Abstract
Unlike pyruvate dehydrogenase complexes (PDCs) from prokaryotes, PDCs from higher eukaryotes have an additional structural component, E3-binding protein (BP), for binding of dihydrolipoamide dehydrogenase (E3) in the complex. Based on the 3-D structure of the subcomplex of human (h) E3 with the di-domain (L3S1) of hBP, the amino acid residues (H348, D413, Y438, and R447) of hE3 for binding to hBP were substituted singly by alanine or other residues. These substitutions did not have large effects on hE3 activity when measured in its free form. However, when these hE3 mutants were reconstituted in the complex, the PDC activity was significantly reduced to 9% for Y438A, 20% for Y438H, and 18% for D413A. The binding of hE3 mutants with L3S1 determined by isothermal titration calorimetry revealed that the binding affinities of the Y438A, Y438H, and D413A mutants to L3S1 were severely reduced (1019-, 607-, and 402-fold, respectively). Unlike wild-type hE3 the binding of the Y438A mutant to L3S1 was accompanied by an unfavorable enthalpy change and a large positive entropy change. These results indicate that hE3-Y438 and hE3-D413 play important roles in binding of hE3 to hBP.
Keywords: Pyruvate dehydrogenase complex, Dihydrolipoamide dehydrogenase, E3-binding protein, Subunit-subunit interaction, Thermodynamics
Introduction
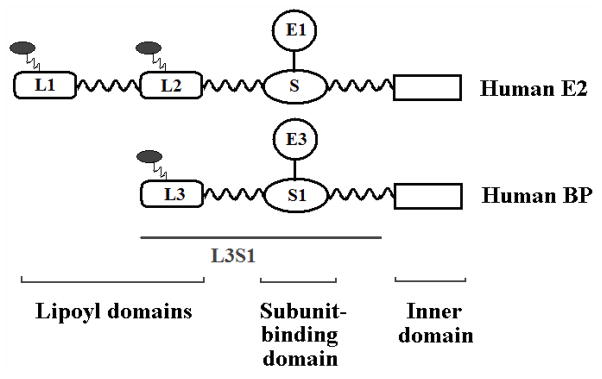
Pyruvate dehydrogenase complex (PDC) plays a pivotal role in the oxidation of pyruvic acid to acetyl-CoA and is composed of multiple copies of three catalytic components: namely pyruvate dehydrogenase (E1), dihydrolipoamide acetyltransferase (E2) and dihydrolipoamide dehydrogenase (E3) [1–3]. Unlike prokaryotic PDCs, PDCs from higher eukaryotes have an additional structural component, E3-binding protein (BP) to bind E3 within PDC. In human (h) PDC multi-copies of hE2 and hBP form the central core of PDC to which are non-covalently bound the other components of the complex [4, 5]. hE2 has three well defined structural domains connected by flexible hinge regions: (i) 2 lipoyl domains, (termed L1, the outer domain and L2, the inner domain), (ii) the subunit-binding domain (S) interacting with E1 only in higher eukaryotes (and with both E1 and E3 in bacteria), and (iii) the inner domain, forming the central core of PDC and carrying out the catalytic reaction of E2 (Fig. 1) [2]. The lipoyl domains forming a “swinging arm” interact with the active sites of E1, E2 and E3. hBP has a domain structure similar to the hE2 structure and is composed of (i) one lipoyl domain (L3), (ii) the E3-binding domain (S1), and (iii) the inner domain. hBP has a structural role in forming the core of PDC by binding hE3 [4, 5]. In contrast to hE2, hBP does not catalyze the acetylation of CoA because it does not have the catalytic histidine residue [6].
Fig. 1.
Structural domains of human E2 and human BP. The di-domain of hBP is indicated as L3S1 [the lipoyl domain (L3), first hinge region, hE3-binding domain (S1) and second hinge region].  , lipoyllysine residue;
, lipoyllysine residue;  , hinge region.
, hinge region.
Knowledge of intra/inter protein-protein interactions is essential to understand the functions of multienzyme complexes. Our understanding of hPDC organization was recently advanced by solving the structure of hE3 bound to the E3-binding domain (S1) of hBP [7], allowing a more detailed investigation of hE3 binding to hBP. A structural analysis of this subcomplex revealed that hE3-Y438 participated in a hydrophobic interaction with hBP-I157 and a hydrogen bond interaction with hBP-R155. hE3-H348 interacted with hBP-R136 and hBP-E140 whereas hE3-D413 bound to hBP-K160 by hydrogen bond interactions. hE3-R447 is also involved in binding to hBP-N137 by a hydrogen bond. In an earlier study, we evaluated several specific residues of hBP for interactions with hE3. Substitution of hBP-I157 with S or R and hBP-R155 with D resulted in significant increases in Kd for binding with WT-hE3, indicating participation of these residues of hBP in binding to hE3 [8]. In the present study we created single-site mutations at H348, Y438, D413, and R447 in hE3. Here we report the activity parameters and isothermal titration calorimetry (ITC) analysis of hE3 mutants to determine specific contributions of these amino acid residues in binding to hBP.
Materials and methods
Construction of singly substituted amino-acid mutants of hE3
Mutagenesis reactions were performed using a double-stranded pPROEX-1 hE3 plasmid [7] with two synthetic mutagenic primers complementary to opposite strands of DNA (Integrated DNA Technologies), and the reagents supplied with the QuikChange site-directed mutagenesis kit (Stratagene) for 16 cycles. The following synthetic 2′-deoxyoligonucleotides and their complements were used as mutagenic primers (mutant codons in boldface type):
5′-GGCTGGTGGTGCTGTGGCCATTGACTACAATTG -3′ for H348A,
5′-GCTGGTGGTGCTGTGCTCATTGACTACAATTG -3′ for H348L,
5′-GCAGAAATCGACAGCCAGAGTACTGGGAG -3′ for D413A,
5′-GGGCAGAAATCGACAAACAGAGTACTGGGAG -3′ for D413N,
5′-GCTCTTGCTTTGGAAGCTGGAGCATCCTGTG -3′ for Y438A,
5′-GCTCTTGCTTTGGAATTTGGAGCATCCTGTGAAG -3′ for Y438F,
5′-CTCTTGCTTTGGAACATGGAGCATCCTGTG -3′ for Y438H,
5′-CCTGTGAAGATATAGCTGCAGTCTGTCATGCACATCC -3′ for R447A,
Expression and purification of WT-hE3 and its mutants and L3S1
Recombinant wild-type (WT) and mutant hE3s were overexpressed by Escherichia coli BL21 cells harboring plasmid pPROEX-1. Expressed E3 proteins were purified using Ni-nitrilotriacetate-agarose chromatography with a linear 30–150 mM imidazole gradient [7]. The resulting hE3s were ~95% pure as judged by densitometry of proteins separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis. L3S1 was expressed as described previously [7], and eluted with a linear gradient of imidazole (25–250 mM) in 50 mM sodium phosphate buffer (pH 8.0). The resulting L3S1 was ~95% pure as judged by densitometry of sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Activity measurements
hE3 specific activities in the forward and reverse directions were measured as reported previously [9]. Additionally, the activities of WT-hE3 and its mutants were measured after reconstitution in the complex with independently expressed WT-hE1 and WT-hE2-BP proteins. The mass ratio of WT-hE1:WT-hE2-BP:hE3 in the reconstituted hPDC was at 1:3:0.5. Pyruvate-dependent reduction of NAD+ was monitored as reported previously [10]. One unit of enzyme activity is defined as 1 μmol of product formed per min per mg of protein at 37º C.
ITC analysis
ITC measurements were carried out using a VP-ITC Microcalorimeter (MicroCal). WT-hE3, its mutants, and WT-L3S1 were dialyzed overnight against 10 mM Hepes buffer (pH 7.4) containing 150 mM NaCl and 2.3 mM EDTA, centrifuged for 10 min and then gently degassed. L3S1 (160 μM) was injected in 10 μL increments into the reaction cell (volume 1.31–1.41 mL) containing 16 μM of hE3 with constant stirring at 25º C. Thermodynamic parameters (Ka, ΔHº, and ΔSº of binding) were determined using the MicroCal ORIGIN version 7.0 software package.
Results and discussion
Kinetic studies of hE3 mutants
BP is a specific PDC component for binding of E3 in PDCs from higher eukaryotes only. The 3-D structural analysis of the subcomplex of hE3 with the di-domain (L3S1) of hBP (Fig. 1) showed a combination of hydrogen bond and hydrophobic interactions [7]. Y438 of hE3 was found to interact with I157 of hBP by a hydrophobic interaction and hE3-D413 was bound to hBP-K160 by a hydrogen bond interaction. Several hydrogen bonds are also involved in binding; hE3-H348 (bound to R136 and E140 of hBP), hE3-Y438 (bound to R155 of hBP) and R447 (bound to N137 of hBP). We created single-site mutants of hE3 based on the 3-D structural information. The specific activities of WT-hE3 and its mutants measured by two different assays are presented in Table 1. In the forward direction hE3 catalyzes the oxidation of dihydrolipoamide to lipoamide with concomitant reduction of NAD+ to NADH. In the reverse direction, hE3 catalyzes the oxidation of NADH in the presence of lipoamide. The effects of substitutions on hE3 specific activities in the forward and reverse directions were minimal [activity ranging from 60%–122%], indicating that these residues are not directly involved in hE3 catalysis.
Table 1.
hE3 and PDC specific activities of WT-hE3 and its mutants. WT- and mutant-hE3 activities were measured in their free forms in both the forward and reverse directions. The PDC activities of hE3s were measured after reconstitution with recombinant WT-hE1 and WT-hE2-BP subcomplex.
| hE3 | hE3 activity(forward reaction) (%) | hE3 activity(reverse reaction) (%) | PDC activity (%) |
|---|---|---|---|
| WT | 100 | 100 | 100 |
| H348A | 60 | 66 | 92 |
| H348L | 65 | 74 | 97 |
| D413A | 85 | 79 | 18 |
| D413N | 119 | 96 | 85 |
| Y438A | 100 | 91 | 9 |
| Y438F | 100 | 112 | 98 |
| Y438H | 99 | 92 | 20 |
| R447A | 110 | 122 | 110 |
WT-hE3 activity (100%) was 617 ± 47 units/mg (n=5) in the forward direction and 195 ± 23 units/mg (n=5) in the reverse direction. Reconstituted PDC activity with WT-hE3 (100%) was 16.5 ± 1.5 units/mg of hE1 (n=5) [9].
The overall PDC activity monitored by NADH production after reconstitution of WT-hE3 or its mutants in the complex was significantly affected by single amino acid substitutions at D413 and Y438; hE3-D413A had 18%, hE3-Y438A 9%, and hE3-Y438H 20% of the activity of WT-PDC [and 85%–110% with the other mutants] (Table 1). The results from kinetic studies indicated that D413 and Y438 of hE3 are involved in the interaction with hBP for the overall activity of PDC.
Binding study of hE3 to L3S1
The enthalpies of interaction of hE3 and hBP di-domain (L3S1) were measured by ITC (Fig. 2). We used L3S1 [the lipoyl domain (L3) and the E3-binding domain (S1), without the inner domain] (Fig. 1) because the recombinant hBP aggregated into non-homogeneous assemblies, preventing detailed binding studies between hBP and hE3 [7]. ITC analysis yielded the binding stoichiometry (N), equilibrium constant for association (Ka) and enthalpy of binding (ΔHº). Comparisons of binding of hE3 mutants with L3S1 are shown in Figs. 2 and 3. Table 2 shows the parameters for the interaction of L3S1 with WT-hE3 or its mutants in 10 mM Hepes buffer, pH 7.4 containing 150 mM NaCl and 2.3 mM EDTA at 25º C. The experiment was performed under the same conditions for different mutants as employed previously in surface Plasmon resonance studies [8]. The Kd (Kd = 1/Ka) value was calculated as 1.6 nM by ITC (Kd = 5.26 nM for WT-hE3 by surface plasmon resonance) [8]. The interaction of WT-hE3 with L3S1 was found to be enthalpically favorable (ΔHº = −6.9 Kcal/mol) and entropically favorable (TΔSº = +5.1 Kcal/mol) by ITC (Fig. 2). The binding stoichiometry for L3S1 and hE3 interaction was a 1:1 ratio (N= 0.956 site/mole), supporting binding of hBP to the hE3 dimer. The free energy of WT-hE3 with L3S1 (−12.0 Kcal/mol) was calculated from the following relationship: ΔGº =−RT lnKa. The binding of kinetics of three hE3 mutants were significantly reduced (1019-fold for hE3-Y438A, 607-fold for hE3-Y438H, and 402-fold for hE3-D413A); other hE3 mutants showed smaller effects (Table 2). The hE3-Y438A mutant showed the largest change in the Ka value. The calculated free energy changes for these three mutants with L3S1 were −7.9 Kcal/mol for hE3-Y438A, −8.2 Kcal/mol for hE3-Y438H, and −8.4 Kcal/mol for hE3-D413A. Interestingly, the binding of hE3-Y438A to hL3S1 was accompanied by an unfavorable enthalpy change (ΔHº = +2.3 Kcal/mol) and large positive entropy change (TΔSº = +10.3 Kcal/mol).
Fig. 2.
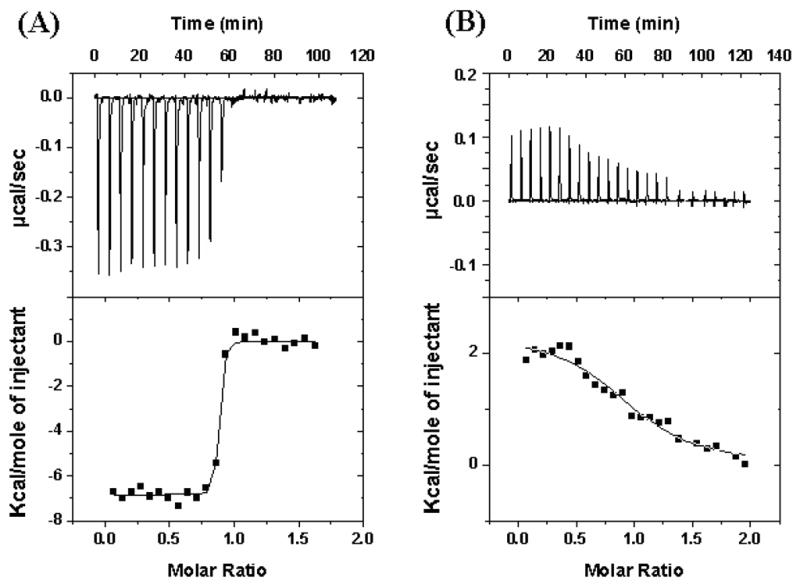
Isothemal titration calorimetry analysis of the interaction of hE3 with L3S1 in 10 mM Hepes buffer, pH 7.4 containing 150 mM NaCl and 2.3 mM EDTA at 25º C. The upper panels are raw data of enthalpy changes, each peak corresponding to each injection of L3S1 into hE3s. The lower panels show the integrated enthalpy plot. The areas under each peak are plotted against the molar ratio. (A) WT-hE3 with L3S1; (B) hE3-Y438A with L3S1. Y axis scales are different for panels A and B.
Fig. 3.
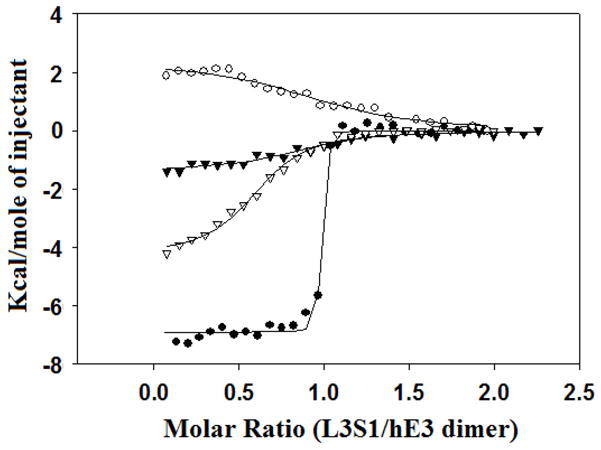
Comparison of the enthalpy integrated plots of WT-hE3 and its mutants (hE3-Y438A, hE3-Y438H and hE3-D413A) with L3S1. closed circle; L3S1/WT-hE3, open circle; L3S1/hE3-Y438A, closed triangle; L3S1/hE3-Y438H, open triangle; L3S1/hE3-D413A. The solid lines are based on a best fit single-site binding model by ORIGIN 7.0.
Table 2.
Binding parameters for WT-hE3 and its mutants with WT-L3S1 obtained by ITC. Ka and ΔGº were determined by ITC (SE < 5%).
| E3 | Ka M−1 (fold) | ΔGo Kcal/mol | ΔΔGo Kcal/mol |
|---|---|---|---|
| WT | 6.43×108 (1) | −12.0 | - |
| H348A | 1.74×108 (4) | −11.2 | 0.8 |
| H348L | 2.09×108 (3) | −11.4 | 0.6 |
| D413A | 1.60×106 (402) | −8.4 | 3.6 |
| D413N | 3.63×108 (2) | −11.7 | 0.3 |
| Y438A | 6.31×105 (1019) | −7.9 | 4.1 |
| Y438F | 5.71×108 (1) | −11.9 | 0.1 |
| Y438H | 1.06×106 (607) | −8.2 | 3.8 |
| R447A | 7.67×108 (1) | −12.1 | −0.1 |
The change in association free energy (ΔΔGº) of the mutants was calculated by the following relationship; ΔΔGº = RT ln (Ka (mutant)/Ka (wt)) [11].
According to a “hot-spot” theory, although protein interaction surfaces are large and complicated, only a few specific regions of the interface are responsible for the majority of the interaction energy [11–13]. A hot-spot is detected as a large change in the association free energy of two proteins (ΔΔGº) [11]. The ΔΔGº values of hE3 mutants with L3S1 were 4.1 Kcal/mol for hE3-Y438A, 3.8 Kcal/mol for Y438H, and 3.6 Kcal/mol for D413A [≤ 0.8 Kcal/mol for other mutants]. The ITC results reveal that hE3-Y438 and hE3-D413 are important residues for binding of hE3 to hBP.
Disease-causing mutants of hE3 were previously analyzed by ITC [14]. The Kd value with WT-hE3 to L3S1 was 0.78 nM in that study (1.6 nM in the present study). The hE3 missense mutation, hE3-R447G, had only 28% of the PDC activity compared with WT-hE3 and had a two orders of magnitude higher Kd than that of WT-hE3 by ITC [14]. Interestingly, in the present study, the substitution of hE3-R447 with A did not cause any perturbation. Since hE3-R447 interacts with hBP-N137 by hydrogen bonding, it can be speculated that the substitution hE3-447 by G interrupted its interaction with hBP-N137 due to its structural conformation.
In summary, the analysis of several amino acid residues in binding of hE3 to hBP indicates that: (i) hE3-D413 and hE3-Y438 are important residues in binding of hE3 to hBP, (ii) hE3-D413 and hE3-Y438 are not directly involved in hE3 catalysis, (iii) Y438 seemed to play a somewhat larger role than D413, and (iv) correct assembly of protein-protein interactions is required for the optimal catalysis by this multi-enzyme complex.
Acknowledgments
This work was supported by National Institutes of Diabetes and Digestive and Kidney Grant DK080748. We thank Dr. Murray Ettinger of this department for critical reading of this manuscript.
Abbreviations
- h
human
- WT
wild-type
- PDC
pyruvate dehydrogenase complex
- E1
pyruvate dehydrogenase
- E2
dihydrolipoamide acetyltransferase
- E3
dihydrolipoamide dehydrogenase
- BP
E3-binding protein
- L3S1 (di-domain)
the lipoyl domain (L3), first hinge region, E3-binding domain (S1) and second hinge region of hBP
- ITC
isothermal titration calorimetry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors maybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Patel MS, Roche TE. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 1990;4:3224–3233. doi: 10.1096/fasebj.4.14.2227213. [DOI] [PubMed] [Google Scholar]
- 2.Patel MS, Korotchkina LG. The biochemistry of the pyruvate dehydrogenase complex. Biochem Mol Biol Educ. 2003;31:5–15. [Google Scholar]
- 3.Holness MJ, Sugden MC. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem Soc Trans. 2003;31:1143–1151. doi: 10.1042/bst0311143. [DOI] [PubMed] [Google Scholar]
- 4.Smolle M, Lindsay JG. Molecular architecture of the pyruvate dehydrogenase complex: bridging the gap. Biochem Soc Trans. 2006;34:815–818. doi: 10.1042/BST0340815. [DOI] [PubMed] [Google Scholar]
- 5.Hiromasa Y, Fujisawa T, Aso Y, et al. Organization of the cores of the mammalian pyruvate dehydrogenase complex formed by E2 and E2 plus the E3-binding protein and their capacities to bind the E1 and E3 components. J Biol Chem. 2004;279:6921–6933. doi: 10.1074/jbc.M308172200. [DOI] [PubMed] [Google Scholar]
- 6.Harris RA, Bowker-Kinley MM, Jeng J, et al. Dihydrolipoamide dehydrogenase-binding protein of the human pyruvate dehydrogenase complex. DNA-derived amino acid sequence, expression, and reconstitution of the pyruvate dehydrogenase complex. J Biol Chem. 1997;272:19746–19751. doi: 10.1074/jbc.272.32.19746. [DOI] [PubMed] [Google Scholar]
- 7.Ciszak EM, Makal A, Hong YS, et al. How dihydrolipoamide dehydrogenase-binding protein binds dihydrolipoamide dehydrogenase in human pyruvate dehydrogenase complex. J Biol Chem. 2006;281:648–655. doi: 10.1074/jbc.M507850200. [DOI] [PubMed] [Google Scholar]
- 8.Patel MS, Korotchkina LG, Sidhu S. Interaction of E1 and E3 components with the core proteins of the human pyruvate dehydrogenase complex. J Mol Catal B Enz. 2009;61:2–6. doi: 10.1016/j.molcatb.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel MS, Hong YS. Lipoic acid as an antioxidant. The role of dihydrolipoamide dehydrogenase. Methods Mol Biol. 1998;108:337–346. doi: 10.1385/0-89603-472-0:337. [DOI] [PubMed] [Google Scholar]
- 10.Korotchkina LG, Patel MS. Site specificity of four pyruvate dehydrogenase kinase isoenzymes toward the three phosphorylation sites of human pyruvate dehydrogenase. J Biol Chem. 2001;276:37223–37229. doi: 10.1074/jbc.M103069200. [DOI] [PubMed] [Google Scholar]
- 11.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 12.Schreiber G, Fersht AR. Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. J Mol Biol. 1995;248:478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 13.Chen CZ, Shapiro R. Site-specific mutagenesis reveals differences in the structural bases for tight binding of RNase inhibitor to angiogenin and RNase A. Proc Natl Acad Sci USA. 1997;94:1761–1766. doi: 10.1073/pnas.94.5.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brautigam CA, Wynn RM, Chuang JL, et al. Structural insight into interactions between dihydrolipoamide dehydrogenase (E3) and E3 binding protein of human pyruvate dehydrogenase complex. Structure. 2006;14:611–621. doi: 10.1016/j.str.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]