Abstract
The proper balance between symmetric and asymmetric stem cell division is crucial both to maintain a population of stem cells and to prevent tumorous overgrowth. Neural stem cells in the Drosophila optic lobe originate within a polarised neuroepithelium, where they divide symmetrically. Neuroepithelial cells are transformed into asymmetrically dividing neuroblasts in a precisely regulated fashion. This cell fate transition is highly reminiscent of the switch from neuroepithelial cells to radial glial cells in the developing mammalian cerebral cortex. To identify the molecules that mediate the transition, we microdissected neuroepithelial cells and compared their transcriptional profile with similarly obtained optic lobe neuroblasts. We find genes encoding members of the Notch pathway expressed in neuroepithelial cells. We show that Notch mutant clones are extruded from the neuroepithelium and undergo premature neurogenesis. A wave of proneural gene expression is thought to regulate the timing of the transition from neuroepithelium to neuroblast. We show that the proneural wave transiently suppresses Notch activity in neuroepithelial cells, and that inhibition of Notch triggers the switch from symmetric, proliferative division, to asymmetric, differentiative division.
Keywords: Neural stem cells, Notch, Drosophila, Optic lobe, Proneural factor
INTRODUCTION
Neurons must be generated in a precisely timed fashion to build functional networks of circuits. This is achieved by controlling the balance between self-renewing neural stem cells and their differentiated progeny. Stem cells can divide symmetrically to generate daughter cells with similar fates, or asymmetrically, to self-renew whilst also producing differentiating daughter cells. During neurogenesis in the mammalian cerebral cortex, neuroepithelial cells initially divide symmetrically to expand the stem cell pool. Later in development, they transform into asymmetrically dividing radial glial cells, which expand the pool of differentiating cells (Farkas and Huttner, 2008; Heins et al., 2002; McConnell, 1995; Miyata et al., 2004; Noctor et al., 2004). The transition from symmetric to asymmetric division must be tightly regulated, as excessive symmetric cell division can lead to tumorous overgrowth, whereas precocious asymmetric division results in premature differentiation and small brains (Cabernard and Doe, 2009; Gauthier-Fisher et al., 2009; Morrison and Kimble, 2006). A sufficiently large pool of undifferentiated stem cells must be maintained so as to generate both early- and later-born neurons, and to repair or regenerate damaged tissue. One way to regulate a symmetrically dividing pool of stem cells is to maintain them in an epithelium. Epidermal, intestinal and neural stem cells are found in mono- or multilayered epithelia that expand laterally and thereby increase the pool of tissue-specific precursor cells (Barker et al., 2008; Blanpain and Fuchs, 2009; Fish et al., 2008).
To date it has been difficult to model cortical development in a simpler invertebrate system. The Drosophila optic lobe provides one such model for studying the progression from symmetric to asymmetric neural stem cell division. The optic lobe is the visual processing centre of the brain. During larval development it consists of a neuroepithelium, in which stem cells divide symmetrically to expand the pool of proliferating precursor cells, and a region of asymmetrically dividing neuroblasts, which produce differentiated neurons (Fig. 1A,B) (Egger et al., 2007; Hofbauer and Campos-Ortega, 1990). Situated between the neuroepithelium and the neuroblasts is a group of cells referred to as ‘the transition zone’ that transiently express the proneural gene lethal of scute (l'sc). l'sc expression sweeps across the epithelium as a ‘proneural wave’, with cells ahead of the wave dividing symmetrically and those behind dividing asymmetrically (Fig. 1B) (Yasugi et al., 2008). Neuroepithelial cells transform into neuroblasts in a highly ordered, sequential manner and the JAK/STAT pathway has recently been implicated in the timing of the transition (Yasugi et al., 2008).
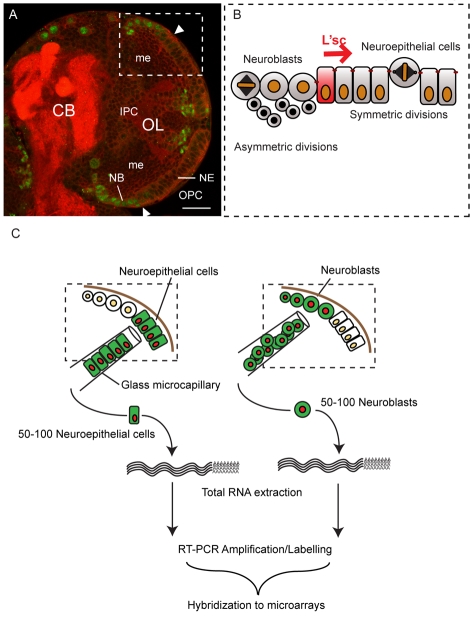
Fig. 1.
Symmetrically dividing neuroepithelial cells give rise to asymmetrically dividing neuroblasts. (A) Single confocal section of a third instar brain lobe showing the central brain (CB) and optic lobe (OL) stained for the neuroblast marker Dpn (green) and Dlg (red) to outline cells and the neuropile, respectively. The lateral half of each brain lobe comprises cells that form the developing optic lobe. Cells of the outer proliferation centre (OPC) give rise to the visual integration centres of the lamina and the outer medulla (me), whereas cells of the inner proliferation centre (IPC) generate the inner medulla, lobula and lobula plate. The OPC harbours two neural stem cell pools: lateral neuroepithelial cells and medial Dpn positive neuroblasts. Scale bar: 20 μm. (B) Optic lobe neuroepithelial cells divide symmetrically to expand the progenitor pool. The most medial neuroepithelial cells upregulate L'sc expression and are transformed into asymmetrically dividing neuroblasts. A wave of L'sc expression sweeps across the epithelium generating progressively more neuroblasts. (C) Genetically labelled neuroepithelial cells or neuroblasts were isolated from third instar larval brains using a glass microcapillary. Total RNA was extracted from 50-100 cells per sample and reverse transcribed, amplified by PCR and fluorescently labelled prior to hybridisation to a full genome microarray.
To identify the genetic switches that underlie the transition from neuroepithelial cells to neuroblasts, we compared the transcriptional profiles of small numbers of optic lobe neuroepithelial cells and neuroblasts. Strikingly, a number of genes encoding members of the Notch signalling pathway are highly expressed in neuroepithelial cells, suggesting that this pathway is active in symmetrically dividing stem cells. We show that Notch is a key regulator in maintaining the balance between symmetrically and asymmetrically dividing stem cell pools. Notch signalling within the neuroepithelium is required to prevent differentiation: clonal Notch mutant neuroepithelial cells are extruded from the epithelial layer and undergo premature neurogenesis. We demonstrate that the proneural gene l'sc transiently represses Notch function to enable the transition from neuroepithelial cell to neuroblast. The timely progression from symmetrically dividing neural precursors in the optic lobe to differentiated neurons closely mirrors that in the vertebrate cerebral cortex, and we propose that the mechanisms regulating neurogenesis in these two systems could be conserved.
MATERIALS AND METHODS
Drosophila strains
Oregon R was used as a control. For cell isolation, we used the GAL4 driver lines GAL4c855a (Manseau et al., 1997) and GAL41407 (an insertion into the inscuteable locus) (Luo et al., 1994) and the responder line w;;UAS-mCD8-GFP, UAS-H2B-mRFP1. To induce MARCM clones, we used the driver line FRT19A, tub-GAL80LL1, hs-FLP1, w; tubP-GAL4, UAS-mCD8-GFPLL5/CyO, act-GFPJMR1 (gift from B. Bello, Biocenter Basel, Switzerland), the responder line w, sn, FRT19A for control clones, and the responder line N[55e11], FRT19A/FM7c (Almeida and Bray, 2005) to induce Notch mutant clones. To induce L'sc misexpression clones, we used the driver line yw, hs-FLP; tubP-GAL4, UAS-mCD8-GFP/CyO, act-GFPJMR1; FRT2A, tub-GAL80LL9 (gift from B. Bello) and the responder w; UAS-l'sc; FRT2A (this study) (Carmena et al., 1995). For the dominant-negative Kuzbanian experiments we used w; UAS-kuz.DN (Pan and Rubin, 1997) driven by w; GAL4c855a; tub-GAL80ts (this study) (McGuire et al., 2003). For the dominant-negative Delta experiments we used w; TM6B/TM3, UAS-Dl.DN (Huppert et al., 1997) driven by GAL4c855a.
Isolation of optic lobe cells and microarray expression profiling
Using a drawn out and bevelled glass capillary, we extracted small groups of cells from dissected third instar larval brains 96 hours ALH. The neuroblast and neuroepithelial cell populations were distinguished under UV light using two different GAL4 lines to drive the expression of membrane-tethered mCD8-GFP and Histone 2B-mRFP1. Neuroepithelial cell samples were collected from flies of genotype w;; GAL4c855a/UAS-mCD8mGFP, UAS-H2B-mRFP1 and neuroblast samples were collected from w; GAL41407/+; UAS-mCD8mGFP, UAS-H2B-mRFP1/+.
Generation of cDNA samples from optic lobe cells
Stock cell lysis buffer was made as follows: 100 μl 10× reaction buffer with MgCl2 (Invitrogen), 10 μl NP-40, 50 μl 0.1 M DTT, 775 μl DEPC H2O. Fresh lysis mix was prepared just prior to cell isolation by adding 1 μl RNase inhibitor mix [1:1 mixture RNasin (Promega) and Prime (Eppendorf)], 1 μl anchored primer (10 ng/μl) and 1 μl 2.5 mM dNTPs to 47 μl lysis buffer. Around 50 cells were isolated per sample and expelled into 3 μl ice-cold lysis mix, and then 2.5 μl of this mix was transferred to a PCR tube. Tubes were incubated at 65°C for 2 minutes to denature RNA and placed immediately on ice. Reverse transcription was carried out by the addition of 2.5 μl of RT mix [0.3 μl Superscript II (Invitrogen), 0.1 μl RNase inhibitor mix, 2.1 μl lysis buffer]. After incubation at 37°C for 90 minutes, the reaction was terminated by heating to 65°C for 10 minutes and then samples were cooled to 4°C. To perform poly(A) tailing of synthesized cDNA, 5 μl of TdT mix [0.15 μl 100 mM dATP, 0.5 μl 10× reaction buffer with MgCl2 (Invitrogen), 3.85 μl DEPC H2O, 0.25 μl TdT (Roche), 0.25 μl RNaseH (Roche)] was added to the reaction mixture. The reaction was incubated for 20 minutes at 37°C then inactivated by heating to 65°C for 10 minutes. PCR amplification of polyadenylated cDNA was then carried out. A 10 μl aliquot of poly(A)-tailed cDNA was mixed with 50 μl PCR reaction solution [5 μl 10× ExTaq buffer (Takara Biochemicals), 5 μl 2.5 mM dNTPs, 1 μl anchored primer (1 μg/μl)], 38.5 μl DEPC H2O, 0.5 μl ExTaq polymerase (Takara Biochemicals)]. The following PCR programme was used: one cycle of 95°C for 1 minute, 37°C for 5 minutes and 72°C for 20 minutes, followed by 32 cycles of 95°C for 30 seconds, 67°C for 1 minute and 72°C for 6 minutes. cDNA was purified using a PCR purification kit (Qiagen), according to the manufacturer's instructions.
Transcriptome analysis
The neuroepithelial and neuroblast cDNA samples were directly compared by hybridisation to microarrays (FL002; Flychip Cambridge Microarray Facility). One microgram of the amplified DNA was labelled with the BioPrime DNA Labelling Kit (Invitrogen) in the presence of fluorescently labelled Cy3- or Cy5-dCTP (GE Healthcare) at 37°C for 3 hours, and the product purified on Sephadex G50 minicolumns. Slide hybridisations were performed for 16 hours at 51°C with a GeneTac hybridisation station (Genomic Solutions). Post-hybridisation washes were performed according to the manufacturer's recommendation. Microarray images were taken with a Genepix 4000B dual laser scanner (Axon), and dapple software was used for spot-finding and quantification (http://www.cs.wustl.edu/~jbuhler/research/dapple/). Log-transformed intensity ratios from four biological replicates (two standard dye configurations plus two swapped dye configurations) were normalized with a Variance Stabilizing Normalization algorithm and averaged. Statistical analysis of the array data was carried out in the R environment using FCAN, an automated microarray analysis pipeline created by FlyChip. Differentially expressed genes were identified by running the normalized data through ‘siggenes’, a computer package for carrying out Significance Analysis of Microarrays (SAM). This involved a two class, unpaired data analysis with modified t-statistic to identify differentially expressed genes and to estimate the False Discovery Rate (FDR), which is presented as a q-value for each gene. We only considered genes with q<0.05.
Immunofluorescence staining, fluorescent in situ hybridisation and imaging procedure
Larval brains were fixed and stained as previously described (Van Vactor et al., 1991). We used the following primary antibodies: mouse anti-NICD (1:50), rat anti-DE-Cad (1:20), mouse anti-Dlg (1:100), mouse anti-Dl[ECD] (1:50), mouse anti-Elav (1:20) (all from Developmental Studies Hybridoma Bank), chicken anti-GFP (1:20, Upstate), rabbit anti-betagalactosidase (1:10000, Cappel), rat anti-Dpn (1:2) (Lee et al., 2006), guinea pig anti-Dpn (1:1000; gift from J. Skeath, Washington University School of Medicine, St Louis, USA), rat anti-L'sc (1:800) (Martin-Bermudo et al., 1991). Fluorescently conjugated secondary antibodies Alexa405, Alexa488, Alexa568 and Alexa633 were used (Molecular Probes, Invitrogen). Fluorescent in situ hybridisation was performed as described previously (Egger et al., 2007). Primers to make the Tom RNA probe are: forward primer, AAATCTCAACAATCCTCAACACAA; reverse primer, CAGTAATACGACTCACTATTATACGAAGACCCTAACAAACAAACA. Primers to make the HLHm5 RNA probe are: forward primer, CAGTTCAGATCAGCTCAGCACATT; reverse primer, CAGTAATACGACTCACTATTATCAATTGGAAGACATGATTCGTTG. Underlined bases indicate T7 polymerase binding site.
Images were acquired with a Leica SP2 confocal microscope and analysed with Imaris 3.2 (Bitplane) and ImageJ. Figures and illustrations were assembled using Adobe Photoshop 8.0. and Adobe Illustrator 11.0.
Clone induction, Kuz-DN, Dl-DN misexpression and larval staging
For MARCM clone induction freshly hatched larvae were collected in a 4 to 6 hour window and staged on cornmeal food. Clones were induced by a double heat shock (5 minutes at 37°C, 5 minutes recovery at 25°C and 30 minutes at 37°C) at first/early second larval instar (21-27 hours after larval hatching). For Notch mutant clone induction, larvae were of the genotype: FRT19A, tub-GAL80, hs-FLP/N[55e11], FRT19A; tubP-GAL4, UAS-mCD8-GFP/ +. For L'sc misexpression clones, larvae were of the genotype: hs-FLP/ +; tubP-GAL4, UAS-mCD8-GFP/UAS-l'sc; FRT2A, tubP-GAL80/FRT2A. For dominant-negative Kuz experiments, larvae of the genotype w; +/UAS-kuz.DN; tub-GAL80ts/GAL4c855a were raised at 25°C. For kuz-DN expression, larvae were shifted to 29°C at 24 hours ALH and shifted back to 25°C at 48 hours ALH to recover for another 24 hours. For all experiments, larvae were dissected and stained mid-third instar (72 hours ALH). For dominant-negative Dl experiments, larvae of the genotype w; +/+; TM3, UAS-Dl.DN/GAL4c855a were raised at 25°C. For all experiments, larvae were dissected and stained at mid- or late-third instar (72 hours or 96 hours ALH).
RESULTS
The Notch signalling pathway is activated in neuroepithelial stem cells
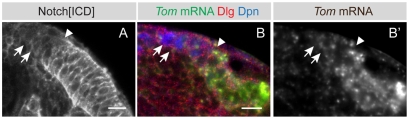
Optic lobe neuroepithelial cells and neuroblasts are clonally related (Egger et al., 2007), suggesting that there might be a limited number of transcriptional changes catalysing the switch from symmetric to asymmetric division. To identify differentially expressed genes between neuroepithelial cells and neuroblasts, we microdissected GFP-labelled neuroepithelial cells or neuroblasts from individual late-third instar larval brains (96 hours after larval hatching, ALH) and extracted total RNA, which was reverse transcribed, amplified, labelled and hybridized to whole transcriptome oligonucleotide arrays. Four samples of each cell type were analysed (Fig. 1C, see Materials and methods). One hundred and ninety genes were differentially expressed between symmetrically dividing neuroepithelial cells and asymmetrically dividing neuroblasts. One hundred and fifty-four genes were upregulated in neuroepithelial cells and 36 in neuroblasts. Strikingly, a number of genes in the Notch signalling pathway (Bray, 2006) (see Fig. S1 in the supplementary material) were expressed in neuroepithelial cells, including negative Delta regulators of the Bearded gene family Bearded (Brd), Twin of m4 (Tom), Ocho, Brother of Bearded A (BobA) and m4, Notch modulators fringe (fng), scabrous (sca) and Gp150 and the direct Notch target gene HLHm5 (see Table S1 in the supplementary material). Notch was expressed throughout the OPC and was reduced in medial neuroblasts (Fig. 2A). In support of our microarray data, in situ hybridisation for the Brd gene Tom and the Enhancer of split gene HLHm5 shows expression in the optic lobe neuroepithelium (Fig. 2B,B′; see also Fig. S2 in the supplementary material). We conclude that Notch signalling is differentially regulated between symmetrically dividing neuroepithelial cells and asymmetrically dividing neuroblasts.
Fig. 2.
The Notch signalling pathway is active in neuroepithelial cells. (A) The intracellular domain of Notch is strongly enriched in neuroepithelial cells (to the right of the arrowhead) and downregulated in neuroblasts (arrows, to the left of the arrowhead). (B,B′) As shown by fluorescent in situ hybridisation, Tom is strongly expressed in neuroepithelial cells (green, grey) and downregulated in Dpn-positive neuroblasts (arrows, blue). Arrowhead indicates the neuroepithelial to neuroblast transition zone. Scale bars: 10 μm.
Notch mutant clones are found at ectopic positions within the medulla cortex
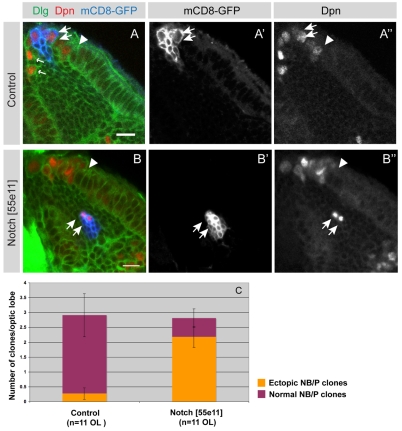
We generated Notch mutant clones using the MARCM (mosaic analysis with a repressible cell marker) system. Wild-type clones and Notch mutant clones (loss of function, N[55e11]), labelled with mCD8-GFP, were induced one day after larval hatching (see Materials and methods; see also Fig. S3 in the supplementary material). At 24 hours ALH, the optic lobe outer proliferation centre (OPC) is comprised primarily of neuroepithelial cells and only a small number of neuroblasts (Egger et al., 2007). As a result, clones induced between 21 and 27 hours ALH originated almost exclusively from within the neuroepithelium. At 72 hours ALH, many of these clones had undergone the transition to asymmetrically dividing neuroblasts (Fig. 3A-A″, arrows).
Fig. 3.
Notch mutant clones differentiate prematurely at aberrant positions. (A-A″) A single mCD8-GFP labelled control clone (blue) that contains several neuroblasts and progeny (A,A′). The clone originated in the neuroepithelium (to the right of the arrowhead) and has transformed into neuroblasts (arrows). Neuroblasts express the transcription factor Dpn (red in A,A″) and generate smaller progeny cells through asymmetric divisions (A,A′, arrows). Wild-type control neuroblasts are localised at the brain surface with the exception of the earliest born neuroblasts that migrate somewhat further into the cortex (A small arrows, A″). (B-B″) A representative mCD8-GFP labelled Notch loss-of-function clone is positioned aberrantly deep within the medulla cortex (B,B′ arrows). Two cells within this clone express Dpn (B,B″ arrows), which marks neuroblasts. Brains are counterstained for Dlg to outline cells. The arrowhead indicates the neuroepithelial to neuroblast transition zone. (C) Quantification of normal and ectopic neuroblast/progeny (NB/P) cell clones at 72 hours ALH. Virtually all control NB/P clones (91%, n=32) are located at the brain surface medial to the OPC neuroepithelium. By contrast, the great majority of Notch loss-of-function NB/progeny clones (77%, n=31) are localised ectopically within the medulla cortex. P<0.001 (unpaired t-test), error bars show s.e.m. Scale bars: 10 μm.
Whereas wild-type neuroblast/progeny (NB/P) clones are found almost exclusively at the medial edge of the neuroepithelium, most of the Notch mutant NB/P clones (24/31) are found within the medulla cortex, underlying the neuroepithelium (Fig. 3B-B″, arrows; Fig. 3C).
These clones might have migrated from the medial surface of the brain, where wild-type neuroblasts are normally found, or they might have dropped out from the overlying neuroepithelium. To distinguish between these two possibilities we characterised the clones in greater detail.
Notch mutant clones are extruded from the neuroepithelium and undergo premature neurogenesis
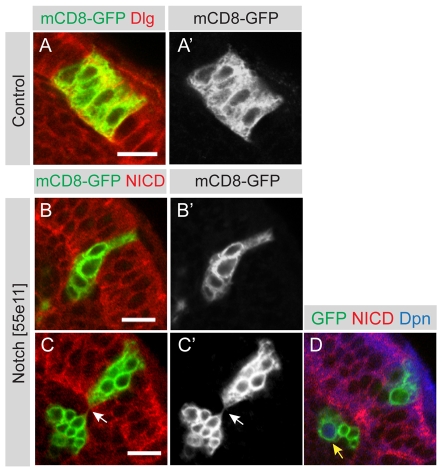
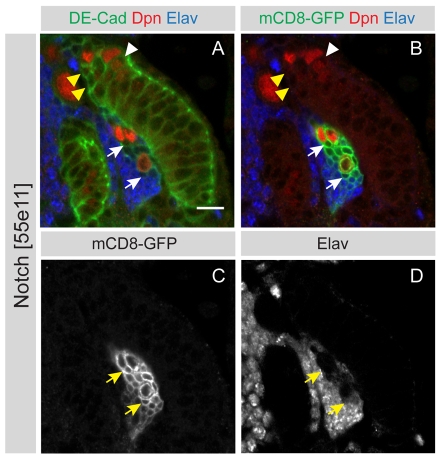
Wild-type neuroepithelial clones contain several cells, indicating that they have divided symmetrically within the epithelium to expand the stem cell pool (Fig. 4A,A′). These cells transform into neuroblasts only at the medial edge of the neuroepithelium, at which point they start to divide asymmetrically. By contrast, we find that Notch loss-of-function clones (n=6) are extruded from the neuroepithelial layer and invade the underlying medulla cortex (Fig. 4B,B′; Fig. 4C,C′). Once the mutant cells have been extruded from the neuroepithelium, a subset of cells begin to express Dpn (one to four cells in each clone; Fig. 4D, arrow). These are associated with a variable number of Dpn-negative progeny, for a total of up to 43 cells per clone. The neuronal marker Elav is absent from newly generated progeny but is induced in more mature progeny, which are found further away from the neuroblasts (Fig. 5A-D and see Fig. S4 in the supplementary material).
Fig. 4.
Notch mutant clones are extruded from the neuroepithelium. (A,A′) A single mCD8-GFP labelled control clone (green) was induced in the OPC, which is labelled by the septate junction protein Dlg (red). The clone contains several neuroepithelial cells generated through symmetric cell division. (B,B′) Shows a single mCD8-GFP labelled Notch loss-of-function clone (green) that is being extruded from the middle of the OPC neuroepithelium. Cells are stained with an antibody against the intracellular domain of Notch (red). (C,C′) A single mCD8-GFP labelled Notch loss-of-function clone (green) that has split in two. A group of cells has been extruded but is still attached by a thin membraneous connection (arrow) to clonally related cells that remain within the epithelium. (D) The same clone as in C, at a deeper section, shows a large cell with nuclear Dpn expression that has budded off smaller progeny cells. Scale bars: 10 μm.
Fig. 5.
Notch mutant clones show a wild-type differentiation pattern. (A-D) A single mCD8-GFP labelled Notch loss-of-function clone that contains Dpn (red) positive neuroblasts aberrantly positioned in the medulla cortex. Newly born progeny close to the neuroblasts do not express the neuronal marker Elav (arrows), whereas more mature progeny that are positioned further away from the neuroblast upregulate Elav. This differentiation pattern is similar to that observed for wild-type neuroblasts at the medial edge (yellow arrowheads). Scale bar: 10 μm.
Therefore, Notch mutant stem cells undergo premature neurogenesis, they express Dpn and switch from symmetric to asymmetric division in a pattern similar to wild-type neuroblasts found at the medial edge of the optic lobe.
L'sc downregulates Notch and is sufficient to transform neuroepithelial cells into neuroblasts
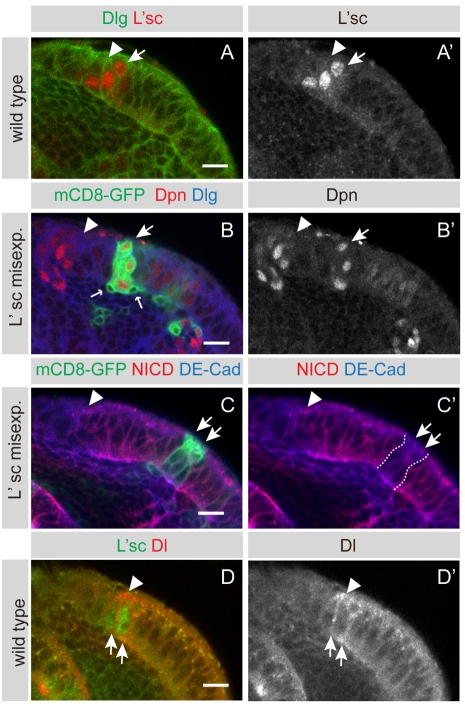
The timely conversion of neuroepithelial cells to neuroblasts in the optic lobe relies on expression of the proneural gene lethal of scute (l'sc) (Yasugi et al., 2008), which is expressed in a small group of neuroepithelial cells flanking the neuroblast region (Fig. 6A,A′). When l'sc is broadly misexpressed, the number of Dpn-positive cells increases markedly (Yasugi et al., 2008). To assess whether L'sc regulates the transition from neuroepithelial cells to neuroblasts via modulation of Notch signalling, we induced l'sc misexpression clones using the MARCM system. Clones were induced at 24 hours ALH and were analysed at 72 hours ALH. Clonal l'sc misexpression led to ectopic expression of the neuroblast marker Dpn within the neuroepithelium (Fig. 6B,B′, arrows; n=7 clones). Some of these cells divided asymmetrically while still embedded within the neuroepithelium (n=6; Fig. 6B, small arrows). Some clones (n=6) delaminated from the epithelium, resulting in ectopic Dpn-positive cells positioned within the medulla cortex (data not shown). Notch loss-of-function, and l'sc gain-of-function, clones resemble each other phenotypically. Notch ICD levels are noticeably reduced in l'sc misexpression clones, as compared with in wild-type neighbouring cells (Fig. 6C,C′, arrows; n=3 clones). We conclude that L'sc induces the switch from symmetric to asymmetric division, at least in part, by downregulation of Notch signalling.
Fig. 6.
L'sc downregulates Notch and is sufficient to induce neuroblasts. (A,A′) The proneural factor L'sc is transiently expressed in the most medial neuroepithelial cells (arrow) that are about to transform into neuroblasts. Cells are outlined by Dlg (green). (B,B′) A single mCD8-GFP labelled clone misexpressing l'sc that has been induced within the neuroepithelium. Cells within the clone ectopically express the neuroblast-specific transcription factor Dpn (larger arrow). The ectopic neuroblasts generate smaller progeny cells (small arrows), indicating a switch to an asymmetric division mode. There is a second larger clone to the right of the white arrow, which also shows upregulated Dpn levels. (C,C′) In clonal cells expressing l'sc (green, arrows), the levels of intracellular Notch (red) are markedly reduced and similar to those observed in the neuroblast zone (to the left of the arrowhead). No change in expression of DE-Cad (blue) was detected. Arrowhead indicates neuroepithelial to neuroblast transition zone. (D,D′) Delta is expressed at low levels throughout the epithelium but expression is highest in the L'sc-positive transition zone (arrows). Delta expression is reduced in the neuroblasts (left from arrowhead). Scale bars: 10 μm.
We next examined where the Notch ligand, Delta, is expressed. We find low levels of Delta throughout the neuroepithelium (Fig. 6D,D′). Delta expression is highest at the transition zone, where it is co-expressed with L'sc at the medial edge of the neuroepithelium (Fig. 6D,D′, arrows).
Notch and Delta function are required within the neuroepithelium to prevent precocious neuroblast formation
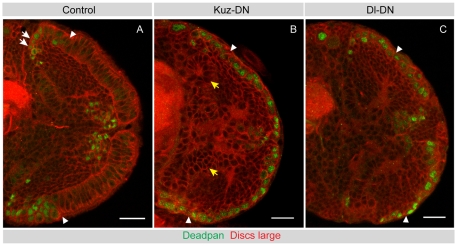
To test whether Delta-Notch signalling within the neuroepithelium is important to regulate neurogenesis, we disrupted Notch signalling in all neuroepithelial cells. Signalling through the canonical Notch pathway can be inhibited by expression of a dominant-negative form of Kuzbanian (Kuz-DN), a metalloprotease that catalyses the S2-cleavage of the Notch receptor and acts primarily in the signal-receiving cell (Delwig and Rand, 2008; Pan and Rubin, 1997; Sotillos et al., 1997). We find that virtually all neuroepithelial cells are transformed into Dpn-positive neuroblasts upon Kuz-DN misexpression (Fig. 7A,B).
Fig. 7.
Disruption of Kuzbanian or Delta function leads to premature neuroblast formation. (A) A mid-third instar control brain is shown, stained for Dpn (green) to label neuroblasts and Dlg to outline cells (red). Dpn-positive medulla neuroblasts form at the medial edges of the OPC neuroepithelium (to the left of the arrowhead). (B) A mid-third instar Kuz-DN brain stained for Dpn (green) and Dlg (red). Disruption of Kuz function during the second larval instar transforms virtually all neuroepithelial cells into Dpn-positive neuroblasts. The IPC epithelium is disrupted and forms loop-like structures (yellow arrows), but exhibits no ectopic Dpn expression. Arrowheads indicate the putative neuroepithelial to neuroblast transition zone. (C) A late-third instar Dl-DN brain stained for Dpn (green) and Dlg (red). Disruption of Dl function during larval instars transforms neuroepithelial cells prematurely into Dpn-positive neuroblasts. Arrowheads indicate the putative neuroepithelial to neuroblast transition zone. Scale bars: 20 μm.
Next, we expressed a dominant-negative form of Delta (Dl-DN) (Huppert et al., 1997) across the entire neuroepithelium. By late-third instar (96 hours ALH), the entire neuroepithelium is transformed into neuroblasts (Fig. 7C). Therefore, the activity of both ligand and receptor are required in neuroepithelial cells, suggesting that Delta-Notch signalling within the neuroepithelium maintains symmetric division and prevents premature differentiation.
DISCUSSION
In the developing cortex, neural stem cells initially divide symmetrically to produce two neural stem cells, thereby increasing the neural precursor pool. The radial glial cells subsequently divide asymmetrically to produce a neural stem cell and either a basal progenitor cell or an immature neuron (Gotz and Huttner, 2005). Most basal progenitor cells divide once more to generate two postmitotic neurons (Haubensak et al., 2004; Miyata et al., 2004; Noctor et al., 2001; Noctor et al., 2008). The Notch signalling pathway is thought to play a role in maintaining the undifferentiated state of neuroepithelial cells, radial glia and basal progenitors, but the downstream signalling cascades activated in these cells might be differentially regulated (de la Pompa et al., 1997; Gao et al., 2009; Mizutani et al., 2007; Yoon and Gaiano, 2005). Neurogenesis is initiated by proneural genes, such as Mash1 and Neurogenin2 (Ngn2) (Bertrand et al., 2002).
We show that this sequence of neurogenic events is remarkably similar to that seen in the development of the optic lobe in Drosophila. Notch is activated in the neuroepithelial cells, which remain undifferentiated. The proneural gene l'sc is expressed within the transition zone, and levels of Delta are increased, while Notch activity is decreased (Fig. 8). Thus neuroepithelial cells ultimately give rise to a variety of differentiated neurons, but only after they have passed through the transition zone.
Fig. 8.
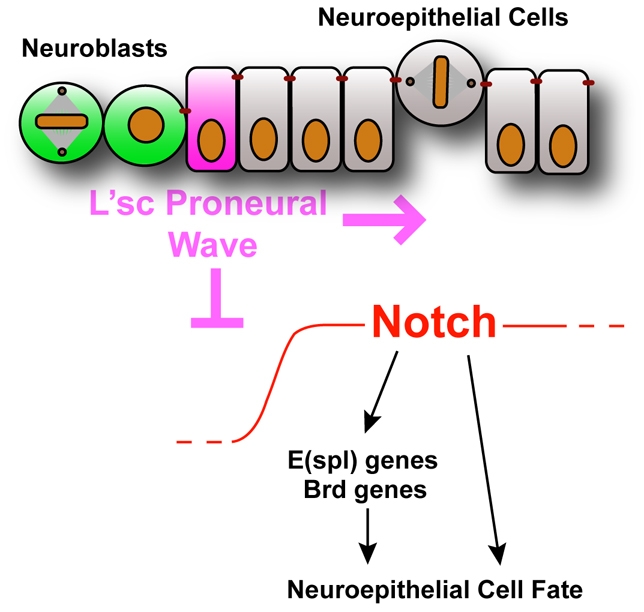
Model for the neuroepithelial to neuroblast transition. A proneural wave sweeps across the neuroepithelium and downregulates Notch to allow the transition from symmetrically dividing neuroepithelial cells to asymmetrically dividing neuroblasts.
We find low levels of Delta expression thoughout the optic lobe neuroepithelium, with increased expression in the transition zone. We also observe expression of several Brd genes within the neuroepithelium. Negative regulation of Delta activity by the Brd proteins (Bardin and Schweisguth, 2006; De Renzis et al., 2006) would be expected to further reduce the level of Delta signalling. This situation might be analogous to the oscillations in Delta and Ngn2 levels observed in vertebrates (Shimojo et al., 2008), and it will be interesting to assess whether the expression of Delta, HLHm5 or proneural genes also oscillate in flies. Strikingly, when we inhibit Delta activity throughout the epithelium, we observe the premature transformation of the entire neuroepithelium into neuroblasts. This suggests that neuroepithelial cells might both send and receive the Notch signal.
We observe higher levels of Delta in the L'sc positive transition zone. High levels of Delta or Serrate can inhibit Notch signalling through cis-inhibition (de Celis and Bray, 1997; Klein et al., 1997; Micchelli et al., 1997), suggesting one possible mechanism for the downregulation of Notch signalling at the transition zone. Interestingly, very recent results suggest that cis-inhibition can create sharp boundaries and this could be the role of the high levels of Delta we observe in the transition zone (Sprinzak et al., 2010).
Epithelial integrity might be important to maintain proliferative cell division. We show that Notch mutant clones are extruded from the neuroepithelium. Furthermore, we find expression in the optic lobe neuroepithelial cells of a number of genes involved in cell adhesion. Notch could regulate cell adhesion molecules at the transcriptional level, or might itself form a complex with adhesion molecules. In either case, Notch loss of function would disrupt cell adhesion and lead to the extrusion of epithelial cells. Subtypes of cadherins, such as DE-Cad, Cad99C, Fat, which we find preferentially expressed in the neuroepithelium (see also Fung et al., 2008), might be activated by Notch to maintain the neuroepithelium, and repressed by L'sc to promote neurogenesis. Notch mutant clones also upregulate expression of the neuroblast transcription factor Dpn (but not of L'sc), and divide asymmetrically only once they have delaminated from the epithelium. In contrast to Notch mutant clones, L'sc misexpression clones upregulate Dpn and switch to asymmetric division whilst still embedded within the neuroepithelium. L'sc acts, at least in part, through repression of Notch signalling, but might also induce neuroblast-specific genes directly.
JAK/STAT signalling negatively regulates the progression of the proneural wave and neurogenesis in the optic lobe (Yasugi et al., 2008). Interestingly, the ability of Notch to maintain radial glial cell fate appears to be largely dependent on functional JAK/STAT signalling (Kamakura et al., 2004). It remains to be seen whether the Notch pathway interacts with JAK/STAT in the Drosophila optic lobe.
Here, we show that the development of the Drosophila optic lobe parallels that of the vertebrate cerebral cortex, suggesting that the pathways regulating the transition from symmetric to asymmetric division might be conserved from flies to mammals. Identifying the effector genes that are regulated by Notch and L'sc, and the links between JAK/STAT and Notch signalling, will yield further insights into the molecular mechanisms that maintain an expanding neural stem cell pool and regulate the timely transition to differentiation.
Supplementary Material
Acknowledgements
We are grateful to B. Fischer and S. Russell from the Flychip microarray facility for help with the microarray experiments. We thank B. Bello, S. J. Bray, Y. Bellaiche, C. Q. Doe, A. Carmena and J. Skeath for fly stocks and reagents. Special thanks go to F. Hirth, S. J. Bray and all members of the Brand lab for fruitful discussions. Thanks also to A. Sossick for help with microscopy. B.E. is supported by a postdoctoral fellowship from the Swiss National Foundation and K.S.G. is a Wellcome Trust 4-year PhD student in Developmental Biology. This work was funded by a Wellcome Trust Programme Grant to A.H.B. Deposited in PMC for release after 6 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.051250/-/DC1
References
- Almeida M. S., Bray S. J. (2005). Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mech. Dev. 122, 1282-1293 [DOI] [PubMed] [Google Scholar]
- Bardin A. J., Schweisguth F. (2006). Bearded family members inhibit Neuralized-mediated endocytosis and signaling activity of Delta in Drosophila. Dev. Cell 10, 245-255 [DOI] [PubMed] [Google Scholar]
- Barker N., van de Wetering M., Clevers H. (2008). The intestinal stem cell. Genes Dev. 22, 1856-1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N., Castro D. S., Guillemot F. (2002). Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517-530 [DOI] [PubMed] [Google Scholar]
- Blanpain C., Fuchs E. (2009). Epidermal homeostasis: a balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 10, 207-217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S. J. (2006). Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678-689 [DOI] [PubMed] [Google Scholar]
- Cabernard C., Doe C. Q. (2009). Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev. Cell 17, 134-141 [DOI] [PubMed] [Google Scholar]
- Carmena A., Bate M., Jimenez F. (1995). Lethal of scute, a proneural gene, participates in the specification of muscle progenitors during Drosophila embryogenesis. Genes Dev. 9, 2373-2383 [DOI] [PubMed] [Google Scholar]
- de Celis J. F., Bray S. (1997). Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development 124, 3241-3251 [DOI] [PubMed] [Google Scholar]
- de la Pompa J. L., Wakeham A., Correia K. M., Samper E., Brown S., Aguilera R. J., Nakano T., Honjo T., Mak T. W., Rossant J., et al. (1997). Conservation of the Notch signalling pathway in mammalian neurogenesis. Development 124, 1139-1148 [DOI] [PubMed] [Google Scholar]
- De Renzis S., Yu J., Zinzen R., Wieschaus E. (2006). Dorsal-ventral pattern of Delta trafficking is established by a Snail-Tom-Neuralized pathway. Dev. Cell 10, 257-264 [DOI] [PubMed] [Google Scholar]
- Delwig A., Rand M. D. (2008). Kuz and TACE can activate Notch independent of ligand. Cell. Mol. Life Sci. 65, 2232-2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B., Boone J. Q., Stevens N. R., Brand A. H., Doe C. Q. (2007). Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2, 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas L. M., Huttner W. B. (2008). The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr. Opin. Cell Biol. 20, 707-715 [DOI] [PubMed] [Google Scholar]
- Fish J. L., Dehay C., Kennedy H., Huttner W. B. (2008). Making bigger brains-the evolution of neural-progenitor-cell division. J. Cell Sci. 121, 2783-2793 [DOI] [PubMed] [Google Scholar]
- Fung S., Wang F., Chase M., Godt D., Hartenstein V. (2008). Expression profile of the cadherin family in the developing Drosophila brain. J. Comp. Neurol. 506, 469-488 [DOI] [PubMed] [Google Scholar]
- Gao F., Zhang Q., Zheng M. H., Liu H. L., Hu Y. Y., Zhang P., Zhang Z. P., Qin H. Y., Feng L., Wang L., et al. (2009). Transcription factor RBP-J-mediated signaling represses the differentiation of neural stem cells into intermediate neural progenitors. Mol. Cell. Neurosci. 40, 442-450 [DOI] [PubMed] [Google Scholar]
- Gauthier-Fisher A., Lin D. C., Greeve M., Kaplan D. R., Rottapel R., Miller F. D. (2009). Lfc and Tctex-1 regulate the genesis of neurons from cortical precursor cells. Nat. Neurosci. 12, 735-744 [DOI] [PubMed] [Google Scholar]
- Gotz M., Huttner W. B. (2005). The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777-788 [DOI] [PubMed] [Google Scholar]
- Haubensak W., Attardo A., Denk W., Huttner W. B. (2004). Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc. Natl. Acad. Sci. USA 101, 3196-3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins N., Malatesta P., Cecconi F., Nakafuku M., Tucker K. L., Hack M. A., Chapouton P., Barde Y. A., Gotz M. (2002). Glial cells generate neurons: the role of the transcription factor Pax6. Nat. Neurosci. 5, 308-315 [DOI] [PubMed] [Google Scholar]
- Hofbauer A., Campos-Ortega J. A. (1990). Proliferation pattern and early differentiation of the optic lobes in Drosophila melanogaster. Roux's Arch. Dev. Biol. 198, 264-274 [DOI] [PubMed] [Google Scholar]
- Huppert S. S., Jacobsen T. L., Muskavitch M. A. (1997). Feedback regulation is central to Delta-Notch signalling required for Drosophila wing vein morphogenesis. Development 124, 3283-3291 [DOI] [PubMed] [Google Scholar]
- Kamakura S., Oishi K., Yoshimatsu T., Nakafuku M., Masuyama N., Gotoh Y. (2004). Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat. Cell Biol. 6, 547-554 [DOI] [PubMed] [Google Scholar]
- Klein T., Brennan K., Arias A. M. (1997). An intrinsic dominant negative activity of serrate that is modulated during wing development in Drosophila. Dev. Biol. 189, 123-134 [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Robinson K. J., Doe C. Q. (2006). Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature 439, 594-598 [DOI] [PubMed] [Google Scholar]
- Luo L., Liao Y. J., Jan L. Y., Jan Y. N. (1994). Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8, 1787-1802 [DOI] [PubMed] [Google Scholar]
- Manseau L., Baradaran A., Brower D., Budhu A., Elefant F., Phan H., Philp A. V., Yang M., Glover D., Kaiser K., et al. (1997). GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev. Dyn. 209, 310-322 [DOI] [PubMed] [Google Scholar]
- Martin-Bermudo M. D., Martinez C., Rodriguez A., Jimenez F. (1991). Distribution and function of the lethal of scute gene product during early neurogenesis in Drosophila. Development 113, 445-454 [DOI] [PubMed] [Google Scholar]
- McConnell S. K. (1995). Constructing the cerebral cortex: neurogenesis and fate determination. Neuron 15, 761-768 [DOI] [PubMed] [Google Scholar]
- McGuire S. E., Le P. T., Osborn A. J., Matsumoto K., Davis R. L. (2003). Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 1765-1768 [DOI] [PubMed] [Google Scholar]
- Micchelli C. A., Rulifson E. J., Blair S. S. (1997). The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development 124, 1485-1495 [DOI] [PubMed] [Google Scholar]
- Miyata T., Kawaguchi A., Saito K., Kawano M., Muto T., Ogawa M. (2004). Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development 131, 3133-3145 [DOI] [PubMed] [Google Scholar]
- Mizutani K., Yoon K., Dang L., Tokunaga A., Gaiano N. (2007). Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 449, 351-355 [DOI] [PubMed] [Google Scholar]
- Morrison S. J., Kimble J. (2006). Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441, 1068-1074 [DOI] [PubMed] [Google Scholar]
- Noctor S. C., Flint A. C., Weissman T. A., Dammerman R. S., Kriegstein A. R. (2001). Neurons derived from radial glial cells establish radial units in neocortex. Nature 409, 714-720 [DOI] [PubMed] [Google Scholar]
- Noctor S. C., Martinez-Cerdeno V., Ivic L., Kriegstein A. R. (2004). Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136-144 [DOI] [PubMed] [Google Scholar]
- Noctor S. C., Martinez-Cerdeno V., Kriegstein A. R. (2008). Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J. Comp. Neurol. 508, 28-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D., Rubin G. M. (1997). Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell 90, 271-280 [DOI] [PubMed] [Google Scholar]
- Shimojo H., Ohtsuka T., Kageyama R. (2008). Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 58, 52-64 [DOI] [PubMed] [Google Scholar]
- Sotillos S., Roch F., Campuzano S. (1997). The metalloprotease-disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development 124, 4769-4779 [DOI] [PubMed] [Google Scholar]
- Sprinzak D., Lakhanpal A., Lebon L., Santat L. A., Fontes M. E., Anderson G. A., Garcia-Ojalvo J., Elowitz M. B. (2010). Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature 465, 86-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vactor D. L., Jr, Cagan R. L., Kramer H., Zipursky S. L. (1991). Induction in the developing compound eye of Drosophila: multiple mechanisms restrict R7 induction to a single retinal precursor cell. Cell 67, 1145-1155 [DOI] [PubMed] [Google Scholar]
- Yasugi T., Umetsu D., Murakami S., Sato M., Tabata T. (2008). Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development 135, 1471-1480 [DOI] [PubMed] [Google Scholar]
- Yoon K., Gaiano N. (2005). Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat. Neurosci. 8, 709-715 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.