Abstract
HLH54F, the Drosophila ortholog of the vertebrate basic helix-loop-helix domain-encoding genes capsulin and musculin, is expressed in the founder cells and developing muscle fibers of the longitudinal midgut muscles. These cells descend from the posterior-most portion of the mesoderm, termed the caudal visceral mesoderm (CVM), and migrate onto the trunk visceral mesoderm prior to undergoing myoblast fusion and muscle fiber formation. We show that HLH54F expression in the CVM is regulated by a combination of terminal patterning genes and snail. We generated HLH54F mutations and show that this gene is crucial for the specification, migration and survival of the CVM cells and the longitudinal midgut muscle founders. HLH54F mutant embryos, larvae, and adults lack all longitudinal midgut muscles, which causes defects in gut morphology and integrity. The function of HLH54F as a direct activator of gene expression is exemplified by our analysis of a CVM-specific enhancer from the Dorsocross locus, which requires combined inputs from HLH54F and Biniou in a feed-forward fashion. We conclude that HLH54F is the earliest specific regulator of CVM development and that it plays a pivotal role in all major aspects of development and differentiation of this largely twist-independent population of mesodermal cells.
Keywords: bHLH proteins, Cell migration, Mesoderm, Specification, Visceral muscles, Drosophila
INTRODUCTION
The Drosophila mesoderm forms from the ventral-most cells of the early embryo that invaginate during gastrulation. The expression and function of two transcription factors, the basic helix-loop-helix (bHLH) protein Twist (Twi) and the zinc-finger protein Snail (Sna), in ventral cells located between ~15 and 85% egg length are essential for their invagination and for subsequent mesodermal tissue development. Although the absence of either twi or sna activity results in similar phenotypes, the molecular roles of the two genes in this pathway differ. twi functions in activating a variety of mesoderm-specific target genes, including several that are known to regulate the invagination, patterning and differentiation of the mesoderm. By contrast, sna is thought to act largely, if not exclusively, as a repressor of a number of neuroectodermal targets, permitting mesoderm formation by restricting the expression of these genes to areas outside the presumptive mesoderm (Kosman et al., 1991; Leptin, 1991; Reuter and Leptin, 1994; Hemavathy et al., 1997; Wakabayashi-Ito and Ip, 2005; Sandmann et al., 2007; Zeitlinger et al., 2007).
Notably, however, there is at least one group of mesodermal cells that requires sna, but not twi, for its initial phase of development. This cell group is located ventrally within the domain of early twi and sna expression, but is restricted to the posterior tip of the mesoderm between ~7.5 and 15% egg length. Because it is fated to develop into the longitudinal muscles of the midgut, it has been termed the caudal visceral mesoderm (CVM) primordium (Nguyen and Xu, 1998; Kusch and Reuter, 1999). In light of the apparent lack of a requirement for twi for the initial development of the CVM (Kusch and Reuter, 1999), it is interesting that the cells of the CVM primordium are marked by the expression of another bHLH-encoding gene, HLH54F (Georgias et al., 1997). This situation raises the possibility that, in the caudal-most portion of the mesoderm, HLH54F instead of twi cooperates with sna to control early CVM development. However, until now, specific mutants for HLH54F have not been available to test this possibility.
The Drosophila midgut musculature consists of syncytial fibers that arise through myoblast fusion between gut muscle founder cells and fusion-competent myoblasts and forms a meshwork of circular and longitudinal muscles around the endodermal layer (reviewed by Lee et al., 2005). The CVM appears to be the sole source of founder cells of the longitudinal midgut muscles (Kusch and Reuter, 1999; San Martin et al., 2001). After their ingression, these cells migrate anteriorly and spread over the future midgut, where they fuse with resident fusion-competent cells to form the multinucleated longitudinal muscle fibers of the midgut (Campos-Ortega and Hartenstein, 1997; Georgias et al., 1997; San Martin et al., 2001). The fusion-competent cells for this event come from a different source, the so-called trunk visceral mesoderm (TVM), which provides a second (and major) contribution of precursors to the musculature of the midgut (San Martin et al., 2001). The primordia of the TVM are arranged bilaterally as 11 metameric cell clusters within the dorsal mesoderm, which subsequently merge with each other into the contiguous band of the TVM. The TVM primordia are marked by the expression of the NK homeodomain gene bagpipe (bap) and the FoxF gene biniou (bin), both of which are essential for TVM formation (reviewed by Lee et al., 2005). Within the TVM, the founder cells of the circular midgut muscles are induced by Jelly belly (Jeb) signals acting through the receptor tyrosine kinase Alk (Englund et al., 2003; Lee et al., 2003). The founder myoblasts from the TVM fuse one-to-one with adjacent fusion-competent myoblasts into binucleated syncytia that form the circular midgut muscles (San Martin et al., 2001; Klapper et al., 2002). Subsequently, after the migrating CVM-derived founder cells have arrived at their destinations, each fuses with multiple fusion-competent cells from the TVM left over from the TVM founder cell fusions (San Martin et al., 2001). It is these multinucleated syncytia that will then differentiate into the longitudinal visceral muscles, which run perpendicularly to the circular muscles along the entire length of the midgut (Klapper et al., 2001). The longitudinal gut muscle fibers from the outer layer of the developing visceral musculature are tightly interwoven with the circular gut muscle fibers from the inner layer (Schröter et al., 2006).
HLH54F is the earliest known marker of the CVM primordia and its expression is maintained throughout the development and differentiation of the longitudinal midgut muscles (Georgias et al., 1997). To test whether HLH54F plays an important role in the development of the CVM and longitudinal midgut musculature, we generated loss-of-function mutations for this gene by imprecise P-excision and EMS mutagenesis screens. We demonstrate that in the absence of HLH54F activity, no longitudinal gut muscle founder cells are formed. The absence of all tested CVM markers and the observed apoptotic death of the cells that would normally be destined to form CVM in HLH54F mutants, show that HLH54F has an essential role in determining the CVM and in specifying the founder cells of the longitudinal gut musculature. This function includes feed-forward regulation and direct binding to target enhancers (e.g. from the Dorsocross genes). We also show that ectopic expression of HLH54F can interfere with normal somatic muscle, cardiac and TVM development. Further, we extend the known pathway of CVM development by showing that the initiation of HLH54F expression is largely independent of twi, but depends critically on the combined activities of sna and terminal patterning genes, particularly the synergistic activities of fork head (fkh) and brachyenteron (byn). Hence, the CVM primordia are determined at the intersection of the domains of these mesodermal and terminal regulators.
MATERIALS AND METHODS
Drosophila strains
The following Drosophila strains were used: twiID, tllL10, byn5, hkbA321R1, sna18, EY06760 (all from Bloomington Stock Center, Indiana University, USA), fkhXT6, byn5 fkhXT6, zfh12 (from R. Reuter, Tübingen University, Germany), Df(3R)Exel9020, Df(2R)14H10W-21 and Df(2R)02B10Y-12 (Mohr and Gelbart, 2002) (from the authors), croc-lacZ (Häcker et al., 1995) and slp m4-lacZ #5 (Lee and Frasch, 2000). For UAS/GAL4-induced ectopic expression, embryos were grown at 28°C using the lines UAS-p35 (Zhou et al., 1997), UAS-bin #35 (Zaffran et al., 2001), byn-GAL4 UAS-GFP (Johansen et al., 2003) (from J. Lengyel, UCLA, USA), bap3-GAL4 (Lee et al., 2003), 2xPE-twi-GAL4; 24B-GAL4 (Reim and Frasch, 2005), UAS-HLH54F-IR (P{KK103428}v103965) and UAS-da-IR (P{GD4440}v51300) (both from VDRC, Vienna, Austria) (Dietzl et al., 2007).
P-excision and EMS mutagenesis screens
The viable P insertion EY06760 was used in an excision screen with Δ2-3-derived transposase. Balanced w− males in F2 were crossed individually to Df(2R)14H10W-21 females (see Fig. 1) (Mohr and Gelbart, 2002). A total of 598 single excision lines were screened over Df(2R)14H10W-21 for semi-lethality. Semi-lethality was confirmed over Df(2R)02B10Y-12. For two semi-lethal lines, HLH54FΔ219 and HLH54FΔ598, the molecular lesions were sequenced using genomic PCR fragments from DNA of homozygous escaper flies.
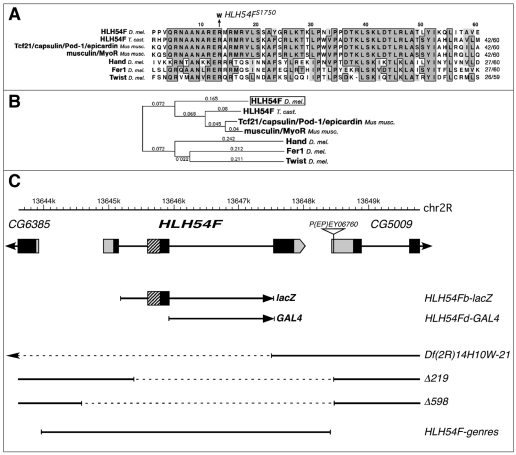
Fig. 1.
HLH54F protein sequence comparison, gene locus and mutations. (A) Alignment of the bHLH domain of HLH54F with the most closely related bHLH domains from mouse, Tribolium and Drosophila. The R-to-W exchange in the predicted protein from EMS allele HLH54FS1750 is indicated. (B) Phylogenetic analysis of data from A. (C) HLH54F gene map showing translated regions (black), the bHLH domain (hatched) and UTRs (gray). Beneath are shown the genomic regions used for the lacZ construct, GAL4 construct and the genomic rescue construct HLH54F-genres, as well as the molecularly defined breakpoints (dashed lines) of Df(2R)14H10W-21 and the P-excision alleles HLH54FΔ219 and HLH54FΔ598.
In addition, we performed an EMS mutagenesis screen for mutants with disrupted specification or migration of the CVM. HLH54Fb-RFP reporter males were mutagenized with 30 mM EMS (Lewis and Bacher, 1968) and balanced with CyO, twi>EGFP. Embryos homozygous for the mutagenized chromosome were screened for the absence, reduction or mis-migration of the RFP-labeled CVM cells. Two lines with absent RFP in the CVM failed to complement the semi-lethality of HLH54FΔ598 and were semi-lethal or lethal, respectively, in trans to larger deficiencies. PCR analysis identified HLH54FS0323 as a deficiency of HLH54F and flanking regions (lethal, with yet undefined breakpoints). Sequencing of genomic DNA amplified from HLH54FS1750 identified a C-to-T transition at position 121 of the open reading frame (ORF) as compared with the parental chromosome.
Construction of HLH54Fb-lacZ and HLH54Fb-GAL4
Genomic HLH54F sequences were cloned into pPelican (upstream sequences, HLH54Fa-lacZ) or pH-Pelican (introns, HLH54Fb-lacZ; downstream sequences, HLH54Fc-lacZ) (Barolo et al., 2000). PCR fragments from yw DNA were obtained with the following primers (5′ to 3′): for the upstream region, primers HLH-UPS-5′ CGGATAACCCATGGTTCAGATTCCTGATAA and HLH-UPS-3′ ACAAGTTGGTGGTATCTCGAGAATCACTTC; for the introns with exon 2, primers HLH-INTRON-5′ TCTCAAAGTGAGTTCGAATTCAATTTATAG and HLH-INTRON-3′ TAATGTGGAATGTGTGGATCCAACAGGCAT; for the downstream region, primers HLH-DS-5′ CTAGCCATGCCCATGGTACCTAAGCGGATA and HLH-DS-3′ TACTTGGGCGTTTTCCTCGAGTTTGTTTCT. Only HLH54Fb-lacZ expressed lacZ.
HLH54Fd-GAL4 was generated from the second intron by inserting a fragment amplified with primers Gal4-2 EcoRI (ATTCGTCCGGTGGAGGGAATTCTAATTTCC) and HLH-INTRON-3′ into the EcoRI site of the p221-GAL4 vector (from C. Klämbt, Münster University, Germany).
Genomic rescue experiments
HLH54F-genres, containing 4.5 kb of sequences from the HLH54F locus, including the intergenic regions up to the 5′ ends of both neighboring genes, was generated by genomic PCR and cloning into pCaSpeR2 (NotI/BamH1). Sequencing confirmed the correct ORF. Rescue of CVM formation was tested in the genetic background of w; HLH54FΔ598 HLH54Fb-lacZ/SM6 eve-lacZ; HLH54F-genres. Upon crossing this line with HLH54FS0323/CyO twi>EGFP, one copy rescued the semi-lethality.
Doc enhancer constructs
The 566 bp DocF4s1 enhancers (3L:9,022,917..9,023,482, vR5.27) were cloned into EcoRI/XhoI of a modified pH-Stinger vector, which has attB sequences inserted into its AvrII site (for nGFP reporters) (H. Jin and M.F., unpublished) and pH-Pelican (for DocF4s1-lacZ) (Barolo et al., 2000). For transformation, the landing site of M{3xP3-RFP.attP'}ZH-22A was used (Bischof et al., 2007) (Bestgene, Chino Hills, CA, USA). In DocF4s1Hmut, the E-boxes CANNTG (positions 227, 293, 417, 436, 508) were mutated to AGNNTG via de novo DNA synthesis (Mr. Gene, Regensburg, Germany). In DocF4s1Bmut, the Bin binding core motifs T/CAAACA (positions 162, 265, 289, 444, 458) were mutated to T/CAGTCA and the CAAACA at position 486 to CGGCCA.
Staining procedures
Antibody stainings and fluorescent double stainings for proteins and mRNAs were performed as described (Azpiazu and Frasch, 1993; Knirr et al., 1999). The following antibodies were used: rabbit anti-Twi (1:2000, from S. Roth, Cologne University, Germany), guinea pig anti-Hb9 (Exex – FlyBase) (1:500, from J. Skeath, Washington University School of Medicine, USA), guinea pig anti-Eve (1:300) (Kosman et al., 1998), rabbit anti-Eve (1:3000, our own production), rabbit anti-Mef2 (1:750, from H. Nguyen, Erlangen University, Germany), rat anti-Tropomyosin (1:400, Babraham Institute, Cambridge, UK), mouse and rabbit anti-Vasa (1:1000, from A. Ephrussi, EMBL Heidelberg, Germany and Ruth Lehmann, NYU Skirball Institute, New York, USA), rabbit anti-GFP (1:2000, Molecular Probes), monoclonal mouse anti-FasIII (7G10, 1:25) and anti-Wg (4D4, 1:10) (Developmental Studies Hybridoma Bank, University of Iowa, USA), guinea pig anti-Doc2/3 (1:500) (Reim et al., 2003), rabbit anti-Zfh1 (1:1000) (Broihier et al., 1998), rabbit anti-β-gal (1:1500, Promega) and mouse anti-β-gal (1:200, Sigma). TUNEL labeling was performed as described (Reim et al., 2003). Rhodamine-conjugated phalloidin (1:50, Molecular Probes) was used as described (Swan et al., 2004). The following digoxigenin-labeled RNA probes were used: HLH54F and bin (Zaffran et al., 2001), beat-IIa (T7 RNA polymerase transcript of EcoRI/XbaI fragment from EST RE17794 in pBluescript KS+), byn [from a PCR-derived fragment using primers byn-5′ CTCATAACTCTCCGTCGCCGACGAATGGAT and byn-3′ ATCACTGCAGCGTGCTCTGTCGCGGAGTGA in the TOPO-II TA vector (Invitrogen)]. Mutant embryos were identified with lacZ or GFP balancers.
RESULTS
HLH54F encodes a conserved bHLH transcription factor and is driven by an intronic enhancer in the CVM
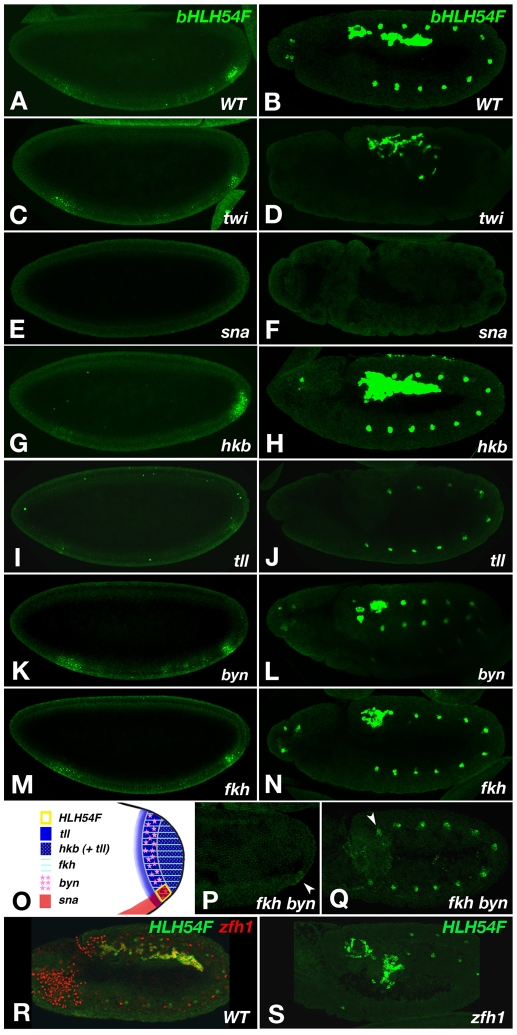
Drosophila HLH54F encodes a bHLH transcription factor and sequence alignments of the bHLH domain show that it is orthologous to a single gene in Tribolium and to the vertebrate paralogs capsulin (also known as Tcf21, Pod-1, epicardin) and musculin (also known as MyoR) (Fig. 1A,B). Within Drosophila, the closest bHLH domains belong to Hand, Fer1 and Twi (Fig. 1A,B). Notably, all of these Drosophila and vertebrate genes, with the exception of neuronally expressed Fer1, are prominently expressed in mesodermal tissues (Georgias et al., 1997; Hidai et al., 1998; Lu et al., 1998; Quaggin et al., 1998; Robb et al., 1998a; Robb et al., 1998b; Lu et al., 1999; Quaggin et al., 1999; Tomancak et al., 2002). To obtain a useful tool for examining the genetic function of HLH54F in the developing CVM of Drosophila embryos, which expresses HLH54F (Georgias et al., 1997), as well as for analyzing the development of this tissue in general, we aimed to generate lacZ reporter constructs that reflect this expression pattern. HLH54Fb-lacZ, which spans the two introns (Fig. 1C), was found to recapitulate the endogenous HLH54F expression pattern in the CVM. Reporter gene activity first appeared during gastrulation in a small domain abutting the posterior-most border of the gastrulation furrow, which reflects the early expression of HLH54F mRNA at the posterior tip of the mesoderm anlage during late blastoderm stages (Fig. 2A,B). This group of cells is known as the CVM primordium (Campos-Ortega and Hartenstein, 1997; Kusch and Reuter, 1999). During germ band elongation, the CVM cells marked by HLH54Fb-lacZ and HLH54F mRNA were internalized and occupied a position within the posterior bend formed by the posterior midgut (PMG) rudiment (stage 9; Fig. 2C,D). Subsequently, the CVM cells from this cluster migrated anteriorly along the dorsal and ventral margins of the TVM and spread along its entire length (Fig. 2E,F; see also below). Eventually the CVM cells fused with fusion-competent cells from the TVM to form multinucleated longitudinal midgut muscles (Fig. 2H; compare with Fig. 2G).
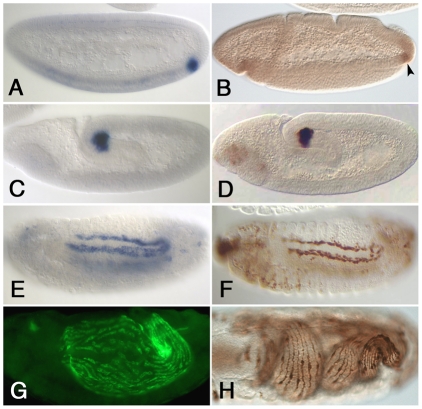
Fig. 2.
An HLH54F-lacZ reporter recapitulates the expression of HLH54F. Wild-type Drosophila embryos hybridized with HLH54F digoxygenin RNA (A,C,E,G) and comparable HLH54Fb-lacZ embryos stained with β-galactosidase (β-gal) antibodies (B,D,F,H). (A,B) Blastoderm stage embryo (stage 5, A) and embryo at early gastrulation (stage 6, B). (C,D) Stage 9 embryos during germ band elongation. (E,F) In stage 13 embryos, the cells expressing HLH54F mRNA (E) and β-gal (F) have migrated anteriorly and spread along either side of trunk visceral mesoderm. (G) Stage 15 embryo with fluorescent detection of HLH54F mRNA, showing newly formed and elongating syncytia distributed over the midgut. (H) Stage 16 embryo showing HLH54F-lacZ-stained longitudinal muscle fibers.
The early steps of cell migration were examined in more detail by stainings of HLH54Fb-lacZ embryos in the context of the neighboring tissue primordia, namely the Twi-labeled trunk mesoderm and the Hb9-labeled endoderm of the PMG rudiment (Thisse et al., 1988; Broihier and Skeath, 2002). At stage 9, the posterior portion of the mesoderm is U-shaped, perhaps because it is being bent around and drawn anteriorly on the inside by the invaginating PMG (Fig. 3A). During gastrulation twi and HLH54F were co-expressed, whereas during germ band elongation twi was downregulated in the HLH54F-expressing CVM cells, which led to mutually exclusive expression domains of the two bHLH-encoding genes after stage 8 (Fig. 3A). During stage 10, the CVM cell cluster became disconnected and moved slightly anteriorly (Fig. 3B), which was accompanied by a split into two bilaterally symmetric clusters (see Fig. 4A). At stage 10, cells became individualized and left the clusters to migrate anteriorly between the trunk mesoderm and the PMG, and ultimately towards more anterior regions of the mesoderm (Fig. 3C,D). A smaller group of CVM cells remained in the posterior portion of the mesoderm (Fig. 3D).
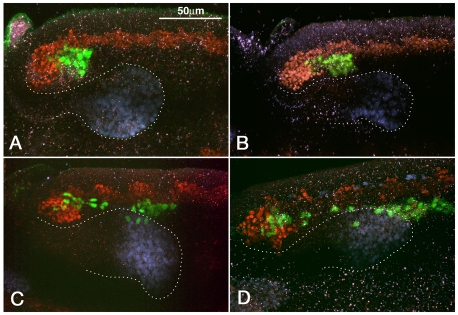
Fig. 3.
Early phase of migration of caudal visceral mesoderm cells. Sagittal views of posterior portion of elongated germ bands of HLH54Fb-lacZ Drosophila embryos stained with antibodies against β-gal (green; CVM), Twist (red; trunk mesoderm) and Hb9 (blue; midgut endoderm). (A) In stage 9 embryos, the posterior portion of the mesoderm bends around internally such that the CVM becomes positioned internally and slightly more anteriorly. (B) At stage 10, the CVM separates from the remaining mesoderm. (C) At late stage 10, individual cells leave the CVM clusters and start migrating anteriorly between posterior midgut (dotted outline) and trunk mesoderm. (D)By the end of stage 11, most CVM cells have surpassed the migrating endoderm anteriorly and are associated with trunk mesoderm.
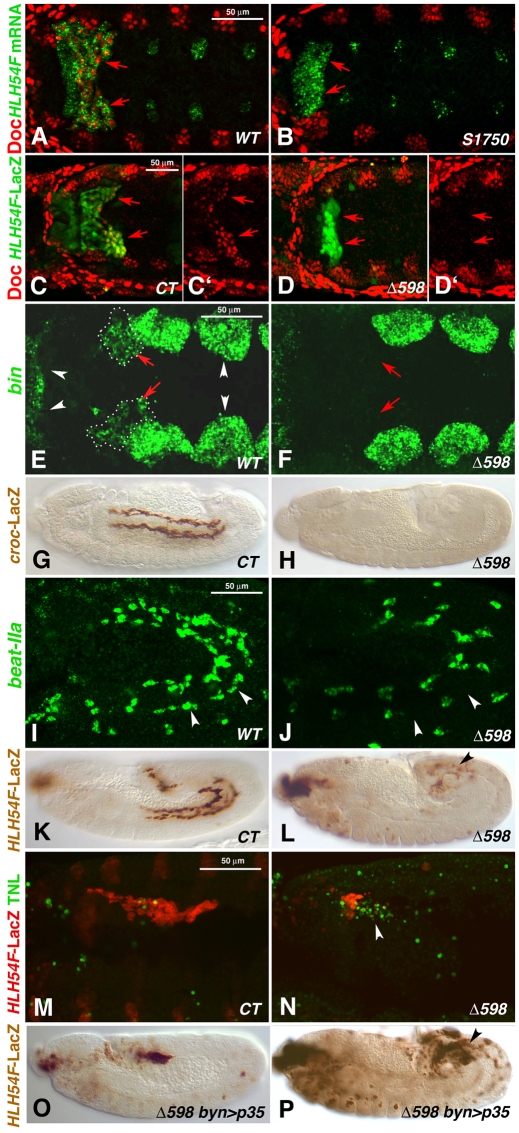
Fig. 4.
Embryonic HLH54F mutant phenotypes. (A,C,C′,E,G,I,K,M) Wild-type (WT) or control (CT) Drosophila embryos and (B,D,D′,F,H,J,L,N-P) HLH54F mutant embryos (alleles as indicated). (A-F) Dorsal views; (G-P) lateral views. (A) Stage 10 WT embryo stained for Doc protein in bilateral CVM clusters (arrows, red) and HLH54F mRNA (green). (B) HLH54FS1750 embryo stained as in A, which lacks Doc signals specifically in the CVM areas marked by mutant HLH54F mRNA (arrows). (C,C′) Stage 10 HLH54Fb-lacZ embryo stained for Doc protein (red, shown singly in C′) and β-gal (green) in CVM (arrows). (D,D′) HLH54FΔ598 embryo with HLH54Fb-lacZ stained as in C is missing Doc expression in β-gal-stained caudal mesoderm. (E) Late stage 10 WT embryo stained for bin mRNA in the CVM (red arrows, dotted outlines) and in TVM and HVM primordia (white arrowheads). (F) HLH54FΔ598 mutant, in which bin mRNA is missing in corresponding areas. (G) Stage 14 WT embryo expressing croc-lacZ in migrating CVM cells. (H) HLH54F; croc-lacZ mutant without croc-lacZ expression. (I) Stage 12 embryo showing beat-IIa mRNA in migrating CVM cells adjacent to the TVM (arrowheads) and in unidentified cells. (J) HLH54FΔ598 mutant embryo without beat-IIa mRNA signals adjacent to the TVM. (K) Stage 12 control embryo expressing HLH54Fb-lacZ in the migrating CVM. (L) Stage 12 HLH54FΔ598 mutant embryo expressing HLH54Fb-lacZ, showing a large decrease in the number of positive cells, which are not migrating (arrowhead). (M) Early stage 12 HLH54Fb-lacZ control embryo stained for β-gal (CVM, red) and by TUNEL assay (green), which shows few apoptotic cells in CVM areas. (N) Stage 12 HLH54FΔ598, HLH54Fb-lacZ mutant embryo with many TUNEL-positive cells (arrowhead). (O) Stage 11 HLH54FΔ598 mutant embryo expressing HLH54Fb-lacZ and byn-GAL4-driven anti-apoptotic p35 with normal numbers of internalized lacZ-positive cells (see Fig. 2D). (P) Stage 12 HLH54FΔ598 mutant embryo with apoptosis inhibition as in O and an increased number of HLH54Fb-lacZ-containing cells as compared with mutants without forced p35 expression (see L), which fail to migrate and differentiate.
HLH54F mutations generated by imprecise P excisions and EMS mutagenesis
To create null alleles of HLH54F a P element, EY06760, located downstream of HLH54F and just inside the 5′UTR of CG5009 (Fig. 1C) was excised. Data from Mohr and Gelbart on deficiencies with defined breakpoints at 54EF indicated that loss of HLH54F may cause semi-lethality (Mohr and Gelbart, 2002). Indeed, two of our excision chromosomes that caused semi-lethality in trans with two of these deficiencies, Df(2R)14H10W-21 and Df(2R)02B10Y-12, carried deletions of HLH54F coding sequences (Fig. 1C, HLH54FΔ219 and HLH54FΔ598). Because the deletions in both alleles also remove a small portion of the 5′UTR of CG5009, we generated a rescue construct encompassing the entire HLH54F locus (Fig. 1C). Our observation that this genomic construct can rescue the semi-lethality to full viability and also rescues the observed loss of CVM (see below) in a HLH54FΔ598 background demonstrate that the phenotypes described herein with this allele are solely due to the loss of the HLH54F gene.
We also obtained two ethyl methane sulfate (EMS)-induced mutations in HLH54F from an ongoing forward genetic screen for mutations affecting the specification and/or migration of the CVM (see Materials and methods). Both alleles, HLH54FS0323 and HLH54FS1750, are semi-lethal in trans to the P-excision allele HLH54FΔ598 and show the same CVM phenotype. Whereas HLH54FS0323 is a deletion of HLH54F and flanking genes, a point mutation in HLH54FS1750 causes a change in a conserved Arg to a Trp within the DNA-binding basic domain (Fig. 1A). The equivalent residue (Arg33) of another bHLH protein, Max, is known to contact the phosphate backbone of the DNA (Ferre-D'Amare et al., 1993). Thus, based upon the molecular and genetic data, HLH54FS1750 and the P-excision alleles appear to be functional nulls. Transheterozygotes of the P-excision alleles with the EMS alleles or with larger deficiencies at the HLH54F locus, in which the effects of any second site mutations are eliminated, occur as adult escapers at less than 10% of the expected frequency and cannot be grown as a stock.
HLH54F is required for proper specification and migration of the CVM
Based on its expression pattern, it was anticipated that HLH54F would play an important role in the development of the CVM and longitudinal midgut muscles. The genes for the Dorsocross T-box factors are normally expressed in the CVM between stages 10 and 12 (Fig. 4A,C,C′) (Reim et al., 2003), but were not turned on in the caudal mesoderm in HLH54F mutant backgrounds (Fig. 4B,D,D′; note that HLH54FS1570 mRNA and HLH54Fb-lacZ were still expressed, which rules out autoregulation and allows identification of the cells that normally become CVM). Likewise, bin, a FoxF gene that is normally expressed in all three types of visceral mesoderm (Fig. 4E) (Zaffran et al., 2001), failed to be expressed in the CVM in HLH54F mutants, whereas it was still expressed in the TVM and hindgut visceral mesoderm (HVM) (Fig. 4F). Other markers of the migrating CVM, such as croc-lacZ, which reflects the CVM expression of the forkhead domain gene crocodile (croc) (Häcker et al., 1995), the Ig domain-encoding gene beaten path IIa (beat-IIa) (Tomancak et al., 2002), the enhancer trap expression of a sloppy paired (slp) reporter (slp-m4-lacZ) (Lee and Frasch, 2000), and expression of byn after stage 10, were also absent in the corresponding areas of HLH54F mutants (Fig. 4G-J; data not shown). The only marker of CVM primordia not affected in HLH54F mutants was the high-level expression of the zinc-finger homeodomain transcription factor Zfh1 (see Fig. S1 in the supplementary material), although, as for byn, zfh1 expression also disappeared prematurely after stage 10. The loss of all tested CVM markers at all stages in the absence of HLH54F (with the exception of early Zfh1) and the observation that the cells that would normally form CVM failed to undergo any rearrangements and movements (Fig. 4B,D,D′; compare with Fig. 4A,C,C′) suggest that HLH54F is required for the specification of the CVM and show that development of the longitudinal visceral muscles is critically dependent on HLH54F function.
It has been observed that the loss of cell fate specification can cause either cell fate transformation or cell death (reviewed by Bonini and Fortini, 1999; Mann and Morata, 2000; Werz et al., 2005). Hence, we used an HLH54Fb-lacZ reporter to address the fate of the caudal mesodermal cells that are no longer identified as CVM in the absence of HLH54F. Following stage 10, we observed a dramatic decrease in the number of HLH54Fb-lacZ-positive cells and a total absence of migrating cells in the HLH54F mutants (compare Fig. 4L with 4K). Apoptosis of these particular cells was demonstrated in a TUNEL assay, which showed TUNEL-positive cells containing traces of lacZ signals in the vicinity of the remaining HLH54Fb-lacZ-positive cells in stage 12 HLH54F mutants (Fig. 4N). This is in contrast to the virtual lack of TUNEL signals in HLH54Fb-lacZ-positive cells in control embryos (Fig. 4M).
To examine whether any possible cell fate transformations or migratory abilities of the HLH54Fb-lacZ-positive cells in HLH54F mutant backgrounds are obscured by their death, we forced the expression of a pan-caspase inhibitor, p35 (BacA\p35), via UAS-p35 and a byn-GAL4 driver in these cells to prevent their apoptosis (Brand and Perrimon, 1993; Hay et al., 1994). Blocking apoptosis specifically in the CVM (along with the presumptive hindgut ectoderm) of HLH54F mutants resulted in a nearly normal-sized HLH54F-lacZ-positive domain (Fig. 4O; compare with Fig. 4C). However, these cells still did not migrate or differentiate into any kind of muscles and, instead, remained as unfused cells randomly distributed in the vicinity of the hindgut (Fig. 4P; data not shown). Thus, we infer that HLH54F is required not only for the survival of the CVM, but also for the initial specification and, in this context, for the correct migratory properties of these cells. In the absence of HLH54F activity, the failure of these cells to be specified apparently causes their entry into apoptosis.
The CVM primordia are thought to form the founder cells for all longitudinal visceral muscles. Apparently as a consequence of the absence of these muscles (see below), midgut constrictions were not formed efficiently and were present to a variable extent in the mutants, with a small fraction of mutant embryos lacking midgut constrictions altogether (Fig. 5A-C).
Fig. 5.
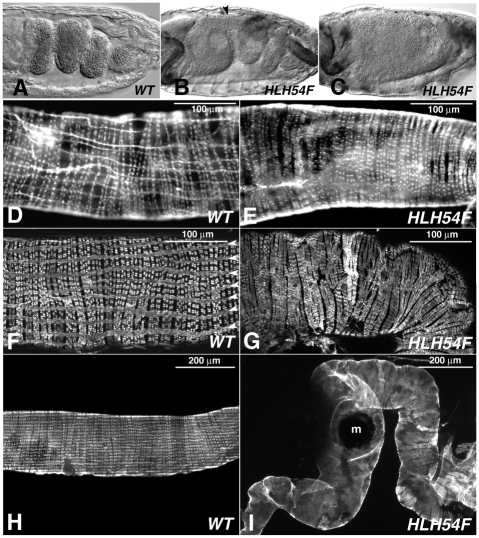
HLH54F mutant phenotypes of embryonic midgut and midgut musculature in larvae and adults. (A) Stage 15 wild-type (WT) Drosophila embryo with constricted and looping midgut. (B) Stage 15 HLH54FS1750/HLH54FΔ598 embryo with incomplete midgut constriction (arrowhead); also stained for GFP reporter for foregut and hindgut muscles, which are unaffected. (C) HLH54F mutant embryo as in B without any midgut constrictions. (D) Section of WT third-instar larval midgut stained with Rhodamine-conjugated phalloidin to visualize both the circular and the longitudinal visceral muscles. (E) Homozygous HLH54FΔ598 third-instar larval gut with absent longitudinal visceral muscles. (F) Section of WT adult midgut stained for Tropomyosin, showing regularly spaced longitudinal and circular muscle fibers. (G) Adult midgut section from a HLH54FS1750/HLH54FΔ589 fly stained for Tropomyosin, showing the lack of longitudinal midgut muscles and circular muscles with irregular morphologies and interruptions. (H,I) WT (H, from F) and mutant (I, from G) adult midguts at lower magnification. Unlike the smooth and linear surface of WT midguts, midguts from mutants have an undulating surface and are curled. m, melanotic mass.
The larval longitudinal midgut muscles persist during metamorphosis and contribute to the longitudinal gut musculature of adult flies (Klapper, 2000). To address the question of whether the CVM is the sole source of founder cells for the longitudinal visceral muscles in larvae and adults, midguts from HLH54F mutant third-instar larvae and adult escapers, as well as from wild-type controls, were stained for F-actin and Tropomyosin. In wild-type animals, these stainings highlight the transverse circular midgut muscles as well as the longitudinal visceral muscles (Fig. 5D,F,H). By contrast, longitudinal visceral muscles were present neither in HLH54F mutant larvae nor in adult flies (Fig. 5E,G,I). At this level of resolution, we did not detect any clear alterations of the larval circular gut muscles and the midgut that would be caused by the absence of the longitudinal gut muscles (Fig. 5E). However, midguts isolated from mutant adults tended to be more curled into a spiral shape and smaller in diameter than their straighter wild-type counterparts (Fig. 5G,I). The adult mutant gut tube exhibited small bulges and frequently showed melanotic masses, similar to hand mutant flies (Lo et al., 2007), which suggests a tendency to rupture in vivo (Fig. 5G,I). In addition to the absence of support by longitudinal muscle fibers, this apparent fragility of adult midguts may be explained by the presence of highly disordered circular muscle fibers that showed frequent interruptions (Fig. 5G).
As HLH54F was isolated originally as a binding partner of the E-protein Daughterless (Da) (Georgias et al., 1997), we sought to obtain indications as to whether these two proteins might functionally interact in vivo during longitudinal muscle development. Indeed, RNAi knockdowns of HLH54F and da via HLH54F-GAL4-driven expression of the respective inverted repeat sequences caused similar phenotypes in the adult longitudinal gut musculatures, consisting of a variable reduction in the number and thickness of fibers as well as their uneven distribution around the midgut. In general, the effects appeared stronger upon knocking down both genes together (see Fig. S2 in the supplementary material). These data are indicative of functional interactions between these two bHLH proteins during longitudinal gut muscle development. Future studies need to address these interactions in more detail and compare them with the repressive effects on myogenesis reported for Da-Twi heterodimers (Castanon et al., 2001).
Based upon alterations in the mesodermal expression pattern of Even-skipped (Eve) and a disrupted Mef2 pattern in myocardial cells in byn mutant embryos, it has been proposed that the CVM plays a non-autonomous role in the normal development of adjacent dorsal muscle and cardiac progenitors during its migration (Kusch and Reuter, 1999). However, with our HLH54F mutants, which cause the specific loss of the CVM, we did not see any of the reported effects on the specification and morphogenesis of the heart and dorsal muscles, which seems to rule out a role of the CVM in the development of these tissues (see Fig. S1 in the supplementary material). It has also been proposed that germ cells require the CVM to direct their early migration from the basal surface of the PMG in anterior directions along the mesoderm (Broihier et al., 1998). Our observations in HLH54F mutants confirmed that the CVM does make the guidance of germ cells towards the gonadal mesoderm more reliable, although it is not absolutely required for this migration (see Fig. S3 in the supplementary material).
HLH54F acts upon a CVM-specific Doc enhancer in combination with its downstream factor Bin
An exhaustive analysis of the Dorsocross (Doc) locus for enhancers (D.S., H. Jin, M.F. and I.R., unpublished) identified a 566 bp element, DocF4s1, that recapitulates the activity of the Doc genes (Fig. 6A,E,I). DocF4s1 contains five E-boxes and six forkhead domain binding motifs that are expected to bind HLH54F and Bin, respectively (Fig. 6I). Ectopic expression of HLH54F via twi-GAL4 caused an expansion of DocF4s1 enhancer activity within the caudal mesoderm as well as into the head mesoderm, and analogous expression of bin caused similar, albeit less pronounced, effects (Fig. 6B,C). Notably, co-expression of HLH54F and bin caused much stronger effects, indicating that the products of the two genes cooperate in activating DocF4s1. The absence of ectopic expression in the trunk mesoderm points to the requirement for additional, as yet unknown activating or repressive factors.
Fig. 6.
HLH54F and Bin as co-regulators of a caudal visceral mesoderm-specific enhancer from the Doc locus. (A-H) lacZ (A-D) or nuclear GFP (E-H) reporter gene expression from DocF4s1 and derivatives is shown in early stage 12 Drosophila embryos as detected with anti-β-gal (diaminobenzidine) and anti-GFP (fluorescence) antibodies (dorsal views). (A) DocF4s1-lacZ in a wild-type background showing expression in bilateral CVM clusters at the beginning of cell migration (right). Additional activity is seen in bilateral clusters of the procephalic neuroectoderm (left). (B-D) DocF4s1-lacZ in embryos with UAS-HLH54F (B), UAS-bin (C) and UAS-HLH54F + UAS-bin (D), driven pan-mesodermally by twi-GAL4. (E) Control DocF4s1-GFP expression. (F) DocF4s1 Hmut-GFP expression (all E-boxes mutated). (G) DocF4s1 Bmut-GFP expression (all Bin binding motifs mutated). (H) DocF4s1 H+Bmut-GFP (all HLH54F and Bin binding motifs mutated). (I) Map of the DocF4s1 enhancer within the Doc locus and of HLH54F (H) and Bin (B) binding motifs within the enhancer.
To test whether the cooperative inputs of HLH54F and Bin upon DocF4s1 require the respective binding motifs for these proteins, we tested mutated DocF4s1 derivatives. Mutation of all five E-boxes caused a delay in the onset of enhancer activity until mid stage 11 (instead of late stage 10; data not shown) and a mild reduction in the number of CVM cells expressing the reporter (Fig. 6F). Likewise, upon mutation of all six Bin binding motifs, reporter gene activity in the CVM was delayed and occurred in even fewer cells and at lower levels (Fig. 6G). Importantly, simultaneous mutation of the binding motifs for both HLH54F and Bin led to a total loss of enhancer activity in the CVM, whereas the unrelated activity in bilateral clusters within the procephalic neurogenic ectoderm was still present (Fig. 6H). Altogether, these data provide strong evidence for a mechanism in which HLH54F and Bin cooperate in activating CVM-specific Doc expression via direct binding to this enhancer in a partially redundant fashion. Because bin activation in these cells requires HLH54F (Fig. 4F), this mechanism represents a feed-forward mode of regulation that might be responsible for the postponed onset of Doc expression in the CVM as compared with HLH54F.
Ectopic expression of HLH54F is insufficient to drive CVM development
To test whether HLH54F is sufficient to turn other mesodermal derivatives into longitudinal midgut muscles or their progenitors, we expressed the gene ectopically under the control of the enhancers of twi and held out wings (how) in all myogenic cells (confirmed by in situ hybridization). However, we did not detect any ectopic longitudinal gut muscles in these embryos and in situ hybridizations for croc, a CVM marker downstream of HLH54F, failed to detect any ectopic croc expression. Notably, there was a reduction in the number of cardioblasts, and most of the somatic muscles were more rounded and often not properly attached to their tendon cells (see Fig. S4 in the supplementary material). In addition, the midguts failed to form any constrictions, which can in part be explained by the loss of Wingless expression in the TVM (see Fig. S4 in the supplementary material). Hence, although HLH54F is not sufficient to transform other mesodermal derivatives into longitudinal midgut muscles, its ectopic presence interferes with the normal development of various muscle tissues.
HLH54F expression is regulated through a combination of posterior and ventral patterning genes
How is the expression of HLH54F in a small quadrant of cells established in the posterior portion of the presumptive mesoderm? Several genes are expressed in overlapping domains in the posterior region of the blastoderm embryo and are essential for the specification of various terminal structures (reviewed by Bronner and Jäckle, 1991; Lengyel and Iwaki, 2002). At the same time, Twi and Sna are expressed in the ventral-most region of the embryo and are required for mesoderm development (Kosman et al., 1991; Leptin, 1991; Ip et al., 1992). There are also regulatory interactions between the posterior gap genes and these ventrally expressed genes; in particular, the repression of sna by the terminal gap gene huckebein (hkb) defines the posterior border of the future mesoderm (Reuter and Leptin, 1994). As all these genes are known to play roles in the development of the CVM (Kusch and Reuter, 1999), we examined whether they act in this pathway via regulating the expression of HLH54F.
Initially, at the blastoderm stage, twi extends to the poles of the embryo and, therefore, overlaps with the CVM primordium. As shown in Fig. 7C, HLH54F mRNA was still expressed in the absence of twi function, albeit at reduced levels (see Fig. 7A). Furthermore, HLH54F expression was still observed in the migrating CVM in the twi mutant embryos (Fig. 7D; compare with Fig. 7B). These results indicate that HLH54F expression is not directly dependent on twi and confirm that the CVM does not require twi for ingression and initial migration [although the trunk mesoderm is required for proper pathfinding (Kusch and Reuter, 1999)]. HLH54F is also expressed in specific somatic muscle progenitors (where we have not detected any phenotype to date) (Fig. 7B) (Georgias et al., 1997). As expected, this expression was missing in twi mutants (Fig. 7D). In contrast to twi mutants, in the absence of sna there was no HLH54F mRNA expression in the CVM primordium at any time (Fig. 7E,F).
Fig. 7.
Regulation of early HLH54F expression. HLH54F mRNA staining of late blastoderm stage Drosophila embryos (A,C,E,G,I,K,M,P) and in stage 11 embryos (B,D,F,H,J,L,N,Q,S,R) of wild type and mutants as indicated. (A,B) Wild-type HLH54F expression. (C,D) Slightly reduced HLH54F expression and mis-migrating CVM cells at stage 11 in twi mutants. (E,F) Complete absence of HLH54F expression in sna mutants. (G,H) Posterior expansion of HLH54F expression at blastoderm stage and increased CVM at stage 11 in hkb mutants. (I,J) Almost complete absence of HLH54F expression at blastoderm and absence of HLH54F-expressing CVM at stage 11 in tll mutants. (K-N) Reduced HLH54F expression domains in byn and fkh mutant blastoderm embryos and reduced HLH54F-labeled CVM at stage 11. (O) Schematic of the expression domains of the tested genes at late blastoderm stage. (P) Early gastrulation stage fkh byn double mutant showing only low, residual levels of HLH54F expression (arrowhead). (Q) Stage 11 fkh byn double mutant with very few HLH54F-expressing cells (arrowhead). (R) Early stage 12 wild-type embryo co-stained for HLH54F mRNA (green) and Zfh1 protein (red) in the migrating CVM. (S) Stage 12 zfh1-deficient embryo stained as in R. HLH54F is expressed in CVM cells, but cell migration is disrupted.
In hkb mutants, HLH54F expression was expanded towards the posterior (Fig. 7G), leading to an increased number of CVM cells as they started to migrate (Fig. 7H; compare with Fig. 7B). These data are in agreement with the known repressive activity of hkb on posterior sna expression and the involvement of sna in the activation of HLH54F expression, as shown above (see Fig. 7O for hkb and sna expression domains).
tailless (tll) is expressed in the posterior ~18% of the embryo (Fig. 7O) and is required for various structures derived from this expression domain, including the anal pads, hindgut, Malpighian tubules and the CVM (Pignoni et al., 1990; Kusch and Reuter, 1999; Lengyel and Iwaki, 2002). Accordingly, in tllL10 (a strong hypomorph), only trace amounts of HLH54F mRNA expression were observed at blastoderm stage, and during elongated germ band stages caudal mesodermal HLH54F expression was lost completely (Fig. 7I,J). tll has been shown to act upstream of byn and fkh. byn encodes a T-box transcription factor that is expressed in a posterior stripe and is required for development of the hindgut, anal pads and Malpighian tubules (Fig. 7O) (Kispert et al., 1994; Singer et al., 1996). This domain of byn expression is established through activation by tll and repression by hkb at the posterior pole, which confines byn expression to within ~7.5-15% egg length (Pignoni et al., 1990; Singer et al., 1996). fkh encodes a winged helix transcription factor that is expressed in a slightly smaller, more posterior domain than tll (0 to ~15% egg length) that will form the future PMG and hindgut (Weigel et al., 1989). byn and fkh mutants showed a similar reduction in size of the domain of HLH54F expression during blastoderm and elongated germ band stages (Fig. 7K-N). In double mutants for byn and fkh, the reduction of HLH54F was much more severe than with the single mutants, indicating that these two genes synergize in activating HLH54F (Fig. 7P,Q).
The zinc-finger homeodomain-encoding gene zfh1 is expressed preferentially in the CVM (and hematopoietic mesoderm) during early mesoderm development (Fig. 7R) (Lai et al., 1991; Broihier et al., 1998). In embryos lacking zfh1, HLH54F was expressed normally, but the migration of the CVM cells was aberrant (Fig. 7S).
Altogether, these data show that HLH54F expression is positively regulated by ventrally active sna (which itself is excluded from the posterior ~7.5% of the embryo by hkb) as well as by the synergistic action of the terminal genes byn and fkh.
DISCUSSION
HLH54F is a key regulator in the CVM, a population of cells in which the bHLH gene twi appears to have only minor functions. Although twi is initially co-expressed with HLH54F in these cells, it makes only a small contribution to activating HLH54F expression, and the expression of both bHLH genes rapidly becomes mutually exclusive. Instead of twi, the activation of HLH54F in the CVM primarily involves the combined activities of mesodermal sna and the terminal genes fkh and byn (Fig. 8). As sna is generally thought to act as a ventral repressor of non-mesodermal genes in early mesoderm development, it will be interesting to determine whether the positive requirement for sna in the activation of HLH54F expression is direct, which would be unique to date. Alternatively, HLH54F might be activated by high levels of nuclear Dorsal and repressed by lateral genes that are repressed by sna ventrally. Along the anteroposterior axis, the posterior border of HLH54F expression is apparently defined by the posterior expression border of sna, which is delineated by the repressive action from hkb. We propose that the anterior border of HLH54F is determined by near-maximal threshold levels of tll, the expression of which declines steeply in the area anterior to the HLH54F domain (Pisarev et al., 2009). However, tll acts largely indirectly, through the combined activities of its downstream genes byn and fkh, in activating HLH54F (Fig. 8). The low residual levels of HLH54F mRNA in fkh byn double mutants suggest the involvement of direct inputs from additional posterior activities, possibly tll or maternal torso. From the data shown herein and elsewhere (Hemavathy et al., 1997; Kusch and Reuter, 1999), it appears that high-level expression of zfh1 in the CVM largely depends on tll and sna, whereas HLH54F and zfh1 do not depend on one another.
Fig. 8.
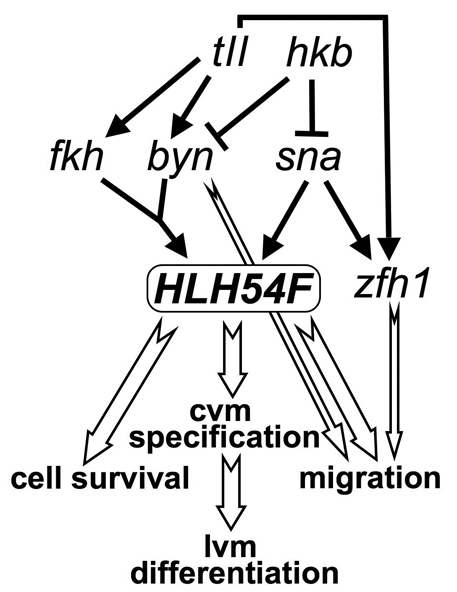
Regulatory interactions during caudal visceral mesoderm development. There is high-level expression of zfh1 in presumptive caudal visceral mesoderm. Although shown separately, the developmental outputs downstream of HLH54F are intimately connected. Black arrows, gene regulation; white arrows, developmental regulation. cvm, caudal visceral mesoderm; lvm, longitudinal visceral muscle.
Notably, neither twi nor HLH54F is required individually for the internalization of the CVM cells during gastrulation (Kusch and Reuter, 1999) (this report), although we cannot exclude a redundant function. We have shown that the posterior portion of the mesoderm, which includes the CVM and portions of the presumptive HVM, bends around during gastrulation to form a second, internal mesodermal layer. It is conceivable that this movement is a passive process brought about by the invagination of the PMG rudiment. However, for subsequent migrations of CVM cells from these positions, the activity of HLH54F, but not twi, is crucial. In addition, byn, zfh1 and fkh are required for normal migration after stage 10 (Kusch and Reuter, 1999) (this report). Whereas their respective functions are likely to be cell-autonomous, the observed requirement of twi for normal pathfinding of CVM cells is likely to be due to the absence of the migration substrate normally formed from the trunk mesoderm.
Our genetic data show that, after the caudal mesodermal cells have ingressed in this manner, they do not develop any further in the absence of HLH54F activity and undergo apoptosis. In the normal situation, HLH54F is needed for the activation of several transcription factor-encoding genes at this stage, including bin, croc and the Doc genes. Although the functions of these genes in CVM development have not been defined, it is likely that they regulate specific aspects of CVM development downstream of, and perhaps in combination with, HLH54F. The data from loss-of-function and ectopic expression analyses of HLH54F show that this gene is essential, but not sufficient, for specification of longitudinal gut muscle founders. Parallel inputs, albeit less pervasive, appear to come from high-level zfh1, which like HLH54F is required for croc/croc-lacZ expression (Kusch and Reuter, 1999). Altogether, we propose that HLH54F is necessary for activating the vast majority of early CVM-specific genes, with one known exception being high-level zfh1, and that zfh1, byn and fkh in various combinations act together with HLH54F to activate certain targets during the specification and early migration of CVM cells.
The continuous expression of HLH54F in the CVM and longitudinal gut muscles suggests that this gene is not only required for specification, but is also directly involved in many other developmental processes, including the continued migration, myoblast fusion and differentiation of the CVM cells. Possible downstream targets of HLH54F in the promotion of proper cell migration include beat-IIa, which encodes an as yet uncharacterized membrane-anchored Ig domain protein, and the FGF receptor-encoding gene heartless (htl), which is known to be required for normal migration (Pipes et al., 2001; Mandal et al., 2004). In this context, it is interesting that the vertebrate orthologs of HLH54F are expressed prominently in specific migrating populations of mesodermal cells as well. For example, musculin is expressed in myoblasts at the myotomal lips that migrate into the developing limbs, and capsulin is expressed in the migrating pro-epicardial cells (e.g. von Scheven et al., 2006). Therefore, it is possible that parts of the regulatory circuit in the control of cell migration have been conserved, even though they occur in different mesodermal cell types. capsulin is also expressed prominently in the splanchnopleura and tissues derived from it, including the developing smooth muscles of the stomach and gut (Hidai et al., 1998; Robb et al., 1998b; von Scheven et al., 2006). Therefore, HLH54F and capsulin might share some functions in the terminal differentiation of the respective gut musculatures in the different systems. Both Capsulin and Musculin have been characterized largely as repressors. However, their activity (and likewise that of HLH54F) as repressors versus activators might well be context specific with respect to the particular enhancer, tissue or developmental stage in question and might depend on the relative concentrations of particular heterodimerization partners (Lu et al., 1999; Miyagishi et al., 2000; Castanon et al., 2001).
Based on the phenotype of byn mutants, a role of the CVM in promoting midgut constrictions has also been proposed (Kusch and Reuter, 1999). The phenotype of HLH54F mutants confirms this effect, although we find that partial constrictions can frequently occur and that the effect is variable. We infer that the physical interactions between developing longitudinal and circular muscle fibers are necessary to provide the full force required for the efficient constriction of the midgut endoderm at the well-defined signaling centers. In the fully developed midgut, scanning electron microscopy images have revealed that the longitudinal fibers are tightly interwoven with the web-shaped circular fibers, which may explain the mechanical strength of this meshwork. Indeed, we find that the mechanical stability and integrity of the midgut, particularly in HLH54F mutant adults, are severely compromised.
In summary, HLH54F appears to sit at the top of the regulatory hierarchy of CVM development and is likely to fulfil additional key roles during the course of development of the CVM and the longitudinal gut muscles. Future efforts need to be directed towards dissecting additional downstream events that regulate the different steps of cell migration, myoblast fusion, morphogenesis and terminal differentiation of the longitudinal midgut musculature.
Supplementary Material
Acknowledgements
We thank the colleagues listed in the Materials and methods section as well as the Bloomington Stock Center (Indiana, USA), VDRC (Vienna, Austria) and DSHB (Iowa, USA) for providing fly stocks and antibody reagents. We gratefully acknowledge funding from the National Institutes of Health (NIDDK and NICHD) to M.F. and Deutsche Forschungsgemeinschaft (DFG) to M.F. and I.R. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.046573/-/DC1
References
- Azpiazu N., Frasch M. (1993). tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 7, 1325-1340 [DOI] [PubMed] [Google Scholar]
- Barolo S., Carver L. A., Posakony J. W. (2000). GFP and β-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29, 726-732 [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312-3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonini N. M., Fortini M. E. (1999). Surviving Drosophila eye development: integrating cell death with differentiation during formation of a neural structure. BioEssays 21, 991-1003 [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415 [DOI] [PubMed] [Google Scholar]
- Broihier H. T., Skeath J. B. (2002). Drosophila homeodomain protein dHb9 directs neuronal fate via crossrepressive and cell-nonautonomous mechanisms. Neuron 35, 39-50 [DOI] [PubMed] [Google Scholar]
- Broihier H., Moore L., Van Doren M., Newman S., Lehmann R. (1998). zfh-1 is required for germ cell migration and gonadal mesoderm development in Drosophila. Development 125, 655-666 [DOI] [PubMed] [Google Scholar]
- Bronner G., Jäckle H. (1991). Control and function of terminal gap gene activity in the posterior pole region of the Drosophila embryo. Mech. Dev. 35, 205-211 [DOI] [PubMed] [Google Scholar]
- Campos-Ortega J. A., Hartenstein V. (1997). The embryonic development of Drosophila melanogaster. Berlin: Springer-Verlag; [Google Scholar]
- Castanon I., Von Stetina S., Kass J., Baylies M. K. (2001). Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development 128, 3145-3159 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151-156 [DOI] [PubMed] [Google Scholar]
- Englund C., Loren C. E., Grabbe C., Varshney G. K., Deleuil F., Hallberg B., Palmer R. H. (2003). Jeb signals through the Alk receptor tyrosine kinase to drive visceral muscle fusion. Nature 425, 512-516 [DOI] [PubMed] [Google Scholar]
- Ferre-D'Amare A. R., Prendergast G. C., Ziff E. B., Burley S. K. (1993). Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363, 38-45 [DOI] [PubMed] [Google Scholar]
- Georgias C., Wasser M., Hinz U. (1997). A basic-helix-loop-helix protein expressed in precursors of Drosophila longitudinal visceral muscles. Mech. Dev. 69, 115-124 [DOI] [PubMed] [Google Scholar]
- Häcker U., Kaufmann E., Hartmann C., Jürgens G., Knöchel W., Jäckle H. (1995). The Drosophila fork head domain protein crocodile is required for the establishment of head structures. EMBO J. 14, 5306-5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay B. A., Wolff T., Rubin G. M. (1994). Expression of baculovirus P35 prevents cell death in Drosophila. Development 120, 2121-2129 [DOI] [PubMed] [Google Scholar]
- Hemavathy K., Meng X., Ip Y. T. (1997). Differential regulation of gastrulation and neuroectodermal gene expression by Snail in the Drosophila embryo. Development 124, 3683-3691 [DOI] [PubMed] [Google Scholar]
- Hidai H., Bardales R., Goodwin R., Quertermous T., Quertermous E. E. (1998). Cloning of capsulin, a basic helix-loop-helix factor expressed in progenitor cells of the pericardium and the coronary arteries. Mech. Dev. 73, 33-43 [DOI] [PubMed] [Google Scholar]
- Ip Y. T., Park R. E., Kosman D., Yazdanbakhsh K., Levine M. (1992). dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev. 6, 1518-1530 [DOI] [PubMed] [Google Scholar]
- Johansen K. A., Iwaki D. D., Lengyel J. A. (2003). Localized JAK/STAT signaling is required for oriented cell rearrangement in a tubular epithelium. Development 130, 135-145 [DOI] [PubMed] [Google Scholar]
- Kispert A., Herrmann B., Leptin M., Reuter R. (1994). Homologs of the mouse Brachyury gene are involved in the specification of posterior terminal structures in Drosophila, Tribolium, and Locusta. Genes Dev. 15, 2137-2150 [DOI] [PubMed] [Google Scholar]
- Klapper R. (2000). The longitudinal visceral musculature of Drosophila melanogaster persists through metamorphosis. Mech. Dev. 95, 47-54 [DOI] [PubMed] [Google Scholar]
- Klapper R., Heuser S., Strasser T., Janning W. (2001). A new approach reveals syncytia within the visceral musculature of Drosophila melanogaster. Development 128, 2517-2524 [DOI] [PubMed] [Google Scholar]
- Klapper R., Stute C., Schomaker O., Strasser T., Janning W., Renkawitz-Pohl R., Holz A. (2002). The formation of syncytia within the visceral musculature of the Drosophila midgut is dependent on duf, sns and mbc. Mech. Dev. 110, 85-96 [DOI] [PubMed] [Google Scholar]
- Knirr S., Azpiazu N., Frasch M. (1999). The role of the NK-homeobox gene slouch (S59) in somatic muscle patterning. Development 126, 4525-4535 [DOI] [PubMed] [Google Scholar]
- Kosman D., Ip Y. T., Levine M., Arora K. (1991). Establishment of the mesoderm-neuroectoderm boundary in the Drosophila embryo. Science 252, 118-122 [DOI] [PubMed] [Google Scholar]
- Kosman D., Small S., Reinitz J. (1998). Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev. Genes Evol. 208, 290-294 [DOI] [PubMed] [Google Scholar]
- Kusch T., Reuter R. (1999). Functions for Drosophila brachyenteron and forkhead in mesoderm specification and cell signalling. Development 126, 3991-4003 [DOI] [PubMed] [Google Scholar]
- Lai Z., Fortini M., Rubin G. (1991). The embryonic expression patterns of zfh-1 and zfh-2, two Drosophila genes encoding novel zinc-finger homeodomain proteins. Mech. Dev. 34, 123-134 [DOI] [PubMed] [Google Scholar]
- Lee H., Frasch M. (2000). Wingless effects mesoderm patterning and ectoderm segmentation events via induction of its downstream target sloppy paired. Development 127, 5497-5508 [DOI] [PubMed] [Google Scholar]
- Lee H. H., Norris A., Weiss J. B., Frasch M. (2003). Jelly belly protein activates the receptor tyrosine kinase Alk to specify visceral muscle pioneers. Nature 425, 507-512 [DOI] [PubMed] [Google Scholar]
- Lee H.-H., Zaffran S., Frasch M. (2005). The development of the visceral musculature of Drosophila. In Drosophila Muscle Development (ed. Sink H.). Georgetown, TX: Landes; [Google Scholar]
- Lengyel J., Iwaki D. (2002). It takes guts: the Drosophila hindgut as a model system for organogenesis. Dev. Biol. 213, 1-19 [DOI] [PubMed] [Google Scholar]
- Leptin M. (1991). twist and snail as positive and negative regulators during Drosophila mesoderm formation. Genes Dev. 5, 1568-1576 [DOI] [PubMed] [Google Scholar]
- Lewis E. P., Bacher F. (1968). Method of feeding ethyl methane sulfonate (EMS) to Drosophila males. Dros. Inf. Serv. 43, 193 [Google Scholar]
- Lo P. C., Zaffran S., Senatore S., Frasch M. (2007). The Drosophila Hand gene is required for remodeling of the developing adult heart and midgut during metamorphosis. Dev. Biol. 311, 287-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Richardson J. A., Olson E. N. (1998). Capsulin: a novel bHLH transcription factor expressed in epicardial progenitors and mesenchyme of visceral organs. Mech. Dev. 73, 23-32 [DOI] [PubMed] [Google Scholar]
- Lu J., Webb R., Richardson J. A., Olson E. N. (1999). MyoR: a muscle-restricted basic helix-loop-helix transcription factor that antagonizes the actions of MyoD. Proc. Natl. Acad. Sci. USA 96, 552-557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal L., Dumstrei K., Hartenstein V. (2004). Role of FGFR signaling in the morphogenesis of the Drosophila visceral musculature. Dev. Dyn. 231, 342-348 [DOI] [PubMed] [Google Scholar]
- Mann R. S., Morata G. (2000). The developmental and molecular biology of genes that subdivide the body of Drosophila. Annu. Rev. Cell Dev. Biol. 16, 243-271 [DOI] [PubMed] [Google Scholar]
- Miyagishi M., Hatta M., Ohshima T., Ishida J., Fujii R., Nakajima T., Fukamizu A. (2000). Cell type-dependent transactivation or repression of mesoderm-restricted basic helix-loop-helix protein, POD-1/Capsulin. Mol. Cell. Biochem. 205, 141-147 [DOI] [PubMed] [Google Scholar]
- Mohr S. E., Gelbart W. M. (2002). Using the P[wHy] hybrid transposable element to disrupt genes in region 54D-55B in Drosophila melanogaster. Genetics 162, 165-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. T., Xu X. (1998). Drosophila mef2 expression during mesoderm development is controlled by a complex array of cis-acting regulatory modules. Dev. Biol. 204, 550-566 [DOI] [PubMed] [Google Scholar]
- Pignoni F., Baldarelli R., Steingrimsson E., Diaz R., Patapoutian A., Merriam J., Lengyel J. (1990). The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell 62, 151-163 [DOI] [PubMed] [Google Scholar]
- Pipes G. C., Lin Q., Riley S. E., Goodman C. S. (2001). The Beat generation: a multigene family encoding IgSF proteins related to the Beat axon guidance molecule in Drosophila. Development 128, 4545-4552 [DOI] [PubMed] [Google Scholar]
- Pisarev A., Poustelnikova E., Samsonova M., Reinitz J. (2009). FlyEx, the quantitative atlas on segmentation gene expression at cellular resolution. Nucleic Acids Res. 37, D560-D566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaggin S. E., Vanden Heuvel G. B., Igarashi P. (1998). Pod-1, a mesoderm-specific basic-helix-loop-helix protein expressed in mesenchymal and glomerular epithelial cells in the developing kidney. Mech. Dev. 71, 37-48 [DOI] [PubMed] [Google Scholar]
- Quaggin S. E., Schwartz L., Cui S., Igarashi P., Deimling J., Post M., Rossant J. (1999). The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development 126, 5771-5783 [DOI] [PubMed] [Google Scholar]
- Reim I., Frasch M. (2005). The Dorsocross T-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Development 132, 4911-4925 [DOI] [PubMed] [Google Scholar]
- Reim I., Lee H. H., Frasch M. (2003). The T-box-encoding Dorsocross genes function in amnioserosa development and the patterning of the dorsolateral germ band downstream of Dpp. Development 130, 3187-3204 [DOI] [PubMed] [Google Scholar]
- Reuter R., Leptin M. (1994). Interacting functions of snail, twist and huckebein during the early development of germ layers in Drosophila. Development 120, 1137-1150 [DOI] [PubMed] [Google Scholar]
- Robb L., Hartley L., Wang C. C., Harvey R. P., Begley C. G. (1998a). musculin: a murine basic helix-loop-helix transcription factor gene expressed in embryonic skeletal muscle. Mech. Dev. 76, 197-201 [DOI] [PubMed] [Google Scholar]
- Robb L., Mifsud L., Hartley L., Biben C., Copeland N. G., Gilbert D. J., Jenkins N. A., Harvey R. P. (1998b). epicardin: a novel basic helix-loop-helix transcription factor gene expressed in epicardium, branchial arch myoblasts, and mesenchyme of developing lung, gut, kidney, and gonads. Dev. Dyn. 213, 105-113 [DOI] [PubMed] [Google Scholar]
- San Martin B., Ruiz-Gomez M., Landgraf M., Bate M. (2001). A distinct set of founders and fusion-competent myoblasts make visceral muscles in the Drosophila embryo. Development 128, 3331-3338 [DOI] [PubMed] [Google Scholar]
- Sandmann T., Girardot C., Brehme M., Tongprasit W., Stolc V., Furlong E. E. (2007). A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 21, 436-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröter R. H., Buttgereit D., Beck L., Holz A., Renkawitz-Pohl R. (2006). Blown fuse regulates stretching and outgrowth but not myoblast fusion of the circular visceral muscles in Drosophila. Differentiation 74, 608-621 [DOI] [PubMed] [Google Scholar]
- Singer J., Harbecke R., Kusch T., Reuter R., Lengyel J. (1996). Drosophila brachyenteron regulates gene activity and morphogenesis in the gut. Development 122, 3707-3718 [DOI] [PubMed] [Google Scholar]
- Swan L. E., Wichmann C., Prange U., Schmid A., Schmidt M., Schwarz T., Ponimaskin E., Madeo F., Vorbruggen G., Sigrist S. J. (2004). A glutamate receptor-interacting protein homolog organizes muscle guidance in Drosophila. Genes Dev. 18, 223-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B., Stoetzel C., Gorostiza-Thisse C., Perrin-Schmitt F. (1988). Sequence of the twist gene and nuclear localization of its protein in endomesodermal cells of early Drosophila embryos. EMBO J. 7, 2175-2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P., Beaton A., Weiszmann R., Kwan E., Shu S., Lewis S., Richards S., Ashburner M., Hartenstein V., Celniker S., et al. (2002). Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3, research0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Scheven G., Bothe I., Ahmed M. U., Alvares L. E., Dietrich S. (2006). Protein and genomic organisation of vertebrate MyoR and Capsulin genes and their expression during avian development. Gene Expr. Patterns 6, 383-393 [DOI] [PubMed] [Google Scholar]
- Wakabayashi-Ito N., Ip Y. T. (2005). Mesoderm formation in the Drosophila embryo. In Drosophila Muscle Development (ed. Sink H.). Georgetown, TX: Landes; [Google Scholar]
- Weigel D., Juergens G., Kuettner F., Seifert E., Jäckle H. (1989). The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell 57, 645-658 [DOI] [PubMed] [Google Scholar]
- Werz C., Lee T. V., Lee P. L., Lackey M., Bolduc C., Stein D. S., Bergmann A. (2005). Mis-specified cells die by an active gene-directed process, and inhibition of this death results in cell fate transformation in Drosophila. Development 132, 5343-5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran S., Küchler A., Lee H. H., Frasch M. (2001). biniou (FoxF), a central component in a regulatory network controlling visceral mesoderm development and midgut morphogenesis in Drosophila. Genes Dev. 15, 2900-2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J., Zinzen R. P., Stark A., Kellis M., Zhang H., Young R. A., Levine M. (2007). Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 21, 385-390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Schnitzler A., Agapite J., Schwartz L. M., Steller H., Nambu J. R. (1997). Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system midline cells. Proc. Natl. Acad. Sci. USA 94, 5131-5136 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.