Abstract
The clustered protocadherins are a subfamily of neuronal cell adhesion molecules that play an important role in development of the nervous systems in vertebrates. The clustered protocadherin genes exhibit complex expression patterns in the central nervous system. In this study, we have investigated the molecular mechanism underlying neuronal expression of protocadherin genes using the protocadherin gene cluster in fugu as a model. By in silico prediction, we identified multiple neuron-restrictive silencer elements (NRSEs) scattered in the fugu protocadherin cluster and demonstrated that these elements bind specifically to NRSF/REST in vitro and in vivo. By using a transgenic Xenopus approach, we show that these NRSEs regulate neuronal specificity of protocadherin promoters by suppressing their activity in non-neuronal tissues. We provide evidence that protocadherin genes that do not contain an NRSE in their 5′ intergenic region are regulated by NRSEs in the regulatory region of their neighboring genes. We also show that protocadherin clusters in other vertebrates such as elephant shark, zebrafish, coelacanth, lizard, mouse and human, contain different sets of multiple NRSEs. Taken together, our data suggest that the neuronal specificity of protocadherin cluster genes in vertebrates is regulated by the NRSE-NRSF/REST system.
INTRODUCTION
Cell adhesion molecules play an important role in animal development, including the formation of complex neural networks in the developing nervous system (1–4). Recently, a gene-rich locus containing three closely-related protocadherin gene subclusters, designated as α, β and γ, has been characterized in mammalian genomes (5–9). Each of these subclusters contains 15–22 ‘variable’ exons, which are driven by independent promoters. Each variable exon is ∼2.4 kb long and encodes an extracellular domain comprising six calcium-binding ectodomain repeats, a transmembrane domain and a short segment of the intracellular domain (8,10). In addition to the variable exons, the 3′ ends of the α and γ (but not β) subclusters contain three ‘constant’ exons each, which are spliced to every individual variable exon in their respective subclusters (8). The constant exons encode the major part of the intracellular domain. Thus, the proteins encoded by each of the α and γ subclusters share an identical cytoplasmic domain, but contain different extracellular domains. This type of gene arrangement can potentially give rise to a large repertoire of cell recognition molecules, which differ from each other by possessing a unique extracellular domain, but contain an identical cytoplasmic domain. Since the β subcluster lacks constant exons, the protocadherins encoded by genes in this subcluster lack the common cytoplasmic domain (8,11).
In mammals, clustered protocadherins are predominantly expressed in neurons and their protein products are highly enriched in synaptic membranes and axons (5,12–14). Gene knockout studies have demonstrated that protocadherins play a crucial role in proper axonal projection, synaptic formation and neuronal survival. Ablation of the protocadherin α subcluster in mice resulted in defects in axonal projection of olfactory sensory neurons to the olfactory bulb (15) and abnormal projection of brainstem serotonergic neuron axons to substantia nigra, hippocampus and caudate-putamen (16). Deletion of the entire protocadherin γ subcluster in mice led to a drastic impairment in synaptic formation and an extensive loss of interneurons in the spinal cord (12,17).
The transcription of protocadherin cluster genes has been shown to be regulated in a complex manner. On the one hand, each protocadherin gene is controlled by individual promoters located adjacent to each of the variable exons. These promoters presumably provide the basic and essential cis-elements for the expression of individual genes (18). On the other hand, the transcription of individual genes seems to be controlled at a higher level of regulation since single neuron RT-PCR experiments have demonstrated that individual Purkinje cells express an overlapping but distinct combination of protocadherin genes. In addition, the expression seems to be allele-selective (18–20). Two long-range tissue-specific enhancer sequences located at the 3′ end of mouse and human protocadherin α subcluster have been identified. One of the enhancer regions in the mouse cluster, designated as HS5-1, was shown to be responsible for the high levels of expression of most protocadherin α subcluster genes in the nervous system (21). However, very little is known about the molecular mechanisms governing the neuronal expression or the complex combinatorial expression of the protocadherin cluster genes. In this study, we have investigated the molecular mechanism responsible for neuronal expression of protocadherin cluster genes using fugu as a model.
At 400 Mb, fugu genome is one of smallest known vertebrate genomes (22). Consequently, the intergenic regions of the fugu protocadherin cluster are very compact (average ∼1 kb compared to more than 10 kb in human) (23). In this study, we have taken advantage of the compact size of the fugu protocadherin cluster and used it as a model for elucidating the molecular mechanism responsible for the neuronal expression of protocadherin cluster genes. We have identified multiple neuron-restrictive silencer elements (NRSEs) scattered in the fugu protocadherin cluster and provided evidence that these elements play a role in regulating neuronal expression of protocadherin genes in the cluster.
MATERIALS AND METHODS
In silico analyses
The genomic sequences of fugu, zebrafish, coelacanth, lizard and elephant shark protocadherin clusters were retrieved from GenBank (accession numbers: DQ986917, DQ986918 for fugu; AC144823, AC144826, AC144828, AC146480, AL929558, AB075928, BX005294, BX957322 for zebrafish; AC150283, AC150284, AC150308–AC150310 for coelacanth; BK006912–BK006917 for lizard and EF693954 for elephant shark). The genomic sequences of human and mouse protocadherin clusters were retrieved from the UCSC Genome Browser (http://genome.ucsc.edu). Intergenic sequences of protocadherin clusters were searched for common sequence motifs by MEME (24). The consensus sequences were aligned and plotted as WebLogo images (http://weblogo.berkeley.edu). Transcription factor binding sites were predicted by TESS (25). Individual NRSEs identified are listed in Supplementary Tables S1 (fugu and zebrafish) and S2 (human, mouse, lizard, coelacanth and elephant shark).
Plasmid constructs
The intergenic regions of fugu protocadherin genes, Fr2α32, α33, α34, α36 and γ6, were amplified by genomic PCR using appropriate primers (Supplementary Table S3). Luciferase and EGFP reporter constructs were prepared by subcloning fugu intergenic regions (‘promoter’ regions) into the promoter-less pGL3-basic (Promega) and pEGFP-1 (Clontech) vectors, respectively. Mutations of NRSEs in Fr2α32, α33 and γ6 promoters were introduced by PCR-directed site-specific mutagenesis. To add an NRSE downstream of the reporter gene in the pEGFP1-Fr2α36 construct, two complementary oligonucleotides (5′-GGCCGTCAGCACCATGGCCAGCGCA-3′ and 5′-GGCCTGCGCTGGCCATGGTGCTGAC-3′) corresponding to the NRSE in Fr2α32 were annealed with a NotI compatible overhang at each end, and inserted into the 3′-end of EGFP coding sequence (at the NotI site). The fugu and rat NRSF/REST expression constructs, pCMVmyc-fNRSF and pCMVmyc-rNRSF, were constructed by subcloning respective NRSF/REST coding sequences into the pCMVmyc vector (Clontech) at SalI and NotI sites in frame with the N-terminal Myc-epitope. The dominant negative isoform of rat NRSF/REST was constructed by subcloning the PCR fragment corresponding to the amino acids 154–296 of rat NRSF/REST into the pCMVmyc in frame with N-terminal Myc-epitope. A mini-cluster system for the fugu protocadherin cluster was generated by stable transfection of HeLa cells with fugu BAC clone b245G6 (http://www.geneservice.co.uk), which contains genomic sequence from Fr2α30 to Fr2γ6. To facilitate selection, a Zeocin resistant cassette was inserted into the fugu BAC vector by Red/ET recombination technology (Gene Bridges) (26). The siRNA knockdown plasmid was constructed by subcloning double-stranded oligonucleotides that encode siRNA corresponding to the human NRSF/REST: 5′-GAAGAACAGTTTGTGCATCACTTGATATCCGGTGATGCACAAACTGTTCTTCCG-3′, into the siRNA delivery vector, pRNAT-CMV3.2-neo (GenScript) at the BamHI and XhoI sites.
Luciferase assay
Mouse Neuro2A cells were cultured in DMEM (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen). Stable cell lines harboring protocadherin promoter-luciferase reporter constructs were generated by co-transfection of the Neuro2A cells with the linearized reporter plasmid and pXJ41neo (for conferring neomycin resistance) DNA using Lipofectamine 2000 method (Invitrogen). Positive colonies were selected by using G418 (Invitrogen) at a final concentration of 600 µg/ml. To determine the effect of NRSF/REST expression on the protocadherin promoter activity, Neuro2A cells harboring various protocadherin promoter-luciferase reporter cassettes were seeded in 6-well plates and transiently transfected with pCMVmyc-rNRSF or the mock plasmid. Parent Neuro2A cells transfected with the same plasmid DNA served as the blank. Cells were harvested at 48 h after transfection and the luciferase assay was performed using the Luciferase Assay System (Promega) according to the manufacturer’s instruction. The luminescence was detected using a Turner Designs TD-20/20 luminometer. The protein content of cell lysates was measured by Bradford reagent (Bio-Rad) and used for normalizing the luciferase activity. The expression of rat NRSF/REST was examined by western blot and immunostaining using anti-Myc antibody (Roche).
Electrophoretic mobility shift assay
Nuclear extracts were prepared by high salt extraction method. Briefly, HEK293 cells transfected with pCMVmyc-fNRSF or the mock plasmids were first lysed in sucrose buffer [0.32 M sucrose, 10 mM Tris–HCl pH 8.0, 3 mM CaCl2, 2 mM Mg(OAc)2, 0.1 mM EDTA, 0.5% NP-40, 1 mM DTT and 0.5 mM PMSF] in a volume of 100 µl per 107 cells. After gently mixing by pipetting, nuclei were collected by centrifugation at 500g for 5 min and resuspended in a low salt hypotonic buffer containing 20 mM HEPES (pH 7.9), 1.5 mM MgCl2, 20 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF and 25% glycerol. The nucleoplasm was extracted by slowly adding an equal volume of a high salt buffer (20 mM HEPES pH 7.9, 1.5 mM MgCl2, 0.8 M KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, 1 µg/ml aprotinin, 1% NP-40 and 25% glycerol). After rotating at 4°C for 45 min, the nuclear extracts were collected by centrifugation at 14 000g for 15 min. Probes were made by annealing two complementary oligonucleotides corresponding to the respective NRSEs. The annealed oligonucleotide duplexes, which contain a 5′ overhang at either end, were labeled with α-32P-dATP by Klenow DNA polymerase. The binding reaction was performed by mixing 2 µg of nuclear extracts with 40 000 cpm (∼0.1 fmol) of the radiolabeled probe in the binding buffer (final concentration: 10 mM Tris–HCl pH 8.0, 150 mM KCl, 0.5 mM EDTA, 0.2 mM DTT, 0.1 mg/ml polydI-dC, 0.1% Triton X-100, 12.5% glycerol) in a volume of 20 µl at room temperature for 30 min. For super-shift assay, 1 µg of anti-Myc antibody (Roche) was added to the reaction mixture. The samples were separated by non-denaturing PAGE and the retarded bands were detected by autoradiography.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed using anti-Myc antibody (for experiments in Figure 2) or anti-NRSF/REST antibody H-290 (#sc25389, Santa Cruz) (for experiments in Figure 4) according to the manufacturer’s instruction. Primers used for the PCR analysis of immunoprecipitation are listed in Supplementary Table S3.
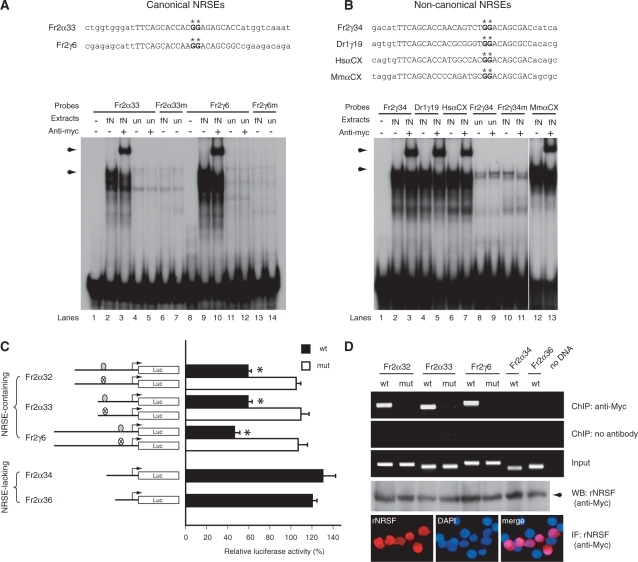
Figure 2.
Functional assay of NRSEs in the fugu protocadherin cluster. EMSA for canonical (A) and non-canonical (B) NRSEs. The NRSE sequences are indicated by upper-case letters whereas the flanking sequences are indicated by lower-case letters. The asterisks indicate the critical guanine dinucleotide that was mutated to adenine nucleotides in NRSE mutants. Arrows in the autoradiography indicate the protein–oligonucleotide complex of the full-length fugu NRSF/REST (A: lanes 2 and 9; B: lanes 2, 4, 6 and 12), or the super-shifted complex in the presence of the anti-Myc antibodies (A: lanes 3 and 10; B: lanes 3, 5, 7 and 13). fN: nuclear extracts from pCMVmyc-fNRSF-transfected HEK293 cells; un: nuclear extracts from mock (pCMVmyc) transfected HEK293 cells. Fr2α33m, Fr2γ6m and Fr2γ34m represent the corresponding mutant NRSE oligonucleotides of Fr2α33, Fr2γ6 and Fr2γ34, respectively. HsαCX and MmαCX represent the NRSEs located in the constant regions of the human and mouse protocadherin α clusters. (C) Relative luciferase activity of Neuro2A stable cell lines harboring individual fugu protocadherin promoter-luciferase reporter cassettes. The solid bars represent the wild-type fugu protocadherin promoters, whereas the open boxes represent the mutant forms. The relative luciferase activity was calculated by the activity of the pCMVmyc-rNRSF-transfected cells over the mock plasmid transfected cells after normalization for the protein concentration. (D) ChIP assay showing that the ectopically-expressed NRSF/REST binds to the wild-type NRSE-containing promoters (Fr2α32, Fr2α33 and Fr2γ6), but not the mutant (Fr2α32mut, Fr2α33mut and Fr2γ6mut) or NRSE-lacking (Fr2α34 and Fr2α36) promoters. The expression of rat NRSF/REST was confirmed by western blot and immunostaining with anti-Myc antibody.
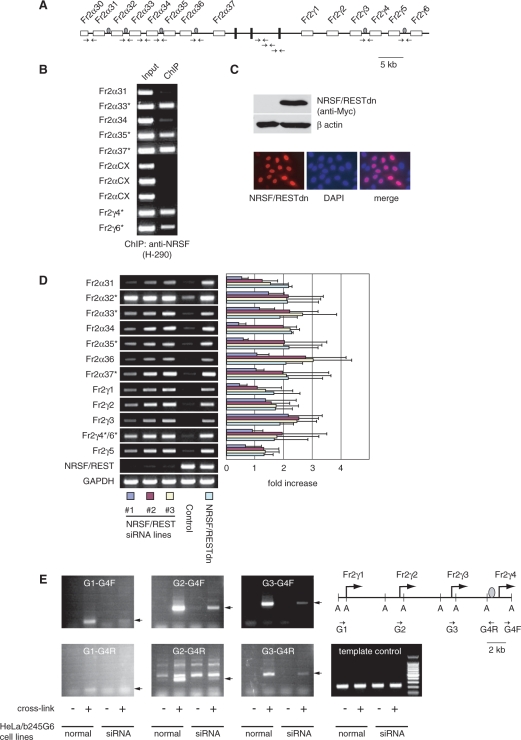
Figure 4.
NRSE regulates protocadherin promoter activity in the multi-gene cluster context. (A) Genomic organization of fugu BAC clone b245G6. Shaded ovals indicate the NRSEs, whereas open boxes and solid vertical bars indicate the variable and constant exons, respectively. The position of PCR primer pairs is shown by arrows. (B) ChIP assay of NRSE occupancy in b245G6-stably-transfected HeLa cells showing that NRSF/REST binds to NRSEs in fugu protocadherin cluster in vivo. Asterisks indicate NRSE-containing protocadherin genes. Input: genomic DNA of the b245G6-stably-transfected HeLa cells. (C) Characterization of the dominant-negative isoform of NRSF/REST (NRSF/RESTdn) in HeLa cells, showing that similar to the full-length NRSF/REST, the NRSF/RESTdn is predominantly expressed in nucleus. (D) Relative expression levels of fugu protocadherin genes in the NRSF/REST knock-down and NRSF/RESTdn-transfected cells. The data shown here (right panel) is average of three independent experiments. Asterisks indicate NRSE-containing protocadherin genes. Primers used for the ChIP assay and the RT-PCR analysis are listed in Supplementary Table S3. (E) Chromosomal spatial organization analysis by 3C assay showing that the promoter regions of Fr2γ1-3 are in proximity to the NRSE of Fr2γ4 in b245G6-stably-transfected HeLa cells. The AseI sites flanking the promoter region are indicated by ‘A’. DNA fragments corresponding to specific PCR products resulting from ligation of the sequence flanking the NRSE and the respective promoter sequences are indicated by arrows (panels 1–6). A fragment of the intergenic region of Fr2γ4 flanking the NRSE site (within the AseI fragment) was amplified by PCR with the Fr2γ4 primers (Supplementary Table S3) to confirm that equal amounts of DNA were used as template (last panel).
RT-PCR analysis of fugu protocadherin expression
The expression profiles of fugu protocadherin genes were determined by real-time RT-PCR. In this experiment, total RNA from various fugu tissues was extracted by the Trizol method (Invitrogen) and reverse-transcribed using SMART RACE cDNA Amplification kit (Clontech). The collective expression level of genes in the fugu Pcdh2α or Pcdh2γ subcluster was quantified by real-time PCR using the LightCycler® Faststart DNA MasterPLUS SYBR Green I kit (Roche) with primers corresponding to the constant exons of fugu Pcdh2α and Pcdh2γ subclusters (Supplementary Table S3). The expression level of actin was determined by the same method with specific primers (Supplementary Table S3) and used for normalizing protocadherin expression levels. The expression of fugu protocadherin in the mini-cluster system was determined by semi-quantitative RT-PCR. In this experiment, the total RNA from cells with various treatments was extracted by the Trizol method and reverse-transcribed into cDNA using the SuperScript First Strand cDNA Synthesis kit (Invitrogen). The relative expression levels of fugu protocadherin genes were determined by density quantification of the PCR products on an agarose gel with the Quantity One software (Bio-Rad) and normalized by GAPDH expression level in HeLa cells.
Transgenic Xenopus
Transgenic Xenopus laevis tadpoles were generated by a modified sperm nuclear injection method (27,28). Briefly, sperm were collected from adult male testis and the nuclei were extracted by digitonin treatment. After incubating with linearized transgene DNA, the sperm nuclei were diluted and injected into de-jellied eggs using the constant flow injector (Harvard Apparatus PHD 2000 Infusion). The fertilized embryos were cultured to 4- or 8-cell stage (∼3 h post-injection). Only tadpoles that developed normally with clear perpendicular arrangements of cleavage furrows were cultured for further analysis. EGFP expression was photographed under fluorescent microscope (Zeiss M2 Bio Quad). For immunohistological examination, tadpoles were fixed overnight in PBS containing 4% paraformaldehyde and embedded in OCT compound (Sakura). The frozen sections were immunostained by TuJ1 antibody. The immunostaining signals and the expression of EGFP were examined and photographed under fluorescent microscopy (Zeiss, Axioskop 40) with an attached Axiocam MRC camera.
3C analysis
The 3C analysis of b245G6-stably-transfected HeLa cells and the NRSF/REST knockdown cells was essentially based on standard protocols (29,30). Briefly, the cells were harvested by trypsinization and treated with 1% formaldehyde at room temperature for 10 min. An equal amount of untreated cells were used as negative control. Cell nuclei were digested by AseI at 37°C overnight, followed by ligation at 16°C for 18 h. The linkage of the genomic DNA fragments was analyzed by PCR with primers specific to each genomic region (Figure 4E, Supplementary Table S3). The identities of PCR fragments were verified by sequencing.
RESULTS
Identification of NRSE-like motifs in fugu protocadherin clusters
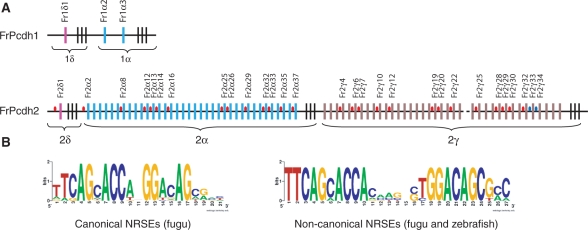
Fugu possesses two unlinked protocadherin clusters, Pcdh1 and Pcdh2, due to a whole genome duplication event in the fish lineage (23). While fugu Pcdh1 is a highly degenerate cluster that has lost the entire β and γ subclusters, and retained only two α and one δ subcluster genes, the fugu Pcdh2 consists of at least 74 genes belonging to the α, γ and δ subclusters (23,31). To identify potential regulatory elements, we searched for common sequence motifs in the intergenic regions of fugu Pcdh1 and Pcdh2 clusters using the motif-finding algorithm MEME (24). Our search identified a common sequence motif ‘TTCAGNACCANGGACAG’ present in the intergenic regions of 27 out of 74 genes in the fugu Pcdh2 cluster (Figure 1A, Supplementary Table S1). This motif does not overlap with the previously identified CGCT element in the mammalian (7,32) and elephant shark (31) protocadherin clusters. We searched for potential transcription factor binding sites in this motif using TESS (25) and discovered that this motif is highly similar to the NRSE, a 21-bp element that was initially identified in the regulatory region of SCG10 (superior cervical ganglion-10, also known as stathmin-2) and type II sodium channel genes (33,34), and was shown to mediate transcriptional repression of these neuronal genes in non-neuronal cells by binding to the transcriptional repressor, NRSF/REST (35,36). The consensus sequence of the fugu elements is highly similar to the consensus sequence of the NRSEs identified in the genomes of mammals (Figure 1B, left panel). No such NRSE-like motif was identified in fugu Pcdh1 cluster.
Figure 1.
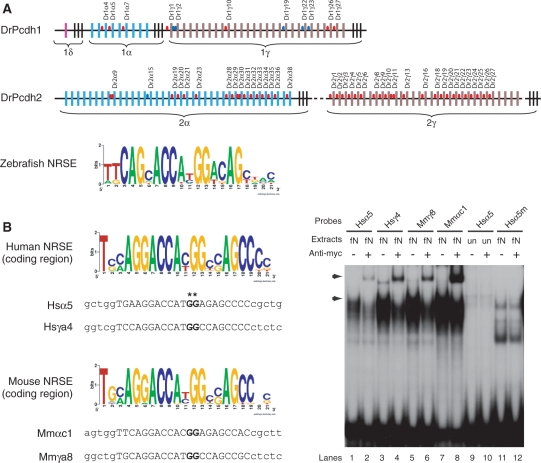
NRSEs in fugu protocadherin clusters. (A) Fugu protocadherin clusters. Colored vertical bars represent individual variable exons, whereas the black vertical bars at the 3′-end of each subcluster represent the constant exons. Red and blue ovals in the intergenic regions represent canonical and non-canonical NRSEs, respectively. The dotted line in the fugu Pcdh2 cluster indicates an undetermined sequence gap. Fr: Fugu rubripes. (B) WebLogo display of the consensus sequence of canonical (left panel) and non-canonical (right panel) NRSEs. Note: the non-canonical NRSE consensus is a combination of both fugu and zebrafish elements.
Recently, genome-wide prediction of NRSEs in mammalian genomes by ChIP assay has identified two types of NRSEs: the canonical and the non-canonical NRSEs (37,38). While the canonical NRSEs contain two non-palindromic halves separated by two spacer-nucleotides, the non-canonical NRSEs contain 5–11 nt in the spacer region. The non-canonical NRSEs nonetheless appear to function similar to the canonical NRSEs (37–39). To determine whether fugu protocadherin clusters also contain non-canonical NRSE, we searched fugu protocadherin clusters using the consensus sequence TYAGMRCCNNGGMSAG with various lengths of spacer sequences (37–41). We identified two non-canonical NRSEs, each with an 8-bp spacer sequence, located in the promoter regions of fugu Fr2γ33 and Fr2γ34 genes (Figure 1B, right panel). Thus, our in silico analysis indicated that 38% (29/77) of fugu protocadherin genes possess a putative NRSE in their regulatory regions.
NRSE-like motifs in fugu protocadherin cluster regulate the promoter activity of protocadherin genes
To determine whether the NRSE-like motifs identified in the fugu protocadherin cluster represent bona fide regulatory elements, we first tested their ability to bind to NRSF/REST in vitro by electrophoretic mobility shift assay (EMSA). Two canonical NRSE-like motifs present in the promoter regions of Fr2α33 and Fr2γ6 were selected for this experiment. Double-stranded oligonucleotide probes corresponding to each of these elements were radiolabeled and incubated with nuclear extracts prepared from fugu NRSF/REST (Myc-tagged at N-terminal) or mock plasmid-transfected HEK293 cells. Electrophoresis showed that both probes can form a protein–oligonucleotide complex with fugu NRSF/REST, and the binding-complex can be further shifted by the anti-Myc antibody (Figure 2A). Previous studies have identified a critical guanine dinucleotide in the NRSE that is essential for the binding activity (42,43). We mutated this dinucleotide in the oligonucleotide probes and found that the mutation completely abolished the association between the oligonucleotide probes and fugu NRSF/REST (Figure 2A, compare lane 2 to lane 6, and lane 9 to lane 13). These results indicate that the binding between the oligonucleotide probes and NRSF/REST is highly specific. In addition to the canonical NRSE-like motifs, we tested the binding activity of non-canonical NRSE-like motifs located in the promoter region of fugu Fr2γ34 gene and found that it can also form a complex with fugu NRSF/REST, similar to the canonical NRSE probes (Figure 2B, compare lanes 2 and 3 to 8–11). These results demonstrate that the NRSE-like motifs identified in the fugu protocadherin cluster are able to bind specifically to NRSF/REST in vitro.
We next examined whether these NRSE-like motifs can regulate the promoter activity of protocadherin genes. To this end, we generated various fugu protocadherin promoter-luciferase reporter constructs and tested whether they respond to ectopic expression of NRSF/REST in cells that lack NRSF/REST expression. Three fugu NRSE-containing (Fr2α32, Fr2α33, Fr2γ6) and two NRSE-lacking (Fr2α34, Fr2α36) protocadherin promoters were selected for this experiment. For each wild-type construct, we also generated a mutant construct in which the critical guanine dinucleotide was substituted with adenine nucleotide. The transcriptional regulation mediated by NRSF/REST requires histone deacetylation and modulation of chromatin structure (44–47). In order to recapitulate these molecular actions of NRSF/REST, we generated stable cell lines for each of these reporter constructs in Neuro2A cells. Neuro2A was chosen for this experiment because these cells express undetectable levels of NRSF/REST and have been widely used for the promoter activity assay of NRSE-containing genes (48,49). As shown in Figure 2C, overexpression of rat NRSF/REST in cells harboring reporter constructs of the wild-type NRSE-containing promoters led to a 40–50% reduction in the luciferase activity, whereas it had little effect on the luciferase activity in cells harboring NRSE-lacking or mutant NRSE-containing promoter constructs. The ectopically expressed NRSF/REST was found to be specifically associated with wild-type NRSEs, but not with mutant NRSEs or the NRSE-lacking promoter sequences (Figure 2D). These results indicate that the NRSE-like motifs in the fugu protocadherin promoter regions can regulate the promoter activity of protocadherin genes.
NRSEs confer neuron-specificity to protocadherin promoters in transgenic Xenopus
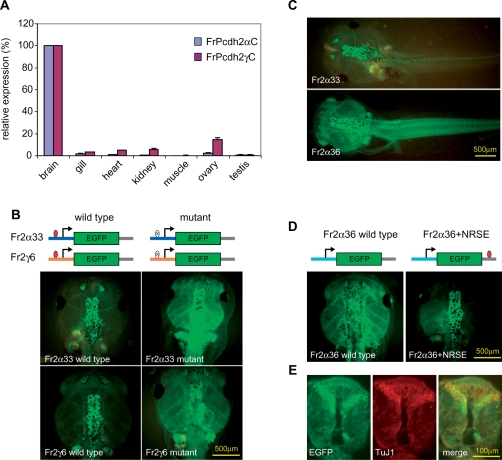
The mammalian protocadherin cluster genes are predominantly expressed in neurons (5,9,13,14,50,51). We examined the expression profile of fugu Pcdh2 cluster genes by real-time RT-PCR using primers specific to the constant exons of α and γ subclusters. Since the constant exons are common to all variable exons in the subcluster, the PCR products amplified with these primers should reflect the collective expression levels of all the protocadherin genes in the subcluster. Our results show that protocadherin genes in both the α and γ subclusters of fugu Pcdh2 are predominantly expressed in the brain (Figure 3A), suggesting that the fugu protocadherin expression is also neuronal. To test whether NRSEs in the fugu protocadherin cluster play a role in regulating neuronal expression of the fugu genes, we generated EGFP reporter constructs for two fugu NRSE-containing promoters, the Fr2α33 and Fr2γ6, and examined their promoter activity in vivo by using a rapid transgenic Xenopus approach described recently (27,28). The transgenic Xenopus approach has been demonstrated as a fast and reliable method for analyzing promoters of fugu and other vertebrate genes in vivo (52,53). We found that both promoters direct high levels of EGFP expression specifically to the nervous system of transgenic Xenopus (Figure 3B, left panels), and the EGFP expression is largely restricted to neurons (Figure 3E). We then mutated NRSEs in the respective promoters by substituting the critical guanine dinucleotide with adenine nucleotide. This mutation led to a significant de-repression of EGFP expression in non-neuronal tissues of transgenic Xenopus, but had little effect on the promoter activity in the nervous system (Figure 3B, right panels). This suggests that the main function of NRSE is to suppress the expression of protocadherin genes in non-neuronal tissues, thereby restricting their expression specifically to neurons.
Figure 3.
NRSEs are responsible for the neuronal specificity of fugu protocadherin promoters. (A) The expression profile of fugu Pcdh2α and Pcdh2γ subcluster genes determined by real-time RT-PCR. The relative protocadherin expression level in each tissue is displayed as a percentage of the expression level in the brain after normalization by actin expression. (B) NRSE-containing protocadherin promoters: red ovals represent the wild-type NRSE, whereas blank ovals with a cross represent the mutant NRSEs. Images of transgenic tadpoles (9–10 days post-fertilization) harboring the wild-type (left panels) and mutant (right panels) protocadherin promoter-EGFP constructs. Upper panels: Fr2α33-EGFP; lower panels: Fr2γ6-EGFP. (C) Comparison of the NRSE-containing and NRSE-lacking protocadherin promoters: the whole body images of transgenic tadpoles (9–10 days post-fertilization) harboring a fugu NRSE-containing (Fr2α33, upper panel) or an NRSE-lacking (Fr2α36, lower panel) protocadherin promoter-EGFP showing that the NRSE-containing promoter directs EGFP expression specifically to the nervous system, whereas the NRSE-lacking promoter directs the transgene expression to both neuronal and non-neuronal tissues. (D) NRSE-lacking protocadherin promoters: images of transgenic tadpoles harboring NRSE-lacking Fr2α36-EGFP transgene in the absence (left panel) and presence (right panel) of an NRSE inserted at the 3′-end of EGFP coding sequence. (E) The transverse section of the brain/spinal cord region of transgenic tadpole harboring the Fr2α33-EGFP showing that the EGFP expression is predominantly neuronal. Left panel: EGFP florescence image; Middle panel: anti-neuron-specific tubulin βIII (TuJ1); Right panel: merged.
Our in silico analysis has shown that more than half of protocadherin genes in the fugu Pcdh2 cluster lack an NRSE in their 5′ intergenic regions. Since these genes are also predominantly expressed in neurons (Figure 3A), we speculated that either they have adopted a different mechanism to exhibit neuronal expression or they are regulated by NRSEs present in the 5′ intergenic regions of their neighboring genes. To test these possibilities, we first examined the tissue-specificity of an NRSE-lacking promoter (Fr2α36) in the transgenic Xenopus system. In contrast to the NRSE-containing promoter (Fr2α33) that directs EGFP expression specifically to the nervous system (Figure 3C, upper panel), this NRSE-lacking promoter is active in both neural and non-neural tissues (Figure 3C, lower panel). This indicates that the NRSE-lacking promoter, by itself, is insufficient to confine its expression to the nervous system. We then investigated whether these promoters could respond to NRSEs located in the 5′-intergenic regions of their downstream genes. To this end, we inserted an NRSE at the 3′-end of the EGFP coding sequence in the Fr2α36 promoter–reporter construct and tested its activity in transgenic Xenopus. Topologically, this NRSE is analogous to the NRSE in the 5′ intergenic region of a downstream protocadherin gene. We found that the insertion of the NRSE was sufficient to repress the promoter activity of Fr2α36 in non-neuronal tissues thereby restricting the EGFP expression only to the nervous system (Figure 3D). This suggests that neuronal expression of protocadherin genes that lack an NRSE in their 5′ intergenic region can be regulated by NRSE elements located in the 5′ intergenic regions of their neighboring genes.
NRSEs in the protocadherin cluster regulate protocadherin gene expression
Having determined the function of NRSEs in individual fugu protocadherin promoters, we next asked whether these elements could interact with NRSF/REST in vivo and regulate protocadherin gene expression in the context of the gene cluster. Unfortunately, due to the lack of a suitable fugu cell line and lack of an antibody that recognizes fugu NRSF/REST, it is difficult to address this question directly in the native fugu system. As an alternative, we generated a mini-cluster system by stably integrating a fugu genomic BAC clone (b245G6, 74.8 kb) in HeLa cells. This clone contains the last eight protocadherin genes (Fr2α30 to Fr2α37) of the α subcluster, and the first six genes (Fr2γ1 to Fr2γ6) of the γ subcluster, which together represents about one-fifth of the genomic sequence of the entire fugu Pcdh2 cluster (Figure 4A). This region contains six NRSEs located one each in the promoter regions of Fr2α32, Fr2α33, Fr2α35, Fr2α37, Fr2γ4 and Fr2γ6 (Figure 4A). After generating the b245G6-stably-transfected cells, we first examined whether these NRSEs interact with the endogenous NRSF/REST expressed in HeLa cells by ChIP assay, with an antibody that recognizes the human NRSF/REST. Our results show that the NRSE sites in the fugu protocadherin cluster are occupied by NRSF/REST expressed by HeLa cells (Figure 4B). We next tested whether this interaction could play a role in regulating fugu protocadherin genes. To this end, we first used a vector-based siRNA delivery system to knockdown the endogenous NRSF/REST expression in the b245G6-stably-transfected cells. RT-PCR experiments showed that in all the three independent NRSF/REST-knockdown cell lines, disruption of NRSF/REST expression resulted in a significant increase (0.5–3-folds) in the expression levels of all the fugu protocadherin genes (Figure 4D). Second, we generated a dominant-negative isoform of NRSF/REST (NRSF/RESTdn), which contains only the DNA-binding domain but lacks the repressor domains at both terminals (35). When expressed in HeLa cells (by transient transfection), the NRSF/RESTdn is predominantly localized in the nucleus (Figure 4C), where it presumably competes with the endogenous NRSF/REST for the occupancy of NRSEs. The RT-PCR experiment showed that over-expression of NRSF/RESTdn in the b245G6-stably-transfected cells led to elevated expression of the fugu protocadherin genes (Figure 4D), similar to that observed in the NRSF/REST-knockdown cell lines. Notably, in both experiments there is no significant difference in the extent of de-repression between the NRSE-containing protocadherin genes (Fr2α32, Fr2α33, Fr2α35, Fr2α37, Fr2γ4 and Fr2γ6, with average of 1.8-folds of increase) and the NRSE-lacking protocadherin genes (Fr2α31, Fr2α34, Fr2α36, Fr2γ1-3 and Fr2γ5, with average of 1.6-folds of increase). This suggests that NRSE-lacking protocadherin genes are also being regulated by the NRSE-NRSF/REST system, presumably by long-range interaction between NRSEs and the promoters of protocadherin genes that do not contain an NRSE in their 5′ intergenic region. To capture such possible long-range interaction, we performed the 3C analysis (54). Our study shows that the promoters of the three NRSE-lacking protocadherin genes (Fr2γ1-3) located at the 5′ end of the fugu Pcdh2γ subcluster are brought in close proximity to the NRSE of a neighboring protocadherin gene (Fr2γ4). Such an association was significantly reduced in NRSF/REST knockdown cells (Figure 4E). These results suggest that there is an interaction between distantly located NRSE-lacking promoters and an NRSE-containing promoter in the protocadherin cluster. Taken together, our results suggest that the expression of fugu protocadherin genes that lack an NRSE in their 5′ intergenic region are regulated by NRSEs located in the 5′ intergenic regions of their neighboring genes.
NRSEs in the protocadherin clusters of other vertebrates
To investigate whether the clustered protocadherin genes in other vertebrates are also regulated by the NRSE-NRSF/REST system, we first performed an in silico analysis of the zebrafish protocadherin clusters. Similar to fugu, zebrafish contains two independent protocadherin clusters, Pcdh1 and Pcdh2, that collectively contain at least 107 genes (6,55–57). However, unlike the highly degenerate fugu Pcdh1 cluster, the zebrafish Pcdh1 cluster contains all the three subclusters (α, γ and δ), with a total of 38 protocadherin genes. Our search revealed that seven out of the 38 genes in zebrafish Pcdh1 cluster and 37 out of the 69 genes in zebrafish Pcdh2 cluster each contain a single canonical NRSE motif in their 5′ intergenic regions, whereas Dr2α9 and Dr2α30 genes contain two such elements each (Figure 5A, Supplementary Table S1). In addition, we identified six non-canonical NRSEs in five zebrafish protocadherin genes (Dr1γ2, Dr1γ19, Dr1γ22, Dr1γ23 and Dr2α15) (Figure 5A, Supplementary Table S1). Thus, the NRSE-containing genes in the zebrafish protocadherin clusters account for about half (49/107) of the total number of genes in the cluster. We performed similar in silico search in the protocadherin clusters of human, mouse, lizard (58), coelacanth (56) and elephant shark (31), using the consensus sequence of the NRSE elements identified in fugu and mammalian genomes (40,41). We found that all of these vertebrates contain different numbers of NRSEs (25 in human, 35 in mouse, five in lizard, 11 in coelacanth and five in elephant shark) in their protocadherin clusters (Supplementary Table S2). To verify if the NRSEs in the protocadherin clusters of these vertebrates are functional, we tested the binding activity of a few selected non-canonical NRSEs by EMSA. We found that all the selected NRSEs can bind to NRSF/REST specifically in vitro (Figure 2B). These experiments indicate that the NRSEs in the protocadherin clusters of zebrafish and mammals are indeed functional regulatory elements and are likely to be involved in regulating neuronal expression of clustered protocadherin genes.
Figure 5.
NRSEs in the protocadherin clusters of zebrafish, human and mouse. (A) Zebrafish protocadherin clusters. The color vertical bars represent individual variable exons, whereas the black vertical bars at the 3′ end of each subcluster represent the constant exons. Red and blue ovals in the intergenic regions of the zebrafish clusters represent the canonical and non-canonical NRSEs, respectively. The dotted lines in the Pcdh2 cluster indicate undetermined sequence gaps. The consensus sequence of zebrafish canonical NRSEs is displayed as a WebLogo plot. Dr: Danio rerio. (B) EMSA for NRSEs located in the protein-coding sequence of human and mouse protocadherin clusters. The consensus sequences are displayed as WebLogo plots. Upper-case letters represent the NRSE sequence, and the lower-case letters represent the flanking sequences. The asterisks indicate the guanine dinucleotide which was mutated to adenine dinucleotide in NRSE mutants. Hsα5m represents the mutant NRSE in the protein-coding sequence of the Hsα5. Hs: Homo sapiens, Mm: Mus muscus. Autoradiography of EMSA: arrows indicate the protein–oligonucleotide complex of the full-length fugu NRSF/REST (lanes 1, 3, 5 and 7) or the super-shifted complex in the presence of the anti-Myc antibodies (lanes 2, 4, 6 and 8). fN: nuclear extracts from pCMVmyc-fNRSF-transfected HEK293 cells; un: nuclear extracts from mock (pCMVmyc) transfected HEK293 cells.
Interestingly, a majority of NRSEs identified in the human and mouse protocadherin clusters (20/25 in human and 19/35 in mouse) are located in the coding sequences of the variable exons. This raised the question whether the occurrence of these sequences is by chance or they are really functional regulatory elements. To verify this, we tested the binding activity of two elements each from the human and mouse protocadherin clusters by EMSA. Our results show that the NRSEs located in the coding region of both human and mouse protocadherin clusters are able to bind to fugu NRSF/REST in vitro, suggesting that they are likely to be functional regulatory elements (Figure 5B). These functional NRSEs are likely to have emerged de novo in the mammalian lineage.
DISCUSSION
The objective of this study was to determine the molecular mechanism underlying neuronal expression of the protocadherin cluster genes in vertebrates. The genes in the protocadherin cluster exhibit a complex expression pattern since neurons of the same kind are known to express an overlapping but distinct combination of these genes (18–20). This implies that individual protocadherin genes are regulated by a complex mechanism. Although NRSE-NRSF/REST system is known to be involved in specifying neuronal expression of a large number of genes in vertebrate genomes, it is not known if the clustered protocadherin genes are also regulated by this system. Using in silico analysis, we identified multiple NRSEs scattered in the protocadherin cluster of fugu and other vertebrates. We then demonstrated that these elements bind to NRSF/REST in vitro and in vivo and are able to suppress the activity of protocadherin promoters in non-neuronal cells, thereby restricting their expression to neuronal cells. Since our in silico analysis showed that only 38% of genes in fugu protocadherin cluster contain an NRSE in their 5′ intergenic regions, we investigated the possibility whether NRSE-lacking genes are regulated by NRSEs located downstream of the gene. We showed that an NRSE placed downstream of a NRSE-lacking protocadherin promoter can restrict the activity of the promoter to neuronal cells in transgenic Xenopus. This finding raised the possibility that an NRSE in the protocadherin cluster may regulate multiple genes that lack an NRSE in their intergenic region. To address this, we used the HeLa cell line that was stably transfected with a part of the fugu protocadherin cluster and investigated the interaction between neighboring protocadherin promoters by 3C assay. Our experiments showed that the promoter of protocadherin genes that lack an NRSE in their intergenic region can be brought in close proximity to an NRSE located downstream of the gene. Moreover, our mini-cluster experiment showed that the expression of both NRSE-containing and NRSE-lacking protocadherin genes can be elevated to a similar extent by ectopic expression of a dominant-negative NRSF/REST or by siRNA-mediated knockdown of the endogenous NRSF/REST expression. Taken together, these results suggest that a single NRSE in the protocadherin cluster can regulates multiple genes including those that do not contain an NRSE in their intergenic regions. Previously, NRSEs located with in introns or in the 3′ non-coding region of neuronal genes (e.g. the L1 cell adhesion molecule and GABAA receptor gamma2 subunit genes) have been shown to be effective in conferring neuronal-specificity to the upstream promoters (59,60). Genome-wide analysis of NRSF/REST chromatin occupancy in mouse TCMK1 kidney cells has also revealed that ∼60% NRSE sites that are occupied by the NRSF/REST are located more than 10 kb away, either up- or down-stream, of the transcription start site of their putative target genes (38). Our study provides further support to the notion that NRSEs can act as long-range repressors. Since the genes in the protocadherin cluster are tightly linked, NRSEs scattered within the cluster may even function like global control regions that are known to regulate expression of multiple genes [e.g. the β globin gene locus (61) and the HoxD gene cluster (62)]. In particular we note that some regions of the fugu Pcdh2 cluster, such as the sequence between Fr2α16 and Fr2α25 (42.7 kb), and the sequence between Fr2γ13 and Fr2α18 (20.5 kb) are virtually devoid of NRSEs (Figure 1A). It is possible that these protocadherin genes are regulated by NRSEs that are located upstream or downstream like a global control region.
Interestingly, in contrast to the fugu Pcdh2 cluster that contains many NRSEs, fugu Pcdh1 cluster does not contain any NRSE. RT-PCR and transgenic Xenopus experiments have shown that similar to the fugu Pcdh2 cluster genes, the three protocadherin genes (Fr1δ1, Fr1α2 and Fr1α3) in the fugu Pchd1 cluster are also predominantly expressed in the nervous system (Li,S.L. and Yu,W.P.Y., unpublished data). It is possible that these genes are regulated by cryptic NRSE elements that could not be identified by the approach used by us. Alternatively, these genes may utilize an NRSE-NRSF/REST independent mechanism to accomplish their neuronal expression, such as by containing tissue-specific enhancers that activate expression specifically in neurons.
In contrast to the multiple NRSEs located exclusively in the intergenic regions of fugu and zebrafish protocadherin clusters, a majority of NRSEs in the human and mouse protocadherin clusters are located in the coding region (Supplementary Table S2). We have provided evidence that at least some of these NRSEs are functional. The presence of regulatory elements in the protein-coding sequences in mammals indicates that during evolution of mammals, the protein-coding sequence of protocadherin genes were co-opted to function as transcriptional regulatory elements. This is a classical example of exaptation whereby protein-coding sequences have been recruited to perform an additional function. Recently, a study aiming to elucidate regulatory mechanisms underlying monoallelic expression of the odorant receptor genes has suggested that some regulatory elements residing within the coding region of the odorant receptor genes may play a critical role in the singular expression of these genes in olfactory sensory neurons (63). Interestingly, like the protocadherin cluster genes, odorant receptor genes have been proposed to play a role in specifying individuality of neurons and the allele-selective expression pattern in neurons. Thus, it would be interesting to determine whether the regulatory elements residing within the coding region of protocadherin cluster are responsible for the allele-selective expression of protocadherin genes.
Recently, two conserved long-range regulatory regions located at the 3′-end of mouse and human protocadherin α cluster have been identified by comparative genomics and DNase hypersensitivity assay (21). Both were found to function as tissue-specific enhancers in reporter assay systems. Furthermore, one of the regulatory regions in the mouse cluster, designated HS5-1 spanning ∼2.5 kb, was shown to be responsible for the high-level expression of most of the protocadherin genes in the mouse α cluster. It was proposed that a competition among promoters of individual variable exons for the enhancer components associated with the HS5-1 may underlie the monoallelic expression of protocadherin α genes. Interestingly, a non-canonical NRSE identified by us is located within this enhancer region (MmαCX, Supplementary Table S2). Our EMSA confirmed that this element binds to NRSF/REST (Figure 2B). This site has also been previously identified by genome-wide ChIP analysis (38). It is therefore likely that this NRSE is responsible for the tissue-specific expression of the long-range enhancer characterized by Ribich et al. (2006). This NRSE is conserved in the human protocadherin cluster and is likely to be involved in the neuron-specific regulation of protocadherin genes in the human α cluster.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biomedical Research Council (BMRC), Singapore (04/1/32/19/352 to WPY). Research work in JLF’s and BV’s laboratories is supported by the Agency for Science, Technology and Research (A*STAR), Singapore. Funding for open access charge: National Medical Research Council (NMRC), Singapore.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ms Qiao Jing Lew, Lei Ling Thia and Janice Tan for their excellent technical assistance. We thank Dr Vydianathan Ravi for his critical reading of the manuscript. We also thank three anonymous reviewers for their critical comments and suggestions that have helped to improve the manuscript. X.J.J. is an attachment post-graduate student from Shandong University, Jinan, Shandong, China.
REFERENCES
- 1.Shapiro L, Colman DR. The diversity of cadherins and implications for a synaptic adhesive code in the CNS. Neuron. 1999;23:427–430. doi: 10.1016/s0896-6273(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 2.Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat. Rev. Neurosci. 2007;8:11–20. doi: 10.1038/nrn2043. [DOI] [PubMed] [Google Scholar]
- 3.Yagi T, Takeichi M. Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev. 2000;14:1169–1180. [PubMed] [Google Scholar]
- 4.Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- 5.Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, Watanabe M, Ishii H, Yasuda M, Mishina M, Yagi T. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 1998;20:1137–1151. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 6.Wu Q. Comparative genomics and diversifying selection of the clustered vertebrate protocadherin genes. Genetics. 2005;169:2179–2188. doi: 10.1534/genetics.104.037606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Q, Zhang T, Cheng JF, Kim Y, Grimwood J, Schmutz J, Dickson M, Noonan JP, Zhang MQ, Myers RM, et al. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 2001;11:389–404. doi: 10.1101/gr.167301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 9.Zou C, Huang W, Ying G, Wu Q. Sequence analysis and expression mapping of the rat clustered protocadherin gene repertoires. Neuroscience. 2007;144:579–603. doi: 10.1016/j.neuroscience.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Sugino H, Hamada S, Yasuda R, Tuji A, Matsuda Y, Fujita M, Yagi T. Genomic organization of the family of CNR cadherin genes in mice and humans. Genomics. 2000;63:75–87. doi: 10.1006/geno.1999.6066. [DOI] [PubMed] [Google Scholar]
- 11.Vanhalst K, Kools P, Vanden EE, van RF. The human and murine protocadherin-beta one-exon gene families show high evolutionary conservation, despite the difference in gene number. FEBS Lett. 2001;495:120–125. doi: 10.1016/s0014-5793(01)02372-9. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Weiner JA, Levi S, Craig AM, Bradley A, Sanes JR. Gamma protocadherins are required for survival of spinal interneurons. Neuron. 2002;36:843–854. doi: 10.1016/s0896-6273(02)01090-5. [DOI] [PubMed] [Google Scholar]
- 13.Phillips GR, Tanaka H, Frank M, Elste A, Fidler L, Benson DL, Colman DR. Gamma-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. J. Neurosci. 2003;23:5096–5104. doi: 10.1523/JNEUROSCI.23-12-05096.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blank M, Triana-Baltzer GB, Richards CS, Berg DK. Alpha-protocadherins are presynaptic and axonal in nicotinic pathways. Mol. Cell Neurosci. 2004;26:530–543. doi: 10.1016/j.mcn.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Hasegawa S, Hamada S, Kumode Y, Esumi S, Katori S, Fukuda E, Uchiyama Y, Hirabayashi T, Mombaerts P, Yagi T. The protocadherin-alpha family is involved in axonal coalescence of olfactory sensory neurons into glomeruli of the olfactory bulb in mouse. Mol. Cell Neurosci. 2008;38:66–79. doi: 10.1016/j.mcn.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 16.Katori S, Hamada S, Noguchi Y, Fukuda E, Yamamoto T, Yamamoto H, Hasegawa S, Yagi T. Protocadherin-alpha family is required for serotonergic projections to appropriately innervate target brain areas. J. Neurosci. 2009;29:9137–9147. doi: 10.1523/JNEUROSCI.5478-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiner JA, Wang X, Tapia JC, Sanes JR. Gamma protocadherins are required for synaptic development in the spinal cord. Proc. Natl Acad. Sci. USA. 2005;102:8–14. doi: 10.1073/pnas.0407931101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Su H, Bradley A. Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 2002;16:1890–1905. doi: 10.1101/gad.1004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, Koide T, Kitsukawa T, Hamada S, Yagi T. Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat. Genet. 2005;37:171–176. doi: 10.1038/ng1500. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko R, Kato H, Kawamura Y, Esumi S, Hirayama T, Hirabayashi T, Yagi T. Allelic gene regulation of Pcdh-alpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. J. Biol. Chem. 2006;281:30551–30560. doi: 10.1074/jbc.M605677200. [DOI] [PubMed] [Google Scholar]
- 21.Ribich S, Tasic B, Maniatis T. Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc. Natl Acad. Sci. USA. 2006;103:19719–19724. doi: 10.1073/pnas.0609445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 23.Yu WP, Yew K, Rajasegaran V, Venkatesh B. Sequencing and comparative analysis of fugu protocadherin clusters reveal diversity of protocadherin genes among teleosts. BMC Evol. Biol. 2007;7:49. doi: 10.1186/1471-2148-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 25.Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. In: Baxevanis AD, editor. Current Protocols in Bioinformatics. John Wiley and Sons; 2003. [DOI] [PubMed] [Google Scholar]
- 26.Muyrers JP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith SJ, Fairclough L, Latinkic BV, Sparrow DB, Mohun TJ. Xenopus laevis transgenesis by sperm nuclear injection. Nat. Protoc. 2006;1:2195–2203. doi: 10.1038/nprot.2006.325. [DOI] [PubMed] [Google Scholar]
- 28.Sparrow DB, Latinkic B, Mohun TJ. A simplified method of generating transgenic Xenopus. Nucleic Acids Res. 2000;28:E12. doi: 10.1093/nar/28.4.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simonis M, Kooren J, de LW. An evaluation of 3C-based methods to capture DNA interactions. Nat. Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 30.Splinter E, Grosveld F, de LW. 3C technology: analyzing the spatial organization of genomic loci in vivo. Methods Enzymol. 2004;375:493–507. doi: 10.1016/s0076-6879(03)75030-7. [DOI] [PubMed] [Google Scholar]
- 31.Yu WP, Rajasegaran V, Yew K, Loh WL, Tay BH, Amemiya CT, Brenner S, Venkatesh B. Elephant shark sequence reveals unique insights into the evolutionary history of vertebrate genes: a comparative analysis of the protocadherin cluster. Proc. Natl Acad. Sci. USA. 2008;105:3819–3824. doi: 10.1073/pnas.0800398105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, Maniatis T. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol. Cell. 2002;10:21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 33.Mori N, Schoenherr C, Vandenbergh DJ, Anderson DJ. A common silencer element in the SCG10 and type II Na+ channel genes binds a factor present in nonneuronal cells but not in neuronal cells. Neuron. 1992;9:45–54. doi: 10.1016/0896-6273(92)90219-4. [DOI] [PubMed] [Google Scholar]
- 34.Kraner SD, Chong JA, Tsay HJ, Mandel G. Silencing the type II sodium channel gene: a model for neural-specific gene regulation. Neuron. 1992;9:37–44. doi: 10.1016/0896-6273(92)90218-3. [DOI] [PubMed] [Google Scholar]
- 35.Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 36.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 37.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 38.Otto SJ, McCorkle SR, Hover J, Conaco C, Han JJ, Impey S, Yochum GS, Dunn JJ, Goodman RH, Mandel G. A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J. Neurosci. 2007;27:6729–6739. doi: 10.1523/JNEUROSCI.0091-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel PD, Bochar DA, Turner DL, Meng F, Mueller HM, Pontrello CG. Regulation of tryptophan hydroxylase-2 gene expression by a bipartite RE-1 silencer of transcription/neuron restrictive silencing factor (REST/NRSF) binding motif. J. Biol. Chem. 2007;282:26717–26724. doi: 10.1074/jbc.M705120200. [DOI] [PubMed] [Google Scholar]
- 40.Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Gottgens B, Buckley NJ. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl Acad. Sci. USA. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mortazavi A, Leeper Thompson EC, Garcia ST, Myers RM, Wold B. Comparative genomics modeling of the NRSF/REST repressor network: from single conserved sites to genome-wide repertoire. Genome Res. 2006;16:1208–1221. doi: 10.1101/gr.4997306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kemp DM, Lin JC, Habener JF. Regulation of Pax4 paired homeodomain gene by neuron-restrictive silencer factor. J. Biol. Chem. 2003;278:35057–35062. doi: 10.1074/jbc.M305891200. [DOI] [PubMed] [Google Scholar]
- 43.Kemp DM, Lin JC, Ubeda M, Habener JF. NRSF/REST confers transcriptional repression of the GPR10 gene via a putative NRSE/RE-1 located in the 5′ promoter region. FEBS Lett. 2002;531:193–198. doi: 10.1016/s0014-5793(02)03502-0. [DOI] [PubMed] [Google Scholar]
- 44.Huang Y, Myers SJ, Dingledine R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 1999;2:867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- 45.Naruse Y, Aoki T, Kojima T, Mori N. Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc. Natl Acad. Sci. USA. 1999;96:13691–13696. doi: 10.1073/pnas.96.24.13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roopra A, Sharling L, Wood IC, Briggs T, Bachfischer U, Paquette AJ, Buckley NJ. Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Mol. Cell Biol. 2000;20:2147–2157. doi: 10.1128/mcb.20.6.2147-2157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl Acad. Sci. USA. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De GS, Houhou L, Oda Y, Corbex M, Pajak F, Thevenot E, Vodjdani G, Mallet J, Berrard S. Is RE1/NRSE a common cis-regulatory sequence for ChAT and VAChT genes? J. Biol. Chem. 2000;275:36683–36690. doi: 10.1074/jbc.M006895200. [DOI] [PubMed] [Google Scholar]
- 49.Uvarov P, Pruunsild P, Timmusk T, Airaksinen MS. Neuronal K+/Cl- co-transporter (KCC2) transgenes lacking neurone restrictive silencer element recapitulate CNS neurone-specific expression and developmental up-regulation of endogenous KCC2 gene. J. Neurochem. 2005;95:1144–1155. doi: 10.1111/j.1471-4159.2005.03434.x. [DOI] [PubMed] [Google Scholar]
- 50.Frank M, Ebert M, Shan W, Phillips GR, Arndt K, Colman DR, Kemler R. Differential expression of individual gamma-protocadherins during mouse brain development. Mol. Cell Neurosci. 2005;29:603–616. doi: 10.1016/j.mcn.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Morishita H, Murata Y, Esumi S, Hamada S, Yagi T. CNR/Pcdhalpha family in subplate neurons, and developing cortical connectivity. Neuroreport. 2004;15:2595–2599. doi: 10.1097/00001756-200412030-00007. [DOI] [PubMed] [Google Scholar]
- 52.Stapleton T, Luchman A, Johnston J, Browder L, Brenner S, Venkatesh B, Jirik FR. Compact intergenic regions of the pufferfish genome facilitate isolation of gene promoters: characterization of Fugu 3′-phosphoadenosine 5′-phosphosulfate synthase 2 (fPapss2) gene promoter function in transgenic Xenopus. FEBS Lett. 2004;556:59–63. doi: 10.1016/s0014-5793(03)01353-x. [DOI] [PubMed] [Google Scholar]
- 53.Zhang T, Tan YH, Fu J, Lui D, Ning Y, Jirik FR, Brenner S, Venkatesh B. The regulation of retina specific expression of rhodopsin gene in vertebrates. Gene. 2003;313:189–200. doi: 10.1016/s0378-1119(03)00680-2. [DOI] [PubMed] [Google Scholar]
- 54.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 55.Tada MN, Senzaki K, Tai Y, Morishita H, Tanaka YZ, Murata Y, Ishii Y, Asakawa S, Shimizu N, Sugino H, et al. Genomic organization and transcripts of the zebrafish Protocadherin genes. Gene. 2004;340:197–211. doi: 10.1016/j.gene.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 56.Noonan JP, Grimwood J, Danke J, Schmutz J, Dickson M, Amemiya CT, Myers RM. Coelacanth genome sequence reveals the evolutionary history of vertebrate genes. Genome Res. 2004;14:2397–2405. doi: 10.1101/gr.2972804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noonan JP, Grimwood J, Schmutz J, Dickson M, Myers RM. Gene conversion and the evolution of protocadherin gene cluster diversity. Genome Res. 2004;14:354–366. doi: 10.1101/gr.2133704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang XJ, Li S, Ravi V, Venkatesh B, Yu WP. Identification and comparative analysis of the protocadherin cluster in a reptile, the green anole lizard. PLoS ONE. 2009;4:e7614. doi: 10.1371/journal.pone.0007614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kallunki P, Edelman GM, Jones FS. Tissue-specific expression of the L1 cell adhesion molecule is modulated by the neural restrictive silencer element. J. Cell Biol. 1997;138:1343–1354. doi: 10.1083/jcb.138.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mu W, Burt DR. Transcriptional regulation of GABAA receptor gamma2 subunit gene. Brain Res. Mol. Brain Res. 1999;67:137–147. doi: 10.1016/s0169-328x(99)00049-2. [DOI] [PubMed] [Google Scholar]
- 61.Mahajan MC, Karmakar S, Weissman SM. Control of beta globin genes. J. Cell Biochem. 2007;102:801–810. doi: 10.1002/jcb.21507. [DOI] [PubMed] [Google Scholar]
- 62.Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 63.Nguyen MQ, Zhou Z, Marks CA, Ryba NJ, Belluscio L. Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell. 2007;131:1009–1017. doi: 10.1016/j.cell.2007.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.