Abstract
Recursive partitioning methods have become popular and widely used tools for non-parametric regression and classification in many scientific fields. Especially random forests, that can deal with large numbers of predictor variables even in the presence of complex interactions, have been applied successfully in genetics, clinical medicine and bioinformatics within the past few years.
High dimensional problems are common not only in genetics, but also in some areas of psychological research, where only few subjects can be measured due to time or cost constraints, yet a large amount of data is generated for each subject. Random forests have been shown to achieve a high prediction accuracy in such applications, and provide descriptive variable importance measures reflecting the impact of each variable in both main effects and interactions.
The aim of this work is to introduce the principles of the standard recursive partitioning methods as well as recent methodological improvements, to illustrate their usage for low and high dimensional data exploration, but also to point out limitations of the methods and potential pitfalls in their practical application.
Application of the methods is illustrated using freely available implementations in the R system for statistical computing.
Scope of This Work
Prediction, classification and the assessment of variable importance are fundamental tasks in psychological research. A wide range of classical statistical methods – including linear and logistic regression as the most popular representatives of standard parametric models – is available to address these tasks. However, in certain situations these classical methods can be subject to severe limitations.
One situation where parametric approaches are no longer applicable is the so called ”small n large p” case, where the number of predictor variables p is greater than the number of subjects n. This case is common, e.g., in genetics, where thousands of genes are considered as potential predictors of a disease. However, even in studies with much lower numbers of predictor variables, the combination of all main and interaction effects of interest – especially in the case of categorical predictor variables – may well lead to cell counts too sparse for parameter convergence. Thus, interaction effects of high order usually cannot be included in standard parametric models.
Additional limitations of many standard approaches include the restricted functional form of the association pattern (with the linear model as the most common and most restrictive case), the fact that ordinally scaled variables, which are particularly common in psychological applications, are often treated as if they were measured on an interval or ratio scale, and that measures of variable importance are only available for a small range of methods.
The aim of this paper is to provide an instructive review of a set of statistical methods adopted from machine learning, that overcome these limitations.
The most important one of these methods is the so called “random forest” approach of Breiman (2001a): A random forest is a so called ensemble (or set) of classification or regression trees (CART, Breiman, Friedman, Olshen, and Stone 1984). Each tree in the ensemble is built based on the principle of recursive partitioning, where the feature space is recursively split into regions containing observations with similar response values. A detailed explanation of recursive partitioning is given in the next section.
In the past years, recursive partitioning methods have gained popularity as a means of multivariate data exploration in various scientific fields, including, e.g., the analysis of microarray data, DNA sequencing and many other applications in genetics, epidemiology and medicine (cf., e.g., Gunther, Stone, Gerwien, Bento, and Heyes 2003; Lunetta, Hayward, Segal, and Eerdewegh 2004; Segal, Barbour, and Grant 2004; Bureau, Dupuis, Falls, Lunetta, Hayward, Keith, and Eerdewegh 2005; Huang, Pan, Grindle, Han, Chen, Park, Miller, and Hall 2005; Shih, Seligson, Belldegrun, Palotie, and Horvath 2005; Diaz-Uriarte and Alvarez de Andrés 2006; Qi, Bar-Joseph, and Klein-Seetharaman 2006; Ward, Pajevic, Dreyfuss, and Malley 2006).
A growing number of applications of random forests in psychology indicates a wide range of application areas in this field, as well: For example, Oh, Laubach, and Luczak (2003) and Shen, Ong, Li, Hui, and Wilder-Smith (2007) apply random forests to neuronal ensemble recordings and EEG data, that are too high-dimensional for the application of standard regression methods. An alternative approach to cope with large numbers of predictor variables would be to first apply dimension reduction techniques, such as principle components or factor analysis, and then use standard regression methods on the reduced data set. However, this approach has the disadvantage that the original input variables are projected into a reduced set of components, so that their individual effect is not longer identifiable. As opposed to that, random forests can process large numbers of predictor variables simultaneously and provide individual measures of variable importance.
Interesting applications of random forests in data sets of lower dimensionality include the studies of Rossi, Amaddeo, Sandri, and Tansella (2005) on determinants of once-only contact in community mental health service and Baca-Garcia, Perez-Rodriguez, Saiz-Gonzalez, Basurte-Villamor, Saiz-Ruiz, Leiva-Murillo, de Prado-Cumplido, Santiago-Mozos, Artes-Rodriguez, and de Leon (2007) on attempted suicide under consideration of the family history. For detecting relevant predictor variables, Rossi et al. (2005) point out that the random forest variable importance ranking proves to be more stable than stepwise variable selection approaches available for logistic regression, that are known to be affected by order effects (see, e.g., Freedman 1983; Derksen and Keselman 1992; Austin and Tu 2004). Moreover, a high random forest variable importance of a variable that was not included in stepwise regression may indicate that the variable works in interactions that are too complex to be captured by parametric regression models. As another advantage, Marinic, Supek, Kovacic, Rukavina, Jendricko, and Kozaric-Kovacic (2007) point out in an application to the diagnosis of posttraumatic stress disorder, that random forests can be used to automatically generate realistic estimates of the prediction accuracy on test data by means of repeated random sampling from the learning data.
Luellen, Shadish, and Clark (2005) explore another field of application in comparing the effects in an experimental and a quasi-experimental study on mathematics and vocabulary performance: When the treatment assignment is chosen as a working response, classification trees and ensemble methods can be used to estimate propensity scores.
However, some of these seminal applications of recursive partitioning methods in psychology also reveal common misperceptions and pitfalls: For example, Luellen et al. (2005) suspect that ensemble methods could overfit (i.e., adapt too closely to random variations in the learning sample, as discussed in detail below) when too many trees are used to build the ensemble – even though recent theoretical results disprove this and indicate that other tuning parameters may be responsible for overfitting in random forests.
More common mistakes in the practical usage and interpretation of recursive partitioning approaches are the confusion of main effects and interactions (see, e.g., Berk 2006) as well as the application of biased variable selection criteria and a significance test for variable importance measures (see, e.g., Baca-Garcia et al. 2007) that has recently been shown to have extremely poor statistical properties.
Some of these pitfalls are promoted by the fact that random forests were not developed in a stringent statistical framework, so that their properties are less predictable than those of standard parametric methods, and some parts of random forests are still “under construction” (cf. also Polikar 2006, for a brief history of ensemble methods, including fuzzy and Bayesian approaches).
Therefore the aim of this paper is not only to point out the potential of random forests and related recursive partitioning methods to a broad scientific community in psychology and related fields, but also to provide a thorough understanding of how these methods function, how they can be applied practically and when they should be handled with caution.
The next section describes the rationale of recursive partitioning methods, starting with single classification and regression trees and moving on to ensembles of trees. Examples are interspersed between the technical explanations and provided in an extra section to highlight potential areas of application. A synthesis of important features and advantages of recursive partitioning methods – as well as important pitfalls – with an emphasis on random forests is given in a later section. For all examples shown here, freely available implementations in the R system for statistical computing (R Development Core Team 2009) were employed. The corresponding code is provided and documented in a supplement as an aid for new users.
The Methods
After the early seminal work on automated interaction detection by Morgan and Sonquist (1963) the two most popular algorithms for classification and regression trees (abbreviated as classification trees in most of the following), CART and C4.5, were introduced by Breiman et al. (1984) and independently by Quinlan (1986, 1993). Their nonparametric approach and the straightforward interpretability of the results have added much to the popularity of classification trees (cf., e.g., Hannöver, Richard, Hansen, Martinovich, and Kordy 2002; Kitsantas, Moore, and Sly 2007, for applications on the treatment effect in patients with eating disorders and determinants of adolescent smoking habits). As an advancement of single classification trees, random forests (Breiman 2001a), as well as its predecessor method bagging (Breiman 1996a, 1998), are so-called “ensemble methods”, where an ensemble or committee of classification trees is aggregated for prediction. This section introduces the main concepts of classification trees, that are then employed as so called “base learners” in the ensemble methods bagging and random forests.
How Do Classification and Regression Trees Work?
Classification and regression trees are a simple nonparametric regression approach. Their main characteristic is that the feature space, i.e. the space spanned by all predictor variables, is recursively partitioned into a set of rectangular areas, as illustrated below. The partition is created such that observations with similar response values are grouped. After the partition is completed, a constant value of the response variable is predicted within each area.
The rationale of classification trees will be explained in more detail by means of a simple psychological example: Inspired by the study of Kitsantas et al. (2007) on determinants of adolescent smoking habits, an artificial data set was generated for illustrating variable and split selection in recursive partitioning.
Our aim is to predict the adolescents’ intention to smoke a cigarette within the next year (binary response variable intention_to_smoke) from four candidate risk factors (the binary predictor variables lied_to_parents, indicating whether the subject has ever lied to the parents about doing something they would not approve of, and friends_smoke, indicating peer smoking of one or more among the four best friends, as well as the numeric predictor variables age, indicating the age in years, and alcohol_per_month, indicating how many times the subject drank alcohol in the past month).
The data were generated such as to resemble the key results of Kitsantas et al. (2007). However, the variables age and alcohol_per_month, that are used only in a discretized form by Kitsantas et al. (2007), were generated as numeric variables to illustrate the selection of optimal cutpoints in recursive partitioning. The generated data set, as well as the R-code used for all examples, are available as supplements.
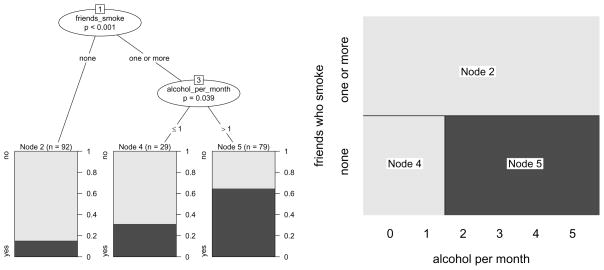
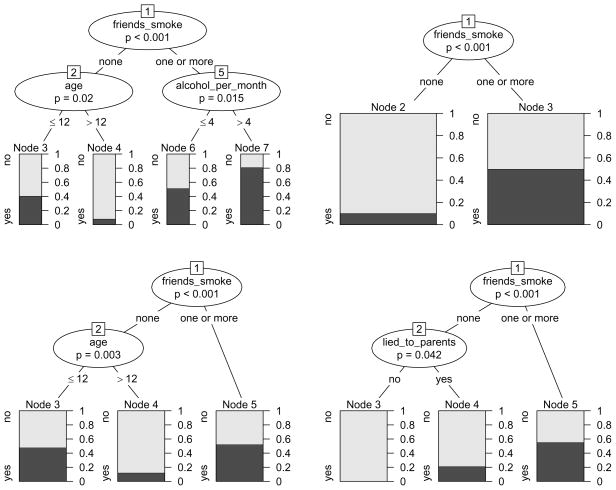
The classification tree derived from the smoking data is illustrated in Figure 1 (left) and shows the following: From the entire sample of 200 adolescents (represented by node 1 in Figure 1 (left), where the node numbers are mere labels assigned recursively from left to right starting from the top node), a group of 92 adolescents is separated from the rest in the first split. This group (represented by node 2) is characterized by the fact that “none” of their four best friends smoke, and that within this group only few subjects intend to smoke within the next year. The remaining 108 subjects are further split into two groups (nodes 4 and 5) according to whether they drank alcohol in “one or less” or “more” occasions in the past month. These two groups again vary in the percentage of subjects who intend to smoke.
Figure 1.
Partition of the smoking data by means of a binary classification tree. The tree representation (left) corresponds to a rectangular recursive partition of the feature space (right). In the terminal nodes of the tree, the dark and light grey shaded areas represent the relative frequencies of “yes” and ”no” answers to the intention to smoke question in each group respectively. The corresponding areas in the rectangular partition are shaded in the color of the majority response.
The model can be displayed either as a tree, as in Figure 1 (left), or as a rectangular partition of the feature space, as in Figure 1 (right): The first split in the variable friends_smoke partitions the entire sample, while the second split in the variable alcohol_per_month further partitions only those subjects whose value for the variable friends_smoke is ”one or more”. The partition representation in Figure 1 (right) is even better suited than the tree representation to illustrate that recursive partitioning creates nested rectangular prediction areas corresponding to the terminal nodes of the classification tree. Details about the prediction rules derived from the partition are given below.
Note that the resulting partition is one of the main differences between classification trees and, e.g., linear regression models: While in linear regression the information from different predictor variables is combined linearly, here the range of possible combinations includes all rectangular partitions that can be derived by means of recursive splitting – including multiple splits in the same variable. In particular, this includes nonlinear and even nonmonotone association rules, that do not need to be specified in advance but are determined in a data driven way.
Of course there is a strong parallel between tree building and stepwise regression, where predictors are also included one at a time in successive order. However, in stepwise linear regression the predictors still have a linear effect on the dependent variable, while extensions of stepwise procedures including interaction effects are typically limited to the inclusion of two-fold interactions, since the number of higher order interactions – that would have to be created simultaneously when starting the selection procedure – is too large.
In contrast to this, in recursive partitioning only those interactions that are actually used in the tree are generated during the fitting process. The issue of including main effects and interactions in recursive partitioning is discussed in more detail below.
Splitting and Stopping
Both the CART algorithm of Breiman et al. (1984) and the C4.5 algorithm (and its predecessor ID3) of Quinlan (1986, 1993) conduct binary splits in numeric predictor variables, as depicted in Figure 1. In categorical predictor variables (of nominal or ordinal scale of measurement) C4.5 produces as many nodes as there are categories (often referred to as “k-ary” or “multiple” splitting), while CART again creates binary splits between the ordered or unordered categories. We concentrate on binary splitting trees in the following and refer to Quinlan (1993) for k-ary splitting.
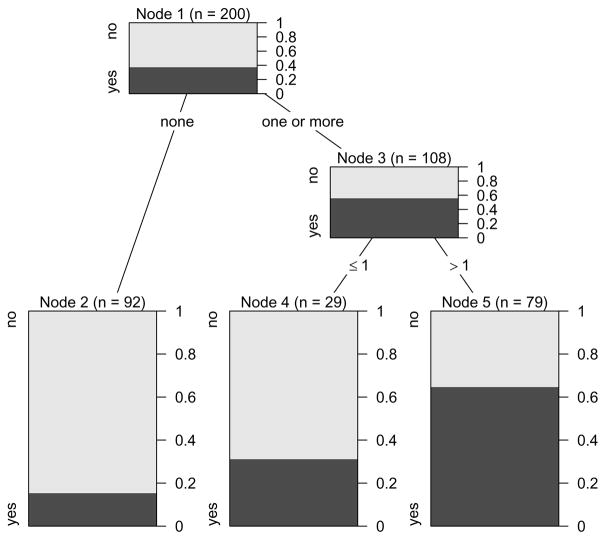
For selecting the splitting variable and cutpoint, both CART and C4.5 follow the approach of impurity reduction, that we will illustrate by means of our smoking data example: In Figure 2 the relative frequencies of both response classes are displayed not only for the terminal nodes, but also for the inner nodes of the tree previously presented in Figure 1. Starting from the root node, we find that the relative frequency of “yes” answers in the entire sample of 200 adolescents is approximately 40%. By means of the first split, the group of 92 adolescents with the lowest frequency of “yes” answers (approximately 15%, node 2) can be isolated from the rest, that have a higher frequency of “yes” answers (almost 60%, node 3). These 108 subjects are then further split to form two groups: one smaller group with a medium (approximately 30%, node 4) and one larger group with a high (more than 60%, node 5) frequency of “yes” answers to the intention to smoke question.
Figure 2.
Relative frequencies of both response classes in the inner nodes of the binary classification tree for the smoking data. The dark and light grey shaded areas again represent the relative frequencies of “yes” and ”no” answers to the intention to smoke in each group respectively.
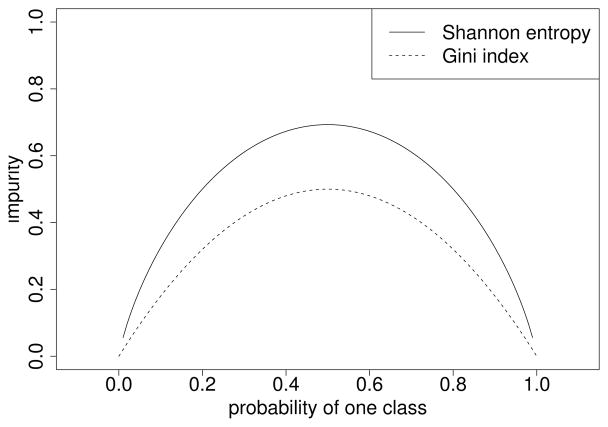
From this example we can see that, following the principle of impurity reduction, each split in the tree building process results in daughter nodes that are more “pure” than the parent node in the sense that groups of subjects with a majority for either response class are isolated. The impurity reduction achieved by a split is measured by the difference between the impurity in the parent node and the average impurity in the two daughter nodes. Entropy measures, such as the Gini Index or the Shannon Entropy, are used to quantify the impurity in each node. These entropy measure have in common that they reach their minimum for perfectly pure nodes with the relative frequency of one response class being zero and their maximum for an equal mixture with the same relative frequencies for both response classes, as illustrated in Figure 3.
Figure 3.
Gini index and Shannon entropy as functions of the relative frequency of one response class. Pure nodes containing only observations of one class receive an impurity value of zero, while mixed nodes receive higher impurity values.
While the principle of impurity reduction is intuitive and has added much to the popularity of classification trees, it can help our statistical understanding to think of impurity reduction as merely one out of many possible means of measuring the strength of the association between the splitting variable and the response. Most modern classification tree algorithms rely on this strategy, and employ the p-values of association tests for variable and cutpoint selection. This approach has additional advantages over the original impurity reduction approach, as outlined below.
Regardless of the split selection criterion, however, in each node the variable that is most strongly associated with the response variable (i.e., that produces the highest impurity reduction or the lowest p-value) is selected for the next split. In splitting variables with more than two categories, that offer more than one possible cutpoint, the optimal cutpoint is also selected with respect to this criterion. In our example, the optimal cutpoint identified within the range of the numeric predictor variable alcohol_per_month is between the values 1 and 2, because subjects who drank alcohol in one or less occasions have a lower frequency of “yes” answers than those who drank alcohol in 2 or more occasions.
After a split is conducted, the observations in the learning sample are divided into the different nodes defined by the respective splitting variable and cutpoint, and in each node splitting continues recursively until some stop condition is reached. Common stop criteria are: split until (a) all leaf nodes are pure (i.e. contain only observations of one class) (b) a given threshold for the minimum number of observations left in a node is reached or (c) a given threshold for the minimum change in the impurity measure is not succeeded any more by any variable. Recent classification tree algorithms also provide statistical stopping criteria that incorporate the distribution of the splitting criterion (Hothorn, Hornik, and Zeileis 2006), while early algorithms relied on pruning the complete tree to avoid overfitting.
The term overfitting refers to the fact that a classifier that adapts too closely to the learning sample will not only discover the systematic components of the structure that is present in the population, but also the random variation from this structure that is present in the learning data due to random sampling. When such an overfitted model is later applied to a new test sample from the same population, its performance will be poor because it does not generalize well. However, it should be noted that overfitting is an equally relevant issue in parametric models: With every variable, and thus every parameter, that is added to the regression model, its fit to the learning data improves, because the model becomes more flexible.
This is evident, e.g., in the R2 statistic reflecting the portion of variance explained by the model, that increases with every parameter added to the model. For example, in the extreme case where as many parameters as observations are available, any parametric model will show a perfect fit on the learning data, yielding a value of R2 = 1, but will perform poorly in future samples.
In parametric models, a common strategy to deal with this problem is to use significance tests for variable selection in regression models. However, one should be aware that in this case significance tests do not work in the same way as in a designed study, where a limited number of hypotheses to be tested are specified in advance. In common forward and/or backward stepwise regression it is not known beforehand how many significance tests will have to be conducted. Therefore, it is hard to control the overall significance level, that controls the probability of falsely declaring at least one of the coefficients as significant.
Advanced variable selection strategies, that have been developed for parametric models, employ model selection criteria such as the AIC and BIC, that include a penalization term for the number of parameters in the model. For a detailed discussion of approaches that account for the complexity of parametric models see Burnham and Anderson (2002) or Burnham and Anderson (2004).
Since information criteria such as the AIC and BIC are, however, not applicable to nonparametric models (see, e.g., Claeskens and Hjort 2008), in recursive partitioning the classic strategy to cope with overfitting is to “prune” the trees after growing them, which means that branches that do not add to the prediction accuracy in cross validation are eliminated. Pruning is not discussed in detail here, because the unbiased classification tree algorithm of Hothorn et al. (2006), that is used here for illustration, employs p-values for variable selection and as a stopping criterion and therefore does not rely on pruning. In addition to this, ensemble methods, that are our main focus here, usually employ unpruned trees.
Prediction and Interpretation of Classification and Regression Trees
Finally a response class is predicted in each terminal node of the tree (or each rectangular section in the partition respectively) by means of deriving from all observations in this node either the average response value in regression or the most frequent response class in classification trees. Note that this means that a regression tree creates a piecewise (or rectangle-wise for two dimensions and cuboid-wise in higher dimensions) constant prediction function.
Even though the idea of piecewise constant functions may appear very inflexible, such functions can be used to approximate any functional form, in particular nonlinear and nonmonotone functions. This is in strong contrast to classical linear or additive regression, where the effects of predictors are restricted to the additive form – the interpretation of which may appear easier, but which may also produce severe artifacts, since in many complex applications the true data generating mechanism is neither linear nor additive. We will see later that ensemble methods, by combining the predictions of many single trees, can approximate functions more smoothly, too.
The predicted response classes in our example are the majority class in each node in Figure 1 (left), as indicated by the shading in Figure 1 (right): Subjects who have not lied to their parents as well as those who have lied to their parents but drank alcohol in one or less occasions are not likely to intend to smoke, while those who have lied to their parents and drank alcohol in 2 or more occasions are likely to intend to smoke within the next year.
For classification problems it is also possible to predict an estimate of the class probabilities from the relative frequencies of each class in the terminal nodes. In our example, the predicted probabilities for answering “yes” to the intention to smoke question would thus be approximately 15%, 30% and 65% in the three groups – which may preserve more information than the majority vote that merely assigns the class with a relative frequency of > 50% as the prediction.
Reporting the predicted class probabilities more closely resembles the output of logistic regression models and can also be employed for estimating treatment probabilities or propensity scores. Note, however, that no confidence intervals are available for the estimates, unless, e.g., bootstrapping is used in combination with refitting to assess the variability of the prediction.
The easy interpretability of the visual representation of classification trees, that we have illustrated in this example, has added much to the popularity of this method, e.g., in medical applications. However, the downside of this apparently straightforward interpretability is that the visual representation may be misguiding, because the actual statistical interpretation of a tree model is not trivial. Especially the notions of main effects and interactions are often used rather incautiously in the literature, as seems to be the case in Berk (2006, p. 272), where it is stated that a branch that is not split any further indicated a main effect. However, when in the other branch created by the same variable splitting continues, as is the case in the example of Berk (2006), this statement is not correct.
The term “interaction” commonly describes the fact that the effect of one predictor variable, in our example alcohol_per_month, on the response depends on the value of another predictor variables, in our example friends_smoke. For classification trees this means that, if in one branch created by friends_smoke it is not necessary to split in alcohol_per_month, while in the other branch created by friends_smoke it is necessary, as in Figure 1 (left), an interaction between friends_smoke and alcohol_per_month is present.
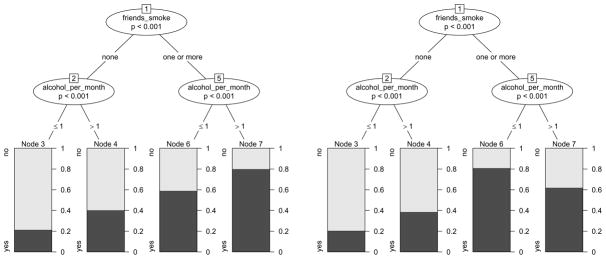
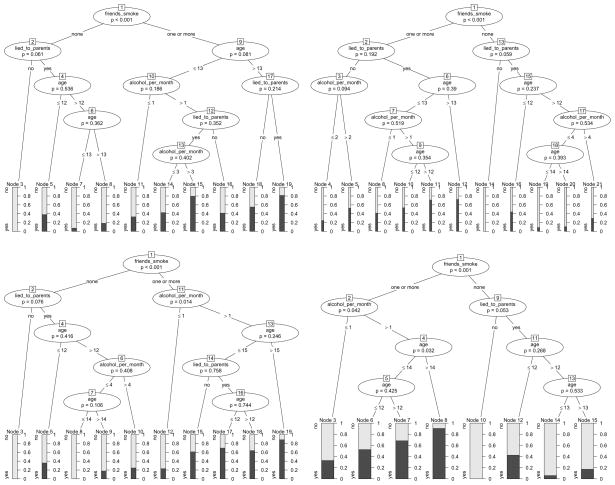
We will further illustrate this important issue and source of misinterpretations by means of varying the effects in our artificial data set. The resulting classification trees are given in Figure 4. Only the left plot in Figure 4, where the effect of alcohol_per_month is the same in both branches created by friends_smoke, represents two main effects of alcohol_per_month and friends_smoke without an interaction: The main effect of friends_smoke shows in the higher relative frequencies of “yes” answers in nodes 6 and 7 as compared to nodes 3 and 4. The main effect of alcohol_per_month shows in the higher relative frequencies of “yes” answers in nodes 4 and 7 as compared to nodes 3 and 6 respectively.
Figure 4.
Classification trees based on variations of the smoking data with two main effects (left) and interactions (right). The tree depicted in Figure 1 based on the original data also represents an interaction.
As opposed to that, both the right plot in Figure 4 and the original plot in Figure 1 represent interactions, because the effect of alcohol_per_month is different in both branches created by friends_smoke. In the right plot in Figure 4 the same split in alcohol_per_month is conducted in every branch created by friends_smoke, but the effect on the relative frequencies of the response classes is different: for those subjects who have no friends that smoke, the relative frequency of a “yes” answer is higher if they drank alcohol in 2 or more occasions (node 4 as compared to node 3), while for those who have one or more friends that smoke, the frequency of a “yes” answer is lower if they drank alcohol in 2 or more occasions (node 7 as compared to node 6). This example represents a typical interaction effect as known from standard statistical models, where the effect of alcohol_per_month depends on the value of friends_smoke.
In the original plot in Figure 1 on the other hand, the effect of alcohol_per_month is also different in both branches created by friends_smoke, because alcohol_per_month has an effect only in the right branch, but not in the left branch.
While this kind of “asymmetric” interaction is very common in classification trees, it is extremely unlikely to actually discover a symmetric interaction pattern as that in Figure 4 (right) or even a main effect pattern as that in Figure 4 (left) in real data.
The reason for this is that, even if the true distribution of the data in both branches was very similar, due to random variations in the sample and the deterministic variable and cutpoint selection strategy of classification trees, it is extremely unlikely that the same splitting variable – and also the exact same cutpoint – would be selected in both branches. However, even a slightly different cutpoint in the same variable would, strictly speaking, represent an interaction. Therefore it is stated in the literature that classification trees cannot (or rather, are extremely unlikely to) represent additive functions that consist only of main effects, while they are perfectly well suited for representing complex interactions.
For exploratory data analysis, further means for illustrating the effects of particular variables in classification trees are provided by the partial dependence plots described in Hastie, Tibshirani, and Friedman (2001, 2009) and the CARTscans toolbox (Nason, Emerson, and Leblanc 2004).
Model-Based Recursive Partitioning
A variant of recursive partitioning, that can also be a useful aid for visual data exploration, is model based recursive partitioning. Here the idea is to partition the feature space not such as to identify groups of subjects with similar values of the response variable, but groups of subjects with similar association patterns, e.g., between another predictor variable and the response.
For example, linear regression could be used to model the dependence of a clinical response on the dose of medication. However, the slope and intersect parameters of this regression may be different for different groups of patients: elderly patients, e.g., may show a stronger reaction to the medication, so that the slope of their regression line would need to be steeper than that of younger patients. In this example, the model of interest is the regression between dose of medication and clinical response – however, the model parameters should be chosen differently in the two (or more) groups defined by the covariate age. Another example and visualization are given in the section “Further application examples”.
The model based recursive partitioning approach of Zeileis, Hothorn, and Hornik (2008) offers a way to partition the feature space in order to detect parameter instabilities in the parametric model of interest by means of a structural change test framework. Similarly to latent class or mixture models, the aim of model based partitioning is to identify groups of subjects for which the parameters of the parametric model differ. However, in model based partitioning the groups are usually not defined by a latent factor, but by combinations of observed covariates, that are searched heuristically. Thus, model based partitioning can offer a heuristic but easy to interpret alternative to latent class – as well as random or mixed effects – models.
An extension of model based partitioning for Bradley-Terry models is suggested by Strobl, Wickelmaier, and Zeileis (2009). An application to mixed models, including the Rasch Model as a special case (as a generalized linear mixed model, see Rijmen, Tuerlinckx, Boeck, and Kuppens 2003; Doran, Bates, Bliese, and Dowling 2007), is currently investigated by Sanchez-Espigares and Marco (2008).
What is Wrong with Trees?
The main flaw of simple tree models is their instability to small changes in the learning data: In recursive partitioning, the exact position of each cutpoint in the partition, as well as the decision which variable to split in, determines how the observations are split up in new nodes, in which splitting continues recursively. However, the exact position of the cutpoint, as well as the selection of the splitting variable, strongly depend on the particular distribution of observations in the learning sample.
Thus, as an undesired side effect of the recursive partitioning approach, the entire tree structure could be altered if the first splitting variable, or only the first cutpoint, was chosen differently due to a small change in the learning data. Due to this instability, the predictions of single trees show a high variability.
The high variability of single trees can be illustrated, e.g., by drawing bootstrap samples from the original data set and investigating whether the trees built on the different samples have a different structure. The rationale of bootstrap samples, where a sample of the same size as the original sample is drawn with replacement (so that some observations are left out, while others may appear more than once in the bootstrap sample) is to reflect the variability inherent in any sampling process: Random sampling preserves the systematic effects present in the original sample or population, but in addition to this it induces random variability. Thus, if classification trees built on different bootstrap samples vary too strongly in their structure, this proves that their interpretability can be severely affected by the random variability present in any data set.
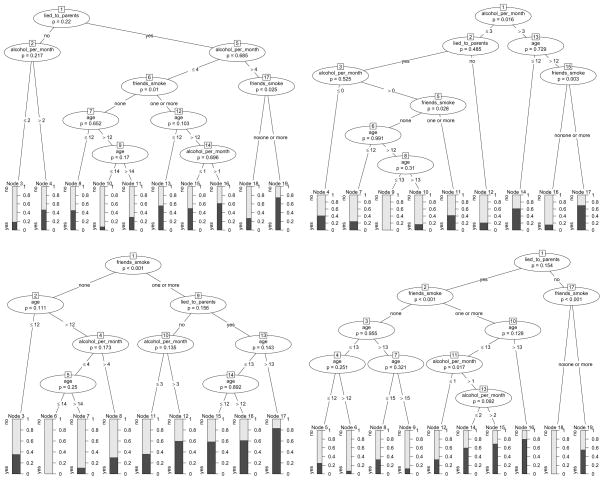
Classification trees built on four bootstrap samples drawn from our original smoking data are displayed in Figure 5. Apparently, the effect of the variable friends_smoke is strong enough to remain present in all four trees, while the further splits vary strongly with the sample.
Figure 5.
Classification trees based on four bootstrap samples of the smoking data, illustrating the instability of single trees.
As a solution to the problem of instability, the average over an ensembles of trees, rather than a single tree, is used for prediction in ensemble methods, as outlined in the following. Another problem of single trees, that is solved by the same model averaging approach, is that the prediction of single trees is piecewise constant and thus may “jump” from one value to the next even for small changes of the predictor values. As described in the next section, ensemble methods have the additional advantage, that their decision boundaries are more smooth than those of single trees.
How Do Ensemble Methods Work?
The rationale behind ensemble methods is to base the prediction on a whole set of classification or regression trees, rather than a single tree. The related methods bagging and random forests vary only in the way this set of trees is constructed.
Bagging
In both bagging and random forests a set of trees is built on random samples of the learning sample: In each step of the algorithm, either a bootstrap sample (of the same size, drawn with replacement) or a subsample (of smaller size, drawn without replacement) of the learning sample is drawn randomly, and an individual tree is grown on each sample. As outlined above, each random sample reflects the same data generating process but differs slightly from the original learning sample due to random variation. Keeping in mind that each individual classification tree depends highly on the learning sample as outlined above, the resulting trees can thus differ substantially.
Another feature of the ensemble methods bagging and random forests is that usually trees are grown very large, without any stopping or pruning involved. As illustrated again for four bootstrap samples from the smoking data in Figure 6, large trees can become even more diverse and include a large variety of combinations of predictor variables.
Figure 6.
Classification trees (grown without stopping or pruning) based on four bootstrap samples of the smoking data, illustrating the principle of bagging.
By combining the prediction of such a diverse set of trees, ensemble methods utilize the fact that classification trees are instable but on average produce the right prediction (i.e. trees are unbiased predictors), which has been supported by several empirical as well as simulation studies (cf., e.g., Breiman 1996a, 1998; Bauer and Kohavi 1999; Dietterich 2000) and especially the theoretical results of Bühlmann and Yu (2002), that show the superiority in prediction accuracy of bagging over single classification or regression trees: Bühlmann and Yu (2002) could show by means of rigorous asymptotic methods that the improvement in the prediction is achieved by means of smoothing the hard cut decision boundaries created by splitting in single classification trees, which in return reduces the variance of the prediction (see also Biau, Devroye, and Lugosi 2008). The smoothing of hard decision boundaries also makes ensembles more flexible than single trees in approximating functional forms that are smooth rather than piecewise constant.
Grandvalet (2004) also points out that the key effect of bagging is that it equalizes the influence of particular observations – which proves beneficial in the case of “bad” leverage points, but may be harmful when “good” leverage points, that could improve the model fit, are downweighted. The same effect can be achieved not only by means of bootstrap sampling as in standard bagging, but also by means of subsampling (Grandvalet 2004), that is preferable in many applications because it guarantees unbiased variable selection (Strobl, Boulesteix, Zeileis, and Hothorn 2007, see also section “Bias in variable selection and variable importance”). Ensemble construction can also be viewed in the context of Bayesian model averaging (cf., e.g., Domingos 1997; Hoeting, Madigan, Raftery, and Volinsky 1999, for an introduction). For random forests, which we will consider in the next section, Breiman (2001a, p. 25) states that they may also be viewed as a Bayesian procedure (and continues: “Although I doubt that this is a fruitful line of exploration, if it could explain the bias reduction, I might become more of a Bayesian.”).
Random Forests
In random forests another source of diversity is introduced when the set of predictor variables to select from is randomly restricted in each split, producing even more diverse trees. The number of randomly preselected splitting variables, termed mtry in most algorithms, as well as the overall number of trees, usually termed ntree, are parameters of random forests that affect the stability of the results and will be discussed further in section “Features and pitfalls”. Obviously random forests include bagging as the special case where the number of randomly preselected splitting variables is equal to the overall number of variables.
Intuitively speaking, random forests can improve the predictive performance even further as compared to bagging, because the single trees involved in averaging are even more diverse. From a statistical point of view, this can be explained by the theoretical results presented by Breiman (2001a), that the upper bound for the generalization error of an ensemble depends on the correlation between the individual trees, such that a low correlation between the individual trees results in a low upper bound for the error.
In addition to the smoothing of hard decision boundaries, the random selection of splitting variables in random forests allows predictor variables, that were otherwise outplayed by a stronger competitor, to enter the ensemble: If the stronger competitor cannot be selected, a new variable has a chance to be included in the model – and may reveal interaction effects with other variables that otherwise would have been missed.
The effect or randomly restricting the splitting variables is again illustrated by means of four bootstrap samples drawn from the smoking data: In addition to growing a large tree on each bootstrap sample, as in bagging, now the variable selection is limited to mtry=2 randomly preselected candidates in each split. The resulting trees are displayed in Figure 7: We find that, due to the random restriction, the trees have become even more diverse; for example the strong predictor variable friends_smoke is no longer chosen for the first split in every single tree.
Figure 7.
Classification trees (grown without stopping or pruning and with a random preselection of 2 variables in each split) based on four bootstrap samples of the smoking data, illustrating the principle of random forests
The reason why even suboptimal splits in weaker predictor variables can often improve the prediction accuracy of an ensemble is that the split selection process in regular classification trees is only locally optimal in each node: A variable and cutpoint are chosen with respect to the impurity reduction they can achieve in a given node defined by all previous splits, but regardless of all splits yet to come.
Thus, variable selection in a single tree is affected by order effects similar to those present in stepwise variable selection approaches for parametric regression (that is also instable against random variation of the learning data, as pointed out by Austin and Tu 2004). In both recursive partitioning and stepwise regression, the approach of adding one locally optimal variable at a time does not necessarily (or rather hardly ever) lead to the globally best model over all possible combinations of variables.
Since, however, searching for a single globally best tree is computationally infeasible (a first approach involving dynamic programming was introduced by van Os and Meulman 2005), the random restriction of the splitting variables provides an easy and efficient way to generate locally suboptimal splits that can improve the global performance of an ensemble of trees. Alternative approaches that follow this rationale by introducing even more sources of randomness are outlined below.
Besides intuitive explanations for “how ensemble methods work”, recent publications have contributed to a deeper understanding of the statistical background behind many machine learning methods: The work of Bühlmann and Yu (2002) provided the statistical framework for bagging, Friedman, Hastie, and Tibshirani (2000) and Bühlmann and Yu (2003) for the related method boosting and, most recently, Lin and Jeon (2006) and Biau et al. (2008) for random forests. In their work Lin and Jeon (2006) explore the statistical properties of random forests by means of placing them in a k-nearest neighbor (k-NN) framework, where random forests can be viewed as adaptively weighted k-NN with the terminal node size determining the size of neighborhood. However, in order to be able to mathematically grasp a computationally complex method like random forests, involving several degrees of random sampling, several simplifying assumptions are necessary. Therefore well planned simulation studies still offer valuable assistance for evaluating statistical aspects of the method in its original form.
Alternative Ensemble Methods
Alternative approaches for building ensembles of trees with a strong randomization component are the random split selection approach of Dietterich (2000), where cutpoints from a set of optimal candidates are randomly selected, and the perfect random trees approach of Cutler (1999) and Cutler (2000), where both the splitting variable and the cutpoint are chosen randomly for each split.
Another very intuitive approach, that resides somewhere in between single classification trees and the ensemble methods we have covered so far, is TWIX (Potapov 2007; Potapov, Theus, and Urbanek 2006). Here the building of the tree ensemble starts in a single starting node but branches to a set of trees at each decision by means of splitting not only in the best cutpoint but also in reasonable extra cutpoints. A data driven approach for selecting extra cutpoints is suggested in Strobl and Augustin (2009).
However, while the approaches involving a strong randomization component manage to overcome local optimality as outlined above, the TWIX approach is limited to a sequence of locally optimal splits. It has been shown to outperform single trees and even to reach the predictive performance of bagging, but in general cannot compete because it becomes computationally infeasible for large sets of trees that are standard in today’s ensemble methods.
Predictions from Ensembles of Trees
In an ensemble of trees the predictions of all individual trees need to be combined. This is usually accomplished by means of (weighted or unweighted) averaging in regression or voting in classification.
The term “voting” can be taken literally here: Each subject with given values of the predictor variables is “dropped through” every tree in the ensemble, so that each single tree returns a predicted class for the subject. The class that most trees “vote” for is returned as the prediction of the ensemble. This democratic voting process is the reason why ensemble methods are also called “committee” methods. Note, however, that there is no diagnostic for the unanimity of the vote. For regression and for predicting probabilities, i.e. relative class frequencies, the results of the single trees are averaged; some algorithms also employ weighted averages. A summary over several aggregation schemes is given in Gatnar (2008). However, even with the simple aggregation schemes used in the standard algorithms, ensembles methods reliably outperform single trees and many other advanced methods (examples of benchmark studies are given in the discussion).
Aside from the issue of aggregation, for bagging and random forests there are two different prediction modes: ordinary prediction and the so called out-of-bag prediction. While in ordinary prediction each observation of the original data set – or a new test data set – is predicted by the entire ensemble, out-of-bag prediction follows a different rationale: Remember that each tree is built on a bootstrap sample, that serves as a learning sample for this particular tree. However, some observations, namely the out-of-bag observations, were not included in the learning sample for this tree. Therefore, they can serve as a “built-in” test sample for computing the prediction accuracy of that tree.
The advantage of the out-of-bag error is that it is a more realistic estimate of the error rate that is to be expected in a new test sample, than the naive and over-optimistic estimate of the error rate resulting from the prediction of the entire learning sample (Breiman 1996b) (see also Boulesteix, Strobl, Augustin, and Daumer (2008) for a review on resampling-based error estimation). The standard and out-of-bag prediction accuracy of a random forests with ntree=500 and mtry=2 for our smoking data example is 74.5% and 71.5% respectively, where the out-of-bag prediction accuracy is more conservative.
In our artificial example, bagging, and even a single tree, would actually perform equally well, because the interaction of friends_smoke and alcohol_per_month, that was already correctly identified by the single tree, is the only effect that was induced in the data.
However, in most real data applications – especially in cases where many predictor variables work in complex interactions – the prediction accuracy of random forests is found to be higher than for bagging, and both ensemble methods usually highly outperform single trees.
Variable Importance
As described in the previous sections, single classification trees are easily interpretable, both intuitively at first glance and descriptively when looking in detail at the tree structure. In particular variables that are not included in the tree did not contribute to the model – at least not in the context of the previously chosen splitting variables.
As opposed to that, ensembles of trees are not easy to interpret at all, because the individual trees in them are not nested in any way: Each variable may appear at different positions, if at all, in different trees, as depicted in Figures 6 and 7, so that there is no such thing as an “average tree” that could be visualized for interpretation.
On the other hand, an ensemble of trees has the advantage that it gives each variable the chance to appear in different contexts with different covariates, and can thus better reflect its potentially complex effect on the response. Moreover, order effects induced by the recursive variable selection scheme employed in constructing the single trees are eliminated by averaging over the entire ensemble. Therefore, in bagging and random forests variable importance measures are computed to assess the relevance of each variable over all trees of the ensemble.
In principle, a possible naive variable importance measure would be to merely count the number of times each variable is selected by all individual trees in the ensemble. More elaborate variable importance measures incorporate a (weighted) mean of the individual trees’ improvement in the splitting criterion produced by each variable (Friedman 2001). An example for such a measure in classification is the “Gini importance” available in random forest implementations. It describes the average improvement in the “Gini gain” splitting criterion that a variable has achieved in all of its positions in the forest. However, in many applications involving predictor variables of different types, this measure is biased, as outlined in section “Bias in variable selection and variable importance”.
The most advanced variable importance measure available in random forests is the “permutation accuracy importance” measure (termed permutation importance in the following). Its rationale is the following: By randomly permuting the values of a predictor variable, its original association with the response is broken.
For example, in the original smoking data, those adolescents who drank alcohol in more occasions are more likely to intend to smoke. Randomly permuting the values of alcohol_per_month over all subjects, however, destroys this association. Accordingly, when the permuted variable, together with the remaining unpermuted predictor variables, is now used to predict the response, the prediction accuracy decreases substantially.
Thus, a reasonable measure for variable importance is the difference in prediction accuracy (i.e. the number of observations classified correctly; usually the out-of-bag prediction accuracy is used to compute the permutation importance) before and after permuting a variable, averaged over all trees.
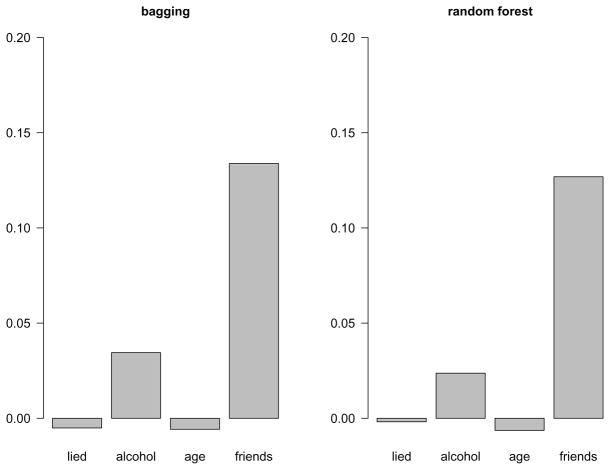
If, on the other hand, the original variable was not associated with the response, it is either not included in the tree (and its importance for this tree is zero by definition), or it is included in the tree by chance. In the latter case, permuting the variable results only in a small random decrease in prediction accuracy, or the permutation of an irrelevant variable can even lead to a small increase in the prediction accuracy (if, by chance, the permutated variable happens to be slightly better suited for splitting than the original one). Thus the permutation importance can even show (small) negative values for irrelevant predictor variables, as illustrated for the irrelevant predictor variables age and lied_to_parents in Figure 8.
Figure 8.
Permutation variable importance scores for the predictor variables of the smoking data from bagging and random forests.
Note also that in our simple example the two relevant predictor variables friends_smoke and alcohol_per_month are correctly identified by the permutation variable importance of both bagging and random forests, even though the positions of the variables vary more strongly in random forests (cf. again Figures 6 and 7). In real data applications, however, the random forest variable importance may reveal higher importance scores for variables working in complex interactions, that may have gone unnoticed in single trees and bagging (as well as in parametric regression models, where modeling high-order interactions is usually not possible at all).
Formally the permutation importance for classification can be defined as follows: Let 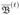 be the out-of-bag sample for a tree t, with t ∈ {1, …, ntree}. Then the importance of variable Xj in tree t is
be the out-of-bag sample for a tree t, with t ∈ {1, …, ntree}. Then the importance of variable Xj in tree t is
| (1) |
where is the predicted class for observation i before and is the predicted class for observation i after permuting its value of variable Xj, i.e. with xi,ψj = (xi,1, …, xi,j−1, xψj(i),j, xi,j+1, …, xi,p). (Note that VI (t)(Xj) = 0 by definition, if variable Xj is not in tree t.) The raw importance score for each variable is then computed as the average importance over all trees
| (2) |
From this raw importance score a standardized importance score can be computed with the following rationale: The individual importance scores VI (t)(xj) are computed from ntree bootstrap samples, that are independent given the original sample, and are identically distributed. Thus, if each individual variable importance VI (t) has standard deviation σ, the average importance from ntree replications has standard error . The standardized or scaled importance, also called “z-score”, is then computed as
| (3) |
When the central limit theorem is applied to the mean importance VI (xj), Breiman and Cutler (2008) argue that the z-score is asymptotically standard normal. This property is often used for a statistical test, that, however, shows very poor statistical properties as outlined in the section on “Features and pitfalls”.
As already mentioned, the main advantage of the random forest permutation accuracy importance, as compared to univariate screening methods, is that it covers the impact of each predictor variable individually as well as in multivariate interactions with other predictor variables. For example Lunetta et al. (2004) find that genetic markers relevant in interactions with other markers or environmental variables can be detected more efficiently by means of random forests than by means of univariate screening methods like Fisher’s exact test.
This, together with its applicability to problems with many predictor values, also distinguishes the random forest variable importance from the otherwise appealing approach of Azen, Budescu, and Reiser (2001) and advanced in Azen and Budescu (2003) for assessing the criticality of a predictor variable, termed “dominance analysis”: The authors suggest employing bootstrap sampling and select the best regression model from all possible models for each bootstrap sample in order to estimate the empirical probability distribution of all possible models. From this empirical distribution for each variable the unweighted or weighted sum of probabilities associated with all models containing the predictor is computed and suggested as an intuitive measure of variable importance. This approach, where for p predictor variables 2p − 1 models are fitted in each bootstrap iteration, has the great advantage that it provides sound statistical inference. However, it is computationally prohibitive for problems with many predictor variables of interest, because all possible models have to be fitted on all bootstrap samples.
In random forests, on the other hand, a tree model is fit to every bootstrap sample only once. Then the predictor variables are permuted in an attempt to mimic their absence in the prediction. This approach can be considered in the framework of classical permutation test procedures (Strobl, Boulesteix, Kneib, Augustin, and Zeileis 2008) and is feasible for large problems, but lacks the sound statistical background available for the approach of Azen et al. (2001). Another difference is that random forest variable importances reflect the effect of a variable in complex interactions as outlined above, while the approach of Azen et al. (2001) reflects the main effects – at least as long as interactions are not explicitly included in the candidate models.
A conditional version of the random forest permutation importance, that resembles the properties of partial correlations rather than that of dominance analysis, is suggested by Strobl et al. (2008).
Literature and Software
Random forests have only recently been included in standard textbooks on statistical learning, such as Hastie et al. (2009) (while the previous edition, Hastie et al. 2001, did not cover this topic yet). In addition to a short introduction of random forests, this reference gives a thorough background on classification trees and related concepts of resampling and model validation, and is therefore highly recommended for further reading. For the social sciences audience a first instructive review on ensemble methods, including random forests and the related method bagging, was given by Berk (2006). We suggest this reference for the treatment of unbalanced data (for example in the case of a rare disease or mental condition), that can be treated either by means of asymmetric misclassification costs or equivalently by means of weighting with different prior probabilities in classification trees and related methods (see also Chen, Liaw, and Breiman 2004, for the alternative strategy of “down sampling”, i.e., sampling from the majority class as few observations as there are of the minority class), even though the interpretation of interaction effects in Berk (2006) is not coherent, as demonstrated above. The original works of Breiman (1996a,b, 1998a,b, 2001a,b), to name a few, are also well suited and not too technical for further reading.
For practical applications of the methods introduced here, several up-to-date tools for data analysis are freely available in the R system for statistical computing (R Development Core Team 2008). Regarding this choice of software, we believe that the supposed disadvantage of command line data analysis criticized by Berk (2006) is easily outweighed by the advanced functionality of the R language and its add-on packages at the state of the art of statistical research. However, in statistical computing the textbooks also lag behind the latest scientific developments: The standard reference Venables and Ripley (2002) does not (yet) cover random forests either, while the handbook of Everitt and Hothorn (2006) gives a short introduction to the use of both classification trees and random forests. This handbook, together with the instructive examples in the following section and the R-code provided in a supplement to this work, can offer a good starting point for applying random forests to your data. Interactive means of visual data exploration in R, that can support further interpretation, are described in Cook and Swayne (2007).
Further Application Examples
For further illustration, two additional application examples of model based partitioning and random forests are outlined. The source code for reproducing all steps of the following analyses as well as the examples in the previous sections in the R system for statistical computing (R Development Core Team 2009) is provided as a supplement.
Model-Based Recursive Partitioning
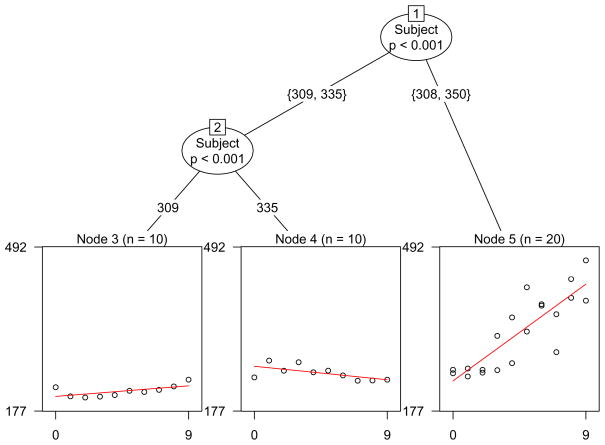
For a reaction time experiment, where the independent variable is sleep deprivation (subset of four exemplary subjects from the sleep-deprived group with measurements for the first 10 days of the study from Belenky, Wesensten, Thorne, Thomas, Sing, Redmond, Russo, and Balkin 2003), it is illustrated in Figure 9 that the effect of sleep deprivation on reaction time differs between subjects.
Figure 9.
Model-based partition for the reaction time data. The model of interest relates the number of days of sleep deprivation to the reaction time.
For each subject, the data set contains ten successive measurements (for days 0 through 9). The subject ID is used as a pseudo-covariate for model based partitioning here: The subject IDs are indicated in the tree structure in Figure 9, where the leftmost node, e.g., includes only the measurements of subject 309, while the rightmost node includes the measurements of subjects 308 and 350. The model of interest in each final node relates the number of days of sleep deprivation (0 through 9, on the abscissa) to the reaction time (on the ordinate).
We find that the reactions to sleep deprivation of subjects 308 and 350 can be represented by one joint model, while the reactions of the other subjects are represented by distinct models. If additional covariates, such as age and gender, rather than only the subject ID, were available for this study, they could be used for partitioning as well, while using the subject ID as a pseudo covariate as in this example resembles a latent class approach for identifying groups of subjects with similar response patterns.
Of course, differences between subjects or groups of subjects could also be modeled by means of, e.g., random effects or latent class models – but again the visual inspection of the model-based partition, that requires no further assumptions, can provide a helpful first glance impression of different response patterns present in the sample or help, e.g., identify groups of non-responders in clinical studies.
Random Forests
When the number of variables is high, as for example in gene expression studies, parametric regression models are not applicable and ensemble methods are often applied for prediction and the assessment of variable importance.
For an exemplary analysis of gene data, we adopted a data set originally presented by Ryan, Lockstone, Huffaker, Wayland, Webster, and Bahn (2006): The data were collected in a case-control study on bipolar disorder including 61 samples (from 30 cases and 31 controls) from the dorsolateral prefrontal cortex cohort. In the original study of Ryan et al. (2006), no genes were clearly found to be differentially expressed, i.e., to have an effect on the disease, in this sample. Therefore, two genes were artificially modified to have an effect, so that we can later control whether these genes are correctly identified.
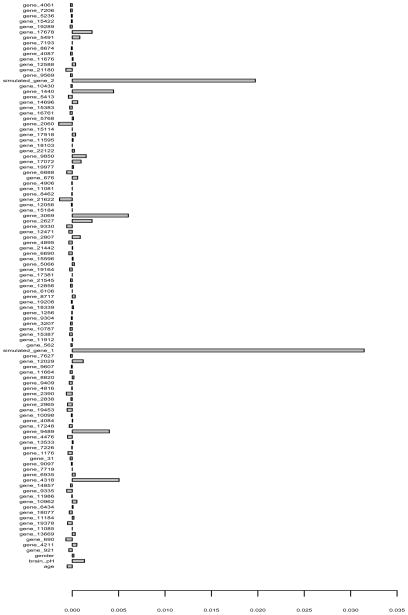
In order to be able to illustrate t’the variable importances in a plot, in addition to the two simulated genes and the three covariates age, gender and brain pH-level, a subset of 100 genes was randomly selected from the 22,283 genes originally present by Ryan et al. (2006) for the example. Note, however, that the application to larger data sets is only a question of computation time.
The permutation importances for all 105 variables are displayed in Figure 10. The effects of the two artificially modified genes can be clearly identified. With respect to the remaining variables, a conservative strategy for exploratory screening would be to include all genes whose importance scores exceed the amplitude of the largest negative scores (that can only be due to random variation) in future studies.
Figure 10.
Variable importances for the original and modified gene data.
A prediction from the random forest can be given either in terms of the predicted response class or the predicted class probabilities, as illustrated in Table 1 for some exemplary subjects, with a mismatch between the true and predicted class for subject 29.
Table 1.
Predicted response class or class probability.
| y | ŷ | p̂(y = 1) | |
|---|---|---|---|
| subject 28 | 1 | 1 | 0.80 |
| subject 29 | 1 | 2 | 0.46 |
| subject 30 | 1 | 1 | 0.64 |
| subject 31 | 2 | 2 | 0.48 |
| subject 32 | 2 | 2 | 0.43 |
For the entire learning sample the prediction accuracy estimate is over-optimistic (90.16%), while the estimate based on the out-of-bag sample is more conservative (67.21%). The confusion matrices in Table 2 display misclassifications separately for each response class.
Table 2.
Confusion matrix with prediction from learning (left) and out-of-bag sample (right).
| ŷ = 1 | ŷ = 2 | |
|---|---|---|
| y = 1 | 28 | 2 |
| y = 2 | 4 | 27 |
| ŷ = 1 | ŷ = 2 | |
|---|---|---|
| y = 1 | 20 | 10 |
| y = 2 | 10 | 21 |
Note that a logistic regression model would not be applicable in a sample of this size, even for the reduced data set with only 102 genes – not even if forward selection was employed and only main effects were considered, disregarding the possibility of interaction effects – because the estimation algorithm does not converge.
In other cases, however, the prediction accuracy of the complex random forests model, involving high-order interactions and nonlinearity, can be compared to that of a simpler, for example linear or logistic regression model including only low-order interactions, to guide the decision whether the simpler, interpretable model would be equally adequate.
To further explore and interpret the effects and interactions of the predictor variables that were found relevant in a random forest, multivariate data visualization tools, such as those described in Cook and Swayne (2007), are strongly suggested.
Features and Pitfalls
The way recursive partitioning methods – in particular the ensemble methods bagging and random forests – work induces some special characteristics, that distinguish them from other (even other nonparametric) approaches. Some of these special features are mostly technical, while others can prove very beneficial in applications, and yet others may pose severe practical problems, that we want to address here.
“Small n Large p” Applicability
The fact that variable selection can be limited to random subsets in random forests make them particularly well applicable in “small n large p” problems with many more variables than observations, and has added much to the popularity of random forests. However, even if the set of candidate predictor variables is not restricted as in random forests, but covers all predictor variables as in bagging, the search is only a question of computational effort: Unlike logistic regression models, e.g., where parameter estimation is not possible (for instance, due to linear constraints in the predictors or perfect separation of response classes in some predictor combinations) when there are too many predictor variables and too few observations, tree-based methods like bagging and random forests only consider one predictor variable at a time, and can thus deal with high numbers of variables sequentially. Therefore Bureau et al. (2005) and Heidema, Boer, Nagelkerke, Mariman, van der A, and Feskens (2006) point out that the recursive partitioning strategy is a clear advantage of random forests as opposed to more common methods like logistic regression in high dimensional settings.
Nonlinear Function Approximation
Classification and regression trees are provably Bayes consistent, i.e. in principle they can approximate any decision boundary, whether linear or highly nonlinear, given a sufficiently large data set and allowed to grow at a proper rate (see, e.g., Devroye, Györfi, and Lugosi 1996). For linear functions, the problem from a practical point of view is that a single tree’s step-function approximation will be rather poor. Ensembles of trees, however, can approximate functions more smoothly by averaging over the single trees’ step-functions.
Therefore, bagging and random forests can be used to approximate any unknown function, even if it is nonlinear and involves complex interactions. An advantage of ensemble methods in this context is that, as compared to other nonlinear regression approaches such as smoothing splines, neither the shape of the function nor the position or number of knots needs to be prespecified (see, e.g., Wood 2006, for knot selection approaches in generalied additive models). On the other hand, the resulting functional shape cannot be interpreted or grasped analytically, and (aside from measures of overall variable importance) can only serve as a “black-box” for prediction. This characteristic of many machine learning approaches has fueled discussions about the legitimacy and usefulness of such complex, nonlinear models (see, e.g., Hand 2006, and the corresponding discussion).
In practice, for a given data set, where nonlinear associations or high-order interactions are suspected, complex approaches like random forests can at least serve as a benchmark predictor: If a linear or other parametric model with a limited number and degree of interaction terms can reach the (cross validated or test sample) prediction accuracy of the more complex model, the extra complexity may be uncalled for and the simpler, interpretable model should be given preference. If, however, the prediction accuracy cannot be reached with the simpler model, and, for example, the high importance of a variable in a random forest is not reflected by its respective parameters in the simpler model, relevant nonlinear or interaction effects may be missing in the simpler model and it may not be suited to grasp the complexity of the underlying process.
In addition to this, a “black-box” method like random forests can be used to identify a small number of potentially relevant predictors from the full feature list, that can then be processed, e.g., by means of a familiar parametric method. This two-stage approach has been successfully applied in a variety of applications (see, e.g., Ward et al. 2006). Note, however, that variable selection should not be conducted before applying another statistical method on the same learning data (Ambroise and McLachlan 2002; Leeb and Pötscher 2006; Boulesteix et al. 2008).
The “XOR”-Problem and Order Effects
In the literature on recursive partitioning, you may come across the so called “XOR”-problem, that describes a situation where two variables show no main effect, but a perfect interaction. In this case, due to the lack of a marginally detectable main effect, none of the variables may be selected in the first split of a classification tree, and the interaction may never be discovered.
In such a perfectly symmetric, artificial XOR problem, a tree would indeed not find a cutpoint to start with. However, a logistic regression model would not be able to identify an effect in any of the variables either, if the interaction was not explicitly included in the logistic regression model – and in that case a tree model, where an interaction effect of two variables can also be explicitly added as a potential predictor variable, would do equally well.
In addition to this, a tree, and even better an ensemble of trees, is able to approximate the XOR problem by means of a sequence of cutpoints driven by random fluctuations that are present in any real data set. In this case, the random preselection of splitting variables in random forests again increases the chance that a variable with a weak marginal effect is still selected, at least in some trees, because some of its competitors are not available.
A similar argument applies to order effects when comparing stepwise variable selection in regression models with the variable selection that can be conducted on the basis of random forest variable importance measures: In both, stepwise variable selection and single trees, order effects are present, because only one variable at a time is considered – in the context of the variables that were already selected, but regardless of all variables yet to come. However, the advantage of ensemble methods, that employ several parallel tree models, is that the order effects of all individual trees counterbalance, so that the overall importance ranking of a variable is much more reliable than its position in stepwise selection (see also Rossi et al. 2005).
Out-of-Bag Error Estimation
It was already mentioned, and used in the application example, that bagging and random forests come with their own “built-in” test sample, the out-of-bag observations, that provide a fair means of error estimation (Breiman 1996b). Of course similar validation strategies, based either on sample splitting or resampling techniques (see, e.g., Hothorn, Leisch, Zeileis, and Hornik 2005; Boulesteix et al. 2008) or ideally even external validation samples (König, Malley, Weimar, Diener, and Ziegler 2007), can and should be applied to any statistical method. However, in many disciplines intensive model validation is not common practice. Therefore a method that comes with a built-in test sample, like random forests, may help sensitize for the issue and relieve the user of the decision for an appropriate validation scheme.
Missing Value Handling
Besides imputation approaches offered by some random forests algorithms, all tree based methods provide another intuitive strategy for missing value handling: This strategy is that, at first, observations that have missing values in the variable that is currently evaluated are ignored in the computation of the impurity reduction for this variable. However, the same observations are included in all other computations, so that the method does not involve cancelation of observations with missing values (which can result in heavy data loss).
After a splitting variable is selected it would be unclear to what daughter node the observations that have a missing values in this variable should be assigned. Therefore a surrogate variable is selected, that best predicts the values of the splitting variable. By means of this surrogate variable the observations can then be assigned to the left or right daughter node (cf., e.g., Hastie et al. 2001). A flaw of this strategy is, however, that currently the permutation variable importance measure is not defined for variables with missing values.
Bias in Variable Selection and Variable Importance
In the classical classification and regression tree algorithms CART and C4.5, variable selection is biased in favor of variables with certain characteristics, even if these variables are no more informative than their competitors. For example, variables with many categories and numeric variables or, even more unintuitively, variables with many missing values are artificially preferred (see, e.g., White and Liu 1994; Kim and Loh 2001; Strobl, Boulesteix, and Augustin 2007).
This bias is carried forward to ensembles of trees: Especially the variable importance can be biased when a data set contains predictor variables of different types (Strobl et al. 2007). The bias is particularly pronounced for the Gini importance, that is based on the biased Gini gain split selection criterion (Strobl et al. 2007), but can also affect the permutation importance. Only when subsamples drawn without replacement, instead of bootstrap samples, in combination with unbiased split selection criteria, are used in constructing the forest, the resulting permutation importance can be interpreted reliably (Strobl et al. 2007).
For applications in R, the functions ctree for classification and regression trees and cforest for bagging and random forests (both freely available in the add-on package party; Hothorn et al. 2006; Hothorn, Hornik, and Zeileis 2008) guarantee unbiased variable selection when used with the default parameter settings, as documented in the supplement to this work.
The functions tree (Ripley 2007) and rpart (Therneau and Atkinson. 2006) for trees and random-Forest (Breiman, Cutler, Liaw, and Wiener 2006; Liaw and Wiener 2002) for bagging and random forests, on the other hand, that resemble the original CART and random forests algorithms more closely, induce variable selection bias and are not suggested when the data set contains predictor variables of different types.
Scaled and Unscaled Importance Measures
For the permutation importance a scaled version, the z-score, is available or even default in many implementations of random forests. The term “scaled” here is somewhat misleading, however, for two reasons: Firstly, the variable importance does not depend on the scaling or variance of the predictor variables in the first place (in fact, the whole method is invariant against the scaling of numeric variables). Therefore it is not necessary to account for the scaling of predictors in the variable importance.
Secondly, for a “scaled” measure one may assume that its values are comparable over different studies – which is not the case for the z-score in random forests, that heavily depends on the choice of tuning parameters, as outlined in the next section.
Therefore, we suggest not to interpret or compare the absolute values of the importance measures, not even the z-scores, but rely only on a descriptive ranking of the predictor variables.
Tests for Variable Importance and Variable Selection
In addition to using variable importance measures as a merely descriptive means of data exploration, different significance tests and schemes for variable selection have been suggested: On the official random forests website, a simple statistical test based on the supposed normality of the z-score is proposed by Breiman and Cutler (2008, date of access), that has been applied in a variety of studies – ranging from the investigation of predictors of attempted suicide (Baca-Garcia et al. 2007) to the monitoring of a large area space telescope on board of a satellite (Paneque, Borgland, Bovier, Bloom, Edmonds, Funk, Godfrey, Rando, Wai, and Wang 2007).
This approach may appear more statistically advanced than a merely descriptive usage of the random forest variable importance. However, it shows such alarming statistical properties that any statement of significance made with this test is nullified (Strobl and Zeileis 2008): The power of this test depends on the number of trees in the ensemble ntree, over which the importance is averaged (cf. Equations 2 and 3 in section “Variable importance”; see also Lunetta et al. 2004). Thus reporting the significance of variable importance scores (like, e.g., Baca-Garcia et al. 2007, who do not even report the parameter settings they use for fitting the random forest) can be highly misleading, because the number of variables whose scores exceed a given threshold for significance depends on the arbitrary choice of a tuning parameter.
In addition to this, all statistical tests and variable selection schemes based on the original permutation importance, such as those suggested by Diaz-Uriarte and Alvarez de Andrés (2006) and Rodenburg, Heidema, Boer, Bovee-Oudenhoven, Feskens, Mariman, and Keijer (2008), suffer from another artifact, that is induced by the way the permutation importance is constructed: the artificial preference of correlated predictor variables. In a permutation test framework Strobl et al. (2008) show that only a conditional permutation scheme reflects the desired null hypothesis, and the resulting conditional importance describes the actual effect of a variable in the presence of correlations more reliably.
For selecting variables for further investigation in an exploratory study, we suggest a conservative decision aid for variable selection, already mentioned in the application example: All variables whose importance is negative, zero or has a small positive value that lies in the same range as the negative values, can be excluded from further exploration. The rationale for this rule of thumb is that the importance of irrelevant variables varies randomly around zero. Therefore positive variation of an amplitude comparable to that of negative variation does not indicate an informative predictor variable, while positive values that exceed this range may indicate that a predictor variable is informative.
Randomness and Stability
One special characteristic of random forests and bagging, that new users are often not entirely aware of, is that they are truly “random” models in the sense that, for the same data set, the results may differ between two computation runs.
The two sources of randomness that are responsible for these possible differences are (a) the bootstrap samples (or subsamples) that are randomly drawn in bagging and random forests and (b) the random preselection of predictor variables in random forests. When When the permutation importance is computed, another source of of variability is the variability is the random permutation of the predictor vectors.
Due to these random processes, a random forest is only exactly reproducible when the random seed, a number that can be set by the user and determines the internal random number generation of the computer, is fixed. Otherwise, the results will vary between two runs of the same code. To illustrate this point, random seeds are set in the supplementary code for the application examples, whenever random sampling is involved.
The differences induced by random variations are, however, negligible – as long as the parameters of a random forest have been chosen such as to guarantee stable results:
The number of trees ntree highly affects the stability of the model. In general, the higher the number of trees the more reliable is the prediction and the interpretability of the variable importance.
-
The number of randomly preselected predictor variables mtry may also affect the stability of the model and the reliability of the variable importance. In general, random forests with random preselection perform better than bagging with no random preselection at all, but small values of mtry do not always prove beneficial: When predictor variables are highly correlated, the results of Strobl et al. (2008) indicate that a higher number of randomly preselected predictor variables is better suited to reflect conditional importance. In addition to that, if the number of randomly preselected predictor variables is very low, interactions of high order may be missed in the tree building process. In situations with few relevant variables, “small mtry results in many trees being built that do not incorporate any of the relevant [variables]” (Diaz-Uriarte and Alvarez de Andrés 2006), which would lead to a decrease in prediction accuracy.
The number of randomly preselected predictor variables can also be chosen such as to optimize prediction accuracy by means of cross validation in some algorithms. Note, however, that the choice of tuning parameters in random forests is not as critical as in other computerintensive approaches, such as support vector machines (Svetnik, Liaw, Tong, and Wang 2004), and random forests often produce good results even “off the shelf” with default settings.
Note that the two tuning parameters, ntree and mtry, also interact: To assess a high number of predictor variables in a data set, a high number of trees or a high number of preselected variables for each split, or ideally both, are necessary so that each variable has a chance to occur in enough trees. Only then its average variable importance measure is based on enough trials to actually reflect the importance of the variable and not just a random fluctuation.
In summary this means: If you observe that, for a different random seed, your prediction results and variable importance rankings (for the top-scoring variables) differ notably, you should not interpret the results but adjust the number of trees and preselected predictor variables.
Do Random Forests Overfit?
The study referred to in Breiman (2001b), where it is stated (and has been extensively cited ever since) that random forests do not overfit, may be a prominent example for a premature conclusion drawn from an unrepresentative sample. A variety of studies exploring the characteristics of machine learning tools such as random forests are based on only a few, real data sets, that happen to be freely available in some data repository. The particular data sets investigated by Breiman (2001b) seem to enhance the impression that random forests would not overfit, but this notion is heavily criticized by Segal (2004).
The theoretical results of Breiman (1996a) do support the fact that ensemble methods do not overfit with an increasing number of trees. However, the real data “case studies” referred to in Breiman (2001b) do not exclude the possibility that they overfit due to other reasons. For further methodological investigations of machine learning algorithms we therefore strongly suggest to employ well designed and controlled simulation experiments, rather than case studies with an unrepresentative selection of real data sets with unknown distributional properties, when analytical results are not feasible.
With respect to the theoretical foundations and practical application of random forests, Segal (2004) implies that the depth of the trees in random forests, rather than the number of trees as suspected, e.g., by Luellen et al. (2005), may regulate overfitting.
While most previous publications have argued that in an ensemble each individual tree should be grown as large as possible and that trees should not be pruned, the recent results of Lin and Jeon (2006) also show that creating large trees is not necessarily the optimal strategy. In problems with a high number of observations and few variables a better convergence rate (of the mean squared error as a measure of prediction accuracy) can be achieved when the terminal node size increases with the sample size (i.e. when smaller trees are grown for larger samples). On the other hand for problems with small sample sizes or even “small n large p” problems, growing large trees usually does lead to the best performance.
Discussion and Conclusion
Recursive partitioning methods have become popular and widely used tools in many scientific fields. Especially random forests have been widely applied in genetics and related disciplines within the past few years. First applications in psychology show that random forests can be of use in a wide variety of applications in this field as well. With this review we hope to have given the necessary background for a successful – yet sensible – use of recursive partitioning methods, in particular of random forests, that have drawn much attention due to their applicability to even high dimensional problems.
Besides the applications to regression and classification problems covered here, the function cforest (Hothorn et al. 2008, 2006) used in the application example can even be applied to survival data with a censored response and can thus serve as a means of data exploration in a broad range of longitudinal studies, too.
Of course other recent statistical learning methods, such as boosting (Freund and Schapire 1997) and support vector machines (cf. Vapnik 1995, for an introduction) can also be applied to the scope of problems we suggested for the application of random forests. The performance of these methods is within a close range with that of random forests, so that in some comparison studies random forests clearly outperform their competitors (cf., e.g., Wu, Abbott, Fishman, McMurray, Mor, Stone, Ward, Williams, and Zhao 2003), while in others they are slightly outperformed (cf., e.g., König, Malley, Pajevic, Weimar, Diener, and Ziegler 2008, for a comparison of several statistical learning methods in a medical example of moderate size, where logistic regression was also applicable).
In summary, one can conclude in accordance with Heidema et al. (2006), that high dimensional data should be approached by several different methods because each single method has its strengths and weaknesses: Boosting, for example, can be employed for variable selection in linear and other additive models (Bühlmann 2006; Bühlmann and Hothorn 2007, for an implementation in R). Similarly, shrinkage approaches like the LASSO (cf., e.g., Hastie et al. 2001), the elastic net (Zou and Hastie 2005) and the recent approach of Candes and Tao (2007) perform variable selection in linear models by means of penalization of the model coefficients. However, in contrast to random forests, for these methods it has to be assumed that the model is linear or additive and that the problem is sparse (meaning that only few predictor variables have an effect). For extremely small sample sizes, on the other hand, exact methods like the multivariate permutation tests described in Mielke and Berry (2001) or Good (2005) may be more suited.
With respect to ease of application, the results of the empirical comparisons between different supervised learning methods conducted by Caruana and Niculescu-Mizil (2006) and Svetnik et al. (2004) indicate that random forests are among the best performing methods even without extra tuning. Therefore random forests can be considered as a valuable “off the shelf” tool for exploring complex data sets, that may in a few years from now become as popular in psychology as it is now in the fields of genetics and bioinformatics.
Supplementary Material
Acknowledgments
The authors would like to thank A. Liaw, Merck & Co. Inc., Whitehouse Station, New Jersey, USA, and A. Zeileis, Wirtschaftsuniversität Wien, Vienna, Austria, for their expert advice on technical issues, as well as the editor and reviewers for their helpful suggestions on the presentation.
Contributor Information
Carolin Strobl, Department of Statistics, Ludwig-Maximilians-Universität, Munich, Germany.
James Malley, Center for Information Technology, National Institutes of Health, Bethesda, MD, USA.
Gerhard Tutz, Department of Statistics, Ludwig-Maximilians-Universität, Munich, Germany.
References
- Ambroise C, McLachlan GJ. Selection bias in gene extraction in tumour classification on basis of microarray gene expression data. Proceedings of the National Academy of Science. 2002;99:6562–6566. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin P, Tu J. Automated variable selection methods for logistic regression produced unstable models for predicting acute myocardial infarction mortality. Journal of Clinical Epidemiology. 2004;57(11):1138–1146. doi: 10.1016/j.jclinepi.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Azen R, Budescu DV. The dominance analysis approach for comparing predictors in multiple regression. Psychological Methods. 2003;8(2):129–48. doi: 10.1037/1082-989x.8.2.129. [DOI] [PubMed] [Google Scholar]
- Azen R, Budescu DV, Reiser B. Criticality of predictors in multiple regression. British Journal of Mathematical and Statistical Psychology. 2001;54:201–225. doi: 10.1348/000711001159483. [DOI] [PubMed] [Google Scholar]
- Baca-Garcia E, Perez-Rodriguez MM, Saiz-Gonzalez D, Basurte-Villamor I, Saiz-Ruiz J, Leiva-Murillo JM, de Prado-Cumplido M, Santiago-Mozos R, Artes-Rodriguez A, de Leon J. Variables associated with familial suicide attempts in a sample of suicide attempters. Progress in Neuro-Psychopharmacology & Biological Psychiatry. 2007;31(6):1312–1316. doi: 10.1016/j.pnpbp.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Bauer E, Kohavi R. An empirical comparison of voting classification algorithms: Bagging, boosting, and variants. Machine Learning. 1999;36(1–2):105–139. [Google Scholar]
- Belenky G, Wesensten N, Thorne D, Thomas M, Sing H, Redmond D, Russo M, Balkin T. Patterns of performance degradation and restoration during sleep restriction and subsequent recovery: A sleep dose-response study. Journal of Sleep Research. 2003;12:1–12. doi: 10.1046/j.1365-2869.2003.00337.x. [DOI] [PubMed] [Google Scholar]
- Berk RA. An introduction to ensemble methods for data analysis. Sociological Methods & Research. 2006;34(3):263–295. [Google Scholar]
- Biau G, Devroye L, Lugosi G. Consistency of random forests and other averaging classifiers. Journal of Machine Learning Research. 2008;9:2015–2033. [Google Scholar]
- Boulesteix AL, Strobl C, Augustin T, Daumer M. Evaluating microarray-based classifiers: An overview. Cancer Informatics. 2008;4:77–97. doi: 10.4137/cin.s408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Bagging predictors. Machine Learning. 1996a;24(2):123–140. [Google Scholar]
- Breiman L. Technical report. Department of Statistics, University of California; Berkeley, CA, USA: 1996b. Out-of-bag estimation. [Google Scholar]
- Breiman L. Arcing classifiers. The Annals of Statistics. 1998;26(3):801–849. [Google Scholar]
- Breiman L. Random forests. Machine Learning. 2001a;45(1):5–32. [Google Scholar]
- Breiman L. Statistical modeling: The two cultures. Statistical Science. 2001b;16(3):199–231. [Google Scholar]
- Breiman L, Cutler A. Random forests – Classification manual. 2008. [Website accessed in 1/2008.]. [Google Scholar]
- Breiman L, Cutler A, Liaw A, Wiener M. R package version 4.5–16. 2006. Breiman and Cutler’s Random Forests for Classification and Regression. [Google Scholar]
- Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and Regression Trees. New York: Chapman and Hall; 1984. [Google Scholar]
- Bühlmann P. Boosting for high-dimensional linear models. Annals of Statistics. 2006;34:559–583. [Google Scholar]
- Bühlmann P, Hothorn T. Boosting algorithms: Regularization, prediction and model fitting. Statistical Science. 2007;22(4):477–505. [Google Scholar]
- Bühlmann P, Yu B. Analyzing bagging. The Annals of Statistics. 2002;30(4):927–961. [Google Scholar]
- Bühlmann P, Yu B. Boosting with the L2 loss: Regression and classification. Journal of the American Statistical Association. 2003;98:324–339. [Google Scholar]
- Bureau A, Dupuis J, Falls K, Lunetta KL, Hayward B, Keith TP, Eerdewegh PV. Identifying SNPs predictive of phenotype using random forests. Genetic Epidemiology. 2005;28(2):171–182. doi: 10.1002/gepi.20041. [DOI] [PubMed] [Google Scholar]
- Burnham K, Anderson D. Model Selection and Multimodel Inference. New York: Springer; 2002. [Google Scholar]
- Burnham K, Anderson D. Multimodel inference. Sociological Methods & Research. 2004;33(2):261–304. [Google Scholar]
- Candes E, Tao T. The Dantzig selector: Statistical estimation when p is much larger than n. The Annals of Statistics. 2007;35(6):2313–2351. [Google Scholar]
- Caruana R, Niculescu-Mizil A. An empirical comparison of supervised learning algorithms. Proceedings of the 23rd International Conference on Machine Learning (ICML 2006); Pittsburgh, PA, USA. New York: ACM Press; 2006. pp. 161–168. [Google Scholar]
- Chen C, Liaw A, Breiman L. Technical Report 666. Department of Statistics, University of California; Berkeley, CA, USA: 2004. Using random forest to learn imbalanced data. [Google Scholar]
- Claeskens G, Hjort N. Model Selection and Model Averaging. Cambridge: Cambridge University Press; 2008. [Google Scholar]
- Cook D, Swayne D. Interactive and Dynamic Graphics for Data Analysis. Berlin: Springer; 2007. [Google Scholar]
- Cutler A. Technical Report. Utah State University, Dept. of Mathematics and Statistics; 1999. Fast classification using perfect random trees. 5/99/99. [Google Scholar]
- Cutler A. Technical Report. Utah State University, Dept. of Mathematics and Statistics; 2000. Voting perfect random trees. 5/00/100. [Google Scholar]
- Derksen S, Keselman H. Backward, forward and stepwise automated subset selection algorithms: Frequency of obtaining authentic and noise variables. British Journal of Mathematical and Statistical Psychology. 1992;45(11):265–282. [Google Scholar]
- Devroye L, Györfi L, Lugosi G. A Probabilistic Theory of Pattern Recognition. New York: Springer; 1996. [Google Scholar]
- Diaz-Uriarte R, Alvarez de Andrés S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietterich TG. An experimental comparison of three methods for constructing ensembles of decision trees: Bagging, boosting, and randomization. Machine Learning. 2000;40(2):139–157. [Google Scholar]
- Domingos P. Why does bagging work? A bayesian account and its implications. Proceedings of the Third International Conference on Knowledge Discovery and Data Mining; Newport Beach, CA, USA. AAAI Press; 1997. pp. 155–158. [Google Scholar]
- Doran H, Bates D, Bliese P, Dowling M. Estimating the multilevel Rasch model: With the lme4 package. Journal of Statistical Software. 2007;20(2) [Google Scholar]
- Everitt B, Hothorn T. A Handbook of Statistical Analyses Using R. Boca Raton: Chapman & Hall/CRC; 2006. [Google Scholar]
- Freedman D. A note on screening regression equations. The American Statistician. 1983;37(2):152–155. [Google Scholar]
- Freund Y, Schapire RE. A decision-theoretic generalization of on-line learning and an application to boosting. Journal of Computer and System Sciences. 1997;55(1):119–139. [Google Scholar]
- Friedman J. Greedy function approximation: A gradient boosting machine. The Annals of Statistics. 2001;29(5):1189–1232. [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Additive logistic regression: a statistical view of boosting. The Annals of Statistics. 2000;28(2):337–407. [Google Scholar]
- Gatnar E. Fusion of multiple statistical classifiers. In: Preisach C, Burkhardt H, editors. Data Analysis, Machine Learning and Applications: Proceedings of the 31st Annual Conference of the German Classification Society (GfKl), Freiburg i. Br; Germany. 2008. pp. 19–28. to appear. [Google Scholar]
- Good P. Permutation, Parametric, and Bootstrap Tests of Hypotheses. 3 New York: Springer Series in Statistics; 2005. [Google Scholar]
- Grandvalet Y. Bagging equalizes influence. Machine Learning. 2004;55(3):251–270. [Google Scholar]
- Gunther EC, Stone DJ, Gerwien RW, Bento P, Heyes MP. Prediction of clinical drug efficacy by classification of drug-induced genomic expression profiles in vitro. Proceedings of the National Academy of Sciences. 2003;100(16):9608–9613. doi: 10.1073/pnas.1632587100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand DJ. Classifier technology and the illusion of progress. Statistical Science. 2006;21(1):1–14. [Google Scholar]
- Hannöver W, Richard M, Hansen NB, Martinovich Z, Kordy H. A classification tree model for decision-making in clinical practice: An application based on the data of the German multicenter study on eating disorders, project TR-EAT. Psychotherapy Research. 2002;12(4):445–461. [Google Scholar]
- Hastie T, Tibshirani R, Friedman JH. The Elements of Statistical Learning. New York: Springer; 2001. [Google Scholar]
- Hastie T, Tibshirani R, Friedman JH. The Elements of Statistical Learning. 2. New York: Springer; 2009. [Google Scholar]
- Heidema AG, Boer JMA, Nagelkerke N, Mariman ECM, van der A DL, Feskens EJM. The challenge for genetic epidemiologists: How to analyze large numbers of SNPs in relation to complex diseases. BMC Genetics. 2006;7:23. doi: 10.1186/1471-2156-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeting JA, Madigan D, Raftery AE, Volinsky CT. Bayesian model averaging: A tutorial. Statistical Science. 1999;14(4):382–417. [Google Scholar]
- Hothorn T, Hornik K, Zeileis A. Unbiased recursive partitioning: A conditional inference framework. Journal of Computational and Graphical Statistics. 2006;15(3):651–674. [Google Scholar]
- Hothorn T, Hornik K, Zeileis A. party: A laboratory for recursive part(y)itioning. R package version 0.9–96 2008 [Google Scholar]
- Hothorn T, Leisch F, Zeileis A, Hornik K. The design and analysis of benchmark experiments. Journal of Computational and Graphical Statistics. 2005;14(3):675–699. [Google Scholar]
- Huang X, Pan W, Grindle S, Han X, Chen Y, Park SJ, Miller LW, Hall J. A comparative study of discriminating human heart failure etiology using gene expression profiles. BMC Bioinformatics. 2005;6:205. doi: 10.1186/1471-2105-6-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Loh W. Classification trees with unbiased multiway splits. Journal of the American Statistical Association. 2001;96(454):589–604. [Google Scholar]
- Kitsantas P, Moore T, Sly D. Using classification trees to profile adolescent smoking behaviors. Addictive Behaviors. 2007;32(1):9–23. doi: 10.1016/j.addbeh.2006.03.014. [DOI] [PubMed] [Google Scholar]
- König I, Malley JD, Pajevic S, Weimar C, Diener H-C, Ziegler A. Patient-centered yes/no prognosis using learning machines. International Journal of Data Minig and Bioinformatics. 2008 doi: 10.1504/ijdmb.2008.022149. To appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König I, Malley JD, Weimar C, Diener HC, Ziegler A. Practical experiences on the necessity of external validation. Statistics in Medicine. 2007;26(30):5499–5511. doi: 10.1002/sim.3069. [DOI] [PubMed] [Google Scholar]
- Leeb H, Pötscher BM. Can one estimate the conditional distribution of post-model-selection estimators? The Annals of Statistics. 2006;34(5):2554–2591. [Google Scholar]
- Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2(3):18–22. [Google Scholar]
- Lin Y, Jeon Y. Random forests and adaptive nearest neighbors. Journal of the American Statistical Association. 2006;101(474):578–590. [Google Scholar]
- Luellen JK, Shadish WR, Clark MH. Propensity scores: An introduction and experimental test. Evaluation Review. 2005;29(6):530–558. doi: 10.1177/0193841X05275596. [DOI] [PubMed] [Google Scholar]
- Lunetta KL, Hayward LB, Segal J, Eerdewegh PV. Screening large-scale association study data: Exploiting interactions using random forests. BMC Genetics. 2004;5:32. doi: 10.1186/1471-2156-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinic I, Supek F, Kovacic Z, Rukavina L, Jendricko T, Kozaric-Kovacic D. Posttraumatic stress disorder: Diagnostic data analysis by data mining methodology. Croatian Medical Journal. 2007;48(2):185–197. [PMC free article] [PubMed] [Google Scholar]
- Mielke PW, Berry KJ. Permutation Methods: A Distance Function Approach. New York: Springer Series in Statitics; 2001. [Google Scholar]
- Morgan JN, Sonquist JA. Problems in the analysis of survey data, and a proposal. Journal of the American Statistical Association. 1963;58(302):415–434. [Google Scholar]
- Nason M, Emerson S, Leblanc M. CARTscans: A tool for visualizing complex models. Journal of Computational and Graphical Statistics. 2004;13(4):1–19. [Google Scholar]
- Oh J, Laubach M, Luczak A. Estimating neuronal variable importance with random forest. Proceedings of the 29th Annual IEEE Bioengineering Conference; Newark, NJ, USA: New Jersey Institute of Technology; 2003. pp. 33–34. [Google Scholar]
- Paneque D, Borgland A, Bovier A, Bloom E, Edmonds Y, Funk S, Godfrey G, Rando R, Wai L, Wang P. Novel technique for monitoring the performance of the LAT instrument on board the GLAST satellite. Proceedings of the First GLAST Symposium; Stanford, CA, USA. American Institute of Physics; 2007. pp. 562–563. [Google Scholar]
- Polikar R. Ensemble based systems in decision making. IEEE Circuits and Systems Magazine. 2006;6(3):21–45. [Google Scholar]
- Potapov S. TWIX: Trees WIth eXtra Splits. R package version 0.2.4 2007 [Google Scholar]
- Potapov S, Theus M, Urbanek S. TWIX: Trees WIth eXtra Splits. Presentation slides from the Third Ensemble Workshop of the Statistical Computing task group of the German Section of the International Biometric Society; Munich, Germany. 2006. [Google Scholar]
- Qi Y, Bar-Joseph Z, Klein-Seetharaman J. Evaluation of different biological data and computational classification methods for use in protein interaction prediction. Proteins. 2006;63(3):490–500. doi: 10.1002/prot.20865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan JR. Induction of decision trees. Machine Learning. 1986;1(1):81–106. [Google Scholar]
- Quinlan JR. C4.5: Programms for Machine Learning. San Francisco: Morgan Kaufmann Publishers Inc; 1993. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- Rijmen F, Tuerlinckx F, Boeck PD, Kuppens P. A nonlinear mixed model framework for item response theory. Psychological Methods. 2003;8(2):185–205. doi: 10.1037/1082-989x.8.2.185. [DOI] [PubMed] [Google Scholar]
- Ripley B. tree: Classification and Regression Trees. R package version 1.0–26 2007 [Google Scholar]
- Rodenburg W, Heidema AG, Boer JM, Bovee-Oudenhoven IM, Feskens EJ, Mariman EC, Keijer J. A framework to identify physiological responses in microarray based gene expression studies: Selection and interpretation of biologically relevant genes. Physiological Genomics. 2008;33(1):78–90. doi: 10.1152/physiolgenomics.00167.2007. [DOI] [PubMed] [Google Scholar]
- Rossi A, Amaddeo F, Sandri M, Tansella M. Determinants of once-only contact in a community-based psychiatric service. Social Psychiatry and Psychiatric Epidemiology. 2005;40(1):50–56. doi: 10.1007/s00127-005-0845-x. [DOI] [PubMed] [Google Scholar]
- Ryan M, Lockstone H, Huffaker S, Wayland M, Webster M, Bahn S. Gene expression analysis of bipolar disorder reveals downregulation of the ubiquitin cycle and alterations in synaptic genes. Molecular Psychiatry. 2006;11:965–978. doi: 10.1038/sj.mp.4001875. [DOI] [PubMed] [Google Scholar]
- Sanchez-Espigares J, Marco L. Rasch model-based recursive partitioning for statistical survey analysis. In: Brito P, editor. Abstract Book of the 18th International Conference on Computational Statistics; Porto, Portugal. 2008. [Google Scholar]
- Segal MR. Technical Report. Center for Bioinformatics & Molecular Biostatistics papers, University of California; San Francisco, CA, USA: 2004. Machine learning benchmarks and random forest regression. [Google Scholar]
- Segal MR, Barbour JD, Grant RM. Relating HIV-1 sequence variation to replication capacity via trees and forests. Statistical Applications in Genetics and Molecular Biology. 2004;3(1):Article 2. doi: 10.2202/1544-6115.1031. [DOI] [PubMed] [Google Scholar]
- Shen KQ, Ong CJ, Li XP, Hui Z, Wilder-Smith E. A feature selection method for multilevel mental fatigue EEG classification. IEEE Transactions on Biomedical Engineering. 2007;54(7):1231–1237. doi: 10.1109/TBME.2007.890733. [DOI] [PubMed] [Google Scholar]
- Shih YS, Seligson D, Belldegrun AS, Palotie A, Horvath S. Tumor classification by tissue microarray profiling: Random forest clustering applied to renal cell carcinoma. Modern Pathology. 2005;18(4):547–557. doi: 10.1038/modpathol.3800322. [DOI] [PubMed] [Google Scholar]
- Strobl C, Augustin T. Adaptive selection of extra cutpoints – an approach towards reconciling robustness and interpretability in classification trees. Journal of Statistical Theory and Practice. 2009;3(1):119–135. [Google Scholar]
- Strobl C, Boulesteix AL, Augustin T. Unbiased split selection for classification trees based on the Gini index. Computational Statistics & Data Analysis. 2007;52(1):483–501. [Google Scholar]
- Strobl C, Boulesteix AL, Kneib T, Augustin T, Zeileis A. Conditional variable importance for random forests. BMC Bioinformatics. 2008;9:307. doi: 10.1186/1471-2105-9-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl C, Boulesteix A-L, Zeileis A, Hothorn T. Bias in random forest variable importance measures: Illustrations, sources and a solution. BMC Bioinformatics. 2007;8:25. doi: 10.1186/1471-2105-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl C, Wickelmaier F, Zeileis A. Technical Report Nr. 54. Department of Statistics, Ludwig-Maximilians-Universität München; Germany: 2009. Accounting for individual differences in Bradley-Terry models by recursive partitioning. [Google Scholar]
- Strobl C, Zeileis A. Danger: High power! – exploring the statistical properties of a test for random forest variable importance. Proceedings of the 18th International Conference on Computational Statistics; Porto, Portugal. 2008. [Google Scholar]
- Svetnik V, Liaw A, Tong C, Wang T. Application of Breiman’s random forest to modeling structure-activity relationships of pharmaceutical molecules. In: Roli F, Kittler J, Windeatt T, editors. Lecture Notes in Computer Science: Multiple Classifier systems. Berlin/Heidelberg: Springer; 2004. pp. 334–343 . [Google Scholar]
- Therneau TM, Atkinson B. rpart: Recursive partitioning. R port by Brian Ripley, R package version 3.1–30 2006 [Google Scholar]
- van Os BJ, Meulman J. Globally optimal tree models. In: Azen S, Kontoghiorghes E, Lee JC, editors. Abstract Book of the 3rd World Conference on Computational Statistics & Data Analysis of the International Association for Statistical Computing; Cyprus, Greece. Matrix Computations and Statistics Group; 2005. p. 79. [Google Scholar]
- Vapnik VN. The Nature of Statistical Learning Theory. New York: Springer; 1995. [Google Scholar]
- Venables W, Ripley B. Modern Applied Statistics with S-Plus. 4. New York: Springer; 2002. [Google Scholar]
- Ward MM, Pajevic S, Dreyfuss J, Malley JD. Short-term prediction of mortality in patients with systemic lupus erythematosus: Classification of outcomes using random forests. Arthritis and Rheumatism. 2006;55(1):74–80. doi: 10.1002/art.21695. [DOI] [PubMed] [Google Scholar]
- White A, Liu W. Bias in information based measures in decision tree induction. Machine Learning. 1994;15(3):321–329. [Google Scholar]
- Wood S. Generalized Additive Models: An Introduction with R. Boca Raton: Chapman & Hall; 2006. [Google Scholar]
- Wu B, Abbott T, Fishman D, McMurray W, Mor G, Stone K, Ward D, Williams K, Zhao H. Comparison of statistical methods for classification of ovarian cancer using mass spectrometry data. Bioinformatics. 2003;19(13):1636–1643. doi: 10.1093/bioinformatics/btg210. [DOI] [PubMed] [Google Scholar]
- Zeileis A, Hothorn T, Hornik K. Model-based recursive partitioning. Journal of Computational and Graphical Statistics. 2008;17(2):492–514. [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society B. 2005;67(2):301–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.