Abstract
A strategy is presented for the live cell imaging of messenger RNA (mRNA) using hairpin DNA-functionalized gold nanoparticles (hAuNP). hAuNP improve upon technologies for studying RNA trafficking by their efficient internalization within live cells without transfection reagents, improved resistance to DNAse degradation, low cytotoxicity, and the incorporation of hairpin DNA molecular beacons to confer high specificity and sensitivity to the target mRNA sequence. Furthermore, the targeted nanoparticle-beacon construct, once bound to the target mRNA sequence, remains hybridized to the target, enabling spatial and temporal studies of RNA trafficking and downstream analysis. Targeted hAuNP exhibited high specificity for glyceraldehyde 3-phosphate dehydrogenase (GADPH) mRNA in live normal HEp-2 cells, and respiratory syncytial virus (RSV) mRNA in live RSV-infected HEp-2 cells with high target to background ratios. Multiplexed fluorescence imaging of distinct mRNAs in live cells and simultaneous imaging of mRNAs with immunofluorescently-stained protein targets in fixed cells was enabled by appropriate selection of molecular beacon fluorophores. Pharmacologic analysis suggested that hAuNP were internalized within cells via membrane-nanoparticle interactions. hAuNP are a promising approach for the real-time analysis of mRNA transport and processing in live cells for elucidation of biological processes and disease pathogenesis.
Keywords: gold nanoparticles, RNA, mRNA, optical imaging, respiratory syncytial virus, molecular beacons, live cell imaging, microscopy, fluorescence
Introduction
Messenger RNA (mRNA) is the blueprint for the cellular production of proteins, and is involved in a number of sorting and trafficking-associated processes thought to enable specific spatial regulatory functions within the cell, such as elongation of axonal growth cones, and guidance of fibroblast migration.1,2 Aberrant mRNA processing and localization play significant roles in several developmental and pathological abnormalities including viral infection,3 disrupted embryonic development,4 altered cell morphology,5 and susceptibility to cellular apoptosis.6,7 While several established techniques for the investigation of mRNA-specific processes are available using lysed or fixed cells, such as polymerase chain reaction (PCR)8 and in situ hybridization (ISH),9,10 the spatial and temporal information of RNA obtainable from these procedures is limited. Approaches for imaging the complex spatial and temporal dynamics of mRNA processes within living cells are needed to investigate the molecular-scale events involved in intracellular processing and trafficking, for enhanced development of diagnostic tools and therapeutic interventions.
Several live cell imaging probes have been utilized to specifically image mRNA within the intracellular milieu in real time, including linear oligonucleotide probes and fluorescence resonance energy transfer (FRET) reporters such as molecular beacons.11–14 While these techniques overcome the limitations and time-consuming RNA processing steps associated with imaging fixed or lysed cells, they require additional procedures for effective delivery of the probe into the cytoplasm, including electroporation,15 chemically-mediated membrane permeabilization,15,16 or surface engineering using cationic cell-penetrating peptides.11,17 Furthermore, these probes are susceptible to nuclease-mediated degradation, and can exhibit high false-positive rates when using certain surface modifications and transfection methods.17–19
Recently, an approach incorporating oligonucleotide-functionalized gold nanoparticulate probes was reported to address these limitations.20,21 Features of using these gold-oligonucleotide complexes for delivery into live cells for mRNA detection include efficient intracellular delivery without the need for transfection or permeabilization reagents, and compared to previous approaches, an increased resistance to nuclease degradation and high target to background ratio of detection.20,21 The reduction in fluorescence background is achieved by taking advantage of gold colloids’ intrinsic dye-quenching properties.22 In this approach, a gold colloid surface functionalized with a probe DNA oligonucleotide sequence targeted toward an intracellular RNA sequence of interest. The surface oligonucleotide is then hybridized to a fluorescently-labeled complementary DNA strand, which is quenched due to proximity with the gold colloid surface. Upon intracellular hybridization of the construct with target RNA, the fluorescent strand is displaced from the nanoparticle-coupled oligonucleotide and is released into the cytoplasm. This event results in emission of in response to the intracellular presence of target RNA. However, since the fluorescent reporter is not hybridized to the target RNA, but is competitively-displaced and diffuses away from the site of hybridization, this strategy cannot be utilized to determine intracellular mRNA localization.
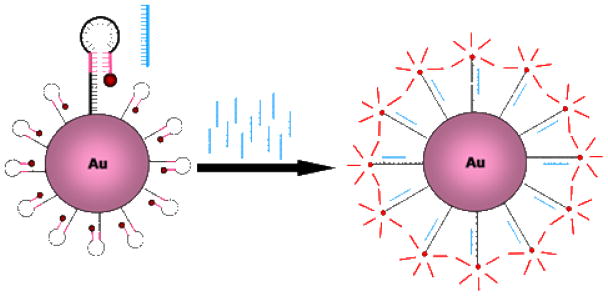
In this work, a new live cell mRNA detection methodology is described for the imaging of mRNA in living cells. This strategy retains the advantages of gold-oligonucleotide constructs, such as intracellular uptake and high specificity, but also exhibits the additional advantage of preserving spatial localization of target mRNA within the cell (Figure 1). In this approach, a modification of a molecular beacon-based strategy,23 a hairpin DNA moiety (h) is coupled to a gold nanoparticle (AuNP) surface. The hairpin DNA structure consists of a 5′ thiol followed by a 10-base polythymine linker sequence and a stem-loop-stem sequence, and is terminated at the 3′ end with a fluorophore. The loop region is designed to hybridize with the target RNA of interest. In the closed state, the hairpin conformation is maintained by the complementarity of the 5′ and 3′ 5 base stem sequences. Additionally, in this closed state, the 5′ thiol group enables linkage to the AuNP surface with sufficient proximity to permit AuNP quenching of fluorescence from the 3′ dye. Specific hybridization of hAuNP with target RNA opens the hairpin DNA sequence, which then separates the 3′ fluorophore from the 5′ AuNP at a sufficient distance to overcome fluorescence quenching. However, the AuNP remains bound to the hairpin DNA in the hybridized state, enabling spatial localization of intracellular target mRNA in live cells.
Figure 1.
Hairpin DNA functionalized gold nanoparticles (hAuNP) for detection of mRNA in live cells. Hairpin DNA is coupled to the gold colloid surface VIA a 5′ thiol, thereby quenching fluorescence emission of the 3′ fluorophore (red). The stem regions (pink) maintain the hairpin conformation through complementary hydrogen bonding. Hybridization between the loop region (black) and the target DNA or RNA sequence (blue) results in opening of the hairpin DNA and fluorescence emission.
In order to demonstrate the utility of this strategy for live cell imaging of mRNA, hAuNP were designed which target specific mRNA sequences of Respiratory Syncytial Virus (RSV), and of the enzyme glyceraldehyde 3-phosphate dehydrogenase (GAPDH). RSV is the leading cause of lower respiratory tract infections in children under age 5, and contributes to over 125,000 hospitalizations per year in the United States.24 RSV is a single-stranded RNA virus of the paramyxovirus family, which includes viruses which cause measles and mumps. A vaccine for preventing RSV infection is not available, and current treatments for RSV infections are only effective if provided early in the course of infection.25 RSV utilizes a library of cell binding (G) and membrane fusion (F) proteins to enter the cell and exploit intracellular machinery to propagate and infect other cells. An important event in this process is translation of viral mRNA using the host cell’s ribosomal apparatus to produce viral proteins. While therapeutics such as pavalizumab (anti-F protein) target these viral proteins to prevent further infection of other cells, understanding the real-time concentration, localization, and trafficking mechanisms of specific viral mRNAs, and targeting RSV at the mRNA level may provide opportunities for earlier, more effective interventions.26 GAPDH is an enzyme involved in glycolysis. Due to its stable, constitutive expression in almost all mammalian cell types, it serves as a useful internal control for normalization in standard quantitative molecular biology assays such as RT-PCR. In this study, hAuNP stability, targeting specificity and sensitivity were evaluated for RSV mRNA in infected mammalian cells, concurrently with hAuNP targeting GAPDH mRNA as an internal control. Also investigated were the cytotoxicity of hAuNP in vitro, the mechanisms underlying hAuNP uptake, and the potential of hAuNP for multiplexed imaging of intracellular fluorescence imaging of mRNA and proteins.
Materials and Methods
Materials
The HEp-2 epithelial cell line was obtained from American Type Culture Collection (ATCC). RSV type A2 virus and anti-F protein monoclonal antibody were a gift from the laboratory of James Crowe, Vanderbilt University Medical Center. Oligonucleotides were synthesized by Biosearch Technologies. Fetal bovine serum, gentamicin/amphotericin B, OptiMEM media, quantum dot nanocrystals, cholera toxin B, dextran, LIVE/DEAD viability kit, Mitotracker Red, and secondary detection antibodies were from Invitrogen Corporation. Lab-Tek II chambered coverglass was from Nunc Inc. Gold nanoparticles were from Ted Pella, Inc. Cytoseal 60 was from Richard Allan Scientific. Goldenhance gold colloid detection kit was from Nanoprobes, Inc. Cytochalasin D and Dynasore were from Tocris Bioscience. Brefeldin A was from Epicentre Biotechnologies. Genistein was from Ascent Scientific. Accutase was purchased from Innovative Cell Technologies. All other chemicals were purchased from Sigma-Aldrich at the highest purity available.
Synthesis and characterization of hAuNP
Hairpin DNA used for coupling to the AuNP surface were synthesized with the following sequences: RSV: 5′-(SH)-TTT TTT TTT TCG ACG AAA AAT GGG GCA AAT ACG TCG-(Cy5)-3′, GAPDH: 5′-(SH)-TTT TTT TTT TCG ACG GAG TCC TTC CAC GAT ACC ACG TCG-(CAL Fluor Red 590)-3′. Bolded regions indicate sequences encoding specificity for target mRNAs, and regions in parenthesis indicate modifications of 5′ thiol and 3′ fluorophore conjugations. The RSV sequence chosen is based on a consensus sequence found in RSV genomic RNA, and has been targeted successfully using antisense therapies directed against RSV replication.27 The GAPDH targeted sequence is based on exon 6 of the GAPDH coding region. Sequences for complementary targets for evaluation of hAuNP hybridization specificity were as follows: RSVcomp: 3′-TTT TTT TAC CCC GTT TAT TTT TT-5′, GAPDHcomp: 3′-TTT TTT GGT ATC GTG GAA GGA CTC TTT TT-5′. A negative control target sequence with similar C-G content of RSVcomp was synthesized as follows: 3′-AGT ATT GGA AGT CGT TCG AAA TTG ATC ACT CGC A-5′.
DNA concentration was verified by UV absorbance at 260 nm. Prior to coupling of hairpin DNA to AuNP, lyophilized DNA was resuspended in 100 mM DTT, 0.2 mM phosphate buffer (pH = 8.3) and incubated for one hour at 25°C to reduce residual 5′ disulfide bonds. A solution of 450 nM DNA and 2.2325 nM 15 nm AuNP was prepared in nuclease-free water and incubated for 4 hours at 25°C protected from light. After 4 hours, the solution was buffered to 10 mM phosphate buffer (pH 7.0), and brought to 0.1 M NaCl, and 0.1% Tween-20. The hAuNPs were then incubated for an additional 4 hours at 25°C. The NaCl concentration was then adjusted to 0.2 M and hAuNP were incubated for an additional 4 hours at 25°C. Finally, the NaCl concentration was adjusted to 0.3 M and incubated for a final 4 hours at 25°C. The solution was then centrifuged at 13,200 rpm for 30 min and hAuNPs were resuspended in PBS. The supernatant was reserved for further analysis. The process of centrifugation and resuspension was repeated thrice.
hAuNP concentration was measured by absorbance at 260 nm and 520 nm, using the extinction coefficient for 15 nm AuNP ε520 = 3.64 × 108 M−1cm−1. Reserved supernatant was also measured for absorbance at 260 nm to determine unbound DNA concentration, enabling calculation of the number of hairpin DNA molecules per AuNP. To determine fluorescence spectra of hAuNP, a solution of 2 μM of RSVcomp, GAPDHcomp, or negative control sequence and 1.4 nM hAuNP specific for RSVcomp or GAPDHcomp was incubated for 1 hour at 70°C and subsequently at 25°C for 12 hours, protected from light. Emission was scanned using a Cary Eclipse fluorescence spectrophotometer from 500–800 nm in 1 nm increments. In a separate experiment, 1.4 nM hAuNP targeting RSV or GAPDH RNA as described above were incubated with increasing concentrations of complementary target RNA (RSVcomp, GAPDHcomp, or negative control sequence) as indicated at 37°C in PBS for 6 hours, and measured for fluorescence emission intensity using a SpectraMax M5 microplate reader (Molecular Devices) configured for hAuNP fluorescence. As a blank, a AuNP surface functionalized with a 10 thymine spacer with identical fluorophore on the 3′ end was used to correct for intrinsic background fluorescence of the quenched hAuNP.
For measurement of hydrodynamic diameter, 0.25 nM solutions of AuNP, or hAuNP specific for RSVcomp or GAPDHcomp were resuspended in nuclease-free water and measured using dynamic light scattering (DLS) on a Malvern Nano ZS.
Resistance of hAuNP to DNAse I degradation was assayed fluorimetrically by incubating a solution of 1.6 nM hAuNP in PBS with 50 mg/mL bovine serum albumin and 0.38 mg/L DNAse I for 120 minutes as previously described.21 Fluorescence emission resulting from degradation of DNA and thus liberation of the fluorophore from the quenching effect of the AuNP surface was monitored throughout the duration of the reaction using a Biotek Synergy HT configured for reading hAuNP-specific emission in 1 min. increments.
Cell culture
HEp-2 cells were cultured at 37°C, 5% CO2 in Opti-MEM supplemented with 2% FBS, 2% L-Glutamine, and 1% gentamicin/amphotericin B. For infection of HEp-2 with RSV, cells in log phase growth were counted using a hemacytometer with trypan blue exclusion. Cells were plated 24 hrs. prior to incubation with virus at varying multiplicities of infection (MOIs) as previously described.26 Incubation of cells with RSV at the appropriate concentration was conducted with OptiMEM for at least 1 hour prior to assays.
Flow cytometric analysis
HEp-2 cells infected at an MOI ranging from 0 to 1 were incubated with 0.5 nM RSV and GAPDH-targeted hAuNP for 6 hours, rinsed thrice with PBS to remove unbound/uninternalized hAuNP, and detached using Accutase. Cells were assayed using a Becton Dickinson LSR II flow cytometer, monitoring emission of hAuNP using appropriate filter settings for CAL Fluor 590 or Cy5. In a separate experiment, cell viability was measured in similarly-treated cells using the LIVE/DEAD single color fixable green kit (Invitrogen Corp.) according to the manufacturer’s instructions. Data was analyzed using FlowJo 7.5 (Treestar Software).
Pharmacologic and environmental inhibition of endocytosis
All solutions for inhibition of HEp-2 endocytosis were sterilized with Pall Supor 0.22 μm filters prior to usage in cell culture assays. HEp-2 cells were seeded in 96-well microplates at a density of 5 × 104 cells per well. A subset of these wells was selected for RSV infection the following day, and cells were incubated for an additional 48 hrs. Cells were rinsed three times in serum-free DMEM, and were either untreated or pretreated with 100 μL volumes of DMEM containing nystatin/progesterone, sodium azide/2-deoxy-D-glucose, chlorpromazine, chloroquine, brefeldin A, dynasore, FBS, nocodazole, heparin, amiloride, genistein, cytochalasin D, or no additives at specified concentrations for 1 hour. The range of working concentrations of reagents was determined using values reported in the literature, or, when possible, empirically using known tracers of endocytosis: transferrin (clathrin-dependent), cholera toxin B (lipid-raft-dependent), or dextran (caveolar-depedent). An acceptable working concentration for hAuNP internalization assays was determined when the reagent inhibited internalization of the tracer by > 15% in HEp-2 cells as measured by microplate spectrophotometry. This concentration was used as the middle concentration in a range of 3 concentrations, to evaluate dose-dependent effects. Media was then aspirated and replaced with DMEM containing the same additives supplemented with RSV or GAPDH-targeted hAuNPs, and cells were incubated for 4 hrs. Following this period, cells were rinsed thrice in HBSS, fixed for 15 min. at room temperature with 4% paraformaldehyde in PBS, and analyzed for hAuNP fluorescence using a Molecular Devices Spectramax M5 microplate reader configured for hAuNP detection. As several solutions used to dissolve endocytosis study reagents were made in DMSO, cells were incubated with hAuNPs suspended in DMEM with the highest concentration of DMSO used (0.5%) to monitor solvent-induced effects. Cells were incubated in parallel with endocytosis study reagents, DMEM, and 1 μM calcein-AM as a viability indicator to ensure reagents did not induce significant (> 5%) cell death in any case as determined by fluorescence microscopy and automated image analysis using Image Pro Plus 5.1.
Confocal microscopy of hAuNP
Uninfected HEp-2 cells or infected HEp-2 cells (5 × 104) in Lab-Tek II chambered coverglass were incubated with 0.5 nM hAuNP for 6 hours. Prior to live cell imaging, cells were rinsed thrice and resuspended in PBS. Imaging was conducted using a Zeiss LSM 510 Meta laser scanning confocal microscope configured for hAuNP-specific emission. SYTO-13 was used as a nuclear counterstain, and in some experiments, Mitotracker Red live cell mitochondrial tracking reagent was used according to the manufacturer’s instructions. Cell viability was monitored in cultured cells processed in parallel using trypan blue to ensure > 95% viability throughout imaging experiments.
For immunofluorescence and immunohistochemical analysis of hAuNP-treated cells, HEp-2 were fixed with 4% paraformaldehyde in PBS for 15 minutes at 25°C, then rinsed thrice with PBS containing 50 mM glycine. Cells were stained for RSV-specific F protein as previously described.26 For detection of gold colloids in fixed cells, the Goldenhance LM gold labeling kit was used according to the manufacturer’s instructions. Samples were mounted in Cytoseal 60 and imaged using confocal microscopy.
Results and Discussion
Characterization of hAuNP
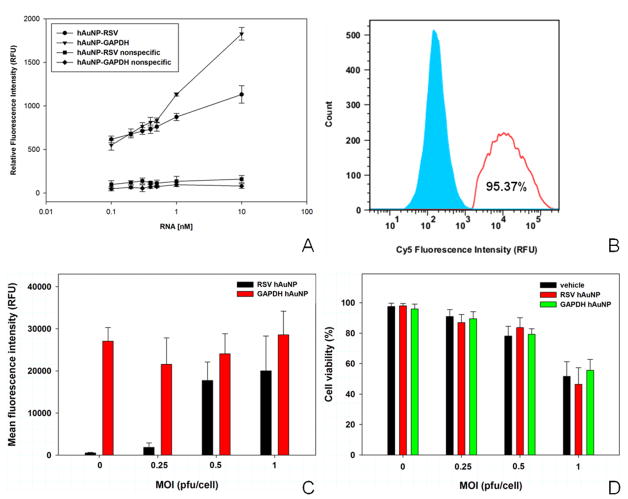
UV - visible absorbance spectra of hAuNP exhibited absorbance peaks at nucleic acid- and AuNP-specific peaks of 260 nm and 520 nm, respectively (Supporting Figure 1), and indicated a coupling efficiency of approximately 40 hairpin DNA moieties per AuNP. DLS analysis indicated an increase in hydrodynamic diameter of 11 nm when AuNP were coupled to RSV-specific hairpin DNA, and an increase of 14 nm when coupled to GAPDH-specific hairpin DNA. hAuNP stability against degradation by DNAse I was quantified using a fluorimetric assay (Supporting Figure 2). The degradation rate of hAuNP targeted toward RSV was 0.072 nmol min−1, and the degradation rate for GAPDH-specific hAuNP was 0.094 nmol min−1. Fluorescence spectrophotometry studies confirmed that hAuNP were specific for target sequences, with emission of fluorescence consistent with the fluorophore coupled to the 3′ end of the hairpin DNA (Figure 2A). hAuNP targeted against RSV or GAPDH both exhibited dose-specific increases in fluorescence intensity in response to complement concentration (Figure 2A). RSV- or GAPDH- hAuNP did not react to an appreciable level when incubated with an concentration of mismatched complementary DNA featuring similar C-G content of RSVcomp (Figure 2A).
Figure 2.
Hybridization specificity, intracellular targeting specificity, and cytotoxicity of hAuNP. (A) RSV-targeted (black) and GAPDH-targeted hAuNP (green) exposed to increasing doses of their target complementary RNA exhibited dose-dependent increase in fluorescence. This effect was not observed when hAuNP were exposed to nonspecific target RNA with similar C–G content (red). (B) Using FACS analysis, RSV-specific hAuNP signal was detected in 95% of RSV-infected cells (MOI = 1, red), but was similar to background fluorescence in uninfected cells (blue). (C) In FACS analysis, RSV-specific hAuNP exhibited increases in fluorescence intensity with increasing RSV infection (MOI), whereas GAPDH-targeted hAuNP emission signal was relatively constant in HEp-2 cells under untreated or RSV-infected conditions. (D) hAuNP did not significantly reduce cell viability in HEp-2 when compared to untreated cells Data presented as n = 3, mean ± S.D..
Characterization data of hAuNP are consistent with previous observations of AuNP-oligonucleotide bioconjugates, with regards to absorbance and emission spectra, size, high specificity, and dose-dependent sensitivity.12,20,21,28 Several observations, however, suggest that hAuNP offer significant advantages in the context of live cell imaging of mRNA. The surface functionalization of up to 40 hairpin DNA moieties may enable high sensitivity detection of low abundance RNA targets within the intracellular milieu. AuNP are efficient quenchers of numerous fluorophores,29 providing a methodology for multiplexed imaging of distinct RNA targets by judicious selection of spectrally-distinct fluorophores. Additionally, hAuNP exhibit improved stability against nuclease degradation compared to previous approaches. Fluorophore-quencher paired molecular beacons have exhibited degradation rates of up to 1.25 nmol−1 min−1,12 and gold-oligonucleotide probes for live cell mRNA detection were degraded at a rate of 0.275 nmol−1 min−1.21 The degradation rate of hAuNP was less than 0.1 nmol−1min−1, which may enable imaging of mRNA with higher target to background ratios.
Flow cytometric analysis
The population of RSV-infected cells (MOI = 1) exhibiting RSV-specific hAuNP fluorescence was 95.3%, which is close to the theoretical fraction of cells that would be infected with RSV at this MOI (> 90%) (Figure 2B). RSV-specific hAuNP fluorescence in infected HEp-2 cells was dependent on the MOI (Figure 2C), consistent with measurements conducted with RSVcomp in buffer. The fluorescence of GAPDH-specific hAuNP, however, was similar in RSV-infected and uninfected cells. Neither RSV- or GAPDH-specific hAuNP were toxic to HEp-2 cells in culture when compared to untreated cells at the same MOI, although cell death attributable to increased RSV infection was observed as expected (Figure 2D).
Flow cytometric analysis collectively indicated that hAuNP were specific for mRNA targets within cells, were efficiently taken up by cells in culture without the use of transfection reagents or cell permeabilization, and were not cytotoxic to cells. Furthermore, hAuNP exhibited target-dependent increases in fluorescence, which may enable the use of hAuNP for the quantitative determination of intracellular mRNA content. The consistent emission signal of GAPDH-targeted hAuNP in RSV-infected and uninfected cells suggests that hAuNP can be used for mRNA normalization and assay standardization as an internal control, similar to how GAPDH primers are utilized in RT-PCR. Furthermore, in addition to its role as an mRNA loading control, GAPDH hAuNP fluorescence emission in uninfected and infected cells serves as a control for the adverse effects of RSV on cells, which in response to infection exhibit marked changes in membrane permeability and integrity, in addition to cell death.26 The consistent GAPDH-targeted hAuNP signal, therefore, also validates that hAuNP efficiently enter cells irrespective of viral infection, which will enable studies of mRNA localization for any molecular biology application which interrogates cellular mRNA dynamics in health and disease.
Mechanisms of hAuNP internalization
An interesting feature of hAuNP and other AuNP-based mRNA detection probes is their nonspecific internalization by mammalian cells,21 but the mechanisms underlying this process are not clear. A systematic investigation of hAuNP internalization in HEp-2 cells under a variety of pharmacologic and environmental conditions which inhibit endocytosis was conducted (Table 1). To eliminate the possibility of aberrant endocytosis under the influence of endocytic inhibitors due to RSV infection, only uninfected cells incubated with GAPDH-targeted hAuNP were studied. As shown in Table 1, compounds which are known to inhibit classical modes of endocytosis, such as clathrin and caveolin-mediated endocytosis, were used to treat cells. Global inhibitors of endocytosis which inhibit cellular metabolism, such as sodium azide/2-deoxy-D-glucose, were also utilized. To investigate the involvement of intracellular trafficking machinery used to sort endosomes on hAuNP uptake, inhibitors of Golgi-ER transport and microtubule-mediated transport were incubated with cells. In addition, treatment of cells with nystatin and progesterone, as well as 4°C treatment, which modulate the fluidity of plasma membrane lipid domains, was also conducted to study the effects of membrane fluidity on hAuNP uptake. Lastly, incubation of cells with hAuNP in the presence of serum (FBS) was conducted to determine whether adsorption of serum proteins on the cellular and/or hAuNP surface was capable of affecting uptake efficiency.
Table 1.
Summary of inhibitors/enhancers of hAuNP uptake in HEp-2 cells. Results are expressed as mean ± S.D. percentage of control fluorescence intensity in microplate spectrophotometry assays. These results are shown for hAuNP-GADPH signal in uninfected HEp-2 cells, although results are representative of those observed for hAuNP-RSV in infected cells.
| Inhibitor | Mechanism of Action | Relative Fluorescence Intensity (% of untreated) |
|---|---|---|
| Nystatin/Progesterone | depletes membrane cholesterol, inhibits lipid rafts and caveolin | 153.40 ± 3.72 |
| Chlorpromazine | inhibits clathrin pit formation | 101.21 ± 3.56 |
| Heparin | competitively inhibits heparin sulfate proteoglycan-mediated endocytosis | 98.03 ± 4.87 |
| Amiloride | inhibits fluid-phase macropinocytosis | 93.94 ± 1.66 |
| Genistein | inhibits tyrosine kinases | 93.60 ± 4.70 |
| Cytochalasin D | inhibits actin-based intracellular transport | 92.14 ± 3.84 |
| Nocodazole | inhibits microtubule-based intracellular transport | 88.60 ± 2.49 |
| Sodium azide/2-deoxyglucose | inhibits cellular metabolism | 87.43 ± 1.42 |
| Dynasore | inhibits dynamin-mediated endocytosis (including clathrin, caveolin) | 87.33 ± 3.37 |
| Chloroquine | inhibits endosomal acidification | 87.13 ± 1.93 |
| Brefeldin A | inhibits late endosomal trafficking (Golgi-ER sorting) | 87.10 ± 1.03 |
| 4°C pretreatment | reduces membrane fluidity | 78.87 ± 8.52 |
| 10% Serum | may inhibit nanoparticle uptake due to adsorption of serum proteins | 69.05 ± 4.48 |
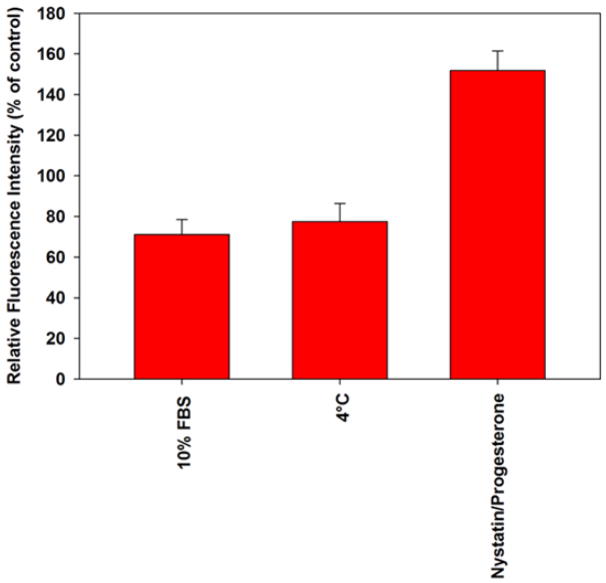
Treatment of HEp-2 cells with known pharmacologic inhibitors of clathrin-mediated endocytosis (chlorpromazine30), dynamin-dependent endocytosis (Dynasore31), and macropinocytosis (amiloride32) did not exhibit dose-dependent effects on hAuNP internalization (Table 1). Furthermore, intracellular endosomal trafficking inhibitors such as brefeldin A (Golgi-ER trafficking33) and nocodazole (microtubule-assisted trafficking34) did not significantly alter hAuNP uptake. Ultimately, not even global inhibition of endocytosis via cellular energy depletion (sodium azide, 2-deoxy-D-glucose35) impacted hAuNP uptake compared to control. These data suggest that energy-dependent translocation mechanisms play a minimal role in the cytosolic delivery of hAuNP. The only treatment which increased the uptake of GAPDH-targeted hAuNP uptake in HEp-2 cells was nystatin/progesterone treatment (Figure 3). Nystatin and progesterone treatment increases membrane fluidity via depletion of cholesterol and inhibits caveolar endocytosis.36 Two treatments, 4°C pretreatment and inclusion of FBS in incubation media, reduced hAuNP intracellular uptake. In separate experiments, nystatin/progesterone treatment did not alter target RNA concentrations as determined by RT-PCR (data not shown). While the data suggest a role of cell membrane-hAuNP interactions in internalization, the multiple types of cellular machinery responsible for uptake of nanomaterials have not been completely elucidated. Therefore, a non-classical mode of endocytosis of hAuNP cannot be ruled out due to lack of extensive methodology in this emerging field.
Figure 3.
Summary of major inhibitors and enhancers of hAuNP transport. Results are expressed as mean ± SD, n = 3, for HEp-2 cells incubated with GADPH mRNA-targeted hAuNP to eliminate artifacts in cellular transport due to RSV infection.
The results of fluorescence-based pharmacologic screening assays indicate that conditions which influence cell membrane fluidity have profound influences on hAuNP internalization. Some inhibitors used in the assay exhibit multiple effects on cellular endocytosis. Specifically, preincubation of cells at 4°C inhibit cellular metabolism and reduce membrane fluidity, and incubation with nystatin compounds inhibit caveolar endocytosis and sequester cholesterol, thus increasing membrane fluidity.36 However, the observation that only 4°C or nystatin/progesterone treatment and not any other specific inhibitors of active endocytosis impaired hAuNP uptake by more than 15% supports the hypothesis that hAuNP uptake is primarily due to lipid membrane-hAuNP interactions. The inclusion of 10% serum in incubation media resulted in reduced internalization of hAuNP. This result could be either due to serum protein adsorption on hAuNP surfaces, thereby conferring steric hindrance or bulkiness upon the construct, or due to competitive inhibition of the attachment protein(s) or lipid domains on the cell membrane by serum components.37 This result is different from previous observations involving AuNP-oligonucleotide complexes, in which serum protein adsorption was hypothesized to promote uptake of AuNP within cells.38 Therefore, uptake mechanisms of AuNP-oligonucleotide complexes may differ substantially based on the configuration of the oligonucleotide on the AuNP surface, and warrants further investigation. Possible explanations for distinct uptake mechanisms may include size dependence, alterations in surface charges, or DNA base pair dependent interactions.
Immunofluorescence analysis
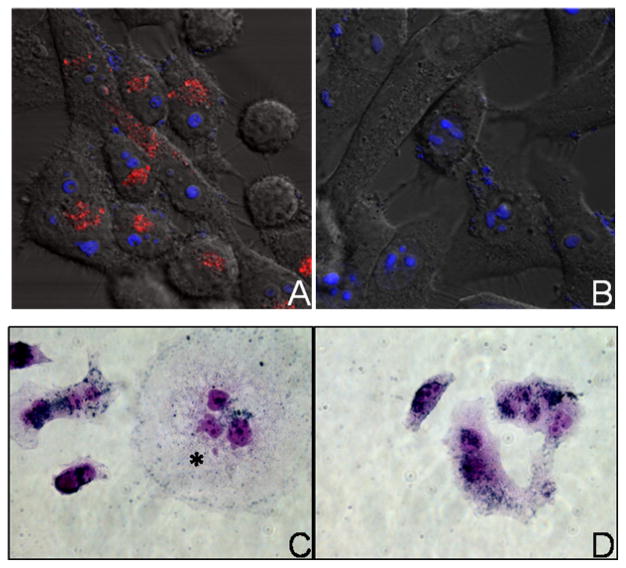
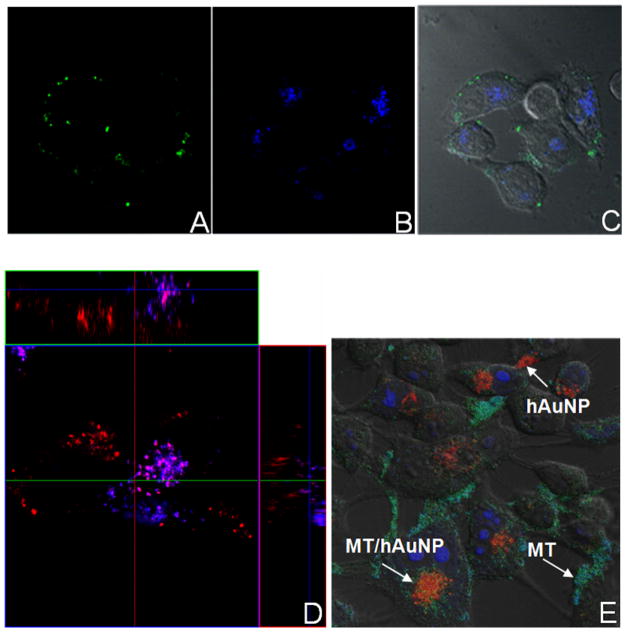
Emission from RSV-specific hAuNP was detected in RSV-infected cells, but not in uninfected cells (Figure 4A-B), consistent with flow cytometric analysis. Distribution of emission was associated with perinuclear patches in the cytoplasm (Figure 4A). Immunohistochemical staining of HEp-2 cells exposed to hAuNP, regardless of their target sequence, indicated that hAuNP were present throughout the cytoplasm in a perinuclear distribution, in RSV syncytia (multinucleated, fused cells) of infected cells (Figure 4C, asterisk), or in normal uninfected cells (Figure 4D). The customized fluorescence emission profiles of hAuNP enabled multiplexed analysis of intracellular mRNAs and proteins (Figure 5), with preservation of mRNA spatial localization afforded by hAuNP. RSV F protein was detected on cell membranes, in areas distinct from RSV mRNA localization (Figure 5A–C), whereas colocalization of RSV mRNA was frequently observed within several regions containing GAPDH mRNA (Figure 5D) and mitochondrial organelles (Figure 5E).
Figure 4.
Intracellular uptake and fluorescence emission of hAuNP. (A) RSV-specific hAuNP emission (red) is observed in RSV-infected cells (MOI = 0.25), but not in uninfected cells (B). Blue: nuclear counterstain. Magnification 400×. (C) Gold nanoparticle immunohistochemical staining indicates that hAuNP are internalized within RSV-infected HEp-2 cells, or uninfected cells (D). Uptake is irrespective of cell infection and the intended target of hairpin DNA.
Figure 5.
Multiplexed confocal analysis of hAuNP-labeled RSV mRNA with other mRNA sequences and subcellular organelles. (A) F protein was detected by monoclonal antibody in RSV-infected HEp-2 (MOI = 0.25) (B) RSV-specific hAuNP emission (C) DIC overlay. (D) Simultaneous imaging of RSV mRNA (red) and GAPDH mRNA (blue) using spectrally-distinct hAuNP indicated areas of distinct localization and colocalization within the cell. (E) RSV-targeted hAuNP were observed to colocalize with Mitotracker(MT)-stained mitochondria, but are also observed in distinct locations within RSV-infected cells.
Studies collectively indicate that hAuNP are nonspecifically-internalized by cells, but exhibit mRNA-specific emission, enabling imaging with high signal to background ratios. Uptake of hAuNP occurs irrespective of the presence or absence of target mRNA. Targeted hAuNP were observed to colocalize in HEp-2 cells with molecular beacons targeted toward the same transcript and introduced into cells using streptolysin O-based reversible membrane permeabilization as previously published (Supplementary Figure 3).14 hAuNP did not penetrate the nuclear compartment following translocation (Supplementary Figure 4). As the design of the hAuNP construct provides for spatial localization of mRNA, new insights into RNA trafficking within the cell may be obtained. By customization of fluorescence emission from hybridized hAuNP, imaging can be extended toward multiple mRNA targets. Furthermore, multiplexed imaging of mRNAs and proteins such as subcellular organelles is possible, using a number of methods such as live cell organelle dye-labeled homing peptides, fluorescent protein encoding, and conventional immunofluorescence techniques.39–42 This strategy for imaging specific RSV-associated sequences simultaneously with an internal control RNA such as GAPDH will enable the quantitation of distinct RSV mRNAs in the cell throughout the time course of RSV infection and replication, for development of targeted therapies and diagnostic tools. In addition, the amenability of hAuNP to customization will enable the imaging of multiple biological and disease-relevant genomic sequences within the cell in other applications. Ongoing work is directed toward the multiplexed imaging of cancer-specific mRNAs in tumor specimens.
Conclusion
hAuNP are a versatile approach for the high specificity and sensitivity imaging of mRNA in living cells without adverse toxicity, and without the need for transfection reagents or cell permeabilization. hAuNP specifically detect specific mRNA sequences for monitoring of viral infections or other biological processes in cell cultures. These constructs are amenable to multiplexed imaging, and due to their preservation of mRNA localization within the cell, mechanisms underlying the specific processing, trafficking, and degradation of specific mRNA sequences can be elucidated using an hAuNP approach. The strategy outlined in this work provides a framework for the detailed study of mRNA-based biological processes, which should enable the discovery of new molecular targets, and early-detection diagnostic strategies.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institutes of Health (R01 EB004537 (DWW)), the Vanderbilt Vision Research Center (P30-EY008126, Core grant in vision research), the Knights Templar Eye Foundation (AJ), the Vanderbilt Institute for Chemical Biology (AJ, DW), the VUMC Cell Imaging Shared Resource, and the Vanderbilt Flow Cytometry Core Facility.
Footnotes
SUPPORTING INFORMATION PARAGRAPH Supplementary figures, including absorbance and fluorescence spectra and hAuNP stability data, as well as confocal micrographs, are available free of charge at http://pubs.acs.org.
References
- 1.Bullock SL. Semin Cell Dev Biol. 2007;18:194. doi: 10.1016/j.semcdb.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Holt CE, Bullock SL. Science. 2009;326:1212. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mallardo M, Schleich S, Krijnse Locker J. Mol Biol Cell. 2001;12:3875. doi: 10.1091/mbc.12.12.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bullock SL, Stauber M, Prell A, Hughes JR, Ish-Horowicz D, Schmidt-Ott U. Development. 2004;131:4251. doi: 10.1242/dev.01289. [DOI] [PubMed] [Google Scholar]
- 5.Hughes JR, Bullock SL, Ish-Horowicz D. Curr Biol. 2004;14:1950. doi: 10.1016/j.cub.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Levadoux M, Mahon C, Beattie JH, Wallace HM, Hesketh JE. J Biol Chem. 1999;274:34961. doi: 10.1074/jbc.274.49.34961. [DOI] [PubMed] [Google Scholar]
- 7.Levadoux-Martin M, Hesketh JE, Beattie JH, Wallace HM. Biochem J. 2001;355:473. doi: 10.1042/0264-6021:3550473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Science. 1985;230:1350. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 9.Zhang G, Taneja KL, Singer RH, Green MR. Nature. 1994;372:809. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
- 10.Bassell GJ, Powers CM, Taneja KL, Singer RH. J Cell Biol. 1994;126:863. doi: 10.1083/jcb.126.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao G, Rhee WJ, Tsourkas A. Annu Rev Biomed Eng. 2009;11:25. doi: 10.1146/annurev-bioeng-061008-124920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santangelo P, Nitin N, Bao G. Ann Biomed Eng. 2006;34:39. doi: 10.1007/s10439-005-9003-6. [DOI] [PubMed] [Google Scholar]
- 13.Santangelo P, Nitin N, LaConte L, Woolums A, Bao G. J Virol. 2006;80:682. doi: 10.1128/JVI.80.2.682-688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santangelo PJ, Nitin N, Bao G. J Biomed Opt. 2005;10:44025. doi: 10.1117/1.2011402. [DOI] [PubMed] [Google Scholar]
- 15.Chen AK, Behlke MA, Tsourkas A. Nucleic Acids Res. 2008;36:e69. doi: 10.1093/nar/gkn331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walev I, Bhakdi SC, Hofmann F, Djonder N, Valeva A, Aktories K, Bhakdi S. Proc Natl Acad Sci U S A. 2001;98:3185. doi: 10.1073/pnas.051429498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nitin N, Santangelo PJ, Kim G, Nie S, Bao G. Nucleic Acids Res. 2004;32:e58. doi: 10.1093/nar/gnh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsourkas A, Behlke MA, Bao G. Nucleic Acids Res. 2003;31:5168. [PubMed] [Google Scholar]
- 19.Tsourkas A, Behlke MA, Rose SD, Bao G. Nucleic Acids Res. 2003;31:1319. doi: 10.1093/nar/gkg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Science. 2006;312:1027. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 21.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. J Am Chem Soc. 2007;129:15477. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubertret B, Calame M, Libchaber AJ. Nat Biotechnol. 2001;19:365. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 23.Santangelo PJ, Nix B, Tsourkas A, Bao G. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weisman LE. Cardiovasc Hematol Agents Med Chem. 2009;7:223. doi: 10.2174/187152509789105471. [DOI] [PubMed] [Google Scholar]
- 25.Pediatrics. 1998;102:531. [Google Scholar]
- 26.Bentzen EL, House F, Utley TJ, Crowe JE, Jr, Wright DW. Nano Lett. 2005;5:591. doi: 10.1021/nl048073u. [DOI] [PubMed] [Google Scholar]
- 27.Player MR, Barnard DL, Torrence PF. Proc Natl Acad Sci U S A. 1998;95:8874. doi: 10.1073/pnas.95.15.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsourkas A, Behlke MA, Xu Y, Bao G. Anal Chem. 2003;75:3697. doi: 10.1021/ac034295l. [DOI] [PubMed] [Google Scholar]
- 29.Dulkeith E, Ringler M, Klar TA, Feldmann J, Munoz Javier A, Parak WJ. Nano Lett. 2005;5:585. doi: 10.1021/nl0480969. [DOI] [PubMed] [Google Scholar]
- 30.Singh RD, Puri V, Valiyaveettil JT, Marks DL, Bittman R, Pagano RE. Mol Biol Cell. 2003;14:3254. doi: 10.1091/mbc.E02-12-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dev Cell. 2006;10:839. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 32.Zaro JL, Rajapaksa TE, Okamoto CT, Shen WC. Mol Pharm. 2006;3:181. doi: 10.1021/mp0500869. [DOI] [PubMed] [Google Scholar]
- 33.Khine AA, Tam P, Nutikka A, Lingwood CA. Glycobiology. 2004;14:701. doi: 10.1093/glycob/cwh085. [DOI] [PubMed] [Google Scholar]
- 34.Fittipaldi A, Ferrari A, Zoppe M, Arcangeli C, Pellegrini V, Beltram F, Giacca M. The Journal of biological chemistry. 2003;278:34141. doi: 10.1074/jbc.M303045200. [DOI] [PubMed] [Google Scholar]
- 35.Foerg C, Ziegler U, Fernandez-Carneado J, Giralt E, Rennert R, Beck-Sickinger AG, Merkle HP. Biochemistry. 2005;44:72. doi: 10.1021/bi048330+. [DOI] [PubMed] [Google Scholar]
- 36.Tagawa A, Mezzacasa A, Hayer A, Longatti A, Pelkmans L, Helenius A. J Cell Biol. 2005;170:769. doi: 10.1083/jcb.200506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Audouy S, Molema G, de Leij L, Hoekstra D. J Gene Med. 2000;2:465. doi: 10.1002/1521-2254(200011/12)2:6<465::AID-JGM141>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 38.Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Nano Lett. 2007;7:3818. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carreon JR, Stewart KM, Mahon KP, Jr, Shin S, Kelley SO. Bioorg Med Chem Lett. 2007;17:5182. doi: 10.1016/j.bmcl.2007.06.097. [DOI] [PubMed] [Google Scholar]
- 40.Daigle N, Ellenberg J. Nat Methods. 2007;4:633. doi: 10.1038/nmeth1065. [DOI] [PubMed] [Google Scholar]
- 41.Miller EW, Zeng L, Domaille DW, Chang CJ. Nat Protoc. 2006;1:824. doi: 10.1038/nprot.2006.140. [DOI] [PubMed] [Google Scholar]
- 42.Sikder S, Reyes JM, Moon CS, Suwan-apichon O, Elisseeff JH, Chuck RS. Photochem Photobiol. 2005;81:1569. doi: 10.1562/2005-06-18-RC-580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.