Abstract
The mutagenic threat of hydrolytic DNA cytosine deamination is met mostly by uracil DNA glycosylases (UDG) initiating base excision repair. However, several sequenced genomes of archaeal organisms are devoid of genes coding for homologues of the otherwise ubiquitous UDG superfamily of proteins. Previously, two possible solutions to this problem were offered by (i) a report of a newly discovered family of uracil DNA glycosylases exemplified by MJ1434, a protein found in the hyperthermophilic archaeon Methanocaldococcus jannaschii, and (ii) the description of TTC0482, an EndoIV homologue from the hyperthermophilic bacterium Thermus thermophilus HB27, as being able to excise uracil from DNA. Sequence homologues of both proteins can be found throughout the archaeal domain of life. Three proteins orthologous to MJ1434 and the family founder itself were tested for but failed to exhibit DNA uracil glycosylase activity when produced in an Ung-deficient Escherichia coli host. Likewise, no DNA uracil processing activity could be detected to be associated with TTC0482, while the protein was fully active as an AP endonuclease. We propose that the uracil processing activities formerly found were due to contaminations with Ung enzyme. Use of Δung-strains as hosts for production of putatively DNA-U processing enzymes provides a simple safeguard.
INTRODUCTION
Base excision repair (BER) is a general and ubiquitous pathway protecting cells from genetic consequences of various forms of chemical damage of DNA nucleobases (1). Repair is initiated by a glycosylase that hydrolyzes the glycosidic bond connecting the damaged base to a DNA sugar moiety; this results in a base-free residue (AP site). The variety of base damage is mirrored by the provision of a number of different DNA repair glycosylases, each having its own characteristic substrate spectrum. Repair glycosylases can be monofunctional, having glycosylase activity only or bifunctional, having an additional (AP lyase) activity (2) that cleaves the DNA strand at the AP site via β-elimination, the reaction product being a 2′,3′-unsaturated aldehyde residue at the 3′-end of the 5′-terminal fragment (3). Subsequently, an AP endonuclease, usually of the ExoIII or EndoIV type, creates a 3′-OH terminus for repair synthesis, either by setting a strand break at the AP site created by a monofunctional glycosylase or by removing the block left behind by a bifunctional glycosylase (4). Repair is completed by functions shared by all individual BER pathways (1).
Hydrolytic deamination of cytosine to uracil residues is a DNA base damage with moderate incidence rate and high mutagenic potential. If not repaired, each pre-mutagenic U/G intermediate gives rise to a C/G→T/A transition in 50% of the progeny. According to the kinetic parameters determined by Frederico et al. (5), about 100 genomic cytosine residues undergo spontaneous hydrolytic deamination everyday in every human cell. Not surprisingly, uracil-specific DNA glycosylases are found throughout all existing life forms. Almost invariably, these enzymes are members of the UDG structural superfamily (6) which consists of families 1 to 5, with Ung (7), TDG/MUG (8,9), SMUG (10), TMUDG (11) and TTUDGB (12) as the respective prototypes.
For fundamental physical reasons, the problem of spontaneous DNA cytosine deamination is greatly accentuated with thermophilic organisms (5). In view of this, it is an apparent paradox that in the complete genome sequences of archaea such as Methanothermobacter thermautotrophicus ΔH (13), Methanocaldococcus jannaschii (14) and Methanopyrus kandleri AV19 (15) no homologues of any of the five UDG families could be detected. Mesophilic archaea, however, are also known to be devoid of UDG genes, e.g. Methanococcus maripaludis S2 (16) and Methanosphaera stadtmanae (17); hence the phenomenon seems to be a speciality of the archaeal domain of life, not strictly linked to high growth temperatures.
The DNA uracil processing glycosylase Mig from M. thermautotrophicus THF, which does not belong to the UDG but rather to the helix–hairpin–helix superfamily (18,19), does not qualify for a general DNA uracil repair enzyme because of its substrate spectrum which is narrowly restricted by sequence context. Its biological role rather seems to be the removal of thymine generated by deamination of 5-methyl-cytosine residues (20–22). In search of the carrier of general DNA-U repair in this organism, our attention was drawn to two gene products: MTH746 and MTH1010, homologues of the thermophile-derived proteins MJ1434 (M. jannaschii) and TTC0482 (Thermus thermophilus HB27), respectively, both of which had been described as DNA uracil processing enzymes (23–25). MJ1434 is a member of the helix–hairpin–helix superfamily of proteins that encompasses many DNA repair glycosylases (2,26,27), TTC0482 is a homologue of Escherichia coli EndoIV, an AP endonuclease (28).
As illustrated below, we have not been able to confirm DNA uracil processing activities to be associated with MJ1434 and TTC0482. On the other hand, the problem of general DNA uracil repair in M. thermautotrophicus ΔH has recently been solved by demonstrating that Mth212, a homologue of E. coli ExoIII, is an endonuclease incising next to DNA-U residues (29) and that this step initiates DNA-U repair in whole-cell extracts (30) and in a minimal in vitro system reconstituted from purified components (31).
MATERIALS AND METHODS
Strains
Escherichia coli DH5α (Invitrogen, Carlsbad, CA, USA), Escherichia coli TOP10 (Invitrogen), Escherichia coli Rosetta-gami B(DE3) (Novagen®, Madison, WI, USA), Escherichia coli BL21_UX (29) [based on BL21-CodonPlus(DE3)-RIL, Stratagene, La Jolla, CA, USA] and Escherichia coli BL21_UXX (30).
Methanothermobacter thermautotrophicus ΔH (DSM 1053), M. kandleri AV19 (DSM 6324), M. maripaludis S2 (DSM 14266), M. jannaschii (DSM 2661) and T. thermophilus HB27 (DSM 7039) were purchased as actively growing cultures from DSMZ (Braunschweig, Germany).
Plasmids
pET_B_001 and pET_B_001_ung (29), pET-21d_ttudga (12), pCR-Blunt II-TOPO vector for cloning of blunt-ended PCR-products was purchased from Invitrogen as part of the ‘Zero Blunt® TOPO’ kit. pET-28a(+) plasmid DNA was acquired from Novagen®. pJET1.2/blunt was purchased from Fermentas (Burlington, Ontario, Canada) as part of the ‘CloneJET™ PCR Cloning Kit’. pET28a-MjUDG (23) was a gift from Ji Hyung Chung (Yonsei University, Seoul, Korea).
Enzymes and chemicals
Restriction enzymes, T4 DNA Ligase and Pfu DNA Polymerase were purchased from Fermentas, thrombin protease from GE Healthcare (Uppsala, Sweden) and Proteinase K from Invitrogen. Chemicals were purchased from either Roth (Karlsruhe, Germany) or Merck (Darmstadt, Germany).
Enzyme substrates
The following 2′-deoxyribooligonucleotides were purchased from Purimex (Grebenstein, Germany) in double HPLC-purified grade. F, fluorescein moiety; U, 2′-d-uridine residue.
40-U (40-mer) 5′ F-GGGTACTTGGCTTACCTGCCCTGUGCAGCTGTGGGCGCAG 3′
35-G (35-mer) 5′ CTGCGCCCACAGCTGCGCAGGGCAGGTAAGCCAAG 3′
Marker_23 (23-mer) 5′ F-GGGTACTTGGCTTACCTGCCCTG 3′
Chung_32_U (32-mer) 5′ F-GGATCCTCTAGAGTCUACCTGCAGGCATGCAA 3′
Chung_32_T (32-mer) 5′ TTGCATGCCTGCAGGTTGACTCTAGAGGATCC 3′
Chung_Marker_15 (15-mer) 5′ F-GGATCCTCTAGAGTC 3′
Single-stranded substrates were prepared by diluting 5 pmol of corresponding fluorescein-labeled oligos in 100 µl 1× SSC (150 mM NaCl, 15 mM trisodiumcitrate) plus 400 µl H2O to a final concentration of 0.01 pmol/µl and stored at −20°C. Preparation of double-stranded substrates was as described earlier (29).
Isolation of genes by PCR and insertion into expression vector
DNA from M. thermautotrophicus ΔH, M. kandleri AV19, M. maripaludis S2, M. jannaschii and T. thermophilus HB27 was prepared by phenol/chloroform extraction and ethanol precipitation from liquid cultures (2 ml) that were previously pelleted, resuspended in 2 ml SCE buffer (1M sorbitol, 100 mM trisodiumcitrate, 60 mM EDTA) and sonicated on ice for 3 min with a Branson Sonifier 250 (microtip, output level 5, duty cycle 50%).
For standard PCR amplification of open reading frames (orfs) from genomic DNAs following primer pairs were used:
Orf mth746-1: 5′ ATGTTTAACCGTCATGATTATTTACC 3′ and 5′ CTGTGAAGTCACTCGAGATCACGTGAACCACCCAG 3′ (BspHI and XhoI sites are underlined)
Orf mth746-2: 5′ CCTATTAATCATGAAGGTGCTCAGGC 3′ and 5′ CTGTGAAGTCACTCGAGATCACGTGAACCACCCAG 3′ (BspHI and XhoI sites are underlined)
Orf mk0193: 5′ GGGCTTCACCATGGTCCCCTCGGTA 3′ and 5′ CACTTCCCTGAGTCGACAGCCTGTT 3′ (NcoI and SalI sites are underlined)
Orf mmp0586: 5′ AGGAAAACCATGGACAATAAAGA 3′ and 5′ CTGAATAATCTCGAGTTTTATTA 3′ (NcoI and XhoI sites are underlined)
Orf mj1434: 5′ CATATGAAAGAGAACAAATTTGAGATG 3′ and 5′ AAGCTTTTACTTTGAGAGGCAGAATTCTTTA 3′ (NdeI and HindIII sites are underlined)
Orf ttc0482: 5′ CATATGCCGCGCTACGGGTTCCAC 3′ and 5′ GTCGACTTAGGCCTCCTCGAGCCAG 3′ (NdeI and SalI sites are underlined)
For sufficient amplification of orf ttc0482, the standard PCR reaction was supplemented with 1.3 M betaine.
Blunt-ended PCR products were gel-purified and cloned into pCR-Blunt II-TOPO vector as described by the supplier. Frames mth746-1, mth746-2, mk0193 and mmp0586 were isolated from pCR-Blunt II by the indicated restriction enzymes and inserted into NcoI- and XhoI-treated pET_B_001. Frame mj1434 was cut out of pCR-Blunt II vector with NdeI and HindIII and cloned into the corresponding restriction sites of pET-28a( + ) resulting in the same plasmid construct as described (23). Frame ttc0482 was cloned into pET-28a( + ) using the restriction sites for NdeI and SalI. Correct sequences of the cloned genes were verified by DNA sequence analyses.
Construction of an mj1434 mutant coding for MJ1434/E132K
For construction of mutant mj1434, two separate PCR amplifications were performed using pET-28a-mj1434 as template. Primers were mj1434-for (5′ CATATGAAAGAGAACAAATTTGAGATG 3′) and E132K-rev (5′ CAACAAAATACTATCAGCTGTTTTCTTTCCCACTCCATTTATTGATA 3′) or mj1434-rev (5′ AAGCTTTTACTTTG AGAGGCAGAATTCTTTA 3′) and E132K-for (5′ TATCAATAAATGGAGTGGGAAAGAAAACAGCTGATAGTATTTTGTTG 3′), respectively, with one primer for each reaction carrying the desired mutation. (NdeI and HindIII sites are underlined and nucleotide exchanges are marked in bold face.) Both PCR products were gel-purified and subjected to a third round of PCR, where the complete frame mj1434/E132K was generated by overlap extension and amplified using primers mj1434-for and mj1434-rev. The blunt-ended PCR product was cloned into pJET1.2/blunt following the supplier’s manual. Frame mj1434/E132K was transferred to pET-28a( + ) using the restriction sites for NdeI and HindIII. Correct sequence of the resulting construct pET-28a-mj1434/E132K was verified by DNA sequence analysis.
Overexpression of cloned genes and purification of proteins
Overexpression of E. coli Ung, MTH746-1, MTH746-2, MK0193 and MMP0586 in BL21_UX and MJ1434 as well as TTC0482 in BL21_UXX, preparation of cleared cell lysate, immobilized metal ion affinity chromatography (IMAC) and heparin column chromatography were essentially as described by us for other proteins (29), with the sole exception of adding the reducing agent dithiothreitole (DTT) to a final concentration of 10 mM to all MJ1434 protein solutions. TTUDGA was produced in BL21_UX and purified as described (12,30).
N-terminal His-tag was cleaved from MJ1434 after heparin column chromatography by incubation with thrombin protease according to the instructor’s manual. Gel filtration to remove His-tag after thrombin incubation was done on a HiLoad 16/60 Superdex 75 prep grade gel filtration column (GE Healthcare/Amersham Biosciences, Fairfield, CT, USA) with 50 mM Tris/HCl pH 7.5, 100 mM KCl as running buffer at a flow rate of 1 ml/min. Elution was monitored by measuring the absorption at 280 and 260 nm simultaneously.
Mutant MJ1434/E132K was produced in BL21_UXX and purified from cleared cell lysate by IMAC in parallel with wild type MJ1434. Fractions containing MJ1434 wild type or mutant protein, respectively, were pooled, concentrated and used directly for enzyme activity assays.
Enzyme activity assays
For glycosylase assays with MTH746-1, MTH746-2, MK0193, MMP0586 and E. coli Ung (control) 0.12 pmol of double- or single-stranded oligonucleotide substrates were pre-incubated in a total volume of 50 µl 10 mM HEPES–KOH, pH 7.6, 100 mM KCl for 10 min at 37°C (Ung, MTH746-1, MTH746-2 and MMP0586) or 65°C (MK0193). Subsequently, 24 pmol of the respective proteins were added and the reactions were incubated for 30 min. To stop the reactions, 5 µl of 1 M NaOH were added and the mixtures were incubated at 95°C for 10 min. Twenty-five microliters loading dye (95% formamide, 20 mM EDTA pH 8.0, 50 mg/ml Dextran-Blue 2000000) were added and samples were kept on ice until gel electrophoretic analysis.
MJ1434 glycosylase assays were performed identically to the protocol of Chung et al. (23) and E. coli Ung was used as a control. Conditions: 5 pmol of protein, 1 pmol of double- or single-stranded oligonucleotide substrates in a total volume of 20 µl 20 mM MES/KOH pH 6.0, 80 mM NaCl, 1 mM DTT, 1 mM EDTA and 3% glycerol at 55°C (for MJ1434) or 37°C (for E. coli Ung) for 20 min. The reactions were stopped and processed as described above.
To test MJ1434 and MJ1434/E132K (IMAC purification grade) in parallel for uracil glycosylase and AP lyase activity, either untreated oligonucleotide substrate or substrate pre-incubated with 0.6 pmol of E. coli Ung were incubated with 2 µg of the respective protein. Pre-incubation with E. coli Ung was performed at 37°C for 20 min, while incubation with MJ1434 wild type or mutant protein was done for 20 min at 55°C. All reactions were performed in a total volume of 60 µl 20 mM MES/KOH pH 6.0, 80 mM NaCl, 1 mM DTT, 1 mM EDTA and 3% glycerol containing 0.12 pmol of oligonucleotide substrate. Subsequently, each sample was split into three equal aliquots. 10 µl of loading dye were added immediately to one aliquot; the second third was post-treated with 2 µl of 1 M NaOH for 10 min at 95°C. Finally, the remaining third was treated with 2 µl 0.5 M EDTA (pH 8.0) containing 5 mg/ml Proteinase K for 20 min at 65°C. After each treatment 11 µl of loading dye were added (not all aliquots from each sample were analysed by gel electrophoresis). Samples were treated with Proteinase K in order to release the oligonucleotides from attaching proteins interfering with subsequent electrophoretic analysis.
Single or combined assays using TTC0482 and TTUDGA were carried out with 0.6 pmol of the respective enzyme and 0.12 pmol of double- or single-stranded oligonucleotide substrates in 50 µl 20 mM Tris/HCl pH 7.5, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT for 30 min at 60°C (enzymes were always added after a 5 min pre-incubation of substrates in reaction buffer at 60°C). When indicated reactions were post-treated with 5 µl of 1 M NaOH for 10 min at 95°C. Subsequently, 25 µl of loading dye were added. Otherwise, reactions were stopped by immediate addition of 25 µl loading dye and kept on ice until gel electrophoretic analysis.
Gel electrophoresis using the Pharmacia A.L.F. sequencer was as described earlier (29). Peak areas were quantified using the software ‘Fragment Manager’ from Pharmacia.
RESULTS
The MJ1434 homologues MTH746, MK0193 and MMP0586 are not able to process uracil residues in DNA
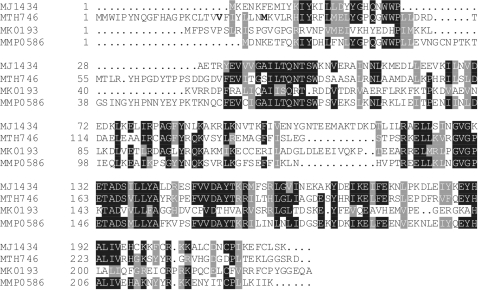
MTH746 shares with MJ1434 38% residue identity and 54% similarity. Figure 1 shows multiple alignment of the two proteins and two additional members of the family, MK0193 from M. kandleri AV19 (15) and MMP0586 from M. maripaludis S2 (16) which were also included in the functional study (compare below). As expected, multiple alignment shows preferential sequence conservation at sites of known functional importance. On the other hand, the four cysteine residues necessary for the formation of a [Fe4S4] cluster, which is wide-spread in the HhH-family of repair glycosylases (2), are present as a complete set only in the case of MJ1434.
Figure 1.
Sequence alignment of deduced amino acid sequences from putative MJ1434 homologues. MJ1434 is from M. jannaschii (NCBI accession number: NP_248438), MTH746 from M. thermautotrophicus ΔH (NP_275889), MK0193 from M. kandleri AV19 (NP_613480) and MMP0586 from M. maripaludis S2 (NP_987706). Sequences were aligned with ClustalW+ from GCG Version 11.1, Accelrys Software Inc., San Diego, CA, USA using default parameters and shaded with BoxShade 3.21 (www.ch.embnet.org/software/BOX_form.html) using a fraction of 0.7 for shading. Bold letters in the N-terminal region of MTH746 point to truncation endpoints of two variants of the protein (see legend of Figure 2 for details).
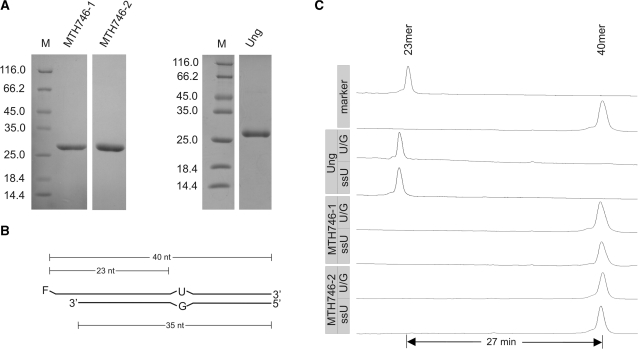
A first attempt to overproduce MTH746 in E. coli from a gene coding for a protein with an N-terminus as annotated (compare Figure 1) and engineered to code for a His6-tag at the C-terminus (preceded by additional amino acids L and E) yielded only insoluble protein. However, soluble protein could be obtained with two N-terminally truncated variants (named MTH746-1 and MTH746-2) constructed as explained in the legend to Figure 2. Proteins were produced in E. coli strain Rosetta-gami B(DE3) and purified to apparent homogeneity (Supplementary Figure S1A) using IMAC, followed by heparin affinity chromatography. At 37°C, both proteins displayed DNA uracil glycosylase activity on both single- and double-stranded DNA substrates (Supplementary Figure S1B and C). However, reduced activity at 65°C and inhibition by the Bacillus subtilis phage PBS2-encoded UDG inhibitor Ugi (32,33) (Supplementary Figure S1D and E, respectively) prompted us to examine whether the observed uracil glycosylase activity might be due to contamination with E. coli Ung, in particular since an analogous case has been reported for E. coli nucleoside diphosphate kinase (34–36).
Figure 2.
Test for uracil DNA glycosylase activity associated with MTH746-1 and MTH746-2 purified from the Ung-deficient E. coli strain BL21_UX. (A) SDS–PAGE analysis (Coomassie stained) of purified proteins after IMAC and heparin affinity chromatography. MTH746 was N-terminally truncated by either omitting the first 19 amino acids (named MTH746-1) or the first 26 amino acids (named MTH746-2). For clarification, the first amino acid post each truncation is indicated in bold in the amino acid sequence of MTH746 depicted in Figure 1. For technical reasons, the N-terminal amino acid residues of MTH746-1 were changed from VF to MI. Relative molecular weights of marker proteins [×10−3] in lane M are indicated to the left. The gel was stained with Coomassie brilliant blue R-250. Ung, uracil-N-glycosylase from E. coli. Calculated relative molecular masses are 28 020 for MTH746-1, 27 040 for MTH746-2 and 26 760 for Ung. For production and purification details, see ‘Materials and Methods’ section. (B) Schematic drawing of the double-stranded U/G mismatched DNA substrate. F, fluorescein moeity. The lengths of the substrate oligonucleotides and the expected products are indicated. For the single-stranded DNA substrate (ssU) the fluorescently labeled oligonucleotide without the complementary strand was used. (C) Track recording of the ALF DNA sequencer after glycosylase assay with E. coli uracil-N-glycosylase Ung (control) or MTH746-1 and MTH746-2, respectively. Incubation was performed with 24 pmol of the respective protein and 0.12 pmol of substrates for 30 min at 37°C. Numbers of minutes refer to run-time differences between marker oligonucleotides. Note that the product in the Ung control is migrating faster than the 23-mer marker oligonucleotide, since it contains a phosphate residue at its 3′-terminus left behind from two rounds of elimination reaction with NaOH. For reaction details refer to ‘Materials and Methods’ section.
In a second round of experiments, MTH746-1 and MTH746-2 were produced in BL21_UX, a Δung-derivative of BL21-CodonPlus(DE3)-RIL (29), and purified as before. In contrast to the control with E. coli Ung, no uracil glycosylase activity could be detected with single-stranded or double-stranded substrate DNA, neither at 37°C (Figure 2) nor at 65°C (data not shown). Homologues MK0193 and MMP0586 (compare Figure 1) were produced and purified in an exactly analogous fashion and subjected to the same test of DNA-U processing activity; no activity was detected (Supplementary Figure S2).
MJ1434 produced in a Δung-strain has no detectable DNA uracil glycosylase activity
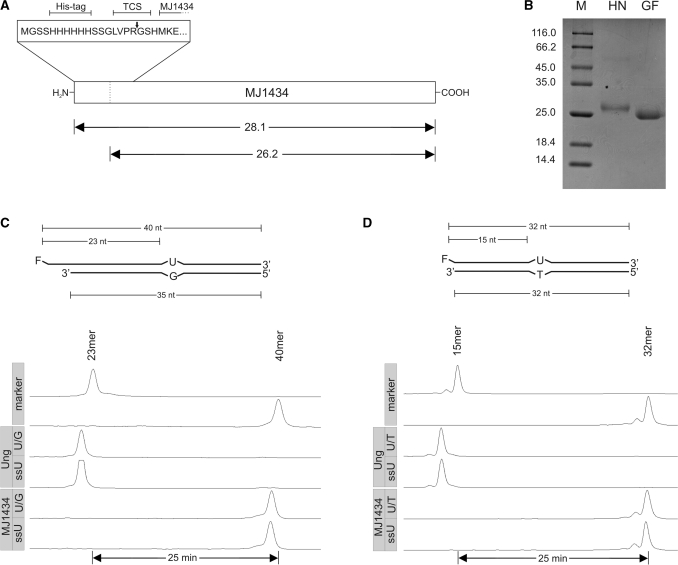
The above findings prompted us to reexamine the activity of the prototype protein MJ1434 described by Chung et al. (23). Their expression plasmid, a pET28a derivative, was reconstructed exactly. From this construct, MJ1434 is produced with an N-terminal His6-tag that can be removed by cleavage with thrombin (Figure 3A).
Figure 3.
Uracil DNA glycosylase assay with MJ1434. (A) Schematic drawing of the expression construct. TCS, thrombin cleavage site. The arrows indicate the expected relative molecular masses [×10−3] of MJ1434 before and after thrombin cleavage. (B) SDS–PAGE analysis of purified MJ1434 after heparin chromatography (HN) and after thrombin cleavage and gel filtration chromatography (GF). Labels as in Figure 2A. (C) Top: schematic drawing of the double-stranded U/G mismatched DNA substrate as in Figure 2B. Bottom: trackings of fluorescence readout as recorded by an ALF DNA sequencer after glycosylase assays with MJ1434 (5 pmol) using double-stranded (U/G) or single-stranded (ssU) uracil containing oligonucleotide substrates (1 pmol each). Ung was used as the positive control. Labels as in Figure 2C. (D) Top: scheme of U/T mismatched substrate. Bottom: readout of glycosylase assay with Ung (control) or MJ1434 as in (C) but with substrates bearing the identical sequence that was used by Chung et al. (23). For details refer to ‘Materials and Methods’ section.
Structural integrity of the gene inserted into the plasmid was verified by DNA sequence analysis. For protein production, Δung-strain BL21_UXX (30) was used. The protein was purified by IMAC, followed by heparin affinity chromatography. The purified protein was treated with thrombin and the liberated His6-tag was removed by gel filtration chromatography. As judged by SDS–PAGE (Coomassie stain), cleavage was complete (Figure 3B). For testing DNA-U glycosylase activity, two double-stranded substrates, one containing a U/G, the other a U/T mispair were prepared (Figure 3C and D). In order to exclude differences brought into the comparison by possible sequence context effects, the U/T substrate was chosen to have the same sequence as the oligonucleotides described by Chung et al. (23). In addition, the fluorescently labeled U-containing oligonucleotides were also tested in single-stranded form. Assays were performed under conditions described by Chung et al. (23); results are displayed in Figure 3C and D. No activity could be detected with either double- or single-stranded DNA. The substrates were, however, completely processed in control reactions with E. coli Ung which confirms that the absence of product formation with MJ1434 was not due to inappropriate substrates or lack of downstream processing of reaction intermediates. MJ1434 produced in Δung-strain BL21_UXX from the original ‘pET28a-MjUDG’ construct (23), made available by Ji Hyung Chung, was likewise devoid of DNA-U glycosylase activity (data not shown).
Mutant protein MJ1434/E132K is an AP lyase but has no DNA uracil glycosylase activity
The results presented in the preceding paragraph certainly do not support the notion of MJ1434 being a DNA uracil glycosylase. At this stage, however, it cannot be stringently ruled out that, in our hands, the activity was lost in the course of protein purification. In a strict sense, this argument applies even to complete lack of activity in the crude cellular extract from which the protein was purified. To clarify this point, some other biochemical function of the protein is required that can be monitored throughout the isolation procedure. In accord with general properties of HhH proteins, this other function could ideally be an independently verified DNA repair glycosylase activity directed towards damaged DNA bases other than uracil. Corresponding experiments, however, are prohibited at present by lack of knowledge what this alternative DNA damage might be (also compare ‘Discussion’ section).
Mutant MJ1434/E132K constructed by Im et al. (24) and described as a combined DNA uracil glycosylase/AP lyase offers a solution of the problem as follows. There are two classes of HhH repair glycosylases, the monofunctional ones exclusively catalyze hydrolysis of the glycosidic bond of their substrate bases, bifunctional ones carry DNA repair one step further by cleavage, via β-elimination, of the damaged DNA strand at the newly produced AP site (2,37). The prototype of the latter (‘glycosylase/AP lyase’) class is E. coli EndoIII and the structural hallmark of the additional AP lyase activity is a lysine residue at a specific location within the active site (K120 in the case of EndoIII). The primary amino function of the lysine side chain forms a transient Schiff base with the aldehyde of the AP site prior to β-elimination (37,38). In vitro, this intermediate can be trapped by saturating the C/N double bond with sodium borohydride or sodium cyanoborohydride (37,39).
Previously, two monofunctional HhH repair glycosylases have been converted to AP lyases by a single amino acid exchange at positions corresponding to K120 of EndoIII, M. thermautotrophicus Mig/Y126K (39) and E. coli MutY/S120K (40). Concomitantly, glycosylase activity was reduced in the latter case and was completely lost in the first. Im et al. (24) correctly point out that residue E132 of MJ1434 aligns with K120 of EndoIII. The construction of mutant MJ1434/E132K reported by Im et al. is therefore directly analogous to the above-quoted work of Begley and Cunningham (39) and of Williams and David (40). Interestingly, albeit not altogether surprisingly, MJ1434/E132K shows AP lyase activity—with the source of the accompanying DNA uracil glycosylase activity still being contentious.
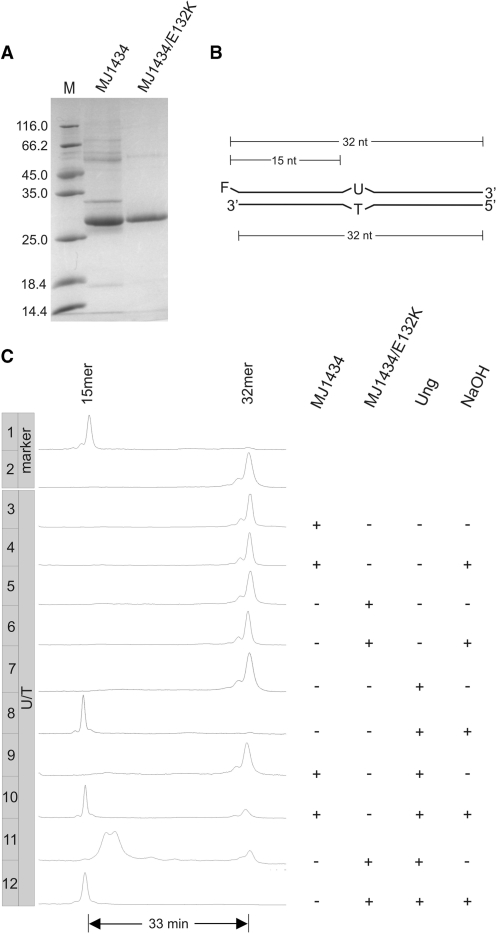
A mutant gene coding for MJ1434/E132K was constructed and the protein purified (by IMAC) in parallel with wild-type MJ1434 from extracts of the same Δung expression host as used before (Figure 4A). Results of DNA uracil glycosylase and AP lyase assays on these protein preparations are displayed in Figure 4C. The necessary substrate containing an AP site was produced from 2′-deoxy-oligonucleotide containing a uracil residue (Figure 4B) by treatment with uracil DNA glycosylase Ung from E. coli.
Figure 4.
DNA-U glycosylase and AP lyase assays with E132K mutant of MJ1434. (A) SDS–PAGE analysis of IMAC purified MJ1434 (wild type) and MJ1434/E132K (for details refer to ‘Materials and Methods’ section). M, molecular weight markers as in Figure 2A. Note that at this state of purification impurities are detectable in the protein preparations. (B) Schematic drawing of the double-stranded U/T mismatched DNA substrate (labels as in Figure 3D). (C) Track recording of the ALF DNA sequencer after incubation of the substrate (0.12 pmol) with 2 µg of MJ1434 (wild type) or mutant E132K or/and 0.6 pmol of E. coli Ung. NaOH: with or without NaOH post-treatment. Samples containing MJ1434 or MJ1434/E132K, if not treated with NaOH, were incubated with Proteinase K prior to gel electrophoresis. For reaction details refer to ‘Materials and Methods’ section.
This treatment yielded a full-length intermediate (Figure 4C, lane 7) which, upon treatment with NaOH, reacted to yield a 5′-fragment with a length of 15 nucleotides (plus a 3′-O-phosphate moiety; Figure 4C, lane 8). Treating the substrate oligonucleotides with NaOH without prior incubation with Ung had no effect (data not shown). Obviously, Ung cleanly converted the DNA uridine residue to an AP site. Upon incubation with the E132K mutant of MJ1434 but not with wild-type MJ1434, this AP-site-containing substrate was almost completely processed to shorter material (Figure 4C, lanes 11 and 9, respectively) largely excluding possible contamination with AP lyase activity from the host. The MJ1434/E132K-specific reaction product (lane 11) migrates slower than the 15mer marker oligonucleotide (lane 1). This is in accord with the well-known AP lyase reaction mechanism: breakage of the DNA strand by β-elimination which leaves behind a 2′,3′-unsaturated aldehyde residue linked to the 3′-end of the 5′-terminal DNA fragment by a phosphodiester bond. Alkali treatment promoted conversion of this intermediate to a faster migrating product (Figure 4C, lane 12, the same as in lane 8)—as expected to result from a second round of elimination leaving behind a 3′-terminal phosphomonoester moiety. Importantly, treatment of substrates with MJ1434/E132K or MJ1434 without prior incubation with Ung had no effect regardless of a post-treatment with NaOH (lanes 3–6).
Clearly, MJ1434/E132K is an AP lyase but, in contrast to its original description, it has no DNA uracil glycosylase activity under the conditions tested. Hence, it can be safely concluded that the DNA uracil glycosylase activity observed by Im et al. (24) in their preparation of MJ1434/E132K was contributed by contaminating E. coli Ung. (Note that the question of possible loss of whichever glycosylase function by introduction of the E132K mutation is irrelevant here since the original claim is that the mutant does carry DNA-U glycosylase activity.)
Together with our findings illustrated above, this leaves little flexibility for how to interpret the nature of the corresponding activity described by Chung et al. (23) as associated with wild-type MJ1434 and by Im et al. (24) with some other MJ1434 mutants (compare below, ‘Discussion’ section). Under the conditions tested (which include the ones used by the authors of the original description), MJ1434 is not a DNA uracil glycosylase.
TTC0482 has AP endonuclease but no DNA uracil processing activity
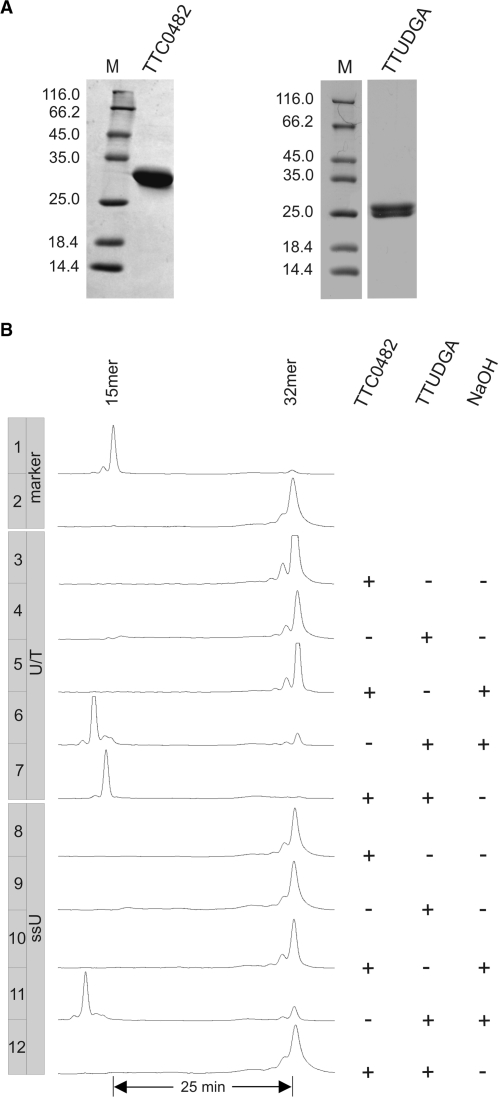
A second candidate for the function of an initiator of DNA-U repair in M. thermautotrophicus ΔH was suggested by work of Back et al. in 2006 (25). These authors characterized the EndoIV homologue from T. thermophilus HB27, TTC0482, as an enzyme capable of processing DNA-U residues in addition to its activity as an AP endonuclease (25). The corresponding homologue in M. thermautotrophicus ΔH is MTH1010. With the hindsight, however, of our experience with MJ1434 and some of its homologues, including MTH746, we decided to first reproduce the findings of Back et al. (25) before embarking on any enzymological characterization of EndoIV homologues from M. thermautotrophicus ΔH or yet other organisms. To this end, we constructed a pET-28a-ttc0482 expression plasmid, produced the protein in BL21_UXX (30) and purified it to near homogeneity by IMAC and heparin affinity chromatography (Figure 5A).
Figure 5.
Assays with TTC0482 for uracil processing and AP endonuclease activities. (A) SDS–PAGE analysis of purified TTC0482 and TTUDGA from T. thermophilus HB27 after IMAC and heparin affinity chromatography (for details refer to ‘Materials and Methods’ section). M, molecular weight markers as in Figure 2A. Calculated relative molecular weights for TTC0482 and TTUDGA are 31 200 and 23 680, respectively. (B) Gel electophoretic analysis of reaction products after incubation of substrates (0.12 pmol) with 0.6 pmol of the EndoIV homologue TTC0482 or/and the uracil DNA glycosylase TTUDGA (12) from T. thermophilus HB27 as indicated. The substrates are identical to the ones used for the glycosylase assay of MJ1434 (Figure 3D). NaOH±: with or without post-reaction NaOH-treatment. Note that the AP-endonuclease reaction product shown in lane 7 migrates as a 15-mer with a 3′-OH terminus (compare marker in lane 1). For reaction details refer to ‘Materials and Methods’ section.
As a member of the EndoIV family of enzymes, TTC0482 is expected to be an AP endonuclease and this property has been confirmed by Back et al. (25). In order to make sure the enzyme at hand is catalytically active, we first assayed our preparation of TTC0482 for this uncontroversial AP endonuclease activity. As in the experiment described in the previous paragraph, substrates containing an AP site were produced by treating corresponding uracil-containing oligonucleotides (Figure 5B) with a uracil DNA glycosylase. For this purpose, we chose TTUDGA (12) from the same organism which can be expected to be compatible with TTC0482 in coupled reactions. TTUDGA-treatment converted the DNA uridine residues of both double- and single-stranded substrates to AP sites which were processed to 15-mer products upon treatment with NaOH (Figure 5, lanes 4, 6 and lanes 9, 11; also compare previous paragraph).
Combining TTUDGA and TTC0482 in the assay mixture led to a 15-mer product with the double-stranded but not with the single-stranded substrate (lanes 7 and 12). This confirmed AP endonuclease activity of TTC0482 and its double-strand specificity as described by Back et al. (25). In contrast, however, no reaction was observed when substrate DNAs were incubated with TTC0482 alone—without or with post-reaction treatment with NaOH (lanes 3, 8 and 5, 10, respectively). Under the conditions tested, TTC0482 clearly is neither a DNA uracil glycosylase nor a DNA uridine endonuclease.
DISCUSSION
Neither MJ1434 nor TTC0482 seems to be a DNA-U processing enzyme (neither a DNA uracil glycosylase nor a DNA uridine endonuclease). This raises the question of how the corresponding misconceptions could have arisen and how similar ones can be avoided in the future. There are several lines of evidence pointing to contaminating Ung protein from the E. coli production host as the most likely source of the activities erroneously assigned to MJ1434 (23,24) and TTC0482 (25).
First, there is precedent. E. coli nucleoside diphosphate kinase (NDPK) was, for a while, thought to be a multifunctional enzyme with DNA-U glycosylase activity (34). However, it was subsequently determined that the activity observed was due to E. coli Ung contaminating the original NDPK preparation (35,36). As elaborated under ‘Results’ section, we experienced similar problems in our early attempts to characterize the enzymological properties of MTH746, the MJ1434 homologue of M. thermautotrophicus ΔH.
In all three studies in question (23–25), Ung-proficient E. coli strains were used as production hosts [BL21 DE3 (23,24) and TB1 (25)]. In the work of Back et al. (25) and Im et al. (24), treatment at 70°C for 10–20 min was used to precipitate or deactivate E. coli proteins. This, however, is not a reliable measure against contamination with Ung, since significant Ung activity was reported recovered after passaging the enzyme through 30 PCR cycles with 94°C denaturing temperature (41).
Several points in the reports by Chung et al. (23), Im et al. (24) and Back et al. (25), however, are not in immediate accord with the notion of E. coli Ung contaminating the MJ1434 and TTC0482 proteins. Most seriously, Chung et al. report their preparation of MJ1434 to be more active at 65°C than at 37°C, an observation clearly against expectation for contaminating Ung. Logically, consistent explanations for this behaviour are at hand, but without detailed knowledge of the pipetting schemes employed, they remain as speculative as complicated and, for the time being, the discrepancy stands out as somewhat of an enigma.
A second point concerns the study of MJ1434 by directed mutagenesis reported by Im et al. (24). Five amino acid replacements were constructed. Two of these showed unchanged and two reduced levels of (apparent) DNA uracil glycosylase activity. A straightforward explanation of these observations is offered by assuming that the different protein preparations were contaminated with E. coli Ung enzyme to varying degrees. The properties of the fifth mutant described by Im et al. (24), MJ1434/E132K, have already been discussed in detail in the ‘Results’ section. As illustrated there, we could verify its AP lyase but not the allegedly concomitant DNA-U glycosylase activity. This not only proves that the mutant protein is an active enzyme, but also makes it quite plausible that, in its wild-type form, MJ1434 is, indeed, a monofunctional DNA repair glycosylase. Interestingly, wild-type M. kandleri homologue MK0193 carries a lysine residue at the position corresponding to E132 of MJ1434 (residue #143; Figure 1) and may thus be a true bifunctional enzyme.
HhH proteins are DNA repair glycosylases prevalent in all domains of life. The superfamily formed by an ancestral structure radiating out into a number of enzyme families having different substrate spectra (26). Each spectrum is defined by a single or a small number of structurally related, chemically damaged nucleobases. On the basis of amino acid sequence, MJ1434 has previously been placed into the MpgII family (26,42) which may hint at methylated DNA purine residues as substrates. However, with no pertinent biochemical data at hand, only one fact about the substrate spectrum of the putative DNA repair glycosylase MJ1434 is presently known for certain: uracil is not part of it.
A final point concerns the apparent DNA-U processing activity of TTC0482 with single-stranded substrate (25). Since the AP endonuclease activity of TTC0482 is double-strand specific, the cleavage observed with a single-stranded, uracil-containing substrate (25, Figure 3B, lane at the extreme right) cannot be explained as resulting from a two-step enzymatic reaction with Ung and TTC0482 working hand in hand. Significantly, however, the product obtained from the single-stranded substrate moves slower in gel electrophoresis than the one obtained from the same DNA as part of a double strand containing a U/G mispair (immediately neighbouring lane). This is the expected behaviour if one assumes that the single-stranded substrate underwent only one round of enzymatic reaction, the removal of the uracil base, and that the oligonucleotide was subsequently cleaved by β-elimination during sample work-up. This point has been discussed repeatedly in the literature (27,43). According to experience of our laboratory, 5 min of heating at 90°C in ‘formamide loading buffer’, as described by the authors, are perfectly apt to bring about this non-enzymatic reaction.
In summary, there are two conclusions to be drawn, one special and one general in nature. First, the case of DNA uracil repair in M. jannaschii has to be re-opened. This organism, like M. thermautotrophicus ΔH, lacks genes for any of the known uracil glycosylases of the UDG superfamily (14). In addition, it is also devoid of an ExoIII-homologue, which excludes the existence of a pathway homologous to the one found in M. thermautotrophicus ΔH (30). Since M. jannaschii certainly depends on DNA-U repair for genome stability, at least one other, still undiscovered mechanism must be at work. Thus, the enzymology of DNA uracil repair in the archaeal domain is even more diverse than what is known to date (30).
Second, given the high activity of E. coli Ung and its (moderate) resistance against thermal denaturation [compare above, (41)] we consider the use of Ung−-strains (preferably Δung) as hosts for the production of putatively DNA-U processing enzymes as the only safe measure to exclude contaminating activity. In view of the widespread use of Ung-proficient strains for that type of work in the past, it cannot be ruled out at present that more cases of erroneously assigned activities are still polluting the scientific record.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Georg-August-Universität Göttingen.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ji Hyung Chung, Yonsei University, Seoul, Korea, for gift of pET28a-MjUDG and Christiane Preiß for expert technical assistance.
REFERENCES
- 1.Friedberg E, Walker G, Siede W, Wood R, Schultz R, Ellenberger T. In DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. Base excision repair; pp. 169–226. [Google Scholar]
- 2.Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997;325(Pt 1):1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dodson ML, Michaels ML, Lloyd RS. Unified catalytic mechanism for DNA glycosylases. J. Biol. Chem. 1994;269:32709–32712. [PubMed] [Google Scholar]
- 4.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 5.Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- 6.Aravind L, Koonin EV. The alpha/beta fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-4-research0007. RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindahl T, Ljungquist S, Siegert W, Nyberg B, Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J. Biol. Chem. 1977;252:3286–3294. [PubMed] [Google Scholar]
- 8.Gallinari P, Jiricny J. A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature. 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- 9.Neddermann P, Jiricny J. Efficient removal of uracil from G.U mispairs by the mismatch-specific thymine DNA glycosylase from HeLa cells. Proc. Natl Acad. Sci. USA. 1994;91:1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haushalter KA, Todd Stukenberg MW, Kirschner MW, Verdine GL. Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr. Biol. 1999;9:174–185. doi: 10.1016/s0960-9822(99)80087-6. [DOI] [PubMed] [Google Scholar]
- 11.Sandigursky M, Franklin WA. Thermostable uracil-DNA glycosylase from Thermotoga maritima a member of a novel class of DNA repair enzymes. Curr. Biol. 1999;9:531–534. doi: 10.1016/s0960-9822(99)80237-1. [DOI] [PubMed] [Google Scholar]
- 12.Starkuviene V, Fritz HJ. A novel type of uracil-DNA glycosylase mediating repair of hydrolytic DNA damage in the extremely thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 2002;30:2097–2102. doi: 10.1093/nar/30.10.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith DR, Doucette-Stamm LA, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J. Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 15.Slesarev AI, Mezhevaya KV, Makarova KS, Polushin NN, Shcherbinina OV, Shakhova VV, Belova GI, Aravind L, Natale DA, Rogozin IB, et al. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl Acad. Sci. USA. 2002;99:4644–4649. doi: 10.1073/pnas.032671499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrickson EL, Kaul R, Zhou Y, Bovee D, Chapman P, Chung J, Conway de Macario E, Dodsworth JA, Gillett W, Graham DE, et al. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 2004;186:6956–6969. doi: 10.1128/JB.186.20.6956-6969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fricke WF, Seedorf H, Henne A, Kruer M, Liesegang H, Hedderich R, Gottschalk G, Thauer RK. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J. Bacteriol. 2006;188:642–658. doi: 10.1128/JB.188.2.642-658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mol CD, Arvai AS, Begley TJ, Cunningham RP, Tainer JA. Structure and activity of a thermostable thymine-DNA glycosylase: evidence for base twisting to remove mismatched normal DNA bases. J. Mol. Biol. 2002;315:373–384. doi: 10.1006/jmbi.2001.5264. [DOI] [PubMed] [Google Scholar]
- 20.Horst JP, Fritz HJ. Counteracting the mutagenic effect of hydrolytic deamination of DNA 5-methylcytosine residues at high temperature: DNA mismatch N-glycosylase Mig.Mth of the thermophilic archaeon Methanobacterium thermoautotrophicum THF. EMBO J. 1996;15:5459–5469. [PMC free article] [PubMed] [Google Scholar]
- 21.Fondufe Y. Doctoral thesis. 1999. Characterization and directed modification of the substrate selectivity of Mig.Mth, a DNA repair glycosylase from the thermophilic archaeon Methanobacterium thermoautotrophicum THF. University of Göttingen. [Google Scholar]
- 22.Starkuviene V. Doctoral thesis. 2001. Identification and characterization of thermostable uracil glycosylases from the archaeon Methanobacterium thermoautotrophicum and the bacterium Thermus thermophilus. University of Göttingen. [Google Scholar]
- 23.Chung JH, Im EK, Park HY, Kwon JH, Lee S, Oh J, Hwang KC, Lee JH, Jang Y. A novel uracil-DNA glycosylase family related to the helix-hairpin-helix DNA glycosylase superfamily. Nucleic Acids Res. 2003;31:2045–2055. doi: 10.1093/nar/gkg319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Im EK, Han YS, Chung JH. Functional changes in a novel uracil-DNA glycosylase determined by mutational analyses. Mikrobiologiia. 2008;77:644–650. [PubMed] [Google Scholar]
- 25.Back JH, Chung JH, Park JH, Han YS. A versatile endonuclease IV from Thermus thermophilus has uracil-excising and 3′-5′ exonuclease activity. Biochem. Biophys. Res. Commun. 2006;346:889–895. doi: 10.1016/j.bbrc.2006.05.187. [DOI] [PubMed] [Google Scholar]
- 26.Denver DR, Swenson SL, Lynch M. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol. Biol. Evol. 2003;20:1603–1611. doi: 10.1093/molbev/msg177. [DOI] [PubMed] [Google Scholar]
- 27.Stierum RH, Croteau DL, Bohr VA. Purification and characterization of a mitochondrial thymine glycol endonuclease from rat liver. J. Biol. Chem. 1999;274:7128–7136. doi: 10.1074/jbc.274.11.7128. [DOI] [PubMed] [Google Scholar]
- 28.Ljungquist S. A new endonuclease from Escherichia coli acting at apurinic sites in DNA. J. Biol. Chem. 1977;252:2808–2814. [PubMed] [Google Scholar]
- 29.Georg J, Schomacher L, Chong JP, Majernik AI, Raabe M, Urlaub H, Muller S, Ciirdaeva E, Kramer W, Fritz HJ. The Methanothermobacter thermautotrophicus ExoIII homologue Mth212 is a DNA uridine endonuclease. Nucleic Acids Res. 2006;34:5325–5336. doi: 10.1093/nar/gkl604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schomacher L, Chong JP, McDermott P, Kramer W, Fritz HJ. DNA uracil repair initiated by the archaeal ExoIII homologue Mth212 via direct strand incision. Nucleic Acids Res. 2009;37:2283–2293. doi: 10.1093/nar/gkp102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schomacher L, Schürer KA, Ciirdaeva E, McDermott P, Chong JP, Kramer W, Fritz HJ. Archaeal DNA uracil repair via direct strand incision: A minimal system reconstituted from purified components. DNA Repair. 2010;9:438–447. doi: 10.1016/j.dnarep.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Mosbaugh DW. Uracil-DNA glycosylase inhibitor of bacteriophage PBS2: cloning and effects of expression of the inhibitor gene in Escherichia coli. J. Bacteriol. 1988;170:1082–1091. doi: 10.1128/jb.170.3.1082-1091.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Mosbaugh DW. Uracil-DNA glycosylase inhibitor gene of bacteriophage PBS2 encodes a binding protein specific for uracil-DNA glycosylase. J. Biol. Chem. 1989;264:1163–1171. [PubMed] [Google Scholar]
- 34.Postel EH, Abramczyk BM. Escherichia coli nucleoside diphosphate kinase is a uracil-processing DNA repair nuclease. Proc. Natl Acad. Sci. USA. 2003;100:13247–13252. doi: 10.1073/pnas.2333230100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennett SE, Chen CY, Mosbaugh DW. Escherichia coli nucleoside diphosphate kinase does not act as a uracil-processing DNA repair nuclease. Proc. Natl Acad. Sci. USA. 2004;101:6391–6396. doi: 10.1073/pnas.0401031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar P, Krishna K, Srinivasan R, Ajitkumar P, Varshney U. Mycobacterium tuberculosis and Escherichia coli nucleoside diphosphate kinases lack multifunctional activities to process uracil containing DNA. DNA Repair. 2004;3:1483–1492. doi: 10.1016/j.dnarep.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Sun B, Latham KA, Dodson ML, Lloyd RS. Studies on the catalytic mechanism of five DNA glycosylases. Probing for enzyme-DNA imino intermediates. J. Biol. Chem. 1995;270:19501–19508. doi: 10.1074/jbc.270.33.19501. [DOI] [PubMed] [Google Scholar]
- 38.Kow YW, Wallace SS. Mechanism of action of Escherichia coli endonuclease III. Biochemistry. 1987;26:8200–8206. doi: 10.1021/bi00399a027. [DOI] [PubMed] [Google Scholar]
- 39.Begley TJ, Cunningham RP. Methanobacterium thermoformicicum thymine DNA mismatch glycosylase: conversion of an N-glycosylase to an AP lyase. Protein Eng. 1999;12:333–340. doi: 10.1093/protein/12.4.333. [DOI] [PubMed] [Google Scholar]
- 40.Williams SD, David SS. A single engineered point mutation in the adenine glycosylase MutY confers bifunctional glycosylase/AP lyase activity. Biochemistry. 2000;39:10098–10109. doi: 10.1021/bi0004652. [DOI] [PubMed] [Google Scholar]
- 41.Thornton CG, Hartley JL, Rashtchian A. Utilizing uracil DNA glycosylase to control carryover contamination in PCR: characterization of residual UDG activity following thermal cycling. Biotechniques. 1992;13:180–184. [PubMed] [Google Scholar]
- 42.Begley TJ, Haas BJ, Noel J, Shekhtman A, Williams WA, Cunningham RP. A new member of the endonuclease III family of DNA repair enzymes that removes methylated purines from DNA. Curr. Biol. 1999;9:653–656. doi: 10.1016/s0960-9822(99)80288-7. [DOI] [PubMed] [Google Scholar]
- 43.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.