Abstract
Antigene RNAs (agRNAs) are small RNA duplexes that target non-coding transcripts rather than mRNA and specifically suppress or activate gene expression in a sequence-dependent manner. For many applications in vivo, it is likely that agRNAs will require chemical modification. We have synthesized agRNAs that contain different classes of chemical modification and have tested their ability to modulate expression of the human progesterone receptor gene. We find that both silencing and activating agRNAs can retain activity after modification. Both guide and passenger strands can be modified and functional agRNAs can contain 2′F-RNA, 2′OMe-RNA, and locked nucleic acid substitutions, or combinations of multiple modifications. The mechanism of agRNA activity appears to be maintained after chemical modification: both native and modified agRNAs modulate recruitment of RNA polymerase II, have the same effect on promoter-derived antisense transcripts, and must be double-stranded. These data demonstrate that agRNA activity is compatible with a wide range of chemical modifications and may facilitate in vivo applications.
INTRODUCTION
The therapeutic potential of gene silencing by small interfering RNAs (siRNAs) is widely recognized (1). siRNA clinical candidates are commonly modified to improve their stability, specificity and potency (2–4). Modified siRNAs have been shown to reduce off-target effects resulting from the miRNA pathway (5,6), the innate immune system (7,8) and loading of the wrong strand (6,9,10). In addition, chemically modified siRNAs have been used to deepen our understanding of the mechanism of RNAi (11–16) and improve siRNA delivery (17,18).
Antigene RNAs (agRNAs) are small duplex RNAs that target gene promoters. Like siRNAs, they are generally 19 bp with 2-nt overhangs at the 3′-ends. Depending on target sequence, cell type and basal level of expression, agRNAs can either silence (19–21) or activate (22–24) gene expression at the transcriptional level. Like gene silencing by siRNAs, modulation of gene expression by agRNAs involves argonaute (AGO) proteins (25,26). However, instead of targeting mRNA, agRNAs target non-coding RNA transcripts (ncRNAs) overlapping gene promoters (27,28). While agRNAs can target either sense or antisense non-coding transcripts, the agRNAs discussed in this study target an antisense transcript overlapping the promoter of the progesterone receptor (PR) gene (28). Thus the guide strand of these agRNA duplexes is the sense strand, in contrast to siRNAs, for which the guide strand is by definition antisense (i.e. complementary to the mRNA).
agRNAs that activate gene expression could be used to modulate expression of disease-associated genes whose upregulation might be beneficial. As with current gene silencing technology, however, it is likely that any development of agRNAs for in vivo use will require chemical modification to improve stability and biodistribution. In this study we examine the effects of modified nucleotides on the function of agRNAs. After examining over 120 duplexes, we identify several chemically modified compounds that retain partial or full activity.
MATERIALS AND METHODS
Double-stranded RNAs
RNAs were synthesized by Alnylam Pharmaceuticals or SIGMA Custom Products using standard protocols, or were purchased from Integrated DNA Technologies. The concentrations of single strands were determined by measuring their absorbance at 260 nm (A260). Complementary single strands were combined in equimolar ratios and diluted to 20 µM stock solutions in 2.5× phosphate-buffered saline. These duplexes were annealed by heating to 95°C and slowly cooled to room temperature in a heating block or thermal cycler. Duplex integrity was tested by a UV thermal melt experiment as described below. Duplexes were stored at −20°C.
Cell culture
T47D and MCF7 cells (American Type Culture Collection) were maintained in RPMI-1640 media (Sigma) supplemented with 10% (v/v) FBS, 0.5 × MEM non-essential amino acids, 0.4 units ml−1 bovine insulin, 10 mM HEPES and 1 mM sodium pyruvate. Cells were cultured at 37°C and 5% (v/v) CO2.
Lipid-mediated transfection
Cells were plated at 110 000–160 000 cells per well in six-well plates (Costar) in RPMI. A higher number of cells was typically seeded for RNA analysis than for protein analysis. Transfections were performed after 1–2 days, when the cells had adhered and attained about 20–30% confluence. Lipofectamine RNAiMAX cationic lipid (1.1 µl/well, Invitrogen) and RNA (31 pmol/well) were individually diluted in Opti-MEM (Invitrogen), then combined and allowed to incubate for 20 min (volume 250 µl/well) before pipetting into cell-culture wells containing 1 ml fresh Opti-MEM (final volume 1.25 ml, 25 nM duplex RNA for all transfections except dose response experiments). The transfection solution was replaced with RPMI after 24 h, and media were changed again 2 days later. For dose response experiments, the ratio of lipid to RNA was kept constant, and the concentration of RNA was varied from 0 to 50 nM.
For competition experiments, the cells were passaged into new plates on Days 2 or 3 after transfection. The second transfection was carried out on Day 4 after the first transfection under identical conditions, and protein was harvested on Days 7 or 8. The negative control duplex Neg1 used in these experiments consisted of oligonucleotides 5′-UCAAGAAGCCAAGGAUAAUdTdT and 5′-AUUAUCCUUGGCUUCUUGAdTdT.
RNA harvest and quantitative real-time PCR
Total RNA from treated MCF7 or T47D cells was harvested 3 days after transfection. After washing with PBS, TRI reagent (1 ml, Sigma) was added to each well, incubated for several minutes, mixed well and transferred to a 1.5-ml microcentrifuge tube. Chloroform (250 µl) was added and the mixture was shaken vigorously for 1 min, then centrifuged at 12 500 rpm for 15 min. The clear aqueous layer was transferred to a new 1.5-ml microcentrifuge tube, avoiding any of the interphase or organic layer. Addition of cold isopropanol (500 µl) followed by shaking, incubation at −20°C (at least 10 min), and centrifugation (12 500 rpm, 15 min, 4°C) gave a pellet, which was washed with 75% ethanol, dissolved in nuclease-free water (20 µl), and quantitated by the A260 of an aliquot (2 µl in 200 µl TE buffer). Samples of 2 µg RNA were removed and treated with DNase I (Worthington). After heat-inactivation of the DNase enzyme (75°C, 10 min), the samples were reverse transcribed using random hexamer primers with the High Capacity cDNA Archive kit (Applied Biosystems). Quantitative real-time PCR (qPCR) was used to measure RNA levels with 30–100 ng of cDNA (carried out in triplicate, and repeated for at least two independent transfections). For analysis of PR mRNA, we used a primer-probe assay (IDT) consisting of primers 5′-CTTACCTGTGGGAGCTGTA-3′ and 5′-GCACTTTCTAAGGCGACATG-3′ as well as probe 5′-CTGCCCTTCCATTGCCCTCTT-3′. For analysis of promoter-derived antisense transcripts, we used SYBRgreen qPCR with primers 5′-GGAGGAGGCGTTGTTAGAAA-3′ and 5′-GAAGGGTCGGACTTCTGCT-3′. Data were normalized to levels of GAPDH mRNA as determined by a TaqMan Gene Expression Assay (Applied Biosystems).
Analysis of protein expression
Protein was generally harvested from treated MCF7 or T47D cells 5 days after transfection. The cells were washed with phosphate-buffered saline (PBS), trypsinized, transferred to 1.5-ml microcentrifuge tubes and centrifuged (3500 rpm, 15 min). The pellets were washed with cold PBS and gently lysed by addition of 50 µl lysis buffer [50 mM Tris–HCl (pH 8.0), 120 mM NaCl, 0.5% Nonidet P40, 1 mM EDTA, 1 mM DTT]. Protease inhibitor cocktail set 1 (Calbiochem) was added to the lysis buffer just before use. After a freeze-thaw cycle to ensure complete lysis, the lysates were centrifuged to pellet insoluble cell debris and cell lysate solutions were transferred to new microtubes. Protein concentrations were calculated using the BCA assay (Thermo Scientific).
Western blots were performed on cell lysates (30 µg protein per lane). Proteins were separated by SDS–PAGE using 7.5% Tris–HCl acrylamide gels, then transferred onto nitrocellulose membranes. After blocking the membrane (5% milk in PBS-T) for 1 h, the upper portion of the membrane was treated with PR monoclonal primary antibody (Cell Signaling, 1:1000 dilution in PBS-T, ∼15 h) and the lower portion was treated with anti–β-actin (Sigma, 1:10 000 dilution, 1 h) as an internal control. Protein was visualized using HRP-conjugated secondary anti-mouse (Jackson Immunolabs, 1:5000 for PR and 1:20 000 for actin, 30–45 min) The membranes were then briefly treated with West Pico SuperSignal developing solution (Thermo Scientific) before exposing them to film. All analogs were tested in at least two independent transfections, of which one representative western blot is shown.
Determination of melting temperature (Tm)
Annealed siRNA duplexes were diluted to 0.8 µM in 0.1 M phosphate buffer, pH 7.4 (final volume 500 µl) and transferred to quartz cuvettes. The solutions were topped with mineral oil (200 µl) and heated from 18 to 99°C at 1°C/min while monitoring changes in A260 using a Cary 100 Bio spectrophotometer. Samples were then re-annealed in the spectrophotometer and the experiment was repeated several times. Tm values were calculated as the first derivatives of the plot of A260 vs temperature, and the results from three experiments were averaged.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed essentially as described (29). Two or three 150-mm dishes of T47D or MCF7 cells were reverse transfected with 25 nM of native agRNA, modified agRNA or luciferase control duplex siGL2 (30).
Media were changed on Day 2 and the cells harvested on Day 3. To crosslink and harvest nuclei, the cells were treated with 1% formaldehyde in PBS (10 ml) for 10 min, then the crosslinking reaction was quenched with glycine (0.125 M final concentration, at least 5 min). Cells were collected and nuclei were isolated by treatment with cold hypotonic lysis buffer (4 ml) containing 10 mM Tris (pH 7.4), 10 mM NaCl, 3 mM MgCl2 and 0.5% Nonidet P40. The pelleted nuclei were lysed with a buffer containing 1% SDS, 10 mM EDTA, 50 mM Tris–HCl (pH 8.1) and 1× complete protease inhibitor cocktail (Roche). DNA was sonicated on ice to an average fragment size of ∼500 bp. The lysate was then centrifuged to remove insoluble cell debris and the supernatant precleared with Protein G Plus/Protein A Agarose suspension (Calbiochem).
An aliquot of the cleared lysate (100 µl) was diluted to 1 ml in buffer [0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris pH 8.1, 167 mM NaCl and 1× complete protease inhibitor cocktail (Roche)]. Mouse anti-RNAPII IgG (Millipore, Catalogue #05-623, 3–6 µg) or normal mouse IgG (Millipore Catalogue #12-371, 3–6 µg) was added and the solution rotated at 4°C overnight. Protein G Plus / Protein A agarose suspension (30–60 µl) was added to precipitate the antibody complexes and the beads were washed as described (29). Complexes were eluted using 2 × 250 µl of NaHCO3 (0.1 M) containing 1% SDS. NaCl was added (final concentration 200 mM) and the samples heated to 65°C for at least 2 h to reverse crosslinks. The samples were then treated with RNase A (1 µl) at 37°C for 30 min, followed by Tris–HCl (pH 7.0, 20 µl), EDTA (0.5 M, 10 µl) and proteinase K (20 µg) at 42°C for 45 min. DNA was isolated by phenol-chloroform extraction and ethanol precipitation. Pellets were diluted to 50 µl in water and 5 µl was used per qPCR reaction. qPCR primers used at the PR promoter were Fwd 5′-CCTAGAGGAGGAGGCGTTGT and Rev 5′-ATTGAGAATGCCACCCACA. Levels of RNAPII at the GAPDH promoter were also measured with primers Fwd 5′-TACTAGCGGTTTTACGGGCG and Rev 5′-TCGAACAGGAGGAGCAGAGAGCGA and were used for normalization. Each sample was normalized to a corresponding 2% input sample.
Statistics, dose response profiles, and curve fitting
Error bars shown on qPCR graphs correspond to standard deviations from triplicate measurements of at least two independent transfections; they include the error from the qPCR measurement as well as from differences between the transfections. For Figures 2–7 we also examined whether RNA expression levels were significantly different from the mismatch control by conducting a two-tailed unpaired Student’s t-test with equal variances, and for this test we considered only biological replicates (using averages of the triplicate qPCR measurements for each). Significance levels of P-values are: *P < 0.05, **P < 0.01 and ***P < 0.005.
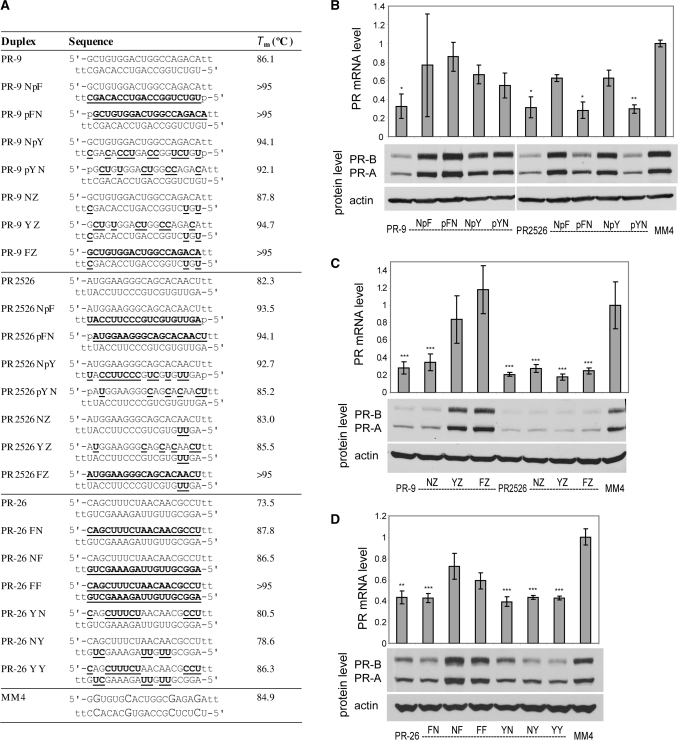
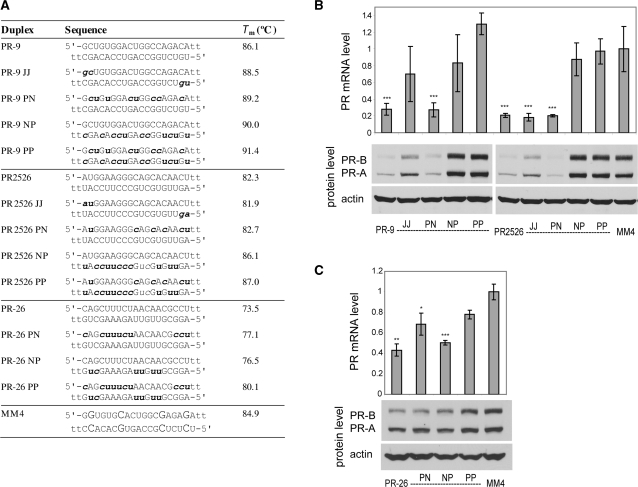
Figure 2.
PR gene expression can be silenced by agRNAs containing 2′F-RNA. (A) Duplex sequences: passenger strands are listed on top in all cases; for siRNA PR2526 this is the sense strand, but for agRNAs PR-9 and PR-26 this is the antisense strand. Modification codes: dna, RNA, 2′F-RNA; p = phosphate. (B) PR-9 analogs and controls containing one strand heavily or fully modified with 2′F-RNA. (C) PR-9 analogs and controls containing a minimally 2′F-RNA-modified guide strand. (D) PR-26 analogs containing one or both strands heavily or fully modified with 2′F-RNA. All data are normalized to mismatch control MM4. RNA levels are the average (±SD) from at least two independent transfections. *P < 0.05, **P < 0.01, ***P < 0.005. Protein levels were also confirmed by at least two independent transfections, from which a typical western blot is shown.
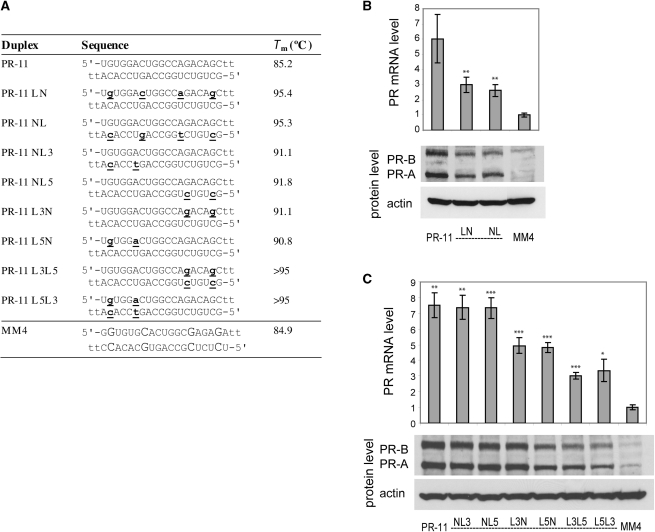
Figure 7.
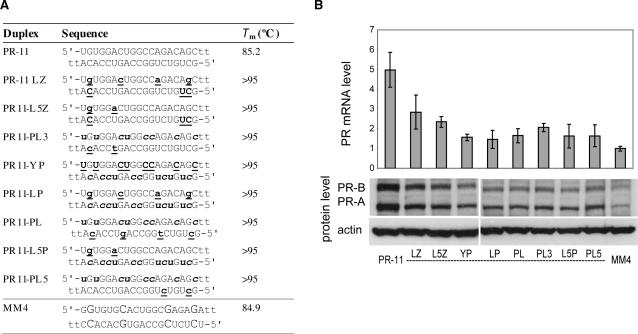
PR gene expression can be activated by agRNAs containing LNA. (A) Duplex sequences: passenger strands are listed on top in all cases; for agRNA PR-11 this is the antisense strand. LNA sequences contain 5-methylcytosine instead of cytosine. Modification codes: dna, RNA, lna. (B) PR-11 analogs containing four evenly distributed LNA units. (C) PR-11 analogs containing LNA at strand ends. All data are normalized to mismatch control MM4. RNA levels are the average (±SD) from at least two independent transfections. *P < 0.05, **P < 0.01, ***P < 0.005. Protein levels were also confirmed by at least two independent transfections, from which a typical western blot is shown.
Dose responses were fit to the model equation y = 100[1 – xm/(nm + xm)], where y is the percent of target (protein or mRNA) remaining, x is the concentration of agRNA in nanomolars. The regression was fit by optimizing m and n using SigmaPlot 11, and n was taken as the IC50 value.
RESULTS AND DISCUSSION
Experimental design
We focused on the PR gene because it is a well-characterized target for agRNAs (20,23,25,28). In T47D breast cancer cells, which express a high level of PR, PR expression can be dramatically reduced by agRNA PR-9 that overlaps the start site from –9 to +10 (20,21). In MCF7 breast cancer cells, which express PR at a lower basal level, its expression can be increased by agRNA PR-11 targeting the region from –11 to +8 (23). PR-11 can also activate gene expression in T47D cells but activation becomes most apparent when the cells are grown under conditions that repress basal expression of PR (23). Whether activation or repression is observed depends primarily on the basal expression level of PR and the target sequence of the agRNA.
While PR-9 and PR-11 are benchmark agRNAs used for most experiments, we also examined other duplexes. For some modifications we tested analogs of silencing agRNA PR-26 (–26 to –8) or activating agRNA PR-22 (–22 to –4) to allow more general insights into gene modulation. To permit comparisons with an RNA that engages the post-transcriptional silencing pathway, we used modified analogs of PR2526, a siRNA that is complementary to PR mRNA from +2526 to +2544. As negative control we used MM4, a duplex containing four mismatches with respect to PR-9. Duplex and single-stranded oligomers were introduced into cells by standard transfection protocols using cationic lipid. Expression was analyzed by western analysis of PR protein and qPCR of PR mRNA.
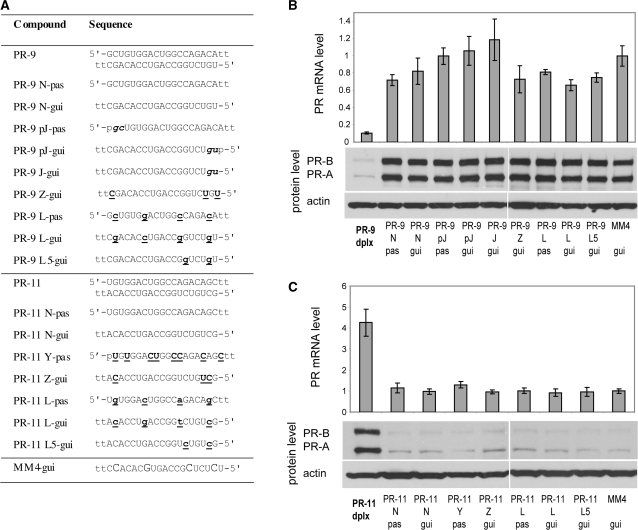
We use descriptive compound identifiers (Table 1). Each type of modified duplex is assigned two uppercase letters. The first letter describes the chemical modification of the passenger strand, while the second letter describes modification of the guide strand. A lowercase ‘p’ indicates that the 5′-end of the strand is phosphorylated. For example, a duplex labeled ‘NpF’ has a native passenger strand and a guide strand that is phosphorylated and fully modified with 2′-fluoro (2′F-RNA) nucleotides. Passenger strands are listed above guide strands throughout.
Table 1.
Descriptive strand names used throughout this work
| Abbreviation | Strand chemistry |
|---|---|
| N | Native RNA strand |
| F | 2′F-RNA, fully modified |
| Y | 2′F-RNA, all pyrimidines |
| Z | 2′F-RNA, contains three units near termini |
| P | 2′-OMe-RNA, all pyrimidines |
| J | 2′-OMe-RNA, two units at the 5′-end |
| L | LNA, contains four units distributed evenly |
| L3 | LNA, contains two units toward the 3′-end |
| L5 | LNA, contains two units toward the 5′-end |
| p | A lower case ‘p’ preceding any strand abbreviation indicates a 5′-phosphate |
Effects of 2′F-RNA modification on gene silencing
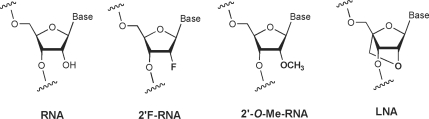
The structure of 2′F-RNA (Figure 1) is similar to that of RNA (31) but confers resistance to endonucleases (32,33). For siRNAs, partial modification with 2′-F RNA is tolerated throughout either strand of the duplex (34–36) and there have been reports of functional siRNAs that are extensively or completely modified with 2′F-RNA (33,37,38). 2′F-RNA has been shown to reduce the immunostimulatory activity of siRNAs (39,40).
Figure 1.
Chemical modifications used in this study.
We tested three types of 2′F-RNA-modified strands: fully modified (F), modified at all of the pyrimidine units (Y), and modified at positions near the termini (Z) (Table 1 and Figure 2A). Relative to unmodified duplex PR-9, we observed reduced potency when either strand was replaced with a fully-fluorinated (PR-9 NpF, PR-9 pFN) or pyrimidine-fluorinated strand (PR-9 NpY, PR-9 pYN) (Figure 2B). siRNA PR2526, by contrast, retained most silencing activity after total or partial modification of its passenger strand (PR2526 pFN, PR2526 pYN) but activity was lost after modification of the guide strand (PR2526 NpF, PR2526 NpY) (Figure 2B). Duplexes based on either PR-9 or PR2526 containing heavy modification in both strands were largely inactive (Supplementary Figure S1).
Given the loss of activity after aggressive modification of PR-9, we tested more limited terminal substitutions. A PR-9 analog containing minimal modification of the guide strand was effective when paired with an unmodified passenger strand (PR-9 NZ) but not when the passenger strand was heavily modified (PR-9 YZ and PR-9 FZ) (Figure 2C). Analogs of PR2526 containing a minimally modified guide strand (PR2526 NZ, PR2526 YZ, PR2526 FZ) were active regardless of the extent of modification of the passenger strand (Figure 2C).
We also examined the effect of 2′F-RNA modification on the activity of silencing agRNA PR-26 in an effort to determine how modification effects might vary depending on the sequence of the agRNA. When the pyrimidines of either or both strands of PR-26 were replaced with 2′F-RNA (PR-26 YN, PR-26 NY, PR-26 YY), most silencing activity was retained (Figure 2D). Taken together, data from PR-9 and PR-26 demonstrate that 2′F-RNA substitutions are compatible with the mechanism for agRNA activity but that the activity of individual patterns of modification can vary between duplexes.
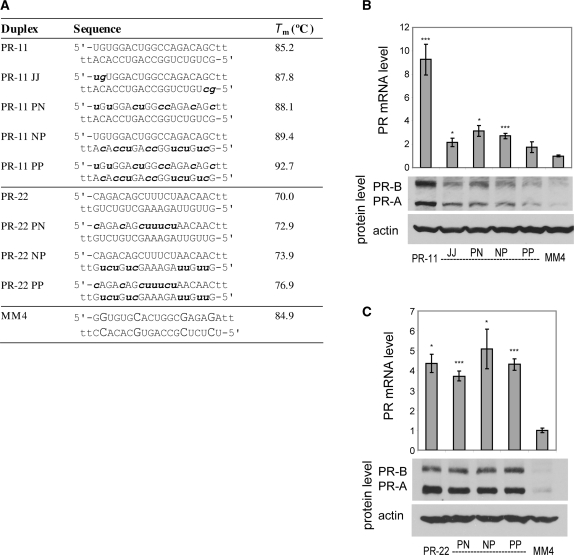
Effect of 2′F-RNA modification on gene activation
We introduced 2′F-RNA into activating agRNA PR-11 (Figure 3A) and observed that the modification was broadly tolerated. Modification of the pyrimidines of the passenger strand (PR-11 pYN), total modification of the passenger strand (PR-11 pFN), or heavy modification of the guide strand (PR-11 NpF and PR11 NpY) all led to gene activation, albeit at levels below those of the unmodified PR-11 duplex (Figure 3B). Combining the unmodified or heavily modified passenger strands with a minimally modified guide strand showed a loss of potency as the degree of modification increased (Figure 3C). Nevertheless, all of these duplexes retained significant activity (2-fold to 6-fold at the RNA level). PR-11 analogs with both strands heavily modified were inactive (Supplementary Figure S2).
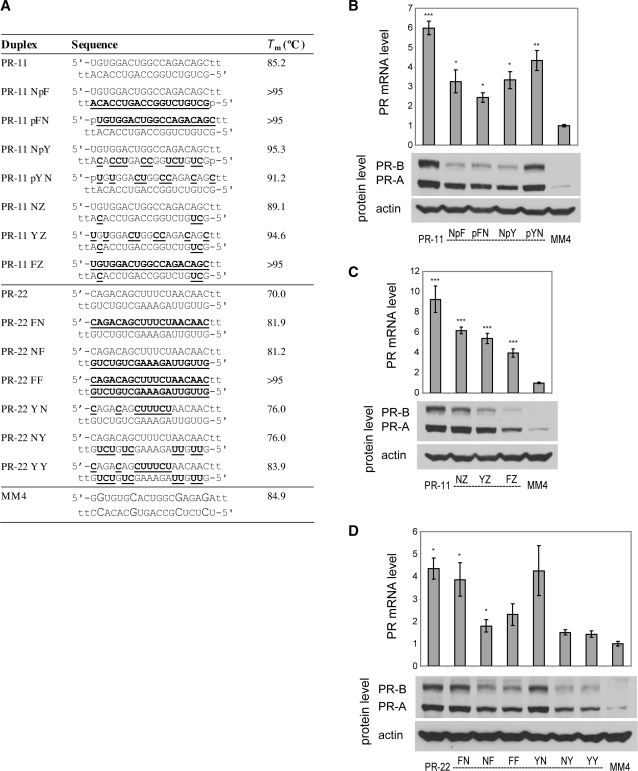
Figure 3.
PR gene expression can be activated by agRNAs containing 2′F-RNA. (A) Duplex sequences: passenger strands are listed on top in all cases; for agRNAs PR-11 and PR-22 this is the antisense strand. Modification codes: dna, RNA, 2′F-RNA; p = phosphate. (B) PR-11 analogs containing one strand heavily or fully modified with 2′F-RNA. (C) PR-11 analogs containing a minimally 2′F-RNA-modified guide strand. (D) PR-22 analogs containing one or both strands heavily or fully modified with 2′F-RNA. All data are normalized to mismatch control MM4. RNA levels are the average (±SD) from at least two independent transfections. *P < 0.05, **P < 0.01, ***P < 0.005. Protein levels were also confirmed by at least two independent transfections, from which a typical western blot is shown.
To assess the generality of 2′F-RNA modification, we also tested a second activating agRNA, PR-22. For PR-22, 2′F-RNA was well-tolerated in the passenger strand and duplexes PR-22 FN and PR-22 YN were able to activate gene expression at both RNA and protein levels without loss of potency (Figure 3D). The guide strand of PR-22 was more sensitive to 2′F-RNA modification, with lower levels of gene activation by agRNAs PR-22 NY and PR-22 YY. Taken together with results from PR-11, these results show that 2′F-RNA substitutions can be well tolerated by activating agRNAs and that chemical modifications on the passenger strand tend to be better tolerated.
Effect of 2′-O-methyl-RNA on gene silencing
2′-O-Methyl-RNA (2′OMe) nucleotides (Figure 1) improve the thermal and nuclease stability of siRNA duplexes (41,42). Inclusion of 2′OMe-RNA units also reduces off-target effects mediated either through the miRNA pathway (5) or the innate immune system (43–45). We tested duplex RNAs containing two 2′OMe ribonucleotides at the 5′-terminus of each strand (J) or at all pyrimidine nucleotides (P) (Figure 4A). Duplexes PR-9 JJ, containing 2′OMe nucleotides at both 5′-termini, and PR-9 PN, containing 2′OMe modification at the pyrimidine nucleotides of the passenger strand, were both effective gene silencing agents (Figure 4B). Duplexes containing pyrimidine modifications on the guide strand (PR-9 NP, PR-9 PP) were inactive, consistent with previous observations that 2′OMe modification is often better tolerated in the passenger strand (41). Variants of PR2526 (PR2526 PN, PR2526 NP and PR2526 PP) had activities similar to the PR-9 variants (Figure 4B).
Figure 4.
PR gene expression can be silenced by agRNAs containing 2′OMe-RNA. (A) Duplex sequences: passenger strands are listed on top in all cases; for siRNA PR2526 this is the sense strand, but for agRNAs PR-9 and PR-26 this is the antisense strand. Modification codes: dna, RNA, 2′Ome-rna. (B) PR-9 analogs and controls containing 2′OMe-RNA. (C) PR-26 analogs containing 2′OMe-RNA. All data are normalized to mismatch control MM4. RNA levels are the average (±SD) from at least two independent transfections. *P < 0.05, **P < 0.01, ***P < 0.005. Protein levels were also confirmed by at least two independent transfections, from which a typical western blot is shown.
We also examined the effects of 2′OMe modification of a second silencing agRNA, PR-26, and identified analogs that inhibited PR expression (Figure 4C). However, for PR-26, modification of the guide strand in PR-26 NP did not prevent gene silencing. These data demonstrate that agRNA-mediated gene silencing can tolerate the introduction of 2′OMe-RNA modifications. The influence of a specific pattern of modification, however, depends on the identity of the parent sequence and can vary between agRNAs.
Effect of 2′-O-methyl-RNA on gene activation
We introduced 2′-O-methyl modifications into two different activating agRNAs, PR-11 and PR-22 (Figure 5A). Substitution of 2′OMe units into PR-11 reduced activation from 8-fold to 2–3-fold (Figure 5B). PR-11 PP with two heavily modified strands were the least activating. By contrast, activation by modified PR-22 duplexes was relatively stable at 4- to 6-fold regardless of modification (Figure 5C). These data suggest that, as observed above for gene silencing, the impact of 2′OMe modifications on gene activation will vary depending on the parent sequence.
Figure 5.
PR gene expression can be activated by agRNAs containing 2′OMe-RNA. (A) Duplex sequences: passenger strands are listed on top in all cases; for agRNAs PR-11 and PR-22 this is the antisense strand. Modification codes: dna, RNA, 2′Ome-rna. (B) PR-11 analogs containing 2′OMe-RNA. (C) PR-22 analogs containing 2′OMe-RNA. All data are normalized to mismatch control MM4. RNA levels are the average (±SD) from at least two independent transfections. *P < 0.05, **P < 0.01, ***P < 0.005. Protein levels were also confirmed by at least two independent transfections, from which a typical western blot is shown.
Effect of LNA on gene silencing
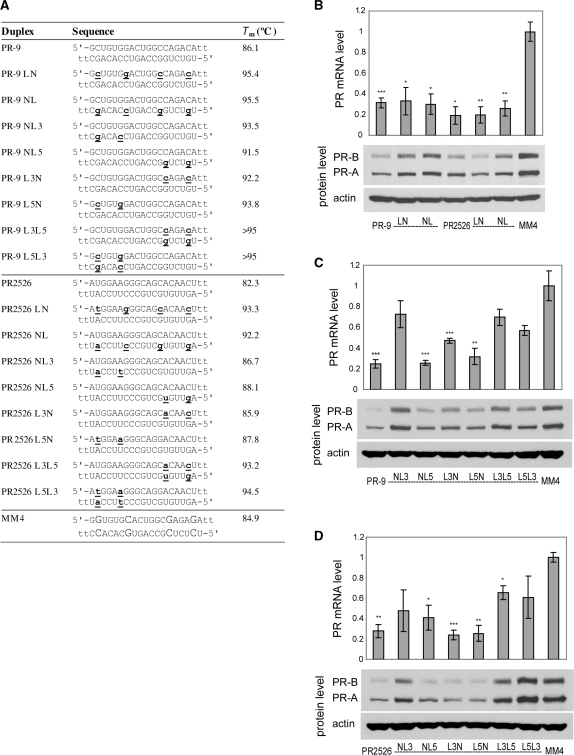
Locked nucleic acid (LNA) (Figure 1) is a rigid nucleotide analog containing a constrained, bicyclic sugar. Introduction of locked nucleotides into DNA or RNA oligomers leads to substantially enhanced affinities for complementary sequences (46–49) and enhanced resistance to digestion by nucleases (49–51). Several studies have also shown that the introduction of LNA nucleotides can improve the properties of siRNAs (9,35,51–53). LNA has been shown to improve loading of the desired strand (51), increase nuclease resistance (51) and lower immune stimulation (53).
We tested a series of duplexes containing four LNA nucleotides evenly distributed across either the passenger or guide strand (Figure 6A). PR-9 LN and PR-9 NL both retained the ability to silence gene expression, albeit to a lesser extent than PR-9 (Figure 6B). Significant silencing activity was maintained regardless of whether the sense or antisense strand was modified.
Figure 6.
PR gene expression can be silenced by agRNAs containing LNA. (A) Duplex sequences: passenger strands are listed on top in all cases; for siRNA PR2526 this is the sense strand, but for agRNA PR-9 this is the antisense strand. LNA sequences contain 5-methylcytosine instead of cytosine. Modification codes: dna, RNA, lna. (B) PR-9 analogs and controls containing four evenly distributed LNA units. (C) PR-9 analogs containing LNA at strand ends. (D) PR2526 analogs containing LNA at strand ends. All data are normalized to mismatch control MM4. RNA levels are the average (±SD) from at least two independent transfections. *P < 0.05, **P < 0.01, ***P < 0.005. Protein levels were also confirmed by at least two independent transfections, from which a typical western blot is shown.
The high binding affinity of LNA has been used to direct loading of the appropriate strand into AGO and bias strand usage (51). We tested a series of thermodynamically biased LNA analogs, to explore whether their activities would differ (Figure 6C). Strand L3 contains LNA at the second and sixth positions from its 3′-end, while strand L5 contains LNA at the second and sixth positions from its 5′-end. Modification of the 3′-end of the guide strand (PR-9 NL3), reduced activity of PR-9 (Figure 6C). All other RNA duplexes containing two LNA substitutions on one strand (PR-9 NL5, PR-9 L3N, PR-9 L5N) were active regardless of which strand was modified or the location of the modification. Some activity was lost when both strands were modified with LNA (PR-9 L3L5, PR-9 L5L3). Variants of PR2526 containing LNA bases yield results that were analogous to LNA-substituted variants of PR-9 (Figure 6B and D).
These results demonstrate that LNA modifications are compatible with gene silencing by agRNAs, but show no evidence for biased strand loading in the context of the PR-9 or PR2526 sequences. The reduction in activity upon modification of the 3′-end of the guide strand (‘NL3’) is consistent with previous observations that LNA modification of the sixth nucleotide from the 3′-end of this strand leads to reduced potency (51). Results were similar for each pattern of modification regardless of whether agRNA PR-9 or siRNA PR2526 was the parent duplex, emphasizing that the response of agRNAs and siRNAs to chemical modification can be similar.
Effect of LNA on gene activation
We tested LNA-modified analogs of PR-11 for gene activation in MCF7 cells (Figure 7A). For activating duplex PR-11, we observed activation (2- to 3-fold versus 6-fold for the unmodified duplex) after modification of either strand with four LNA nucleotides (PR-11 LN, PR-11 NL) (Figure 7B). The potency was reduced to the same extent whether the guide or passenger strand was modified. For activation by PR-11, all of the thermodynamically biased LNA analogs maintained significant activity (Figure 7C). Modification of either end of the guide strand (PR-11 NL3, PR-11 NL5) gave activity indistinguishable from that of the native PR-11. Passenger strand modification (PR-11 L3N, PR-11 L5N) gave a slight reduction in activation, and modification of both strands (PR-11 L3L5, PR-11 L5L3) led to a further reduction in activation. Nonetheless, even these duplexes modified in both strands gave >3-fold activation at the RNA level and clear activation at the protein level. Together with results of 2′F and 2′-OMe substitution, these data suggest that agRNA-mediated gene activation is broadly tolerant of chemical modification.
Effect of combining modifications on gene silencing
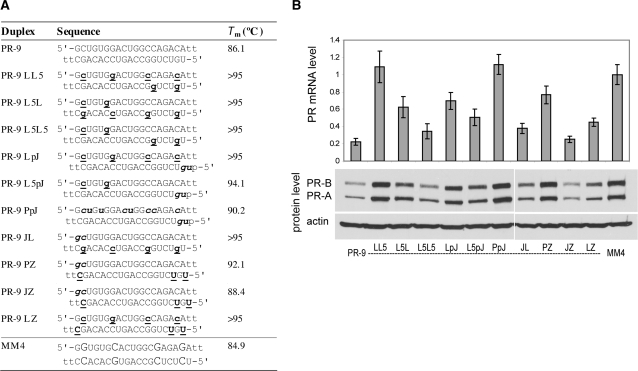
Many examples exist of potent and stable siRNAs containing multiple types of chemical modifications (38,54–56). In some cases, combination of multiple modifications led to potency and stability unattainable by the individual modifications alone (54,56). We set out to explore whether combination agRNA duplexes (Figure 8A) would yield better gene inhibition or activation in their respective cell lines. Even if existing potency is only maintained, the combination of different chemistries could afford better stability and other desirable drug properties.
Figure 8.
PR gene expression can be silenced by agRNAs containing combinations of modifications. (A) Duplex sequences: passenger strands are listed on top in all cases; for agRNA PR-9 this is the antisense strand. LNA sequences contain 5-methylcytosine instead of cytosine. Modification codes: dna, RNA, 2′F-RNA, 2′Ome-rna, lna; p = phosphate. (B) PR-9 analogs containing combinations of previously identified active strands. All data are normalized to mismatch control MM4. RNA levels are the average (±SD) from at least two independent transfections. Protein levels were also confirmed by at least two independent transfections, from which a typical western blot is shown.
Our criteria for choosing combinations was to select modified strands from PR-9 and PR-11 analogs showing the highest activity and recombine them in new ways. For PR-9 analogs, this led to the identification of several potent new duplexes (Figure 8B). Duplexes including PR-9 L5L5, PR-9 JL, PR-9 JZ and PR-9 LZ retained substantial activity. Duplexes PR-9 LL5 and PR-9 PpJ lost most activity. The effects appear complex and cannot simply be characterized by considering additive effects from the individual modifications. Nonetheless, it appears that terminal 2′F-RNA modification (Z) on the guide strand and LNA modification at the 5′-end of the passenger strand (L5) are generally well-tolerated, whereas 2′OMe modification of all pyridimines (P) on the passenger strand tends to disrupt the activity.
It is noteworthy that two of the most active combination duplexes also had Tm values that were too high for us to measure (PR-9 L5L5 and PR-9 JL). It is sometimes assumed that high Tm values prevent the strand separation required for siRNA/agRNA activity, but our findings confirm that the relationship between Tm and activity is more complex. This result is consistent with our previous observation that the limited number of agRNA duplexes tested to date do not rigidly adhere to published guidelines for predicting the activity of duplex RNAs (23).
We combined modified strands from active PR-11 analogs to give a new series of activating agRNAs (Figure 9A). All the combination duplexes based on PR-11 lost significant activity with respect to the unmodified control. Duplexes containing LNA in the passenger strand and 2′F-RNA at the termini of the guide strand (PR-11 LZ and PR-11 L5Z) were the best of this series, but only showed 2- to 3-fold activation at the RNA level (Figure 9B).
Figure 9.
PR gene expression can be activated by agRNAs containing combinations of modifications. (A) Duplex sequences: passenger strands are listed on top in all cases; for agRNA PR-11 this is the antisense strand. LNA sequences contain 5-methylcytosine instead of cytosine. Modification codes: dna, RNA, 2′F-RNA, 2′Ome-rna, lna. (B) PR-11 analogs containing combinations of previously identified active strands. All data are normalized to mismatch control MM4. RNA levels are the average (±SD) from at least two independent transfections. Protein levels were also confirmed by at least two independent transfections, from which a typical western blot is shown.
No evidence that single-strands mediate agRNA activity
There have been reports that single-stranded antisense RNA can lead to inhibition via the RNAi pathway (12,57,58). In addition, previous work from our lab (59–61) and others (62–65) has shown that single-stranded oligonucleotides including peptide nucleic acid (PNA) and LNA–DNA chimeras were able to bind chromosomal DNA and repress gene expression at the transcriptional level.
To explore whether the single-stranded RNAs contained within agRNAs can mediate such effects, we transfected T47D and MCF7 cells with various native and modified single strands from functional silencing or activating agRNA duplexes (Figure 10A). Transfections were carried out under the same conditions and at the same concentration (25 nM) as duplex RNAs. Results from both qPCR and western blots demonstrate that single-stranded agRNA, when transfected alone, had no effect on silencing or activation of PR gene expression (Figure 10B and C). These data demonstrate that potent modulation of gene expression by anti-PR agRNAs requires the presence of intact duplex RNA regardless of chemical modification. Our data indicate that gene silencing or activation by the agRNA in our study is not due (i) to slight excess of one strand, (ii) to degradation of one strand (prior to incorporation into AGO), or (iii) to single-strand-mediated off-target effects. The requirement for RNA duplexes also provides further evidence that both native and chemically modified agRNAs make use of the RNAi machinery.
Figure 10.
Single strands of native or modified agRNAs are inactive. (A) Sequences: passenger strands are given 5′-3′, guide strands are given 3′-5′ as shown. For these agRNAs the passenger strand is the antisense strand. Modification codes: dna, RNA, 2′F-RNA, 2′Ome-rna, lna; p = phosphate. (B) PR-9 duplex and native or modified single-strands. (C) PR-11 duplex and native or modified single-strands. All data are normalized to the guide strand of mismatch control MM4. RNA levels are the average (±SD) from at least two independent transfections. Protein levels were also confirmed by at least two independent transfections, from which a typical western blot is shown.
Concentration dependence of modified duplexes
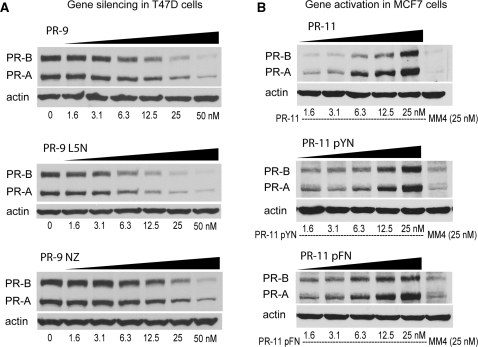
To understand the effect of chemical modification on the potency of agRNAs we examined the dependence of gene silencing or activation on the concentration of selected RNAs. We observed that potencies of gene silencing by modified agRNAs PR-9 L5N and PR-9 NZ were similar to parent agRNA PR-9 (Figure 11A, see also Supplementary Figures S3 and S4). Likewise, potencies of gene activation by PR-11 pYN and PR-11 pFN were similar to gene activation by parent agRNA PR-11 (Figure 11B). Thus it is possible to achieve potent, dose-dependent gene modulation by both activating and inhibitory chemically modified agRNAs. For silencing agRNAs based on PR-9, we calculated IC50 values for the native duplex and several modified analogs (Table 2) (Supplementary Figures S3 and S4). At both the protein and RNA levels, some of the modified analogs were of comparable potency to PR-9 itself (e.g. PR-9 NL). Others, while still active, showed reduced potency (e.g. PR-9 NZ).
Figure 11.
Sample dose response experiments for both (A) silencing and (B) activation. More dose response experiments for PR-9 analogs are shown in Supplementary Figures S3 and S4.
Table 2.
IC50 values for PR-9 and modified analogs in T47D cellsa
| Silencing agRNA | Type of chemistry | IC50 (RNA) (nM) | IC50 (protein) (nM) |
|---|---|---|---|
| PR9 | native | 5.6 | 12.2 |
| PR9 NZ | 2′F | 18.6 | 19.6 |
| PR9 JpJ | 2′OMe | 9.4 | 10.2 |
| PR9 PN | 2′OMe | 12.9 | 12.6 |
| PR9 NL | LNA | 6.8 | 13.2 |
| PR9 L5N | LNA | 13.3 | 7.4 |
aPlots used to derive these data are shown in Supplementary Figures S3 and S4.
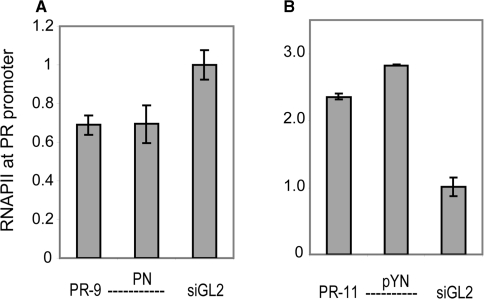
Modified duplexes affect recruitment of RNA polymerase II
We have previously used ChIP of RNA polymerase II (RNAPII) [see the supporting information of ref. (28)] and nuclear run-on assays (25) to show that both silencing and activating agRNAs affect PR transcription. To investigate whether chemically modified agRNAs operate through the same mechanism as their native counterparts, we carried out RNAPII ChIP using two active modified duplexes (Figure 12). In T47D cells, treatment with either PR-9 or its modified analog PR-9 PN led to reduced occupancy of RNAPII at the PR promoter relative to cells treated with the luciferase control duplex siGL2 (Figure 12A). In MCF7 cells, treatment with either PR-11 or the modified analog PR-11 pYN increased levels of RNAPII at the PR promoter (Figure 12B). The finding that native and modified duplexes behave similarly supports the hypothesis that they act through similar mechanisms.
Figure 12.
ChIP of RNA Polymerase II (RNAPII) shows that both native and modified agRNAs operate at the transcriptional level. (A) Treatment of T47D cells with PR-9 or PR-9 PN leads to reduced occupancy of RNAPII at the PR promoter relative to cells treated with control duplex siGL2. (B) Treatment of MCF7 cells with PR-11 or PR-11 pYN leads to increased occupancy of RNAPII at the PR promoter relative to cells treated with control duplex siGL2.
Native and modified duplexes have the same effect on antisense transcript levels
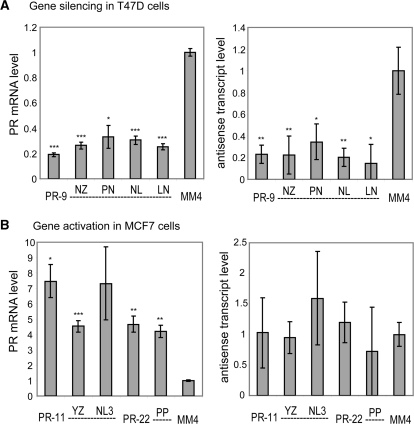
We have previously observed that both activating and inhibitory agRNAs bind to an antisense transcript that overlaps the PR promoter (28). This transcript originates within PR mRNA and extends 70 000-bp upstream. It is spliced, and the spliced product contains target sequences for all the agRNAs used in this study. When biotinylated agRNAs are transfected into cells they bind the antisense transcript. RNA immunoprecipitation shows that both activating and silencing agRNAs recruit AGO to the antisense transcript. The antisense transcript, therefore, appears to play a central role in the mechanism of agRNAs.
We explored the effect of chemically modified agRNAs on levels of the promoter-associated antisense transcript. We previously observed that treatment of T47D cells with silencing agRNA PR-9 led to a reduction of antisense transcript levels, while treatment of MCF7 cells with activating agRNA PR-11 led to no significant change in antisense transcript levels (28). We repeated these experiments using both the native duplexes and several of the most active modified analogs. We monitored expression of PR mRNA and the antisense transcripts that span the agRNA target region using qPCR.
Native and modified duplexes gave comparable results. PR-9 and modified PR analogs reduced expression of both PR mRNA and the promoter-derived antisense transcript similarly (Figure 13A). Conversely, PR-11 and modified analogs increased expression of PR mRNA, but caused no significant change in antisense transcript levels (Figure 13B). The finding that native and modified duplexes have the same effect on levels of the antisense transcript supports the conclusion that they act through similar mechanisms.
Figure 13.
Native and modified duplexes have the same effect on antisense transcript levels. (A) Treatment of T47D cells with PR-9 and modified duplexes leads to a significant downregulation of both PR mRNA (left) and the promoter-associated antisense transcript (right). (B) Treatment of MCF7 cells with PR-11 and modified duplexes leads to a significant upregulation of PR mRNA (left) but no significant change in levels of the promoter-associated antisense transcript (right). *P < 0.05, **P < 0.01, ***P < 0.005.
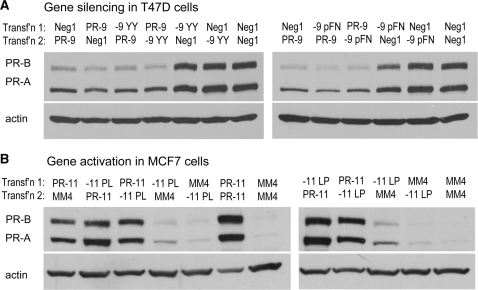
Inactive modified duplexes do not compete for target sites
It is interesting to speculate as to why some patterns of modification cause reduced activity. Inactive agRNAs could be failing at any of a number of steps: (i) loading into the RNA-induced silencing complex (RISC), (ii) cleavage or dissociation of the passenger strand, (iii) import into the nucleus, (iv) binding of the RNA target, or (v) execution of the effect (activation or silencing). We previously observed that inactive native duplexes such as PR-8 (targeting –8 to +11) and PR-12 (–12 to +7) could compete for target sites with the active duplex PR-11 (23). Thus, treating MCF7 cells with inactive duplex PR-12 prevented gene activation by PR-11 in a subsequent transfection several days later. On the other hand, initial treatment with PR-11 followed later by PR-12 gave activation comparable to PR-11 alone (23). This experiment showed that the order of addition is crucial and the duplexes compete for the same target sites. Duplexes PR-8 and PR-12 failed at step (v) above—they were loaded into RISC and even bound the correct RNA target, but failed to execute an agRNA effect. Therefore, since shifting the duplex–target interaction by 1–3 bp is sufficient to eliminate activity, the geometry of interaction at the promoter may be crucial for induction of gene activation.
To test where inactive chemically modified agRNAs might be deficient we carried out a similar experiment using two inactive modified duplexes each for silencing and activation. In T47D cells we used the inactive duplexes PR-9 YY and PR-9 pFN. In both cases, levels of PR expression were identical whether the inactive duplexes were transfected before or after the active duplex PR-9 (Figure 14A). Similarly, treatment of MCF7 cells with PR-11 gave gene activation regardless of whether the duplexes were transfected before or after inactive modified duplexes PR-11 PL and PR-11 LP (Figure 14B). These data suggest that the modified duplexes did not compete for target sites with native duplexes. Since the inactive modified duplexes are not binding the target site, they must be failing at one of the first four steps in the process: RISC loading, passenger strand removal, nuclear import or target binding. Furthermore, some of the inactive modified duplexes contain native guide strands, thus it seems clear that they are failing at one of the first two steps—RISC loading or passenger strand removal—since after this point they would be identical with native duplexes. Therefore in most cases, the challenges causing modified agRNAs to fail are the same as those faced by chemically modified siRNAs that are complementary to mRNA.
Figure 14.
Inactive modified duplexes do not compete for target sites. Western analysis showing no competition between native agRNAs and inactive modified duplexes applied to cells in two transfections 4 days apart. (A) Native PR-9 silences gene expression equally effectively whether it is transfected before or after an inactive chemically modified duplex (or negative control). (B) Native PR-11 activates gene expression equally effectively whether it is transfected before or after an inactive chemically modified duplex.
agRNAs and chemical modifications
Double stranded RNAs that are complementary to mRNA are currently being tested in clinical trials (1–4). Given the success of siRNAs in the laboratory and progress in the clinic, why would agRNAs enhance the potential for in vivo application of double-stranded RNA? One reason is that activating gene expression would open up a wider range of therapeutic targets and may provide new opportunities for development. For example, if more of a protein product is needed to treat a disease, upregulating expression using agRNAs might prove advantageous, especially if the protein’s site of action is intracellular and cannot be accessed by systemically administered protein. Even in the case of gene silencing, where existing siRNA technology often gives superb results, it is possible that silencing at the transcriptional level may be advantageous for some targets. It is possible to envision that agRNAs may be more potent in some cases, produce longer lasting effects, or produce different modulation of isoform expression. Finally, understanding the effect of modifications will also important for future studies of agRNA function in cultured cells. Potential applications for promoter-targeted duplex RNAs have been summarized in a recent review (66).
Our data have several implications for the mechanism of chemically modified agRNAs: (i) As with any structure–function analysis, the most important finding is that some designs yield active compounds whereas others do not; (ii) Two strands are necessary for efficient action of agRNAs, we observe no silencing or activation by single strands at the highest concentrations used in this study; (iii) Activation and silencing show a regular, predictable dose dependence; (iv) As with siRNAs, optimal modifications need to be determined empirically for each sequence; (v) As with siRNAs, both guide and passenger strands can be modified, but modifications are generally better tolerated on the passenger strand; (vi) IC50 values and maximal efficacies of modified and unmodified agRNAs are similar, consistent with the expectation that they operate by a common mechanism; (vii) Both native and modified duplexes affect recruitment of RNAPII, also consistent with a common mechanism; (viii) Addition of modified and unmodified agRNAs leads to almost identical changes in antisense transcript levels, again consistent with a shared mechanism; and (ix) Inactive duplexes do not compete with active duplexes, suggesting that the inactive duplexes fail at a step prior to association with the antisense transcript.
The finding that agRNAs containing 2′F-RNA, 2′OMe-RNA, or LNA retain the ability to silence or activate gene expression is significant because it demonstrates that agRNAs tolerate the introduction of modifications known to be powerful tools for improving in vivo properties. For duplex RNAs that are complementary to mRNA, it is commonly observed that different sequences tolerate different chemical modifications or different patterns of the same chemical modification. The interplay of chemical modifications with each particular sequence is the ultimate determinant of duplex potency and duplexes with optimal properties must be determined empirically. No universal rules of siRNA modification have been developed because our understanding of the nature of this interplay is not yet sufficient.
Our data on agRNAs suggest the same conclusion. We have tested two different silencing agRNAs, PR-9 and PR-26, and two different activating agRNAs, PR-11 and PR-22. While both tolerate a wide range of modifications, the relative potencies of analogous modified duplexes differ. Like siRNAs, we conclude that empirical testing will be needed to identify agRNAs with the best balance of properties. While empirical testing may be necessary, we found that a wide range of modifications were compatible with gene silencing or activation by an agRNA-mediated mechanism and we predict that it will not be difficult to develop active chemically modified agRNAs for genes of interest where a parent unmodified agRNA has already been identified.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIGMS 77253); Robert A. Welch Foundation (I-1244); Fonds québécois de la recherche sur la nature et les technologies (postdoctoral fellowship to J.K.W.); Alnylam Pharmaceuticals. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.de Fougerolles A, Vornlocher H-P, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Corey DR. Chemical modification: the key to clinical application of RNA interference? J. Clin. Invest. 2007;117:3615–3622. doi: 10.1172/JCI33483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA: tools and applications. Drug Discov. Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ui-Tei K, Naito Y, Zenno S, Nishi K, Yamato K, Takahashi F, Juni A, Saigo K. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008;36:2136–2151. doi: 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Judge A, MacLachlan I. Overcoming the innate immune response to small interfering RNA. Hum. Gene Ther. 2008;19:111–124. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- 8.Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 9.Bramsen JB, Laursen MB, Damgaard CK, Lena SW, Ravindra Babu B, Wengel J, Kjems J. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res. 2007;35:5886–5897. doi: 10.1093/nar/gkm548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen PY, Weinmann L, Gaidatzis D, Pei Y, Zavolan M, Tuschl T, Meister G. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA. 2008;14:263–274. doi: 10.1261/rna.789808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parrish S, Fleenor J, Xu S, Mello C, Fire A. Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol. Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- 12.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 13.Schwarz DS, Hutvágner G, Haley B, Zamore PD. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell. 2002;10:537–548. doi: 10.1016/s1097-2765(02)00651-2. [DOI] [PubMed] [Google Scholar]
- 14.Chiu Y-L, Rana TM. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol. Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- 15.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Leuschner PJF, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Paula D, Bentley MVLB, Mahato RI. Hydrophobization and bioconjugation for enhanced siRNA delivery and targeting. RNA. 2007;13:431–456. doi: 10.1261/rna.459807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 19.Morris KV, Chan SWL, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 20.Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy D, Shames DS, Minna JD, Corey DR. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat. Chem. Biol. 2005;1:216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 21.Janowski BA, Hu J, Corey DR. Silencing gene expression by targeting chromosomal DNA with antigene peptide nucleic acids and duplex RNAs. Nat. Protocols. 2006;1:436–443. doi: 10.1038/nprot.2006.64. [DOI] [PubMed] [Google Scholar]
- 22.Li L-C, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl Acad. Sci. USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 24.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat. Struct. Mol. Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 26.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 27.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc. Natl Acad. Sci. USA. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA. Antisense transcripts are targets for activating small RNAs. Nat. Struct. Mol. Biol. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardy DB, Janowski BA, Corey DR, Mendelson CR. Progesterone receptor plays a major antiinflammatory role in human myometrial cells by antagonism of nuclear factor-kB activation of cyclooxygenase 2 expression. Mol. Endocrinol. 2006;20:2724–2733. doi: 10.1210/me.2006-0112. [DOI] [PubMed] [Google Scholar]
- 30.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 31.Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, Cook PD. Uniformly modified 2′-deoxy-2′-fluoro-phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem. 1993;36:831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 32.Layzer JM, McCaffrey AP, Tanner AK, Huang ZAN, Kay MA, Sullenger BA. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrissey DV, Blanchard K, Shaw L, Jensen K, Lockridge JA, Dickinson B, McSwiggen JA, Vargeese C, Bowman K, Shaffer CS, et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology. 2005;41:1349–1356. doi: 10.1002/hep.20702. [DOI] [PubMed] [Google Scholar]
- 34.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 36.Harborth J, Elbashir SM, Vandenburgh K, Manninga H, Scaringe SA, Weber K, Tuschl T. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 37.Blidner RA, Hammer RP, Lopez MJ, Robinson SO, Monroe WT. Fully 2′-deoxy-2′-fluoro substituted nucleic acids induce RNA interference in mammalian cell culture. Chem. Biol. Drug Des. 2007;70:113–122. doi: 10.1111/j.1747-0285.2007.00542.x. [DOI] [PubMed] [Google Scholar]
- 38.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotech. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 39.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann K-K, Schlee M, et al. 5′-Triphosphate RNA Is the Ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 40.Zamanian-Daryoush M, Marques JT, Gantier MP, Behlke MA, John M, Rayman P, Finke J, Williams BRG. Determinants of cytokine induction by small interfering RNA in human peripheral blood mononuclear cells. J. Interferon Cytokine Res. 2008;28:221–233. doi: 10.1089/jir.2007.0090. [DOI] [PubMed] [Google Scholar]
- 41.Czauderna F, Fechtner M, Dames S, Aygun H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collingwood MA, Rose SD, Huang L, Hillier C, Amarzguioui M, Wiiger MT, Soifer HS, Rossi JJ, Behlke MA. Chemical modification patterns compatible with high potency dicer-substrate small interfering RNAs. Oligonucleotides. 2008;18:187–200. doi: 10.1089/oli.2008.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Judge AD, Bola G, Lee ACH, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 44.Robbins M, Judge A, Liang L, McClintock K, Yaworski E, MacLachlan I. 2′-O-Methyl-modified RNAs act as TLR7 antagonists. Mol. Ther. 2007;15:1663–1669. doi: 10.1038/sj.mt.6300240. [DOI] [PubMed] [Google Scholar]
- 45.Cekaite L, Furset G, Hovig E, Sioud M. Gene expression analysis in blood cells in response to unmodified and 2′-modified siRNAs reveals TLR-dependent and independent effects. J. Mol. Biol. 2007;365:90–108. doi: 10.1016/j.jmb.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 46.Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 47.Obika S, Nanbu D, Hari Y, Andoh J-i, Morio K-i, Doi T, Imanishi T. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides. Tetrahedron Lett. 1998;39:5401–5404. [Google Scholar]
- 48.Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 2003;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 49.Braasch DA, Corey DR. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 50.Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hokfelt T, Broberger C, Porreca F, Lai J, Ren K, et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl Acad. Sci. USA. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, Wahren B, Liang Z, Urum H, Koch T, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mook OR, Baas F, de Wissel MB, Fluiter K. Evaluation of locked nucleic acid-modified small interfering RNA in vitro and in vivo. Mol. Cancer Therap. 2007;6:833–843. doi: 10.1158/1535-7163.MCT-06-0195. [DOI] [PubMed] [Google Scholar]
- 53.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 54.Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 55.Koller E, Propp S, Murray H, Lima W, Bhat B, Prakash TP, Allerson CR, Swayze EE, Marcusson EG, Dean NM. Competition for RISC binding predicts in vitro potency of siRNA. Nucleic Acids Res. 2006;34:4467–4476. doi: 10.1093/nar/gkl589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dande P, Prakash TP, Sioufi N, Gaus H, Jarres R, Berdeja A, Swayze EE, Griffey RH, Bhat B. Improving RNA interference in mammalian cells by 4′-thio-modified small interfering RNA (siRNA): effect on siRNA activity and nuclease stability when used in combination with 2′-O-alkyl Modifications. J. Med. Chem. 2006;49:1624–1634. doi: 10.1021/jm050822c. [DOI] [PubMed] [Google Scholar]
- 57.Holen T, Amarzguioui M, Babaie E, Prydz H. Similar behaviour of single-strand and double-strand siRNAs suggests they act through a common RNAi pathway. Nucleic Acids Res. 2003;31:2401–2407. doi: 10.1093/nar/gkg338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Janowski BA, Kaihatsu K, Huffman KE, Schwartz JC, Ram R, Hardy D, Mendelson CR, Corey DR. Inhibiting transcription of chromosomal DNA with antigene peptide nucleic acids. Nat. Chem. Biol. 2005;1:210–215. doi: 10.1038/nchembio724. [DOI] [PubMed] [Google Scholar]
- 60.Beane RL, Ram R, Gabillet S, Arar K, Monia BP, Corey DR. Inhibiting gene expression with locked nucleic acids (LNAs) that target chromosomal DNA. Biochemistry. 2007;46:7572–7580. doi: 10.1021/bi700227g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu J, Corey DR. Inhibiting gene expression with peptide nucleic acid (PNA)-peptide conjugates that target chromosomal DNA. Biochemistry. 2007;46:7581–7589. doi: 10.1021/bi700230a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 63.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 64.Giovannangeli C, Diviacco S, Labrousse V, Gryaznov S, Charneau P, Helene C. Accessibility of nuclear DNA to triplex-forming oligonucleotides: the integrated HIV-1 provirus as a target. Proc. Natl Acad. Sci. USA. 1997;94:79–84. doi: 10.1073/pnas.94.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Larsen HJ, Nielsen PE. Transcription-mediated binding of peptide nucleic acid (PNA) to double-stranded DNA: sequence-specific suicide transcription. Nucleic Acids Res. 1996;24:458–463. doi: 10.1093/nar/24.3.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morris KV. RNA-directed transcriptional gene silencing and activation in human cells. Oligonucleotides. 2009;19:299–306. doi: 10.1089/oli.2009.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.