Abstract
Despite the critical role of pre-mRNA splicing in generating proteomic diversity and regulating gene expression, the sequence composition and function of intronic splicing regulatory elements (ISREs) have not been well elucidated. Here, we employed a high-throughput in vivo Screening PLatform for Intronic Control Elements (SPLICE) to identify 125 unique ISRE sequences from a random nucleotide library in human cells. Bioinformatic analyses reveal consensus motifs that resemble splicing regulatory elements and binding sites for characterized splicing factors and that are enriched in the introns of naturally occurring spliced genes, supporting their biological relevance. In vivo characterization, including an RNAi silencing study, demonstrate that ISRE sequences can exhibit combinatorial regulatory activity and that multiple trans-acting factors are involved in the regulatory effect of a single ISRE. Our work provides an initial examination into the sequence characteristics and function of ISREs, providing an important contribution to the splicing code.
INTRODUCTION
Post-transcriptional gene regulatory mechanisms play central roles in programming the complexity of biological systems. One such process is alternative splicing, a mechanism that produces multiple protein isoforms from a single gene by altering the ways in which exons are joined (1). Splicing patterns are regulated by the interplay between auxiliary cis-acting elements that include exonic and intronic splicing enhancers (ESEs and ISEs, respectively) and silencers (ESSs and ISSs, respectively) and the trans-acting factors that modulate them, leading to a ‘splicing code’ (2). The elucidation of this splicing code is of great interest given that >90% of human genes are alternatively spliced (3) and up to 50% of disease-causing mutations affect splicing (4).
Recent genome-wide studies have made significant progress in identifying the splice isoforms for all human transcripts (5–7). In addition, experiments coupling high-throughput sequencing with cross-linking immunoprecipitation (CLIP) have led to the identification of functional protein-RNA interactions in vivo for a small subset of trans-acting splicing factors (8,9). However, these experimental tools have not allowed for a systematic examination of nucleotide sequences that can confer changes in splicing patterns. High-throughput functional screens that rely on changes in alternatively spliced transcript levels coupled with bioinformatic analyses are needed to support such studies.
Bioinformatic and experimental analyses have identified several RNA motifs that regulate splicing; however, much of this effort has been directed toward the functional characterization of cis-acting exonic regulatory sequences. Specifically, in vivo, in vitro and in silico strategies have been implemented to screen for ESEs and ESSs from small randomized libraries (10–15) or genomic sequence data (15,16). Selected ESSs are able to control alternative 5′ and 3′ ss recognition when placed between competing sites (17). ESEs and ESSs also play critical roles in directing splicing to consensus splice sites rather than decoy sites (18). Several properties of intronic splicing regulatory elements (ISREs) have complicated their functional characterization. Recently, ISSs and ISEs have been identified near alternatively spliced exons and exhibit antagonistic activities (19), suggesting that they behave in a combinatorial manner (20). ISSs may inhibit exon inclusion by recruiting one or more repressors that directly antagonize splicing factor binding (21). The activities of several intronic elements have been shown to be context dependent (3). Despite the widespread importance of ISREs, knowledge regarding their sequence composition, the mechanisms through which they regulate splicing and the regulatory networks of trans-acting splicing factors by which they are bound (splicing regulatory networks, SRNs) is limited (22).
We have developed an in vivo screening strategy for ISREs, which we call SPLICE (Screening PLatform for Intronic Control Elements). SPLICE was used to begin to generate a functional definition of ISREs by identifying sequences adjacent to the 3′ splice site (ss) that regulate the inclusion of an alternatively spliced exon that triggers rapid transcript decay. Our approach combines a systematic screening strategy, genome-wide bioinformatic analyses and experimental characterization to identify ISRE consensus motifs and characterize the SRNs associated with these regulatory elements. Our results indicate that cis-acting intronic regulatory sequences function through combinatorial effects from multiple elements and trans-acting factors, and that the immediate transcript context has a dominant effect on ISRE activity.
MATERIALS AND METHODS
Base SPLICE constructs
Plasmids were constructed using standard molecular biology techniques (23). All enzymes, including restriction enzymes and ligases, were obtained through New England Biolabs unless otherwise noted. DNA synthesis was performed by Integrated DNA Technologies, Inc. Ligation products were electroporated into Escherichia coli DH10B (Invitrogen) using a GenePulser XP system (BioRAD), and clones verified through colony polymerase chain reaction (PCR) and restriction mapping. All cloned constructs were sequence verified through Laragen. Primer sequences and plasmid descriptions are available in Supplementary Tables S1 and S2, respectively.
The green fluorescent protein (GFP)-SMN1 mini-gene fusion construct was constructed through a PCR assembly and site-directed mutagenesis strategy. A region encompassing exons 6 through 8 of the SMN1 mini-gene was amplified through PCR from template pCISMNxΔ6-wt (24) with primers Ex6 and Ex8 and PfuUltra high-fidelity DNA polymerase (Stratagene). The GFP gene was amplified from the template pKW430 (25) with primers GFP1 and GFP2. The GFP-SMN1 gene fusion was constructed by performing PCR assembly on the resulting purified products (Qiagen) as templates and flanking primers GFP1 and Ex8. The resulting gene fusion product was digested with XhoI and KpnI and ligated into the corresponding restriction sites of the mammalian expression vector pcDNA5/FRT (Invitrogen), resulting in the positive control vector pCS238. A PTC (TAA at position +1 in exon 7) and ISRE insertion sites EcoRV/ClaI in intron 6 (positions –62 and –51 from 3′ ss of exon 7, respectively) were introduced by site-directed mutagenesis with primers ECmutF1/ECmutR1 using a Quickchange II Kit (Stratagene) according to manufacturer’s instructions, resulting in the base nonsense-mediated decay (NMD) reporter construct pCS516. ISRE sequences were digested and ligated into the EcoRV and ClaI restriction sites within intron 6 of the base NMD construct.
ISRE library and ISS controls construction
A random 15-nt ISRE library was generated through PCR using a 47-nt template (ISStemp) with primers Lib1 and Lib2. The library PCR was conducted for 12 cycles in a 100 µl reaction containing 20 pmol DNA template, 300 pmol each Lib1 and Lib2, 200 µM each dNTPs, 1.6 mM MgCl2 and 10 U Taq DNA polymerase (Roche). ISS and negative controls were constructed by replacing the random 15-nt region in the above template with previously characterized ISS sequences and scrambled sequences, respectively. The resulting ISRE library, ISS control and negative control fragments were digested with EcoRV and ClaI and ligated into the corresponding restriction sites within intron 6 of pCS516. Control ISS sequences correspond to previously characterized binding sites for U2AF65 (TTTTTTTTTCCTTTTTTTTTCCTTTT; pCS668) (26); hnRNP H (TAAATGTGGGACCTAGA; pCS669) (27), PTB(1) (TAGCATCAGCCTGG TGCCTACCTTCGGCCCC; pCS670) (26); PTB(2) (TCTTCTCTTCTCTTCTCTTC; pCS667) (28). In addition, 15 scrambled sequences were examined in place of the 15-nt random region as negative control constructs. The base random 15-nt sequence ACCTCAGGCTCTGAA (pCS517) was subsequently used as the negative control for all FACS experiments.
Cell culture, transfections, stable cell lines and FACS
HEK-293 FLP-In cells (Invitrogen) were cultured in D-MEM supplemented with 10% fetal bovine serum (FBS) and 100 μg/ml Zeocin at 37°C in 5% CO2. HeLa cells were cultured in MEM media supplemented with 10% FBS. Transfections for all cell lines were carried out with Fugene (Roche) according to the manufacturer’s instructions. All cell culture media was obtained from Invitrogen.
HEK-293 FLP-In stable cell lines were generated by co-transfection of the appropriate SMN1 mini-gene construct with a plasmid encoding the Flp recombinase (pOG44) in growth medium without Zeocin according to the manufacturer’s instructions (Invitrogen). The library stable selections were carried out in 225 cm2 flasks containing ∼4 × 107 HEK-293 FLP-In cells where 37 µg of pOG44 and 3.7 µg of the SMN1 ISRE plasmid library (10:1 ratio) were co-transfected. Fresh medium was added to the cells 24 h after transfection. The cells were expanded by a 1:4 dilution and Hygromycin B was added to a final concentration of 200 μg/ml 48 h after transfection. In total, ∼450 000 stable transformants were pooled from 60 transfections. Clones were harvested by trypsinization, pooled and analyzed on a FACS Aria (Becton Dickinson Immunocytometry Systems) 10–14 days after transfection. GFP fluorescence was excited at 488 nm and emission was measured with a FITC filter. Detailed sorting procedures are presented in Supplementary Figure S1. In the first screening round, positive cells were bulk sorted into 96-well plates, where no more than 25 000 cells were collected into a single well. After ∼1–2 weeks of growth, positives were re-sorted into three fractions (A, B and C) based on varying fluorescence levels (Supplementary Figure S1B). Positive cells were bulk sorted in the second screening round as described for the first round. Total genomic DNA from bulk sorted cells was purified using the DNeasy Blood & Tissue total DNA purification kit (Qiagen) according to the manufacturer’s instructions and used as a template for amplification of recovered ISRE sequences with primers Lib3 and Lib4. The recovered ISRE fragments were then digested, ligated into the corresponding sites of pCS516 and sequenced verified by Functional Biosciences, Inc.
For transient transfection studies, HEK-293 and HeLa cells were seeded in 12-well plates at ∼5 × 104 cells per well 16–24 h prior to transfection. Cell lines were transfected with 625 ng of the appropriate GFP-SMN1 mini-gene constructs. The cells were harvested by trypsinization, pooled and analyzed on a FACS Aria 48 h after transfection. Experiments were carried out on different days and transfections were completed in duplicate, where the mean GFP fluorescence of the transfected population and the average error between samples is reported. A comparison of FACS gating procedures used in transient and stable assays is presented in Supplementary Figure S2. Cell lines harboring PTC-containing transcripts tend to increase in fluorescence at higher passage numbers (>10), whereas the GFP-SMN1 cell line does not. As such, the fluorescence levels of the enriched cell populations at the time of sorting (Figure 1G) do not directly match the expression levels for individual recreated clones (Figure 2B). To minimize differences in expression due to such instabilities, the analysis of all stable cell lines was performed at an identical, early passage.
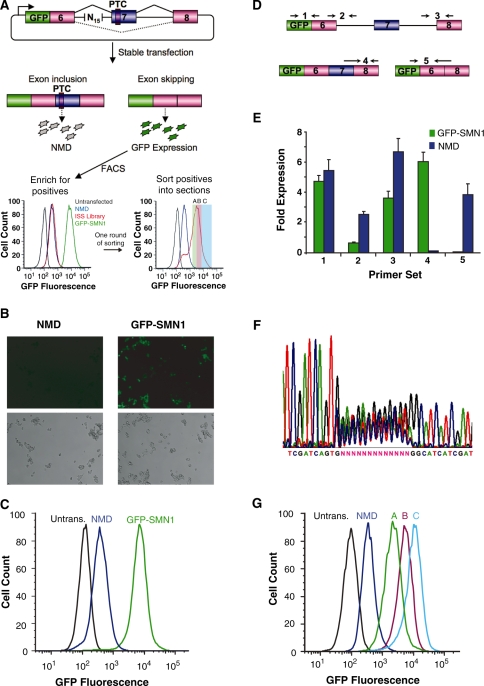
Figure 1.
A Screening PLatform for Intronic Control Elements (SPLICE) provides a generalizable in vivo screening strategy for ISREs. (A) SPLICE couples an exon inclusion event in a mini-gene (SMN1) to the expression level of a fluorescent protein (GFP) through a NMD-based reporter system. A random nucleotide library cloned upstream of the 3′ ss is screened for ISREs by sorting cells based on fluorescence levels. The enriched cells are sorted into sections (A, B, C) in a second screening round. (B) Microscope images of stable cell lines expressing the negative (NMD) and positive (GFP-SMN1) control constructs. (Upper panels) GFP fluorescence, lower panels: phase contrast images. (C) Flow cytometry histograms of stable cell lines expressing the control constructs. An untransfected HEK-293 FLP-In cell population (Untrans.) was also analyzed for reference. (D) Schematic representing the relative locations of primer set binding for transcript isoform analysis by qRT-PCR. Primer sets were used to quantify levels of total transcript (set 1), intron 6 retention (set 2), intron 7 retention (set 3), exon 7 included isoform (set 4) and exon excluded isoform (set 5). Primer binding locations within the SMN1 mini-gene are presented in Supplementary Figure S3. (E) qRT-PCR analysis of the NMD and GFP-SMN1 control cell lines supports decay of the PTC harboring isoform. Expression levels were normalized to the levels of HPRT (hypoxanthine-guanine phosphoribosyltransferase). Data presented is the mean expression of duplicate PCR samples ± the average error. (F) DNA sequencing analysis of purified genomic DNA from HEK-293 cell lines harboring the library constructs. (G) The enriched cell populations maintain the fluorescence levels of the sorted sections (A, B, C). Following the second round of sorting, the fluorescence levels of expanded populations were re-analyzed through flow cytometry to confirm maintenance of expression levels.
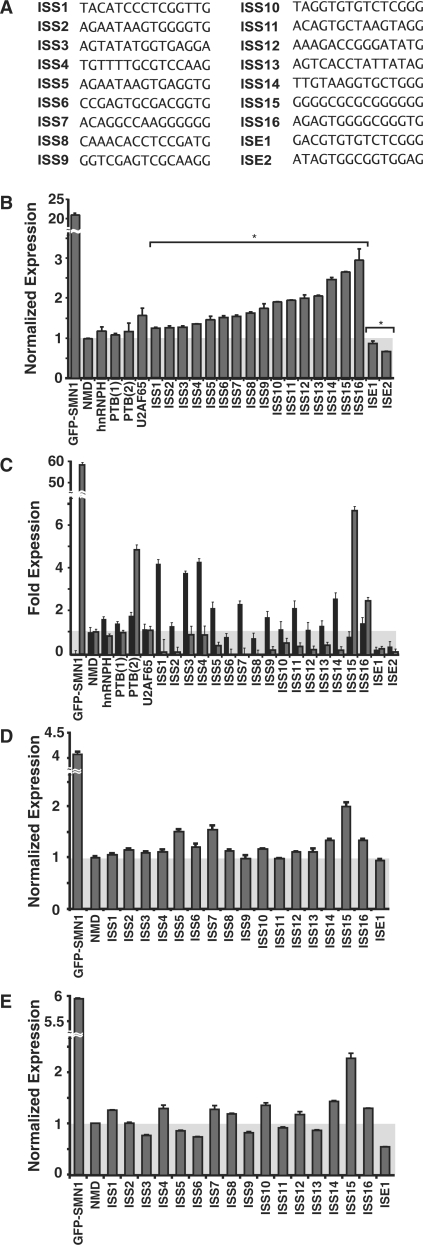
Figure 2.
Functional analysis of recovered ISRE sequences. (A) Recovered ISRE sequences examined for regulatory activity. (B) Flow cytometry analysis of HEK-293 FLP-In stable cell lines generated for recovered ISRE sequence and control constructs. For all reported activities, the mean GFP levels from two independent experiments were determined and normalized to the NMD control. Normalized expression and average error are reported. ISRE sequences are labeled according to function. P-values derived from the Student’s t-test are as follows: *P < 0.05. (C) qRT-PCR analysis of the recovered ISRE sequences and control constructs with primer sets specific for exon 7 excluded (primer set 5, black bars) and included (primer set 4, gray bars) products. Fold expression data is reported as the mean expression for each sample divided by the mean NMD expression value ± the average error. (D, E) Flow cytometry analysis of recovered ISRE sequences and control constructs transiently transfected in (D) HEK-293 and (E) HeLa cells.
Quantitative reverse transcriptase real-time PCR
Total cellular RNA was purified from stably transfected HEK-293 Flp-In cells using GenElute mammalian total RNA purification kit (Sigma) according to the manufacturer’s instructions, followed by DNase treatment (Invitrogen). cDNA was synthesized using a gene-specific primer for the pcDNA5/FRT vector (SMN1cDNA) and Superscript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. qRT-PCR analysis was performed using isoform-specific primers (Supplementary Tables S3 and S4) where each reaction contained 1 µl template cDNA, 10 pmol of each primer and 1X iQ SYBR green supermix (BioRAD) to a final volume of 25 µl. Reactions were carried out using a iCycler iQ system (BioRAD) for 30 cycles (95°C for 15 s, 72°C for 30 s). The purity of the PCR products was determined by melt curve analysis. Data analysis was completed using the iCycler IQ system software v.3.1.7050 (BioRAD). Isoform-specific relative expression was calculated using the ΔCt (change in cycling threshold) method (29). Expression levels were normalized to the levels of HPRT (hypoxanthine-guanine phosphoribosyltransferase). Fold expression data are reported as the mean expression for each sample divided by the mean NMD expression value ± the average error.
siRNA-mediated silencing of trans-acting splicing factors
siRNAs targeting hnRNP H (GAUCCACCACGAAAGCUUA), hnRNP A1 (CAACUUCGGUCGUGGAGGA), PTB (CGUCAAAGGAUUCAAGUUC), CUG-BP1 (GAGCCAACCUGUUCAUCUA) and SF2/ASF(CGUGGAGUUUGUACGGAAA) and a mock control siRNA were purchased from Dharmacon. All duplexes were resuspended in 1X PBS to a concentration of 20 µM. Briefly, HEK-293 FLP-In cells were plated at ∼2 × 105 cells per well in six-well plates. After 24 h, the cells were transfected with individual siRNA duplexes to a final concentration of 50 nM using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. Cells were collected for RNA isolation and western blotting 48 h after transfection.
Western blot analysis
Whole-cell extracts were prepared from harvested cells using M-PER mammalian protein extraction reagent (Pierce) and equal amounts of protein (50 µg) were resolved on 4–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels (Invitrogen) and transferred onto Protran nitrocellulose membranes (Whatman) using the Trans-Blot SD semi-dry transfer cell (BioRAD). After blocking with 5% BSA in TBST, the membranes were incubated with the specified antibodies overnight at 4°C. After incubation, the membranes were washed with TBST and then incubated with the corresponding secondary antibody conjugated with HRP. Signals were detected using the ECL western blotting substrate (Thermo Scientific) according to the manufacturer’s protocol. The primary antibody dilutions were 1 : 500 for goat anti-hnRNP H (N-16), 1 : 1000 for goat anti-Actin (I-19), 1 : 200 for goat anti-hnRNP A1 (Y-15), 1 : 200 for mouse anti-PTB (SH54), 1 : 200 for mouse anti-SF2/ASF (96) and 1 : 200 for mouse anti-CUG-BP1 (3B1). The secondary antibody dilutions were 1 : 10 000 for donkey anti-goat IgG-HRP (sc-2020) and 1 : 10 000 for goat anti-mouse IgG-HRP (sc-2005). All of the antibodies were purchased from Santa Cruz Biotechnology Inc. The relative band intensities were measured by densitometry analyses using Quantity One software (BioRAD).
Discovery of sequence motifs enriched in ISRE sequences
A sliding-window count of all n-mers (4–6 nt) within the nonredundant sample set of 125 sequences was performed. Two nucleotides flanking the 5′- and 3′-ends of the random region were included to account for bias due to the constant sequences. A similar sliding-window count on a set of 450 000 computer generated sequences containing a uniformly random 15-nt region flanked by the same constant nucleotides was performed to calculate the maximum likelihood probabilities for expected occurrences. For both data sets, the counts were transformed into probabilities and the enrichment was determined according to the binomial confidence interval method (30). Sequence motifs were constructed from the significantly enriched (P < 0.1) n-mers using the graph clustering method and software (GCCS) (30) with the following parameters: minimum cluster size = 4, rounds of clustering = 5, minimum substring length = 5 (rounds 1–3) and 4 (rounds 4 and 5). GCCS uses the MCL algorithm (31) to find clusters. Parameters were set as follows: MCL inflation = 3 and MCL scheme = 4. The other MCL parameters were set to default values. To validate the enrichment of ISRE motifs, the GCCS analysis was repeated using five sets of 125 random 15-mers with the same constant flanking bases. The average number of significantly enriched n-mers observed in the random samples (RS) was only 91 and each yielded an average of 11 clusters.
Overlap of ISRE sequences with known splicing regulatory elements
The set of pentamers enriched in the ISRE sequences were compared to previously compiled lists of ESEs (15,16), ESSs (14,16) and ISEs (32) (http://www.snl.salk.edu/∼geneyeo/stuff/papers/supplementary/ISRE/). These data sets were originally reported as hexamers, such that pentameric equivalents were created by extracting all pentamers that occurred at least one time within the original data sets. The ISRE enriched pentamers were also compared to ISREs (33), conserved intronic sequences (CISs) (30) and motifs enriched upstream of weak polypyrimidine (PY) tracts (34). These data sets were composed of various length n-mers and were adjusted to pentameric equivalents to achieve independent sampling by extracting all pentamers that occurred at least once. Lastly, the ISRE pentamers were compared to conserved pentamers enriched in intronic regions of exons excluded in neural progenitor (NP) cells (35).
The significance of overlap between data sets was determined using a 2 × 2 Chi-test of association. Each pentamer was classified according to which of the four ways it could be distributed: (i) in both sets, (ii) in set A but not set B, (iii) not in set A but in set B and (iv) in neither set. The counts for each distribution were then used to calculate the likelihood that this arrangement could have occurred randomly (according to the Chi-distribution with one degree of freedom).
Statistical analysis
Student’s t-test and analysis of variance (ANOVA) analyses were performed using Microsoft Excel. P < 0.05 were taken to be significant. P-values derived from the Student’s t-test are as follows: *P < 0.05 and **P < 0.01, unless otherwise noted.
RESULTS
SPLICE: a Screening PLatform for Intronic Control Elements
SPLICE is a high-throughput in vivo screen for ISRE function based on a construct encoding the GFP fused 5′ of a three-exon, two-intron mini-gene (Figure 1A). The alternatively spliced middle exon harbors a premature termination codon (PTC) that triggers the NMD pathway (36), such that cells with higher levels of exon inclusion will display lower fluorescence. We positioned a random 15-nt library 45-nt upstream of the 3′ ss in the first intron (33-nt upstream of the branch point, Supplementary Figure S3) within the acceptor intronic region, as auxiliary splicing elements are normally located close to splice sites and have been shown to vary between 10- and 30-nt in length (37). Library placement was based on previous results from computational studies, suggesting that ISREs are prevalent in the –64 to –15 upstream region of exons (38) and starting from intron position –41 (32). For the initial screen, we used a cassette exon mini-gene construct based on part of the SMN1 gene because the splicing elements within this gene region have been well characterized. In contrast to previous in vivo screening strategies that require modification to select for enhancers and silencers (13,14), by coupling NMD to splicing efficiency both ISSs and ISEs can be selected within the same construct as cells exhibiting increased levels of fluorescence harbor putative ISSs and those exhibiting lower levels of fluorescence harbor putative ISEs. In addition, our construct design allows for relatively easy modification of the reporter gene and mini-gene sequences and for the screening of ISREs in the context of natural exon–intron sequences, as opposed to previous strategies that are based on chimeric coupling of exons and introns from different transcripts (13,14,39).
We built a NMD-based reporter construct (NMD control), containing a 15-nt control insert and a PTC in exon 7, and a construct lacking a PTC (GFP-SMN1 control) as negative and positive controls, respectively. Constructs were stably transfected into HEK-293 FLP-In cells to generate isogenic cell lines. Flow cytometry (Figure 1A and C) and fluorescence microscopy analyses (Figure 1B) reveal that the fluorescence levels between the GFP-SMN1 and NMD controls differ by ∼22-fold. Transcript isoform analysis indicates that the level of exon 7 inclusion in the NMD control is ∼60-fold less than the GFP-SMN1 control (Figure 1D and E), supporting that observed differences in fluorescence are due to differences in exon inclusion. The high level of exon 7 inclusion for the GFP-SMN1 control is in line with previous observations for the splicing of the SMN1 mini-gene (40). The elevated levels of exon 7 exclusion in the NMD control compared to the GFP-SMN1 control suggest that the PTC may have a secondary effect of increasing the levels of the exon excluded transcript. Such observations have been previously observed and may be the result of nonsense-associated altered splicing (41).
A library of synthetic oligonucleotides containing a random 15-nt region (∼1 × 109 sequences) was ligated into the NMD control construct and transformed into E. coli. Library constructs were purified from ∼1 × 106 transformants, representing ∼0.1% of possible sequences, and transfected into HEK-293 FLP-In cells, generating ∼450 000 stable transformants (a sampling of ∼0.045% of the library, see ‘Materials and Methods’ section). Minimal sequence bias was observed before and after transfection (Figure 1F), indicating that the ISRE library represents an essentially random pool. Flow cytometry analysis indicated that ∼0.1% of the cell population exhibits fluorescence levels greater than the NMD control, corresponding to putative ISSs. The top ∼0.1% of the cell population was bulk sorted, grown 2–3 weeks in culture and re-analyzed by flow cytometry. The round-one pool exhibits an ∼6-fold increase in mean fluorescence compared to the NMD control (Figure 1A) and was sorted into groups (A, B and C) based on fluorescence levels to further enrich the population and select for sequences varying in activity (Figure 1A and Supplementary Figure S1). Sorted section A fell within a fluorescence region slightly below and above the NMD control. As putative ISEs are expected to exhibit fluorescence levels lower than the NMD control, sorted section A may harbor sequences exhibiting ISE activity. The mean fluorescence levels of the enriched populations correlated well with their sorted sections (Figure 1G).
Recovered ISRE sequences enable tuning of alternative splicing
We identified 125 unique sequences with enhanced fluorescence from 226 sequenced isolates derived from the three sorted sections (Supplementary Table S5). A subset of these sequences was individually cloned into the NMD-based reporter and stable cell lines harboring these constructs were generated to validate the activity of the recovered sequences. We used fluorescence and qRT-PCR assays to analyze 18 randomly selected 15-mers (Figure 2A) and four known ISS sequences: an hnRNP H (27), two PTB (26,28) and a U2AF65 binding sites (26). Of the known ISSs, only the U2AF65 element demonstrates significant silencing activity relative to the NMD control through flow cytometry analysis, exhibiting an ∼1.5-fold higher fluorescence level (Figure 2B). This result is in line with previous studies demonstrating that the activities of several characterized splicing regulatory elements (SREs) are context dependent (3). In contrast, 16 of the selected sequences display significant silencer activity and two exhibit enhancer activity relative to the NMD control (Figure 2B). Similar trends were observed from an additional 12 sequences (Supplementary Figure S4). The activities of the majority of tested sequences, grouped as low (ISS1-5), medium (ISS6-10) and high (ISS11-16), correlated with sectioned populations (Figure 1G), supporting that regulatory activity is related to sequence.
The regulatory activity of the selected sequences and ISS controls were confirmed through transcript isoform analysis by qRT-PCR. The total transcript levels (primer set 1, Figure 1D) and the levels of intron retention (primer sets 2 and 3) for the examined ISS and control sequences were similar to the NMD control, while these levels for the selected ISEs differed from the NMD control (Supplementary Figure S5A–D). The GFP-SMN1 control exhibits a low level of the skipped exon isoform (primer set 5, Figure 1D) compared to the NMD control (Figure 2C). Most of the recovered and control ISS sequences exhibit significantly higher levels of the skipped exon isoform than the NMD control (P < 0.05), with the exception of ISS8 and ISS15. In addition, the ISE sequences exhibited lower levels of the skipped exon isoform relative to the NMD control (P < 0.05).
All constructs except for the GFP-SMN1 control are expected to exhibit low levels of the exon 7 included isoform, as this isoform should be rapidly degraded through NMD. The GFP-SMN1 control exhibits a high level of exon 7 inclusion (99.7%) (primer set 4, Figure 1D). Exon inclusion levels for the ISS controls do not differ from the NMD control (P > 0.35), with the exception of PTB(2), which had a higher level of exon inclusion (P < 0.05). Exon inclusion levels for 14 of the 16 recovered ISS sequences (ISS1-14) and the ISE sequences range from 2- to 20-fold less than the NMD control (P < 0.05), whereas ISS15 and ISS16 exhibited increased levels of the exon included isoform (P < 0.05).
To confirm that the recovered sequences maintain splicing regulatory function independent of triggering decay through the NMD pathway, we inserted several sequences (ISE1, ISS5 and ISS14-ISS16) into a GFP-SMN1 non-NMD based reporter. Transcript isoform analysis by qRT-PCR demonstrates that the activity of the tested sequences in the NMD reporter construct match those in the non-NMD reporter construct (Supplementary Figure S5D). As noted above, the transcript isoform levels of ISS15 and ISS16 do not correlate with measured fluorescence levels from the NMD reporter system, indicating that these two sequences may enable the transcript to somehow evade decay through the NMD pathway and show up as false positives from the screen. It is also possible that certain sequences may result in alternative 3′ ss processing leading to enhanced exon inclusion without triggering NMD. However, gel analysis of qRT-PCR products run with a primer set for exon included isoforms (forward primer set 1, Figure 1D and Supplementary Figure S3B) and a unique primer within exon 7 (Supplementary Table S1) indicates that the exon included spliced products (detected in ISS15 and ISS16) are of a length corresponding to processing at the expected 3′ ss (Supplementary Figure S6). While these data suggest that the sequences do not lead to alternative 3′ ss processing, we cannot rule out the possibility of a minor change at the 3′ ss due to aberrant splicing that would alter the reading frame of the PTC. Overall, these results demonstrate that the majority of the sequences obtained from SPLICE, exhibit transcript isoform levels that correlate with measured fluorescence levels, confirming the utility of this screening system. Moreover, the splicing regulatory activity of the recovered sequences is maintained independent of coupling to the NMD pathway.
ISRE sequences function in a different cell type
The relative levels and activities (42) of trans-acting splicing factors vary across different cell types, which can result in cis-acting sequences exhibiting different regulatory activities. We examined the activity of ISS1-16, ISE1 and the NMD and GFP-SMN1 controls in transient transfection assays in HEK-293 and HeLa cell lines. The qualitative activity of 15 of the recovered ISREs was maintained in HEK-293 and 11 sequences exhibited significantly increased expression (P < 0.05) (Figure 2D). The majority of examined sequences (12 of 16) maintain the same trend in activity in the two cell lines and ANOVA analysis of the activities shows a strong correlation (P < 0.01). However, four of the sequences (ISS3, 5, 6 and 13) exhibit enhancer activity relative to the NMD control in HeLa cells (Figure 2E), which may be due to differences in levels of trans-acting factors, total RNA levels or the rate of NMD between the cell lines. The results support that most sequences recovered from SPLICE retain function in a second cell line and may represent global splicing regulators.
GCCS clustering of recovered ISREs identifies conserved motifs
We utilized Graph Clustering by Common Substrings (GCCS) to identify conserved motifs in the SPLICE-generated sequences (30). Since RNA binding proteins typically recognize short sequence motifs, we restricted our analysis to n-mers from 4-6-nt. We determined the enrichment of n-mers in a 19-nt region of the set of 125 sequences using a confidence interval for the binomial distribution based on probabilities expected for 19-nt sequences containing 15-nt of random bases flanked by 2 constant bases present in the experimental system. In the data set, 241 n-mers consisting of 39 4-mers, 93 5-mers and 109 6-mers were significantly enriched (α1-tailed = 0.1; Supplementary Figure S7; Supplementary Table S6). The GCCS analysis grouped 80.1% of the statistically enriched n-mers into 30 consensus motif clusters that we grouped into three classes based on sequence (Figure 3A and Supplementary Tables S7 and S8).
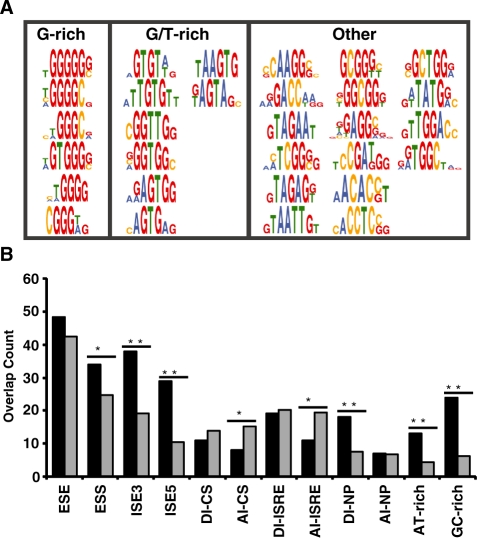
Figure 3.
Enriched motifs derived from recovered ISRE sequences map to known and unknown splicing factors and overlap with SREs. (A) Motifs are grouped into three classes: G-rich, GT-rich and other elements (classes 1–3, respectively). (B) Observed (black) and expected overlap (gray) between data sets. The set of pentamers enriched in the ISRE sequences were compared to previously compiled lists of ESEs (15,16), ESSs (14,16), ISEs (32) ISREs (33), CISs (30), motifs enriched upstream of weak PY tracts (34) and motifs enriched in intronic regions of exons excluded in NP cells (35). P-values derived from the chi-squared test of association are as follows: *P < 0.05 and **P < 0.0001.
Many of the consensus motifs (19 out of 30) identified by the GCCS analysis resemble known binding sites for trans-acting splicing factors (Figure 3A and Supplementary Table S9), supporting the possible functional role of selected ISREs. The first class of enriched motifs are G-rich (contain at least three continuous G nucleotides, class 1). Class 1 motifs resemble binding sites for the splicing factors hnRNP A1 (TAGGG) (43) and hnRNP F/H (GGGGG) (44).
The second class of enriched motifs are GT-rich (contain at least one GT dinucleotide, class 2). The motifs GTGT, TGTG and GGTT resemble known binding sites for the CELF/Bruno-like family, which regulates splicing patterns by binding sequences that contain CTG repeats and exhibits a higher affinity for GT repeats (45). Four motifs in class 2 contain an AGT core element, similar to the hnRNP G binding motif (AAGT) (46).
Half of the GCCS identified motifs do not fall into the G-rich or GT-rich classes (class 3). Three motifs resemble known binding sites for the SR protein family whose members act as general splicing factors. The motifs CAAGG, GACC and TAGAA share three or more nucleotides with binding sites for SRp40 (ACAAG) (10), 9G8 (GACC) (47) and SF2/ASF (GAAGAA) (48), respectively. Although the examples of SR protein involvement in splicing repression are limited, the enrichment of these motifs in our screen may suggest a role for this protein family in intronic regulation. Elements in class 3 may also represent weak binding sites for characterized splicing factors. For example, the motif TCGG[G/C] shares up to 80% sequence identity to a hnRNP A1 binding site. Other elements in this class may represent previously uncharacterized or novel regulatory elements.
Enriched n-mers resemble known splicing regulatory elements
We examined the number of pentamer motifs identified in our enriched n-mer data set (Supplementary Table S6) that are identical to pentamers in published sets of SREs (see ‘Materials and Methods’ section). We analyzed data corresponding to four SRE classes: ESEs (15,16), ESSs (14,16), ISEs (32) and computationally identified conserved intronic elements, which includes CISs (30), ISREs (33), motifs enriched in intronic regions of excluded exons in NP cells (35) and motifs enriched upstream of weak PY tracts in AT- and GC-rich introns (34). Significant overlap exists between the enriched pentamers and ESSs, ISEs, donor intronic (DI, 5′ ss region) elements in NP cells and motifs enriched upstream of weak PY tracts (Figure 3B). The dominant motifs that overlap between SPLICE-generated pentamers and ISEs and weak PY elements are G-rich and GT containing elements. The results suggest that the selected elements may function as ESSs and intronic modulators of splicing depending on their context across various cell types and are likely regulated by general splicing factors. The observed overlap between enriched pentamers and conserved acceptor intronic elements (AI, 3′ ss region) for CIS and ISRE datasets is far less than expected (P < 0.05), suggesting that SPLICE selected against these elements. Elements within clusters CGA, TAGAG, AAGG, GGTT and GCGG do not overlap with known SREs (Supplementary Table S10), indicating that SPLICE may also generate novel regulatory elements.
Analysis of enriched hexamers confirms independent and combinatorial function
Most of the recovered ISRE sequences contain several enriched n-mers, where 88% of all extended 15-mers (plus 2-nt flanking region) contain at least one enriched hexamer which comprise the largest group of n-mers in our data set (Supplementary Table S11). ISS5 contains seven enriched hexamers resembling binding sites for the CELF and SF2/ASF proteins, which cluster into three main regions within the sequence (Figure 4A). We introduced two point mutations within each region and in combination and assessed their activity through transient transfection assays. Mutations within each region decrease expression to levels comparable to the NMD control, supporting that each region has regulatory activity. Simultaneous mutations to regions 2 and 3 resulted in expression levels comparable to or slightly higher than the individual mutations, indicating that the regulatory function of ISS5 is likely not due to combinatorial recognition of motifs.
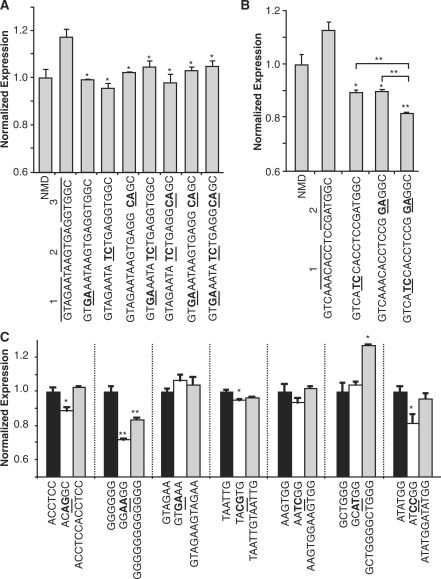
Figure 4.
Enriched ISRE hexamers demonstrate silencer activity. (A) Mutational analysis of ISS5 supports the silencing activity of individual hexamer regions. The combined and individual activity of hexamer regions within the context of ISS5 was examined by introducing 2 point mutations into 3 hexamer regions in combination and separately. Sequences were characterized in transient transfection assays in HEK-293 cells. For all reported data, silencing activity was assessed by flow cytometry analysis, where the mean GFP levels from two independent experiments were normalized to the NMD control. Normalized expression and average error are reported. P-values derived from the Student’s t-test are as follows: *P < 0.05 and **P < 0.01. (B) Mutational analysis of ISS8 supports the silencing activity of combined hexamer regions. The combined and individual activity of hexamer regions within the context of ISS8 was examined by introducing 2 point mutations into 2 hexamer regions in combination and separately. (C) Individual hexamer analysis supports the silencing activity of enriched hexamer sequences. Hexamers and corresponding mutant and duplicate sequences were characterized in transient transfection assays in HEK-293 cells. The mean GFP levels from two independent experiments were normalized to the wild-type hexamer construct.
In contrast, the extended ISS8 sequence contains eight enriched hexamers resembling the preferred binding sites for the hnRNP L protein over two regions and a novel element that overlaps regions 1 and 2. A similar analysis of ISS8 shows that the individual mutations within each region disrupt silencer activity to levels below the NMD control, resulting in an ∼18% decrease in activity (Figure 4B). Mutations to both regions in combination result in an ∼25% decrease in silencer activity, suggesting that the hexamer regions in ISS8 work in combination to effect silencer activity. Therefore, the enriched hexamers within the recovered sequences can exhibit regulatory function independently and in combination with other hexamers, but the effects are context dependent and may depend on the specific trans-acting factors involved.
We examined the silencer activities of representative hexamers from the GCCS cluster motifs through transient assays to confirm the activity of individual motifs outside the context of a selected 15-mer. Hexamers were chosen at random from each class (Figure 3A), and silencing activity was investigated by comparing expression levels of the hexamer alone, in duplicate, and with double point mutations (Figure 4C). A majority of the mutated hexamers (ACCTCC, GGGGGG, TAATTG and ATATGG) exhibits significant loss of function (classes 1–3). The GTAGAA hexamer lacked silencer activity by itself, but displayed function in the context of ISS5 (Figure 4A), suggesting that the regulatory activity of this hexamer is context dependent. Only one of the hexamers (GCTGGG) displays an increase in silencer activity when present in duplicate. The results indicate that while individual hexamers exhibit silencing activity and likely represent core ISREs, they do not generally behave in an additive manner. This may be due to context and spacing requirements, as the duplicate GGGGGG hexamer does not exhibit increased silencing, whereas ISS15, which differs by three cytosine residues that may provide critical spacing between the G-rich hexamers, exhibits strong silencer activity (Figure 2B).
Splicing factor depletion alters splicing of endogenous genes containing ISREs
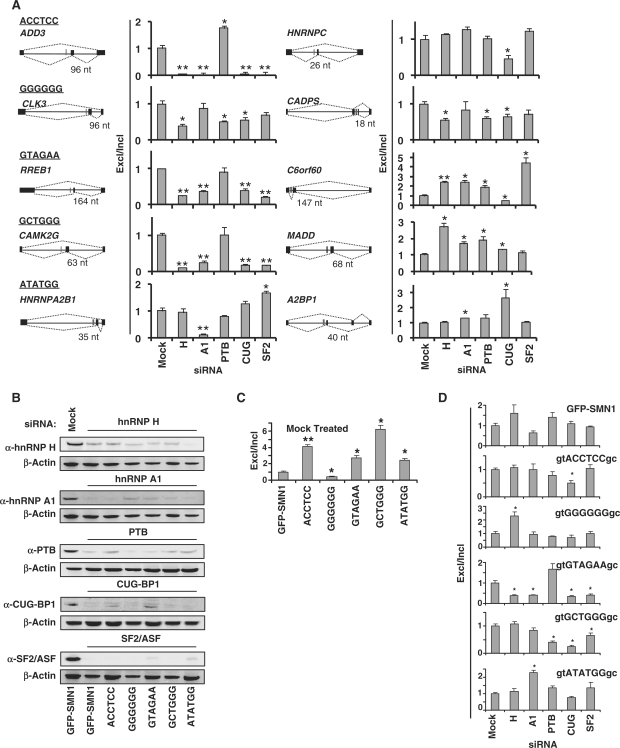
We performed a genome-wide analysis to determine the occurrence of enriched motifs in the region 80-nt upstream of the AI regions flanking skipped exons (30). Several motifs significantly associate with alternative (2 of 30; class 2) and constitutive splicing (10 of 30; classes 1–3) (Supplementary Figure S7). Using results from this association analysis, we screened a panel of siRNAs targeting known splicing regulators (hnRNP H, hnRNP A1, PTB, CUG-BP1 and SF2/ASF) for effects on the splicing patterns of 10 alternatively spliced endogenous genes containing conserved intronic hexamers (Figure 5A and Supplementary Table S4). Hexamers were chosen from each class (ACCTCC, GGGGGG, GTAGAA, GCTGGG and ATATGG) and harbor potential binding sites for the selected trans-acting factors. RNAi-mediated silencing of each gene resulted in a substantial reduction (>70%) of the targeted protein (Figure 5B) and displayed minimal effects on other examined splicing factors (Supplementary Figure S9A). We observed significant changes in the alternative splicing patterns of all genes upon depletion of one or more splicing factors by qRT-PCR, where half of the genes showed decreased exon exclusion (ADD3, CLK3, RREB1, CAMK2G, HNRNPC and CADPS) and two displayed higher levels (MADD and A2BP1) of exon exclusion. These results highlight potential members of the SRNs associated with these alternatively spliced exons; however, the intronic and exonic regions of the genes likely contain additional regulatory motifs which precludes the clear determination of factors associated with selected ISREs within the context of endogenous genes.
Figure 5.
The effects of in vivo depletion of splicing factors on the splicing patterns of endogenous and synthetic genes containing conserved intronic hexamers. (A) qRT-PCR analysis of the siRNA treated GFP-SMN1 control cell lines with primer sets specific for exon included (primer set 4, Figure 1D) and excluded (primer set 5) products of 10 endogenous genes. The splicing patterns of each gene are diagrammed where black bars represent exons and red bars represent the location of conserved ISRE hexamer motifs. Data is reported as the ratio of the mean expression of the exon excluded isoform to the exon included isoform normalized to the ratio for the GFP-SMN1 control ± the average error. P-values derived from the Student's t-test are as follows: *P < 0.05 and **P < 0.01. (B) Western blot analysis of total cell lysates prepared from the GFP-SMN1 control and ISRE hexamer cell lines treated with siRNAs targeted to trans-acting splicing factors and a mock siRNA negative control. β-Actin was used as a loading control for all blots. The results of the GFP-SMN1 mock treated lysate is representative of all mock-treated cell lines. (C) qRT-PCR analysis of the mock-treated ISRE hexamer and GFP-SMN1 control cell lines with primer sets specific for exon 7 included (primer set 4) and excluded (primer set 5) products. Data is reported as the ratio of the mean expression of the exon excluded isoform to the exon included isoform normalized to the ratio for the GFP-SMN1 control ± the average error. (D) qRT-PCR analysis of the siRNA treated ISRE hexamer and GFP-SMN1 control cell lines with primer sets specific for exon 7 included and excluded products (primer sets 4 and 5, respectively). Data is reported as the ratio of the mean expression of the exon excluded isoform to the exon included isoform normalized to the ratio for the mock siRNA treated cell line control ± the average error.
Splicing factor depletion influences ISRE regulated splicing in vivo
To more cleanly investigate the functional significance of the GCCS-identified motifs and the associated trans-acting factors, we screened the same panel of siRNAs for effects on the splicing patterns of selected hexamers in our synthetic SMN1 mini-gene system in stable cell lines. This system allows for the controlled insertion of selected hexamers within the context of constant and well-characterized exonic and intronic regions. Hexamers and a random control were subjected to the RNAi-based screen using a mini-gene lacking a PTC to avoid any siRNA-mediated effects on the NMD pathway.
We analyzed the splicing patterns of the hexamer and control constructs through transcript analysis by qRT-PCR. Four of the hexamers exhibit silencing activity and one (GGGGGG) exhibits enhancer activity relative to the GFP-SMN1 control in the presence of the mock siRNA (Figure 5C and Supplementary Figure S9B). Splicing patterns of all hexamer-containing constructs were significantly affected by the depletion of at least one trans-acting factor (Figure 5D and Supplementary Figure S9C). In contrast, depletion of the trans-acting factors had statistically insignificant effects on the splicing pattern of the GFP-SMN1 control (P > 0.05). Therefore, observed changes in the splicing patterns of hexamer-containing constructs in response to trans-acting factor depletion are specific to the introduced hexamer sequences.
The splicing patterns of three hexamer constructs exhibit significant changes in response to the depletion of one trans-acting factor relative to the mock control. The GGGGGG motif is identical to the hnRNP F/H binding site (44). Depletion of hnRNP H leads to a 2.3-fold increase in exon exclusion, demonstrating that this factor enhances the recognition of the 3′ ss, likely through direct binding to the hexamer. The novel motif ACCTCC is similar to the CT-rich PTB binding site; however, depletion of PTB leads to a marginal decrease in exon exclusion, whereas depletion of CUG-BP1 leads to a 2-fold decrease in exon exclusion. Although PTB has been shown to act antagonistically to CELF proteins (19), it is unlikely that CUG-BP1, which binds CTG and GT-rich motifs, directly binds to the ACCTCC hexamer, suggesting it may be recruited through interactions with other regulatory proteins. Depletion of hnRNP A1 leads to an ∼2.3-fold increase in exon exclusion levels from the ATATGG hexamer construct. The hexamer and flanking regions contain a GGG motif that may be a weak binding site for hnRNP A1. However, any direct binding of hnRNP A1 likely competes with other regulatory factors since its depletion leads to an increase in exon exclusion. Alternatively, modulation of hnRNP A1 levels may affect the levels of other trans-acting factors that play a role in the splicing regulatory effect of the hexamer.
Two hexamer constructs exhibit significant changes in their splicing pattern in response to depletion of multiple factors. The GTAGAA motif resembles the SF2/ASF SELEX-derived binding site (GAAGAA) (48), the TAGA motif may be a weak binding site for hnRNP A1 and the hexamer and flanking regions contain two GT repeats, which may serve as binding sites for CUG-BP1. Depletion of hnRNP H, hnRNP A1, CUG-BP1 and SF2/ASF led to a 2.5-fold or greater reduction in exon exclusion levels for the construct. One possible mechanism is that SF2/ASF, CUG-BP1 and hnRNP A1 directly compete for binding to the hexamer and hnRNP H acts positively in their recruitment. hnRNP H and CUG-BP1 can form an RNA-dependent suppressor splicing complex (49), suggesting that several of these factors may be involved in an inhibitory complex that aids in the recruitment of factors that directly bind to the transcript. The GCTGGG motif and flanking regions contain GT and TG dinucleotides and a CTG element that resemble CUG-BP1 binding sites. Depletion of CUG-BP1 results in a 4-fold decrease in exon exclusion of the construct. Depletion of PTB and SF2/ASF also cause significant decreases in exon exclusion, although the preferred binding sites of these factors do not resemble motifs within the hexamer and flanking regions. The results suggest that CUG-BP1 may be directly involved in binding to the GCTGGG hexamer, and PTB or SF2/ASF may be recruited by CUG-BP1 or other factors.
Comparing results from the endogenous gene and synthetic SMN1 mini-gene depletion studies, we observed significant changes in the alternative splicing patterns of all endogenous genes upon depletion of the splicing factor that showed the greatest effect on the splicing patterns of each hexamer within the context of the SMN1 mini-gene (Figure 5A). Half of the genes responded to depletion of the splicing factor similarly to that observed in the SMN1 mini-gene studies, suggesting that hexamers ACCTCC, GTAGAA, GCTGGG and ATATGG may function as ISSs within the context of endogenous genes. These results suggest that our SMN1 mini-gene depletion studies identified key trans-acting regulators of the examined hexamers. The significant changes in splicing of the synthetic mini-gene and endogenous genes containing conserved hexamers upon splicing factor depletion support the functional role of SPLICE-identified ISREs and highlight key members of the SRNs associated with these elements.
DISCUSSION
To gain insight into the function and sequence composition of ISREs, we developed a novel strategy to screen for ISREs in vivo (SPLICE), using an NMD-based alternative splicing reporter system. Our screen produced 125 unique 15-mer sequences where the majority of examined sequences displayed significant regulatory activity in subsequent characterization studies. Control studies indicated a low frequency of false positives were generated by the fluorescence-based screen, most likely due to rare sequences that enable the transcript to evade the NMD pathway. Importantly, examined sequences displayed similar splicing regulatory activity in NMD- and non-NMD-based reporter constructs, suggesting that recovered sequences maintain splicing regulatory function independent of triggering decay through the NMD pathway. Therefore, this work validates SPLICE as an effective screening platform for ISREs.
The ISREs obtained from our in vivo selection provide a diverse composition of functional sequences, which enable tuning of alternative splicing, and reveal motifs that resemble binding sites for known and previously unidentified trans-acting factors that are enriched in the acceptor intronic regions of genes throughout the human genome. Conserved motifs were identified by SPLICE and characterization supports the properties of context-dependent, combinatorial and independent regulation. The functional activity of the sequences harboring novel motifs was validated in subsequent characterization assays, where eight of the 28 examined ISREs harbored previously unidentified motifs (Supplementary Table 10). In addition, the activities of two novel hexamer motifs (ACCTCC and ATATGG) were validated in the RNAi and hexamer characterization studies. Although shorter functional motifs (6-nt) were identified from the recovered sequences, we were able to observe the enhanced regulatory effects of combinatorial regulation from multiple motifs by screening a larger ISRE length (15-nt). Future studies can examine the functional sequences elucidated here in different mini-genes to gain insight into the relative activities of the identified sequences in the context of intron–exon sequences that exhibit different basal splicing efficiencies.
The results from our RNAi silencing study highlight some of the key splicing factors and the complexity of the SRNs associated with ISRE function, suggesting a role for multiple splicing factors influencing regulation at a single motif. These studies revealed that simply changing the enriched hexamer in the context of the SMN1 mini-gene modifies splicing patterns and the SRNs involved with the transcript, supporting the possibility that most RNA-binding proteins do not strictly act as enhancers or suppressors, but instead have a context dependency. Results from these studies also reveal that the splicing of the endogenous transcripts containing conserved hexamers is influenced similarly to the SMN1 experimental constructs, further supporting the biological relevance of the motifs. Our work sets the stage for large-scale characterization studies of the identified ISREs and associated trans-acting factors, which will further elucidate ISRE regulatory activity and mechanism, including the role of combinatorial control and sequence context in the function of these elements.
Significant advances in our understanding of the mechanisms that guide splice site selection and the distributions of regulatory elements has aided the formulation of an early version of a ‘splicing code’ (14). Currently missing from this draft is a thorough understanding of the sequence characteristics and function of ISREs. Our results indicate that a dominant factor guiding function of ISREs is the immediate transcript context in which the splicing factors bind. Therefore, refinements of the splicing code will benefit from elucidation of co-regulatory sequences that may be identified in functional screens examining pair-wise or combinatorial motifs. The identified motifs offer a rich data set to expand the splicing code, determine the extent of single nucleotide polymorphisms (SNPs) that modulate splicing through ISREs and refine bioinformatic search algorithms for genome-wide identification of intronic regulators, which will facilitate the diagnosis and treatment of disease.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Caltech Joseph Jacobs Institute for Molecular Engineering for Medicine (grant to C.D.S.); National Science Foundation (grant MCB-0616264, J.A.B.); National Institutes of Health (fellowship to K.G.H., grant to C.D.S.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Diamond and D. Perez for FACS assistance and expert technical advice, A. Krainer for the pCISMNxΔ6-wt construct.
REFERENCES
- 1.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 3.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 5.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson M. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat. Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 8.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct. Mol. Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian H, Kole R. Strong RNA splicing enhancers identified by a modified method of cycled selection interact with SR protein. J. Biol. Chem. 2001;276:33833–33839. doi: 10.1074/jbc.M102957200. [DOI] [PubMed] [Google Scholar]
- 12.Schaal TD, Maniatis T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulter LR, Landree MA, Cooper TA. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell. Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 16.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol. Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang XH, Kangsamaksin T, Chao MS, Banerjee JK, Chasin LA. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol. Cell. Biol. 2005;25:7323–7332. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charlet BN, Logan P, Singh G, Cooper TA. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 20.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–88. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 21.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell. Biol. 2005;6:386398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 22.Venables JP. Downstream intronic splicing enhancers. FEBS Lett. 2007;581:4127–4131. doi: 10.1016/j.febslet.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 24.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 25.Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 26.Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 27.Chen CD, Kobayashi R, Helfman DM. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez I, Lin CH, McAfee JG, Patton JG. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Voelker RB, Berglund JA. A comprehensive computational characterization of conserved mammalian intronic sequences reveals conserved motifs associated with constitutive and alternative splicing. Genome Res. 2007;17:1023–1033. doi: 10.1101/gr.6017807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeo G, Hoon S, Venkatesh B, Burge CB. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl Acad. Sci. USA. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeo GW, Nostrand EL, Liang TY. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007;3:e85. doi: 10.1371/journal.pgen.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray JI, Voelker RB, Henscheid KL, Warf MB, Berglund JA. Identification of motifs that function in the splicing of non-canonical introns. Genome Biol. 2008;9:R97. doi: 10.1186/gb-2008-9-6-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeo GW, Xu X, Liang TY, Muotri AR, Carson CT, Coufal NG, Gage FH. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput. Biol. 2007;3:1951–1967. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green RE, Lewis BP, Hillman RT, Blanchette M, Lareau LF, Garnett AT, Rio DC, Brenner SE. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics. 2003;19(Suppl. 1):i118–i121. doi: 10.1093/bioinformatics/btg1015. [DOI] [PubMed] [Google Scholar]
- 37.Ladd AN, Cooper TA. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-11-reviews0008. reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang XH, Leslie CS, Chasin LA. Dichotomous splicing signals in exon flanks. Genome Res. 2005;15:768–779. doi: 10.1101/gr.3217705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Liu G, Yu J, Cao W, Lobo VG, Xie J. In vivo selection of kinase-responsive RNA elements controlling alternative splicing. J. Biol. Chem. 2009;284:16191–16201. doi: 10.1074/jbc.M900393200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 42.Hanamura A, Caceres JF, Mayeda A, Franza BR, Jr, Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 43.Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, Osborne HB. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem. J. 2006;400:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nasim MT, Chernova TK, Chowdhury HM, Yue BG, Eperon IC. HnRNP G and Tra2beta: opposite effects on splicing matched by antagonism in RNA binding. Hum. Mol. Genet. 2003;12:1337–1348. doi: 10.1093/hmg/ddg136. [DOI] [PubMed] [Google Scholar]
- 47.Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paul S, Dansithong W, Kim D, Rossi J, Webster NJ, Comai L, Reddy S. Interaction of muscleblind, CUG-BP1 and hnRNP H proteins in DM1-associated aberrant IR splicing. EMBO J. 2006;25:4271–4283. doi: 10.1038/sj.emboj.7601296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.