Abstract
In recent years, increasingly sophisticated computational and bioinformatics tools have evolved for the analyses of protein structure, function, ligand interactions, modeling and energetics. This includes the development of algorithms to recursively evaluate side-chain rotamer permutations, identify regions in a 3D structure that meet some set of search parameters, calculate and minimize energy values, and provide high-resolution visual tools for theoretical modeling. Here we discuss the interdependency between different areas of bioinformatics, the evolution of different algorithm design approaches, and finally the transition from theoretical models to real-world design and application as they relate to Ca2+-binding proteins. Within this context, it has become evident that significant pre-experimental design and calculations can be modeled through computational methods, thus eliminating potentially unproductive research and increasing our confidence in the correlation between real and theoretical models. Moving from prediction to production, it is anticipated that bioinformatics tools will play an increasingly significant role in research and development, improving our ability to both understand the physiological roles of Ca2+ and other metals and to extend that knowledge to the design of function-specific synthetic proteins capable of fulfilling different roles in medical diagnostics and therapeutics.
Keywords: Calcium-binding, EF-hand, prediction, algorithm, design, statistics, graph theory, machine learning
1. INTRODUCTION
The importance of Ca2+ in biological systems has been well-established, yet research continues to identify new and diverse functions associated with this metal. Calcium’s biological role is mainly fulfilled by its interaction with different classes of Ca2+-binding proteins (CaBPs) [1–3]. Intracellularly, calcium binding to trigger proteins such as calmodulin and troponin C with helix-loop-helix motifs results in Ca2+-induced conformational change which in turn mediates the activity of different cellular processes [4]. Buffer proteins such as calbindin D9k and parvalbumin are essential to maintain proper calcium homeostasis. Conversely, many extracellular CaBPs, such as metabotropic glutamate receptors (mGluRs) and calcium sensing receptors, have weak Ca2+-binding affinities (~mM) yet are essential for extracellular calcium signaling and cell-cell communication [5, 6].
Currently the availability of sophisticated computational methods and resources has become indispensible as research tools. These methods may be directed at the prediction and identification of Ca2+-binding sites; determining the function of CaBPs in a biological system; tracing or predicting the evolutionary path of CaBPs; analyzing the effects of competing, toxic metals on CaBP functions; and engineering binding sites for Ca2+ or other metal ions either for pure research or for various biotechnological applications, including medical imaging, drug delivery and cancer therapies.
As with any computational approach, the analyses of known data provide an important foundation for developing input parameters. For studies of CaBPs, this includes the structural, physical and chemical properties associated with Ca2+-binding proteins in both the bound (holo) and unbound (apo) states. Fortunately, the increasing availability of bioinformatics tools such as databases, programs and algorithms have provided researchers with the means to statistically evaluate large samples [7–10].
In this review we begin with a discussion of current knowledge related to (1) structural parameters associated with Ca2+-binding (i.e., ligand number and type, bond angles, geometric configurations and charge distributions) and (2) dynamic properties including conformational change and charge interactions. Other important dynamic properties, specifically affinity, selectivity, energy minimization and molecular dynamics, have been excluded from this review due to space limitations.
As the data presented here were largely obtained through extensive statistical analyses or mathematical models, we then discuss their significance with respect to the development of algorithms designed to predict or identify either Ca2+-binding proteins or specific binding sites, followed by an overview and comparison of the algorithms. Finally, we discuss specific examples detailing experimental results that were obtained as a result of pre-experimental application of algorithms to design or predict Ca2+-binding sites.
2. DEFINING CA2+-BINDING THROUGH STATISTICAL ANALYSES
a. Distribution of Ligand Number and Types
In broad terms, metal-binding sites are characterized by a central shell of hydrophilic ligands to chelate the ion, with a surrounding shell of hydrophobic residues [11, 12]. The centers of high hydrophobic contrast, where ions are located, are typically solvent inaccessible, low-dielectric cavities that enhance electrostatic metal-ligand interactions [13–15]. Binding ligand atoms for common biologically-important metals (e.g. − Fe3+, Mg2+, Zn2+, Ca2+, Mn2+) are mainly oxygen, nitrogen and sulfur from sidechain groups, and oxygen from mainchain carbonyls [13, 14, 16–23]. Although nitrogen is frequently considered a potential Ca2+ ligand based on small molecule interactions [21–23], it is apparent that interactions between sulfur and nitrogen from proteins with Ca2+ are negligible. According to an explanation of the orbital chemistry relevant to ligand binding, as well as an understanding of how metal-ligand binding relates to polarizability of different classes of Lewis acid metals, the preference of Ca2+ for oxygen can be understood from its hard Lewis acid definition describing less covalent interaction with a more rigid ionic radius [24].
The coordination of Ca2+-binding utilizes various types of oxygen atoms from sidechain carboxyl groups (Asp, Glu), carboxamide groups (Asn, Gln), hydroxyl groups (Ser, Thr), mainchain carbonyl oxygen atoms from most residues, cofactors, and water molecules. The majority of all Ca2+-binding ligands originate from turn/loop regions, rather than helix and sheet regions [12, 25, 26]. This distribution persists even after EF-Hand sites are deliberately excluded [26].
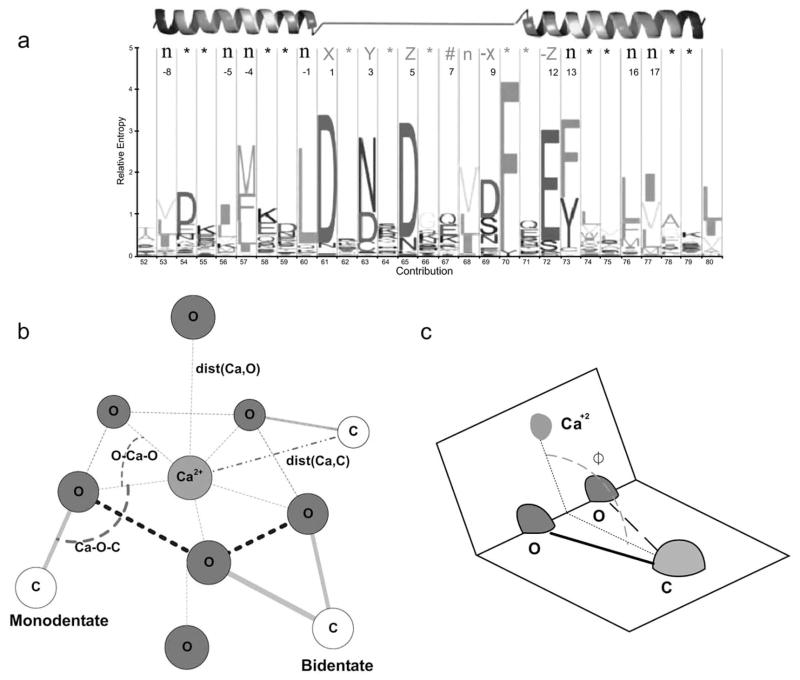
The coordination number is frequently reported in the range of 3–8 ligand atoms [27–29]. However, certain classes of Ca2+-binding sites exhibit preferences with respect to residue type and coordination number. In EF-Hand binding sites [26, 30–32], certain residues appear consistently at specific positions within the loop (Fig. (1a)), with some variations apparent based on the extent of binding sites analyzed. Collectively these include: Asp at position 1, Asp or Asn at position 3, Gly at position 6, Ile, Val or Leu at position 8, and Glu at position 12 [33–35].
Fig. 1.
(a) Canonical EF-Loop binding site. (b) Illustration of key EF-Hand structural characteristics. The physical relationships between the Ca2+ ion (Ca), the ligand oxygen (O), and the ligand oxygen atoms covalently-bound carbon (C) are defined by the angle Ca-O-C and distances dist(Ca,C) and dist(Ca,O). (c) Dihedral angle of bidentate ligands between two planes formed by O-Ca-O and O-C-O.
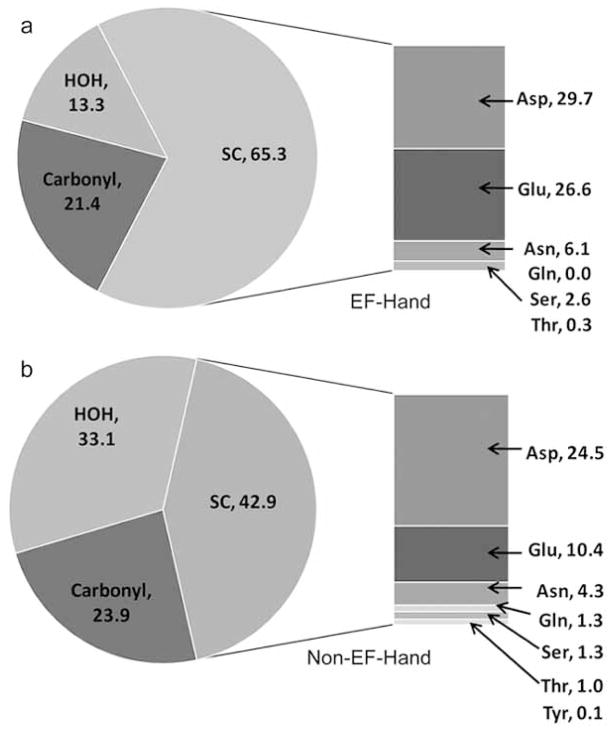
A recent statistical analysis of the structural characteristics of both EF-Hand and Non-EF-Hand proteins conducted in our laboratory revealed significant structural diversity in Non-EF-Hand proteins [28]. The dataset used for our analysis was comprised of 1605 binding sites from 558 Protein Data Bank (PDB) files with resolution R ≤ 2.0 Å, based on a cutoff distance of 3.5 Å as the maximum ligand distance for oxygen and nitrogen, and following removal of all structures with 90% or greater sequence homology. The coordination numbers reported were similar to previous studies indicating an average between 6–7 ligands [26, 32]. Non-EF-Hand sites utilize lower coordination numbers (6 ± 2 vs. 7 ± 1), fewer protein ligands (4 ± 2 vs. 6 ± 1) and more water ligands (2 ± 2 vs. 1 ± 0) than EF-Hand sites. The orders of ligand preference for Non-EF-Hand and EF-Hand sites, respectively, were H2O (33.1%) > sidechain Asp (24.5%) > mainchain carbonyl (23.9%) > sidechain Glu (10.4%), and sidechain Asp (29.7%) > sidechain Glu (26.6%) > mainchain carbonyl (21.4%) > H2O (13.3%).
The Non-EF-Hand proteins exhibit increased solvent interaction although the data for EF-Hand sites follow previously-reported trends (carboxylates > carbonyls > water > hydroxyl atoms) [14]. Reducing the cutoff to 2.9 Å to compare distributions of water oxygen ligands (Table 1) revealed only a modest difference from previous studies for Non-EF-Hand sites [15, 29], although a more significant difference was observed with the 13.3% reported for EF-Hand sites, where all water ligands fell within 2.9 Å of the ion. Additionally, while the results obtained with respect to coordination number for Non-EF-Hand binding sites are consistent with those reported by Pidcock [26], they differed significantly from 6.7 and 7.0 reported in earlier studies which did not exclude EF-Hand sites [29, 36].
Table 1.
Comparison of Ligand Distributions as Percentages of Total Ligand Numbers
For Non-EF-Hand sites, the higher than expected percentage of Asp residues and lower than expected percentage of Glu residues involved in calcium binding sites may be due to the more common occurrence of Asp in Asx turns, or a preference by Ca2+ for less sterically bulky sidechains [26]. However, the more equivalent distribution of Asp and Glu in EF-Hand sites is less readily understood. Canonical EF-Hand sites typically include 2–3 Asp and 1 Glu residues that bind Ca2+ with carboxyl oxygen atoms. Glu residues are of strategic importance in EF-Hand binding sites as bidentate, anchoring ligands for Ca2+ [29, 37–40] and exhibit strong propensity towards helix formation which is consistent with its observed distribution in EF-Hand binding sites (Fig. (2)).
Fig. 2.
Distribution of ligand residues between water, carbonyl and sidechain (SC) residue oxygen ligands for (a) EF-Hand and (b) Non-EF-Hand.
b. Binding Site Geometry
In EF-Hand binding sites, the ligand oxygen atoms, usually six to seven, exhibit bipyramidal/monopyramidal pentagonal geometry (Fig. (1b)). Five oxygen atoms of the pentagon and Ca2+ lie nearly on the same plane, with variations [29, 37, 41]. The O-Ca-O angles in the pentagonal plane (Fig. (1b)) are approximately 72°. In monopyramidal geometry, the extraplanar peak oxygen atom is usually provided by an Asp/Asn sidechain, and the line between it and the calcium ion is nearly vertical with the pentagonal plane. In the bipyramidal geometry, the second extraplanar oxygen atom is typically from water. Other Non-EF-Hand Ca2+-binding sites usually do not have similar, well-defined geometrical structures.
The mean Ca-O ligand binding distance has been reported, from different studies, as either 2.4 ± 0.1 Å or 2.42 Å with a range of 2.01 – 3.15 Å [2, 14, 24, 42–45]. The mean Ca–O distance values reported for EF-Hand and Non-EF-Hand indicate very little difference between carbonyl and side-chain oxygens, and between the different classes for each ligand type. However, bidentate ligand distances are slightly longer for both EF-Hand (2.5 ± 0.2 Å) and Non-EF-Hand (2.6 ± 0.3 Å) than for the carbonyl and side-chain ligands in their respective classes [28]. A more pronounced change was observed for the bidentate mean Ca–C distances, which were 0.5–0.6 Å shorter than the distances found for carbonyl and side-chain ligand oxygen atoms, resulting in overlap between the Ca–O and Ca–C shells [28].
The mean Ca–O–C angles (Fig. (1b)) are observed to be different among carbonyl, side-chain, and bidentate oxygen ligands. The Ca–O–C angles were largest for carbonyl (151.5 ± 15.8° and 159.8 ± 12.5°), followed by side chain (140.4 ± 15.2° and 136.7 ± 16.0°) and bidentate (93.6 ± 11.3° and 92.9 ± 6.8°) for Non-EF-Hand and EF-Hand, respectively. It was observed that a Gaussian distribution of Ca–O–C angle values is associated with Non-EF-Hand ligands, and the range values for both classes are nearly identical for carbonyl, side chain, and bidentate.
For the bidentate ligands, the dihedral angle is the angle between the plane formed by the side-chain carboxyl group (–COO), and the plane formed by the two carboxyl oxygen atoms and the Ca2+ ion (Fig. (1c)). For Non-EF-Hand and EF-Hand the mean and standard deviation values for dihedral angles were found to be 168.1 ± 9.7° and 170.6 ± 7.1°, indicating that they almost lie in the same plane.
This analysis presented a variety of flexible coordination schemes associated with Ca2+-binding and tremendous structural diversity, consistent with earlier conclusions by Martin [46].
c. Charge in the Binding Site Microenvironment
Formal charge (FC) by site is typically simplified to account only for negatively charged side-chain carboxyl groups (−1) from Glu and Asp [28, 34]. Mean negative formal charge values of 1 ± 1 and 3 ± 1 have been reported for Non-EF-Hand and EF-Hand sites, respectively. However, only a small percentage of Non-EF-Hand sites exhibit a negative charge greater than 2. While binding affinity may be enhanced by an increase in the number of negatively-charged ligands, there appears to be an optimal level for this [47, 48] which varies based on ion charge (e.g., trivalent cations are better charge acceptors than divalent cations) and the presence of bidentate ligands in the binding site [13–15]. However, recent work in our laboratory has demonstrated that increasing the number of negatively-charged ligands from three to five in an engineered Ca2+-binding site increases the binding affinity for both Ca2+ and La3+ [49]. Additionally, removal of charges distant from the binding site may lead to significant decreases in binding affinity [50] due to reduction in the calcium kinetic on-rate [51]. Ca2+-binding sites with high negative formal charges are likely located in flexible loop regions of the protein [52]. Binding sites of zero formal charge were identified within EF-Hand sites, such as the protein calprotectin (1xk4.pdb) [53], and Non-EF-Hand proteins, although this was predictably more evident in Non-EF-Hand (20%) than EF-Hand (4%) sites. Detailed structural analyses of the protein environments of these charge-deficient sites reveal that charge–charge stabilization beyond the chelated metal ion can lead to the exclusion of available negatively charged side-chain residues and facilitate the binding of Ca2+ with carbonyl oxygen atoms [28].
Increasing the number of bidentate ligands is energetically favorable, and bidentate binding may allow for an increase in the number of charged ligands in the binding site. This subsequently leads to better protection for the cation from other intracellular anions. The carboxlyate binding mode is highly-significant with respect to Ca2+-binding, as the conserved bidentate anchor in EF-Hand proteins is instrumental in conformational changes associated with protein function [54, 55].
d. Calcium-Induced Conformational Change
The comparative analyses of static properties may be further extended to evaluate dynamic aspects of Ca2+-binding. A significant body of research is available related to the mechanistic and functional aspects of protein conformational change associated with binding [7, 55–60]. In general, these conformational analyses can be divided into two categories: the analyses of conformational changes of individual proteins and proteins on a database scale.
Vertical studies (i.e., between native and metal-complexed structures of the same protein) for individual proteins have compared changes in interatomic distances and angles, and hydrophobicity or β-factor changes associated with Ca2+-binding. Zhang et al. reported the Ca2+-induced conformational transition observed by comparing NMR structures of apo- and holo-calmodulin [55]. Chrysina et al. compared the X-ray structures of apo- and holo-α-lactalbumin with respect to Ca2+-induced protein folding [59]. In addition, protein conformational changes have been investigated using simulations (e.g., Molecular Dynamics, Monte Carlo simulations) [61–63]. In a different approach, Dudev and Lim [15] used ab initio calculations, continuum dielectric methods (CDM) representing different dielectric media, and density functional theory (DFT) to compute thermodynamic values related to the exchange of metal-bound water molecules with small molecule analogs for Asp and Glu (formate); Asn, Gln and backbone peptide (forma-mide); and His (imidazole). For Ca2+ sites, which tend to have higher coordination numbers (CN) than Mg2+ or Zn2+, the presence of more charged ligands leads to higher charge repulsion in the binding site, thus increasing the cavity size and contributing to the selectivity of Ca2+ over the more abundant Mg2+.
Serial studies (i.e., between datasets of different proteins) for comparison of protein conformational changes on a database scale have evaluated different calcium-binding pockets for common features. These analyses are dependent upon (1) the availability of empirical and experimental data deposited in the PDB, and (2) the methodologies and measurements used to analyze the dataset. A database-scaled study by Babor et al. analyzed 58 apo/holo pairs of calcium binding sites from the PDB to evaluate conformational change based on comparing changes in Root Mean Square Deviation (RMSD), distance, angle, β-factor and solvent accessibility as a result of Ca2+-binding [64]. Babor et al. evaluated conformational changes between the apo/holo forms, where conformational change was defined based on side-chain dihedral angles that were required to differ by a minimum of 40° between structures to qualify as a conformational change. Results of this study suggested that approximately 20% of Ca2+-binding sites undergo backbone rearrangements upon Ca2+ binding, while only a small percentage of binding sites (~5%) exhibits significant rotation in more than two side-chains if the backbone is not rearranged upon calcium-binding.
Conversely, Eyal et al., analyzing conformational change associated with point mutations, considered a 60° change as a cutoff for defining conformational change [9]. Similarly, Zhao et al. evaluated multiple paired PDB protein structures that did not involve metal-protein complex formation and concluded that the residue angle differences in terms of one paired entry are intrinsic properties of the individual residues, and that inaccurate conclusions may be drawn based on arbitrary selection of a single cutoff value for all residues [65].
Future comparison of the holo- and apo- protein forms of NMR structures on a database scale will provide additional important information on dynamic conformational changes in solution.
e. Methodologies to Identify Ca2+-Binding Sites
While statistical analyses identify structural, chemical and energetic characteristics necessary for the characterization of Ca2+-binding sites, computational methods are required to exploit these data to identify or predict the location of Ca2+-binding sites in the protein structure. These methodologies include: homology analyses against known Ca2+-binding sequences or structures; energy or pseudo-energy calculations using Ca2+-binding ligands (e.g., oxygen clusters) or rotamer libraries of Ca2+-binding residues; grid functions; Bayesian statistical methods or scoring functions based on Ca2+-binding characteristics (e.g., charge, distance); and graph theory applications identifying Ca2+-binding ligand clusters. Many current applications combine two or more of these methods with statistical parameters as filters. Details and examples of these methodologies are discussed in the next section.
3. ALGORITHMS: EVOLUTION, DESIGN AND APPLICATIONS
a. Primary Sequence Analysis and Structure Prediction
Ca2+-binding sites may be predicted or identified based on primary sequence or three-dimensional tertiary structure. Software is also available to predict or identify secondary structure, including PDBSum (http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/). Prediction is dependent on matrices that evaluate existing databases to provide predictions based on known sequence and structure data, which may include static structural information (e.g., ligand number, ligand type, bond angles, etc.) and dynamic parameters (e.g., binding affinity).
Significant achievements have been made in the prediction or identification of Ca2+-binding sites utilizing programs which align primary sequences with related conserved regions. Examples of these would include the MacVector package (Oxford Molecular Group), ClustalW (http://www.ebi.ac.uk/clustalw/), FASTA (http://www.ebi.ac.uk/Tools/fasta/index.html) and BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Similarly, programs such as CALM [66] and CaPS [35] apply pattern matching to identify target motifs in a sequence based on derived flexible patterns. In canonical EF-Hand proteins first analyzed by Kretsinger et al. [67], Ca2+-binding sites were found to be highly conserved with respect to a helix-loop-helix motif and, to a certain degree, by the relative locations of certain residues and residue ligand preference within the motif regions. Variants of the EF-Hand, including the lesser conserved pseudo EF-Hand and EF-Hand like motifs, also demonstrate less pronounced but recognizable primary sequence patterns. Patterns describing these motifs are usually generated by first conducting multiple sequence alignments and identifying highly-conserved residues. Accordingly, the sequence pattern search method is a simple and efficient way to predict EF-Hand and EF-Hand like sites in proteins.
Using the sequence searching software CALM [66] which was designed to search for continuous calcium binding sites, Bertrand and coworkers [68] identified five calcium binding amino acid sequences which possessed significant sequence homology with EF-Hand III loop of calmodulin in the N-terminal domain of neuronal nicotinic receptors. All of these sequences were rich in hydrophilic and acidic amino acids and included a terminal glutamate. The site-directed mutation of all the Glu to Gln residues in the five sequences reduced the Ca2+ binding affinity, thus providing confirmation of the predicted binding ligands.
Similarly, Egmond and coworkers [69] reported identification of a calcium binding site in Staphylococcus hyicus Lipase (SHL) by sequence alignment with other staphylococcal lipases. Sequence alignment was conducted by MacVector package (Oxford Molecular Group) [70]. Replacing predicted Ca2+-binding ligands generated a Ca2+-insensitive staphylococcus hyicus lipase.
Zhou et al. developed new PROSITE-like motifs for the prediction of EF-Hand and EF-Hand-Like Ca2+-binding sites by first conducting extensive sequence analyses across various databases and then utilizing these results (Fig. (1a)) as the basis for sequential patterns [35]. These patterns were then applied to bacterial proteins on a database scale. A total of 467 canonical EF-Hand and zero pseudo EF-Hand sites were identified, which suggested a later phylogenetical evolution of pseudo EF-Hand compared with canonical EF-Hand motif. An online server for CaPS is available at http://lithium.gsu.edu/faculty/Yang/Calciomics.htm. Zhou et al. later applied CaPS to identify a putative EF-Hand calcium binding motif within the rubella virus [71]. Similar prediction results were obtained using the programs PSIPRED [72], JPRED [73], and PredictProtein [74] to analyze the helix-loop-helix structure of the predicted calcium binding sequence. Parallel evidence came from similar prediction results obtained using the GG algorithm [75] based on a homology model of the rubella virus nonstructural pro-tease generated by the program SWISS-MODEL [76]. Experimental validation was obtained when the predicted EF-Hand motif from rubella virus was grafted onto a scaffold protein CD2, the wild-type form of which exhibits no Ca2+-binding capability [77]. This grafting approach allowed for analysis of the intrinsic Ca2+-binding capability of an isolated Ca2+-binding site. Site-directed mutagenesis of presumed Ca2+-binding ligands was conducted, and subsequent affinity studies validated the predicted site, which is believed to be required for the stability of rubella virus nonstructural protease under physiological conditions [71].
b. Prediction Based on Structural Data: Algorithm Design, Parameter Definition
Currently, most efforts to predict Ca2+-binding sites may more accurately be described as the identification of known Ca2+-binding sites using some computational methods or combination of methods to accurately locate a documented Ca2+ ion when little or no significant conformational change is observed. Conversely, true prediction will be achieved when the location of a Ca2+ binding site in the holo (loaded) form of the protein can be determined in silico based on only the apo (free) protein structure or the sequence alone. Many prediction algorithms employ structural parameters as distinguishing factors between Ca2+-binding and nonbindingsites. This includes parameters related to ligand number and type and binding geometry restrictions related to distance and angle values between Ca2+ and its ligands as well as between the ligands with each other. Frequently, multiple algorithms are combined, typically using some type of scoring function with a grid system or other method of spatial analysis (e.g., graph theory) to evaluate physicochemical characteristics systematically and identify either binding ligands or binding sites based on comparative scoring. It is relevant to note that these functions are dependent on a priori statistical analyses to define or constrain function parameters based on structural, chemical or thermodynamic data. Prediction algorithms discussed in this section are summarized in Table 2.
Table 2.
Summary of Prediction Algorithms
| Name | Input | Output | Method | Applicability | |
|---|---|---|---|---|---|
| a Zhou | CaPS | Primary sequence | Binding pattern & region | Sequence pattern search | Identify EF-Hand and EF-like Ca2+– binding sites in proteins |
| b Yamashita | --- | PDB file | Ca2+ coordinates | Hydrophobicity contrast function | Predict metal (including Ca2+) binding sites of insignificant conformational changes |
| c Nayal | --- | PDB file | Ca2+ coordinates | Valence function | Predict Ca2+– binding sites of insignificant conformational changes |
| d Wei | Feature | PDB file | Ca2+ coordinates & physiochemical properties | Probability scoring function | Predict Ca2+– binding sites of insignificant conformational changes |
| e Lin | SVMProt | Primary sequence | Functional families of the input sequence | Machine learning | Assign functional families (including Ca2+ and metal binding) to the input sequence |
| f Schymkowitz | --- | PDB file | Ca2+ coordinates & estimated binding affinity | Fold-X force field | Predict metal (include Ca2+) binding sites of insignificant conformational changes |
| g Deng | GG | PDB file | Ca2+ coordinates & ligand oxygen atoms | Graph theory | Predict Ca2+–binding sites with minor conformational changes |
| h Wang | MUG | PDB file | Ca2+ coordinates & ligand oxygen atoms | Graph theory | Predict Ca2+–binding sites with minor conformational changes |
| i Babor | CHED | PDB file | Binding residues | Rotamer | Predict transition metal–binding sites with possible side-chain movement |
| j Hellinga | DEZYMER | PDB file | Binding residues & possible mutations | Rotamer | Suggest mutations for Ca2+-binding based on a PDB file |
| k Dahiyat | ORBIT | Backbone coordinates | Sequence appropriate for the backbone fold | Rotamer library & optimization | De novo design of protein (including CaBP) sequence based on a desired backbone fold |
HYDROPHOBICITY CONTRAST FUNCTION
One of the first programs to predict calcium binding sites in three-dimensional structures was based on a hydrophobicity contrast function, which identified centers of high hydrophobic contrast as the metal-binding sites [11]. To quantitatively evaluate the correlation between points of high hydrophobicity expressed by the function and metal-binding sites, this program examined the structures of 10 metalloproteins and 13 metal-host molecules (five with documented Ca2+-binding sites) with metal ions deleted. By embedding the structure into 0.5 Å grids and then computing at each grid the value of the function, the program was able to verify whether grid points returning high values were indeed near the metal-binding sites.
COMBINATORY ALGORITHMS
An early FORTRAN algorithm for predicting Ca2+-binding sites, which closely followed the approach reported by Yamashita [11], combined a fine grid system with a scoring function to describe points of valency based on electrostatic contribution of ligand atoms [27]. The scoring function parameters for the prediction of Ca2+-binding were determined based on a priori analysis of the 62 available Ca2+-binding proteins deposited in the PDB at that time which revealed that 371 of the 376 inner coordination shell atoms (i.e., within 2.7 Å) were oxygen. Based on these data, this algorithm required that Ca2+ be ligated by at least three protein oxygen atoms, where the metal-ligand distance must be smaller than 3.4 Å but larger than the van der Waals radius between the two. Each point in the grid system was then evaluated to determine if the oxygen coordination requirements were met and then a valence score was assigned to that point. An identification-removal procedure was used to iteratively eliminate all valence points within 3.5 Å of the highest valence point identified, leaving only the highest valence point in that region as the Ca2+-binding site. Results demonstrated that 58/62 Ca2+-binding sites had valence greater than or equal to 1.4 and that 87% of the high valence points fell within 1.0 Å of the documented Ca2+ ion.
The Fold-X algorithm combines geometric pattern matching and energy calculations based on Fold-X empirical force field [78, 79], where Ca2+-binding sites are constrained to four to six ligand oxygen atoms. In the preprocess stage, the algorithm first identifies canonical positions [80] of Ca2+ with respect to each type of oxygen atom. The canonical ions are then superimposed onto a given protein where conflicting canonical ions are either removed or fused together depending on their relative proximities. Finally, the Fold-X force field [35, 79] is used for energy calculation and consequently for the optimization of the position of the predicted calcium ion. This algorithm can also coarsely predict binding affinity based on energy-optimized placement of the Ca2+ ion to distinguish between low and high affinity sites, as well as high affinity Ca2+- and Mg2+-binding sites. Additionally, this approach was applicable to predicting Mg2+, Ca2+, Zn2+, Mn2+, and Cu2+ and water molecules associated with the structure. Although these results indicate higher prediction rates than previous efforts, this method was less successful at identifying either multiple binding sites or sites with lower coordination numbers (CN<4).
GRAPH THEORY-BASED ALGORITHMS
The program GG (Graph theory and Geometry) presented a new approach to prediction based on a combination of graph theory and geometric parameters [75]. Based on the established parameters associated with Ca2+-binding, the GG algorithm was designed to identify oxygen clusters as the basis for predicting Ca2+-binding sites. In this method a graph G(V, E) was first constructed with oxygen atoms as its vertices and edges between two vertices if their spatial distance was no more than 6.0 Å. Oxygen clusters corresponding to cliques in G(V, E) consisting of exactly four vertices were identified, and the calcium center was determined at an equidistant center within each cluster if the distance ranged from 1.8 to 3.0 Å. If no such qualified center was located, that cluster was abandoned; otherwise, the center was the predicted calcium position and the atoms in the cluster were the predicted ligand atoms. This algorithm did not include a procedure to merge overlapping, predicted Ca2+ locations, resulting in multiple, adjacent predicted sites near the documented Ca2+-binding site. Nonetheless, this algorithm produces rapid results, mainly due to the absence of a grid algorithm, and achieves approximately 90% site sensitivity and 80% site selectivity on testing datasets. These results are comparable to those reported by other algorithms. GG was implemented in C++. An online server is available at http://chemistry.gsu.edu/faculty/Yang/Calciomics.htm.
To fully utilize known geometric properties such as distance, angles and dihedral angles of Ca2+-binding sites, which are essential in pinpointing Ca2+ position, Wang et al. developed the MUG (MUltiple Geometries) program, which is able to predict Ca2+–binding sites with different coordination numbers in proteins with atomic resolution. MUG requires that potential calcium-binding sites consist of at least four oxygen atoms [82] although MUG further allows oxygen atoms from either water or cofactors in addition to amino acid residues. Spatial analysis is achieved based on graph theory modeling as well as imposition of a grid system. After first identifying all possible oxygen clusters by finding maximal cliques, a calcium center (CC) for each cluster, corresponding to the potential Ca2+ position, is located to maximally regularize the structure of the (cluster, CC) pair. The structure is then inspected by geometric filters. An unqualified (cluster, CC) pair is further handled by recursively removing oxygen atoms and relocating the CC until its structure is either qualified or contains fewer than four ligand atoms. Ligand coordination is then determined for qualified structures. MUG also has the capability of predicting both low coordination sites and multiple Ca2+-binding sites that share a common ligand. The MUG algorithm was implemented in Java and is available for use through an online server at http://chemistry.gsu.edu/faculty/Yang/Calciomics.htm.
MACHINE LEARNING TECHNIQUES
Machine learning techniques, which are general approaches to any classification problem, are particularly effective at classifying ligands as binding residues and determining whether the target atom is close enough to the ion for binding [81, 85, 86]. In this method, a window including neighboring residues slides through the sequence so that each successive residue is viewed as a target residue in turn, to identify binding residues. The training procedures include applying the same learning machine to a training dataset consisting of sequences with known metal-binding residues. Classification features include location in the primary sequence, secondary structure, metal-binding propensity, and solvent accessibility. Additional information on machine learning methods for predicting functional classes can be found through the online server SVMProt [87] at http://jing.cz3.nus.edu.sg/cgi-bin/svmprot.cgi.
TOWARDS APO STRUCTURE PREDICTION
Wei and Altman [25] reported the development of the FEATURE program (a machine learning system) that utilizes a Bayesian approach to classify potential Ca2+-binding sites based on positive identification of predefined Ca2+-binding characteristics. Following identification of the distributions of these features using FEATURE, the query protein structure was embedded in a 3D-grid of 2 Å grid cubes, and a score was computed for each grid (e.g., query site) based on a scoring function that uses Bayes’ Rule to combine prior known distributions and observed frequencies (e.g., derived from a training dataset and the query site) to indicate the likelihood of Ca2+-binding in the queried site. An online server WebFEATURE is available at http://feature.stanford.edu/webfeature/. Besides being applied to static X-ray crystal structures, FEATURE has recently been applied to multiple conformations of protein parvalbumin β generated from molecular dynamics (MD) simulation [88]. For MD generated conformations of both holo and apo forms, FEATURE correctly identifies the calcium-binding sites and non-sites with some interesting fluctuations in score. The combination of machine learning techniques such as FEATURE with MD simulations may provide new insights into Ca2+-binding affinity, persistence and the stability of calcium-binding sites.
More recently, Babor et al. demonstrated related progress towards the prediction of metal (Zn, Fe, Cu, Co, Ni and Mn ions) binding sites for given protein apo structures [89] based on previous statistical analysis results, indicating that transition metal-binding residues were almost exclusively limited to Cys, His, Asp and Glu (CHED) residues (Table 2) [64]. An online server related to this method is available at http://ligin.weizmann.ac.il/ched.
Currently the MUG algorithm developed by Wang et al. [82] has been modified to accurately prediction protein calcium-binding sites including multiple-binding sites that undergo local conformational change or side chain rotations upon calcium-binding. Given an apo (unbound) protein structure, this new approach (MUGSR) conducts a side chain rotation procedure according to a rotamer library [90] to find binding sites with possible side-chain movements (local conformational changes) caused by calcium binding. Given an apo protein structure, MUGSR is able to capture calcium binding sites that may undergo local conformational changes, which is validated by comparison with the corresponding holo protein structure sharing more than 98% sequence similarity with the apo protein. For the test dataset consisting of 47 documented binding sites in the holo structures, MUGSR was able to predict 44/47 sites using the corresponding apo structures (manuscript in preparation).
a. Application of Algorithms for the Prediction or Design of Ca2+-Binding Proteins
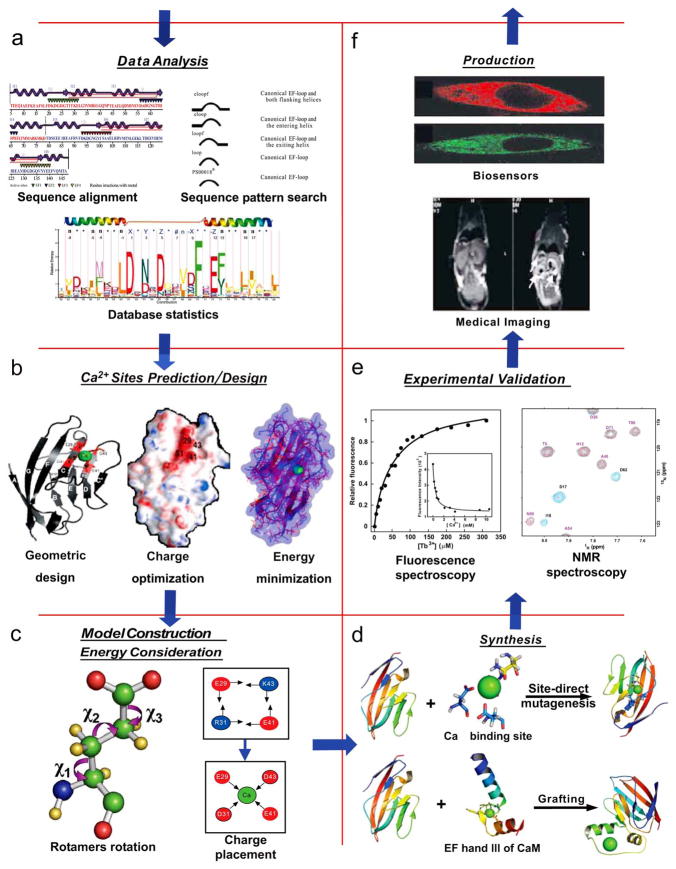
Pre-experimental, computational design of Ca2+-binding proteins facilitates the transition from prediction and theory to empirical analysis and application. Various computational or prediction methods have been utilized for the rational design of proteins, the objective being to identify target sequences or structures with increased probability of obtaining some desired characteristics. This design phase may include sequential and statistical analysis, structural modeling, charge analysis, rotamer analysis and energy minimization as precursors to synthesis of novel proteins (Fig. (3)).
Fig. 3.
Schematic illustrating rational design of a Ca2+-binding protein for biotechnological applications. (a) Calcium pattern search results and definition of Ca2+-binding motifs, reproduced from Zhou et al. [35]. (b) Geometric design and charge optimization, and (c) charge placement for energy optimization, reproduced from Li et al. [100]. (d) General strategy for construction of Ca2+-binding site on protein scaffold either by mutating existing residues (site-directed mutagenesis) or by inserting new residues (grafting). (e) Fluorescence spectrum (left) for identification of Ca2+-binding site in Rubella virus, reproduced from Zhou [71] and NMR spectrum for designed CaBP, reproduced from Yang et al. [97]. (f) Application of designed proteins in medical imaging and as biosensors, reproduced from Yang et al. [105] and Zou et al. [104], respectively.
Previous work by Yang and coworkers [37] has shown that different classes of naturally evolved calcium binding sites can be re-identified based on local geometric properties using DEZYMER [83] which utilizes a two-step process to generate one or more potential ligand binding sites in proteins (apo or holo) of known structure.
The subsequent modification/engineering of Ca2+-binding proteins may be accomplished by either site-directed mutagenesis or through a grafting approach [77, 91–93]. For the study of Ca2+-binding proteins, these well-established methods usually involve restructuring the charge environment in the binding site [94, 95] which may alter the binding affinity. Both design and grafting have been applied to overcome the complexities encountered in cooperative, multi-site binding associated with natural Ca2+-binding proteins, where novel proteins have been engineered with a single Ca2+-binding site in order to dissect the key structural factors that control Ca2+-binding affinity, conformational change and cooperativity, as well as to function as novel reagents for research. Several de novo calcium-binding sites have been designed into a non-calcium binding protein CD2 with strong metal selectivity based on the local geometric properties and common chemical features of calcium binding sites in proteins [96]. The NMR solution structure of a novel protein (1t6w.pdb) revealed that Ca.CD2 binds Ca2+ at the intended site with the designed arrangement, which validated a general strategy for designing de novo Ca2+-binding proteins [97].
Ye et al. reported the first estimation of the intrinsic Ca2+ affinities of the four EF-Hand loops of calmodulin and a better estimation of the cooperativity of coupled EF-Hand proteins based on a designed grafting approach [77]. This approach has also been successfully applied to obtain site-specific calcium binding affinity of a predicted single EF-Hand motif in the non-structural protein of rubella virus [35] and Non-EF-Hand continuous calcium binding sites in GPCR, pumps and channels.
Shifman et al. observed a method of rational design to modify CaM for domain limited binding of only two Ca2+ ions (either C-terminal or N-terminal) to circumvent cooperativity problems with CaM-Ca2+ complexes and their interaction with protein kinase II [98]. The program ORBIT [84] was applied to minimize the energy of a given protein by optimizing the sequence, which was based on empirical atom-based force field calculation and a fast side-chain selection algorithm. Far-UV circular dichroism was then applied to evaluate correct folding of the engineered CaMs. Their results indicated that CaM, bound with only two Ca2+ ions in the C-terminal could both bind to CaMKII and partially activate the kinase.
Recent studies based on rational design have shown that calcium and lanthanide binding affinities and protein stability can be systematically tuned by their arrangement in the coordination shell and its protein environment [49, 99]. Jones et al. analyzed electrostatic interactions and engineered a novel Ca2+-binding site on CD2, which was found to increase its CD48 binding affinity [99]. In addition, Li et al. further designed by site-directed mutagenesis a calcium-dependent CD2.trigger the conformation of which can be reversibly switched upon calcium binding without using coupled EF-Hand motifs [97, 100]. The extensive in silico design of this protein variant based on pre-experimental analysis of structure, charge interactions and energy minimizations, provides an excellent example illustrating the progression from prediction and modeling to the creation of a desired product. This designed Ca2+ binding protein is very useful since it serves not only as a model system for understanding Ca2+ dependent cell adhesion but also as a basis for designing Ca2+-dependent trigger proteins for the control of protein structures and conformations by metal binding.
Bunick et al. reported efforts to design a variant of calbindin D9k capable of responding to Ca2+-binding with conformational changes similar to CaM in order to understand how differences in the amino acid sequence lead to differences in the Ca2+-binding response [101]. Calbindin D9k was re-engineered with 15 mutations based on comparative analyses of sequence and structures combined with model building using MODELLER [102] to generate calbindo-modulin (CBM-1), which did not exhibit any non-native-like molten globule properties despite the large number, and, in some cases, the nonconservative nature, of the mutations. Ca2+-induced changes in CD intensity and in the binding of the hydrophobic probe, ANS, implied that CBM-1 did undergo Ca2+ sensor-like conformational changes. The X-ray crystal structure of Ca2+-CBM-1 determined at 1.44 Å resolution revealed the anticipated increase in hydrophobic surface area relative to the wild-type protein.
Huang et al. recently identified several calcium binding sites in the extracellular domain of calcium sensing receptors using algorithms based on the geometric description and surface electrostatic potentials [5]. Ca2+-sensing receptors (CaSRs) represent a class of receptors that respond to changes in the extracellular Ca2+ concentration ([Ca2+]o) and activate multiple signaling pathways. A major barrier to advancing our understanding of the role of Ca2+ in regulating CaSRs is the lack of adequate information regarding the locations of their Ca2+-binding sites due to the absence of a solved three-dimensional structure and rapid off rates related to low Ca2+-binding affinities. In the absence of a solved CaSR structure, Huang et al. used SWISS-MODEL [76] and MODELLER to generate a putative CaSR structure model that was based on its homology with the previously-solved mGluR structure. Mutation of the predicted ligand residues in the full-length CaSR caused abnormal responses to [Ca2+]o, similar to those observed with naturally occurring activating or inactivating mutations of the CaSR, supporting the essential role of these predicted Ca2+-binding sites in the sensing capability of the CaSR.
In a later related work, Huang et al. employed a similar computational strategy to probe the intrinsic Ca2+-binding properties of predicted CaSR Ca2+-binding sites [103]. For this study, two predicted continuous Ca2+-binding sequences were individually engineered into a scaffold protein provided by a non-Ca2+-binding protein, CD2. Metal-binding affinities of these predicted sites in the CaSR were calculated by monitoring aromatic-sensitized Tb3+ fluorescence energy transfer. Removal of the predicted Ca2+-binding ligands resulted in the loss or significant weakening of cation binding. The potential Ca2+-binding residues were shown to be involved in Ca2+/Ln3+ binding by high resolution NMR and site-directed mutagenesis, further validating predictions of Ca2+-binding sites within the extracellular domain of the CaSR. To further evaluate the potential calcium binding capabilities of predicted noncontinuous Ca2+-binding sites in the ECD, the intact CaSR was dissected into three globular subdomains, each of which contained two to three predicted Ca2+-binding sites. This approach allowed for analysis of the mechanisms underlying the binding of multiple metal ions to extended polypeptides derived from a location within the ECD of the CaSR, which would be anticipated to more closely mimic the structure of the native CaSR ECD. Studies on these subdomains suggested the existence of multiple metal-binding sites and metal-induced conformational changes that might be responsible for the switching on and off of the CaSR by the transition between its open inactive form and closed active form [103].
Application of these methods has also been used to develop novel, protein-based reagents. A rational grafting approach was used to develop calcium sensors capable of real-time quantitative Ca2+ concentration measurements in specific sub-cellular environments without using natural Ca2+ binding proteins such as calmodulin [104]. These engineered Ca2+ sensors have been shown to exhibit large ratiometric fluorescence and absorbance changes upon Ca2+ binding with affinities corresponding to the Ca2+ concentrations found in the ER (Kd values ranging from 0.4 to 2 mM). The developed Ca2+ sensor was successfully targeted to the ER of mammalian cell lines to monitor Ca2+ changes occurring in this organelle in response to stimulation with agonists. Similarly, computational design was utilized in the recent development of a new class of protein-based MRI contrast agents with significantly improved longitudinal and transverse relaxation [35, 105].
CONCLUSIONS AND PERSPECTIVES
The evolution of bioinformatics has led to an incredibly diverse set of algorithms, databases and tools that are rapidly becoming indispensible to all scientific inquiry in fields related to biology, chemistry and medicine. The availability of structured data collections, prediction algorithms and software for modeling structural and energetic characteristics has facilitated an important new phase in research, allowing for pre-experimental evaluation of hypotheses and the rational design of research methodologies. Due to the scope of the field, this review was restricted to a concentrated focus with respect to Ca2+-binding as a central theme; however, it should be clear that the studies presented here can be extended beyond CaBPs as summarized in this work. The progression from prediction to development illustrates not only the increasing sophistication of bioinformatics tools, but their relative applicability to move beyond theoretical models and provide precise data for real-world design and development. Within the context of Ca2+-binding research, our ability to make assumptions regarding the structure and function of CaBPs has advanced in step with the evolution of algorithms capable of recognizing or predicting binding sites, and the development of tools to predict binding sites in apo-structures may become routine in the near future. Using prediction as a basis for rational design, it is anticipated that the engineering of proteins with precise affinity and metal selectivity will significantly improve our ability to provide function-specific proteins for therapeutic, drug-delivery and diagnostic purposes.
Acknowledgments
We would like to thank Dan Adams for his critical review of this manuscript. This work is supported in part by the following sponsors: Georgia State University Molecular Basis of Disease Fellowships, NIH GM081749, NIH GM 62999 and NSF MCB-0092486 to JJY, and; DMS-0500951 to GTC.
ABBREVIATIONS
- CaBP
Calcium-Binding Protein
- CN
Coordination Number
- FC
Formal Charge
- PDB
Protein Data Bank
References
- 1.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–9. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 2.Herzberg O, Moult J, James MN. A model for the Ca2+-induced conformational transition of troponin C: a trigger for muscle contraction. J Biol Chem. 1986;261:2638–44. [PubMed] [Google Scholar]
- 3.Mann KG, Nesheim ME, Church WR, Haley P, Krishnaswamy S. Surface-dependent reactions of the vitamin K-dependent enzyme complexes. Blood. 1990;76:1–16. [PubMed] [Google Scholar]
- 4.Nelson MR, Chazin WJ. An interaction-based analysis of calcium-induced conformational changes in Ca2+ sensor proteins. Protein Sci. 1998;7:270–82. doi: 10.1002/pro.5560070206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Y, Zhou Y, Yang W, et al. Identification and dissection of Ca(2+)-binding sites in the extracellular domain of Ca(2+)-sensing receptor. J Biol Chem. 2007;282:19000–10. doi: 10.1074/jbc.M701096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hofer AM, Curci S, Doble MA, Brown EM, Soybel DI. Intercellular communication mediated by the extracellular calcium-sensing receptor. Nat Cell Biol. 2000;2:392–8. doi: 10.1038/35017020. [DOI] [PubMed] [Google Scholar]
- 7.Bower MJ, Cohen FE, Dunbrack RL., Jr Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: a new homology modeling tool. J Mol Biol. 1997;267:1268–82. doi: 10.1006/jmbi.1997.0926. [DOI] [PubMed] [Google Scholar]
- 8.Gunasekaran K, Nussinov R. How different are structurally flexible and rigid binding sites? Sequence and structural features discriminating proteins that do and do not undergo conformational change upon ligand binding. J Mol Biol. 2007;365:257–273. doi: 10.1016/j.jmb.2006.09.062. [DOI] [PubMed] [Google Scholar]
- 9.Eyal E, Najmanovich R, Edelman M, Sobolev V. Protein side-chain rearrangement in regions of point mutations. Proteins. 2003;50:272–82. doi: 10.1002/prot.10276. [DOI] [PubMed] [Google Scholar]
- 10.Najmanovich R, Kuttner J, Sobolev V, Edelman M. Side-chain flexibility in proteins upon ligand binding. Proteins. 2000;39:261–8. doi: 10.1002/(sici)1097-0134(20000515)39:3<261::aid-prot90>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita MM, Wesson L, Eisenman G, Eisenberg D. Where metal ions bind in proteins. Proc Natl Acad Sci USA. 1990;87:5648–52. doi: 10.1073/pnas.87.15.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bagley SC, Altman RB. Characterizing the microenvironment surrounding protein sites. Protein Sci. 1995;4:622–35. doi: 10.1002/pro.5560040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudev T, Lim C. Effect of carboxylate-binding mode on metal binding/selectivity and function in proteins. Acc Chem Res. 2007;40:85–93. doi: 10.1021/ar068181i. [DOI] [PubMed] [Google Scholar]
- 14.Dudev T, Lim C. Principles governing Mg, Ca, and Zn binding and selectivity in proteins. Chem Rev. 2003;103:773–87. doi: 10.1021/cr020467n. [DOI] [PubMed] [Google Scholar]
- 15.Dudev T, Lin YL, Dudev M, Lim C. First-second shell interactions in metal binding sites in proteins: a PDB survey and DFT/CDM calculations. J Am Chem Soc. 2003;125:3168–80. doi: 10.1021/ja0209722. [DOI] [PubMed] [Google Scholar]
- 16.Babu CS, Dudev T, Casareno R, Cowan JA, Lim C. A combined experimental and theoretical study of divalent metal ion selectivity and function in proteins: application to E. coli ribonuclease H1. J Am Chem Soc. 2003;125:9318–28. doi: 10.1021/ja034956w. [DOI] [PubMed] [Google Scholar]
- 17.Chakrabarti P. Geometry of interaction of metal ions with histidine residues in protein structures. Protein Eng. 1990;4:57–63. doi: 10.1093/protein/4.1.57. [DOI] [PubMed] [Google Scholar]
- 18.Chakrabarti P. Geometry of interaction of metal ions with sulfur-containing ligands in protein structures. Biochemistry. 1989;28:6081–5. doi: 10.1021/bi00440a052. [DOI] [PubMed] [Google Scholar]
- 19.Chakrabarti P. Interaction of metal ions with carboxylic and carboxamide groups in protein structures. Protein Eng. 1990;4:49–56. doi: 10.1093/protein/4.1.49. [DOI] [PubMed] [Google Scholar]
- 20.Glusker JP, Lewis M, Rossi M. Crystal structure analysis for chemists and biologists. New York: VCH; 1994. [Google Scholar]
- 21.Harding MM. Geometry of metal-ligand interactions in proteins. Acta Crystallogr D Biol Crystallogr. 2001;57:401–11. doi: 10.1107/s0907444900019168. [DOI] [PubMed] [Google Scholar]
- 22.Harding MM. The geometry of metal-ligand interactions relevant to proteins. Acta Crystallogr D Biol Crystallogr. 1999;55:1432–43. doi: 10.1107/s0907444999007374. [DOI] [PubMed] [Google Scholar]
- 23.Harding MM. The geometry of metal-ligand interactions relevant to proteins. II. Angles at the metal atom, additional weak metal-donor interactions. Acta Crystallogr D Biol Crystallogr. 2000;56:857–67. doi: 10.1107/s0907444900005849. [DOI] [PubMed] [Google Scholar]
- 24.Glusker JP. Structural aspects of metal liganding to functional groups in proteins. Adv Protein Chem. 1991;42:1–76. doi: 10.1016/s0065-3233(08)60534-3. [DOI] [PubMed] [Google Scholar]
- 25.Wei L, Altman RB. Pac Symp Biocomput World Scientific. 1998. Recognizing protein binding sites using statistical descriptions of their 3D environments. [PubMed] [Google Scholar]
- 26.Pidcock E, Moore GR. Structural characteristics of protein binding sites for calcium and lanthanide ions. J Biol Inorg Chem. 2001;6:479–89. doi: 10.1007/s007750100214. [DOI] [PubMed] [Google Scholar]
- 27.Nayal M, Di Cera E. Predicting Ca(2+)-binding sites in proteins. Proc Natl Acad Sci USA. 1994;91:817–21. doi: 10.1073/pnas.91.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirberger M, Wang X, Deng H, Yang W, Chen G, Yang JJ. Statistical analysis of structural characteristics of protein Ca(2+)-binding sites. J Biol Inorg Chem. 2008;13:1169–81. doi: 10.1007/s00775-008-0402-7. [DOI] [PubMed] [Google Scholar]
- 29.McPhalen CA, Strynadka NC, James MN. Calcium-binding sites in proteins: a structural perspective. Adv Protein Chem. 1991;42:77–144. doi: 10.1016/s0065-3233(08)60535-5. [DOI] [PubMed] [Google Scholar]
- 30.Hill E, Broadbent ID, Chothia C, Pettitt J. Cadherin superfamily proteins in Caenorhabditis elegans and Drosophila melanogaster. J Mol Biol. 2001;305:1011–24. doi: 10.1006/jmbi.2000.4361. [DOI] [PubMed] [Google Scholar]
- 31.Truong K, Ikura M. The cadherin superfamily database. J Struct Funct Genomics. 2002;2:135–43. doi: 10.1023/a:1021352716284. [DOI] [PubMed] [Google Scholar]
- 32.Yang JJ, Yang W. Encyclopedia of inorganic chemistry. New York: Wiley; 2005. [Google Scholar]
- 33.Rigden DJ, Galperin MY. The DxDxDG motif for calcium binding: multiple structural contexts and implications for evolution. J Mol Biol. 2004;343:971–84. doi: 10.1016/j.jmb.2004.08.077. [DOI] [PubMed] [Google Scholar]
- 34.Marsden BJ, Shaw GS, Sykes BD. Calcium binding proteins. Elucidating the contributions to calcium affinity from an analysis of species variants and peptide fragments. Biochem Cell Biol. 1990;68:587–601. doi: 10.1139/o90-084. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Y, Yang W, Kirberger M, Lee HW, Ayalasomayajula G, Yang JJ. Prediction of EF-hand calcium-binding proteins and analysis of bacterial EF-hand proteins. Proteins. 2006;65:643–55. doi: 10.1002/prot.21139. [DOI] [PubMed] [Google Scholar]
- 36.Katz AK, Glusker JP, Beebe SA, Bock CW. Calcium ion coordination: a comparison with that of beryllium, magnesium, and zinc. J Am Chem Soc. 1996;118:5752–63. [Google Scholar]
- 37.Yang W, Lee HW, Hellinga H, Yang JJ. Structural analysis, identification, and design of calcium-binding sites in proteins. Proteins. 2002;47:344–56. doi: 10.1002/prot.10093. [DOI] [PubMed] [Google Scholar]
- 38.Kawasaki H, Kretsinger RH. Calcium-binding proteins 1: EF-hands. Protein Profile. 1995;2:297–490. [PubMed] [Google Scholar]
- 39.Falke JJ, Drake SK, Hazard AL, Peersen OB. Molecular tuning of ion binding to calcium signaling proteins. Q Rev Biophys. 1994;27:219–90. doi: 10.1017/s0033583500003012. [DOI] [PubMed] [Google Scholar]
- 40.Marsden BJ, Hodges RS, Sykes BD. 1H-NMR studies of synthetic peptide analogues of calcium-binding site III of rabbit skeletal troponin C: effect on the lanthanum affinity of the interchange of aspartic acid and asparagine residues at the metal ion coordinating positions. Biochemistry. 1988;27:4198–206. doi: 10.1021/bi00411a043. [DOI] [PubMed] [Google Scholar]
- 41.Strynadka NC, James MN. Crystal structures of the helix-loop-helix calcium-binding proteins. Annu Rev Biochem. 1989;58:951–98. doi: 10.1146/annurev.bi.58.070189.004511. [DOI] [PubMed] [Google Scholar]
- 42.Einspahr H, Bugg C. Calcium and its role in biology. In: Sigel H, editor. Crystal Structure Studies of Calcium Complexes and Implications for Biological Systems. New York: M. Dekker; 1984. pp. 52–97. [Google Scholar]
- 43.Harding MM. Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr D Biol Crystallogr. 2006;62:678–82. doi: 10.1107/S0907444906014594. [DOI] [PubMed] [Google Scholar]
- 44.Swain AL, Kretsinger RH, Amma EL. Restrained least squares refinement of native (calcium) and cadmium-substituted carp parvalbumin using X-ray crystallographic data at 1.6-A resolution. J Biol Chem. 1989;264:16620–8. [PubMed] [Google Scholar]
- 45.Vyas MN, Jacobson BL, Quiocho FA. The calcium-binding site in the galactose chemoreceptor protein. Crystallographic and metal-binding studies. J Biol Chem. 1989;264:20817–21. [PubMed] [Google Scholar]
- 46.Martin R. Bioinorganic chemistry of calcium. New York: Marcel Dekker; 1984. [Google Scholar]
- 47.Drake SK, Lee KL, Falke JJ. Tuning the equilibrium ion affinity and selectivity of the EF-hand calcium binding motif: substitutions at the gateway position. Biochemistry. 1996;35:6697–705. doi: 10.1021/bi952430l. [DOI] [PubMed] [Google Scholar]
- 48.Drake SK, Zimmer MA, Miller CL, Falke JJ. Optimizing the metal binding parameters of an EF-hand-like calcium chelation loop: coordinating side chains play a more important tuning role than chelation loop flexibility. Biochemistry. 1997;36:9917–26. doi: 10.1021/bi9703913. [DOI] [PubMed] [Google Scholar]
- 49.Maniccia AW, Yang W, Li SY, Johnson JA, Yang JJ. Using protein design to dissect the effect of charged residues on metal binding and protein stability. Biochemistry. 2006;45:5848–56. doi: 10.1021/bi052508q. [DOI] [PubMed] [Google Scholar]
- 50.Linse S, Helmersson A, Forsen S. Calcium binding to calmodulin and its globular domains. J Biol Chem. 1991;266:8050–4. [PubMed] [Google Scholar]
- 51.Martin SR, Linse S, Johansson C, Bayley PM, Forsen S. Protein surface charges and Ca2+ binding to individual sites in calbindin D9k: stopped-flow studies. Biochemistry. 1990;29:4188–93. doi: 10.1021/bi00469a023. [DOI] [PubMed] [Google Scholar]
- 52.Cheng Y, Sequeira SM, Malinina L, Tereshko V, Sollner TH, Patel DJ. Crystallographic identification of Ca2+ and Sr2+ coordination sites in synaptotagmin I C2B domain. Protein Sci. 2004;13:2665–72. doi: 10.1110/ps.04832604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korndorfer IP, Brueckner F, Skerra A. The crystal structure of the human (S100A8/S100A9)2 heterotetramer, calprotectin, illustrates how conformational changes of interacting alpha-helices can determine specific association of two EF-hand proteins. J Mol Biol. 2007;370:887–98. doi: 10.1016/j.jmb.2007.04.065. [DOI] [PubMed] [Google Scholar]
- 54.Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A. Solution structure of calcium-free calmodulin. Nat Struct Biol. 1995;2:768–76. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 55.Zhang M, Tanaka T, Ikura M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat Struct Biol. 1995;2:758–67. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
- 56.Davis IW, Arendall WB, 3rd, Richardson DC, Richardson JS. The backrub motion: how protein backbone shrugs when a sidechain dances. Structure. 2006;14:265–74. doi: 10.1016/j.str.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 57.Esposito L, De Simone A, Zagari A, Vitagliano L. Correlation between omega and psi dihedral angles in protein structures. J Mol Biol. 2005;347:483–7. doi: 10.1016/j.jmb.2005.01.065. [DOI] [PubMed] [Google Scholar]
- 58.Dunbrack RL, Jr, Karplus M. Conformational analysis of the backbone-dependent rotamer preferences of protein sidechains. Nat Struct Biol. 1994;1:334–40. doi: 10.1038/nsb0594-334. [DOI] [PubMed] [Google Scholar]
- 59.Chrysina ED, Brew K, Acharya KR. Crystal structures of apo- and holo-bovine alpha-lactalbumin at 2.2-A resolution reveal an effect of calcium on inter-lobe interactions. J Biol Chem. 2000;275:37021–9. doi: 10.1074/jbc.M004752200. [DOI] [PubMed] [Google Scholar]
- 60.Nelson MR, Chazin WJ. Structures of EF-hand Ca(2+)-binding proteins: diversity in the organization, packing and response to Ca2+ binding. Biometals. 1998;11:297–318. doi: 10.1023/a:1009253808876. [DOI] [PubMed] [Google Scholar]
- 61.Iyer LK, Qasba PK. Molecular dynamics simulation of alpha-lactalbumin and calcium binding c-type lysozyme. Protein Eng. 1999;12:129–39. doi: 10.1093/protein/12.2.129. [DOI] [PubMed] [Google Scholar]
- 62.Shesham RD, Bartolotti LJ, Li Y. Molecular dynamics simulation studies on Ca2+ -induced conformational changes of annexin I. Protein Eng Des Sel. 2008;21:115–20. doi: 10.1093/protein/gzm094. [DOI] [PubMed] [Google Scholar]
- 63.Richardson RC, King NM, Harrington DJ, Sun H, Royer WE, Nelson DJ. X-Ray crystal structure and molecular dynamics simulations of silver hake parvalbumin (Isoform B) Protein Sci. 2000;9:73–82. doi: 10.1110/ps.9.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Babor M, Greenblatt HM, Edelman M, Sobolev V. Flexibility of metal binding sites in proteins on a database scale. Proteins. 2005;59:221–30. doi: 10.1002/prot.20431. [DOI] [PubMed] [Google Scholar]
- 65.Zhao S, Goodsell DS, Olson AJ. Analysis of a data set of paired uncomplexed protein structures: new metrics for side-chain flexibility and model evaluation. Proteins. 2001;43:271–9. doi: 10.1002/prot.1038. [DOI] [PubMed] [Google Scholar]
- 66.Haiech J, Sallantin J. Computer search of calcium binding sites in a gene data bank: use of learning techniques to build an expert system. Biochimie. 1985;67:555–60. doi: 10.1016/s0300-9084(85)80276-5. [DOI] [PubMed] [Google Scholar]
- 67.Kretsinger RH, Nockolds CE. Carp muscle calcium-binding protein. II. Structure determination and general description. J Biol Chem. 1973;248:3313–26. [PubMed] [Google Scholar]
- 68.Galzi JL, Bertrand S, Corringer PJ, Changeux JP, Bertrand D. Identification of calcium binding sites that regulate potentiation of a neuronal nicotinic acetylcholine receptor. EMBO J. 1996;15:5824–32. [PMC free article] [PubMed] [Google Scholar]
- 69.Simons JW, van Kampen MD, Ubarretxena-Belandia I, et al. Identification of a calcium binding site in Staphylococcus hyicus lipase: generation of calcium-independent variants. Biochemistry. 1999;38:2–10. doi: 10.1021/bi981869l. [DOI] [PubMed] [Google Scholar]
- 70.Rastogi PA. In: MacVector: Integrated Sequence Analysis for the Macintosh, in Bioinformatics Methods and Protocols. Misener S, Krawetz SA, editors. Humana Press; Totowa, N.J: 2000. [DOI] [PubMed] [Google Scholar]
- 71.Zhou Y, Tzeng WP, Yang W, et al. Identification of a Ca2+-binding domain in the rubella virus nonstructural protease. J Virol. 2007;81:7517–28. doi: 10.1128/JVI.00605-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–5. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 73.Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–3. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 74.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–6. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deng H, Chen G, Yang W, Yang JJ. Predicting calcium-binding sites in proteins - a graph theory and geometry approach. Proteins. 2006;64:34–42. doi: 10.1002/prot.20973. [DOI] [PubMed] [Google Scholar]
- 76.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–5. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ye Y, Lee HW, Yang W, Shealy S, Yang JJ. Probing site-specific calmodulin calcium and lanthanide affinity by grafting. J Am Chem Soc. 2005;127:3743–50. doi: 10.1021/ja042786x. [DOI] [PubMed] [Google Scholar]
- 78.Schymkowitz JW, Rousseau F, Martins IC, Ferkinghoff-Borg J, Stricher F, Serrano L. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc Natl Acad Sci USA. 2005;102:10147–52. doi: 10.1073/pnas.0501980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guerois R, Serrano L. The SH3-fold family: experimental evidence and prediction of variations in the folding pathways. J Mol Biol. 2000;304:967–82. doi: 10.1006/jmbi.2000.4234. [DOI] [PubMed] [Google Scholar]
- 80.Pitt WR, Goodfellow JM. Modelling of solvent positions around polar groups in proteins. Protein Eng. 1991;4:531–7. doi: 10.1093/protein/4.5.531. [DOI] [PubMed] [Google Scholar]
- 81.Lin HH, Han LY, Zhang HL, et al. Prediction of the functional class of metal-binding proteins from sequence derived physico-chemical properties by support vector machine approach. BMC Bioinformatics. 2006;7 (Suppl 5):S13. doi: 10.1186/1471-2105-7-S5-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang X, Kirberger M, Qiu F, Chen G, Yang JJ. Towards predicting Ca(2+)-binding sites with different coordination numbers in proteins with atomic resolution. Proteins. 2008;75:787–98. doi: 10.1002/prot.22285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hellinga HW, Richards FM. Construction of new ligand binding sites in proteins of known structure. I. Computer-aided modeling of sites with pre-defined geometry. J Mol Biol. 1991;222:763–85. doi: 10.1016/0022-2836(91)90510-d. [DOI] [PubMed] [Google Scholar]
- 84.Dahiyat BI, Mayo SL. De novo protein design: fully automated sequence selection. Science. 1997;278:82–7. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 85.Sodhi JS, Bryson K, McGuffin LJ, Ward JJ, Wernisch L, Jones DT. Predicting metal-binding site residues in low-resolution structural models. J Mol Biol. 2004;342:307–20. doi: 10.1016/j.jmb.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 86.Lin CT, Lin KL, Yang CH, Chung IF, Huang CD, Yang YS. Protein metal binding residue prediction based on neural networks. Int J Neural Syst. 2005;15:71–84. doi: 10.1142/S0129065705000116. [DOI] [PubMed] [Google Scholar]
- 87.Cai CZ, Han LY, Ji ZL, Chen X, Chen YZ. SVM-Prot: web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acids Res. 2003;31:3692–7. doi: 10.1093/nar/gkg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Glazer DS, Radmer RJ, Altman RB. Combining molecular dynamics and machine learning to improve protein function recognition. Pac Symp Biocomput. 2008;13:332–343. [PMC free article] [PubMed] [Google Scholar]
- 89.Babor M, Gerzon S, Raveh B, Sobolev V, Edelman M. Prediction of transition metal-binding sites from apo protein structures. Proteins. 2008;70:208–17. doi: 10.1002/prot.21587. [DOI] [PubMed] [Google Scholar]
- 90.Dunbrack RL, Jr, Cohen FE. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 1997;6:1661–81. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ye Y, Lee HW, Yang W, et al. Metal binding affinity and structural properties of an isolated EF-loop in a scaffold protein. Protein Eng. 2001;14:1001–13. doi: 10.1093/protein/14.12.1001. [DOI] [PubMed] [Google Scholar]
- 92.Toma S, Campagnoli S, Margarit I, et al. Grafting of a calcium-binding loop of thermolysin to Bacillus subtilis neutral protease. Biochemistry. 1991;30:97–106. doi: 10.1021/bi00215a015. [DOI] [PubMed] [Google Scholar]
- 93.Ye Y, Lee HW, Yang W, Yang JJ. Calcium and lanthanide affinity of the EF-loops from the C-terminal domain of calmodulin. J Inorg Biochem. 2005;99:1376–83. doi: 10.1016/j.jinorgbio.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 94.Wilkins AL, Ye Y, Yang W, Lee HW, Liu ZR, Yang JJ. Metal-binding studies for a de novo designed calcium-binding protein. Protein Eng. 2002;15:571–4. doi: 10.1093/protein/15.7.571. [DOI] [PubMed] [Google Scholar]
- 95.Falke JJ, Snyder EE, Thatcher KC, Voertler CS. Quantitating and engineering the ion specificity of an EF-hand-like Ca2+ binding. Biochemistry. 1991;30:8690–7. doi: 10.1021/bi00099a029. [DOI] [PubMed] [Google Scholar]
- 96.Yang W, Jones LM, Isley L, et al. Rational design of a calcium-binding protein. J Am Chem Soc. 2003;125:6165–71. doi: 10.1021/ja034724x. [DOI] [PubMed] [Google Scholar]
- 97.Yang W, Wilkins AL, Ye Y, et al. Design of a calcium-binding protein with desired structure in a cell adhesion molecule. J Am Chem Soc. 2005;127:2085–93. doi: 10.1021/ja0431307. [DOI] [PubMed] [Google Scholar]
- 98.Shifman JM, Choi MH, Mihalas S, Mayo SL, Kennedy MB. Ca2+/calmodulin-dependent protein kinase II (CaMKII) is activated by calmodulin with two bound calciums. Proc Natl Acad Sci USA. 2006;103:13968–73. doi: 10.1073/pnas.0606433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jones LM, Yang W, Maniccia AW, Harrison A, van der Merwe PA, Yang JJ. Rational design of a novel calcium-binding site adjacent to the ligand-binding site on CD2 increases its CD48 affinity. Protein Sci. 2008;17:439–49. doi: 10.1110/ps.073328208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li S, Yang W, Maniccia AW, et al. Rational design of a conformation-switchable Ca2+- and Tb3+-binding protein without the use of multiple coupled metal-binding sites. FEBS J. 2008;275:5048–61. doi: 10.1111/j.1742-4658.2008.06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bunick CG, Nelson MR, Mangahas S, et al. Designing sequence to control protein function in an EF-hand protein. J Am Chem Soc. 2004;126:5990–8. doi: 10.1021/ja0397456. [DOI] [PubMed] [Google Scholar]
- 102.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 103.Huang Y, Zhou Y, Castiblanco A, Yang W, Brown EM, Yang JJ. Multiple Ca(2+)-binding sites in the extracellular domain of the Ca(2+)-sensing receptor corresponding to cooperative Ca(2+) response. Biochemistry. 2009;48(2):388–98. doi: 10.1021/bi8014604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zou J, Hofer AM, Lurtz MM, et al. Developing sensors for real-time measurement of high Ca2+ concentrations. Biochemistry. 2007;46:12275–88. doi: 10.1021/bi7007307. [DOI] [PubMed] [Google Scholar]
- 105.Yang JJ, Yang J, Wei L, et al. Rational design of protein-based MRI contrast agents. J Am Chem Soc. 2008;130:9260–7. doi: 10.1021/ja800736h. [DOI] [PMC free article] [PubMed] [Google Scholar]