Abstract
We developed a powerful expression system to produce aptamers and other types of functional RNA in yeast to examine their effects. Utilizing the intron homing process, the aptamer-coding sequences were integrated into hundreds of rRNA genes, and the aptamers were transcribed at high levels by RNA polymerase I without any additional promoter being introduced into the cell. We used this system to express an aptamer against the heat shock factor 1 (HSF1), a conserved transcription factor responsible for mobilizing specific genomic expression programs in response to stressful conditions such as elevated temperature. We observed a temperature sensitive growth retardation phenotype and specific decrease of heat shock gene expression. As HSF1 enables and promotes malignant growth and metastasis in mammals, and this aptamer binds yeast HSF1 and its mammalian ortholog with equal affinity, the results presented here attest to the potential of this aptamer as a specific and effective inhibitor of HSF1 activity.
INTRODUCTION
Due to their high affinity and specificity, RNA aptamers offer advantages over several other types of reagents for studying biological processes (1,2). Their application as ‘intramers’ is particularly useful to manipulate and control protein function in the context of living cells or organisms (3,4). However, their intracellular delivery is not as straightforward. A tactic employed to circumvent this problem is to deliver RNA aptamers as synthetic genes (5). In mammalian cells, RNA polymerase III (Pol III) promoters are often used to express aptamers and observe their effects (6); but the same type of promoter when used in yeast was less successful (7). To address this issue, we have developed an expression system to achieve high levels of aptamer accumulation in the baker’s yeast Saccharomyces cerevisiae. This system utilizes the process of group I intron homing to insert an aptamer-coding sequence into multiple copies of yeast rDNA, so the aptamers can be produced through transcription by RNA polymerase I (Pol I) without the requirement of additional promoters. Here, we report the use of this system to produce an aptamer against heat shock factor 1 (HSF1) to knock down the activity of this protein.
HSF1 is a major transcription regulator of the stress response, and it is highly conserved among eukaryotes (8,9). Under unfavorable conditions, and especially at elevated temperatures, HSF1 activates the expression of heat shock (HS) genes, producing HS proteins with chaperonin activity to repair cellular damage and boost cellular resistance to further injury (10,11). Intriguingly, although it presumably evolved to enhance cellular survival, this mechanism can impede organismal survival under certain circumstances. In particular, HSF1 can affect diverse pathways as a powerful supporter of malignant transformation (12). Therefore, in mammalian species, blocking the activity of HSF1 may hold promise as a therapeutic modality for the treatment of cancer. However, while HSF1 is the dominant HS regulator in mammals and is responsible for the maintenance of cancerous phenotype, HSF has multiple isoforms that appear to have somewhat specialized functions, which complicates the analysis of HSF1 function (13–15). In addition, the diversity of genetic backgrounds among various cancers makes it difficult to elucidate the mode of action of a potential HSF-targeting therapeutic agent. In contrast, HSF activity in yeast is encoded by a single gene, and the HSF1 protein is essential for viability and vegetative growth, making yeast a convenient and inexpensive vehicle to screen for reagents capable of inhibiting HSF1 activity to a sufficient degree to generate a systemic effect (16,17).
Like many other transcription factors, HSF1 possesses a DNA-binding domain and an activation domain. Additionally, it has a trimerization domain and a flexible linker that collaborates with flanking domains to allow the homotrimer of HSF to bind avidly to a Heat Shock Element (HSE) (8). Based on this molecular configuration, an effective strategy for decoupling the transcriptional response to HSF1 from its activation by a HS stimulus is to mask the DNA binding and adjacent domains, thereby preventing HSF from binding to the HSEs. To this end, we generated an RNA aptamer for HSF that competes with the HSE for binding to HSF1 (18). This aptamer, named AptHSF-RA1, binds yeast and mammalian HSF1 equally well through interaction with the DNA-binding domain and the flanking linker region. An in vitro transcription assay demonstrated the ability of this aptamer to inhibit HS gene transcription (18). When the HSF aptamer was expressed using the system described here, we observed growth retardation of the yeast cells under HS conditions. Consistent with this cellular phenotype, we also observed a specific decrease in the expression of genes activated by the HSF1.
MATERIALS AND METHODS
Plasmids and cloning
The group I intron trans-integration system is composed of two plasmids, pCPIPpo (19), which carries the Physarum homing endonuclease I-PpoI, and pRSTtLSU1-ClaI (20), which carries the Tetrahymena intron TtLSU1 (Tth.L1925) flanked by yeast rDNA sequence. pRSTtLSU1-ClaI has a unique ClaI site resulting from the mutation of four nucleotides in the P1 loop, which is used for the insertion of the aptamer AptHSF-RA1 in either monomeric or dimeric form. The monomer insert was generated through bi-directional extension of an overlapping pair of oligonucleotide primers purchased from Integrated DNA Technologies. The dimeric insert was synthesized by GenScript. The sequences of these ClaI inserts are given below.
RA-1(M): 5′-ATCGATGCGGCCGCGAATTCAACTGCC TTCGGGCATCGCGATACAAAATTAAGTTGAACGCGA GTTCGCGGCCGCATCGAT-3′. RA-1(D): 5′-ATCGATGCGGCCGCGTGACGTTAATT CAACTGCCTTCGGGCATCGCGATACAAAATTAAG TTGAACGCGAGTTTTCGTCATACTCCTT GGCATCGCGATACAAAATTAAGTTGAACG CGAGTTCTTCGGAATTCAACTGCCTT GGAGGCGCGGCCGCATCGAT-3′. The ligation product was transformed into the DH5α Escherichia coli strain. Single colonies containing inserts were confirmed using polymerase chain reaction (PCR) and verified by sequencing.
Yeast strains and intron homing
The parental strains for intron homing were W303-1A (MATa; leu2-3,112; trp1-1; can1-100; ura3-1; ade2-1; his3-11,15), YPH500 (MATα; ura3-52; lys2-801_amber; ade2-101_ochre; trp1-Δ63; his3-Δ200; leu2-Δ1) and BY4741(MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0). The two plasmids for trans-integration were co-transformed into the parental strains using the LiAc/ssDNA/PEG method (21). Transformants carrying both plasmids were identified on plates with SD—Ura–His media. To induce intron homing, these transformants were cultured on SGal–Ura–His plates and subcultured several times. Samples were collected at several stages for colony PCR to monitor the progress of homing until the process was complete. The primers used in this assay have the following sequences: E1 (a.k.a. JL83, #1803–1824 of yeast 25S rDNA; downstream), 5′-TCACCCCGGAATTGGTTTATCC-3′; E2 (a.k.a. JL84, #2772–2753 of yeast 25S rDNA; upstream), 5′-CGAATGGGACCTTGAATGC-3′; I (R1), 5′-CAATTTGACGGTCTTGCC-3′. Northern blot analysis was performed according to a standard protocol (22) with the following probes end-labeled using [γ-32P] adenosine triphosphate (ATP) (GE Healthcare) and T4 polynucleotide kinase (NEB): probe for rRNA, 5′-CGAATGGGACCTTGAATGCTAGAACGTGGAAAATGAATTCC-3′; probe for aptamer, 5′-ATCGATGCGGCCGCGAACTCGCGTTC AACTTAATTTTGTATCGCGATGCCCGAAG-3′. RNA samples were run on a 1.67% agarose gel containing 2.2 M formaldehyde in 1× MOPS buffer. Hybridization was performed at 55°C overnight. The membrane was washed with 0.1× saline-sodium citrate (SSC) containing 0.5% sodium dodecyl sulfate (SDS) at 65°C.
Yeast growth under normal and stressful conditions
Yeast strains were grown continuously at 30°C on SGal–Ura–His plates. To examine their phenotypes under different conditions, strains were inoculated using several serial dilutions into the following media and cultured at the temperature indicated: SGal–Ura–His media at 22°C, SGal–Ura–His media at 30°C, SGal–Ura–His media at 37°C, SGal–Ura–His media supplemented with 5 mM CuSO4 at 30°C, SGal–Ura–His media with 0.3 mM H2O2 at 30°C and SGal–Ura–His media with 1 M sorbitol at 30°C. The growth of the cell patches was compared and recorded after 3–4 days’ incubation.
Reverse transcription (RT) and PCR
Liquid yeast culture grown at 22°C in log phase (O.D. 600 = 0.5–1.0.) was split into two equal parts. The first half was subjected to HS at 39°C for 20 min, while the other half remained at 22°C as the non-HS (NHS) control. Both HS and NHS yeast cells were immediately spun down and broken open with glass beads. Total RNA was extracted using hot phenol (23). For RT, 4 pmol of each primer was hybridized with 100 ng of yeast total RNA for each 10-µl reaction. The RNA was then reverse-transcribed with SuperScript III reverse transcriptase (Invitrogen) at 37°C for 30 min and 50°C for 30 min, followed by RNase cocktail treatment to eliminate the remaining RNA. The product was diluted 200-fold, and 2 µl of the diluted product was taken for further qPCR. The qPCR was performed according to a protocol described elsewhere (24). Serial dilutions of the input RNAs were used to plot the standard curves for each gene tested. NHS genes such as U6 and ADH1 were used as internal controls to normalize the RNA levels of the HS genes and the aptamer being tested. The sequences of the primers used are given below: SSA3F, 5′-AGGGAGGCAGAACGAGTTCAGG-3′; SSA3R, 5′-CTCCAGGACCTGCGCCGGCACC-3′; HSP82F, 5′-TCTGGGAATCCAACGCTGGTGG-3′; HSP82R, 5′-CAGAATGTCTCTTGATAACTTCC-3′; SSE2F, 5′-CAATAACTCAGTACTAGCAGTTGCC-3′; SSE2R, 5′-ACCGCCGAACTCCACTTCCACAC-3′; HSP12F, 5′-TGAAGCCAGACTCTCAAAAGTC-3′; HSP12R, 5′-CTTCTTGGTTGGGTCTTCTTCACC-3′; ADH1DNF, 5′-GGACATTGTCGGTGCTGTTCTAAAGG-3′; ADH1DNR, 5′-ATTTCTGGCAAGGTAGACAAGCCG-3′; U6F, 5′-GTTCGCGAAGTAACCCTTCGTG-3′; and U6R, 5′-GAAATAAATCTCTTTGTAAAACGG-3′.
RESULTS
Conceptual design of an indirect aptamer delivery system
When aptamers are used inside cells as ‘intramers’ to modulate protein function, it is important to balance the expression levels of the aptamer and the target protein to achieve the desired change in target function and cellular phenotype. Several additional issues also need to be addressed, including the rate of aptamer production, correctness of folding, stability and subcellular localization. Our expression system aims primarily at generating a high level of aptamer accumulation, so that the effect of aptamers can be easily observed and evaluated. It also ensures reasonable stability of correctly folded aptamers and retains the aptamers within the nucleus, the organelle that harbors the protein targets of our aptamers. This system comprises a set of molecular constructs that ferries the aptamer-coding DNA to a specified location in the middle of each rRNA gene. The strategy exploits unique features of the nucleolar rRNA genes to achieve the goals set above: in yeast, rRNAs are transcribed at a high level by Pol I from about 150 copies of rDNA. Since each rRNA gene contains an aptamer, the high-capacity transcription of rRNA results in the production of a large amount of aptamers as byproducts.
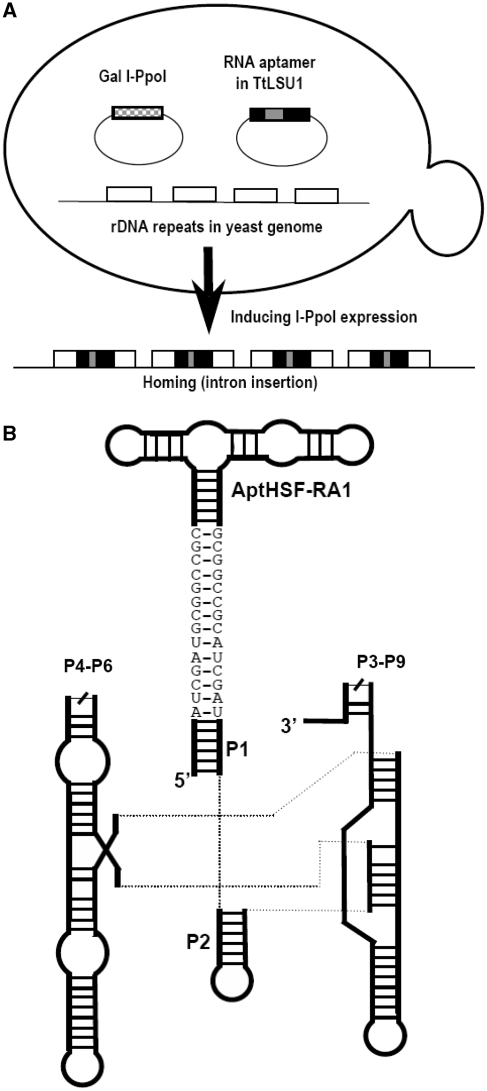
Utilization of the power of Pol I transcription in this scheme requires the capability of inserting a heterologous DNA fragment into all copies of the rDNA and ensuring that normal rRNA production and maturation are not affected by this insertion. These problems were solved by using a group I intron trans-integration process. In our adaptation of this system, depicted in Figure 1A, we combined components from three different species. The yeast S. cerevisiae served as the host, and harbored a group I intron, TtLSU1, from the ciliate Tetrahymena thermophila (25), and a homing endonuclease, I-PpoI, from the slime mold Physarum polycephalum (26). Homing of the group I intron into yeast rDNA was driven in trans by I-PpoI, which was part of the intron PpLSU3 (Ppo.L1925). This endonuclease recognizes a 15-bp site in each of the rDNA repeats on chromosome 12 and cleaves the DNA (27). PpLSU3 and TtLSU1 are closely related. The Tetrahymena intron was chosen because its self-splicing ribozyme is far more active then that of the Physarum intron (28), and we found that it is able to accommodate an inserted sequence and retain the ability to splice quickly and accurately (20). The aptamer-coding sequence receives a ‘piggyback ride’ as an insert in the group I intron, which integrates into these rDNA repeats at the cleaved I-PpoI site. Using this system the intron RNA accumulation can reach 2% of the total RNA (19). As depicted in Figure 1B, we have designed a construct in which the aptamer is introduced as an extension of the P1 stem of the intron. At the interface between the P1 stem and the aptamer, we used a ‘GC clamp’ (29) as a structural insulator between the intron and the aptamer, so that the splicing activity of the intron would not be compromised and the aptamer would be well exposed.
Figure 1.
The indirect aptamer delivery system. (A) Trans-integration of a group I intron. The three components, the host and the two plasmids, are depicted. The arrow shows the transition of rDNA to the intron-homed state caused by I-PpoI cleavage of the rDNA and insertion of the intron-aptamer. (B) Schematic diagram of the intron-aptamer. P4–P6 and P3–P9 signify the two catalytic domains of the group I intron. The sequences shown as bases depict the ClaI sites and the GC clamp extending from the P1 stem.
Construction of yeast strains and confirmation of aptamer expression
To demonstrate the general utility of the method, we tested it in three yeast strains commonly used in laboratories, W303-1A, YPH500 and BY4741. To form the aptamer-coding construct, a segment of DNA coding for a minimized version of AptHSF-RA1 was inserted into the sequence of the group I intron TtLSU1. The plasmid pRSTtLSU1-RA1(M), carrying the gene for this intron-aptamer, was co-transformed into the parental yeast strains together with the plasmid pCPIPpo, which carries the homing endonuclease I-PpoI under Gal 1,10 promoter control. In this arrangement, homing of the intron was controlled by the induced expression of the homing endonuclease. Since the insertion of the intron would abolish the I-PpoI site in rDNA, the homing process proceeded until all copies of rDNA had acquired an intron.
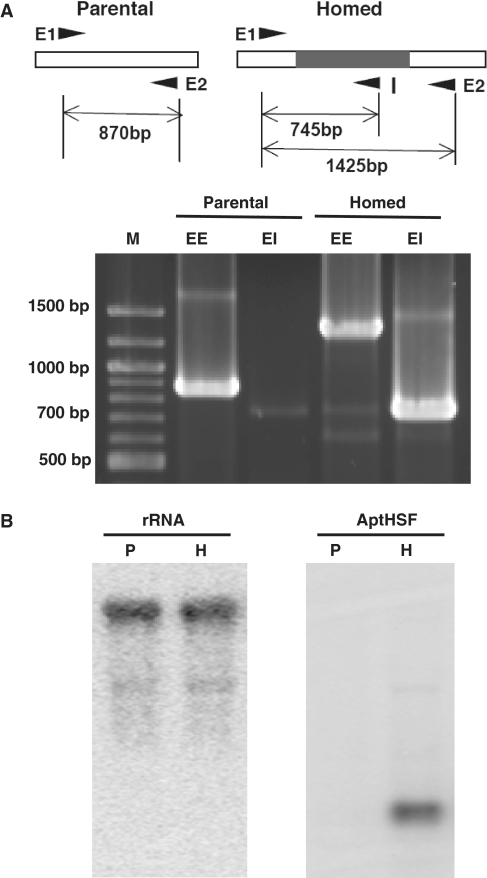
To observe this process and to assess our aptamer delivery system at the DNA level, we subcultured the transformants several times and performed PCR with two exon-specific primers (see Figure 2A, lanes labeled EE) or an exon-specific primer paired with an intron-specific primer (lanes labeled EI). Intron homing would result in increased size of the EE fragment (from 870 to 1425 bp). The 745 bp EI fragment should only be produced by rDNA with a homed intron. In Figure 2A, we show the results of one completely homed clone compared to one parental clone. Although the data shown were generated using the W303-1A parental strain, this experiment was performed in all three genetic backgrounds with identical results.
Figure 2.
Functionality of the aptamer delivery system. (A) Intron homing to rDNA as confirmed by PCR. The primer annealing sites on rDNA before and after homing is given at the top. The homing of the intron to all rDNA is shown for one clone in the W303-1A parental strain. EE designates bands produced by the two exonic primers. EI designates bands produced by the exonic primer E1 and the intronic primer I. ‘M’ indicates molecular weight markers. (B) RNA expression as monitored by Northern blot analysis. ‘P’ indicates the parental strain. ‘H’ indicates the homed strain. W303-1A was used for this experiment.
To confirm that this aptamer delivery system functioned properly, two more tests at the RNA level were required. First, the presence of the aptamer should not affect the expression of rRNA (the splicing of the group I intron from rRNA transcripts should leave the mature rRNA intact). Second, the level of intron-aptamer accumulation should be sufficiently high as to be comparable to the rRNA level.
For these purposes, we prepared RNA from the homed and parental strains, and measured the expression of the rRNA and the aptamer using Northern blot analysis. An oligonucleotide 41 bases in length covering the sequence of the downstream exon at the PCR primer-annealing site was used to detect the rRNA. To visualize the aptamer, we used a probe 55 bases in length complementary to the aptamer sequence. As shown in Figure 2B, the rRNA probe revealed a band with identical mobility and intensity in both parental (P) and homed (H) strains. The aptamer probe detected a band in the homed strain only, and the intensity of the band detected is close to that of the rRNA band.
Cellular phenotypes caused by the HSF aptamer
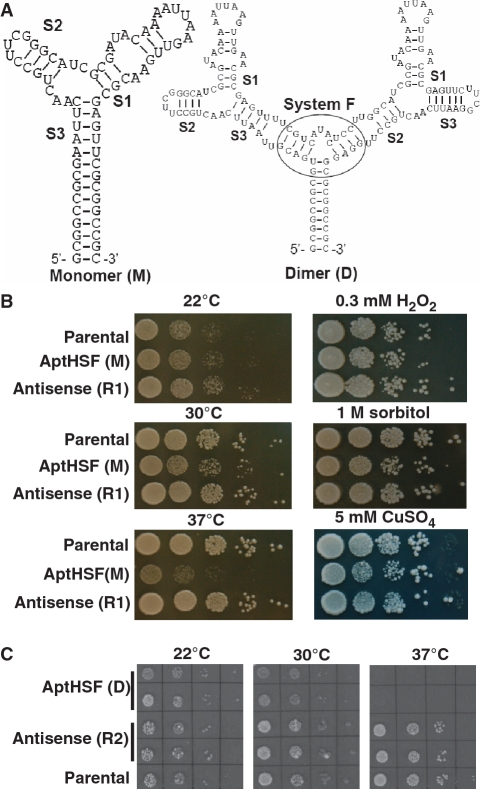
The minimized ‘Core’ of the AptHSF-RA1 comprises a three-way junction. Of the three stems connected to the junction, Stem 1 contains an apical loop and an internal loop, which are required for the aptamer’s activity. Stems 2 and 3 are short and both can serve as a point of integration with other structural or functional elements. As shown in Figure 3A, in the TtLSU1-RA1(M) construct, we connected Stem 3 with the P1 stem of the intron and grafted a UUCG tetra-loop to the tip of Stem 2 to maintain strand continuity, as this arrangement was closer to the original full length version. Having successfully tested the expression system using the monomeric construct, we also created a dimeric construct. To ensure correct folding, an additional three-way junction was used to join the two aptamers and the intron together. This three-way junction was previously shown to be stable among other variants (30) and used to present multiple aptamers in molecular composites (31). As shown in Figure 3A, we used either Stem 3 or 2 to connect the aptamer to this additional three-way junction to form the dimer RA1(D). This longer and more complex sequence also helped demonstrate the general utility of the aptamer expression system.
Figure 3.
Cellular phenotype of the inhibitory HSF aptamer. (A) Predicted secondary structure of the monomer and the dimer being expressed in yeast. The sequence between the two ClaI sites is shown. Different stems are indicated. The additional three-way junction used to present the dimer is encircled. (B) Growth of the monomer (M)-expressing strain and control strains cultured on different solid media and under different temperatures. ‘Antisense (R1)’ is a strain in which the antisense sequence of the monomer was expressed. (C) Growth of the dimer (D)-expressing strain and control strains cultured under different temperatures. ‘Antisense (R2)’ is a strain in which the antisense sequence of the dimer was expressed. In both (B) and (C), the parental strain was W303-1A. Patches in both (B) and (C) were inoculated by 10-fold serial dilutions of yeast stock.
By preventing HSF1 from binding to its cognate DNA sites, the aptamer AptHSF-RA1 was anticipated to compromise yeast survival at elevated temperatures. Prompted by this hypothesis, we examined the growth rate of yeast strains harboring the homed intron-aptamer at different temperatures. As shown in Figure 3B and C, the growth rate of the homed strain and the parental strain did not differ significantly when they were cultured at 22°C. As the temperature increased, we observed moderate growth retardation for the homed strain at 30°C. The most severe phenotype was observed when the temperature was raised to 37°C: the AptHSF(M) homed strain grew very slowly, and the AptHSF(D) homed strain was not able to grow under this HS condition. As an additional control, we also tested homed strains with an intron carrying the antisense sequence of the aptamer (R1 for the monomer and R2 for the dimer). Growth of these strains was normal at all three temperatures.
In addition to heat treatment, we also examined the growth of the aptamer-expressing yeast under several other stressful conditions. Because responses to these non-heat stresses are not mediated by HSF1, these conditions served as tests of aptamer specificity. As shown in Figure 3B, exposure to hydrogen peroxide and hyper-osmotic shock did not reveal any difference between the aptamer-expressing strains and the antisense and parental controls, although the growth of aptamer-expressing strains appeared to be slightly affected by CuSO4. These results strongly indicated a causal relationship between aptamer expression and a compromised response to thermal stress.
Molecular phenotypes caused by the HSF aptamer
HSF1 plays a predominant role in the induction of HS genes. The temperature sensitive phenotype of the aptamer-expressing yeast strains described above led us to predict that the HSF aptamer interfered with HSF activation of at least some HS genes. To investigate the mechanism underlying the systemic effect of the aptamer, we examined the mRNA level of three representative HS genes before and after heat treatment using W303-1A stains expressing the AptHSF in either monomeric (M) or dimeric (D) forms, and compared them with the strain harboring a control antisense version (R1 or R2) of each construct. These three typical HS genes, SSA3, HSP83 and SSE2, all have HSEs in the promoter and are inducible by heat. As a control, we used the HSP12 gene, which is inducible by heat or other stresses but does not contain typical HSEs in its promoter. If the aptamer were effective as an antagonist of HSF1, we would expect that the levels of transcripts in aptamer-expressing strains would be lower after HS than those in strains expressing the antisense sequence.
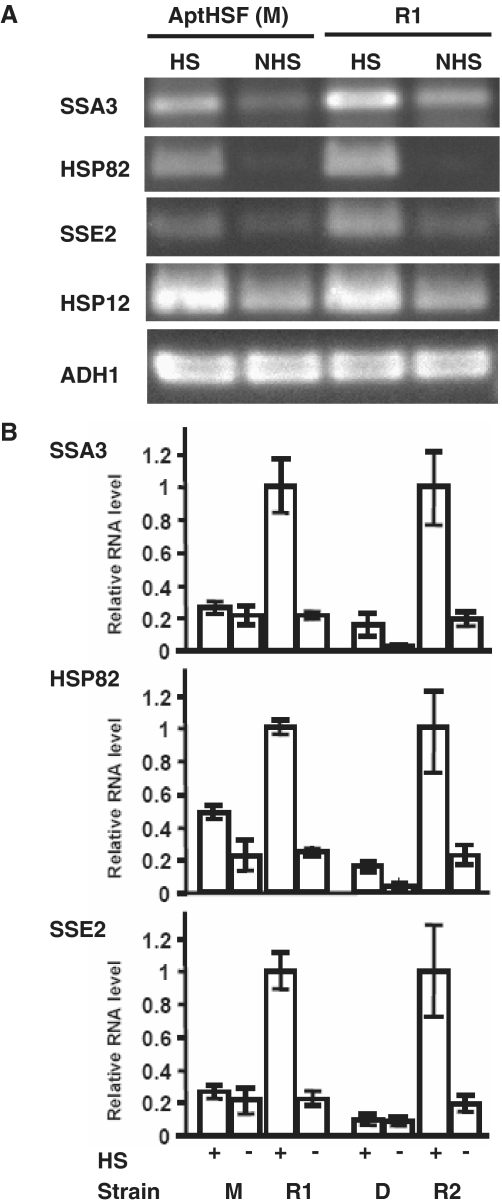
As shown in Figure 4A, using RT–PCR, we observed a significant decrease in heat-induced expression of all three typical HS genes in the strain expressing the AptHSF monomer. The difference in ratios of mRNA levels under HS and NHS conditions for the aptamer and antisense strains for each gene analyzed indicated a downregulation of HSF1 activity by the aptamer. In contrast, the expression level of HSP12 was not changed under either HS or NHS conditions. To confirm this result and compare the efficacy of the monomer and dimer, we used quantitative PCR (qPCR) to measure the mRNA level of the same three HS genes in both monomer- and dimer-expressing strains. Figure 4B shows a more pronounced decrease in the level of these mRNAs in the dimer-expressing strain than in the monomer-expressing strain under both HS and NHS conditions. Notably, the aptamer did not entirely inhibit HSF function, as in some cases an increase in transcription still occurred after HS. However, the resulting level of transcription apparently did not produce sufficient proteins to enable survival of the HS. In this experiment, we used the RNA polymerase II-driven ethanol-inducible gene ADH1 and the Pol III-driven constitutive gene U6 to normalize the data. These genes were not induced by HS and were not inhibited by the aptamer.
Figure 4.
Molecular phenotype of the inhibitory HSF aptamer. (A) Effect of aptamer monomer (M) on the level of HS genes measured by conventional RT–PCR. ‘R1’ is a strain expressing the antisense sequence of the aptamer monomer. HS (20 min at 39°C). (B) Effect of both aptamer monomer (M) and dimer (D) on the level of HS genes measured by RT–qPCR. ‘R2’ is a strain expressing the antisense sequence of the aptamer dimer. The RNA level for each gene is presented as the ratio to the full HS induction level in the antisense control strain, which is set to 1. The expression level for each gene is normalized to that of ADH1. (We also used U6 to normalize the data, and the data sets were consistent with each other.) The error bars show the standard error from RT–qPCR experiments using three independently heat-shocked yeast RNA preparations from the same strain. In both panels the parental strain used was W303-1A.
DISCUSSION
HSF1 is a transcription factor involved in a wide variety of biological processes, including stress responses. It is also a master regulator of a cancer-enabling gene network and a potential target for therapeutic inhibition in mammalians. Interfering with DNA binding of a transcription factor is an efficient way to downregulate genes controlled by this factor, a strategy demonstrated elegantly by an aptamer for NF-κB (6,32,33). The aptamer used in the present study was previously demonstrated to recognize the DNA-binding domain and the adjacent linker region of HSF with high affinity (18). However, without an exhaustive global proteomic assay, it is impossible to completely rule out off-target effects when an aptamer is used in an assay involving cell extract or in living cells. Yeast exhibits a general response of gene expression to stress as well as specific genomic expression patterns for particular environmental conditions (34). Here, we showed that the growth of aptamer-expressing yeast was severely impeded by elevated temperature, but not by exposure to non-heat stresses whose responses are not mediated by HSF1. In parallel experiments, downregulation was observed in the aptamer-expressing yeast for HS genes having HSEs in their promoter but not for the control gene without HSE. The congruence of cellular and molecular phenotypes indicated that the specific knockdown of HS genes in these strains was responsible for the systemic effect. Interestingly, the AptHSF-RA1 is also able to recognize mammalian HSF1 (35), thus providing us with the exciting prospect of further developing it as a potential anticancer drug lead.
The forced expression of high levels of functional synthetic RNAs in vivo requires the use of powerful promoters to drive the transcription of genes encoding these RNAs. To avoid shunting the transcripts into a pre-mRNA pathway, Pol I- or Pol III-driven promoters are often used for this purpose. However, a previous study using the Pol III-driven promoter of the RNase P RNA gene yielded aptamers equivalent to only ∼0.3% of endogenous U6 RNA (7). Perhaps the high level of transcription of a small number of housekeeping genes by these RNA polymerases suggests that these transcription systems are operating at close to full capacity, leaving less room to accommodate additional genes. If this conjecture were true, introducing large numbers of additional promoters into the cell not only would produce insufficient amount of the synthetic RNA, but also might impede the production of native transcripts by ‘soaking up’ the limiting factors, thereby creating a situation in which the real effect of the synthetic RNA would be difficult to assess. In this study, we designed and constructed an expression system in which the total number of Pol I transcription initiation events in the cell remained unchanged, which ensured that potentially limiting Pol I transcription factors were not consumed by additional promoters.
We achieved a high level of aptamer accumulation by using a specific homing endonuclease to insert a self-splicing intron containing the aptamer sequence into every copy of the ∼150 highly expressed rRNA genes. In general, homing endonucleases recognize sequences 14–40 bp in length (36) and are being developed as powerful and precise tools to insert therapeutic genes into a chosen location to circumvent the hazards of random insertion technology (37–39). Our aptamer expression system provided an innovative demonstration of their utility. Using this approach, presumably other aptamers or small RNAs could be delivered into host cells with minimal collateral disruption. Performance of this system may be affected by several factors, including efficiency of homing and splicing of the intron as well as activity of the aptamer. We have tested different introns and different points of integration between the intron and the aptamer to find the optimal condition for homing as described herein. But additional care should be taken to design each intron-aptamer construct, as success hinges on the functionality of both the aptamer and the intron in the composite. In principle, both components should be correctly folded and the aptamer should be well exposed. For these purposes, we introduced a long ‘GC-clamp’ (29) between the aptamer and the intron. In the dimeric construct, we further introduced a structurally stable three-way junction (30) to enforce the folding modularity. These design features can be used in other expression constructs.
FUNDING
This work was supported by grants from the National Institutes of Health [GM40918 and CA140730]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Mr. E. Koman for technical assistance and Dr. K. Nishikawa for comments on the manuscript.
REFERENCES
- 1.Gold L, Polisky B, Uhlenbeck O, Yarus M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 2.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 3.Famulok M, Blind M, Mayer G. Intramers as promising new tools in functional proteomics. Chem. Biol. 2001;8:931–939. doi: 10.1016/s1074-5521(01)00070-9. [DOI] [PubMed] [Google Scholar]
- 4.Famulok M, Mayer G. Intramers and aptamers: applications in protein-function analyses and potential for drug screening. Chembiochem. 2005;6:19–26. doi: 10.1002/cbic.200400299. [DOI] [PubMed] [Google Scholar]
- 5.Shi H, Hoffman BE, Lis JT. RNA aptamers as effective protein antagonists in a multicellular organism. Proc. Natl Acad. Sci. USA. 1999;96:10033–10038. doi: 10.1073/pnas.96.18.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mi J, Zhang X, Rabbani ZN, Liu Y, Su Z, Vujaskovic Z, Kontos CD, Sullenger BA, Clary BM. H1 RNA polymerase III promoter-driven expression of an RNA aptamer leads to high-level inhibition of intracellular protein activity. Nucleic Acids Res. 2006;34:3577–3584. doi: 10.1093/nar/gkl482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas M, Chedin S, Carles C, Riva M, Famulok M, Sentenac A. Selective targeting and inhibition of yeast RNA polymerase II by RNA aptamers. J. Biol. Chem. 1997;272:27980–27986. doi: 10.1074/jbc.272.44.27980. [DOI] [PubMed] [Google Scholar]
- 8.Wu C. Heat shock transcription factors: structure and regulation. Annu. Rev. Cell Dev. Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 9.Birch-Machin I, Gao S, Huen D, McGirr R, White RA, Russell S. Genomic analysis of heat-shock factor targets in Drosophila. Genome Biol. 2005;6:R63. doi: 10.1186/gb-2005-6-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindquist S, Craig EA. The heat-shock proteins. Annu. Rev. Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 11.Kregel KC. Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J. Appl. Physiol. 2002;92:2177–2186. doi: 10.1152/japplphysiol.01267.2001. [DOI] [PubMed] [Google Scholar]
- 12.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rabindran SK, Giorgi G, Clos J, Wu C. Molecular cloning and expression of a human heat shock factor, HSF1. Proc. Natl Acad. Sci. USA. 1991;88:6906–6910. doi: 10.1073/pnas.88.16.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarge KD, Zimarino V, Holm K, Wu C, Morimoto RI. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 1991;5:1902–1911. doi: 10.1101/gad.5.10.1902. [DOI] [PubMed] [Google Scholar]
- 15.Schuetz TJ, Gallo GJ, Sheldon L, Tempst P, Kingston RE. Isolation of a cDNA for HSF2: evidence for two heat shock factor genes in humans. Proc. Natl Acad. Sci. USA. 1991;88:6911–6915. doi: 10.1073/pnas.88.16.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiederrecht G, Seto D, Parker CS. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell. 1988;54:841–853. doi: 10.1016/s0092-8674(88)91197-x. [DOI] [PubMed] [Google Scholar]
- 17.Sorger PK, Pelham HR. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988;54:855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- 18.Zhao X, Shi H, Sevilimedu A, Liachko N, Nelson HC, Lis JT. An RNA aptamer that interferes with the DNA binding of the HSF transcription activator. Nucleic Acids Res. 2006;34:3755–3761. doi: 10.1093/nar/gkl470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin J, Vogt VM. I-PpoI, the endonuclease encoded by the group I intron PpLSU3, is expressed from an RNA polymerase I transcript. Mol. Cell. Biol. 1998;18:5809–5817. doi: 10.1128/mcb.18.10.5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suran R. Ph.D. Dissertation. Ithaca, NY: Cornell University; 2009. Expression of proteins from chimeric Tetrahymena introns integrated in ribosomal RNA gene of Saccharomyces cerevisiae. [Google Scholar]
- 21.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Meth. Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Russell DW. Molecular Cloning: A laboratory Manual. 3rd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 23.Kohrer K, Domdey H. Preparation of high molecular weight RNA. Meth. Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- 24.Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brehm SL, Cech TR. Fate of an intervening sequence ribonucleic acid: excision and cyclization of the Tetrahymena ribosomal ribonucleic acid intervening sequence in vivo. Biochemistry. 1983;22:2390–2397. doi: 10.1021/bi00279a014. [DOI] [PubMed] [Google Scholar]
- 26.Ellison EL, Vogt VM. Interaction of the intron-encoded mobility endonuclease I-PpoI with its target site. Mol. Cell. Biol. 1993;13:7531–7539. doi: 10.1128/mcb.13.12.7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flick KE, Jurica MS, Monnat RJ, Jr, Stoddard BL. DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI. Nature. 1998;394:96–101. doi: 10.1038/27952. [DOI] [PubMed] [Google Scholar]
- 28.Zhang F, Ramsay ES, Woodson SA. In vivo facilitation of Tetrahymena group I intron splicing in Escherichia coli pre-ribosomal RNA. RNA. 1995;1:284–292. [PMC free article] [PubMed] [Google Scholar]
- 29.Cassiday LA, Maher LJ., 3rd Yeast genetic selections to optimize RNA decoys for transcription factor NF-kappa B. Proc. Natl Acad. Sci. USA. 2003;100:3930–3935. doi: 10.1073/pnas.0736013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diamond JM, Turner DH, Mathews DH. Thermodynamics of three-way multibranch loops in RNA. Biochemistry. 2001;40:6971–6981. doi: 10.1021/bi0029548. [DOI] [PubMed] [Google Scholar]
- 31.Xu D, Shi H. Composite RNA aptamers as functional mimics of proteins. Nucleic Acids Res. 2009;37:e71. doi: 10.1093/nar/gkp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lebruska LL, Maher LJ., 3rd Selection and characterization of an RNA decoy for transcription factor NF-kappa B. Biochemistry. 1999;38:3168–3174. doi: 10.1021/bi982515x. [DOI] [PubMed] [Google Scholar]
- 33.Huang DB, Vu D, Cassiday LA, Zimmerman JM, Maher LJ, 3rd, Ghosh G. Crystal structure of NF-kappaB (p50)2 complexed to a high-affinity RNA aptamer. Proc. Natl Acad. Sci. USA. 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao X. Ph.D. Dissertation. Ithaca, NY: Cornell University; 2007. A synthetic non-coding RNA that inhibits an extremely strong macromolecular interaction in vivo. [Google Scholar]
- 36.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 37.Chevalier BS, Kortemme T, Chadsey MS, Baker D, Monnat RJ, Stoddard BL. Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell. 2002;10:895–905. doi: 10.1016/s1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
- 38.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosen LE, Morrison HA, Masri S, Brown MJ, Springstubb B, Sussman D, Stoddard BL, Seligman LM. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 2006;34:4791–4800. doi: 10.1093/nar/gkl645. [DOI] [PMC free article] [PubMed] [Google Scholar]