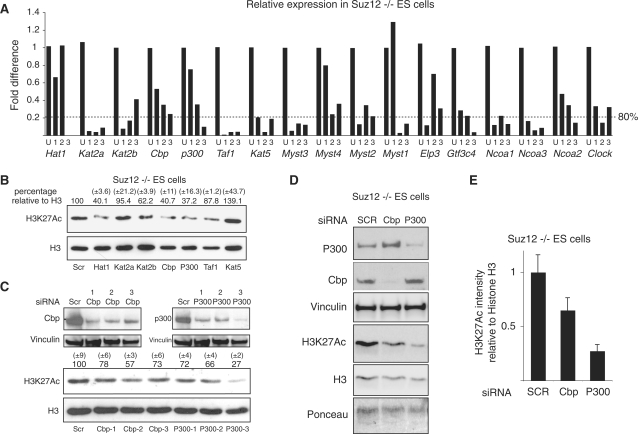
In mammals, 17 different HATs have been characterized so far and several of these have been reported to acetylate different lysine residues of histone H3 (43). To identify the HAT that could be involved in H3K27 acetylation, we generated a library containing three different siRNA oligonucleotides for each of the 17 HATs. First, we tested the efficiency of the different oligonucleotides to reduce the expression of each gene by real-time qPCR analysis of RNA extracted from Suz12 KO ES cells transfected with control (scrambled) or the specific siRNA oligonucleotide for 48 h. As shown in Figure 5A, the qPCR analysis showed that at least one oligonucleotide per gene reduced the expression of the target gene by at least 80%. Next, we picked the most efficient siRNA oligonucleotide for each gene and tested the effects of siRNA knockdown on H3K27Ac by western blot analysis. An example is presented in Figure 5B showing that siRNAs to Hat1, Kat2b, Cbp and p300 led to a significant reduction of H3K27Ac. While we were unable to further validate the effects of Hat1 and Kat2b downregulation (data not shown), independent experiments using different oligonucleotides to Cbp and p300 led to a loss of H3K27Ac levels in Suz12 KO cells (Figure 5C and D) as further confirmed by the quantification presented in Figure 5E. Importantly, the siRNA oligonucleotide that induced the most efficient downregulation of p300 correlates with the strongest reduction of H3K27Ac (Figure 5C). Taken together, these results suggest that p300 and Cbp are the major H3K27 HATs in ES cells.
Figure 5.
p00 and Cbp are required for efficient H3K27 acetylation in Suz12 KO ES cells. (A) qPCR expression analyses of the indicated genes in Suz12 KO ES cells transfected for 48 h with the indicated siRNA oligos. ‘U’ indicates the control siRNA oligo carrying a scrambled oligoribonucleotide sequence. (B) Western blot analyses of histones purified from Suz12 KO ES cells transfected with the indicated siRNA oligos using the indicated antibodies. H3 is presented as loading control. Quantification of the H3/H3K27Ac signal is indicated above each lane. A scrambled siRNA oligo (SCR) was used as negative control. (C and D) Western blot analyses of protein extracts and of purified histones from Suz12 KO ES cells transfected with the indicated siRNA oligos using the indicated antibodies. Vinculin, Ponceau staining and H3 are presented as loading controls. A scrambled siRNA oligo (SCR) was used as negative control. Quantification of the H3/H3K27Ac signal of western blots presented in ‘C’ is indicated above each lane. (E) Average quantification of the H3/H3K27Ac signals between the two independent siRNA experiments presented in C and D.
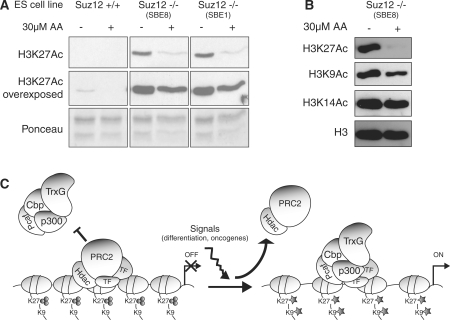
To obtain independent evidence that Cbp and p300 are regulating H3K27 acetylation, we took advantage of the ability of anacardic acid (AA) to inhibit the in vitro and in vivo acetyltransferase activity of p300 and CBP (44,45). Thus, we analyzed the H3K27Ac levels in Suz12 KO ES cells cultured in the presence of AA for 72 h by western blot analysis. As shown in Figure 6A, the treatment of two independent Suz12 KO ES cell lines with AA led to a strong reduction of H3K27Ac. Moreover, overexposure of the same western blots showed that, in WT ES, the physiological levels of H3K27Ac are also reduced upon AA treatment. Importantly, while H3K27Ac was strongly reduced upon AA treatment, the acetylation of other histone H3 lysine residues was only mildly (H3K9) or not affected (H3K14) (Figure 6B). Although we cannot exclude that AA treatment could also inhibit the activity of other acetyltransferases, these data, together with the siRNA results presented in Figure 5, strongly support that p300 and Cbp are H3K27 acetyltransferases and further suggest a competition between PRC2, p300 and Cbp in the posttranslational modification of H3K27.
Figure 6.
Inhibition of p300 and Cbp activities leads to downregulation of H3K27 acetylation. (A) Western blot analyses of histones purified from WT and two independent Suz12 KO ES cell lines cultured in the presence (+) or absence (−, DMSO) of AA using the indicated antibodies. Ponceau staining of histones is presented as loading control. (B) Western blot analyses of histones purified from Suz12 KO ES cell lines cultured in the presence (+) or absence (−, DMSO) of AA using the indicated antibodies. H3 is presented as loading control. (C) Schematic representation of a model for the competition between PcG and p300/Cbp activities during ES cell differentiation and HSPC immortalization. TF, transcription factor.
DISCUSSION
By using an unbiased approach, we have identified and characterized a switch between the acetylation and the trimethylation of H3K27 that correlates with the transcriptional activation and repression of PcG target genes, respectively. We have shown that such changes in H3K27 posttranslational modifications occur both during ES cell differentiation and upon MLL-AF9 transformation. Finally, we have provided evidence that H3K27 acetylation is (at least in part—see long exposure in Figure 6A) controlled by the activity of p300 and Cbp.
In addition to preventing the acetylation of H3K27, our results suggest that the association of the PcG proteins and H3K27 methylation also inhibit the ‘spreading’ of acetylation to other H3 residues (Figures 2D and 4B). The activation and the recruitment of Cbp/p300 may involve Trithorax (Trx)-like factors that, by competing with PcG repressive activity, are required for the maintenance of active transcription of target genes (1,2). The fact that loss of PRC2 activity leads to a global increase of H3K27Ac and that re-expression of Suz12 in the KO ES cells restores WT levels of H3K27Ac suggests that the HAT activity (likely P300 and Cbp) could be limiting in these conditions. In agreement with this model, genome-wide localization analysis have shown that the binding sites for p300 and PRC2 do not significantly overlap (46).
The Trx-like activity may involve the mixed lineage leukemia proteins (MLL1-5), the mammalian orthologues of Drosophila Trx. The MLL proteins are histone methyltransferases (HMTs) catalyzing di- and trimethylation of H3K4 and exist in large multiprotein complexes (47). Several chromatin-modifying enzymatic activities are associated with the MLL proteins, including the H3K4-specific HMT ASH2 and the H3K27me3/me2 histone demethylase UTX (33). However, the switch from H3K27me3 to H3K27Ac might not require de novo methylation of H3K4. In ES cells, H3K27me3 often coexists with H3K4me3 generating so-called ‘bivalent domains’ (48). Although bivalent domains are not confined to ES cells (35), the fact that H3K4me3 does not exclude H3K27me3 suggests that the H3K4 methylation is independent of the MLLs. Thus, the major function of the MLLs could be to recruit HATs to PcG target promoters. In this context, it is interesting that MLL has also been shown to bind directly to Cbp (49). Taken together with our results this may suggest that MLL is mediating the recruitment of Cbp, and due to their functional association also p300 and PCAF/GCN5 (50), to PcG-regulated promoters.
Our results are in agreement with recent work from the Harte laboratory (51). Like in mouse ES cells, these authors have shown that loss of H3K27me3 in Drosophila melanogaster results in a global increase of H3K27Ac, and that this requires the activity of Cbp (the Drosophila orthologue of mammalian Cbp and p300). Moreover, the authors showed that Trx is essential for Cbp-mediated H3K27Ac. Together with our data, these data support the notion that H3K27Ac is controlled by Cbp and p300 throughout evolution and therefore, suggests that the mechanism of transcriptional regulation is conserved between distantly related species. In our experiments, we find a more modest effect on H3K27 acetylation by siRNA-mediated knockdown of p300 and Cbp than reported for Drosophila Cbp. We do not know the reason for this difference; however, it could be due to the redundant functions of Cbp and p300 in mammalian cells as well as siRNA-based experiments that do not lead to a complete knockdown of the targeted gene. This assumption is further supported by the results presented in Figure 5C showing that the H3K27Ac levels are dependent on the efficiency of p300 downregulation. Of note, we have tried to knockdown Cbp and p300 simultaneously in ES cells; however, this is very toxic for the ES cells and we have therefore not been able to analyze the H3K27 acetylation levels in such cells. Nevertheless, inhibition of Cbp and p300 acetyltransferase activity by AA treatment in Suz12 KO ES cells led to a strong decrease of H3K27Ac, further supporting the siRNA-based data.
The mechanism by which the PcG and TrxG proteins are recruited in mammalian cells is still not well understood. In flies, PcG and TrxG proteins are recruited to Polycomb responsive elements (PRE) often located several kilo bases from transcription start sites (TSS) (52). Different DNA-binding transcription factor mediates the recruitment of PcG and TrxG proteins at PREs in flies (52). These factors, with the exception of YY1 (the mammalian orthologue of Pho), are not conserved in mammalian cells. In mammalian cells, several proteins have been suggested to participate in PcG promoters. Recently, we and others have shown that the DNA-binding protein Jarid2 is part of the PRC2 complex and that Jarid2 is required for recruitment of PcG proteins to most of their target genes in mouse ES cells (53–55). However, as in flies, PcG recruitment most likely involves the combined presence of several transcription factors to provide the necessary combinatorial diversity to regulate the several thousand PcG target genes in stem cells and during differentiation. Independently of these mechanisms our findings, together with the data obtained in D. melanogaster (51), suggest that both the competing mechanisms of PcG and TrxG recruitment to the same target sites and the antagonistic enzymatic activities that regulate transcription are conserved throughout evolution.
The correlation between the increased levels of H3K27Ac, the loss of H3K27me3 and the reactivation of genes during ES cell differentiation suggest that the switch in H3K27 posttranslational modification could play a role in the correct regulation of gene expression during development. Consistent with this, similar to PcG proteins (10–13), KO mice for either p300 or Cbp are embryonic lethal between E8.5–11.5 highlighting the essential role of these proteins in the regulation of embryonic development (56,57). Moreover, also similar to the PRC2 PcG proteins, p300 and Cbp are not required for ES cell proliferation, but are essential for the correct in vitro differentiation of ES cells (58).
The model proposed in Figure 6C is also in agreement with the observation that MLL-AF9 immortalization of HSPCs increase the expression of Hoxa9 and that this correlates with loss of H3K27me3 and increased levels of H3K27Ac at the Hoxa9 promoter. Hoxa9 expression plays an important role in the development of leukemia. Moreover, the displacement of PcG proteins mediated by MLL-AF9 expression further supports the idea of a competition between PcG and TrxG proteins at the same target gene. It is likely that H3K27Ac involves the activity of CBP and p300 also in this context, and it is therefore tempting to speculate that MLL-AF9 mediates the recruitment of p300 and CBP to the Hoxa9 promoter and that this activity could be important for the transformation process. In support of this, both CBP and p300 are found directly fused to MLL in several leukemia patients, and it has been shown that the HAT activity of both proteins is essential for the oncogenic potential of these fusion proteins (59–61).
It is important to mention that H3K27 is not the only substrate of p300 and Cbp. Several reports have shown that p300 and Cbp can acetylate different lysine residues of histone H3 as well as other non-histone proteins (62,63). Nevertheless, our data suggest that PcG binding and H3K27me3 are sufficient to prevent acetylation of unmodified lysine residues of histone H3. Moreover, accessibility of H3K27 followed by the displacement of PcG proteins might be the triggering event that, starting from H3K27Ac, leads to the hyperacetylation of other H3K27 surrounding lysines (Figure 6C). In summary, our findings provide novel important insights into the mechanisms of PcG- and TrxG-mediated regulation of gene expression required for proper cellular differentiation and leukemogenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Post-doctoral fellowship from Danish Medical Research Council (to D.P.); post-doctoral fellowship from NordForsk (Nordic Union to J.W.); Danish National Research Foundation (to K.H. and O.N.J.); Danish Cancer Society, Novo Nordisk Foundation, Danish Medical Research Council (to K.H.). Funding for open access charge: Danish National Research Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the Helin lab for discussions, technical advice and support.
REFERENCES
- 1.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 3.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 4.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 5.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 7.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 8.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr. Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 11.O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl Acad. Sci. USA. 2003;100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell. Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 18.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl Acad. Sci. USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koontz JI, Soreng AL, Nucci M, Kuo FC, Pauwels P, van Den Berghe H, Cin PD, Fletcher JA, Sklar J. Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proc. Natl Acad. Sci. USA. 2001;98:6348–6353. doi: 10.1073/pnas.101132598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 21.Dietrich N, Bracken AP, Trinh E, Schjerling CK, Koseki H, Rappsilber J, Helin K, Hansen KH. Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. EMBO J. 2007;26:1637–1648. doi: 10.1038/sj.emboj.7601632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gil J, Bernard D, Martinez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat. Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- 23.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 25.Bryja V, Bonilla S, Arenas E. Derivation of mouse embryonic stem cells. Nat. Protoc. 2006;1:2082–2087. doi: 10.1038/nprot.2006.355. [DOI] [PubMed] [Google Scholar]
- 26.Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell. Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 28.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 29.Lamers CHJ, van Elzakker P, van Steenbergen SCL, Sleijfer S, Debets R, Gratama JW. Retronectin-assisted retroviral transduction of primary human T lymphocytes under good manufacturing practice conditions: tissue culture bag critically determines cell yield. Cytotherapy. 2008;10:406–416. doi: 10.1080/14653240801982961. [DOI] [PubMed] [Google Scholar]
- 30.Martin ME, Milne TA, Bloyer S, Galoian K, Shen W, Gibbs D, Brock HW, Slany R, Hess JL. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer Cell. 2003;4:197–207. doi: 10.1016/s1535-6108(03)00214-9. [DOI] [PubMed] [Google Scholar]
- 31.Bruno L, Hoffmann R, McBlane F, Brown J, Gupta R, Joshi C, Pearson S, Seidl T, Heyworth C, Enver T. Molecular signatures of self-renewal, differentiation, and lineage choice in multipotential hemopoietic progenitor cells in vitro. Mol. Cell. Biol. 2004;24:741–756. doi: 10.1128/MCB.24.2.741-756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung HR, Pasini D, Helin K, Jensen ON. Quantitative mass spectrometry of histone H3.2 and H3.3 in Suz12 deficient mouse ES cells reveals distinct, dynamic post-translational modifications at K27 and K36. Mol. Cell Proteomics. 2010 doi: 10.1074/mcp.M900489-MCP200. February 11. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villa R, Pasini D, Gutierrez A, Morey L, Occhionorelli M, Vire E, Nomdedeu JF, Jenuwein T, Pelicci PG, Minucci S, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11:513–525. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 37.Ford HL. Homeobox genes: a link between development, cell cycle, and cancer? Cell Biol. Int. 1998;22:397–400. doi: 10.1006/cbir.1998.0329. [DOI] [PubMed] [Google Scholar]
- 38.Thorsteinsdottir U, Mamo A, Kroon E, Jerome L, Bijl J, Lawrence HJ, Humphries K, Sauvageau G. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99:121–129. doi: 10.1182/blood.v99.1.121. [DOI] [PubMed] [Google Scholar]
- 39.Daser A, Rabbitts TH. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev. 2004;18:965–974. doi: 10.1101/gad.1195504. [DOI] [PubMed] [Google Scholar]
- 40.Kumar AR, Hudson WA, Chen W, Nishiuchi R, Yao Q, Kersey JH. Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood. 2004;103:1823–1828. doi: 10.1182/blood-2003-07-2582. [DOI] [PubMed] [Google Scholar]
- 41.Moore MA, Chung KY, Plasilova M, Schuringa JJ, Shieh JH, Zhou P, Morrone G. NUP98 dysregulation in myeloid leukemogenesis. Ann. N. Y. Acad. Sci. 2007;1106:114–142. doi: 10.1196/annals.1392.019. [DOI] [PubMed] [Google Scholar]
- 42.Corral J, Lavenir I, Impey H, Warren AJ, Forster A, Larson TA, Bell S, McKenzie AN, King G, Rabbitts TH. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 43.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 44.Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK. Small molecule modulators of histone acetyltransferase p300. J. Biol. Chem. 2003;278:19134–19140. doi: 10.1074/jbc.M301580200. [DOI] [PubMed] [Google Scholar]
- 45.Eliseeva ED, Valkov V, Jung M, Jung MO. Characterization of novel inhibitors of histone acetyltransferases. Mol. Cancer Ther. 2007;6:2391–2398. doi: 10.1158/1535-7163.MCT-07-0159. [DOI] [PubMed] [Google Scholar]
- 46.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 47.Tenney K, Shilatifard A. A COMPASS in the voyage of defining the role of trithorax/MLL-containing complexes: linking leukemogensis to covalent modifications of chromatin. J. Cell. Biochem. 2005;95:429–436. doi: 10.1002/jcb.20421. [DOI] [PubMed] [Google Scholar]
- 48.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol. Cell. Biol. 2001;21:2249–2258. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giles RH, Peters DJ, Breuning MH. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 51.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 53.Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 54.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanaka Y, Naruse I, Hongo T, Xu M, Nakahata T, Maekawa T, Ishii S. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech. Dev. 2000;95:133–145. doi: 10.1016/s0925-4773(00)00360-9. [DOI] [PubMed] [Google Scholar]
- 57.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 58.Roth JF, Shikama N, Henzen C, Desbaillets I, Lutz W, Marino S, Wittwer J, Schorle H, Gassmann M, Eckner R. Differential role of p300 and CBP acetyltransferase during myogenesis: p300 acts upstream of MyoD and Myf5. EMBO J. 2003;22:5186–5196. doi: 10.1093/emboj/cdg473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lavau C, Du C, Thirman M, Zeleznik-Le N. Chromatin-related properties of CBP fused to MLL generate a myelodysplastic-like syndrome that evolves into myeloid leukemia. EMBO J. 2000;19:4655–4664. doi: 10.1093/emboj/19.17.4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohnishi H, Taki T, Yoshino H, Takita J, Ida K, Ishii M, Nishida K, Hayashi Y, Taniwaki M, Bessho F, et al. A complex t(1;22;11)(q44;q13;q23) translocation causing MLL-p300 fusion gene in therapy-related acute myeloid leukemia. Eur. J. Haematol. 2008;81:475–480. doi: 10.1111/j.1600-0609.2008.01154.x. [DOI] [PubMed] [Google Scholar]
- 61.Taki T, Sako M, Tsuchida M, Hayashi Y. The t(11;16)(q23;p13) translocation in myelodysplastic syndrome fuses the MLL gene to the CBP gene. Blood. 1997;89:3945–3950. [PubMed] [Google Scholar]
- 62.Hermanson O, Glass CK, Rosenfeld MG. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol. Metab. 2002;13:55–60. doi: 10.1016/s1043-2760(01)00527-6. [DOI] [PubMed] [Google Scholar]
- 63.Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.